Abstract
Active self-assembly, in which non-thermal energy is consumed by the system to put together building blocks, allows the creation of non-equilibrium structures and active materials. Microtubule spools assembled in gliding assays are one example of such non-equilibrium structures, capable of storing bending energies on the order of 105 kT. Although these structures arise spontaneously in experiments, the origin of microtubule spooling has long been debated. Here, using a stepwise kinesin gradient, we demonstrate that spool assembly can be controlled by the surface density of kinesin motors, showing that pinning of microtubules due to dead motors plays a dominant role in spool initiation.
Introduction
Self-assembly is traditionally viewed as assembly via spontaneous, thermodynamically driven processes, e.g. crystal formation.1–3Adding energy to the system via molecular motors, which couple the system to a store of chemical energy, results in greater transport speed for larger building blocks. This makes it possible to accelerate the self-assembly process, which is especially important for the assembly of larger building blocks that move slowly by diffusive transport. The additional energy also enables the creation of non-equilibrium structures and active materials.4 Thus, the design space for nanodevices and materials can be greatly expanded by active self-assembly.1
Studies in active self-assembly often utilize the kinesin motor protein and its associated filament the microtubule.5–10 In in vitro experiments, a surface is coated with kinesin motors, which move microtubules along the surface while consuming ATP. By functionalizing the microtubule with biotin, streptavidin– with its four biotin-binding sites– can be used to cross-link microtubules. In such assays, microtubules have been observed to form “bundles,” “wires,” and “spools” (Fig 1).5, 7, 10–15Spools are especially interesting because they are non-equilibrium structures, storing on the order of 105kT per spool of bending energy (the persistence length of a microtubule is on the order of millimetres).5, 16
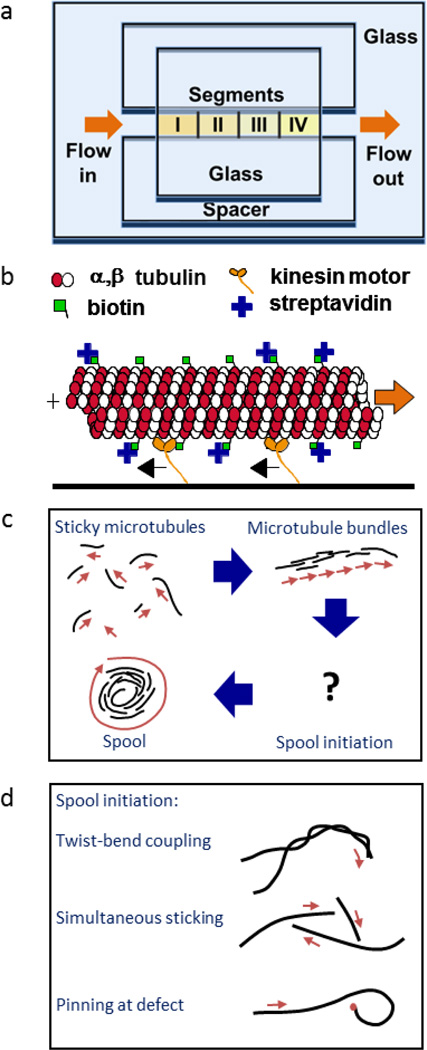
Figure 1.
Experimental set-up. (a) The flow cell is constructed with glass coverslips and double-sided tape. Kinesin is flowed in section by section, resulting in a stepwise gradient. (b) Kinesin motors attach to the surface and move the biotinylated microtubule around. Reproduced from Hess, et al.5 with permission from the Americal Chemical Society. (c) The biotin and streptavidin allow the microtubules to crosslink to one another forming microtubule bundles and spools. Reproduced from Luria, et al.20 with permission from the Royal Society of Chemistry. (d) There are several theories on how spools are initiated. Twist-bend coupling occurs as a result of the microtubule structure; simultaneous sticking of three or more microtubules is dependent on the microtubule surface density; and pinning events are dependent on the kinesin surface density. Adapted from Luria, et al.20 with permission from the Royal Society of Chemistry.
At present, three mechanisms of spool formation have been proposed. The first mechanism is that spools emerge as a result of the intrinsic microtubule structure, thus making spool size independent of kinesin and microtubule density.7, 8 The second mechanism proposes that spools arise when three or more microtubules collide and cross-link into a closed structure. This mechanism is primarily dependent on the surface microtubule density. The third mechanism proposes that spools are formed when the microtubule is pinned at the leading end by a defective motor or other obstacle and forced to buckle. In this case, both spool size and spool density are dependent on kinesin density.
The first mechanism is motivated by the observation that some microtubules polymerized in vitro have an inherent supertwist. During polymerization, tubulin dimers form long chains called protofilaments which in turn assemble into the hollow cylindrical structure of the microtubule. While 13 protofilaments form a straight cylinder, microtubules polymerized in vitro may have anywhere from 8 to 19 protofilaments.17, 18Thesenon-13 protofilament microtubules have an inherent supertwist, which kinesin motors follow. Thus, these microtubules rotate when being propelled forward.18If in a gliding assay, one non-13 protofilament microtubule encounters another microtubule and cross-linking occurs, the two microtubules may twist around one another forming a helical structure. Microtubule complexes involving multiple microtubules wrapped around each other have been observed via electron microscopy.8It has been shown that for such helical structures, stress relaxation results in out-of-plane buckling when an external compressive load is applied.19This twist-bend coupling may result in curved trajectories of the microtubules, which in turn may lead to spool formation (Fig 1d).
The second mechanism postulating that spools are formed at microtubule intersections was explored by Crenshaw et al. by computer simulation.20, 21 It was discovered that when three or more microtubules cross paths and cross-link together, a closed polygon forms, which then relaxes into a ring-like shape over time (Fig 1d).20The distribution of spool circumferences generated by the simulation was in good agreement to experimental results.20 This theory is further supported by the fact that even in the absence of streptavidin and biotin cross-linkers, high microtubule densities also lead to loop formation, something which is not noted at lower microtubule densities.22
The third mechanism postulates that spools are formed due to the presence of non-functional motors or defects on the surface, which pin part of the microtubule in place.20 Since the rest of the microtubule is still propelled by other attached functional motors, the microtubule buckles and eventually forms a loop which initiates spooling (Fig 1d). Thus, spool formation would be dependent on the kinesin motor density, which affects both the buckling force and the density of dead motors. Such pinning events have been previously observed in both actin filaments as well as microtubules, and it has been shown that pinning can result in filament curvature and even spiralling.23Furthermore, it has been shown that applying compressive forces to microtubule bundles does lead to buckling and ring formation in experiments done on vesicle-encased microtubule bundles loaded via micropipette aspiration.16
Out of the three mechanisms presented, the first two postulate that spool formation is independent of kinesin motor density; the third postulates that both the frequency of spool initiation and spool size are dependent on the motor density. Here, we prepare surfaces with step-wise variations in kinesin density and show that spool density and circumference are both dependent on kinesin motor density, supporting the third mechanism which posits that spool formation is a result of pinning due to dead motors.
Experimental
Microtubule preparation
Microtubules were prepared by polymerizing 20 µg of biotin-labeled tubulin (Cytoskeleton Inc., Denver, CO, T333P) in 6.25 µL of growth solution containing 4 mM of MgCl2, 1 mM of GTP, and 5% DMSO (v/v) in BRB80 buffer (80 mM piperazine diethanesulfonic acid, 1 mM magnesium chloride (MgCl2), and 1 mM ethylene glycol tetra acetic acid, titrated to pH 6.9 with potassium hydroxide (KOH)) for 30 min at 37°C. The microtubules were then diluted 100-fold and stabilized in 10 µM paclitaxel in BRB80 (Sigma, Saint Louis, MO).
Kinesin preparation
For the spooling experiments, a kinesin construct consisting of the wild-type, full-length Drosophila melanogaster kinesin heavy chain and a C-terminal His-tag was expressed in Escherichia coli and purified using a Ni-NTA column. Based on landing rate measurements,24the concentration of this kinesin solution is 730±180nM in this stock solution.
Flow cell preparation
A 10 µL drop of casein solution (2 mg/mL in BRB80) was sandwiched between a 22 mm × 22 mm coverslip and a 50 mm × 35 mm coverslip. The casein (purified from bovine milk and containing all casein subunits; Sigma, Saint Louis, MO, C7078) was allowed to adsorb for 5 min before the two coverslips were disassembled and allowed to air dry. With the two casein-coated sides facing each other, a ~100 µm × 5 mm × 22 mm flow cell was assembled using two pieces of double-sided Scotch tape as spacers. Segments of ~5 mm in length were demarcated prior to fluid injection into the flow cell (Fig 1a).
Kinesin solution (25 nM kinesin, 0.5 mg/mL casein, and 0.02 mM ATP in BRB80) was flowed into the flow cell incrementally. Enough kinesin solution was flowed in to meet the first demarcation (approximately 2.5 µL), covering the first segment of the flow cell, and was allowed to adsorb for 2 min before more kinesin solution (approximately 2.5 µL) was flowed in to cover the second segment. The solution was allowed to sit for another 2 min before kinesin solution was added to cover the third segment. Again the kinesin was allowed to adsorb for 2 min before additional kinesin solution was added into the flow cell. After another 2 min elapsed, the excess kinesin was washed out with anti fade solution (20 mM D-glucose, 20 µg/mL glucose oxidase, 8 µg/mL catalase, 10 mM dithiothreitol, and 0.02 mM ATP in BRB80). We used a kinesin solution resulting in a surface density of 740 ± 240 µm−2 after 5 minutes of adsorption, calculated from the landing rate experiments done on the stock kinesin solution.24, 25Based on landing rate experiments done on the kinesin gradient, the kinesin densities relative to the first section are estimated to be 0.55 ± 0.09, 0.17 ± 0.03, and 0.05 ± 0.02, implying that 36 ± 5% of the kinesin motors adsorb over the course of two minutes(SI). Thus, the kinesin densities can be estimated to be 1600 ± 580 µm−2, 870 ± 350µm−2, 270 ± 110 µm−2, and 90 ± 40 µm−2 for the first, second, third, and fourth sections, respectively.
The antifade solution was immediately exchanged with motility solution (3 µg/mL tubulin in 10 µM of paclitaxel and an antifade system made up of 20 mM of D-glucose, 20 µg/mL of glucose oxidase, 8 µg/mL of catalase, 10 mM of dithiothreitol, and 0.02 mM of ATP in BRB80). The microtubules were allowed to adsorb for 5 min before 15 µL of antifade solution was used to wash out the excess microtubules. Alexa488-labeled streptavidin (0.16 µM with 0.5 mg/mL casein and 10 µM paclitaxel in BRB80) was then introduced to the system and allowed to incubate for 5 min. Excess streptavidin was then washed out with antifade solution. It was observed that the initial density of microtubules was roughly equal in each section of the flow cell. Since the exchange of solutions takes about 20 seconds, based on the diffusion coefficient for a 1 µm long microtubule, it can be calculated that less than 5% of the microtubules are expected to settle onto the surface during solution addition or exchange.
To evaluate the stability of the kinesin gradient surface over time, a different flow cell was created following the same protocol as mentioned above. However, instead of immediately adding the motility solution after the first antifade wash, the motility solution was added after 2 hr had elapsed. The flow cell was kept in a humidified environment to prevent evaporation of the solution for the 2 hrs between flows. The subsequent solutions were flowed through as described above, and spooling behaviour was observed in each quadrant after 2 hrs. All experiments were performed at 20° C.
Imaging and analysis
For the spooling experiments, flow cells were imaged using an epifluorescence microscope (Nikon TE2000) equipped with an X-cite 120 lamp (EXFO, Ontario, Canada) and an iXON DU885LC EMCCD camera (Andor Technology, South Windsor, CT). Image sequences of 3 different fields of view were taken using a 40× air objective (N.A. 0.75) for each section of the flow cell approximately once every 15 minutes for 2 hours (0.5 fps for 20s with 0.5s exposure time). In experiments for the evaluation of the gradient stability, the flow cells were only imaged 2 hrs after the motility solution was added.
Data analysis was conducted using Image J imaging software (available at http://rsbweb.nih.gov/ij/).
Results and Discussion
The assembly process is completed within 90 minutes for all kinesin densities (see SI). The spool population was distinctly different in the four sections of decreasing kinesin densities (Fig 2a–d). The regions in between the sections were characterized by an approximately 200 µm wide stripe (one field of view) in which very few spools or gliding microtubules were seen. Instead, there were many stationary disordered microtubule aggregates and short microtubules which appeared as speckles. Evaporation of the fluid within the flow cell during the 2 min between subsequent additions of kinesin solution may have disabled adhered motors causing this distinct boundary between sections.
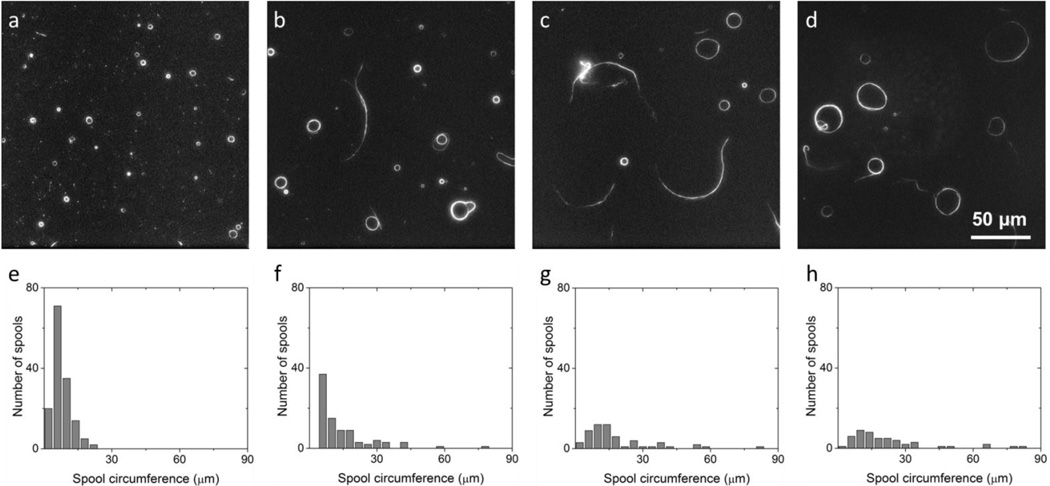
Figure 2.
Sample images and spool circumference distributions taken at 2 hours after the final solution exchange in the flow cell for sections I (a and e), II (b and f), III (c and g), and IV (d and h). As kinesin density decreases, average spool circumference decreases while spool density increases.
Microtubule movement was observed in the central region of each segment throughout the full 2 hours of the experiment. The average gliding velocities of the microtubules in each section of the flow cell stayed around 0.1 µm/s throughout the experiment. In control experiments, similar differences in spool circumferences and densities between sections were obtained, even after waiting 2 hr before the addition of the microtubule solution. Thus, we concluded that the differences in kinesin density between various sections were stable over the course of the 2 hours. Thus, in the flow cell, only the kinesin density differed between sections while all other variables were constant (i.e. initial microtubule population and density, streptavidin concentration, and antifade solution.)
The section with the highest kinesin density was characterized by the presence of many smaller spools. As the kinesin density decreased, fewer spools were created, and more non-spool structures (i.e. bundles and aggregates) were observed (Fig. 2a–d). The distribution of spool sizes was recorded for all kinesin densities (Fig 2e–h). Lowering the kinesin density results in a heavier distribution tail and a right shift in the peak.
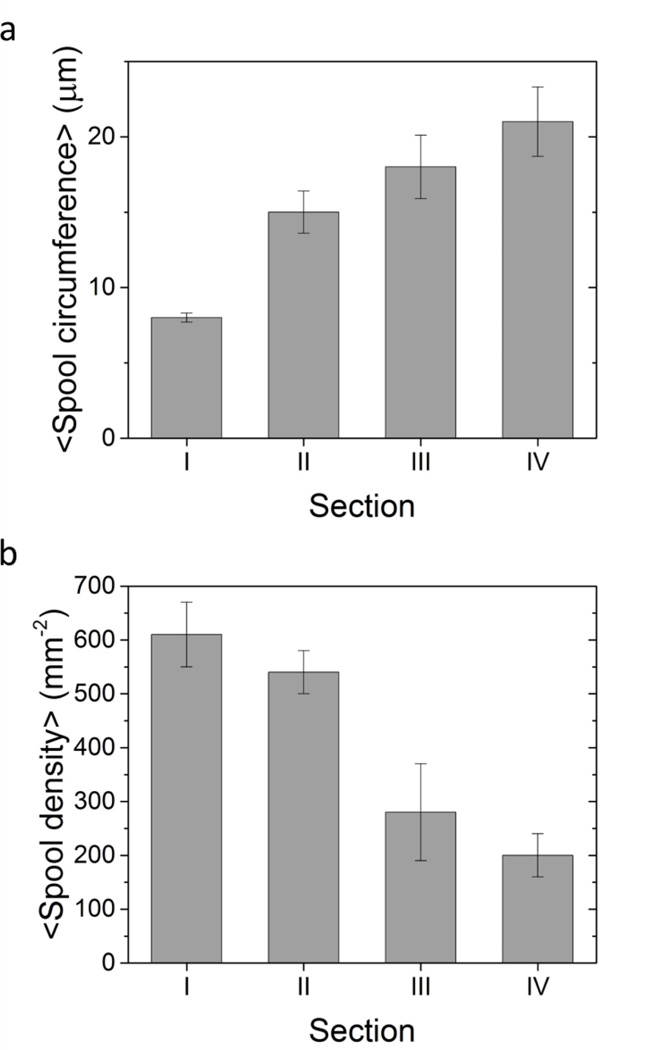
These results are predicted by the model described by Luria et al.20 In this model, the probability that a spool of a specified circumference is formed is given by the product of the probability that the microtubule forming the spool has a sufficient length and the probability that the microtubule has a sufficient number of kinesin motors attached to provide enough force to bend the microtubule. The first is a function of the distribution of microtubule bundle lengths while the second is dependent on the stiffness of the tip of the microtubule bundle, the force exerted per motor, length of the microtubule/microtubule bundle, and the surface density of kinesin motors. Since neither the properties of individual microtubules and kinesins nor the initial distribution of microtubule lengths should change between different sections of the flow cell, the kinesin density is the only parameter which changes the distribution. However, because the absolute kinesin densities are unknown, we cannot use this model to generate the expected spool distributions. The highest spool density was observed in the section with the highest kinesin density, and corresponded with the lowest average spool circumference. Conversely, the lowest spool density corresponded with the highest average spool circumference in the section with the lowest kinesin density (Fig 3). These results are consistent with the results of assays run with GFP-kinesin (SI).
Figure 3.
Average spool circumferences (a) and spool densities (b) for each of the four sections and their standard errors. As kinesin density decreases, average spool circumference increases while average spool density decreases, i.e., higher kinesin densities lead to a greater number of spools and smaller spools.
For the flow cell section with the highest kinesin density, there was a much greater number of very short microtubules, which appeared as specks on the field of view (Fig. 2a). The high density of kinesin is capable of both providing enough force to break bent microtubules and of sustaining gliding of the shortened microtubules. This may change the length distribution of the microtubules prior to assembly, and lead to the preferential formation of smaller spools seen in Fig. 2h.26
It was also noted after two hours that the longest microtubule bundle (230 µm) was formed in the section with the lowest kinesin density. Disordered structures (i.e. aggregates of cross-linked microtubules which have no alignment with respect to one another) were also found in this section, and a number of microtubules drifted in and out of focus. Such floating structures were not counted as spools, even though some of them did form closed loops. These sorts of structures and behavior were not noted in the areas with higher kinesin densities.
If spools were generated only due to the relaxation of the strain rooted in the supertwist of the microtubule, it would be expected that spool size would not be dependent on motor density. Because a kinesin motor always takes 8 nm steps regardless of how many motors are attached to the microtubule, the amount of twisting experienced over the trajectory would be solely determined by the number of protofilaments in the microtubule. Previous studies have found that the rotation of microtubule spools is dependent on the handedness of the helical structure, with right-handed and left-handed helices resulting in clockwise and counter-clockwise rotation, respectively.8, 27 However, here, we have demonstrated that there is a significant difference in the average spool circumferences and the number of spools formed at different kinesin densities; thus, the supertwist of the microtubule cannot be the main origin of spooling.
Similarly, the initiation of spooling due to microtubule intersections does not depend on kinesin density, and instead is only dependent on the microtubule density. While for short adsorption times, higher kinesin densities result in higher initial microtubule densities on the surface, for the fixed adsorption time used for this experiment, the initial microtubule density should only be dependent on the concentration of microtubules in solution.25 The images taken closest to the initial time points for each section show that the density of microtubules is roughly constant throughout the flow cell, as expected from calculations based on the diffusion constant of the microtubule. If the intersection of microtubules were the sole reason spools form, then likewise, the spool distribution should be similar across all sections. As it is not, this too cannot be the main mechanism which initiates spooling.
If spools were generated by pinning, it would be expected that both spool density and size would be dependent on kinesin density. A higher density of kinesin means that dead motors are spaced closer to one another, thus initiating a greater density of spools. Furthermore, higher kinesin densities lead to increased force on pinned microtubules allowing for smaller spool circumferences. Both of these effects have been observed. While we cannot rule out that the supertwist or that multiple intersections are the origin of some spools, it is clear that pinning of the microtubule plays a dominant role in spool initiation.
Conclusions
Understanding the origins of spooling is the first step to being able to better control the structures which arise from the active self-assembly of microtubules. Here we have shown that increasing kinesin density increases the spool density and decreases spool circumference. This clear dependence on kinesin density shows that neither the supertwist of the microtubules nor the intersection of microtubules is the primary cause of spool formation; rather, pinning of microtubules by dead motors plays a dominant role in spool initiation.
This demonstrates that forces acting upon the building blocks are important factors for determining the size of the assembled structure. By changing these parameters, it may be possible to better determine structures that arise from active self-assembly processes of micro- to mesoscale building blocks, and build systems which capitalize on these properties to assemble non-equilibrium structures in a more uniform and controlled manner.
We have also demonstrated a proof-of-concept method for making a kinesin gradient based solely on its diffusion constant and ability to attach to a surface. Though our flow cells only contained four distinct regions with relatively sharp boundaries in kinesin surface density, it would be feasible to divide the flow cells into increasingly smaller segments and ultimately create a smooth gradient. To prevent evaporation for slower flow rates, it would be necessary to keep the flow cell in a humidified environment. This adds to the existing methods for creating protein gradients,28, 29 and using this simple method, it may be possible to pursue further studies on protein movement, adsorption, and assembly.
Supplementary Material
Acknowledgements
Financial support through NSF grant DMR1063771 is gratefully acknowledged.C.C. thanks the Swiss National Science Foundation for a postdoctoral fellowship.
This work was performed, in part, at the Center for Integrated Nanotechnologies, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Los Alamos National Laboratory (Contract DE-AC52-06NA25396) and Sandia National Laboratories (Contract DE-AC04-94AL85000).
TIRF images were collected in the Confocal and Specialized Microscopy Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University, supported by NIH grant #P30 CA013696. The confocal microscope was purchased with NIH grant #S10 RR025686.
references
- 1.Whitesides GM, Grzybowski B. Science. 2002;295:2418–2421. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- 2.Lehn J-M. Proceedings of the National Academy of Sciences. 2002;99:4763–4768. doi: 10.1073/pnas.072065599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitesides GM, Boncheva M. Proceedings of the National Academy of Sciences. 2002;99:4769–4774. doi: 10.1073/pnas.082065899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez T, Chen DTN, DeCamp SJ, Heymann M, Dogic Z. Nature. 2012;491:431-+. doi: 10.1038/nature11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hess H, Clemmens J, Brunner C, Doot R, Luna S, Ernst K-H, Vogel V. Nano Letters. 2005;5:629–633. doi: 10.1021/nl0478427. [DOI] [PubMed] [Google Scholar]
- 6.Kawamura R, Kakugo A, Osada Y, Gong JP. Nanotechnology. 2010;21:145603. doi: 10.1088/0957-4484/21/14/145603. [DOI] [PubMed] [Google Scholar]
- 7.Bachand M, Trent AM, Bunker BC, Bachand GD. Journal of Nanoscience and Nanotechnology. 2005;5:718–722. doi: 10.1166/jnn.2005.112. [DOI] [PubMed] [Google Scholar]
- 8.Liu HQ, Spoerke ED, Bachand M, Koch SJ, Bunker BC, Bachand GD. Advanced Materials. 2008;20:4476–4481. [Google Scholar]
- 9.Choi DS, Byun KE, Hong S. Small. 2011;7:1755–1760. doi: 10.1002/smll.201002267. [DOI] [PubMed] [Google Scholar]
- 10.Kawamura R, Kakugo A, Osada Y, Gong JP. Langmuir. 2010;26:533–537. doi: 10.1021/la902197f. [DOI] [PubMed] [Google Scholar]
- 11.Idan O, Lam A, Kamcev J, Gonzales J, Agarwal A, Hess H. Nano Letters. 2012;12:240–245. doi: 10.1021/nl203450h. [DOI] [PubMed] [Google Scholar]
- 12.He SH, Lam AT, Jeune-Smith Y, Hess H. Langmuir. 2012;28:10635–10639. doi: 10.1021/la302034h. [DOI] [PubMed] [Google Scholar]
- 13.Schaller V, Weber CA, Hammerich B, Frey E, Bausch AR. Proceedings of the National Academy of Sciences. 2011 doi: 10.1073/pnas.1107540108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue D, Kabir AMR, Mayama H, Gong JP, Sada K, Kakugo A. Soft Matter. 2013;9:7061–7068. [Google Scholar]
- 15.Tamura Y, Kawamura R, Shikinaka K, Kakugo A, Osada Y, Gong JP, Mayama H. Soft Matter. 2011;7:5654–5659. [Google Scholar]
- 16.Elbaum M, Kuchnir Fygenson D, Libchaber A. Physical Review Letters. 1996;76:4078–4081. doi: 10.1103/PhysRevLett.76.4078. [DOI] [PubMed] [Google Scholar]
- 17.Howard J. Mechanics of Motor Proteins and the Cytoskeleton. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- 18.Ray S, Meyhofer E, Milligan RA, Howard J. Journal of Cell Biology. 1993;121:1083–1093. doi: 10.1083/jcb.121.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De La Cruz EM, Roland J, McCullough BR, Blanchoin L, Martiel JL. Biophysical Journal. 2010;99:1852–1860. doi: 10.1016/j.bpj.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luria I, Crenshaw J, Downs M, Agarwal A, Seshadri SB, Gonzales J, Idan O, Kamcev J, Katira P, Pandey S, Nitta T, Phillpot SR, Hess H. Soft Matter. 2011;7:3108–3115. [Google Scholar]
- 21.Crenshaw JD, Liang T, Hess H, Phillpot SR. Journal of Computational and Theoretical Nanoscience. 2011;8:1999–2005. [Google Scholar]
- 22.Liu L, Tuzel E, Ross JL. Journal of Physics-Condensed Matter. 2011;23 doi: 10.1088/0953-8984/23/37/374104. [DOI] [PubMed] [Google Scholar]
- 23.Bourdieu L, Duke T, Elowitz MB, Winkelmann DA, Leibler S, Libchaber A. Physical Review Letters. 1995;75:176–179. doi: 10.1103/PhysRevLett.75.176. [DOI] [PubMed] [Google Scholar]
- 24.Katira P, Agarwal A, Fischer T, Chen H-Y, Jiang X, Lahann J, Hess H. Advanced Materials. 2007;19:3171–3176. [Google Scholar]
- 25.Agarwal A, Luria E, Deng XP, Lahann J, Hess H. Cellular and Molecular Bioengineering. 2012;5:320–326. [Google Scholar]
- 26.Pampaloni F, Lattanzi G, Jonas A, Surrey T, Frey E, Florin E-L. Proceeding of the National Academy of Sciences. 2006;103:10248–10253. doi: 10.1073/pnas.0603931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawamura R, Kakugo A, Shikinaka K, Osada Y, Gong JP. Biomacromolecules. 2008;9:2277–2282. doi: 10.1021/bm800639w. [DOI] [PubMed] [Google Scholar]
- 28.Karnik R, Castelino K, Duan C, Majumdar A. Nano Letters. 2006;6:1735–1740. doi: 10.1021/nl061159y. [DOI] [PubMed] [Google Scholar]
- 29.Ionov L, Stamm M, Diez S. Nano Letters. 2005;5:1910–1914. doi: 10.1021/nl051235h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.