Abstract
Purpose
MYC-amplified medulloblastomas are highly lethal tumors. BET bromodomain inhibition has recently been shown to suppress MYC-associated transcriptional activity in other cancers. The compound JQ1 inhibits BET bromodomain-containing proteins, including BRD4. Here we investigate BET bromodomain targeting for the treatment of MYC-amplified medulloblastoma.
Experimental Design
We evaluated the effects of genetic and pharmacological inhibition of BET bromodomains on proliferation, cell cycle, and apoptosis in established and newly generated patient- and GEMM-derived medulloblastoma cell lines and xenografts that harbored amplifications of MYC or MYCN. We also assessed the effect of JQ1 on MYC expression and global MYC-associated transcriptional activity. We assessed in vivo efficacy of JQ1 in orthotopic xenografts established in immunocompromised mice.
Results
Treatment of MYC-amplified medulloblastoma cells with JQ1 decreased cell viability associated with arrest at G1 and apoptosis. We observed down-regulation of MYC expression and confirmed inhibition of MYC-associated transcriptional targets. Exogenous expression of MYC from a retroviral promoter reduced the effect of JQ1 on cell viability, suggesting that attenuated levels of MYC contribute to the functional effects of JQ1. JQ1 significantly prolonged survival of orthotopic xenograft models of MYC-amplified medulloblastoma (p<0.001). Xenografts harvested from mice after five doses of JQ1 had reduced expression of MYC mRNA and a reduced proliferative index.
Conclusion
JQ1 suppresses MYC expression and MYC-associated transcriptional activity in medulloblastomas, resulting in an overall decrease in medulloblastoma cell viability. These preclinical findings highlight the promise of BET bromodomain inhibitors as novel agents for MYC-amplified medulloblastoma.
Keywords: BET-bromodomain, JQ1, MYC, MYCN, MYCL1, medulloblastoma
INTRODUCTION
Medulloblastoma is the most common malignant brain tumor of childhood(1). Patients with local disease receive surgical resection, radiation therapy and chemotherapy, with five-year overall-survival exceeding 80%(2). These treatments cause significant therapy-related morbidity, including disabling cognitive deficits(3), growth failure, and increased risk of secondary malignancies(4). However, despite intensive chemotherapy and radiotherapy, the overall survival of these “high-risk” patients remains dismal, with 10-year overall-survival rates as low as 20%(5). Thus, tremendous impetus exists for the development of more effective treatments, based on known molecular targets, in medulloblastoma.
Medulloblastoma is a genetically heterogeneous disease, comprised of molecular subtypes characterized by differing transcriptional signatures, genomic alterations, and clinical courses(6–9). The current consensus is of at least four distinct subtypes including Wingless (WNT), Sonic Hedgehog (SHH), Groups 3 and 4(10). Group 3 medulloblastomas have the worst prognosis, are commonly metastatic and refractory to standard therapy, with 10-year overall-survival rates of 39%(5, 6, 10). Amplifications of one of three members of the MYC family of genes (MYC, MYCN, MYCL1) are found in several subtypes. Group 3 tumors are often associated with amplification of MYC(11), which is the most frequently observed amplification observed across multiple cancer types(12). Group 3 tumors without MYC amplification are often characterized by over-expression of MYC(6) or amplification of MYCN(11). MYCL1 amplifications have been reported in a few SHH tumor cases while SHH and Group 4 tumors are enriched with amplifications of MYCN(11).
MYC and other transcription factors complicit in cancer are poor targets for small molecule inhibition. Alternative strategies target MYC through epigenetic modulation of MYC transcription itself or of MYC target genes(13, 14). In particular, bromodomain and extra terminal (BET)-containing proteins, which recognize and engage side-chain acetylated lysine on open chromatin to facilitate gene transcription(15), have been identified as novel targets for small molecule development(13). Evidence that transcription of MYC and MYCN and subsequent activation of their downstream transcriptional programs can be targeted by BET bromodomain inhibition(13, 16, 17) presents a novel therapeutic strategy for patients with MYC-amplified medulloblastoma.
METHODOLOGY
Ethics statement
Ethics approval was granted by the relevant human IRB and/or animal ethics IACUC research committees of Dana-Farber Cancer Institute and Stanford University.
Cell lines and Culture
D283, D425, D458 and D556 were generously provided by Dr. Darrell Bigner (Duke University). Daoy cells were obtained from the American Type Culture Collection. Cell lines were maintained in Dulbecco modified Eagle medium (Gibco) supplemented with 10% FBS (100106, Benchmark) and 1% penicillin–streptomycin with 1% glutamine (Gibco). UW228, R256, R262 and R308 were a kind gift from Michael Bobola (University of Washington, Seattle). MB002 cells were derived from an autopsy specimen of the leptomeningeal compartment from a child with metastatic, treatment-refractory (chemotherapy only) medulloblastoma. The MB002 primary tumor displayed histological features of large cell medulloblastoma and gene expression markers consistent with Group 3 medulloblastoma (Supplemental Figure 1A)(11). MB004 cells were derived from the primary surgical resection of a tumor in a child whose tumor recurred after therapy. The MB004 primary tumor displayed focal anaplasia and gene expression markers consistent with Group 3 and Group 4 medulloblastoma (see Supplemental Figure 1)(6). MYC-amplification in the MB002 and MB004 cells were confirmed with NanoString nCounter v2 Cancer CN Codeset, which estimates copy-number of 86 genes commonly amplified or deleted in cancer (Supplemental Table 1). Human neural stem cells were derived from subventricular zone tissue surgically excised during a functional hemispherectomy in a child with refractory seizures. MB002 and MB004 cells were maintained in culture media with 1:1 Dulbecco modified Eagle medium (Gibco) and neural stem cell media (Gibco) supplemented with B27 (Gibco) EGF (02653, Stem Cell), FGF (GF003, Millipore), Heparin (07980, Stem Cell) and LIF (LIF1010, Millipore). The SVZ-derived neural stem cells were maintained similarly with the exception of LIF supplement. The MYC and MYCN-driven medulloblastoma GEMM cell lines were derived and cultured as previously described(18, 19). Briefly, for Myc amplified GEMM lines, cerebellar stem cells infected with Myc and DNp53 retroviruses were transplanted into cerebella of NOD-SCID-IL2Rgammanull (NSG) mice. When mice became symptomatic, tumors were harvested and dissociated into single cell suspensions.
Patient-derived cell lines MB002 and MB004 were authenticated using STS fingerprinting. Cell lines obtained from the Bigner and Babola labs were authenticated using SNP250k or SNP6.0 arrays, which revealed copy-number alterations consistent with previously published karyotypes(12, 20).
shRNA suppression
Lentiviral vectors encoding shRNAs specific for BRD4, MYC, and the control LACZ were obtained from The RNAi Consortium (Clones and sequence: shBRD4 TRCN0000021426, 5’ CGTCCGATTGATGTTCTCCAA; shMYC TRCN0000039640, 5’ CAGTTGAAACACAAACTTGAA; and shLacZ TRCN0000231726, 5’ TGTTCGCATTATCCGAACCAT). Lentivirus was produced by transfection of 293T cells with vectors encoding each shRNA (2 µg) with packaging plasmids encoding PSPAX2 and VSVG using Lipofectamine (Invitrogen, 56532). Lentivirus-containing supernatant was collected 48 hours and 72 hours after transfection, pooled, and stored at −80 °C. Cells were infected (ratio of 1:4 virus media) in polybrene-containing media (2.5µg/ml), and incubated overnight. Cells were selected in puromycin (2.0 µg/ml) starting 48 hrs after infection.
Over-expression of MYC in D283 for MYC rescue experiments
293T cells were transfected with 2 µg retroviral pBabe expression vectors (empty vector or pBabeMYC) with packaging plasmids encoding gag-pol and VSVG using Lipofectamine. Retro-virus containing supernatant was collected at 48 hours and 72 hours after transfection, pooled and stored at −80 °C. D283 ells were infected (ratio of 1:4 virus media) in polybrene-containing media (2.5µg/ml). Treatment with 1µM JQ1R or JQ1S was commenced at six hours post infection. Cell viability was assessed at 24 hours post treatment with JQ1.
Cell viability assays following treatment with JQ1 or shRNA suppression
To assess responsiveness to JQ1, 1000 cells were plated in 96-well plates in serial dilutions of either JQ1R or JQ1S, in triplicate. Cell viability was measured by assessing ATP content at 0, 24, 48, 72, 96 and 120 hours using Cell Titre-Glo (Promega) according to the manufacturer’s instructions. Mean ± standard deviation (SD) was calculated. Non-linear dose response curves were applied to the data using GraphPad Prism.
To assess dependence of cells on BRD4 or MYC, cells were infected with lentiviral plasmids encoding shRNA. Forty-eight hours after infection, 1000 cells were plated in each well of 96-well plates, in triplicate, in media containing puromycin(1µg/ml). Cell viability was measured by assessing ATP content using Cell Titre-Glo (Promega), and results were normalized to baseline. Mean ± SD was calculated.
Flow Cytometry
Cell cycle analysis was performed by measuring DNA content by Propidium Iodide staining in cells treated with 1µM JQ1R or JQ1S for 72 hours. Apoptosis was measured with annexin V/PI staining. The annexin V was labeled with Alexa Fluor (A13201, Invitrogen) and flow cytometry was performed per manufacturer’s guidelines.
Protein extraction and immunoblotting
MYC amplified cells were lysed in boiling RIPA lysis buffer containing protease and phosphatase inhibitors, and centrifuged at 13,000 × g for 10 minutes. For MYCN-amplified lines, western blot analysis was performed as previously described(19) with the following modification: Lysis buffer with 1% SDS. Supernatant was mixed with 4× SDS sample buffer, boiled for 10 minutes, and subjected to SDS-PAGE on 4–12% gradient gels. Blots were probed with antibodies against BRD4 (12183, Cell Signaling), MYC (sc-764, Santa Cruz), MYCN (ab-16898, Abcam), β-tubulin(MAB 3408, Millipore) and actin (sc-1615, Santa Cruz).
RNA extraction and Real-Time RT-PCR
RNA was extracted with the RNeasy kit (Qiagen). cDNA was synthesized from 1µg RNA using High Capacity RNA to cDNA kits (Applied Biosystems). Real-time RT-PCR was performed using SYBR Green master mix (Applied Biosytems). Cycling was performed as follows: 50°C for two minutes and 95°C for ten minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 30 seconds. This was followed with a dissociation stage of 95°C for 15 seconds, 60°C for 30 seconds, and 95°C for 15 seconds. Primers for BRD4, MYC and β-actin are listed in Supplemental Table 2. Samples were amplified in triplicate and data analyzed using the ΔΔCT method.
Copy-number analysis
Relative copy-number estimates were generated from published Affymetrix SNP 6.0 data for 1073 tumors(11) using comparison data from 131 normal samples and an analytic pipeline described in detail elsewhere (Tabak et al, in preparation). Briefly, signal intensities for each probe were normalized to uniform intensity values and merged to form probeset-level values using SNPFileCreator, a Java implementation of dChip(21, 22). Marker-level intensities were calibrated to DNA copy-number levels using Birdseed for SNP markers(23) and using the results of experiments with cell lines with varied copy-numbers of the X chromosome for copy-number probes (Tabak et al., in preparation). Regions of frequent germline copy-number variation were identified using a large bank of normal tissue samples and excluded from the data (Tabak et al, in preparation). Noise was reduced by applying Tangent normalization(12) followed by Circular Binary Segmentation(24, 25). Data were mean-centered for each sample. Amplifications were defined as greater than a relative copy-number of 2.4. For samples with a relative copy-number of 2.4 to 3, we applied the ABSOLUTE algorithm(26) and confirmed that each of these samples had an absolute copy-number of greater than three copies.
Genome-wide expression analysis
Previously published microarray expression and copy number data(11) were obtained from the Gene Expression Omnibus (GSE37385 and GSE37382). The expression data were obtained using the GEOImporter module in GenePattern. Z scores of gene expression values of genes within samples were calculated.
For analyses of gene sets enriched among samples exhibiting high expression of MYC family members (z score>1 for MYC, z score>0.85 for MYCL1 and z score>1.5 for MYCN), gene set enrichment analysis (GSEA)(27, 28) was performed using the C2 canonical pathway (CP) gene sets and seven additional gene sets from MSigDB that represent MYC activation signatures (Supplemental Table 2). Gene sets with a nominal p-value less then 0.05 were considered significant.
To examine the effect of JQ1 on global gene transcription, cell lines were treated with JQ1R or JQ1S (1µM for 24 hours) and RNA was extracted. Gene expression profiles were assayed using Affymetrix Human Gene 1.0 ST microarrays (Affymetrix, Santa Clara, CA). CEL files were normalized using RMA(29). Expression-array data have been deposited in the Gene Expression Omnibus portal under the accession number GSE51020. Comparative marker selection analysis(30) between JQ1S- and JQ1R-treated cells was performed in GenePattern using the default settings.
The recently-described JQ1 consensus signature(16) was applied to the gene expression profiles using the 52 genes identified as being significantly differentially expressed following treatment with JQ1. Agglomerative hierarchical clustering was performed using pairwise complete-linkage and a Pearson correlation metric across both samples and genes.
To identify gene sets differentially expressed following treatment with JQ1, GSEA was performed using the same customized C2 (CP) gene sets (MSigDB) with the seven additional MYC activation gene sets. Gene sets with a nominal p-value ess then 0.05 were considered significant.
In vivo experiments
In vivo efficacy studies were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee at Stanford University. Briefly, MB002 cells were transduced with a GFP-luciferase lentiviral expression construct and FACS sorted to obtain 30,000 GFP-luciferase positive cells that were then injected with stereotaxic guidance into the cerebella of 4–6 week old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (The Jackson Laboratories). To confirm engraftment, mice were administered D-luciferin (75 mg/kg; Promega) and imaged on a Xenogen IVIS2000 (Perkin-Elmer) 14 days after injection. Mice were randomized into treatment and control groups (n=5 mice per group) and administered JQ1-S (50 mg/kg in 1:10 solution of DMSO:10% cyclodextrin) or vehicle alone (1:10 solution of DMSO:10% cyclodextrin), daily via intraperitoneal injection, until euthanasia was required. Tumor growth was monitored by IVIS imaging at 14 and 21 days of treatment. Statistical significance for Kaplan-Meier analysis was determined by log-rank (Mantel–Cox) test.
For immunohistochemistry analysis of xenografted medulloblastomas, 4–6 week old NSG mice received intracerebellar injections of MB002 cells (30,000 cells) and were administered JQ1-S (50mg/kg twice daily; n=3) or vehicle (n=3), for five doses and then euthanized. Brains were carefully dissected and either frozen in RNAlater (Qiagen) or preserved in 4% paraformaldehyde and subsequently embedded in paraffin. RNA was extracted from frozen cerebellum using the RNeasy kit (Qiagen) as per manufacturers instructions.
Immunohistochemistry
JQ1- and vehicle-treated MB002 xenografts were harvested, rinsed in PBS and fixed in 4% paraformaldehyde overnight at 4°C. Five-micron thick sections were mounted on poly-D-lysine-coated slides and treated with xylene followed by several changes of graded alcohol. Antigen retrieval was performed by application of citrate buffer pH 6.00 for 20 minutes. Slides were then incubated with anti-Ki67 (Labvision, SP6 RM-9106-S, lot 9106S1210D) overnight at 4°C. Cells were washed with several changes of PBS, and secondary antibody conjugated to HRP was applied and detected using the Dako Envision kit for DAB.
Immunofluorescence
Primary medulloblastoma cells (MB002) were cultured in 12-well plates at density of 1 × 105 cells/well and treated with vehicle, (S)-JQ1 (500nM and 1µM for 6 and 12 hours). Cells were centrifuged at 1000 rpm for 5 minutes, washed in PBS, and mechanically dissociated for 5 minutes at 37 °C. Single cell suspensions were transfered to coverslips pre-coated with Poly-L-Lysine (10µg/ml in ddH2O, catalog number P6516, Sigma-Aldrich) and then fixed with 4% PFA for 15 minutes. Immunolabeling was carried out with the antibody anti-BAD(C-7) (1:400, SC8044, Santa Cruz), detected by Cy3-conjugated secondary Donkey anti-mouse antibody (1:200, JacksonImmuno Research) and visualized by confocal fluorescence microscopy (Leica DM5500 B, Leica microsystem).
For statistical analysis, p values were calculated using Fisher’s, T-tests or Pearson’s as appropriate. ANOVA with correction was used for comparison of multiple groups.
RESULTS
Medulloblastomas exhibit several indicators of MYC pathway activation
We evaluated indicators of MYC pathway activation using integrated datasets of genome-wide copy-number estimates from 1071 medulloblastomas and corresponding gene expression data for 282 medulloblastomas(11). These tumors comprised 79 Wnt subgroup medulloblastomas, 265 Sonic Hedgehog subgroup tumors, 168 Group 3 medulloblastomas, and 313 Group 4 tumors, as determined by exon array or nanoString analysis(11). Subtype designation was not available for 245 tumors. We evaluated copy-numbers and expression of all three MYC family members (MYC, MYCN, and MYCL1) and nine signatures of MYC pathway activation obtained from the ‘Gene Set Enrichment Database’.
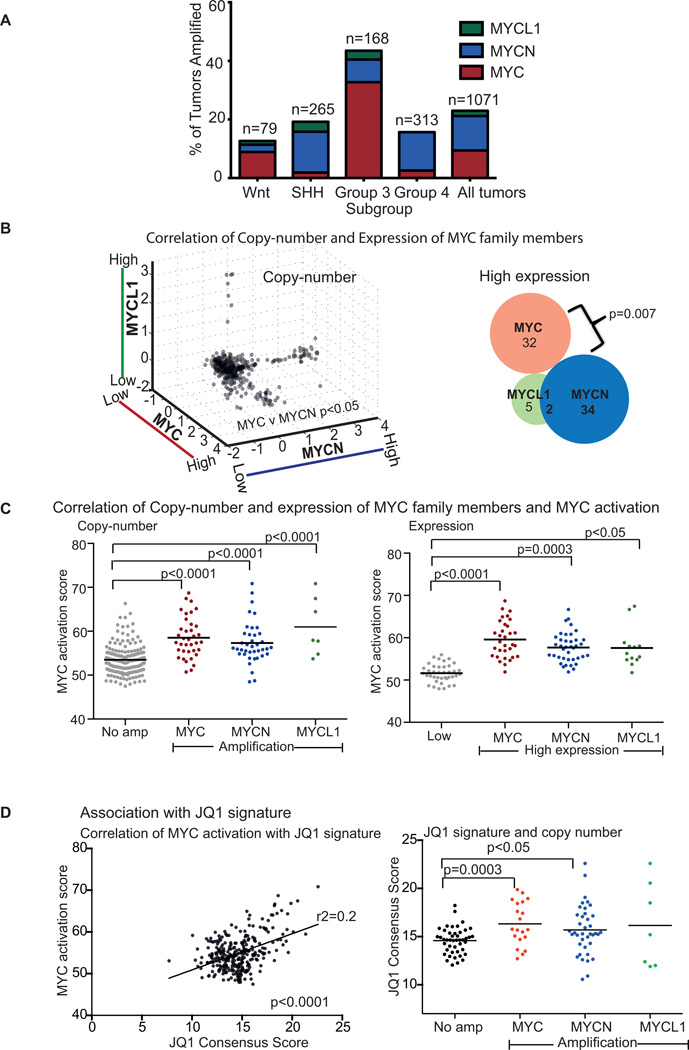
We found that 23% of all medulloblastomas exhibit amplifications of one or more of MYC, MYCN, or MYCL1 (9%, 12%, and 2% of all tumors, respectively; Figure 1A). Group 3 tumors exhibit amplifications of MYC family members approximately three times as frequently (44% of cases) as the other subtypes (13%, 19%, and 16% among Wnt, Sonic Hedgehog and Group 4 tumors, respectively). This enrichment was most profound for MYC; Group 3 tumors accounted for 70% of medulloblastomas exhibiting MYC amplifications. Amplifications of MYCN were observed slightly more often in SHH and Group 4 tumors (38% and 35% of all MYCN amplifications, respectively) than Group 3 tumors (10%).
Figure 1. Amplifications of the MYC isoforms MYC, MYCN, or MYCL1 are common and mutually exclusive in medulloblastoma.
A, Fractions of tumors in the Wnt, Sonic Hedgehog, Group 3 and Group 4 subtypes of medulloblastoma with amplifications of MYC, MYCN, or MYCL1 in 1071 medulloblastomas.
B, Scatterplot (Left) depicting copy-numbers of different MYC isoforms in 1071 medulloblastoma samples. p value depicts significant anti-correlation between MYC and MYCN. Venn diagram (Right) showing medulloblastomas with high expression of MYC (z score >1), MYCN (z score >1.5) or MYCL1 (z score >0.85). p value depicts significant anti-correlation between MYC and MCYN.
C, Correlation of MYC activation scores with copy-numbers of MYC isoforms (Left) and correlation of MYC activation scores with gene expression of MYC isoforms. p values depict significant differences in MYC activation scores. ANOVA p value <0.0001.
D, Association between MYC activation and JQ1 consensus score (Left). p value depicts significant correlation as determined by Pearson’s Correlation test. Association between JQ1 consensus score and MYC amplifications (Right). p values depict significant differences in JQ1 scores. ANOVA p value <0.05.
Amplifications of MYC and MYCN were anti-correlated (p < 0.05; Figure 1B), suggesting that they have similar functional effects. Anti-correlated genetic alterations often indicate functional redundancies(31–33) because redundant alterations are not required by the same tumor. Although there was a trend towards anti-correlation between amplification of MYCL1 and either MYC or MYCN, this did not reach significance (p=0.8 and 0.8, respectively), perhaps because MYCL1 amplifications were observed so infrequently.
Amplifications of each MYC family member were also associated with increased expression of its gene transcript (p<0.0001 in all cases). Indeed, high expression of MYCN or MYCL1 tends to be found exclusively in medulloblastomas with amplification of those genes. High MYC expression, however, is often present in samples without MYC amplification, suggesting alternative mechanisms for increased MYC expression. Increased expression of MYC and MYCN were anti-correlated (p=0.007; Figure 1B), as were MYC and MYCN amplifications. Increased expression of MYCL1 trended towards a similar anti-correlation with the other MYC family members, but did not reach statistical significance.
Amplification and over-expression of each MYC family member was also associated with increased expression of genes up-regulated by MYC (p<0.05 in all cases; Figures 1C). We assessed MYC pathway activation by summing the expression levels of 68 previously published genes known to be up-regulated by MYC (14). We obtained similar results with GSEA analysis using nine signatures of MYC activation (Supplemental Table 2) and comparing tumors with high expression of any MYC to tumors with low expression of all MYCs. Among the nine MYC activation signatures present in our gene sets, six were significantly associated with high MYC expression, five with MYCN and four with MYCL1, respectively (Supplemental Table 3).
Medulloblastomas that exhibited indicators of MYC activation also exhibited high expression of genes observed to be down-regulated with treatment with the BET bromodomain inhibitor JQ1(16). Genes down-regulated by JQ1 were previously identified in tumors from multiple lineages, including multiple myeloma, leukemia and neuroblastoma(16). We found a positive correlation between the expression of genes that are targeted by JQ1 (and thus opposite to the signature following treatment with JQ1) and MYC activation signatures in medulloblastoma (Figure 1D). We also observed positive correlations between expression of genes targeted by JQ1 and amplifications of MYC (p value 0.003) and MYCN (p value <0.05). The finding that medulloblastomas with indicators of MYC activation exhibit gene expression profiles that are opposite to the signature of JQ1 is not surprising, because the JQ1 signature has already been shown to reflect down-regulation of MYC activity(16). Nevertheless, these findings raise the hypothesis that treatment with JQ1 may limit MYC activity and suppress proliferation of MYC-driven medulloblastomas.
JQ1 reduces cell proliferation in MYC- and MYCN-amplified medulloblastoma cells
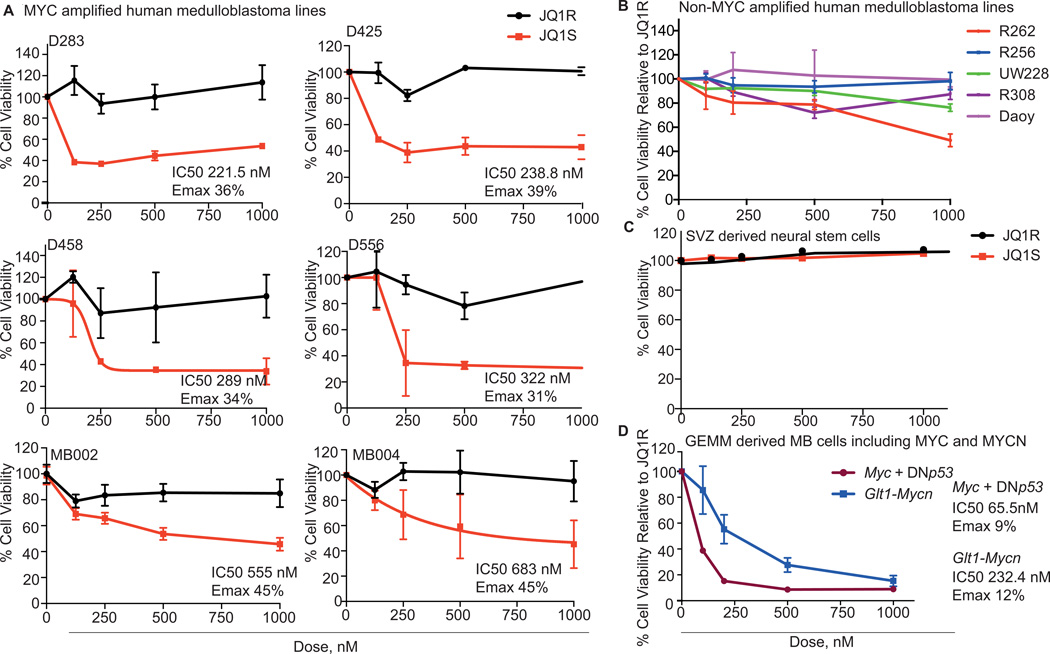
We examined the efficacy of treatment with JQ1S in six patient-derived medulloblastoma cell lines documented to have MYC-amplification relative to five non-MYC amplified medulloblastoma lines, and human neural stem cells(34, 35). Western immunoblotting of the patient-derived cell lines confirmed increased expression of MYC in lines with MYC-amplification (Supplemental Figure 1B). No patient derived MYCN-amplified medulloblastoma cell lines have been generated to date, therefore we evaluated JQ1 activity in the setting of Mycn-amplification using tumor cells derived from recently developed mouse models of group 4 MYNC-amplified medulloblastomas(18, 36). We also evaluated JQ1 activity in a murine model of group 3 MYC-driven medulloblastoma. The activity of JQ1 was initially assessed by comparing proliferation rates in the presence of the active stereoisomer of JQ1 (JQ1S) to proliferation rates in the presence of an inactive stereoisomer, JQ1R(13).
In all MYC-amplified patient-derived cell lines, treatment with JQ1S for 48 hours at doses less than 1µM resulted in 57–69% reduction in cell viability compared to treatment with JQ1R (Figure 2A). Each MYC-amplified line had an IC50 of less than 1µM. In contrast, cell viability of non-MYC amplified cell lines (Figure 2B) and neurons derived from the subventricular zone (SVZ) (Figure 2C) was not substantially altered by treatment with either JQ1S or JQ1R. Cells with MYC-amplification exhibited up to five-fold increases in cell number by day four of treatment with JQ1R, but showed no increase in cell numbers with JQ1S treatment over the same time period (Supplemental Figure 2A).
Figure 2. JQ1 reduces cell proliferation in MYC-amplified medulloblastoma cell lines and murine models of MYC and MYCN amplified medulloblastoma.
A, Dose-response curves of patient derived MYC-amplified medulloblastoma cell lines treated with JQ1S and JQ1R for 48 hours. % Cell viability is relative to untreated cells. Values shown represent mean ± SD.
B, Dose-response curves of patient derived non MYC-amplified medulloblastoma cell lines treated with JQ1S for 72 hours. % Cell viability is relative to untreated cells. Values shown represent mean ± SD.
C, Dose-response curves of neural stem cells derived from the subventricular zone treated with JQ1S and JQ1R. Values shown represent mean ± SD.
D, Dose-response curves of cancer cells from genetically engineered mice models of Group 3 and 4 medulloblastoma with Myc or Mycn overexpression, after treatment with JQ1S. Values shown represent mean ± SD.
Treatment with JQ1S also reduced proliferation of cells derived from a murine model of group 3–4 medulloblastoma with MYCN over-expression. In these cells, treatment with JQ1 reduced viability by 75% compared to treatment with JQ1R (Figure 2D). Medulloblastoma cells from a mouse model with MYC over-expression exhibited even greater (91%) sensitivity to treatment with JQ1S(Figure 2D). Taken together, these data show that JQ1 is efficacious in reducing cell viability in medulloblastoma cell lines driven by MYC and also in cells from a murine model of MYCN-driven medulloblastoma, with minimal effect observed in non-MYC-amplified medulloblastoma cells or neural stem cells.
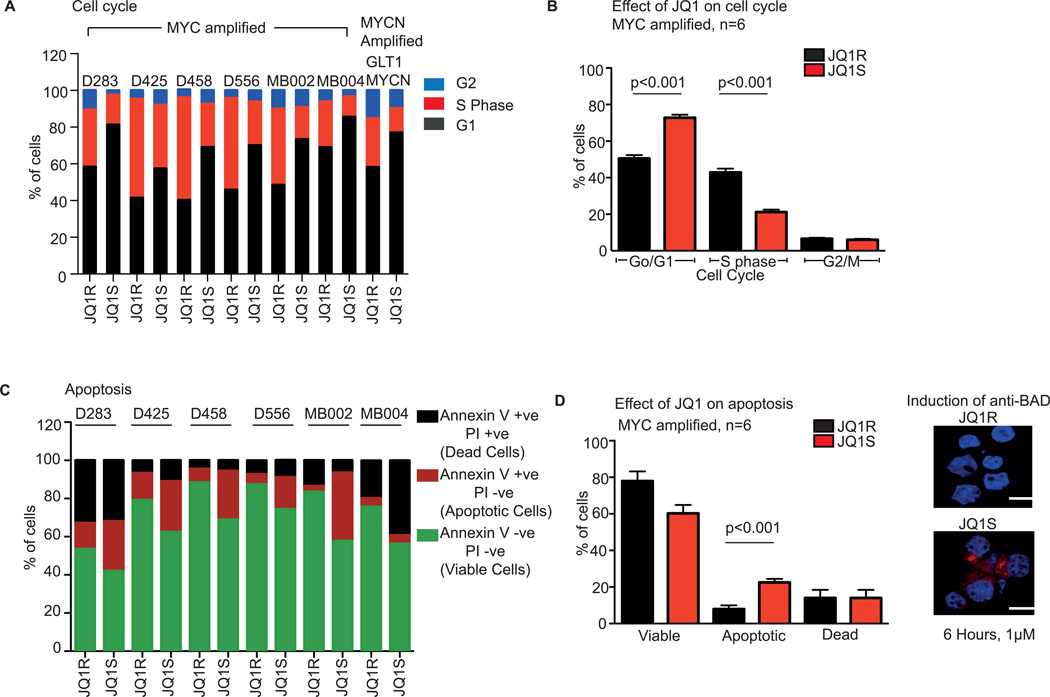
JQ1 reduces cell viability by inducing G1 arrest and apoptosis in MYC-amplified medulloblastoma cell lines
Treatment with JQ1S significantly affected cell cycling of the MYC-driven cell lines. We profiled cell cycling by flow-cytometry measurement of propidium iodide (PI) stained cells treated for 72 hours with either 1 µM of JQ1S or JQ1R. In six patient derived MYC-amplified cell lines, and one Mycn-driven GEMM derived cell line, we observed treatment with JQ1S resulted in G1 arrest, with an increased percentage of cells in G1 and reduction of cells in S phase (Figure 3A). Across all MYC-amplified lines, treatment with JQ1S reduced the number of cells in S phase by approximately 50% compared to JQ1R (21 ± 3% vs 43 ± 5%, p value <0.001) and increased the number of cells in G1 (73 ± 4% vs 51 ± 6%; p<0.001) (Figure 3B).
Figure 3. JQ1 induces G1 arrest and apoptosis in patient-derived MYC-amplified medulloblastoma cell lines.
A Cell cycle analysis of MYC amplified patient medulloblastoma cell lines and one MYCN amplified cell line derived from a GEMM treated with 1µM of JQ1S and JQ1R for 72 hours
B Pooled cell cycle analysis of MYC amplified medulloblastoma cell lines treated with 1µM of JQ1S and JQ1R for 72 hours. Values represent mean ± SD. ANOVA p value<0.001.
C Annexin V/PI apoptosis assays of MYC amplified medulloblastoma cell lines treated with 1µM of JQ1S and JQ1R for 72 hours. Viable cells are negative for annexin V and PI, apoptotic cells are positive for Annexin V and negative for PI and dead cells are positive for both Annexin V and PI.
D, Pooled Annexin V/PI apoptosis analysis of the six MYC-amplified medulloblastoma cell lines following treatment with 1µM of JQ1S and JQ1R for 72 hours. Values represent mean ± SD. Images on right shows induction of the apoptotic protein, BAD, in MB002 cell line, six hours following treatment with 1µM of JQ1S. Scale shown represents 20 micron.
Treatment with JQ1S also induced apoptosis in patient-derived MYC-amplified cells. We assessed induction of apoptosis in cells treated with 1 µM JQ1S or JQ1R for 72 hours by flow cytometry analysis of Annexin V/PI staining (Figure 3C). MB004 was observed to have an increased in the number of necrotic cells while in all other cell lines there was a clear increase in apoptosis noted. When the results of all cell lines were pooled (including MB004), JQ1S almost tripled the number of apoptotic cells (8 ± 2% vs 22.5 ± 4%, p=02; Figure 3D). As an independent correlation of induction of apoptosis we treated MB002 cells with JQ1 and observed induction of the apoptotic protein, BAD, within six hours of treatment (Figure 3D. right panel). These data suggest that JQ1S reduces cell viability of MYC-driven medulloblastoma cell lines by inducing G1 arrest and apoptosis.
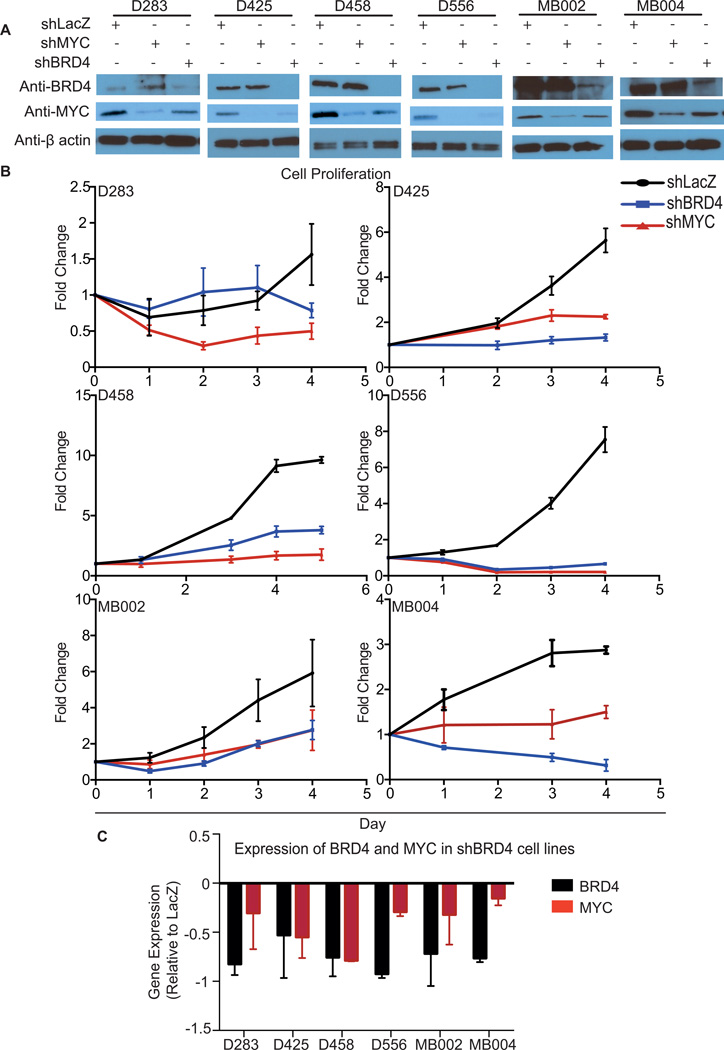
BRD4 suppression attenuates expression of MYC in medulloblastoma cells
We hypothesized that the effects of JQ1 in the medulloblastoma cells were mediated in large part by decreased activity of MYC through inhibition of BRD4. BRD4 has been shown to be a cofactor for MYC-dependent transcription in multiple cell types(13), and JQ1 has previously been shown to have higher affinity for binding domains of BRD4 than other bromodomain-containing proteins(13).
We therefore suppressed expression of either MYC or BRD4 in each of the six MYC-amplified patient derived cell lines using MYC- and BRD4-directed shRNAs. We measured proliferation and compared the results to controls treated with LacZ-targeted shRNAs. Suppression of MYC and BRD4 protein levels was confirmed by immunoblotting (Figure 4A and Supplemental Figure 3).
Figure 4. Suppression of BRD4 in patient-derived MYC-amplified medulloblastoma cell lines suppresses proliferation and MYC expression.
A, Immunoblots of BRD, MYC, and β-actin loading controls following infection with shRNAs targeting BRD4, MYC, or LacZ controls.
B, Cell viability following infection with shRNAs targeting BRD4, MYC, or LacZ. Cell proliferation of MYC amplified medulloblastoma cell lines following suppression of BRD4, MYC or LacZ. Error bars represent the mean ± SD of 3 replicates per condition.
C, BRD4 and MYC mRNA levels determined 72 hours after BRD4 suppression in the indicated cell lines. Values shown are the mean ± SD of 3 replicates per condition, normalized to expression in LacZ controls.
In all cell lines, suppression of either MYC or BRD4 resulted in greater than 50% reductions in proliferation relative to cells treated with LacZ-targeted shRNAs (Figure 4B). We observed absolute decreases in cell numbers among four lines with both MYC suppression and BRD4 suppression.
In all cell lines, suppression of BRD4 was also associated with a reduction of MYC mRNA, by an average of 45% relative to the LacZ-suppressed controls (p<0.0001; Figure 4C). We also observed a reduction in MYC protein levels in cells following suppression of BRD4 (Figure 4A and Supplemental Figure 3). This was most pronounced in the D425 and D556 cell lines. Suppression of BRD4 has previously been shown to decrease MYC expression in other cell types(13). The finding that BRD4 suppression resulted in attenuation of MYC mRNA and protein suggested that JQ1 also exerted its effects in part through suppression of MYC.
JQ1 suppresses MYC activation pathways and expression of MYC and MYCN themselves
To determine the transcriptional effects of JQ1 treatment, we performed genome-wide expression profiling of five patient-derived MYC-amplified cell lines treated with 1 µM of either JQ1S or JQ1R for 24 hours. We identified 43 genes that were significantly down-regulated by JQ1S (FDR<0.1; Supplemental Table 4). No genes were significantly up-regulated at this significance threshold.
The JQ1S-treated cells exhibited significant enrichment of a signature of JQ1 treatment derived from multiple myeloma, leukemia and neuroblastoma cells (16) (p<0.0001; Figure 5A), and significant attenuation of MYC activity (p<0.05 in all cases; Figure 5B). An unbiased screen of pathways altered by treatment with JQ1S (using the Gene Set Enrichment Algorithm and the C2 (CP) set of signatures) indicated significant alteration of 46 pathways (p value<0.05; Supplementary Table 5). Four of these 46 pathways represented MYC activation signatures (Figure 5B), and were down-regulated following treatment with JQ1S (compared to cells treated with JQ1R).
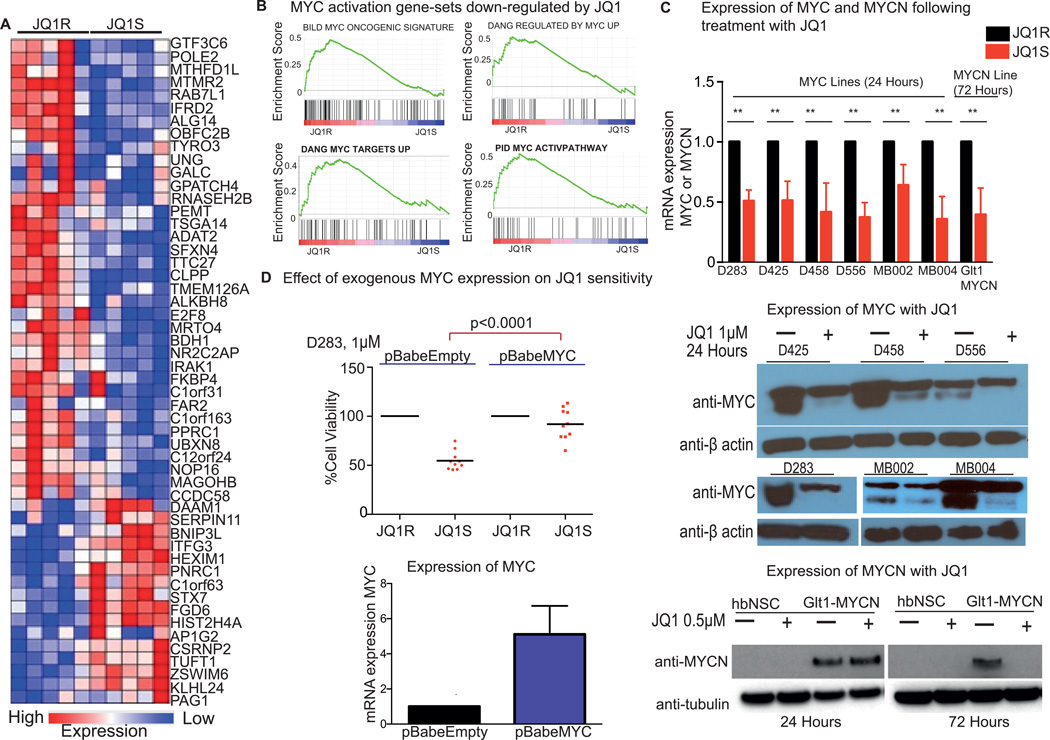
Figure 5. JQ1 treatment down-regulates transcription of MYC, MYCN and MYC activating pathways.
A, Expression of genes in the JQ1 consensus signature in MYC-amplified medulloblastoma cell lines following 24-hour treatment with 1µM JQ1R or JQ1S. Red indicates increased gene expression and blue reduced gene expression.
B, GSEA analysis of gene sets down-regulated among genes in medulloblastoma cell lines treated with JQ1S.
C, Expression of MYC and MYCN mRNA and protein following treatment with JQ1R or JQ1S. MYC amplified cells were treated with 1µM of JQ1R or JQ1S for 24 hours and MYCN derived GEM lines were treated with 0.5 µM JQ1S over the indicated time course. mRNA expression values are normalized to housekeeping control and expression is depicted relative to JQ1R controls. Values shown represent mean ± SD of 3 replicates per condition. ** indicates p<0.05. Immunoblots for MYC and β-actin loading controls following 24-hour treatment with 1µM of JQ1S in the indicated cell lines or for MYCN and β-tubulin loading controls treated with 0.5 µM JQ1S over the indicated time course are shown. Lysates from hindbrain neural stems cells are shown as a negative control for MYCN expression.
D, Cell viability of D283 infected with pBabeEmpty or pBabeMYC which were treated with 1µM of JQ1R or JQ1S six hours after infection. Cell viability is shown at 24 hours following treatment with JQ1R or JQ1S. Bars depict mean of ten replicates (individual values shown as scatterplot). qPCR for mRNA expression of MYC in D283 cells infected with retrovirus for pBabeEmpty or pBabeMYC is shown on the right. Expression is shown relative to β-actin controls, and normalized to expression for pBabeEmpty. Values depict mean ± SD of six replicates.
In all patient derived MYC amplified cell lines, treatment with 1µM JQ1S also led to decreased expression of MYC itself. Levels of MYC mRNA declined by an average of 46% (range 29–78%; p<0.0001; Figure 5C), and these changes were associated with decreased expression of MYC protein. We also examined expression of MYC mRNA in three cell lines treated with lower doses of JQ1S, and found attenuation of MYC mRNA expression at doses of JQ1S as low as 125 nM (Supplemental Figure 2B). Treatment of Mycn over-expressing cells derived from tumors from the Glt1 Mycn GEMM model with 0.5 µM JQ1 for 72 hours was associated with suppression of Mycn mRNA and protein expression (Figure 5C).
We next over-expressed MYC from an exogenous retroviral promoter in D283 cells and treated them with JQ1 to assess whether forced expression of MYC could rescue the cells from the effects of JQ1 (Figure 5D). In cells infected with the empty pBabe retroviral vector we observed reduced cell viability following treatment with JQ1S. In contrast, in cells infected with pBabe MYC, we observed minimal effect on cell viability following treatment with JQ1S.
Taken together, these results confirm BET bromodomain inhibition with JQ1 results in down-regulation of MYC and MYC activation pathways, resulting in reduced cell proliferation. JQ1 treatment also reduces MYC and MYCN expression in patient-derived MYC-amplified medulloblastoma cell lines and in cells derived from a MYCN-driven medulloblastoma mouse model. Forced expression of MYC from an exogenous promoter rescues D283 cells from the effects of JQ1S.
Treatment with JQ1 prolongs survival of MYC-amplified medulloblastoma xenografts
We examined the efficacy of JQ1 in vivo, using orthotopic xenografts generated by intra-cerebellar injections of MB002 cells. Compared to vehicle control, mice undergoing daily treatment with JQ1 (50 mg/kg/day) exhibited significantly prolonged survival (Figure 6A) with a slower rate of tumor growth at 14 and 21 days post-injection as indicated by bioilluminescence (Figure 6B). This was statistically significant on day 14 (p value 0.03). A cohort of mice were sacrificed following five doses of JQ1 or vehicle treatment. We extracted RNA from cerebellum of vehicle or JQ1 treated mice and assessed expression of MYC mRNA (Figure 6C). We observed significant down-regulation of MYC mRNA expression following treatment with JQ1. Ki67 immunostaining confirmed a reduced proliferative index in tumors treated with JQ1 compared to vehicle controls (82% vehicle vs 27% JQ1 treated, p value <0.0001) (Figure 6D).
Figure 6. JQ1 prolongs survival of xenograft models of Group 3 medulloblastoma.
A, Kaplan-Meier curves of mice with intracranial xenograft injections of MB002 treated with JQ1 or vehicle.
B, Rate of tumor growth in NSG mice with orthotopic MB002 xenografts following treatment with JQ1 as monitored by bioilluminescence detection
C, qPCR of MYC mRNA levels (normalized to β-actin) in cerebellar xenografts of MB002 following treatment with either vehicle control or JQ1. Values represent mean ± SD of six replicate measurements.
D, Representative images of Ki67 staining of MB002 xenografts treated with vehicle control and JQ1 (Top panel). % Ki67 positivity in tumors from mice treated with either vehicle control (n=2) or JQ1(n=3). Values represent mean ± SD of three replicate measurements for each animal.
DISCUSSION
Our data provide the first preclinical evidence for a potential therapeutic role of BET bromodomain inhibition for MYC-amplified medulloblastoma. Further we show redundancy between amplification of the different MYC genes. We observed responsiveness to JQ1 therapy of all patient cell lines harboring MYC amplifications, and in a murine cell line of a Mycn-driven medulloblastoma model.
MYC-amplified medulloblastomas are characteristically resistant to conventional treatments including radiation therapy and chemotherapy. In this study, the use of JQ1 as a single agent was able to reduce cell proliferation, tumor growth and prolong survival of mice with intracranial xenografts. Thus, our data provides rationale for the development of clinical trials to assess the role of these agents in the treatment of patients with medulloblastoma.
Ongoing work, however, is required to determine optimal strategies to incorporate these agents into upfront therapy for children with newly diagnosed MYC-amplified medulloblastoma. Specifically, further work is required to assess strategies to combine BET-bromodomain inhibition with other treatment modalities such as radiation therapy and chemotherapy. Specific predictors of sensitivity and resistance to JQ1 remain to be elucidated.
The anti-correlation between amplifications of MYC and MYCN, and the correlations between these amplifications (and amplifications of MYCL1) and expression of genes up-regulated by MYC, indicate that these amplifications are associated with increased MYC activity. However, some samples without MYC amplification had high expression of MYC or of other genes up-regulated by MYC. This observation suggests that other factors are also involved in regulation of MYC activation pathways, and that some tumors without MYC amplification may benefit from therapeutics targeting MYC, including BET-bromodomain inhibition.
Although we observed minimal responsiveness of the non-MYC amplified medulloblastoma cell lines to JQ1, it is possible that these lines may not represent the full spectrum of non-MYC-amplified medulloblastomas. For example, none of our non-MYC amplified lines included either the Wnt or SHH sub-types. We are unable to speculate whether these subtypes may also have sensitivity to treatment with BET-bromodomain inhibition.
Importantly, inhibition of MYC family members and activation pathways may result in down-regulation of other cell signaling pathways in tumors that do not harbor amplification of MYC isoforms. This is particularly relevant in the SHH subtype. Both MYC(37) and MYCN(38–40) have been reported to interact with and regulate transcription factors involved in SHH signaling, raising the possibility that inhibition of MYC activation may also result in inhibition of the SHH pathway, and other pathways important in the pathogenesis of medulloblastoma.
We show that MYC-amplified medulloblastomas can be targeted by epigenetic modulation of oncogenes. BRD4 is an epigenetic reader that binds to ε-N-lysine acetylation motifs(41). It is increasingly recognized that genomic alterations may result in global epigenetic dysregulation in pediatric brain tumors(42–44). Indeed, Group 3 medulloblastomas have been found to harbor somatic copy number alterations of genes involved in chromatin modeling(45, 46). These alterations frequently affect genes of modifiers of H3K27 methylation(46). Thus, it is possible that targeting other chromatin modifiers may also have activity in this group of tumors. Further work is required to define the interplay between methylation and acetylation of these histone marks to help determine whether the combination of different classes of epigenetic modulators may improve their efficacy.
We were unable to obtain any patient derived medulloblastoma cell lines that harbor amplifications of either MYCN or MYCL1. Our integrative analysis of copy number and expression profiles suggest that both isoforms are associated with increased expression of MYC activation pathways, suggesting that BET-bromodomain inhibition may be effective in tumors that harbor these amplifications. Indeed, BET-bromodomain inhibition has been shown to suppress MYCN expression in patient derived neuroblastoma cell lines (16). We have shown efficacy of JQ1 treatment in a cell line generated from a MYCN-driven mouse model, however ideally this should be validated in patient-derived MYCN-amplified medulloblastoma cell lines.
In summary, we show that BET bromodomain inhibition results in suppression of MYC activation pathways in MYC-driven medulloblastoma models, resulting in reduced cell viability, induction of G1 arrest and apoptosis with prolongation of survival in xenograft models. These data provide a rationale for early phase clinical trials for BET bromodomain inhibitors for children with this aggressive disease.
Supplementary Material
Statement of translational relevance.
Collectively, MYC, MYCN and MYCL1 are the most commonly amplified oncogenes in medulloblastoma, and are associated with a dismal prognosis. The recent development of strategies to block MYC activity through inhibition of BET bromodomain proteins represents a possible novel therapeutic strategy for these tumors. Here we report that JQ1, a potent inhibitor of BET bromodomain proteins, results in both reduced cell proliferation and prominent apoptosis using in vitro models of MYC-amplified medulloblastoma, and prolongs survival in xenograft models. We confirm effective down-regulation of MYC-related pathways with JQ1 and suppression of expression of MYC. We also show reduced cell proliferation with JQ1 treatment of cells derived from MCYN driven tumors harvested from a genetically engineered mouse model. BET bromodomain inhibition therefore represents a novel therapeutic strategy for children with MYC-amplified medulloblastoma. These data support further evaluation in early phase clinical trials.
ACKNOWLEDGEMENTS
We gratefully acknowledge the following funding sources: St. Baldrick’s Foundation Scholar Award (YJC), Beirne Faculty Scholar Endowment (YJC), NIH U01-CA176287 (YJC, WAW), Stanford Center for Children’s Brain Tumors (YJC, SC), Pediatric Low-Grade Astrocytoma Foundation (PB, GB, MWK, RB), Friends of DFCI (PB), the Sontag Foundation (RB), Gray Matters Foundation (RB), Men’s Collaborative for Women’s Cancers (JEB and RB), Stop&Shop Pediatric Brain Tumor Program (PB and MWK), the Mill Foundation for Kids (MWK), Damon-Runyon Cancer Research Foundation (JQ and JEB), Nuovo-Soldati Foundation (GB), Philippe Foundation (GB), R01-CA159859 (WAW, RWR), NIH R01-CA148699 (WAW), R01-CA133091(WAW), Swedish Childhood Cancer Foundation (SB, FJS), the Swedish Cancer Society (SB, FJS), the Swedish Research Council (SB, FJS), the Swedish Society of Medicine (SB, FJS), the Swedish Brain Foundation (SB, FJS), Åke Wiberg’s Foundation (SB, FJS), the Association for International Cancer Research (SB, FJS). RWR is the recipient of a Leadership Award (LA1-01747) from the California Institute of Regenerative Medicine. We would also like to thank Vida Shokoohi and John Coller in the Stanford Functional Genomics Core; Christopher DeHeer and Karen Krantz at NanoString Technologies, Inc.; Steve Avolicino at Histo-Tec Laboratory; and John Daley and Suzan Lazo-Kallanian in the DFCI Flow Cytometry Core Facility for their technical assistance. We also gratefully acknowledge Dr Scott Pomeroy and Dr Tenley Archer, Boston Children’s Hospital, for their assistance and sharing of reagents.
Footnotes
Conflict of Interest: The Dana-Farber Cancer Institute has licensed drug-like derivatives of JQ1 prepared in the Bradner laboratory to Tensha Therapeutics for clinical translation as cancer therapeutics. Dana-Farber and Dr. Bradner have been provided minority equity shares in Tensha. Dr Qi has a consultant agreement with Tensha Therapeutics.
References
- 1.Kieran MW, Walker D, Frappaz D, Prados M. Brain tumors: from childhood through adolescence into adulthood. J Clin Oncol. 2010;28:4783–4789. doi: 10.1200/JCO.2010.28.3481. [DOI] [PubMed] [Google Scholar]
- 2.Packer RJ, Gajjar A, Vezina G, Rorke-Adams L, Burger PC, Robertson PL, et al. Phase III study of craniospinal radiation therapy followed by adjuvant chemotherapy for newly diagnosed average-risk medulloblastoma. J Clin Oncol. 2006;24:4202–4208. doi: 10.1200/JCO.2006.06.4980. [DOI] [PubMed] [Google Scholar]
- 3.Mulhern RK, Palmer SL, Merchant TE, Wallace D, Kocak M, Brouwers P, et al. Neurocognitive consequences of risk-adapted therapy for childhood medulloblastoma. J Clin Oncol. 2005;23:5511–5519. doi: 10.1200/JCO.2005.00.703. [DOI] [PubMed] [Google Scholar]
- 4.Packer RJ, Zhou T, Holmes E, Vezina G, Gajjar A. Survival and secondary tumors in children with medulloblastoma receiving radiotherapy and adjuvant chemotherapy: results of Children's Oncology Group trial A9961. Neuro Oncol. 2013;15:97–103. doi: 10.1093/neuonc/nos267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123:473–484. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29:1424–1430. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;3:e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 10.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488:49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raeder MB, Birkeland E, Trovik J, Krakstad C, Shehata S, Schumacher S, et al. Integrated genomic analysis of the 8q24 amplification in endometrial cancers identifies ATAD2 as essential to MYC-dependent cancers. PLoS One. 2013;8:e54873. doi: 10.1371/journal.pone.0054873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filippakopoulos P, Knapp S. The bromodomain interaction module. FEBS Lett. 2012;586:2692–2704. doi: 10.1016/j.febslet.2012.04.045. [DOI] [PubMed] [Google Scholar]
- 16.Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3:308–323. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrmann H, Blatt K, Shi J, Gleixner KV, Cerny-Reiterer S, Mullauer L, et al. Small-molecule inhibition of BRD4 as a new potent approach to eliminate leukemic stem- and progenitor cells in acute myeloid leukemia AML. Oncotarget. 2012;3:1588–1599. doi: 10.18632/oncotarget.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pei Y, Moore CE, Wang J, Tewari AK, Eroshkin A, Cho YJ, et al. An animal model of MYC-driven medulloblastoma. Cancer Cell. 2012;21:155–167. doi: 10.1016/j.ccr.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swartling FJ, Savov V, Persson AI, Chen J, Hackett CS, Northcott PA, et al. Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer Cell. 2012;21:601–613. doi: 10.1016/j.ccr.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weeraratne SD, Amani V, Teider N, Pierre-Francois J, Winter D, Kye MJ, et al. Pleiotropic effects of miR-183~96~182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol. 2012;123:539–552. doi: 10.1007/s00401-012-0969-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-8-research0032. RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 25.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 26.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 29.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 30.Gould J, Getz G, Monti S, Reich M, Mesirov JP. Comparative gene marker selection suite. Bioinformatics. 2006;22:1924–1925. doi: 10.1093/bioinformatics/btl196. [DOI] [PubMed] [Google Scholar]
- 31.Ciriello G, Cerami E, Sander C, Schultz N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 2012;22:398–406. doi: 10.1101/gr.125567.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaske CJ, Benz SC, Sanborn JZ, Earl D, Szeto C, Zhu J, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26:i237–i245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandin F, Upfal E, Raphael BJ. Algorithms for detecting significantly mutated pathways in cancer. J Comput Biol. 2011;18:507–522. doi: 10.1089/cmb.2010.0265. [DOI] [PubMed] [Google Scholar]
- 34.Friedman HS, Burger PC, Bigner SH, Trojanowski JQ, Wikstrand CJ, Halperin EC, et al. Establishment and characterization of the human medulloblastoma cell line and transplantable xenograft D283 Med. J Neuropathol Exp Neurol. 1985;44:592–605. doi: 10.1097/00005072-198511000-00005. [DOI] [PubMed] [Google Scholar]
- 35.Aldosari N, Wiltshire RN, Dutra A, Schrock E, McLendon RE, Friedman HS, et al. Comprehensive molecular cytogenetic investigation of chromosomal abnormalities in human medulloblastoma cell lines and xenograft. Neuro Oncol. 2002;4:75–85. doi: 10.1093/neuonc/4.2.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swartling FJ, Grimmer MR, Hackett CS, Northcott PA, Fan QW, Goldenberg DD, et al. Pleiotropic role for MYCN in medulloblastoma. Genes Dev. 2010;24:1059–1072. doi: 10.1101/gad.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon JW, Gallant M, Lamm ML, Iannaccone S, Vieux KF, Proytcheva M, et al. Noncanonical Regulation of the Hedgehog Mediator GLI1 by c-MYC in Burkitt Lymphoma. Mol Cancer Res. 2013;11:604–615. doi: 10.1158/1541-7786.MCR-12-0441. [DOI] [PubMed] [Google Scholar]
- 38.Colvin Wanshura LE, Galvin KE, Ye H, Fernandez-Zapico ME, Wetmore C. Sequential activation of Snail1 and N-Myc modulates sonic hedgehog-induced transformation of neural cells. Cancer Res. 2011;71:5336–5345. doi: 10.1158/0008-5472.CAN-10-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kenney AM, Cole MD, Rowitch DH. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- 40.Oliver TG, Grasfeder LL, Carroll AL, Kaiser C, Gillingham CL, Lin SM, et al. Transcriptional profiling of the Sonic hedgehog response: a critical role for N-myc in proliferation of neuronal precursors. Proc Natl Acad Sci U S A. 2003;100:7331–7336. doi: 10.1073/pnas.0832317100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM, Bouffet E, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012;124:439–447. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 45.Jones DT, Northcott PA, Kool M, Pfister SM. The role of chromatin remodeling in medulloblastoma. Brain Pathol. 2013;23:193–199. doi: 10.1111/bpa.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dubuc AM, Remke M, Korshunov A, Northcott PA, Zhan SH, Mendez-Lago M, et al. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol. 2013;125:373–384. doi: 10.1007/s00401-012-1070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.