Abstract
Memory programming of CTLs by inflammatory cytokines can be regulated by mTOR. We have shown that inhibition of mTOR during CTL activation leads to the enhancement of memory, but the molecular mechanisms remain largely unknown. Using high-throughput RNA-Seq, we identified genes and functions in mouse CTLs affected by mTOR inhibition through rapamycin. Of the 43,221 identified transcripts, 184 transcripts were differentially expressed after rapamycin treatment, corresponding to 128 annotated genes. Of these genes, 114 were downregulated and only 14 were upregulated. Most importantly, 50 of them are directly related to cell death and survival. In addition, several genes such as CD62L are related to migration. Furthermore, we predicted downregulation of transcriptional regulators based on the total differentially expressed genes, as well as the subset of apoptosis related genes. Quantitative PCR confirmed the differential expressions detected in RNA-Seq. We conclude that the regulatory function of rapamycin may work through inhibition of multiple genes related to apoptosis and migration, which enhance CTL survival into memory.
Keywords: CTLs, memory, mTOR, rapamycin, RNA-Seq
INTRODUCTION
Generation of functional memory CTLs holds promise for vaccines against some important chronic infections. However, effective strategies have not yet become available (Mescher et al. 2006). Activation of naïve CTLs requires three signals: the antigen derived from the proliferating pathogen, a co-stimulation provided by dendritic cells, and a third signal provided by inflammatory cytokines (Curtsinger et al. 2003; Mescher et al. 2006). Deficiency of signaling to IL-12 and IFNα abolishes memory CTL formation during infections, further supporting the critical role of inflammatory cytokines in memory CTL generation (Xiao et al. 2009). In addition, IL-12 can program memory CTLs in in vitro activation (Rao et al. 2010; Xiao et al. 2009), suggesting that programming of memory CTLs may take place during early activation, as demonstrated in some infections (Badovinac et al. 2004).
mTOR is a critical regulator of cells’ response to environmental factors, such as energy levels, insulin and other growth factors (Thomson et al. 2009). Recently, inhibition of mTOR in mice was reported to enhance memory CTL generation during infections through suppressing mTORC1 or modulating cell metabolisms (Araki et al. 2009; Pearce et al. 2009). Inhibition of mTOR using rapamycin during CTL activation leads to increased memory programming (Li et al. 2011; Rao et al. 2010), which is related to increased memory CTL precursors and their survival (Rao et al. 2010), and increased expansion (Li et al. 2011). However, how rapamycin regulates this memory CTL programming has not been evaluated on a global level.
RNA-Seq is a recently developed approach using high-throughput sequencing technologies. It allows global characterization and quantification of transcriptomes, which provide a precise measurement of transcripts (Ozsolak and Milos 2011). RNA-Seq has generated exciting results in the study of cancers and immune related diseases (Ceol et al. 2011). To determine the regulatory mechanisms of rapamycin on memory CTLs, we examined the differential gene expression in CTLs with rapamycin treatment using high-throughput RNA-Seq. Among 43,221 transcripts, 183 transcripts were found to be differentially expressed in rapamycin treatment as compared to the control, corresponding to 128 annotated genes in Ensembl database. Of these genes, many were directly implicated in immune function and cell survival. Most of the differentially expressed genes were downregulated, including genes associated with cell death and survival. The predicted transcriptional regulators were inhibited by rapamycin based on the total differentially expressed genes, as well as genes related to apoptosis and migration. Therefore, we consider that rapamycin may enhance memory CTL programming through promoting survival and regulating migration.
2. Results
RNA-Seq analysis of CTLs
We have shown that IL-12 can program memory CTLs during in vitro activation (Xiao et al. 2009). This memory programming can be further enhanced by inhibition of mTOR by rapamycin (Fig. 1). Rapamycin can increase the memory formation (Fig. 1A), and promote a more central memory phenotype (CD62L hi/KLRG1 lo) (Fig. 1B), consistent with recent reports (Li et al. 2011; Rao et al. 2010). To further study the underlying molecular mechanisms of rapamycin in this regulation, naïve OT-I cells were purified and subsequently stimulated by 3 signals (antigen, costimulation and interleukin-12) in the presence or absence of rapamycin, as described previously (Li et al. 2011). Stimulated cells were harvested 3 days after in vitro stimulation, and RNA was purified from these samples. After construction of cDNA libraries, whole transcriptome RNA-Seq was performed to determine the differences in gene expression between rapamycin treated and control CTLs, as demonstrated in Fig. S1. Sequence statistics were analyzed with FASTQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc website). Base sequence qualities and proportion of bases per cycle are shown in Fig. S2.
Figure 1.
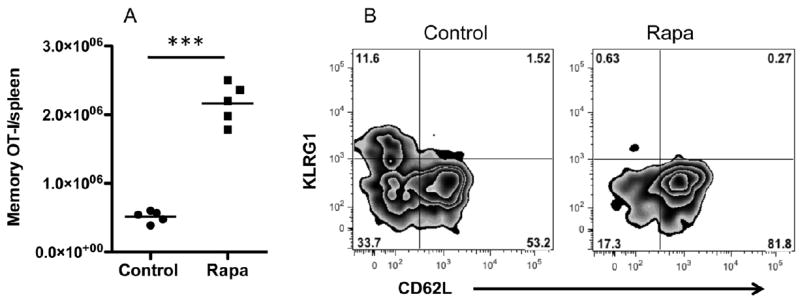
Enhancement of CTL memory by rapamycin. Purified OT1 cells were stimulated with antigen+B7+IL-12 in the presence of rapamycin at 250 ng/ml as (Li et al. 2011). Programmed CTLs were harvested at 72 hrs, and transferred into naïve B6 mice at 106/mouse through tail injection (Li et al. 2011). (A) Memory OT1 cells in spleen at day 40 after transfer. (B) Representative staining of spleen samples from (A). ***, P<0.001.
Based on the initial FastQC report, we trimmed the first 13 bp from all reads due to their low quality. Analysis of the trimmed reads was done using Tophat software for alignment with the reference genome (NCBI37/mm9.0). Of the total sequenced fragments, 91.20% and 90.98% were mapped to the reference genome, respectively (Table S1). In addition, the number of reads was comparable between rapamycin treated and control (Fig. S3). The high quality reads were then used for the downstream Tophat-Cufflink pipeline analysis.
Identification of differentially expressed genes
The application of RNA-Seq to immunology facilitates the global analysis of gene differential expression in response to rapamycin treatment in CTLs. Overall, we tested a total of 43,221 transcripts. Of these, 184 were determined by Cuffdiff method to be significantly differentially expressed after Benjamini-Hochberg correction for multiple-testing (Fig. 2) corresponding to 128 genes (FDR < 0.05). As seen in Fig. 3, of these 128 differentially expressed genes, 14 genes were upregulated by rapamycin, and 114 were found to be downregulated (Table S2).
Figure 2.
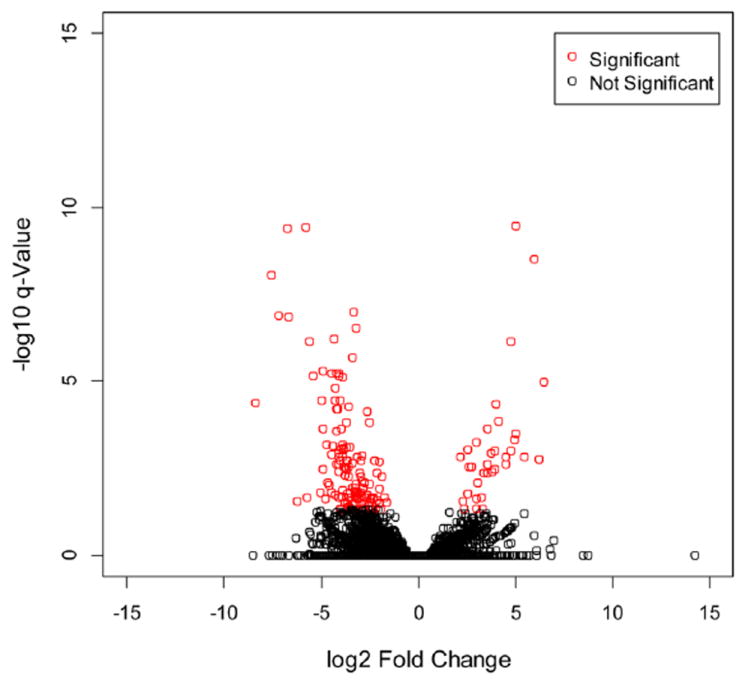
Volcano plot of differentially expressed transcripts by rapamycin. The 183 significantly differentially expressed protein-coding transcripts (represented in red), as determined by Cuffdiff default settings with normalization. X-axis values are log2 (fold change) and y-axis values are the −log10 of q-value. q-values indicate FDR-adjusted p-value for multiple-testing.
Figure 3.
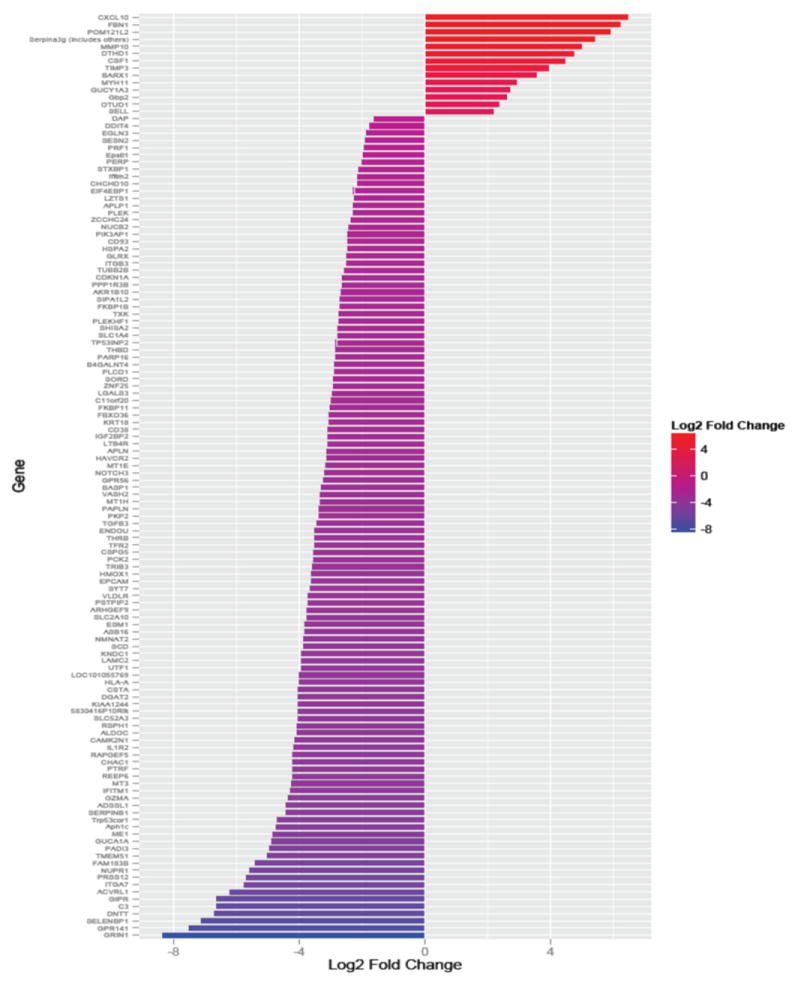
Significantly differentially expressed genes identified by Cuffdiff with differential fold change.
To investigate the molecular and cellular functions modulated by rapamycin, the significantly differentially expressed genes were submitted to Ingenuity Pathway Analysis (IPA) version 16542223 (Ingenuity Systems, Inc., Redwood City, CA). The first function affected by ramycin was Cell Death and Survival, which involves 50 genes, followed by Cellular Function and Maintenance (42 genes), Small Molecule Biochemistry (39 genes), Cellular Movement (35 genes) and Cell-to Cell Signaling and Interaction (37 genes) (Table 1). This suggests that regulation of cell death and survival may be an important function of rapamycin during memory programming. To further investigate the functions, differentially expressed genes were separated as downregulated (114) and upregualted (14), and imported into IPA as individual group. Cell Death and Survival and Cellular Function and Maintenance were the top functions in the downregulated genes, whereas Cellular Movement was the top function in upregulated genes (Table 2). Therefore, rapamycin may suppress apoptosis of CTLs by affecting multiple genes.
Table 1.
Molecular and Cellular Functions of differential expressed genes
| Functions | Genes | P value |
|---|---|---|
| Cell Death and Survival | 50 | 5.66E-07 – 6.81E-03 |
| Cellular Function and Maintenance | 42 | 2.81E-08 – 6.81E-03 |
| Small Molecule Biochemistry | 39 | 2.28E-06 – 6.81E-03 |
| Cellular Movement | 35 | 6.03E-06 – 6.81E-03 |
| Cell-to Cell Signaling and Interaction | 37 | 8.26E-08 – 6.81E-03 |
Table 2.
Molecular and Cellular Functions of gens downregulated or upregulated by rapamycin
| Function | Genes | P-value |
|---|---|---|
| Down regulated | ||
| Cell Death and Survival | 46 | 1.99E-06 – 1.08E-02 |
| Cellular Function and Maintenance | 46 | 1.05E-05 – 1.05-02 |
| Protein Synthesis | 18 | 9.55E-06 – 8.37E-03 |
| Cellular Assembly and Organization | 21 | 2.98E-05 – 1.05E-02 |
| Cellular Compromise | 14 | 2.98E-05 – 1.03E-02 |
| Up-regulated | ||
| Cellular Movement | 8 | 9.33E-07 – 1.04-02 |
| Cell-To-Cell Signaling and Interaction | 4 | 7.07E-05 – 1.04-02 |
| Cellular Development | 7 | 2.16E-04 – 1.03-02 |
| Cell Growth and Proliferation | 9 | 2.16E-04 – 1.04-02 |
| Cellular Function and Maintenance | 4 | 2.19E-04 – 4.68-03 |
Apoptosis is one of the major mechanisms for CTL contraction, during which about 90 to 95% of effector CTLs disappear, and the remaining small fraction becomes memory cells (Kaech et al. 2002; Wherry et al. 2003). Several of these genes are involved in apoptosis in CTLs, such as perforin and granzyme (Badovinac et al. 2004; Mescher et al. 2006), but most of them have not been directly studied in CTLs (Tables 3 and S3). Rapamycin generally regulated apoptotic genes through inhibitionof genes that promote apoptosis. For example, cyclin-dependent kinase inhibitor 1A (CDKN1A) is involved in the genes suppression by tumor protein p53 (TP53), so inhibition of CDKN1A by rapamycin will reduce the suppression of P53-responsive genes like survivin (Lohr et al. 2003). Transforming growth factor beta 3 (TGFB3) is involved in the apoptosis of cells in intestinal mucosa (Dunker et al. 2002), and TGFB3 may be required in FasL-Fas-Caspase pathway (Huang et al. 2011). TIMP metallopeptidase inhibitor 3 (TRIMP-3) promotes apoptosis in lung carcinoma (Kallio et al. 2011). Both TGFB3 and TIMP3 were inhibited by rapamycin, which should enhance CTL survival. On the other hand, the genes promoting survival were upregulated. For instance, Colony Stimulating Factor 1 (CSF-1) decreases apoptosis (Menke et al. 2009), and its expression was increased by rapamycin. Therefore, rapamycin may enhance the memory CTL programming by suppressing apoptosis and enhancing survival.
Table 3.
Representative apoptosis related genes regulated by rapamycin
| Gene | Fold Change | Type(s) | Location | Entrez Gene Name |
|---|---|---|---|---|
| CDKN1A | -2.649 | Kinase | Nucleus | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
| CSF1 | 4.473 | Cytokine | Extracellular Space | Colony stimulating factor 1 (macrophage) |
| DAP | -1.642 | Transcription regulator | Cytoplasm | Death-associated protein |
| DDIT4 | -1.775 | Other | Cytoplasm | DNA-damage-inducible transcript 4 |
| EIF4EBP1 | -2.261 | Translation regulator | Cytoplasm | Eukaryotic translation initiation factor 4E binding protein 1 |
| GRIN1 | -8.378 | Ion channel | Plasma Membrane | Glutamate receptor, ionotropic, N-methyl D-aspartate 1 |
| GZMA | -4.380 | Peptidase | Cytoplasm | Granzyme A (granzyme 1, cytotoxic T-lymphocyte-Gssociated serine esterase 3) |
| C3 | -6.671 | Ehemical - endogenous mammalian | unknown | |
| NOTCH3 | -3.219 | Transcription regulator | Plasma Membrane | Notch 3 |
| NUPR1 | -5.601 | Transcription regulator | Nucleus | Nuclear protein, transcriptional regulator, 1 |
| PERP | -2.031 | Other | Plasma Membrane | PERP, TP53 apoptosis effector |
| PRF1 | -1.976 | Other | Cytoplasm | Perforin 1 (pore forming protein) |
| TGFB3 | -3.483 | Growth factor | Extracellular Space | Transforming growth factor, beta 3 |
| TIMP3 | 3.947 | Other | Extracellular Space | TIMP metallopeptidase inhibitor 3 |
There were 14 genes upregulated by rapamycin (Table S2), including CD62L (Araki et al. 2011; Li et al. 2011), which is an important marker for central memory CTL and directs the migration of immune cells to secondary lymphoid tissues (Obar and Lefrancois 2010). Another adhesion molecule, CXCL10, was also enhanced by rapamycin. CXCL10 (another name IP-10) is a chemokine mainly produced by macrophages and dendritic cells during infections (Liu et al. 2011), and can also be produced by mucosal CD4 and NK cells (Singh et al. 2008). This chemokine is critical for recruitment of immune cells, including CTLs, to the site of infection to perform immune function (Liu et al. 2011). Therefore, the regulation of rapamycin on CTLs may include two aspects: to enhance their survival, thus forming more memory, and also to take an active route to circulating both inside secondary lymphoid tissues and original site of infection.
IPA predicted that the cytotoxicity of the cells in the rapamycin treated group was overall decreased as compared to control (Fig. S4). This determination was based on the downregulation of genes such as granzyme and perforin, which are the key cytotoxic proteins used by CTLs to kill their targets (Curtsinger et al. 2003; Mescher et al. 2006). A serine protease inhibitor Spi2a (Serpina3g), which has been implicated in aiding memory CTLs formation (Liu et al. 2004), was upregulated. This means that rapamycin may also regulate memory related genes like Spi2a directly. The decrease in these cytotoxic genes, along with the increase in expression of Serpina3g may also be one of the mechanisms contributing to greater memory formation.
Network analysis of apoptosis related genes regulated by rapamycin
There were 45 apoptosis related genes regulated by rapamycin (Table 3 and Table S3). By running through IPA program, 27 of them were directly connected in a network (Fig. 4). This suggests that there may be common upstream molecules responsible for their regulation. The analysis of upstream molecules identified some important transcriptional regulators linked to these 45 apoptosis-related genes (Fig. S5). STAT4 is a critical transcription factor involved in IL-12 signaling (Watford et al. 2004), and the genes regulated by rapamycin suggested that STAT4 function was inhibited. In addition, tumor protein p63 (TP63) shares high structure similarity with TP53, and interacts with TP53 and tumor protein p73 (TP73) in tumorigensis (Graziano and De Laurenzi 2011). TP63 induces apoptosis in addition to its critical role in development and epithelial differentiation (Dotsch et al. 2010), therefore the inhibition of TP63 may suppress apoptosis, thus enhancing CTL survival to become memory. Interestingly, three upstream transcription regulators regulated by rapamycin were related to hyopoxic conditions: Aryl hydrocarbon receptor nuclear translocator 2 (ARNT2) (Garritano et al. 2013), Hypoxia-inducible factor 1-alpha (HIF-1) (Barbi et al. 2013) and single-minded homolog 1 (SIM1) (Michaud et al. 1998). This suggests that rapamycin may induce a hypoxia-like response in CTLs. Interestingly, HIF-1 is required for Th17 generation (Barbi et al. 2013).
Figure 4.
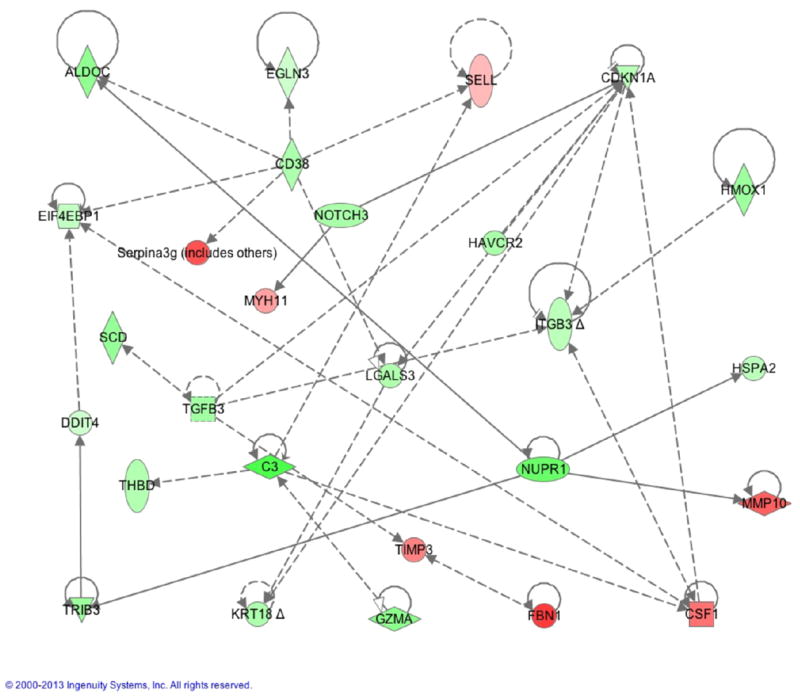
Network analysis of apoptosis related genes regulated by rapamycin. Network was composed through IPA. Green: significantly downregulated. Red: significantly upregulated.
Quantitative RT-PCR Validation of the RNA-Seq Results
To validate the precision of our RNA-Seq data, we randomly selected 15 candidate genes (10 downregulated, 5 upregulated) to perform real-time RT-PCR. These genes cover immune function, migration and apoptosis. Real time PCR found that the expression levels were consistent with the results detected in RNAseq, all 10 downregulated genes were downregulated in real time PCR, whereas upregulated genes were also enhanced by rapamyin in real time PCR (Fig. 5A). To compare the expression levels directly in both RNA-Seq and real time PCR, we performed Spearman rank correlation. These two data sets were nicely correlated (Fig. 5B), with Pearson correlation value rho = 0.9285714, and p value -value < 2.2e-16. All the PCR products were confirmed by sequencing. Thus, RNA-Seq gave reliable gene expression measurements, which were validated by quantitative PCR.
Figure 5.
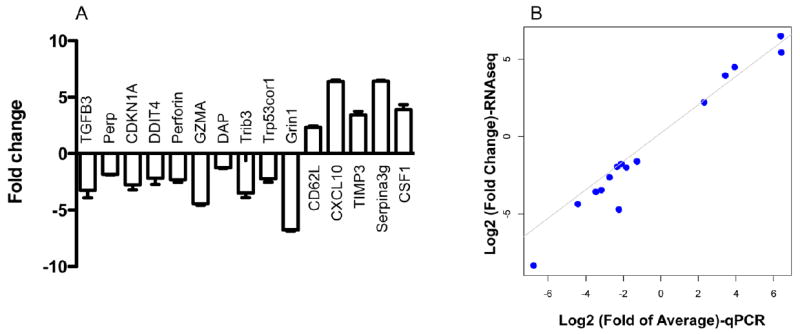
Validation of RNA-Seq data using quantitative RT-PCR. (A) qRT-PCR analysis of 10 selected downregulated genes and 5 upregulated genes. Fold change (y-axis) were generated by comparing rapamycin treated sample to control. The data represent mean of three individual samples plus SEM. The experiment was repeated twice with similar results. (B) The Log2 Fold of Average qRT-PCR (x-axis) were plotted against Log2 Fold Change values of RNA sequencing(y-axis), (Pearson correlation values >0.92)
3. Discussion
Rapamycin enhances memory CTL programming (Li et al. 2011; Rao et al. 2010), which could be related to mTORC1(Zeng et al. 2013). Using high throughput sequencing, we were able to find only 128 genes that were significantly altered by rapamycin, and most of them were inhibited, which is consistent with the function of mTOR in cell growth and proliferation (Araki et al. 2009; Chi 2012; Hara et al. 2002). Of them, 45 genes are directly related to apoptosis (Table 3), and most of these genes are somewhat connected (Fig. 4). This suggests that effects of the regulation of mTOR by rapamycin on memory CTL programming may be related to regulation of survival for activated CTLs.
Survival signals for CTLs can be provided by stimulation from TCR, costimulation and cytokines (Kaech et al. 2003; Williams and Bevan 2007), presumably through regulation of prosurvival and proapoptotic molecules, such as Bcl-2 and Bim in Bcl-2 family (Gillings et al. 2009). Many transcription factors are known to be involved in Bcl-2 regulation including cyclic AMP response element (CRE)-binding protein (CREB) (Xiang et al. 2006) and STAT5 (Hand et al. 2010). In addition, transcription factors traditionally belonging to other pathways participate in the regulation of cell survival. T cell factor 1 in canonical Wnt signaling (Staal et al. 2008) modulates memory CTLs through regulation of Bcl-2 expression and IL-2/IL-15 signaling (Zhou et al. 2010). Similarly, proapoptotic molecules can also be regulated by transcription factors. Forkhead box O transcription factor 1 (FOXO1) regulates expression of FAS and Bim (Dejean et al. 2010; Kerdiles et al. 2009). Because survival is a key for memory CTLs, it is possible that the regulation of critical transcription factors is one of the major molecular mechanisms for rapamycin’s regulation on memory programming. We found that most of genes related to apoptosis were inhibited by rapamycin. Although further functional study is warranted, we postulate that this apoptosis regulation by rapamycin may be not single gene dependent, but rather a general feature of regulation of multiple genes.
Treatment with rapamycin modifies the effects of IL-12 on 3SI CD8 T-cells. The addition of rapamycin decreased genes related to effector killing functions such as granzyme and perforin, as opposed to 3SI (antigen, Costimulation plus IL-12) treatment without rapamycin which upregulated those genes compared to 2SI (only antigen and costimulation). Agarwal et al. found that CD62L (SELL) was down regulated in the 3SI treated CD8 T cells compared to 2SI (Agarwal et al. 2009), but was upregulated in the 3SI + Rapamycin treatment (Fig. 5). Some apoptosis related genes were also upregulated in 3SI treatment versus 2SI (Agarwal et al. 2009), but were down regulated in the rapamycin treated 3SI cells (Table 3 and S3). These include PERP, TRIB3, CDKN1A, EIF4EBP1, and DDIT4, and TGF-β related genes. TGFB3 was down regulated by rapamycin, while 3SI up regulated TGFB3 compared to 2SI (Agarwal et al. 2009). Furthermore, ACVRL1, a type I cell-surface receptor for TGF-β, was upregulated by IL-12, but down regulated by 3SI + rapamycin treatment. Therefore, rapamycin may enhance memory CTL programming by modulating some of the effects of IL-12.
CXCL10 is critical for the induction of protective CTLs in intracellular parasite infections (Majumder et al. 2012). CXCL10 is induced by IFNg/TNFa/IL-1 (Liu et al. 2011), and recently is reported to be induced by type I IFNa during LCMV infection (Sung et al. 2012). The receptor for CXCL10 is CXCR3, which also binds to CXCL9 and CXCL11 (Singh et al. 2008). Agarwal et al. reported that CXCL10 is upregulated in CTLs by IFNα, but not IL-12 during in vitro stimulation (Agarwal et al. 2009). This gene was drastically upregulated by rapamycin (Fig. 5). Interestingly, CXCR3, the receptor for CXCL10 was also enhanced by rapamycin (data not shown), and is regulated by IL-12 ((Agarwal et al. 2009). This means that rapamycin is regulating the expression of a pair of adhesion molecules, CXCL10 and CXCR3. It is possible that this paired regulation enables activated CTLs to migrate to the site of infection more efficiently as a group. Further experiments are needed to determine if this is functionally significant.
The upregulation of CXCL10 and CD62L by rapamycin suggests that the memory programming, is not only regulating survival, but also directing the CTLs where to migrate, which is consistent with previous reports (Li et al. 2011; Rao et al. 2010). The central memory phenotype of the rapamycin regulated memory CTLs may keep most of them in circulating the secondary lymphoid tissues (Obar and Lefrancois 2010), whereas the effector memory CTLs reside in the tissues (Jameson and Masopust 2009). This TCM resides close to the entry channel in draining lymph nodes, which makes them accessible to the draining pathogen through the tunnel (Kastenmuller et al. 2013; Sung et al. 2012). The residence of TCM may be directed by different chemokines in different infections (Kastenmuller et al. 2013; Sung et al. 2012). Therefore, we postulate that the regulatory function of rapamycin on memory programming is through an integrated process: regulation of apoptosis and cell migration as a general pattern of biological process.
4. Materials and Methods
Animal
OT-I mice, gifts from MF Mescher, University of Minnesota, MN, express a transgenic TCR specific for H-2Kb OVA257–26. The mice were housed in specific pathogen free conditions at the University of Maryland. These studies have been reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Maryland (protocol ID R-12–23). All conjugated fluorescent Abs were purchased from BD Biosciences (San Diego, CA), eBioscience (San Diego, CA) or Biolegend (San Diego, CA).
T-Cell Purification
This was performed in the same way we reported previously [16]. In brief, inguinal, axillary, brachial, cervical, and mesenteric lymph nodes (LNs) were harvested from WT OT-I mice. The nodes were then pooled and disrupted to obtain a single cell suspension necessary for CD8+ CD44lo cell enrichment. The enrichment was performed using negative selection with MACS magnetic beads (Milteny Biotec). This involved coating the cells with FITC-labeled antibodies specific for CD4, B220, I-Ab, and CD44. Anti-FITC magnetic MicroBeads (Miltenyi Biotech) were then added and the suspension passed through separation columns attached to a MACS magnet. Cells that did not bind were collected, and were >95% CD8+ and <0.5% CD44hi.
In vitro stimulation of naïve OT-I T cells
Naïve OT-I CD8 cells were purified as described above and stimulated for 3 days in vitro in flat-bottom microtiter wells stimulated with antigen (DimerX H-2Kb:Ig fusion protein loaded with OVA257-264 peptide; BD Pharmingen), recombinant B7-1/Fc chimeric protein (R&D Systems) and 2U/ml of murine rIL-12 (R&D Systems) as previously described (Li et al. 2011; Xiao et al. 2009). 3 × 105 cells in 1.5 ml Allos media were placed in each well and 2.5U/ml IL-2 added to all wells (24-well plate). For rapamycin treated CTLs, cultures were supplemented with rapamycin at 250 ng/ml. Rapamycin was purchased from EMD (Gibbstown, NJ).
RNA preparation
RNA samples were extracted using RNeasy Micro kit (Qiagen) following the manufacturer’s instruction. The isolated RNA samples were stored at -80 degree upon to use.
RNA-Seq construct preparation
cDNA libraries were prepared from 1 microgram of RNA per sample using Illumina TruSeq RNA Sample Preparation v2 kit following the manufacturer’s instructions. Sequencing libraries were created and sequenced with Illumina’s HiSeq 1000 at the University of Maryland IBBR Sequencing facility. In brief, samples were tested for quality with the Agilent Bioanalyzer chip before Illumina adapters were added after PCR amplification. Fifteen cycles of PCR were used to amplify the cDNA library with Illumna adaptors. qPCR was also performed for quantity check after the Illumina cDNA libraries were prepared. Finally, Cluster generation was performed using Illumina cBot at 15pM starting concentration for each library. The libraries were barcoded, multiplexed, and sequenced in one HiSeq 1000 lane on the Illumina’s HiSeq 1000.
Pre-process and analyzing RNA SEQ reads using Tophat-cufflink
The raw quality of the reads of rapamycin treated and control samples were checked using FastQC(http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Based on the FastQC summarized statistics, we trimmed and the first 13 bp of low quality reads using fastx_trimmer in the Fastx toolkit(http://hannonlab.cshl.edu/fastx_toolkit/index.html). The final reads were mapped to the reference Mouse genome (mm 9, NCBI Build 37) using Tophat (v-2.0.4.Linux_x86_64) (Kim et al. 2013). Cufflinks (v-2.0.2) was employed to assemble transcript models from RNA-Seq alignments and to estimate transcript and abundances (Trapnell et al. 2010). Differential expression of annotated genes was identified using Cuffdiff (Trapnell et al. 2013), a program in the Cufflinks, the upper quartile normalization option was used to improve differential expression tests for less abundant transcripts. Genes with a false discovery rate (FDR) < 0.05 were considered significant. Otherwise, the default options were used in these software packages in our analyses (Trapnell et al. 2012).
Enriched pathway analysis of differentially expressed genes
To further identify canonical signaling pathways, molecular networks, and biological functions, 128 genes determined to be significantly differentially expressed between rapamycin treatment and control by TopHat/Cufflinks were uploaded into the Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems, http://www.ingenuity.com). The IPA database is maintained and edited by humans, and contains genes, proteins, and RNA in order to not only find associations between expression data and canonical pathways, but also build new networks. The significance of associations were computed using the right-tailed Fisher exact test. All signaling pathways identified by IPA with a P value ≤0.05 have a statistically significant, nonrandom association.
Real-time RT-PCR
cDNA was synthesizes from purified RNA using QuantiTech Reverse Transcription kit (Qiagen). The synthesized cDNAs were further 1:20 diluted with Rnase and Dnase free water. Quantitation PCR was performed using 2 microlitter of diluted cDNA as a template in a 20 ul of SybrGreen (Biorad) surpermix reaction on a MyiQ™ Single-Color Real-Time PCR Detection System (Bio-Rad). Details of the real-time PCR conditions used are available upon request. Primer sequences are listed in Table S4.
Supplementary Material
Acknowledgments
We thank Mr. Ken Class for technical assistance. This work was partially supported by National Institutes of Health Grants R21AI095715A (to X. Z.) and Startup from UMD (to X. Z.). This work was supported in part by AFRI grant No. 2011-67015-30183 from USDA NIFA (to GEL). Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. The USDA is an equal opportunity provider and employer.
References
- Agarwal P, Raghavan A, Nandiwada SL, Curtsinger JM, Bohjanen PR, Mueller DL, Mescher MF. Gene regulation and chromatin remodeling by IL-12 and type I IFN in programming for CD8 T cell effector function and memory. J Immunol. 2009;183:1695–704. doi: 10.4049/jimmunol.0900592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Ellebedy AH, Ahmed R. TOR in the immune system. Curr Opin Cell Biol. 2011;23:707–15. doi: 10.1016/j.ceb.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–12. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badovinac VP, Porter BB, Harty JT. CD8+ T cell contraction is controlled by early inflammation. Nat Immunol. 2004;5:809–17. doi: 10.1038/ni1098. [DOI] [PubMed] [Google Scholar]
- Barbi J, Pardoll D, Pan F. Metabolic control of the Treg/Th17 axis. Immunol Rev. 2013;252:52–77. doi: 10.1111/imr.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, Fritsch L, Lin WM, Hollmann TJ, Ferre F, Bourque C, Burke CJ, Turner L, Uong A, Johnson LA, Beroukhim R, Mermel CH, Loda M, Ait-Si-Ali S, Garraway LA, Young RA, Zon LI. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–7. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–38. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtsinger JM, Lins DC, Mescher MF. Signal 3 determines tolerance versus full activation of naive CD8 T cells: dissociating proliferation and development of effector function. J Exp Med. 2003;197:1141–51. doi: 10.1084/jem.20021910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejean AS, Hedrick SM, Kerdiles YM. Highly specialized role of Forkhead box O transcription factors in the immune system. Antioxid Redox Signal. 2010;14:663–74. doi: 10.1089/ars.2010.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotsch V, Bernassola F, Coutandin D, Candi E, Melino G. p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol. 2010;2:a004887. doi: 10.1101/cshperspect.a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker N, Schmitt K, Schuster N, Krieglstein K. The role of transforming growth factor beta-2, beta-3 in mediating apoptosis in the murine intestinal mucosa. Gastroenterology. 2002;122:1364–75. doi: 10.1053/gast.2002.32991. [DOI] [PubMed] [Google Scholar]
- Garritano S, Inga A, Gemignani F, Landi S. More targets, more pathways and more clues for mutant p53. Oncogenesis. 2013;2:e54. doi: 10.1038/oncsis.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillings AS, Balmanno K, Wiggins CM, Johnson M, Cook SJ. Apoptosis and autophagy: BIM as a mediator of tumour cell death in response to oncogene-targeted therapeutics. FEBS J. 2009;276:6050–62. doi: 10.1111/j.1742-4658.2009.07329.x. [DOI] [PubMed] [Google Scholar]
- Graziano V, De Laurenzi V. Role of p63 in cancer development. Biochim Biophys Acta. 2011;1816:57–66. doi: 10.1016/j.bbcan.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Hand TW, Cui W, Jung YW, Sefik E, Joshi NS, Chandele A, Liu Y, Kaech SM. Differential effects of STAT5 and PI3K/AKT signaling on effector and memory CD8 T-cell survival. Proc Natl Acad Sci U S A. 2010;107:16601–6. doi: 10.1073/pnas.1003457107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–89. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- Huang X, Yokota T, Iwata J, Chai Y. Tgf-beta-mediated FasL-Fas-Caspase pathway is crucial during palatogenesis. J Dent Res. 2011;90:981–7. doi: 10.1177/0022034511408613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson SC, Masopust D. Diversity in T cell memory: an embarrassment of riches. Immunity. 2009;31:859–71. doi: 10.1016/j.immuni.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4:1191–8. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol. 2002;2:251–62. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- Kallio JP, Hopkins-Donaldson S, Baker AH, Kahari VM. TIMP-3 promotes apoptosis in nonadherent small cell lung carcinoma cells lacking functional death receptor pathway. Int J Cancer. 2011;128:991–6. doi: 10.1002/ijc.25404. [DOI] [PubMed] [Google Scholar]
- Kastenmuller W, Brandes M, Wang Z, Herz J, Egen JG, Germain RN. Peripheral prepositioning and local CXCL9 chemokine-mediated guidance orchestrate rapid memory CD8+ T cell responses in the lymph node. Immunity. 2013;38:502–13. doi: 10.1016/j.immuni.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerdiles YM, Beisner DR, Tinoco R, Dejean AS, Castrillon DH, DePinho RA, Hedrick SM. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol. 2009;10:176–84. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Garcia K, Sun Z, Xiao Z. Temporal regulation of rapamycin on memory CTL programming by IL-12. PLoS ONE. 2011;6:e25177. doi: 10.1371/journal.pone.0025177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Guo S, Hibbert JM, Jain V, Singh N, Wilson NO, Stiles JK. CXCL10/IP-10 in infectious diseases pathogenesis and potential therapeutic implications. Cytokine Growth Factor Rev. 2011;22:121–30. doi: 10.1016/j.cytogfr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Phillips T, Zhang M, Wang Y, Opferman JT, Shah R, Ashton-Rickardt PG. Serine protease inhibitor 2A is a protective factor for memory T cell development. Nat Immunol. 2004;5:919–26. doi: 10.1038/ni1107. [DOI] [PubMed] [Google Scholar]
- Lohr K, Moritz C, Contente A, Dobbelstein M. p21/CDKN1A mediates negative regulation of transcription by p53. J Biol Chem. 2003;278:32507–16. doi: 10.1074/jbc.M212517200. [DOI] [PubMed] [Google Scholar]
- Majumder S, Bhattacharjee S, Paul Chowdhury B, Majumdar S. CXCL10 is critical for the generation of protective CD8 T cell response induced by antigen pulsed CpG-ODN activated dendritic cells. PLoS ONE. 2012;7:e48727. doi: 10.1371/journal.pone.0048727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke J, Iwata Y, Rabacal WA, Basu R, Yeung YG, Humphreys BD, Wada T, Schwarting A, Stanley ER, Kelley VR. CSF-1 signals directly to renal tubular epithelial cells to mediate repair in mice. J Clin Invest. 2009;119:2330–42. doi: 10.1172/JCI39087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mescher MF, Curtsinger JM, Agarwal P, Casey KA, Gerner M, Hammerbeck CD, Popescu F, Xiao Z. Signals required for programming effector and memory development by CD8+ T cells. Immunol Rev. 2006;211:81–92. doi: 10.1111/j.0105-2896.2006.00382.x. [DOI] [PubMed] [Google Scholar]
- Michaud JL, Rosenquist T, May NR, Fan CM. Development of neuroendocrine lineages requires the bHLH-PAS transcription factor SIM1. Genes Dev. 1998;12:3264–75. doi: 10.1101/gad.12.20.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obar JJ, Lefrancois L. Early signals during CD8(+) T cell priming regulate the generation of central memory cells. J Immunol. 2010;185:263–72. doi: 10.4049/jimmunol.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–7. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 2010;32:67–78. doi: 10.1016/j.immuni.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh UP, Singh S, Singh R, Cong Y, Taub DD, Lillard JW., Jr CXCL10-producing mucosal CD4+ T cells, NK cells, and NKT cells are associated with chronic colitis in IL-10(-/-) mice, which can be abrogated by anti-CXCL10 antibody inhibition. J Interferon Cytokine Res. 2008;28:31–43. doi: 10.1089/jir.2007.0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal FJ, Luis TC, Tiemessen MM. WNT signalling in the immune system: WNT is spreading its wings. Nat Rev Immunol. 2008;8:581–93. doi: 10.1038/nri2360. [DOI] [PubMed] [Google Scholar]
- Sung JH, Zhang H, Moseman EA, Alvarez D, Iannacone M, Henrickson SE, de la Torre JC, Groom JR, Luster AD, von Andrian UH. Chemokine guidance of central memory T cells is critical for antiviral recall responses in lymph nodes. Cell. 2012;150:1249–63. doi: 10.1016/j.cell.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson AW, Turnquist HR, Raimondi G. Immunoregulatory functions of mTOR inhibition. Nat Rev Immunol. 2009;9:324–37. doi: 10.1038/nri2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watford WT, Hissong BD, Bream JH, Kanno Y, Muul L, O’Shea JJ. Signaling by IL-12 and IL-23 and the immunoregulatory roles of STAT4. Immunol Rev. 2004;202:139–56. doi: 10.1111/j.0105-2896.2004.00211.x. [DOI] [PubMed] [Google Scholar]
- Wherry EJ, Teichgraber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, Ahmed R. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol. 2003;4:225–34. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- Williams MA, Bevan MJ. Effector and memory CTL differentiation. Annu Rev Immunol. 2007;25:171–92. doi: 10.1146/annurev.immunol.25.022106.141548. [DOI] [PubMed] [Google Scholar]
- Xiang H, Wang J, Boxer LM. Role of the cyclic AMP response element in the bcl-2 promoter in the regulation of endogenous Bcl-2 expression and apoptosis in murine B cells. Mol Cell Biol. 2006;26:8599–606. doi: 10.1128/MCB.01062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Casey KA, Jameson SC, Curtsinger JM, Mescher MF. Programming for CD8 T cell memory development requires IL-12 or type I IFN. J Immunol. 2009;182:2786–94. doi: 10.4049/jimmunol.0803484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Yang K, Cloer C, Neale G, Vogel P, Chi H. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 2013;499:485–90. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity. 2010;33:229–40. doi: 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


