Abstract
The transformation of 4-hydroxyphenylpyruvate to homogentisate, catalyzed by 4-hydroxyphenylpyruvate dioxygenase (HPPD), plays an important role in degrading aromatic amino acids. As the reaction product homogentisate serves as aromatic precursor for prenylquinone synthesis in plants, the enzyme is an interesting target for herbicides. In this study we report the first x-ray structures of the plant HPPDs of Zea mays and Arabidopsis in their substrate-free form at 2.0 Å and 3.0 Å resolution, respectively. Previous biochemical characterizations have demonstrated that eukaryotic enzymes behave as homodimers in contrast to prokaryotic HPPDs, which are homotetramers. Plant and bacterial enzymes share the overall fold but use orthogonal surfaces for oligomerization. In addition, comparison of both structures provides direct evidence that the C-terminal helix gates substrate access to the active site around a nonheme ferrous iron center. In the Z. mays HPPD structure this helix packs into the active site, sequestering it completely from the solvent. In contrast, in the Arabidopsis structure this helix tilted by about 60° into the solvent and leaves the active site fully accessible. By elucidating the structure of plant HPPD enzymes we aim to provide a structural basis for the development of new herbicides.
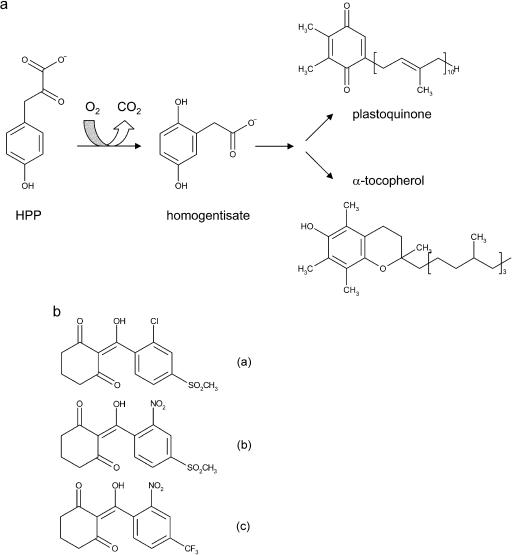
Dioxygenases play a key role in the degradation of aromatic compounds. 4-hydroxyphenylpyruvate dioxygenase (HPPD; EC 1.13.11.27) is an important enzyme in both Tyr and Phe catabolism of most organisms and in the biosynthesis of plastoquinones and tocopherols in plants that starts with the reaction product homogentisate. HPPD catalyzes a complex reaction involving oxidative decarboxylation of the 2-keto acid side chain of 4-hydroxyphenylpyruvate (HPP), accompanied by hydroxylation of the aromatic ring, 1,2-migration of the carboxymethyl group, and consumption of one molecule of dioxygen (Jefford and Cadby, 1981; Fig. 1A). Steady state kinetic experiments indicate an ordered bi bi mechanism in which HPP is the first substrate to bind and carbon dioxide is the first product to dissociate (Rundgren, 1977). Therefore, it seems that HPP is an activating effector for the reaction of HPPD with molecular oxygen, leading to formation of the first oxygenated intermediate (Johnson-Winters et al., 2003). Similar mechanisms have been proposed for other Fe2+-dependent oxygenases (Arciero et al., 1985; Harpel and Lipscomb, 1990; Brown et al., 1995; Shu et al., 1995; Roach et al., 1997; Solomon et al., 2000). The shift of the decarboxylated side chain is still poorly understood but the migration of smaller substituents is observed in other aromatic oxygenases (Sono et al., 1996) and may involve an arene oxide intermediate (Forbes and Hamilton, 1994).
Figure 1.
a, Reaction catalyzed by HPPD. b, Structures of HPPD inhibitors sulcotrione (a), mesotrione (b) and NTBC (c), which are based on a triketone backbone and may mimic an HPPD reaction coordinate transition state or intermediate.
HPPD belongs to the α-keto acid-dependent group of dioxygenases; however, in contrast to other enzymes of this class, HPPD, catalyzes the incorporation of both atoms of molecular oxygen into a single substrate. Moreover, the typical α-keto acid cosubstrate, α-ketoglutarate, such as in prolyl hydroxylase, cephalosporin synthase, and clavaminate synthase, forms part of the substrate HPP (Que and Ho, 1996; Solomon et al., 2000). Therefore, it is not surprising that the sequences of all HPPD genes available to date reveal only low homology to other α-keto acid dependent dioxygenases (Prescott, 1993). In addition, the HPPDs from prokaryotes and eukaryotes show a limited sequence identity of only about 30%, which is also reflected by their different oligomerization state. All studied mammalian HPPDs behave as homodimers of 43- to 49-kD subunits (Wada et al., 1975; Lindblad et al., 1977b; Roche et al., 1982; Endo et al., 1992; Rüetschi et al., 1993), whereas the Pseudomonas fluorescens enzyme is a homotetramer of 41-kD subunits (Lindstedt and Odelhög, 1987) as confirmed by structural analysis (Serre et al., 1999). Nevertheless, all HPPD enzymes purified and studied so far contain nonheme Fe2+ as an essential cofactor (Wada et al., 1975; Lindblad et al., 1977b; Roche et al., 1982; Endo et al., 1992; Rüetschi et al., 1993; Kim and Petersen, 2002; Johnson-Winters et al., 2003) with a mononuclear iron binding motif in the C-terminal half of the protein in which the Fe2+ ion is coordinated by two His and one carboxylic acid residues.
Due to its central role in the metabolism of aromatic amino acids in mammals and quinone synthesis in plants, the inhibition of HPPD has recently become the focus of considerable research interest. In plant tissues the HPPD reaction product homogentisate (2,5-dihydroxyphenylacetate) is the aromatic precursor of plastoquinones and tocopherols, two major classes of lipid-soluble quinone compounds in higher plant chloroplasts. The first is known for its role as an electron carrier between PSII and the cytochrome b6/f complex and as an electron carrier for NAD(P)H-plastoquinone oxidoreductase (Berger et al., 1993). Plastoquinone is also a cofactor for phytoene desaturase (Pallett et al., 1998), which is involved in the biosynthesis of photosynthetic pigments. Tocopherol functions as a membrane-associated antioxidant and structural component of membranes. Interestingly, in Synechocystis sp. PCC 6803—which is presumably related to the common plastid ancestor—HPPD is only required for the synthesis of tocopherols but not for the synthesis of plastoquinone as demonstrated by a gene-disruption experiment (Dähnhardt et al., 2002). The corresponding mutant experiments in Arabidopsis showed that HPPD is indispensable for the formation of both compounds (Norris et al., 1995). In nonphotosynthetic bacteria HPPD contributes to the degradation of Phe and Tyr (Durand and Zenk, 1974; Fernández-Cañón and Peñalva, 1995).
Defects of Tyr catabolism in humans range from relatively mild symptoms in type II tyrosinemia (Huhn et al., 1998; Phornphutkul et al., 2002) to fatal, if untreated, type I tyrosinemia (Russo and O'Regan, 1990; Russo et al., 2001). In type I tyrosinemia a dearth of active fumarylacetoacetase results in the accumulation of fumarylacetoacetate in the liver where it is either saturated or decarboxylated to succinylacetoacetone or succinylacetone, respectively (Lindblad et al., 1977a). They cause the early onset of liver cirrhosis and primary liver cancer as well as inhibition of heme synthesis at porphobiligen synthase, resulting in severe anemia (Kavana and Moran, 2003).
Different classes of HPPD inhibitors have been developed including sulcotrione, mesotrione, and 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione (NTBC) (Fig. 1) that are based on a triketone backbone and may mimic a reaction coordinate transition state or intermediate (Kavana and Moran, 2003). They are used both for clinical purposes and in agriculture as bleaching herbicides (Schulz et al., 1993). The bleaching effect in plants is associated with an accumulation of the carotenoid precursor phytoene explained by an indirect inhibition of phytoene desaturase that results from the absence of plastoquinone, an essential cofactor for the desaturase (Schulz et al., 1993; Norris et al., 1995). As a result, the photosynthetic apparatus can no longer be stabilized by the carotenoid level and chlorophyll molecules are destroyed by excessive light energy.
To date, HPPDs of only a few plant sources have been sequenced and partially cloned, including the enzymes of Coleus blumei, Hordeum vulgare, Daucus carota, and Arabidopsis (Bartley et al., 1997; Garcia et al., 1997; Kim and Petersen, 2002). The only structural information available is from the distantly related tetrameric enzyme from the chemo-organotrophic aerobic gram-negative prokaryote P. fluorescens (Serre et al., 1999).
Here, we present the first crystal structures of plant HPPDs from Zea mays and Arabidopsis at a resolution of 2.0 and 3.0 Å, respectively, which reveal the dimeric architecture of the plant enzymes. As an enzyme from an anabolic pathway, HPPD is important for plant growth and therefore an interesting target for the development of new inhibitors used as herbicides.
RESULTS AND DISCUSSION
Cloning and Expression of Plant HPPDs
Plant HPPD enzymes differ significantly at their N termini from both the bacterial and mammalian enzymes as they have extensions of at least 30 amino acids. These residues may serve as subcellular targeting signals, e.g. chloroplast import signal as HPPD could contribute to both anabolic synthesis of prenylquinones in chloroplasts and to catabolism of aromatic amino acids in the cytosol (Fiedler et al., 1982). The subcellular localization and the existence of isoforms of HPPD in the plant cell have not been investigated extensively. For cultured carrot cells it was shown that HPPD activity is restricted to the cytosol (Lenne et al., 1995; Garcia et al., 1997). Meanwhile, Norris et al. (1998) analyzed a 17-bp deletion in the single copy HPPD gene of Arabidopsis (Garcia et al., 1999), which causes a complete loss of enzyme activity in the cytosol, indicating the contribution of the cytosolic enzyme to both degradation of amino acids and prenylquinone synthesis.
In order to determine the shortest protein construct for crystallography we purified the native protein from Z. mays seedlings grown for 6 d. Protein sequencing yielded Ala-18 as native N terminus. In contrast, the first codon of the cloned cDNA encoded Phe-34 obtained with primer sequences based on the consensus sequence of Oryza sativa expressed sequence tags, H. vulgare, D. carota, and Arabidopsis (Linden, 2000). The Z. mays HPPD variant starting with Phe-34 yielded only insoluble protein; therefore, we added residues Ala-18 to Asn-33 according to the sequence of Maxwell et al. (1997). Soluble expression of the resulting protein was improved but still irregular. However, it could be significantly improved by lowering the temperature of expression to 20°C and the pH to about 5.0 prior to induction. The recombinant Z. mays HPPD protein used in our studies comprised amino acids Ala-18 to Glu-435 preceded by the sequence H6SSGLVPRGSHM originating from the cloning. The His-tag was not removed for crystallization.
To clone Arabidopsis HPPD, total mRNA was isolated from seedlings. Primer sequences for cDNA synthesis were deduced from the deposited gene sequence of Bartley (1997). The amplified DNA was cloned in pET14b. Arabidopsis HPPD could be expressed as soluble protein in Escherichia coli under less stringent conditions than Z. mays HPPD. The N-terminal His-tag was cleaved off for structural studies and the recombinant protein comprised residues Gly-2 to Gly-445 preceded by Gly-Ser-His-Met from the cleavage site.
Structure Analysis and Quality of the Models
HPPDs from Z. mays and Arabidopsis have been crystallized in the orthorhombic space group P212121 and the tetragonal space group P41212, respectively. Crystals diffracted to 2.0 Å and 3.0 Å, respectively (Table I). The structure of Z. mays HPPD has been solved by single isomorphous replacement and 4-fold noncrystallographic symmetry averaging and refined at 2.0 Å resolution (Table I). The model comprises residues Phe-37 to Leu-431 for all four asymmetric monomers, one iron atom per monomer, and 847 solvent molecules. In addition, monomer A shows density for Arg-36, monomers B and C also for Phe-34 at the N terminus, whereas Glu-432 is visible in monomer B, leaving the C-terminal residues Ala-433 to Gln-435 disordered in all monomers. No electron density could be found for the N-terminal residues Ala-18 to Asn-33 and residues Glu-249 to Ser-256 in all four monomers, indicating their high flexibility. Residues Lys-397 to Glu-403 located in the insertion including β-strands F and G have weak electron density in all monomers.
Table I.
Summary of data collection and atomic model refinement statistics
| Data Set | Arabidopsis HPPD | Z. mays HPPD (Native) | Z. mays HPPD (Mercury) |
|---|---|---|---|
| Space group | P41212 | P212121 | P212121 |
| Unit cell dimensions | |||
| a/b/c (Å) | 95.2/95.2/185.7 | 89.0/110.9/174.8 | 89.0/111.3/174.6 |
| Resolution limits (Å) | 16.83–3.0 | 19.84–2.0 | 34.1–3.0 |
| Reflections, unique | 16,577 | 115,602 | 33,799 |
| Reflections, measured | 67,542 | 377,176 | 136,536 |
| Completeness, overall (%) | 82.0 | 98.6 | 95.6 |
| Completeness, last shell (%) | 76.4 | 96.2 | 88.9 |
| Rmerge (%) | 11.2 | 8.2 | 11.2 |
| Rmerge (%), last shell | 42.3 | 49.3 | 24.4 |
| No. of residues | 803 | 2,440 | |
| No. of nonhydrogen protein atoms | 5,743 | 11,813 | |
| No. of solvent molecules | 0 | 847 | |
| No. of metal ions | 2 | 4 | |
| R-value (%)/Rfree (%) | 23.7/31.2 | 27.5/32.4 | |
| rms deviation | |||
| Bond length (Å) | 0.008 | 0.010 | |
| Bond angles (°) | 1.43 | 1.81 | |
| between monomers/dimers in the asymmetric unit | 0.60 | 0.26 | |
| B-Factors (Å2) | |||
| From Wilson scaling | 53.6 | 30.3 | |
| Overall anisotropic B-factor (B11, B22, B33) | 5.3, 5.3, −10.7 | 14.2, −23.3, 9.1 | |
| Main chain | 21.84 | 40.03 | |
| Side chain | 21.82 | 41.29 | |
| Solvent | - | 48.91 | |
| Metal | 28.16 | 60.53 | |
| Number of sites | - | - | 6 |
| Phasing power (acentric/centric) | - | - | 1.04/0.78 |
| Isomorphous difference (%) | - | - | 24.1 |
| Mean figure of merit | - | - | 0.23 |
Rmerge = ∑(I−〈I〉)/∑I. R = ∑(|Fobs| − |Fcalc|)/∑|Fobs|.
Rfree was calculated from 5% of measured reflections omitted from refinement.
The structure of HPPD from Arabidopsis was solved by the molecular replacement method using the physiological dimer of the Z. mays structure and refined at 3.0 Å resolution. The model of Arabidopsis HPPD is composed of residues Val-33 to Phe-428 in monomer A and Val-33 to Thr-437 in monomer B. The iron ions were clearly visible in the Fo-Fc difference electron density map. There was no electron density for residues Gly-2 to Phe-32 and residues located in loop structures including Leu-107 to Thr-116, Ala-194 to Glu-201, Asp-211 to Phe-215 in both molecules, for residues Glu-252 to Glu-262 in monomer A, and Ala-255 to Glu-262 in molecule B.
The Ramachandran plot (Ramachandran and Sasisekharan, 1968) of Z. mays HPPD shows 89.1% of the residues in the most favored regions and 10.5% in the additional allowed regions; no residues are found in the disallowed regions. The Arabidopsis HPPD model has acceptable geometry for a 3.0 Å structure with 81.7% in the most favored, 17.7% in additional allowed, and less 0.7% of the amino acid residues in generously allowed regions but again none in disallowed regions of the Ramachandran plot.
Overall Fold of Plant HPPD
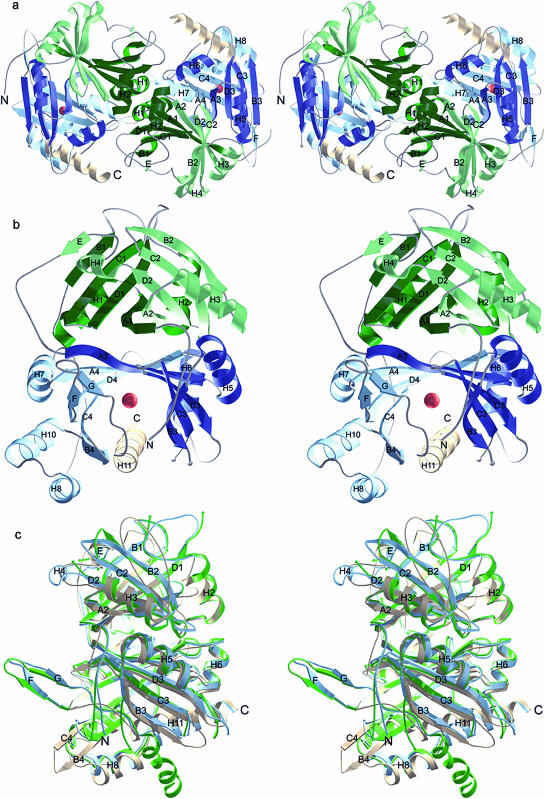
The asymmetric unit of the Z. mays HPPD crystals of space group P212121 contains two dimers of 44.8-kD subunits (Fig. 2A). Crystals of Arabidopsis HPPD of space group P41212 contain one dimer per asymmetric unit. As both the Z. mays and Arabidopsis dimers show an identical dimerization mode we conclude that these dimers also reflect the physiological oligomeric state present in solution. The inspection of lattice contacts indicates no formation of higher oligomers in the crystals. For the Arabidopsis enzyme gel permeation chromatography and dynamic light scattering showed a Mr of 90 kD and 102 kD, respectively, in accordance with a calculated Mr of 97.6 kD (Linden, 2000). These findings confirm the result of the previous characterization of the oligomeric state of eukaryotic HPPD enzymes (Wada et al., 1975; Lindblad et al., 1977b; Roche et al., 1982; Endo et al., 1992; Rüetschi et al., 1993; Lenne et al., 1995; Garcia et al., 1997). The two dimers found in the asymmetric unit of Z. mays HPPD crystals are virtually identical (root mean square [rms] deviations for 769 Cα positions = 0.26 Å), whereas there are some minor differences between molecules of Arabidopsis HPPD dimer, which have rms deviations of 0.60 Å for 359 Cα atoms. The rms difference between the dimers of Z. mays and Arabidopsis that share 66% identical residues is approximately 1.0 Å for 695 main chain atoms, which reflects also the similarity in dimer formation for both plant enzymes.
Figure 2.
Structure of HPPD. a, Ribbon representation of the Z. mays HPPD dimer. The view is down the 2-fold axis. b, The Z. mays HPPD monomer shows two open β-barrel domains (green and blue), which are both built up by two similar modules (light and dark colors). The catalytic iron atom in the C-terminal domain is shown as a red sphere. c, Superimposition of HPPD monomers of Z. mays (blue), Arabidopsis (green), and P. fluorescens (light brown).
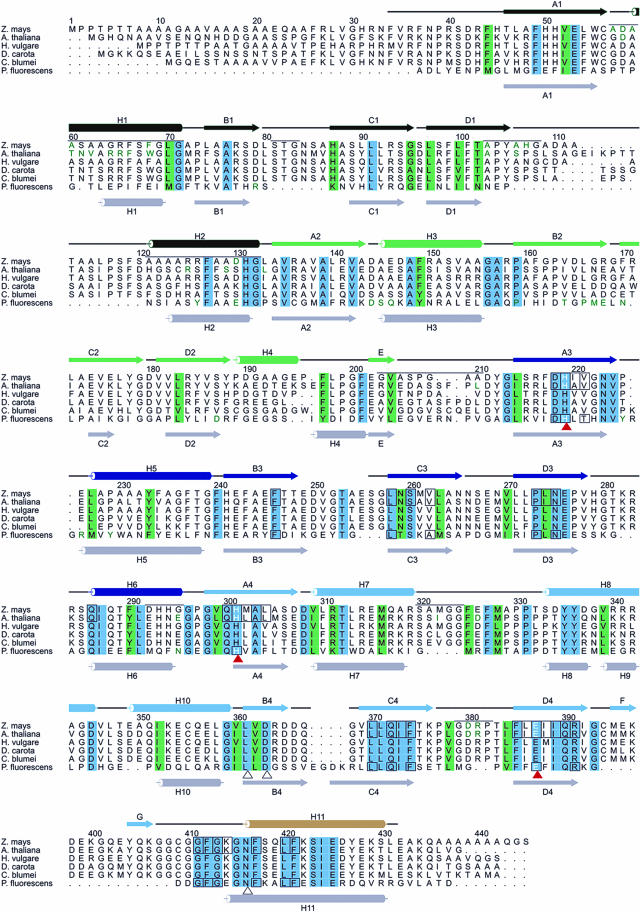
As both plant structures are quite similar to each other we will confine the description to the Z. mays HPPD. Figure 2 shows ribbon diagrams of the Z. mays monomer and dimer, and a structure based sequence alignment and assignment of secondary structure elements of the plant and bacterial enzymes is provided in Figure 3.
Figure 3.
The alignment of different plant HPPD sequences is structure based on the sequences of Z. mays, Arabidopsis, and P. fluorescens. Numbering is given for Z. mays HPPD according to Maxwell et al., (1997). Secondary structures are color coded like in Figure 2 and were numbered according to Serre et al. (1999). The secondary structure assignment of P. fluorescens HPPD is depicted in light gray. Black lines indicate loop regions, whereas disordered residues of Z. mays HPPD are indicated without the corresponding lines. Structural elements mediating dimerization are marked by a black line above the Z. mays sequence with residues directly involved in dimer contacts shown in green. Amino acids highlighted in blue are invariant, conservative exchanges are depicted in light-green. The iron coordinating residues are marked by red triangles. Active site residues are boxed. The residues building the rigid cavity of the articular are marked by a black triangle.
The structures reveal that the monomers of both enzymes are folded into two structural domains that are arranged as an N- and C-terminal open β-barrel of eight β-strands each. The N-terminal domain (residues Arg-36 to Asp-211 in Z. mays and residues Val-33 to Asp-218 in Arabidopsis) has apparently no direct catalytic function, whereas the C-terminal domain (Tyr-212 to Glu-432 in Z. mays and Tyr-219 to Thr-437 in Arabidopsis) harbors the iron binding site (His-219, His-301, and Glu-387) in the center of the β-barrel, the surrounding catalytic residues, and the α-helix H11 that probably functions as a gate controlling substrate access to the active site.
Both barrels pack against each other related by a pseudo-2-fold axis and form an extended β-sandwich in the center of the monomer. This core is flanked by 10 α-helices in total whereby α-helices H2 and H11 partly pack into the groves formed by the exposed sides of the open β-barrels. Each β-barrel is composed of two topologically similar modules that comprise four β-strands in the order A-D-C-B with an up-up-down-up topology. Both modules form an antiparallel β-sheet between strands A, which results in the overall structure of each domain as an eight-stranded β-barrel. β-strands A and B of each module are connected by an α-helix (H1, H3, H5, and H7, respectively) that pack against the exposed side of the module. Modules 1 and 2 are connected by α-helix H2. The link between modules 2 and 3, that is also the link between both domains, is formed by a sequence stretch that contacts the first module of the N-terminal domain and includes a rather irregular helix H4 and short β-strand E that runs antiparallel to β-strand B1 on the outer edge of module 1. Modules 3 and 4 are connected by α-helix H6. The C-terminal module 4 shows larger deviations from this general scheme by the insertion of α-helices H8 and H10 preceding β-strand B4 and a relatively well ordered two-stranded β-sheet (β-strands F and G) that point into the solvent without contacts to the remaining molecule. The comparison of all four modules shows a topology of βαβββα with larger variations at the second α-helix which is not surprising as it contains the module-connecting sequences. Despite the mechanistic relation to α-keto acid dependent dioxygenases, HPPD shares these structural features with one class of extradiol ring-cleaving dioxygenases that includes catechol-2,3-dioxygenase (Kita et al., 1999).
Comparison of Plant and Bacterial HPPD
The monomer of the bacterial HPPD from P. fluorescens shares the overall fold with plant HPPDs. The superposition of the Cα atoms of the HPPD monomers of Z. mays and P. fluorescens and Arabidopsis and P. fluorescens result in higher rms deviations of about 1.5 Å in both cases for 264 and 242 atoms, respectively (Fig. 2C). This reflects not only the lower sequence identity of about 30% between plant and prokaryotic enzyme but also some prominent structural deviations.
These deviations mainly relate to the different oligomerization modes (see below) and include two prominent insertions into the N-terminal domain from Ser-81 to Ala-86 and Ala-106 to Ser-120 of six and 15 residues, respectively. Interestingly, the Arabidopsis HPPD has a longer insertion of 21 residues in total at the latter position. However, the added residues in the Z. mays sequence that are located at an outer edge of the dimer contact are partly disordered. This large insertion induces a drastic shift of β-strands B2 and C2 with the largest effect in the connecting turn. This region is involved in an important contact in the P. fluorescens tetramer. The second surface participating in bacterial tetramerization shows smaller deviations in the backbone orientation but nevertheless shows the deletion of two residues after Pro-225 and an insertion of two residues after Thr-281. Therefore, not only does the surface involved in the formation of the plant dimer deviate significantly from the bacterial enzyme but also those responsible for bacterial tetramer formation are altered significantly. In addition, the region connecting both domains (Gly-203 to Ala-210) has a completely different backbone trace including a deletion of two residues that shifts it toward the dimer contact.
α-Helix H4 in the N-terminal domain of P. fluorescens (Serre et al., 1999) has only a rather distorted counterpart in the plant structures. The hairpin-loop formed from the short β-sheet between β-strands F and G of the plant enzymes in the C-terminal domain points out into the solvent representing an insertion of 15 amino acids with respect to the P. fluorescens enzyme that is unique to the plant enzymes. It is stabilized by a disulphide bond between the conserved cysteines Cys-394 and Cys-409 at its base.
Dimer Formation
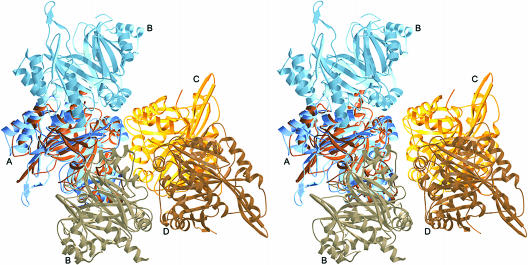
In contrast to the dimeric plant enzymes, the bacterial P. fluorescens HPPD behaves as a tetramer of 222 point symmetry with the shape of a flat parallelepiped and tetramerization is dominated by N terminal-N terminal contacts with C-terminal domains interacting only with one neighboring domain (Serre et al., 1999). This arrangement contains three potential dimers but only two bury an extensive portion of the molecular surface. Interestingly, tetramerization involves only loops connecting secondary structural elements and mainly nonconserved residues including Tyr-167 that forms a prominent stacking interaction between monomers, which is a Pro in mammalian and most plant HPPDs.
Surprisingly, the plant HPPDs dimerize in a completely different mode using an orthogonal molecular surface (Figs. 2A and 4). Dimer formation buries an average of 1,355 Å2 in Arabidopsis HPPD and 1,544 Å2 in Z. mays HPPD in each monomer and involves both domains compared to 2,600 Å2 in P. fluorescens HPPD. However, the majority of contributions come from N-terminal-N-terminal domain interactions and no contacts are formed between C-terminal domains of neighboring subunits. These involve secondary structural elements (Ala-57 to Gly-71) including α-helix H1 in the center of the dimer where it packs against the neighboring α-helix H1 with both helix axes tilted by about 20° with respect to the 2-fold axis. In addition, α-helix H2 (Ala-121 to Gly-132) packs against H2 perpendicular to the 2-fold axis. Other parts of the N-terminal domain involve the loops including Ser-81 to Gly-83 and Ala-103 to Gly-108. The latter region serves as abutment of the C terminus of the neighboring α-helix H11. The C-terminal domain makes a smaller contribution to the dimer involving only secondary structure connecting elements. The most prominent are the loop regions including Ser-320 to Gly-324 and Gly-379 to Pro-382 which contact parts of the neighboring α-helix H1. The loop around Asp-292 to Gly-295, like that around Ser-320 to Gly-324, also contacts the region around Ala-103 to Gly-108.
Figure 4.
Quaternary structure of bacterial and plant HPPDs. The Z. mays HPPD dimer (blue) uses an orthogonal molecular surface compared to the dimer contacts present in the P. fluorescens tetramer (brown to orange) of 222 point symmetry.
It should be noted that the largest differences between bacterial and plant enzymes are associated with dimer formation. They affect the N-terminal half of helix H1, the region including Asp-79 to His-87 and Ala-103 to Gly-108 that are part of insertions and the region around Gly-202 to Ala-210 that has a strongly deviating backbone orientation (Fig. 2C). Differences in the C-terminal domain involve insertion from Ala-321 to Gly-324 and around Asp-380. As a consequence of the large insertion from Tyr-105 to Ser-120, the β-sheet between has to shift up to 6.3 Å for Gly-169 compared to the topologically corresponding Met-109 in P. fluorescens.
Intersubunit contacts are primarily hydrophilic and involve the side chains of Asp-58, Arg-65, Tyr-105, Asp-380, and Arg-381 that are strictly conserved among plant HPPDs and the nonconserved residues Ser-61, which can also be asparagines, and His-107, which can also be Asn or Pro. A prominent hydrophobic stacking interaction occurs between Phe-68 and its symmetry mate corresponding to Trp-66 in Arabidopsis HPPD (Glu-31 in P. fluorescens). An aromatic residue at that position is strictly conserved among plant HPPDs. This contact bears some resemblance to the stacking interaction between Tyr-167 in P. fluorescens HPPD and its symmetry mate. In addition, Met-322 at the edge of the contact surface packs into a small hydrophobic pocket formed from Pro-73 and Ala-210 of the neighboring molecule that are not well conserved. Taken together, the high homology in the dimer-forming regions and the strict conservation of key residues involved in dimer formation further support the view that the dimerization mode observed for the Z. mays and Arabidopsis HPPD is common to all plant HPPDs. The only major difference between both plant sequences is an insertion of six residues after Ala-112 that does not influence dimerization.
Gating to the Active Site
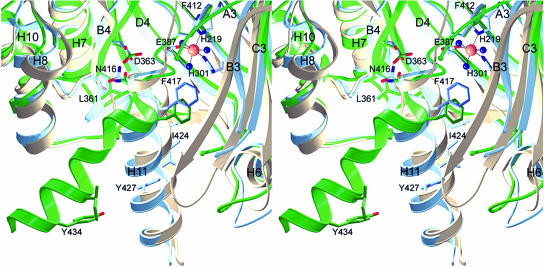
The C-terminal α-helix H11 sequesters the active site barrel completely from the surrounding solvent. Based on the structure of P. fluorescens HPPD it has been postulated that α-helix H11 has to move out of the active site to allow substrate entrance. The present structure analyses provide four copies of the Z. mays HPPD and two copies of the Arabidopsis HPPD. Whereas the C-terminal helices block the active site entrance in both dimers of Z. mays HPPD, this helix is tilted out of the cleft in the Arabidopsis structure of monomer B, leaving the substrate entrance widely open. A symmetry related molecule is found in the vicinity of the C-terminal helix. There are three contacts between the side chains of Ile-431 and Glu-150′, Tyr-434 and Ser-154′, and Thr-437 and Ala-158′ at a distance of 4.2, 3.5, and 3.6 Å, respectively. Taking the small number and the rather weak nature of these interactions into account, we conclude that the C-terminal helix is not locked in an open state by crystal packing. However, it cannot be ruled out that the open state is composed of additional conformations that are accessible to the C-terminal helix. In Arabidopsis monomer A the helix is completely disordered.
This movement can be best described as swinging out of α-helix H11 by about 60° around the Cα position of Asn-416 that functions as a rigid pivot point. The side chain of Asn-416 is fixed between the side chains of Leu-361 and Asp-363 of β-strand B4 and points orthogonal onto the corresponding β-sheet, a highly conserved region that can be compared to the rigid cavity of an articular. The rotation around the Cα position of Asn-416 results in a shift of the entire following α-helix H11, starting with Phe-417, as its secondary structure is essentially unaltered. On one side of the active site entrance α-helix H8 and the preceding residue form a rigid boundary against which the α-helix H11 can back both in the open (Thr-332 to Gly-338) or closed (Met-328 to Pro-331) state (Fig. 5). At the other side the entrance is lined by a more flexible part of the structure comprising the turn region connecting β-strands B3 and C3, which includes the disordered region from Glu-249 to Glu-255. In P. fluorescens HPPD the latter region is shorter by three residues (Fig. 3), completely ordered, and embraces the C-terminal α-helix H11, fixing it in the active site cleft. It should be noted that the hinge around Asn-416 and the articular cavity around Leu-361 and Asp-363 are strictly conserved in all HPPDs (Fig. 3). Therefore, we conclude that the gating mechanism revealed by the plant structures is universal to all HPPDs.
Figure 5.
Gating to the active site. Z. mays HPPD is shown in blue, Arabidopsis in green, and P. fluorescens in light brown. The highly conserved Asn-416 forms the hinge region around which the C-terminal α-helix H11 rotates to open the active site, whereas Leu-361 and Asp-363 function as rigid articular cavity. The region around α-helix H8 remains rigid, whereas the loop connecting β-strands B3 and C3 is flexible and presumably adopts to the movement of the gating α-helix H11.
The region preceding the hinge from Cys-409 to Phe-412 is almost identical in both plant enzymes but deviates slightly from the backbone position in the P. fluorescens structure, probably influenced by the insertion of the FG hairpin-loop in the plant enzymes. Nevertheless, this region is important as it is located in direct vicinity of the iron center including a hydrogen bond (2.9–3.1 Å) between the backbone carbonyl of Phe-412 and a water ligand of the iron atom. Although the metal ligands are not resolved at 3.0 Å in the Arabidopsis structure, the almost unaltered position of the corresponding Phe-419 suggests that a similar interaction with the coordination sphere of iron is maintained also in the open conformation. In the closed conformation α-helix H11 contributes only hydrophobic side chains (Phe-412, Phe-417, Leu-420, Ile-424, and Tyr-427) to the active site compartment.
Active Site Architecture
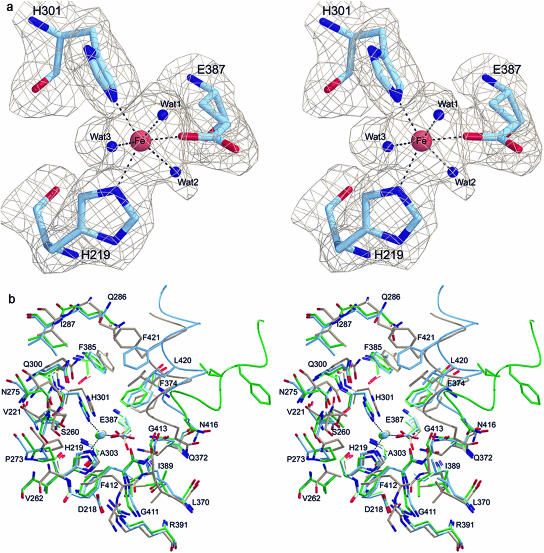
The active site of HPPD is located in the C-terminal domain formed by an eight-stranded highly twisted half-open β-barrel that surrounds a pronounced cavity that is between 8 and 14 Å in width. In the active form of the enzyme an Fe2+-ion is bound in the center of this cavity to three strictly conserved residues, His-219, His-301, and Glu-387, located on β-strands A3, A4, and D4, respectively (Fig. 6A). The temperature factors for all four iron atoms of Z. mays HPPD are between 40 and 76 Å2, which matches for only one monomer quite well with the ligand atoms, suggesting that the iron sites are not fully occupied in the other three subunits. The Arabidopsis enzyme shows temperature factors of the iron atoms of 24 and 32 Å2, which matches quite well with the surrounding protein atoms and suggests a high occupancy. Oxidation of Fe2+ under aerobic conditions during purification and crystallization cannot be excluded and we have no experimental evidence on the oxidation state of both plant HPPD enzymes, although we find that storage under aerobic conditions does not inactivate the enzymes. However, electron paramagnetic resonance spectroscopy demonstrated a high spin Fe3+ for the P. fluorescens HPPD (Serre et al., 1999); furthermore, Johnson-Winters et al. (2003) showed that the holoenzyme from Streptomyces avermitilis, both in complex with HPP and in its substrate free form, reacts with molecular oxygen. Likewise, the 2,3-dihydroxybiphenyl-1,2-dioxygenase (DHBD) structure was originally solved with Fe3+ as an inactive enzyme that could be reactivated with Fe2+ and ascorbate prior the anaerobic crystallization that allowed the characterization of dioxygen and product complexes (Uragami et al., 2001).
Figure 6.
Active site architecture. a, Stereo picture showing the 2Fo-Fc electron density at the 1σ level at the active site of Z. mays HPPD. The active site iron shows an octahedral coordination sphere of three amino acid ligands and three water molecules. b, Superimposition of the active site structures of Z. mays (blue), Arabidopsis (green), and P. fluorescens (light brown) HPPD. The view includes residues approximately 14 Å around the catalytically active iron atom. Ile-220, Leu-274, Met-302, Leu-304, Ile-373, Leu-386, Ile-388, and Lys-414 in the front were omitted for clarity. The C-terminal gating helices are shown as coils.
The structure of Z. mays HPPD, analyzed at 2.0 Å, clearly shows an octahedral coordination sphere that is comprised of three protein ligand atoms and three water molecules. The ligand sphere of the Arabidopsis HPPD was not resolved at 3.0 Å; however, there was clearly additional electron density at the iron atom that could not be satisfactorily explained by a single water molecule. The active site geometry of HPPD is very similar to that of the structurally related extradiol-cleaving catechol dioxygenase metapyrocatechase (Kita et al., 1999) and the biphenyl-cleaving extradiol dioxygenase DHBD (Han et al., 1995; Senda et al., 1996) although the latter enzymes have open active sites without a gating mechanism. All three enzymes share two His and a Glu residue as iron ligands. The previous structural analysis of the bacterial P. fluorescens HPPD solved at 2.4 Å demonstrated a larger ligand density that was interpreted as a monodentate acetate; metapyrocatechase showed a bound acetone molecule and DHBD a t-butanol, all of which originate from crystallization additives. Therefore, the coordination sphere of iron in these enzymes is described as a distorted tetrahedron or square pyramid as in DHBD. Interestingly, the acetone and t-butanol ligands are not close enough to the iron to serve as direct metal ion ligands. The substrate complex of P. fluorescens HPPD was modeled (Serre et al., 1999) based on the 2,3-dihydroxybiphenyl complex with oxidized inactive DHBD (Senda et al., 1996). The coordination sphere is likewise octahedral for the end-on dioxygen complex of BphC (Uragami et al., 2001). Similar to the Z. mays HPPD, other α-keto acid dependent oxygenases like cephalosporin synthase and clavaminate synthase, which split their cosubstrate 2-oxoglutarate oxidatively, show a coordination sphere of the Fe2+ ion in an almost perfect octahedral geometry, with two His residues, one acidic residue, and three solvent molecules (Valegard et al., 1998; Zhang et al., 2000).
The iron center of HPPD is surrounded by an almost strictly conserved environment (Fig. 6B) that is clearly dominated by 21 hydrophobic residues in comparison to four hydrophilic and only three charged residues. The polar side chains within a radius of 5 to 8 Å around the iron are Gln-372, Asn-275, and Gln-300. With the exception of Phe-412 on the loop between β-strand G and helix H11 and Leu-420, Ile-424, and Phe-421 on the C-terminal α-helix H11, all residues are found in rigid secondary structure elements and nearly all of them are strictly conserved among all known HPPD sequences.
CONCLUSION
The crystal structures of Z. mays and Arabidopsis HPPD enzymes presented here provide for the first time to our knowledge structural insight into the plant representatives of these ubiquitous enzymes. As expected from the sequence homology, especially in the C-terminal half, the monomers of plant and bacterial enzymes share the overall three-dimensional fold but differ in their oligomeric state. We could demonstrate that the plant and the bacterial enzymes use nonoverlapping, orthogonal parts of the monomer surface for dimer and tetramer formation, respectively. Prominent insertions into the plant enzymes in the N-terminal domain that were not clear from previous sequence alignments are involved in the dimer contacts. In addition, the structures provide direct evidence for the substrate gating to the active site by the C-terminal α-helix H11 and reveal its hinge-like rotation around Asn-416. Despite the dissimilar quaternary structures of plant and bacterial enzymes their active site architecture is remarkably well conserved, which very likely reflects steric requirements for catalysis in the multi-step reaction. Clearly, ligand binding studies will be necessary to elucidate details of the enzyme mechanism.
MATERIALS AND METHODS
Cloning
Total Zea mays RNA was prepared from ethiolated seedlings grown for 6 d by using the RNeasy Mini Kit (Qiagen, Hilden, Germany). The first strand of the cDNA was obtained by reverse transcription with displayTHERMO-RT (Display Systems Biotech, Vista, CA) and an oligo(dT) primer according to the instructions of the manufacturer. Because of the high content of predicted secondary structure elements in the mRNA of Z. mays HPPD the reverse transcription reaction had to be performed with a thermostable reverse transcriptase. The HPPD gene coding for amino acids Phe-34 to Gln-435 was amplified by modified PCR introducing NdeI and BamHI restriction sites. The 5′-primer (5′-GGAATTCCTATGGCATCAGCAGCGG-3′) was deduced from the N terminus of native purified Z. mays HPPD (Linden, 2000), and the 3′ primer (5′-GCGGATCCTTATTGCTTGGCTTCAAGG-3′) was synthesized based on the C-terminal consensus sequence of plant HPPD from Daucus carota, Hordeum vulgare, and Arabidopsis. The amplification was carried out in a 50-μL reaction volume using 2.5 μL cDNA, 1 μm forward and reverse primer, respectively, containing 5 μL Q-solution (Qiagen), 1mm dNTPs, 5 μL Pwo polymerase buffer, and 2.5 units Pwo DNA polymerase (Roche, Mannheim, Germany). The PCR was started with an initial denaturation step at 94°C for 3 min, followed by 35 cycles (at 94°C for 45 s, 50°C for 60 s, and 72°C for 10 min) and a final extension step at 72°C for 7 min. The purified PCR product was first cloned into the pCR 2.1-TOPO vector (Invitrogen, Groningen, Netherlands) and for protein expression recloned into vector pET14b (Novagen, Madison, WI). This construct resulted in the expression of insoluble protein. Therefore, the gene was extended at the N terminus by 16 amino acids (Ala-18 to Asn-33) using the primer (5′-GGAATTCCATATGGCATCAGCAGCGGAGCAAGCGGC-3′) whose sequence was deduced from the native purified protein, affirmed by the published sequence (Maxwell et al., 1997) and recloned into the pET14b vector.
Arabidopsis mRNA was isolated from seedlings, the cDNA synthesized by reverse transcription, and the gene amplified via PCR in a similar way. Primer sequences (5′-GGAATTCCTATGGGCCACCAAA-3′ and 5′GCGGATCCTTATTGCTTGGCTTCA-AGG-3′) were deduced from the known cDNA sequence (GenBank accession no. U89267; Bartley et al., 1997). For protein expression the gene was also cloned in the vector pET14b. The amino acid sequences corresponding to the cloned genes are given in Figure 3.
Expression, Purification, and Crystallization
Z. mays and Arabidopsis HPPDs were expressed in BL21(DE3) E. coli cells. All cultures were grown in the presence of 100 μg mL−1 carbenicillin. Isolated plasmid DNA of individual colonies was restricted and analyzed on SDS-PAGE to identify clones carrying the HPPD gene. Positive clones were used to inoculate 50 mL of Luria-Bertani (LB) broth. Successful expression of Z. mays HPPD could be monitored by a red-brownish coloration of the medium even without induction. The precultures were used to inoculate 1 L of LB broth and grown with vigorous shaking at 37°C to an OD600 of 0.6 to 0.8. Isopropylthio-β-galactoside was added to a final concentration of 1 mm and incubation was continued for 16 h.
The gain of soluble Z. mays HPPD could be increased by direct inoculation of the preculture immediately after transformation without spreading on LB plates. The precultures were used to inoculate 1 L LB broth. When bacterial growth was equivalent to an OD600 of 2.0, the pH was lowered to 5.0 with 37% HCl, the medium cooled down to 20°C, and α-lactose was added to a final concentration of 0.75% (w/v) to induce the protein expression. The incubation was continued for 24 h at room temperature.
Cells were harvested by centrifugation at rcf = 5,005g* at 4°C for 20 min. The pellets were resuspended in 50 mm KPi, pH 8.0 and 500 mm NaCl (Arabidopsis HPPD) and 50 mm Tris-HCl, pH 8.0 (Z. mays HPPD), respectively, and the cells were disrupted by sonification (Branson, Heinemann, Schwäbisch Gmünd, Germany). The crude extracts were centrifuged at rcf = 75,398g* for 20 min to yield a cell-free supernatant.
Arabidopsis HPPD was purified in two chromatographic steps. The supernatant was loaded onto a Co2+ Talon metal chelating column (Clontech, Palo Alto, CA), equilibrated with cell disruption buffer. Arabidopsis HPPD was eluted with 100 mm HEPES, pH 7.0, 200 mm NaCl, and 250 mm imidazole. The fractions containing HPPD were pooled, concentrated, and the buffer exchanged for 10 mm HEPES, pH 7.0 by ultrafiltration in Ultrafree filter devices (Millipore, Bedford, MA). The N-terminal His-tag was cut by incubation with thrombin (Novagen, Madison, WI) for 16 h at 4°C (0.5 unit thrombin/mg of protein). To remove thrombin and His-tag an anion exchange chromatography was carried out on Resource Q (Amersham-Pharmacia Biotech, Freiburg, Germany) in 10 mm HEPES, pH 7.0. Elution of the recombinant HPPD was done in a linear gradient from 0 to 300 mm NaCl. HPPD-containing fractions were pooled and the buffer was exchanged to 10 mm HEPES, pH 7.0 by ultrafiltration.
The soluble fraction with recombinant Z. mays HPPD was applied to a Co2+ affinity column (Clontech), equilibrated with 50 mm Tris, pH 8.0 and the protein was purified to almost homogeneity by elution using a linear imidazole gradient (50 mm Tris, pH 7.0, 500 mm imidazole). The protein was concentrated and a buffer exchange to 10 mm HEPES, pH 7.0 was carried out in an Ultrafree filter device (Millipore). The purity of the recombinant proteins was checked by SDS-PAGE stained with Coomassie Brilliant Blue (Sambrook et al., 1989). The enzymatic activity of the proteins was tested with modifications using the photometric assay described by Lindstedt and Rundgren (1982) that monitors the decrease of the HPP enol form at 308 nm. A total of 250 μm HPP was incubated for 10 min at 20°C to equilibrate keto and enol form in a volume of 900 μL containing 0.8 m KH2PO4, 0.4 m H3BO3, 0.2 m Tris/HCl, pH 7.6, 5.7 mm ascorbate, and 1,200 units catalase. The reaction was started by addition of 100 μL containing 10 mm HEPES/NaOH pH 7.0, 1 mm FeSO4, and 1–5 μg HPPD. The reaction was performed for 15 min at 25°C. The specific activity of the soluble protein extract was in the range 32 to 27 nmol of HPP consumed/min/mg of protein with Arabidopsis HPPD and 60 nmol of HPP consumed/min/mg of protein with Z. mays HPPD, respectively (data not shown).
For crystallization the proteins were concentrated to 10 mg/mL (Z. mays) and 12 mg/mL (Arabidopsis), respectively. Crystals of HPPD were grown from active protein by the sitting drop vapor diffusion method. HPPD from Arabidopsis crystallized from 50% (v/v) 2-methyl-2,4-pentanediol and 6% (m/v) PEG200 in 25 mm sodium citrate, pH 5.6. Crystals grew within 4 to 6 weeks to a size of 200 × 200 × 300 μm3 at 20°C. Z. mays HPPD crystallized from 28% (v/v) 2-methyl-2,4-pentanediol, 4.9% (m/v) PEG 8000, 0.025% (v/v) dichlormethane, and 0.7 m cacodylate, pH 6.5. At 16°C crystals of Z. mays HPPD took 3 weeks to reach a size of 200 μm × 100 μm × 500 μm.
Data Collection, Structure Solution, and Refinement
Inhouse diffraction data were collected on a MAR Research 345 imaging plate detector system mounted on a Rigaku RU-200 rotating anode operated at 50 mA and 100 kV with λ = CuKα = 1.542 Å. The high resolution native data set of Z. mays HPPD was collected at beam line BW6 at the Deutsches Elektronen Synchrotron in Hamburg, Germany employing a MAR Research CCD detector at a wavelength (λ) of 1.050 Å. All data collections were carried out under cryogenic conditions at 100 K using an Oxford Cryosystem with the mother liquor serving as a cryoprotectant.
The structure of Z. mays HPPD was solved by the SIRAS method. A heavy atom derivative was prepared by soaking a crystal at 16°C in 1 mm thiomersal (C9H9HgNaO2S) in mother liquor for 12 h. The data sets were integrated and scaled using the HKL suite (Otwinowski and Minor, 1997). The CCP4 suite (Collaborative Computational Project, Number 4, 1994) was used for scaling of native and derivative data. Six heavy atom positions were determined with SHELXS (Sheldrick, 1990) and heavy atom parameters refined with MLPHARE (Otwinowski, 1991) using both the isomorphous and anomalous contributions at 3 Å resolution. Phases were improved by solvent flattening and density modifications implemented in DM of the CCP4 suite (Cowtan and Main, 1993). The heavy atom sites and partial models were used to identify the NCS operators, which were improved with inosine-5′-monophosphate (Kleywegt and Jones, 1994). The electron density was 4-fold averaged with AVE and masks were generated with MAMA (Kleywegt and Jones, 1994). Model building was done with the program MAIN (Turk, 1992). The structure was refined with the program CNS (Brünger et al., 1998) with 5% of the data in the test set to calculate the free R-factor. Water molecules were added to the protein model with CNS. All water molecules were checked manually. NCS-restraints on atomic positions and temperature factors were applied in the initial stages of refinement. After adding water molecules the NCS-restraints were gradually decreased. In the final round of refinement no NCS-restraints were applied. The distances between the catalytic iron atom and its ligands were restrained to 2.1 Å.
The crystal structure of Arabidopsis HPPD was determined by molecular replacement with the program MOLREP (Collaborative Computational Project, Number 4, 1994). Reflections in the resolution range from 16 Å to 5 Å were used with the structure of Z. mays HPPD monomer as search model. The first monomer was positioned with a correlation of 0.22 and a crystallographic R-factor of 50%. Positioning of the second monomer increased the correlation to 0.45 and decreased the R-factor to 35.7%. The structure was refined as described above, however, maintaining NCS-restraints throughout refinement taking the lower resolution into consideration.
The stereochemistry of the refined structures was analyzed with the program PROCHECK (Laskowski et al., 1993). Data collection statistics and statistics of crystallographic refinement are reported in Table I. Figures were prepared using MOLSCRIPT (Kraulis, 1991), Raster3D (Merritt and Bacon, 1997), and ALSCRIPT (Barton, 1993). Secondary structures were assigned with the program STRIDE (Frishman and Argos, 1995).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.034082.
References
- Arciero DM, Orville AM, Lipscomb JD (1985) [17O] water and nitric-oxide binding by protocatechuate 4,5-dioxygenase and catechol 2,3-dioxygenase – evidence for binding of exogenous ligands to the active-site Fe2+ of extradiol dioxygenases. J Biol Chem 260: 14035–14044 [PubMed] [Google Scholar]
- Bartley GE, Maxwell CA, Wittenbach VA, Scolnik PA (1997) Cloning of an Arabidopsis thaliana cDNA for p-hydroxyphenylpyruvate dioxygenase. Plant Physiol 113: 1465–1468 [Google Scholar]
- Barton GJ (1993) ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng 6: 37–40 [DOI] [PubMed] [Google Scholar]
- Berger S, Ellersiek U, Westhoff P, Steinmüller K (1993) Studies on the expression of NDH-H, a subunit of the NAD(P)H-plastoquinone-oxidoreductase of higher plant chloroplasts. Planta 190: 25–31 [Google Scholar]
- Brown CA, Pavlosky MA, Westre TE, Zhang Y, Hedman B, Hodgson KO, Solomon EI (1995) Spectroscopic and theoretical description of the electronic structure of S=3/2 iron-nitrosyl complexes and their relation to O-2 activation by non-heme iron enzyme active sites. J Am Chem Soc 117: 715–732 [Google Scholar]
- Brünger AT, Adams PD, Clore GM, Delano WI, Gros P, Grosse-Kustleve RW, Jiang JS, Kuszewski J, Nilges N, Pannu NS, Read RJ, Rice LM, et al (1998) Crystallography and NMR systems (CNS): A new software system for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Cowtan K, Main P (1993) Improvement of macromolecular electron-density maps by the simultaneous application of real and reciprocal space constraints. Acta Crystallogr D 49: 148–157 [DOI] [PubMed] [Google Scholar]
- Dähnhardt D, Falk J, Appel J, van der Kooij TAW, Schulz-Friedrich R, Krupinska K (2002) The hydroxyphenylpyruvate dioxygenase from Synechocystis sp. PCC 6803 is not required for plastoquinone synthesis. FEBS Lett 523: 177–181 [DOI] [PubMed] [Google Scholar]
- Durand R, Zenk MH (1974) The homogentisate ring-cleavage pathway in the biosynthesis of acetate-derived naphthoquinones of Droseraceae. Phytochemistry 13: 1483–1492 [Google Scholar]
- Endo F, Awata H, Tanoue A, Ishiguro M, Eda Y, Titani K, Matsuda I (1992) Primary structure deduced from complementary DNA sequence and expression in cultured cells of mammalian 4-hydroxyphenylpyruvic acid dioxygenase: evidence that the enzyme is a homodimer of identical subunits homologous to rat liver-specific alloantigen F. J Biol Chem 267: 24235–24240 [PubMed] [Google Scholar]
- Fernández-Cañón JM, Peñalva MA (1995) Molecular characterization of a gene encoding a homogentisate dioxygenase from Aspergillus nidulans and identification of its human and plant homologues. J Biol Chem 270: 21199–21205 [DOI] [PubMed] [Google Scholar]
- Fiedler E, Soll J, Schultz G (1982) The formation of homogentisate in the biosynthesis of tocopherol and plastoquinone in spinach chloroplasts. Planta 155: 511–515 [DOI] [PubMed] [Google Scholar]
- Forbes BJR, Hamilton GA (1994) Mechanism and mechanism-based inactivation of 4-hydroxyphenylpyruvate dioxygenase. Bioorg Chem 22: 343–361 [Google Scholar]
- Frishman D, Argos P (1995) Knowledge-based protein secondary structure assignment. Proteins 23: 566–579 [DOI] [PubMed] [Google Scholar]
- Garcia I, Rodgers M, Lenne C, Rolland A, Sailland A, Matringe M (1997) Subcellular localization and purification of a p-hydroxyphenylpyruvate dioxygenase from cultured carrot cells and characterization of the corresponding cDNA. Biochem J 325: 761–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia I, Rodgers M, Pepin R, Hsieh TF, Matringe M (1999) Characterization and subcellular compartmentation of recombinant 4-hydroxyphenylpyruvate dioxygenase from arabidopsis in transgenic tobacco. Plant Physiol 119: 1507–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Eltis LD, Timmis KN, Muchmore SW, Bolin JT (1995) Crystal structure of the biphenyl-cleaving extradiol dioxygenase from a PCB-degrading pseudomonad. Science 270: 976–980 [DOI] [PubMed] [Google Scholar]
- Harpel MR, Lipscomb JD (1990) Gentisate 1,2-dioxygenase from pseudomonas – substrate coordination to active-site Fe2+ and mechanism of turnover. J Biol Chem 265: 22187–22196 [PubMed] [Google Scholar]
- Huhn R, Stoermer H, Klingele B, Bausch E, Fois A, Farnetani M, Di Rocco M, Boue J, Kirk JM, Coleman R, Scherer G (1998) Novel and recurrent tyrosine aminotransferase gene mutations in tyrosinemia type II. Hum Genet 102: 305–313 [DOI] [PubMed] [Google Scholar]
- Jefford CW, Cadby PA (1981) Evaluation of the models for the mechanism of action of 4-hydroxyphenylpyruvate dioxygenase. Experientia 37: 1134–1137 [DOI] [PubMed] [Google Scholar]
- Johnson-Winters K, Purpero VM, Kavana M, Nelson T, Moran GR (2003) (4-hydroxyphenyl)pyruvate dioxygenase from Streptomyces avermitilis: the basis for ordered substrate addition. Biochemistry 42: 2072–2080 [DOI] [PubMed] [Google Scholar]
- Kavana M, Moran GR (2003) Interaction of (4-hydroxyphenyl)pyruvate dioxygenase with the specific inhibitor 2-[2-nitro-4-(trifluoromethyl)benzoyl]-1,3-cyclohexanedione. Biochemistry 42: 10238–10245 [DOI] [PubMed] [Google Scholar]
- Kim KH, Petersen M (2002) cDNA-cloning and functional expression of hydroxyphenylpyruvate dioxygenase from cell suspension cultures of Coleus blumei. Plant Sci 163: 1001–1009 [Google Scholar]
- Kita A, Kita S, Fujisama I, Inaka K, Ishida T, Horrike K, Nozaki M, Miki K (1999) An archetypical extradiol-cleaving catecholic dioxygenase: the crystal structure of catechol 2,3-dioxygenase (metapyrocatechase) from Pseudomonas putida mt-2. Structure 7: 25–34 [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA (1994) Halloween: masks and bones. In S Bailey, R Hubbard, D Waller, eds, From First Map to Final Model. Science and Engineering Research Council, Daresbury Laboratory, Warrington, United Kingdom, pp 59–66
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24: 946–950 [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Lenne C, Matringe M, Roland A, Sailland A, Pallett KE, Douce R (1995) Partial purification and localization of p-hydroxyphenylpyruvate dioxygenase activity from cultured carrot cells. In P Mathis, ed, Photosynthesis: From Light to Biosphere, Vol 5. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 285–288
- Lindblad B, Lindstedt S, Steen G (1977. a) Enzymic defects in hereditary tyrosinemia. Proc Natl Acad Sci USA 74: 4641–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad B, Lindstedt G, Lindstedt S, Rundgren M (1977. b) Purification and some properties of human 4-hydroxyphenylpyruvate dioxygenase. J Biol Chem 252: 5073–5084 [PubMed] [Google Scholar]
- Linden L (2000) Isolierung, Klonierung, Expression, Reinigung und Kristallisation eukaryotischer 4-Hydroxyphenylpyruvat Dioxygenasen – Röntgenstrukturanalyse von Inhibitorkomplexen der löslichen anorganischen Pyrophosphatase aus Saccharomyces cerevisiae. PhD Thesis. Technische Universität München, München, Germany
- Lindstedt S, Rundgren M (1982) Blue color, metal content, and substrate binding in 4-hydroxyphenylpyruvate dioxygenase from Pseudomonas sp. strain P. J. 874. J Biol Chem 257: 11922–11931 [PubMed] [Google Scholar]
- Lindstedt S, Odelhög B (1987) 4-Hydroxyphenylpyruvate dioxygenase from Pseudomonas. Method Enzymol 142: 143–148 [DOI] [PubMed] [Google Scholar]
- Maxwell CA, Scolnik PA, Wittenbach VA, Gutteridge S, inventors. December 31, 1997. Plant gene for p-hydroxyphenylpyruvate dioxygenase. International Application No. PCT/US97/11295
- Merritt EA, Bacon DJ (1997) Raster3D photorealistic molecular graphics. Method Enzymol 277: 505–524 [DOI] [PubMed] [Google Scholar]
- Norris SR, Barrette TR, DellaPenna D (1995) Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell 7: 2139–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris SR, Shen XH, DellaPenna D (1998) Complementation of the Arabidopsis pds1 mutation with the gene encoding p-hydroxyphenylpyruvate dioxygenase. Plant Physiol 117: 1317–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z (1991) Proceedings of the CCP4 study weekend. In W Wolf, PR Evans, AGW Leslie, eds, Isomorphous Replacement and Anomalous Scattering. Science and Engineering Research Council, Daresbury Laboratory, Warrington, United Kingdom, pp 80–86
- Otwinowski Z, Minor W (1997) Processing of x-ray diffraction data collected in oscillation mode. Method Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pallett KE, Little JP, Sheekey M, Veerasekaran P (1998) The mode of action of isoxaflutole I. Physiological effects, metabolism, and selectivity. Pestic Biochem Phys 62: 113–124 [Google Scholar]
- Phornphutkul C, Introne WJ, Perry MB, Bernardini I, Murphey MD, Fitzpatrick DL, Anderson PD, Huizing M, Anikster Y, Gerber LH, Gahl WA (2002) Natural history of alkaptonuria. N Engl J Med 347: 2111–2121 [DOI] [PubMed] [Google Scholar]
- Prescott AG (1993) A dilemma of dioxygenases (or where biochemistry and molecular biology fail to meet). J Exp Bot 44: 849–861 [Google Scholar]
- Que LJ, Ho RYN (1996) Dioxygen activation by enzymes with mononuclear non-heme iron active sites. Chem Rev 96: 2607–2624 [DOI] [PubMed] [Google Scholar]
- Ramachandran GN, Sasisekharan V (1968) Conformation of polypeptides and proteins. Adv Protein Chem 23: 283–437 [DOI] [PubMed] [Google Scholar]
- Roach PL, Clifton IJ, Hensgens CM, Shibata N, Schofield CJ, Hajdu J, Baldwin JE (1997) Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation. Nature 387: 827–830 [DOI] [PubMed] [Google Scholar]
- Roche PA, Moorehead TJ, Hamilton GH (1982) Purification and properties of hog liver 4-hydroxyphenylpyruvate dioxygenase. Arch Biochem Biophys 216: 62–73 [DOI] [PubMed] [Google Scholar]
- Rüetschi U, Dellsen A, Sahlin P, Stenman G, Rymo L, Lindstedt S (1993) Human 4-hydroxyphenylpyruvate dioxygenase, primary structure and chromosomal localization of the gene. Eur J Biochem 213: 1081–1089 [DOI] [PubMed] [Google Scholar]
- Rundgren M (1977) Steady-state kinetics of 4-hydroxyphenylpyruvate dioxygenase from human liver.3. J Biol Chem 252: 5094–5099 [PubMed] [Google Scholar]
- Russo P, O'Regan S (1990) Visceral pathology of hereditary tyrosinemia type I. Am J Hum Genet 47: 317–324 [PMC free article] [PubMed] [Google Scholar]
- Russo PA, Mitchell GA, Tanguay RM (2001) Tyrosinemia: a review. Pediatr Dev Pathol 4: 212–221 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) In N Ford, C Nolan, M Ferguson, eds, Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schulz A, Oswald O, Beyer P, Kleinig H (1993) SC-OO51 a 2-benzoyl-cyclohexane-1,3-dione bleaching herbicide, is a potent inhibitor of the enzyme p-hydroxyphenylpyruvate dioxygenase. FEBS Lett 318: 162–166 [DOI] [PubMed] [Google Scholar]
- Senda T, Sugiyama K, Narita H, Yamamoto T, Kimbara K, Fukuda M, Sato M, Yano K, Mitsui Y (1996) Three-dimensional structures of free form and two substrate complexes of an extradiol ring-cleavage type dioxygenase, the BphC enzyme from Pseudomonas sp. Strain KKS102. J Mol Biol 255: 735–752 [DOI] [PubMed] [Google Scholar]
- Serre L, Sailland A, Sy D, Boudec P, Rolland A, Pebay-Peyroula E, Cohen-Addad C (1999) Crystal structure of Pseudomonas fluorescens 4-hydroxyphenylpyruvate dioxygenase: an enzyme involved in the tyrosine degradation pathway. Structure 7: 977–988 [DOI] [PubMed] [Google Scholar]
- Sheldrick G (1990) Phase annealing in SHELX90: direct methods for larger structures. Acta Crystallogr A 46: 467–473 [Google Scholar]
- Shu L, Chiou YM, Orville AM, Miller MA, Lipscomb JD, Que L Jr (1995) X-ray-absorption spectroscopic studies of the Fe(II) active-site of catechol 2,3-dioxygenase – implications for the extradiol cleavage mechanism. Biochemistry 34: 6649–6659 [DOI] [PubMed] [Google Scholar]
- Solomon EI, Brunold TC, Davis MI, Kemsley JN, Lee SK, Lehnert N, Neese F, Skulan AJ, Yang YS, Zhou J (2000) Geometric and electronic structure/function correlations in non-heme iron enzymes. Chem Rev 100: 235–350 [DOI] [PubMed] [Google Scholar]
- Sono M, Roach MP, Coulter ED, Dawson JH (1996) Heme-containing oxygenases. Chem Rev 96: 2841–2888 [DOI] [PubMed] [Google Scholar]
- Turk D (1992) Weiterentwicklung eines Programms für Molekülgraphik und Elektronendichte-Manipulation und seine Anwendung auf verschiedene Protein-Strukturaufklärungen. PhD Thesis. Technische Universität München, München, Germany
- Uragami Y, Senda T, Sugimoto K, Sato N, Nagarajan V, Masai E, Fukuda M, Mitsui Y (2001) Crystal structures of substrate free and complex forms of reactivated BphC, an extradiol type ring-cleavage dioxygenase. J Inorg Biochem 83: 269–279 [DOI] [PubMed] [Google Scholar]
- Valegard K, Terwisscha van Scheltinga AC, Lloyd MD, Hara T, Ramaswamy S, Perrakis A, Thompson A, Lee HJ, Baldwin JE, Schofield CJ, Hajdu J, Andersson I (1998) Structure of a cephalosporin synthase. Nature 394: 805–809 [DOI] [PubMed] [Google Scholar]
- Wada GH, Fellman JH, Fujita FS, Roth E (1975) Purification and properties of avian liver p-hydroxyphenylpyruvate hydroxylase. J Biol Chem 250: 6720–6726 [PubMed] [Google Scholar]
- Zhang Z, Ren J, Stammers DK, Baldwin JE, Harlos K, Schofield CJ (2000) Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase. Nat Struct Biol 7: 127–133 [DOI] [PubMed] [Google Scholar]