Abstract
Signaling through Toll-like receptors (TLRs), crucial molecules in the induction of host defense responses, requires adaptor proteins that contain a Toll/interleukin-1 receptor (TIR) domain. The pathogen Staphylococcus aureus produces several innate immune-evasion molecules that interfere with the host's innate immune response. A database search analysis suggested the presence of a gene encoding a homologue of the human TIR domain in S. aureus MSSA476 which was named staphylococcal TIR domain protein (TirS). Ectopic expression of TirS in human embryonic kidney, macrophage and keratinocyte cell lines interfered with signaling through TLR2, including MyD88 and TIRAP, NF-κB and/or mitogen-activated protein kinase pathways. Moreover, the presence of TirS reduced the levels of cytokines MCP-1 and G-CSF secreted in response to S. aureus. The effects on NF-κB pathway were confirmed using S. aureus MSSA476 wild type, an isogenic mutant MSSA476ΔtirS, and complemented MSSA476ΔtirS +pTirS in a Transwell system where bacteria and host cells were physically separated. Finally, in a systematic mouse infection model, TirS promoted bacterial accumulation in several organs 4 days postinfection. The results of this study reveal a new S. aureus virulence factor that can interfere with PAMP-induced innate immune signaling in vitro and bacterial survival in vivo.
Key Words: Staphylococcus aureus, Staphylococcal TIR domain protein, TirS, TLR signaling, Mitogen-activated protein kinase, NF-κB, Bacterial TIR domain
Introduction
The human Toll-like receptors (TLRs) TLR1-TLR10 play important roles in the induction of host innate immune responses [1, 2]. They recognize a wide range of conserved bacterial structures [2, 3], collectively called pathogen-associated molecular patterns, PAMPs [1]. TLRs and interleukin-1 receptors (IL-1Rs) all have a conserved cytoplasmic region of approximately 200 amino acids known as the Toll/IL-1 receptor (TIR) domain [1, 4]. The intracellular TIR domain can recruit a variety of TIR-containing adaptor protein(s) including myeloid differentiation factor 88 (MyD88), MyD88 adaptor-like protein (Mal, also known as TIR-associated protein or TIRAP), TIR domain-containing adaptor protein including interferon-β (TRIF), TRIF-related adaptor molecule, TRAM, and sterile adaptor α- and armadillo-motif-containing protein (SARM). TIRAP in particular acts as a bridging molecule for MyD88 in the context of TLR2 and TLR4 activation [1, 2]. The ligand-receptor-induced recruitment of TIR-containing adaptors attracts IL-1R-associated kinases, IRAKs, which then transmit the signal via a downstream signaling pathway. A critical outcome of this process is activation of mitogen-activated protein kinase (MAPK) cascades and the transcription factor NF-κB, resulting in induction of a pro-inflammatory host response [1].
The importance of TLR signaling in human immune defense is implicated by the impact of naturally occurring genetic mutations or polymorphisms in innate immune genes. Pediatric patients and mice with MyD88 or IRAK4 deficiencies are predisposed to bacterial infections caused by Gram-positive pathogens [5, 6, 7]. Also, more subtle changes such as certain polymorphisms in human genes that encode TLRs or TLR-intracellular signaling components affect susceptibility to infection. For example, humans with TLR2 Arg753Gln show increased susceptibility to infection with Mycobacterium tuberculosis and Gram-positive bacteria, while individuals with TLR4 Asp299Gly or Thr399Ile are hyporesponsive to lipopolysaccharide resulting in increased susceptibility to Gram-negative bacterial infections [8].
Many bacteria use molecular mimicry of host proteins to perturb the host immune system and establish a critical population size in vivo [9]. An initial report on Escherichia coli described a TIR-containing protein that suppressed innate immunity by interfering with TLR signaling [10]. This inhibition is based on structural mimicry with the TIR domains of the host receptors and their adaptors [11, 12]. Subsequently, TIR-containing proteins have been reported in a wide range of human non-pathogenic and pathogenic bacteria [10, 12, 13, 14, 15, 16, 17, 18], as well as fungi, archaea, viruses and eukaryotes [17, 19]. Molecular studies on bacterial TIR-containing proteins have been conducted for several Gram-negative bacteria including Salmonella enterica (TIR-like protein A, TlpA) [13], Brucella spp. (TIR domain-containing protein B, TcpB, also called Brucella TIR-protein 1, Btp1) [10, 16], uropathogenic E. coli (TIR-containing protein C, TcpC) [10, 20], Yersinia pestis (Y. pestis TIR domain protein, YpTdp) [18] and Paracoccus dentrificans (P. dentrificans TIR-like protein, PdTLP) [14]. As a common theme, these studies show that bacterial TIR-containing proteins can negatively interfere with TLR signaling [10, 13, 15, 16].
Comparison of amino acid sequences of TIR domains in eukaryotic TIR-containing proteins reveal some common amino acid sequence motifs, called box 1, box 2 and box 3, where boxes 1 and 2 are of special importance in mediating signaling [21]. The structure of the TIR domain of human TLR1 consists of a five-stranded parallel β-sheet (βA-βE) surrounded by five helices (αA-αE) connected by loops. The functionally relevant BB loop connects strand βB and αB, and is located within box 2. Most amino acid sequence variations among TIR domains are found in helices αB and αD and loops BB, CD and DD. The diversity is suggested to be crucial for the specificity of signal transduction [22]. The BB-loop of bacterial TIR proteins was found to be of particular importance in the suppressive effect on host signaling [16, 23].
S. aureus is an important nosocomial and community-acquired pathogen. Increased antibiotic resistance among hospital-acquired strains is a global concern and continuing challenge for public health [24]. MSSA476 belongs to a main global lineage associated with invasive community-acquired disease and contains a new type of staphylococcal cassette chromosome (SCC) element, SCC476, which is merged at the same site on the chromosome as SCCmec elements in methicillin-resistant S. aureus, MRSA [25]. The presence of a microbial TIR domain in the methicillin-susceptible S. aureus strain MSSA476 has been suggested [10] but never pursued experimentally. The aim of this study was to confirm the presence of a putative TIR domain-containing protein in S. aureus strain MSSA476, and to investigate its possible interference with TLR signaling and influence on bacterial virulence.
Methods and Materials
Bacterial Strains, Mammalian Cell Lines and Plasmids
S. aureus subsp. aureus Rosenbach MSSA476 was purchased from LGC standard AB (ATCC-BAA-1721; Boras, Sweden). S. aureus 61010305 (not containing tirS), spa type t186, was obtained from the Tromsø Staph and Skin Study, Norway [26]. HEK293 cells, a human embryonic kidney cell line, were purchased from the European Collection of Cell Cultures (Porton Down, UK) while HaCaT cells, a human keratinocyte cell line, were purchased from PromoCell (Heidelberg, Germany). RAW264.7 cells, a mouse macrophage cell line, were a kind gift from N. Seredkina. The plasmids and primers are described in table 1.
Table 1.
Plasmids and primers
| Plasmids/primers | Properties/sequence | Use | Origin |
|---|---|---|---|
| pCMV-Myc-TLR2 | Eukaryotic expression plasmid encoding TLR2 | Transfection | [43] |
| AU1-MyD88 full | Eukaryotic expression plasmid encoding the full-length human MyD88 | Transfection | [44] |
| DN-AU1-MyD88 | Eukaryotic expression plasmid encoding TIR domain (152–296 aa) of human MyD88; acts as dominant negative on MyD88 signaling | Transfection | [44] |
| NF-κB-luc | Reporter of the activity of transcription factor NF-κB | Transfection | Stratagene |
| Myc-DDK-TIRAP | Eukaryotic expression plasmid encoding the adaptor protein TIRAP | Transfection | OriGene |
| pEGFP-TirS | Eukaryotic expression plasmid encoding TirS | Transfection | This work |
| pEGFP-C2 | Eukaryotic expression vector | Transfection | BD Biosciences |
| pCMV | Eukaryotic expression vector | Transfection | Clontech |
| pcDNA3 | Eukaryotic expression vector | Transfection | Invitrogen |
| PKOR1 | S. aureus shuttle vector | Making isogenic mutant | [45] |
| pCM29 | GFP fluorescence expression vector | Reporter | [46] |
| tirS prev For | 5′-CAGTCTTACCTGCTCGATTC-3′ | RT-PCR | This work |
| tirS prev Rev | 5′-CTTACGCACATCAATAACGA-3′ | RT-PCR | This work |
| gyrA prev For | 5′-GTCAAAATCTGCAAAAATAGCTAG-3′ | RT-PCR | This work |
| gyrA prev Rev | 5′-GTCAAAATCTGCAAAAATAGCTAG-3′ | RT-PCR | This work |
| tirS EcoRI For | 5′-AATCTAGAATTCGAGGTATTATATGTCAG-3′ | Cloning | This work |
| tirS BamHI Rev | 5′-ATTGTTCTCGGATCCTTCCCTCTTTGCTTTTAAAG-3′ | Cloning | This work |
| Up For tirS + attB1 | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCGCTAGGTTG GGCTTTTCCACAT-3′ | Generating fusion construct | This work |
| Up Rev tirS | 5′-AAAGTAGATAACCAATACTATTAATAATACCTCGCTTTTT ATAATC-3′ | Generating and sequencing fusion construct | This work |
| Down For tirS | 5′-GATTATAAAAAGCGAGGTATTATTAATAGTATTGGTTATC TACTTT-3′ | Generating and sequencing fusion construct | This work |
| Down Rev tirS + attB2 | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCAGTTACTCC CGCTTCTGTTAATG-3′ | Generating fusion construct | This work |
| pKOR1 For | 5′-AGCTCCAGATCCATATCCTTC-3′ | Sequencing fusion construct | This work |
| pKOR1 Rev | 5′-CACACAGGAAACAGCTATGAC-3′ | Sequencing fusion construct | This work |
| tirS KO confirm For | 5′-GCTTCGAGAGTGGTTAGAC-3′ | Isogenic mutant confirm | This work |
| tirS-CL-For | 5′-GCGGAATTCATGTCAGTATTAGAAACTAAATTAAAAAGTC-3′ | Complementation | This work |
| tirS-CL-Rev | 5′-GCGAAGCTTCTAATTCTTAGAATTAACGATTACTTG-3′ | Complementation | This work |
| pDC123 | 5′-GCGGAATTCATGTCAGTATTAGAAACTAAATTAAAAAGTC-3′ | Shuttle vector used for complementation | [45] |
| tirS KO confirm Rev | 5′-GGTTATCATCAAATGAGCTACCTG-3′ | Isogenic mutant confirm | This work |
| tirS Int For | 5′-TTAATCTTCAAAAAGAGCAGTCTAGG-3′ | Isogenic mutant confirm | This work |
| tirS Int Rev | 5′-GGGTTGTATGCACGTACATCTTCAAC-3′ | Isogenic mutant confirm | This work |
| tirS-prom For | 5′-CGCCTGCAGGCTCCACAGTTTGTTCATCCTGAT-3′ | Generating reporter construct | This work |
| tirS-prom Rev | 5′-CGCGGTACCCGTTTGTTCCCTTTTATATTGTATATCTTATC-3′ | Generating reporter construct | This work |
Cloning of tirS in a Eukaryotic Expression Vector
Bacterial genomic DNA was extracted as previously described [26]. The tirS gene was amplified by PCR of S. aureus MSSA476 using the tirS EcoRI For + tirS BamHI Rev primers (table 1). The PCR product was digested with EcoRI and BamHI (New England Biolabs, Hitchin, UK), and ligated to the corresponding sites of pEGFP-C2. The presence of tirS in the pEGFP-C2 vector was confirmed by sequencing.
Targeted Mutagenesis and Complementation Vector Construction
Markerless precise allelic replacement of tirS was performed in S. aureus MSSA476 using previously described methods [27] with minor changes. Briefly, DNA fragments 1,029 bp upstream and 1,016 bp downstream of tirS were amplified using Up For tirS + attB1, Up Rev tirS, Down For tirS and Down Rev tirS + attB2 primers (table 1). The Up Rev TirS and Down For TirS primers were constructed with ∼25 bp 5′ overhangs for the opposite flanking region. The upstream and downstream PCR products were fused in a second round of PCR using primers Up For tirS + attB1 and Down Rev tirS + attB2. The fusion construct was subcloned into the temperature-sensitive pKOR1 (table 1) using Gateway BP Clonase II Enzyme mix (Invitrogen, Carlsbad, Calif., USA). The BP reaction product was transformed into DC10b ultracompetent cells. The resulting plasmid pKOR1ΔtirS was confirmed by fusion construct sequencing primers (table 1) and transformed into S. aureus MSSA476 through electroporation (100 Ω resistance, 25 µF capacitance and 2.5 kV voltage). Precise allelic replacement of tirS was established by temperature shifting and antisense counter selection. Deletion of tirS was confirmed by PCR using tirS KO confirm For + tirS KO confirm Rev and tirS Int For + tirS Int Rev primers (table 1). For complementation analysis, tirS was PCR-amplified from S. aureus MSSA476 genome using tirS-CL-F and tirS-CL-R primers (table 1) and cloned into shuttle expression vector pDC123, yielding plasmid pTirS. The resulting construct was transformed into S. aureus MSSA476ΔtirS through electroporation as described previously, yielding S. aureus MSSA476ΔtirS +pTirS.
tirS Expression Profile
Expression of TirS in S. aureus MSSA476 was assessed by three independent methods: semiquantitative reverse transcriptase PCR (RT-PCR), use of a tirS-GFP reporter construct and by immunoblot analysis using TirS antibodies (see below).
To assess expression of TirS in broth cultures, an overnight culture of S. aureus MSSA476 was diluted 1:100 in brain-heart infusion broth (BHI; Sigma Aldrich, Munich, Germany) and incubated at 37°C under shaking conditions. Samples were harvested at an optical density at 600 nm (OD600) of 0.3, 0.6, 0.9 and 1.2, and resuspended in RNA protect solution (Qiagen, Hilden, Germany). RNA extraction was carried out using the RNeasy mini kit (Qiagen) with a prolonged initial lysis step in TE-buffer containing 50 μg/ml lysostaphin (Sigma Aldrich). On-column DNase treatment (Qiagen) and HL-dsDNase digestion (ArcticZymes, Tromso, Norway) was performed according to the manufacturer's instructions. RNA integrity and quantity was determined by checking the 260/280 ratio by Nanodrop and 16S/23S by Experion Automated Electrophoresis Station (Bio-Rad, Hercules, Calif., USA). Reverse transcription of the total RNA was performed on 100 ng of RNA using High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, Calif., USA) according to the manufacturer's recommendations. PCR was conducted using tirS detection primers tirS prev For and tirS prev Rev and housekeeping gene gyrA primers (table 1). In each assay, a no template control and a no reverse transcriptase control were included. PCR cycling conditions were 25 cycles of 95°C for 1 min, 55°C for 1 min and 72°C for 1 min. PCR products were visualized on 1% agarose gel.
To assess expression of tirS in the presence of eukaryotic cells, we first generated a tirS-GFP reporter construct. The tirS promoter region was amplified by PCR from genomic DNA of S. aureus MSSA476 using tirS prom For and tirS prom Rev primers (table 1). The PCR product (333 bp) was digested with PstI and KpnI (Roche, Indianapolis, Ind., USA), ligated to the corresponding sites of pCM29 (table 1) upstream of GFP and transformed into DC10b ultracompetent cells. The presence of the tirS promoter in pCM29 was confirmed by sequencing. The construct was transformed into S. aureus MSSA476 by electroporation (100 Ω resistance, 25 μF capacitance and 2.5 kV voltage). Visual assessment of GFP was carried out by using a blue LED for excitation of GFP. The generated S. aureustirS-GFP reporter strain was subsequently used to assess tirS expression in the presence of HaCaT cells. 2 × 104 HaCaT or HEK293 cells/well were seeded into a 96-well microtiter plate (Corning Inc., Corning, N.Y., USA) in DMEM 10% FBS and infected with S. aureustirS-GFP reporter strain. For this purpose, an overnight culture of S. aureus MSSA476 harboring tirS-reporter construct was diluted 1:100 into pre-warmed TSB medium, incubated at 37°C under shaking conditions, and harvested at an OD600 of 1.2. The bacteria were pelleted, washed in phosphate-buffered saline (PBS), and diluted to the appropriate CFU/ml in DMEM 10% FBS. Fluorescence was measured using a Synergy H1 Hybrid Reader (BioTek, Winooski, Vt., USA) with excitation/emission of 488/520 nm. A control sample of non-transformed S. aureus MSSA476 was included for background correction. Cell viability was examined by Trypan blue staining.
To assess whether release of TirS requires physical contact with host cells, Transwell chambers with a 0.4-μm pore size polyester membrane (Corning) were used. 1 × 105 HEK cells/well were seeded in the upper compartment of a 12-well Transwell plate. Twenty-four hours later, cells were washed and fresh antibiotic-free DMEM 10% FBS was added to each well. 3 × 107S. aureus MSSA476 or S. aureus MSSA476ΔtirS or just DMEM were added to the lower compartment. The media from the lower and upper compartments were harvested 5.5 h later, and concentrated using Amicon® Ultra 30K centrifugal filter device (Millipore Corp., Billerica, Mass., USA). The expression of TirS and gyrase A in the supernatants was evaluated by immunoblot using antibodies against the indicated proteins.
Cell Stimulation Practices
Overnight cultures of S. aureus 61010305, S. aureus MSSA476 and S. aureus MSSA476ΔtirS (depending on the experimental plan) were diluted 1:100 in BHI (Sigma Aldrich) and incubated at 37°C under shaking conditions. Bacterial growth was monitored by OD600. The bacteria were pelleted, washed in PBS and diluted to the appropriate CFU/ml in DMEM (Sigma Aldrich) supplemented with 10% (v/v) heat-inactivated FBS (Invitrogen).
The mammalian cells were stimulated for the indicated time periods by addition of an appropriate number of S.aureus cells/well in DMEM media, 100 ng/ml of synthetic triacylated lipoprotein Pam3CSK4 (Invitrogen) or 30 ng/ml of human tumor necrosis factor (TNF)-α (Bionordika, Lysaker, Norway). When indicated, the mammalian cells were incubated with 2 µg/well staphylococcal superantigen-like protein 3 (SSL3) [28] for 1 h at 37°C before stimulation with the TLR2 ligand Pam3CSK4.
Transfection and Luciferase Assay
HEK293, HaCaT and RAW264.7 cells were maintained in DMEM 10% (v/v) FBS (Invitrogen), penicillin (100 units/ml) and streptomycin (100 µg/ml; Sigma Aldrich) at 37°C with 5% CO2. 2 × 105 cells/well or 1 × 105 cells/well were seeded in 6-well plates or in the upper compartment of a 12-well transwell plate, respectively. After 24 h, transfection was carried out using METAFECTENE® PRO (Biontex, San Diego, Calif., USA) for HEK293 and RAW264.7, and Attractene (Qiagen) for HaCaT cells according to the manufacturer's instructions. The total amount of DNA used in transfection was kept constant by adding CT-DNA (Calf thymus DNA; Invitrogen). After stimulation, the cells were washed twice with PBS (Biochrom, Berlin, Germany) and harvested in 100 µl of Tropix® solution (Applied Biosystems) containing 0.05% 1 M dithiothreitol (Invitrogen). Cell lysates were cleared by centrifugation at 14,000 g for 7 min at 4°C and luciferase activity was measured in 20 µl lysate using Promega E4550 kit (Promega Corp., Madison, Wisc., USA) on a Luminoscan RT luminometer (Labsystems, Helsinki, Finland) according to the manufacturer's instructions. Co-expression of Renilla luciferase or β-galactosidase was avoided as our stimuli influenced the reporters [unpubl. data]. When required, cells were starved overnight by replacing standard culture medium. Transfection with pEGFP-C2 was done for monitoring transfection efficiency in each experiment. Transfection efficiency was >80% for HEK cells and 40–50% for RAW264.7 and HaCaT cells.
Immunoblot Analysis
Immunoblot analysis was carried out on cell lysates as described previously [29], using antibodies against EGFP (Roche, Basel, Switzerland), phospho-SAPK/JNK (Thr183/Tyr 185; Cell Signaling Technology Inc., Danvers, Mass., USA), DNA gyrase A (Abcam, Cambridge, UK) and TirS. The affinity purified TirS antibody was generated (Eurogentec, Seraing, Belgium) by immunizing rabbits with the peptides RINKKRKPTSSNIRD and NQKKLSSMLDKNTKG.
To confirm equal protein loading, membranes were washed in 0.2 M NaOH for 5 min, rinsed with PBST (PBS containing 0.1% Tween 20; Sigma Aldrich), blocked with blocking buffer containing 5% non-fat dry milk (Nestlé, Frankfurt, Germany) and 0.1% Tween 20, and reprobed with ERK2 (C-14): sc-154 (Santa Cruz Biotechnology, Santa Cruz, Calif., USA). Densitometric analysis was performed using ImageJ.
Cytokine Assay
2 × 105 HEK293 cells/well were seeded into 12-well plates in DMEM 10% FBS. The next day, cells were transfected with TLR2 (500 ng/ml) and pEGFP-TirS (100 ng/ml) or pEGFP-C2 (100 ng/ml) as described previously. Twenty-four hours post-transfection, the cells were washed and new antibiotic-free DMEM 10% FBS was added. Cells were stimulated with 1.5 × 107S. aureus 61010305 cells/well and culture supernatants were collected 8 h postinfection and cleared by centrifugation. MCP-1 (monocyte chemoattractant protein) and G-CSF (granulocyte colony-stimulating factor) were measured in the culture supernatants using MILLIPLEX® MAP kit (Millipore Corp.) based on Luminex technology and according to the manufacturer's instructions.
Growth Curves of S. aureus MSSA476 Wild-Type and MSSA476ΔtirS
S. aureus MSSA476 wild-type and MSSA476ΔtirS bacteria were grown overnight in Todd-Hewitt broth (THB). The next day, bacteria were diluted and grown to OD600 = 0.6 in THB, washed, and resuspended in different media, including RPMI 1640 medium supplemented with 1% casamino acids, RPMI-CA, DMEM medium supplemented with 1% casamino acids, DMEM-CA, DMEM supplemented with 10% FCS, DMEM-FCS, THB, or BHI. 1 × 105 CFU were added to 100-well Bioscreen Honeycomb plates (Growth Curves, Piscataway, N.J., USA) in a total volume of 200 µl. Growth was monitored by measuring OD600 every 30 min for 20 h under shaking conditions using a Bioscreen C MBR machine (Growth Curves).
Murine Model of Subcutaneous and Intravenous Infection
We used established models of bacteremia and skin abscess formation to determine the difference in virulence between S. aureus MSSA476 wild type and MSSA476ΔtirS. For systemic infection, 8-week-old female C57BL/6 mice (Charles River, Wilmington, Mass., USA) were infected intravenously with 5 × 106 CFU early exponential-phase bacteria (OD600 = 0.25) by tail vein injection. Bacterial load (log CFU/g organ) in the kidneys (paired), spleen, liver, heart, brain and blood was quantified at 4 days postinfection. Mice that died 3 days postinfection were excluded from the experiment.
For the skin abscess model, 8-week-old female C57BL/6 mice (Charles River) were infected subcutaneously with 2 × 108 CFU early exponential-phase bacteria (OD600 = 0.25) on a shaved flank. Daily area measurements (mm2) of lesions (as dermonecrosis) and abscess (as dermonecrotic area) were carried out for 3 consecutive days. On day 3, abscesses were excised and bacterial loads (CFU/abscess) were quantified.
Ethical Approval, Animal Care and Compliance Statement
The Tromsø Staph and Skin Study has been approved by the Regional Committee for Medical Research Ethics, North Norway (Ref. 200605174-12/IAY/400), the Norwegian Data Inspectorate (Ref. 07/00886-2/CAO), and is in the Biobank Registry (Ref. 2397). Mouse infection studies were performed under approved protocol S00227M of the University of California San Diego Institutional Animal Care and Use Committee.
Statistical Analysis and Data Validation
The data are expressed as mean ± standard deviation (SD) of an individual experiment except for figure 3c, where the data are expressed as mean ± SD of three independent pooled experiments, and figure 3d, which presents mean ± SD of pooled data of two independent experiments. Results were validated by performing most experiments in triplicate and repeating at least three times. Statistical analysis was carried out on the data of pooled experiments. One-way ANOVA or Student's t test in Excel was used for the determination of statistically significant differences between groups (p < 0.05). Excel or GraphPad Prism was used for generating graphs.
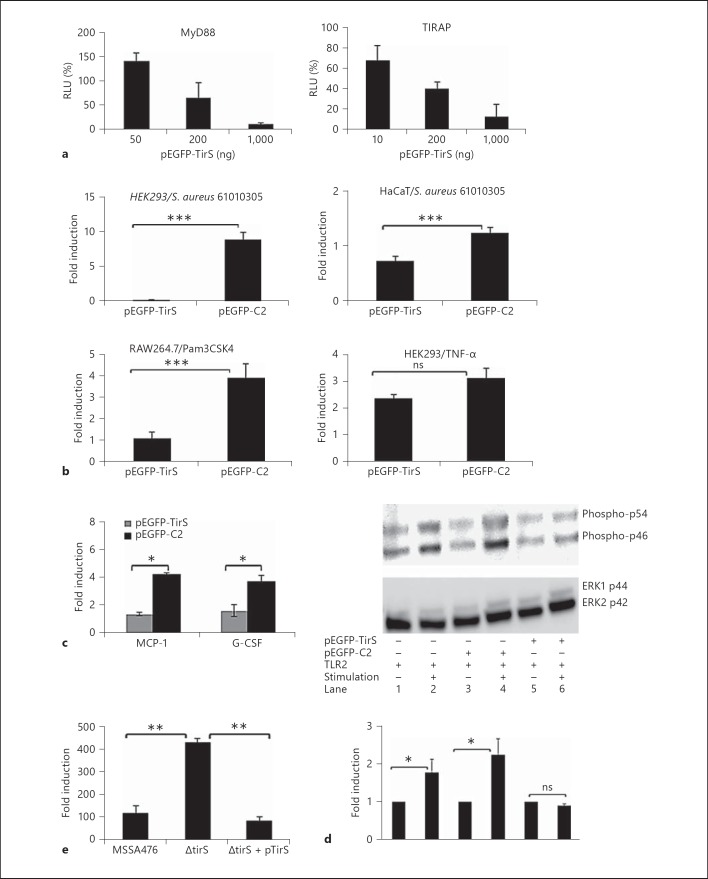
Fig. 3.
TirS inhibits NF-κB activation and MAPK signaling in different cell lines. a Expression of TirS interferes with MyD88 or TIRAP-induced NF-κB activation in HEK293 cells. pEGFP-C2-transfected cells were arbitrarily set as 100%, and the luciferase activity in the pEGFP-TirS is represented as the percentage of mock transfection. b TirS (pEGFP-TirS) inhibits TLR2-induced activation of NF-κB-luciferase reporter in HEK293, HaCaT and RAW264.7 cells, but not TNF-α-induced NF-κB activation in HEK293. The untreated control cells were arbitrarily set as 1, and the luciferase activity in the treated cells is represented as fold induction. c Expression of MCP-1 and G-CSF from TLR2-transfected HEK293 cells is reduced by co-expression TirS (pEGFP-TirS), but not by control plasmid (pEGFP-C2). Cells were left untreated or stimulated with TirS-negative S. aureus 61010305 for 8 h. Cytokine secretion in treated cells is represented as fold induction compared to cytokine secretion by untreated control cells, which was arbitrarily set as 1. d Expression of TirS (pEGFP-TirS) but not control plasmid (pEGFP-C2) suppresses activation of SAPK/JNK activation in TLR2-transfected HEK293 as determined by Western blotting. Cells were left untreated or stimulated with TirS-negative S. aureus 61010305 for 3 h and cell lysates were used for Western blot analysis using phospho-SAPK/JNK (Thr183/Tyr185) antibody (upper panel). Equal loading was verified by ERK2 (C-14) antibody (middle panel). The ratio of phospho-JNK/ERK2 was determined by densitometry analysis on two membranes. The ratio obtained in untreated cells was arbitrarily set to 1, and the values obtained for the stimulated cells are expressed as fold induction (lower panel). e Loss of TirS from S. aureus MSSA476 results in increased activation of NF-κB in TLR2-transfected HEK293 cells as determined by NF-κB-luciferase activity. Fold induction was calculated as described in b. ns = Not significant. * p < 0.05; ** p < 0.01; *** p < 0.001.
Results
Genomic Localization, Similarity to Other TIR-Containing Proteins and Predicted Tertiary Protein Structure of TirS
S. aureus was suggested to possess a TIR-containing coding region with homology to E. colitcpC[10]. We identified the putative ORF (annotated as SAS0038) in the S. aureus MSSA476 genome by BLAST search (accession number BX571857.1; [25]) and named it tirS. From the annotation the ORF was located on the 22.8-kb staphylococcal cassette chromosome SCC476 element present in MSSA476 (fig. 1a). Further BLAST searches revealed the presence of tirS in nine additional genome-sequenced S. aureus strains (online suppl. table 1; for all online suppl. material, see www.karger.com/doi/10.1159/000357618).
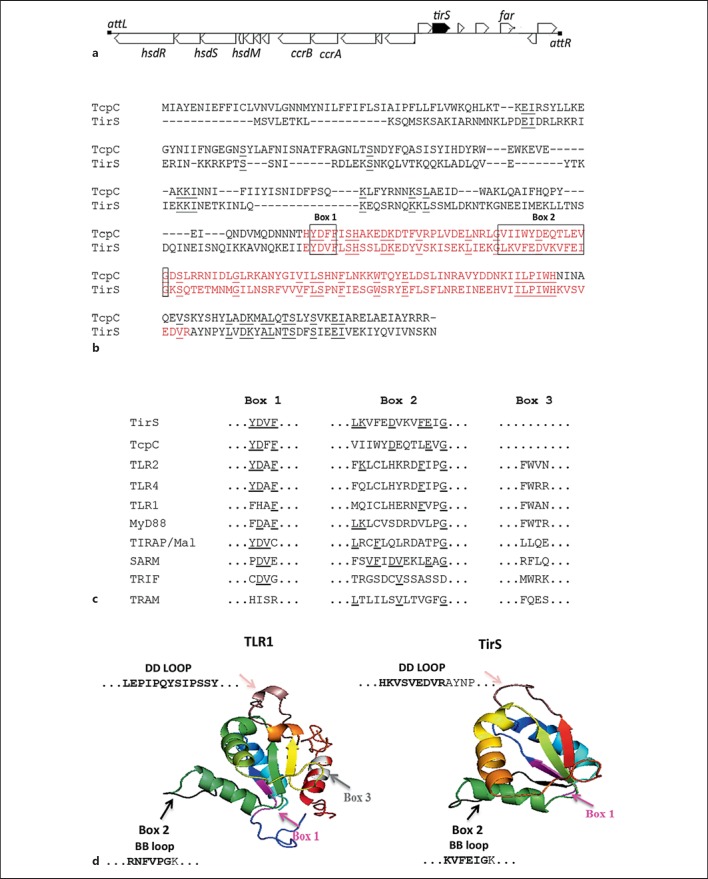
Fig. 1.
Localization and bioinformatic characterization of TirS in S. aureus. a MSSA476tirS is located within the mobile genetic element SCC476 between ORFs encoding hypothetical proteins. The annotation is from KEGG Genome map and Holden et al. [25]. hsdRSM = Restriction modification system; ccrAB = cassette chromosome recombinase A and B; far = fusidic acid resistance protein; attL/attR = attachment site left/right, respectively. b Comparison of the amino acid sequences of full-length TirS and E. coli TcpC. The TIR domain sequences are marked in red. The identical residues are underlined. c Comparison of the amino acid sequence within signature motifs boxes 1, 2 and 3 of the TIR domains of TirS, TcpC, TLR2, TLR4, TLR1, MyD88, TIRAP, SARM, TRIF and TRAM. The identical residues are underlined. d The predicted three-dimensional structure was generated using Phyre server [47] and figures were generated by PyMol. 100% of the TirS TIR domain (amino acids 142-245) was modeled with 99.9% confidence by the single highest scoring template as TIR domain of TLR1 (PDB FYVA). Box 1 (purple), box 2 (black), box 3 (light grey) and the DD loop (dark pink) are indicated within the TIR domain. The BB loop (bold amino acid sequences are part of box 2) and DD loop (bold amino acid sequences are part of the TIR domain) amino acid sequences in both TLR1 and TirS are shown.
The ORF tirS was predicted to encode a protein of 280 amino acids containing a conserved TIR domain 103 amino acids in length (fig. 1b). Comparison of the full-length amino acid sequence of TirS and E. coli TcpC revealed that the TIR domain was located at the C-terminal part of both proteins, and shared 62% amino acid similarity (fig. 1b). The amino acid sequence within box 1, box 2 and box 3 motifs of TirS were compared to eukaryotic TIR domains and the prokaryotic TcpC. S. aureus TirS showed particular sequence homology to TcpC, TLR2 and TLR4 in box 1 (fig. 1c). Comparison of the secondary structure of TirS with TLR1 TIR domain using ESPript (Easy Sequencing in Postscript) [30] suggested additional structural homologies in the BB loop (fig. 1d), which is one of the key residues within the TIR domain. The predicted TIR domain structure of TirS suggested that it belongs to the flavodoxin-like fold according to the Structural Classifications of Proteins, SCOP, which is also found for the other TIR structures [19]. The functionally important BB loop, which is part of the box 2 motif, is positioned away from the main core. The amino acid sequence and predicted structure does not support the existence of a box 3 motif in TirS (fig. 1c, d).
Expression and Release of tirS from S. aureus
We analyzed the expression profile of tirS in regular growth medium (BHI) by RT-PCR and in DMEM in the presence or absence of eukaryotic cells using a tirS-specific GFP reporter construct. The tirS gene was expressed during different growth phases in BHI (fig. 2a). Furthermore, tirS expression was strongly induced in MSSA476 when the bacteria were grown in the presence of HaCaT and HEK293 cells compared to growth in DMEM alone (fig. 2b and data not shown).
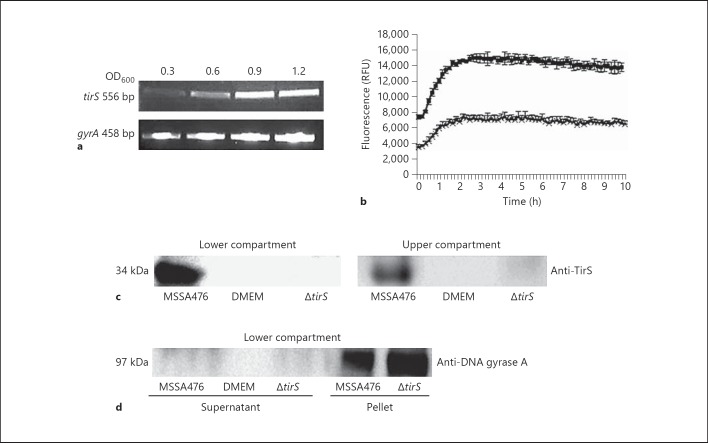
Fig. 2.
Expression profile of tirS. a Expression of MSSA476 tirS in BHI was evaluated at OD600 0.3, 0.6, 0.9 and 1.2 by RT-PCR. b Expression of MSSA476 tirS in DMEM in the absence (×) or presence (◼) of HaCaT cells using S. aureus MSSA476 harboring tirS-GFP reporter construct. c Presence of TirS in both the lower (bacteria only) and upper compartment (HEK293 only) of a Transwell system as detected by immunoblot. d Presence of the cytoplasmic protein gyrase A in bacteria but not the supernatant of the lower compartment as assessed by immunoblot and gyrA-specific antibody.
To evaluate whether TirS is expressed and released by the bacteria without physical contact to HEK293 cells, we employed a Transwell system followed by immunoblot of supernatants using a TirS-specific polyclonal antibody. TirS was detected in the media of the lower compartments inoculated with MSSA476, but not MSSA476ΔtirS or DMEM media alone (fig. 2c). Bacterial cells were unable to cross the membrane to the upper compartment, as verified by plating of the supernatant for bacterial counts (data not shown). However, TirS could still be detected in the upper compartment associated with the host cells (fig. 2c, right section), indicating passive diffusion of the proteins released by the bacterial cells localized to the lower compartment. Importantly, the cytoplasmic marker gyrA was not detected in the supernatant of the lower compartment, indicating that TirS is released by a mechanism other than bacterial cell death (fig. 2d).
The Effect of TirS on TLR2 Signaling
Stimulation of TLRs results in the recruitment and activation of TIR-containing adaptor proteins like MyD88 and TIRAP [3]. Given the homology between the TIR domains of TirS and eukaryotic TLRs/adaptor proteins, we hypothesized that TirS could interfere with TLR intracellular signaling. Overexpression of adaptors like MyD88 results in constitutive activation of the TLR pathway [31]. Therefore, HEK293 cells were transfected with expression plasmids encoding MyD88 or TIRAP in combination with increasing levels of pEGFP-TirS or pEGFP-C2 to assess the effect of TirS on NF-κB activation. TirS significantly suppressed both MyD88- and TIRAP-induced NF-κB activation in a dose-dependent manner (fig. 3a).
Next, the effect of ectopic TirS expression on stimuli-induced activation was addressed. HEK293 and HaCaT cells were transfected with a NF-κB-reporter and expression plasmid encoding TLR2 in combination with expression plasmids encoding pEGFP-TirS or pEGFP-C2. Transfected cells were stimulated with S. aureus 61010305 (not containing tirS) for 6 h (fig. 3b). S. aureus-stimulated mock-transfected cells induced 10- and 1.5-fold induction of the luciferase reporter in HEK293 and HaCaT cells, respectively. In contrast, pEGFP-TirS transfected cells resulted in reduced NF-κB reporter activation in both cell lines (fig. 3b). Similarly, presence of TirS significantly inhibited the Pam3CSK4-induced NF-κB activation in the mouse monocyte macrophage cell line RAW264.7 (p < 0.001; fig. 3b) and in HEK293 cells (data not shown). Furthermore, the inhibition of Pam3CSK4-induced NF-κB activation by TirS was similar to inhibition observed in the presence of DN-hMyD88 or when TLR2 stimulation was blocked by SSL3 (online suppl. fig. 1). The presence of TirS was checked by Western blot in most of these experiments, and its presence was confirmed (data not shown). Another stimulus that induces NF-κB activation is TNF-α, which acts independently of TLR2 and MyD88 [32]. TirS did not interfere with TNF-α-induced NF-κB activation (fig. 3b) which demonstrated that TirS specifically interferes with TLR2-mediated signaling.
We also assessed whether ectopic TirS expression interfered with S. aureus-induced host cytokine secretion. HEK293 cells were transfected with expression plasmids encoding TLR2 and pEGFP-TirS or control pEGFP-C2. The cells were then left untreated or stimulated with tirS-negative S. aureus prior to Milliplex analysis of secreted cytokines. In pEGFP-C2-transfected HEK293, presence of S. aureus induced 4.1- and 3.6-fold increases of secreted MCP-1 and G-CSF, respectively (fig. 3c). In contrast, the levels of secreted MCP-1 and G-CSF were significantly reduced in S. aureus-stimulated cells transfected with pEGFP-TirS (p < 0.05; fig. 3c).
In addition to NF-κB activation, stimulation of TLRs can also activate the MAPK pathway [1]. Therefore, we tested the effect of TirS on the MAPK signaling pathway. HEK293 cells were transfected with expression plasmids encoding TLR2 and pEGFP-TirS or pEGFP-C2 and starved overnight. Cells were left untreated or stimulated with tirS-negative S. aureus and cell lysates were harvested for immunodetection of phospho-SAPK/JNK (Thr183/Tyr185). Equal loading was verified using the ERK2 antibody. Both phospho-p54 and phospho-p46, representing SAPK/JNK, were phosphorylated upon stimulation with S. aureus (fig. 3d, compare lanes 2 and 4 with lane 1). Co-transfection with the TirS-expressing plasmid negatively interfered with S. aureus-induced phosphorylation of JNK (fig. 3d, compare lane 6 with lanes 4 and 2), indicating that TirS interferes with the SAPK/JNK signaling pathway.
Finally, to confirm the results generated by ectopic expression of TirS, the effect on TLR2-mediated NF-κB activation was assessed using MSSA476 wild-type, MSSA476ΔtirS and the complemented strain MSSA476ΔtirS +pTirS in the Transwell system where the host cells and bacteria are physically separated. The MSSA476ΔtirS mutant resulted in an increased activation of the NF-κB reporter in comparison to the wild type and complemented strains (fig. 3e).
Taken together, these results indicate that TirS attenuates TLR-induced activation of NF-κB and MAPK signaling pathways as well as induction of the proinflammatory cytokines G-CSF and MCP-1.
Presence of TirS Increases S. aureus Load in Multiple Organs in a Murine Intravenous Infection Model
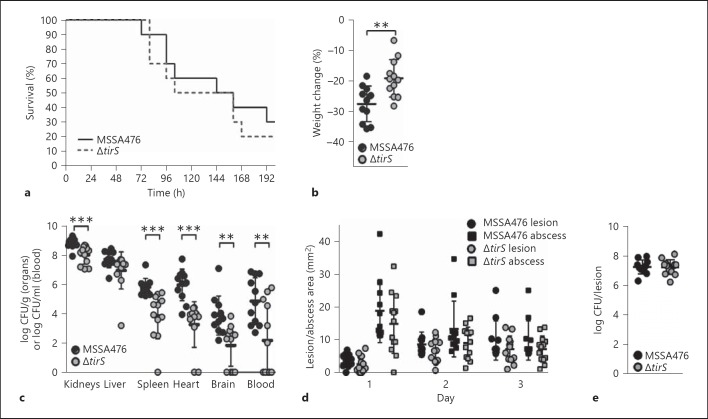
Finally, we evaluated whether the observed effects of TirS on host immune signaling in vitro could influence S. aureus pathogenesis in vivo. First, we confirmed that bacterial growth was not affected by deletion of TirS by assessing growth in five different media including THB medium (online suppl. fig. 2). Thereafter, mice were injected intravenously with the two strains. The survival rates of wild-type and ΔtirS-infected mice were similar (fig. 4a); however, percentage weight loss was significantly higher in mice infected with wild-type MSSA476 (fig. 4b). Strikingly, disruption of the tirS gene resulted in significantly decreased S. aureus loads in the kidneys, heart, brain, blood and spleen (fig. 4c). In contrast, the effect of TirS upon S. aureus virulence was not apparent in a localized abscess/lesion formation skin infection model. No significant differences in lesion or abscess sizes were detected upon infection with MSSA476 wild-type or MSSA476ΔtirS over a course of 3 days (fig. 4d). In addition, bacterial loads in excised abscesses were similar for both strains on day 3 (fig. 4e).
Fig. 4.
TirS improves bacterial survival in a bacteremia but not in a skin abscess model in C57BL/6 mice. Percentage survival (a), percentage weight change (b) and bacterial numbers (c) in the kidneys, spleen, liver, heart and brain upon systemic infection with S. aureus MSSA476 and S. aureus MSSA476ΔtirS (10 mice per group). Skin lesion size (⚪) and abscess size (◻) over the course of infection (d), and bacterial load per abscess 3 days after subcutaneous infection with S. aureus MSSA476 or S. aureus MSSA476ΔtirS (e). ** p < 0.01; *** p < 0.001.
Discussion
In this study, the expression of a gene encoding a 280 amino acid TIR-containing protein, tirS, within S. aureus MSSA476 was confirmed. The tirS gene is localized in the staphylococcal cassette chromosome SCC476 element (fig. 1a), which is classified as a mobile genetic element [25]. Several other bacterial TIR-containing genes have been identified, and these are localized in genomic regions of phage origins, or close to genes of phage integrase, site-specific recombinases and transposases. This pattern strongly suggests that these genes are transmitted by lateral transfer [19]. tirS is currently identified in other sequenced S. aureus strains (online suppl. table 1). Whether or how tirS is spread among S. aureus strains remains to be investigated.
Sequence conservation among TIR domains has been reported to be in the 20–30% range and the diversity has been suggested to be crucial for the specificity in signal transduction [4]. Comparison of the primary sequence of several bacterial TIR proteins has revealed that there is diverse domain architecture in the sequence surrounding the TIR domain [19]. Also, the N-terminal part of TirS is highly divergent in comparison to TcpC (fig. 1b) and TcpB [16]. This suggests that various bacterial TIR-containing proteins may have species-specific function(s) in addition to the effect on intracellular signaling.
Here we demonstrated that TirS attenuates stimuli-induced TLR2-mediated NF-κB activation, JNK phosphorylation and secretion of MCP-1 and G-CSF (fig. 3). Moreover, inhibition of TLR-induced cellular activation was not only observed upon ectopic expression of TirS in eukaryotic cells, but also in infection experiments where NF-κB activation was significantly increased as a consequence of co-culturing MSSA476ΔtirS with host cells in comparison to MSSA476 wild-type bacteria and MSSA476ΔtirS +pTirS (fig. 3e). Our results are in line with previous publications reporting negative interference with stimuli-induced TLR signaling in vitro by TIR-containing proteins from various Gram-negative bacteria, including S. enterica [13], Brucella spp. [10, 16, 33], uropathogenic E. coli [10, 20] and Y. pestis[18].
Interference with TLR-dependent NF-κB signaling requires intracellular localization of the bacterial effector. Effectors from Gram-negative bacteria can be directly injected into the host cell by the type III or type IV secretion systems (T3SS or T4SS) [34, 35], or secreted into the medium and subsequently taken up by the host cells [10]. Using the Transwell system, where host cells and bacterial cells are separated by a 0.4-μm pore size polyester membrane, we could demonstrate that TirS is released from S. aureus through an as yet unidentified mechanism (fig. 2b, c). Furthermore, the TirS effect on signaling does not require S. aureus invasion into host cells to exert an effect on the NF-κB reporter (fig. 3e). Production of membrane-derived vesicles by S. aureus during infection and their role in the delivery of virulence factors to the host cells has been reported previously [36]. Alternatively, TirS could be endocytosed by host cells. However, the mechanism by which TirS enters the host cells remains to be elucidated.
To study whether TirS affects S. aureus virulence, we compared the pathogenicity of MSSA476 wild-type versus an isogenic mutant MSSA476ΔtirS in two different mouse infection models. Loss of TirS reduced weight loss and bacterial load in multiple organs upon systemic infection suggesting that TirS increases bacterial survival in the host. Similar results have been obtained for Gram-negative bacteria. The E. coli TIR-containing protein TcpC, which inhibits the TLR4/MyD88 and TLR2/MyD88 pathway, has a direct effect on the pathogenesis of acute pyelonephritis in mouse urinary tract infection models. Moreover, the prevalence of tcpC among urine isolates from humans with asymptomatic bacteriuria or severe kidney or bladder infections suggested that tcpC is associated with bacterial virulence [10]. A similar association with severity of infection was observed when the prevalence of tcpC was examined among 302 E. coli isolates from the urinary tract, skin and soft tissue infections and commensal E. coli strains [37]. Furthermore, increased virulence was seen for strains containing the TIR proteins TlpA or TcpB [13, 16], while YpTdp did not have a role in the virulence of Y. pestis in a mouse model of bubonic plague [17]. Interestingly, both patients and mice with genetic defects in TLR signaling pathways, such as TLR2, CD36, TIRAP, MyD88 or IRAK4 deficiencies, are highly susceptible to infection with Gram-positive bacteria [6, 7, 38, 39]. In mouse infection models, the number of S. aureus cells in the blood, spleen and kidney were increased in TLR2- and MyD88-deficient mice compared to the wild type [6, 38, 39]. These findings suggest that the MyD88/IRAK-4 pathway is of particular importance for defense against Gram-positive organisms including S. aureus. Therefore, interfering with TLR-mediated cellular activation is an effective immune evasion strategy of S. aureus to avoid immune clearance. This is also underlined by a recent paper demonstrating that S. aureus-secreted SSL3 inhibits the TLR2 pathway by blocking the TLR2 ligand-binding domain [28, 40].
MSSA476ΔtirS was more attenuated than the wild type strain in the intravenous infection model (fig. 4a-c), but exhibited comparable virulence in the skin abscess model (fig. 4d, e). The discrepancy in the contribution of TirS in the systemic versus the skin infection model is probably multifactorial. First of all, the tirS gene belongs to the S. aureus accessory genome (fig. 1; online suppl. table 1), and MSSA476 was originally isolated from a severe invasive infection in an immune-competent child [25], which suggests a capacity for effective bloodstream dissemination. Mice deficient in TLR2 or MyD88 are highly susceptible to systemic S. aureus infection [6] and our in vitro results clearly show that TirS inhibits signaling from TLR2 and MyD88 (fig. 3). Although not directly proven in vivo, this may explain the increase of virulence in the bacteremia model (fig. 4c). Finally, during skin abscess formation in mice, IL-1R/MyD88-, but not TLR2/MyD88-, signaling is of importance [41]. Although we show that TirS can inhibit signaling downstream of MyD88 in vitro (fig. 3a), TirS may also target TLR2 directly. Indeed, many bacterial effectors often exert their influence at several stages in a signaling pathway [e.g. YopJ; for a review see [42]]. Therefore, if the main contribution of TirS is to block TLR2 and not MyD88 in mice, this may explain the lack of contribution of TirS in the abscess model. However, the full natures of the differential virulence manifestations based on the route of administration remain to be investigated.
In summary, our study suggests a new intracellular immune evasion mechanism of S. aureus. The TLR-MyD88 signaling pathway, especially through TLR2, is important for clearance of infection, and tirS increases bacterial survival in the host. The tirS-gene is located on SCC476 together with fusidic acid resistance gene and other putative virulence factors [25], which may enhance bacterial fitness. The localization of these genes on a mobile genetic element may enable horizontal gene transfer to other strains, resulting in an increased prevalence of tirS among S. aureus isolates in the future.
Supplementary Material
Supplementary data
Supplementary data
Acknowledgments
This work was supported by the Research Council of Norway (grant 191264/V50) and the Northern Norway Regional Health Authority (Helse Nord RHF) grants Toppforskning (2004-2009), SFP877-09 and Miljøstøtte MIL963-10 (2010-2012), NIH/NIAID/GLRCE research grant AI057153 (VN), and Marie Curie International Incoming Fellowship (FP7; NMvS). We thank Carsten J. Kirschning, Caroline Jefferies and Reindert Nijland for providing pCMV-Myc-TLR2, AU1-MyD88 and pCM29 constructs, respectively. We thank Natalya Seredkina for providing RAW264.7 cells. We gratefully acknowledge the assistance of Renate Slind Olsen and Kristin Hansen.
References
- 1.Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 3.O'Neill LAJ, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 4.Xu YW, Tao X, Shen BH, Horng T, Medzhitov R, Manley JL, Tong L. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408:111–115. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- 5.von Bernuth H, Picard C, Puel A, Casanova JL. Experimental and natural infections in MyD88- and IRAK-4-deficient mice and humans. Eur J Immunol. 2012;42:3126–3135. doi: 10.1002/eji.201242683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeuchi O, Hoshino K, Akira S. Cutting edge: TLR2-deficient and MyD88-deficient mice are highly susceptible to Staphylococcus aureus infection. J Immunol. 2000;165:5392–5396. doi: 10.4049/jimmunol.165.10.5392. [DOI] [PubMed] [Google Scholar]
- 7.Picard C, von Bernuth H, Ghandil P, Chrabieh M, Levy O, Arkwright PD, McDonald D, Geha RS, Takada H, Krause JC, Creech CB, Ku CL, Ehl S, Marodi L, Al-Muhsen S, Al-Hajjar S, Al-Ghonaium A, Day-Good NK, Holland SM, Gallin JI, Chapel H, Speert DP, Rodriguez-Gallego C, Colino E, Garty BZ, Roifman C, Hara T, Yoshikawa H, Nonoyama S, Domachowske J, Issekutz AC, Tang M, Smart J, Zitnik SE, Hoarau C, Kumararatne DS, Thrasher AJ, Davies EG, Bethune C, Sirvent N, de Ricaud D, Camcioglu Y, Vasconcelos J, Guedes M, Vitor AB, Rodrigo C, Almazan F, Mendez M, Arostegui JI, Alsina L, Fortuny C, Reichenbach J, Verbsky JW, Bossuyt X, Doffinger R, Abel L, Puel A, Casanova JL. Clinical features and outcome of patients with IRAK-4 and MyD88 deficiency. Medicine. 2010;89:403–425. doi: 10.1097/MD.0b013e3181fd8ec3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpenter S, O'Neill LA. How important are Toll-like receptors for antimicrobial responses? Cell Microbiol. 2007;9:1891–1901. doi: 10.1111/j.1462-5822.2007.00965.x. [DOI] [PubMed] [Google Scholar]
- 9.Stebbins CE, Galan JE. Structural mimicry in bacterial virulence. Nature. 2001;412:701–705. doi: 10.1038/35089000. [DOI] [PubMed] [Google Scholar]
- 10.Cirl C, Wieser A, Yadav M, Duerr S, Schubert S, Fischer H, Stappert D, Wantia N, Rodriguez N, Wagner H, Svanborg C, Miethke T. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat Med. 2008;14:399–406. doi: 10.1038/nm1734. [DOI] [PubMed] [Google Scholar]
- 11.Chan SL, Low LY, Hsu S, Li S, Liu T, Santelli E, Le Negrate G, Reed JC, Woods VL, Jr, Pascual J. Molecular mimicry in innate immunity: crystal structure of a bacterial TIR domain. J Biol Chem. 2009;284:21386–21392. doi: 10.1074/jbc.C109.007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cirl C, Miethke T. Microbial Toll/interleukin 1 receptor proteins: a new class of virulence factors. Int J Med Microbiol. 2010;300:396–401. doi: 10.1016/j.ijmm.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Newman RM, Salunkhe P, Godzik A, Reed JC. Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infect Immun. 2006;74:594–601. doi: 10.1128/IAI.74.1.594-601.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Low LY, Mukasa T, Reed JC, Pascual J. Characterization of a TIR-like protein from Paracoccus denitrificans. Biochem Biophys Res Commun. 2007;356:481–486. doi: 10.1016/j.bbrc.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salcedo SP, Marchesini MI, Lelouard H, Fugier E, Jolly G, Balor S, Muller A, Lapaque N, Demaria O, Alexopoulou L, Comerci DJ, Ugalde RA, Pierre P, Gorvel JP. Brucella control of dendritic cell maturation is dependent on the TIR-containing protein Btp1. PLoS Pathog. 2008;4:e21. doi: 10.1371/journal.ppat.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radhakrishnan GK, Yu Q, Harms JS, Splitter GA. Brucella TIR domain-containing protein mimics properties of the Toll-like receptor adaptor protein TIRAP. J Biol Chem. 2009;284:9892–9898. doi: 10.1074/jbc.M805458200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spear AM, Loman NJ, Atkins HS, Pallen MJ. Microbial TIR domains: not necessarily agents of subversion? Trends Microbiol. 2009;17:393–398. doi: 10.1016/j.tim.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Rana RR, Simpson P, Zhang MH, Jennions M, Ukegbu C, Spear AM, Alguel Y, Matthews SJ, Atkins HS, Byrne B. Yersinia pestis TIR-domain protein forms dimers that interact with the human adaptor protein MyD88. Microb Pathog. 2011;51:89–95. doi: 10.1016/j.micpath.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, Zmasek CM, Cai XH, Godzik A. TIR domain-containing adaptor SARM is a late addition to the ongoing microbe-host dialog. Dev Comp Immunol. 2011;35:461–468. doi: 10.1016/j.dci.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yadav M, Zhang J, Fischer H, Huang W, Lutay N, Cirl C, Lum J, Miethke T, Svanborg C. Inhibition of TIR domain signaling by TCPC: MyD88-dependent and independent effects on Escherichia coli virulence. PLoS Pathogens. 2010;6:e1001120. doi: 10.1371/journal.ppat.1001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slack JL, Schooley K, Bonnert TP, Mitcham JL, Qwarnstrom EE, Sims JE, Dower SK. Identification of two major sites in the type I interleukin-1 receptor cytoplasmic region responsible for coupling to pro-inflammatory signaling pathways. J Biol Chem. 2000;275:4670–4678. doi: 10.1074/jbc.275.7.4670. [DOI] [PubMed] [Google Scholar]
- 22.Rana RR, Zhang MH, Spear AM, Atkins HS, Byrne B. Bacterial TIR-containing proteins and host innate immune system evasion. Med Microbiol Immun. 2013;202:1–10. doi: 10.1007/s00430-012-0253-2. [DOI] [PubMed] [Google Scholar]
- 23.Spear AM, Rana RR, Jenner DC, Flick-Smith HC, Oyston PCF, Simpson P, Matthews SJ, Byrne B, Atkins HS. A Toll/interleukin (IL)-1 receptor domain protein from Yersinia pestis interacts with mammalian IL-1/Toll-like receptor pathways but does not play a central role in the virulence of Y. pestis in a mouse model of bubonic plague. Microbiology. 2012;158:1593–1606. doi: 10.1099/mic.0.055012-0. [DOI] [PubMed] [Google Scholar]
- 24.Foster TJ. Colonization and infection of the human host by staphylococci: adhesion, survival and immune evasion. Vet Dermatol. 2009;20:456–470. doi: 10.1111/j.1365-3164.2009.00825.x. [DOI] [PubMed] [Google Scholar]
- 25.Holden MTG, Feil EJ, Lindsay JA, Peacock SJ, Day NPJ, Enright MC, Foster TJ, Moore CE, Hurst L, Atkin R, Barron A, Bason N, Bentley SD, Chillingworth C, Chillingworth T, Churcher C, Clark L, Corton C, Cronin A, Doggett J, Dowd L, Feltwell T, Hance Z, Harris B, Hauser H, Holroyd S, Jagels K, James KD, Lennard N, Line A, Mayes R, Moule S, Mungall K, Ormond D, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Sharp S, Simmonds M, Stevens K, Whitehead S, Barrell BG, Spratt BG, Parkhill J. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci USA. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sangvik M, Olsen RS, Olsen K, Simonsen GS, Furberg AS, Sollid JU. Age- and gender-associated Staphylococcus aureus spa types found among nasal carriers in a general population: the Tromso Staph and Skin study. J Clin Microbiol. 2011;49:4213–4218. doi: 10.1128/JCM.05290-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Bardoel BW, Vos R, Bouman T, Aerts PC, Bestebroer J, Huizinga EG, Brondijk THC, van Strijp JAG, de Haas CJC. Evasion of Toll-like receptor 2 activation by staphylococcal superantigen-like protein 3. J Mol Med. 2012;90:1109–1120. doi: 10.1007/s00109-012-0926-8. [DOI] [PubMed] [Google Scholar]
- 29.Johannessen M, Delghandi MP, Rykx A, Dragset M, Vandenheede JR, Van Lint J, Moens U. Protein kinase D induces transcription through direct phosphorylation of the cAMP-response element-binding protein. J Biol Chem. 2007;282:14777–14787. doi: 10.1074/jbc.M610669200. [DOI] [PubMed] [Google Scholar]
- 30.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 31.Gay NJ, Gangloff M, O'Neill LAJ. What the myddosome structure tells us about the initiation of innate immunity. Trends Immunol. 2011;32:104–109. doi: 10.1016/j.it.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 32.Hayden MS, Ghosh S. NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radhakrishnan GK, Splitter GA. Biochemical and functional analysis of TIR domain containing protein from Brucella melitensis. Biochem Biophys Res Commun. 2010;397:59–63. doi: 10.1016/j.bbrc.2010.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ge J, Xu H, Li T, Zhou Y, Zhang Z, Li S, Liu L, Shao F. A Legionella type IV effector activates the NF-κB pathway by phosphorylating the IκB family of inhibitors. Proc Natl Acad Sci USA. 2009;106:13725–13730. doi: 10.1073/pnas.0907200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newton HJ, Pearson JS, Badea L, Kelly M, Lucas M, Holloway G, Wagstaff KM, Dunstone MA, Sloan J, Whisstock JC, Kaper JB, Robins-Browne RM, Jans DA, Frankel G, Phillips AD, Coulson BS, Hartland EL. The type III effectors NleE and NleB from enteropathogenic E. coli and OspZ from Shigella block nuclear translocation of NF-κB p65. PLoS Pathog. 2010;6:e1000898. doi: 10.1371/journal.ppat.1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gurung M, Moon DC, Choi CW, Lee JH, Bae YC, Kim J, Lee YC, Seol SY, Cho DT, Kim SI, Lee JC. Staphylococcus aureus produces membrane-derived vesicles that induce host cell death. PloS One. 2011;6:e27958. doi: 10.1371/journal.pone.0027958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erjavec MS, Jesenko B, Petkovsek Z, Zgur-Bertok D. Prevalence and associations of tcpC, a gene encoding a Toll/interleukin-1 receptor domain-containing protein, among Escherichia coli urinary tract infection, skin and soft tissue infection, and commensal isolates. J Clin Microbiol. 2010;48:966–968. doi: 10.1128/JCM.01227-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoebe K, Georgel P, Rutschmann S, Du X, Mudd S, Crozat K, Sovath S, Shamel L, Hartung T, Zahringer U, Beutler B. CD36 is a sensor of diacylglycerides. Nature. 2005;433:523–527. doi: 10.1038/nature03253. [DOI] [PubMed] [Google Scholar]
- 39.Stuart LM, Deng JS, Silver JM, Takahashi K, Tseng AA, Hennessy EJ, Ezekowitz RAB, Moore KJ. Response to Staphylococcus aureus requires CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. J Cell Biol. 2005;170:477–485. doi: 10.1083/jcb.200501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yokoyama R, Itoh S, Kamoshida G, Takii T, Fujii S, Tsuji T, Onozaki K. Staphylococcal superantigen-like protein 3 binds to the Toll-like receptor 2 extracellular domain and inhibits cytokine production induced by Staphylococcus aureus, cell wall component, or lipopeptides in murine macrophages. Infect Immun. 2012;80:2816–2825. doi: 10.1128/IAI.00399-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller LS, O'Connell RM, Gutierrez MA, Pietras EM, Shahangian A, Gross CE, Thirumala A, Cheung AL, Cheng G, Modlin RL. MyD88 mediates neutrophil recruitment initiated by IL-1r but not TLR2 activation in immunity against Staphylococcus aureus. Immunity. 2006;24:79–91. doi: 10.1016/j.immuni.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 42.Johannessen M, Askarian F, Sangvik M, Sollid JE. Bacterial interference with canonical NFκB signalling. Microbiology. 2013;159:2001–2013. doi: 10.1099/mic.0.069369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirschning CJ, Wesche H, Merrill Ayres T, Rothe M. Human Toll-like receptor 2 confers responsiveness to bacterial lipopolysaccharide. J Exp Med. 1998;188:2091–2097. doi: 10.1084/jem.188.11.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jefferies C, Bowie A, Brady G, Cooke EL, Li XX, O'Neill LAJ. Transactivation by the p65 subunit of NF-κB in response to interleukin-1 (IL-1) involves MyD88, IL-1 receptor-associated kinase 1, TRAF-6, and Rac1. Mol Cell Biol. 2001;21:4544–4552. doi: 10.1128/MCB.21.14.4544-4552.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Sorge NM, Beasley FC, Gusarov I, Gonzalez DJ, von Kockritz-Blickwede M, Anik S, Borkowski AW, Dorrestein PC, Nudler E, Nizet V. Methicillin-resistant Staphylococcus aureus bacterial nitric-oxide synthase affects antibiotic sensitivity and skin abscess development. J Biol Chem. 2013;288:6417–6426. doi: 10.1074/jbc.M112.448738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Surewaard BG, de Haas CJ, Vervoort F, Rigby KM, Deleo FR, Otto M, van Strijp JA, Nijland R. Staphylococcal alpha-phenol soluble modulins contribute to neutrophil lysis after phagocytosis. Cell Microbiol. 2013;15:1427–1437. doi: 10.1111/cmi.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelley LA, Sternberg MJE. Protein structure prediction on the web: a case study using the Phyre server. Nature Protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data