Abstract
Our current knowledge of recombination hot spot activity in mammalian systems implicates a role for both the primary DNA sequence and the nature of the chromatin domain around it. In mice, the only recombination hot spots mapped to date have been confined to a cluster within the major histocompatibility complex (MHC) region. We present a high resolution analysis of a new recombination hot spot in the mouse genome which maps to mouse chromosome 8 C-D. Haplotype diversity analysis across 40 different strains of mice has enabled us to map recombination breakpoints to a 1-kb interval. This hot spot has a recombination intensity that is 10- to 100-fold above the genome average and has a mean gene conversion tract length of 371 bp. This meiotically active locus happens to be flanked by a transcribed region encoding a non-protein-coding RNA polymerase II transcript and the previously characterized repair site. Many of the primary DNA sequence features that have been reported for the mouse MHC hot spots are also shared by this hot spot locus and in addition, along with three other MHC hot spot loci, we show a new parallel feature of association of the crossover sites with the nuclear matrix.
Most of the advances in our understanding of meiosis and recombination have come from studies of lower eukaryotes, in particular Saccharomyces cerevisiae. Recombination initiates via programmed double-strand breaks at the leptotene interval which are resected and undergo strand exchange and finally give rise to double Holliday junctions by the mid-pachytene interval. Mature crossover products follow around the end of pachytene (14, 24). It is generally believed that a similar process operates in higher eukaryotes, including mammals. The rate of recombination is not a uniform function of the physical distance between two markers, leading to the concept of recombination hot spots and cold spots (36). In mice and humans, regions are generally termed as hot spots if their recombination frequency significantly exceeds 1 centimorgan (cM)/Mb. There are three methods that have been used to map hot spots in mammals. The classical method is to correlate physical distances with genetic distances established by standard pedigree-based linkage analysis. The resolution of this method is limited by the density of markers, the number of individuals examined, and the number of recombination events that occur in a typical pedigree. Methods based on linkage disequilibrium (LD), which refers to the nonrandom association of alleles at linked sites, take into account ancestral recombination events, which would be far greater than those observed from any pedigree-based linkage study, leading to a finer mapping of recombination hot spots. Most measures of LD quantify the degree of association between pairs of markers which decreases as a function of physical distance between them due to increased probability of recombination between the markers. However, both of these techniques do not have the resolution to characterize crossovers at a molecular level. The technique of mapping by sperm typing uses PCR to examine alleles present in haploid sperms and offers the highest possible resolution and characterization of hot spots, due to the availability of unlimited number of sperms, each of which is a product of a meiotic event (2). However, sperm typing does not give information on recombination rates in females, is technically demanding, and requires a large number of heterozygous markers in the region being studied. Hence, population genetic methods based on LD can be very useful to infer recombination rates from patterns of genetic variation among DNA sequences (49).
Several recombination hot spots have been identified in humans and mice. In humans this has been possible because of the availability of high resolution physical and genetic maps, while analysis of recombinants in crosses between laboratory strains have helped identify the hot spots in mice. Among the few hot spots that have been mapped to a fine resolution, the human hotspots include (i) minisatellite-associated hot spot MS 32 (20), (ii) the CMT1A/HNPP hot spot associated with hereditary neuropathies (39), and (iii) the major histocompatibility complex (MHC)-associated TAP2 and DNA2 hot spots (19, 21). In the mouse, a total of eight hotspots have been identified so far (56), and the ones that have been characterized to a high degree of resolution include (i) the Lmp2/Psmb9 hot spot (32), (ii) the hot spot located in the Eb second intron (57), (iii) the Pb hot spot (18), and (iv) the hot spot located in the Ea fourth intron (22). All the known mouse recombination hot spots have been identified in the MHC region by pedigree analysis, hence not much is known about the hot spot morphology and the recombination processes operating within them, except for the recently published study of the mouse MHC Eb hot spot showing crossover asymmetry and the resulting biased gene conversion of markers (56). More importantly, little is known about the recombinational activity of DNA outside these mouse MHC hot spots.
The primary DNA sequence features associated with known recombination hot spots are long terminal repeat (LTR) elements, transposable sequences, hypervariable minisatellite sequences (HVMS), and GT repeats (29, 46, 53). It is now becoming clear that increased accessibility of DNA within the chromatin to the recombination machinery is an important prerequisite for initiating recombination. Open chromatin structure can manifest because of the intrinsic nature of the DNA sequence (bent DNA sequence, scaffold/matrix-associated regions [S/MARs]) that makes it exclude nucleosomes (β hot spots) or as a result of transcription factor binding and transcriptional activity in the region (α hot spots) (34, 36). The mechanistic relationship between meiotic recombination and transcription has been substantiated by the global mapping of recombination hotspots in S. cerevisiae (13).
S/MARs are DNA elements 300 bp to several thousand base pairs long which mediate attachment of the chromatin to the nuclear scaffold or matrix and are separated by loops of approximately 60 to 100 kb. Important processes like replication and transcription have been shown to take place at macromolecular complexes located on the nuclear matrix (30). Actively transcribed genes are closely associated with the nuclear matrix, although the actual DNA attachment sites are within the noncoding regions. S/MARs are expected to be recombinogenic due to their significant potential for alternate secondary structure and the presence of curved sequences, which promote base unpairing under superhelical stress (5). This expectation is supported by the observations that transfected S/MAR constructs integrate at much higher copy numbers than their S/MAR-free counterparts and that they are the dominant integration sites for retroviruses (5). S/MARs have therefore been shown to be associated with illegitimate and site-specific recombination, though nothing much is known about their association with meiotic recombination.
The pachytene interval of meiotic prophase is also known to exhibit high levels of DNA repair synthesis (35). DNA polymerase β has been shown to immunolocalize to discrete sites along homologous chromosomes during zygotene and pachytene intervals (37). More recently, a novel DNA polymerase, polymerase λ, has been reported which is expressed only in late pachytene spermatocytes (12), suggesting distinct functions for DNA polymerases β and λ during meiotic recombination. Earlier work from our laboratory, while characterizing the DNA repair sites of pachytene spermatocytes, had resulted in the isolation of a 1.3-kb DNA fragment from the rat genome, which contained several recombination potentiating sequences (38), that is highly conserved in most mammalian species (unpublished data). We have now isolated a larger 17.2-kb fragment from the mouse genome flanking the mouse 1.3-kb sequence and used haplotype and LD analysis to provide evidence that the 17.2-kb locus in the mouse genome does define a recombination hot spot. We also provide the first estimate of the recombination parameter R for any mouse hot spot locus and the recombination intensity and average size of gene conversion tracts for this locus. A close correlation between hot spot activity and the presence of S/MAR elements was observed for the 17.2-kb locus as well as for the three other previously mapped mouse MHC hotspots. Our results therefore suggest S/MAR elements may define an important new molecular feature of meiotic hotspot activity in the mouse genome. We also find a novel 2.4-kb non-protein-coding RNA polymerase II transcript located at the 5′ end of the crossover domain in the 17.2-kb hot spot locus.
MATERIALS AND METHODS
Cloning of the mouse 17.2-kb fragment.
The mouse 1.3-kb fragment was amplified from genomic DNA by using primers 1 and 2 (Table 1) designed based on the sequence of the rat 1.3-kb fragment, and the sequence identity was confirmed. The 564-bp sequence (118 to 682 bp within the 1.3-kb repair fragment) devoid of repeat elements, and thus representing a unique sequence, was amplified using primers 3 and 4 and used to screen the mouse genomic library. We screened 3 × 105 plaques of a mouse (Mus musculus 129/SvEvTacfBR) genomic library constructed in Lambda FIX II vector (Stratagene) with the 564-bp radioactive probe. The 17.2-kb insert was released from the only positive clone obtained by NotI digestion, and its identity was confirmed by Southern blot analysis (42). Four smaller contigs of 3.4, 4.0, 4.5, and 5.3 kb were generated from the 17.2-kb fragment following restriction with NotI and BamHI. The 4.0- and 4.5-kb contigs were cloned into the BamHI site of plasmid pUC 18, and the 3.4- and 5.3-kb contigs were cloned into the NotI/BamHI sites of Bluescript vector pBS SK+. The clones were sequenced by the primer-walking method, and the individual sequence reads were assembled together to generate the entire sequence of the 17.2-kb fragment by using the fragment assembly program of GCG software.
TABLE 1.
Sequence of primers used in the present study
| Primer 1 (sense) | 5′ CCGGAATTCCTGCTACCTGCC 3′ |
| Primer 2 (antisense) | 5′ ACGGAATTCTTCAGCAGTCTA 3′ |
| Primer 3 (sense) | 5′ CGCCCCTCTGTATAAATACACCCTAATCAGC 3′ |
| Primer 4 (antisense) | 5′ CCGCTTGGTCAAATTATCATTAATACAC 3′ |
| Primer 5 (sense) | 5′ ACACACTCCCCTCTGTTATTCC 3′ |
| Primer 6 (antisense) | 5′ GAAGACCCCAAACCCCACTACAC 3′ |
| Primer 7 (sense) | 5′ CTCGAGCAATTTTGGGAA 3′ |
| Primer 8 (antisense) | 5′ CTGTTGACCTAATTTTGTGATTAC 3′ |
| Primer 9 (sense) | 5′ CAGGACAGACAGGTCAGGACAGAGAA 3′ |
| Primer 10 (antisense) | 5′ CCAATGTCCAAATGAACAGAGCA 3′ |
| Primer 11 (sense) | 5′ GTGACTTGCTCTTCATTAGAT 3′ |
| Primer 12 (antisense) | 5′ GGTATTTTGTTCCCAATCTAAGG 3′ |
| Primer 13 (antisense) | 5′ ACTGTTTTATAATCTCTTGCCTTG 3′ |
| Primer 14 (sense) | 5′ GGTCAGATTGATGAATGGGAGATGTTG 3′ |
| Primer 15 (antisense) | 5′ TGACACATATCCTAGATGATGC 3′ |
| Primer 16 (sense) | 5′ GCCCACCTTGATAAATACCTA 3′ |
| Primer 17 (antisense) | 5′ TGGGGTAGTCTCTGGATAGTC 3′ |
| Primer 18 (sense) | 5′ TCTCCTGTTAACACTATGTAGCCAG 3′ |
| Primer 19 (antisense) | 5′ GCAAGAGGAAAGCATTATATTGAG 3′ |
| Primer 20 (sense) | 5′ GAGACTGACAGATGTGTGAATG 3′ |
| Primer 21 (antisense) | 5′ GGAGGTCACAGTAAGAACTGAG 3′ |
| Primer 22 (sense) | 5′ CACCCTGCTCAACTGCAGCCA 3′ |
| Primer 23 (antisense) | 5′ GTCTGCCAGCCCTTGTAAGC 3′ |
| Primer 24 (sense) | 5′ GCTGCAGTTACAGGTGACAGGG 3′ |
| Primer 25 (antisense) | 5′ CAGAATCACTGTGTGGCTGTG 3′ |
| Primer 26 (sense) | 5′ GAGGTAAAATCTACAGCCAGC 3′ |
| Primer 27 (antisense) | 5′ ATAGTTGCCTTTGGCTAGGG 3′ |
| Primer 28 (sense) | 5′ CAGAAGTCAGAATTGGGAGGTATCGGT 3′ |
| Primer 29 (antisense) | 5′ CATAGATGCCAACAAGCTGGTTC 3′ |
| Primer 30 (sense) | 5′ GCACTCATGAAGAGGAAACAGATGG 3′ |
| Primer 31 (antisense) | 5′ GTCAGCTGCTATTAGGACCAATG 3′ |
In silico sequence analysis.
Analysis for the presence of exons, promoter elements, poly(A) sites, and CpG islands and BLAST searches of the DNA, protein, and EST databases were done using the NIX suite of programs run on the HGMP bioinformatics server at http://menu.hgmp.mrc.ac.uk/menu-bin/Nix/Nix.pl and the CpG island searcher program at http://ccnt.hsc.usc.edu/cpgislands. Gene models were also constructed by the NIX suite of programs by pattern recognition of different types of exons, promoters, splice sites, and poly(A) signals followed by dynamic programming that puts them together in the most optimal combination. The programs Repeat Masker (http://ftp.genome.washington.edu/RM/RepeatMasker.html), MARFINDER (http://www.futuresoft.org), THERMODYN (http://wings.buffalo.edu/gsa/dna/dk/WEBTHERMODYN), and mfold (http://bioweb.pasteur.fr/seqanal/interfaces/mfold.html) were used for identifying genome-wide repeats, S/MARs, DNA unwinding elements (DUEs), and RNA secondary structure, respectively.
Chromosome localization by mapping with radiation hybrid panels.
DNA samples from 96 mouse-hamster cell lines from the T31 radiation hybrid panel were purchased from Research Genetics. Approximately 50 ng of DNA from each of these samples was used for PCR analysis. We sequenced the 1.3-kb fragment from the hamster genome, and due to the conserved nature of the mouse and hamster 1.3-kb sequences, we used primers 5 and 6, which amplify an 800-bp product specifically from the mouse genome, from a region adjacent to the mouse 1.3-kb position in the 17.2-kb fragment. As an internal control we used primers 7 and 8, which amplify a 260-bp product from both the mouse and hamster 1.3-kb locus. Each of the 96 DNA samples from the radiation hybrid panel was scored for the presence of the 800-bp product specific to the mouse 17.2-kb fragment, as well as for a 260-bp product that amplifies from both the mouse and hamster 1.3-kb sequences. The Jackson Laboratory RH database server was used for interpretation of the data from which the chromosomal location of the 17.2-kb locus was identified.
Northern blotting, RT-PCR, and primer extension.
Poly(A)+ RNA was isolated from mouse liver and testis by standard methods and electrophoretically separated (2 μg/lane) on a 1% agarose gel containing 2.2 M formaldehyde. After transferring to nylon membranes, the blot was probed with each of the 32P-labeled contigs. For multiple-tissue Northern analysis, the mouse MessageMap blot (Stratagene) containing electrophoretically separated poly(A)+ RNA was probed with the 900-bp reverse transcription (RT)-PCR product amplified with primers 9 and 10. β-Actin cDNA was used as an internal control. RT-PCR to amplify the 2.4-kb transcript was performed using total RNA prepared from mouse testis, which was reverse transcribed into cDNA using avian myeloblastosis virus reverse transcriptase (Invitrogen) with the antisense primer 12 at 52°C, followed by PCR amplification with primers 11 and 12 by standard techniques. The transcription start site for the 2.4-kb transcript was determined by primer extension on mouse testis total RNA by using primer 13 end-labeled with γ-32P. The extension products were run on a 6% polyacrylamide gel containing 8.3 M urea and were autoradiographed.
Haplotype diversity analysis.
Genomic DNA for the M. musculus species was obtained from Jackson Laboratories for all of the 33 strains except for Swiss, BALB/c, CD1, C3H, C57, and FVB/n. For these laboratory strains, DNA was isolated from the tails of mice by standard procedures. For strain 129/SvEvTacfBR, the genomic library in λ FIX II vector was available. For haplotype analysis, a 1-kb region from 13.3 to 14.3 kb was PCR amplified using the primer pair 18 and 19. The PCR products were gel eluted and sequenced on an ABI 377 sequencer. The sequences from all of the 40 strains were aligned using the CLUSTALW program. The DnaSP version 3.5 software (41) was used for the analysis of polymorphism and LD between parsimony informative sites (sites where the low-frequency variant is present at least two times) as well as to perform the Tajima's D test (50) on the unphased diploid genotype data. The Tajima's D test is used for testing the hypothesis that all mutations or polymorphisms seen in a DNA sequence are selectively neutral. Pairwise LD values were plotted against the physical map using MATLAB software. Recombination events were also identified by the four gamete test (17). Coalescent simulations to simulate the history of our sample for different values of 4Nr were performed using DnaSP. The GENECONV program (44) was used to determine gene conversion tracts (http://www.math.wustl.edu/∼sawyer). This program looks for aligned segments (after removal of monomorphic sites) for which a pair of sequences are sufficiently similar to be suggestive of past gene conversion events. The similarity is judged based on an unusually long pairwise identity or an unusually high score (matches count as +1 with a penalty for mismatches) for that pair of sequences.
Association of the 17.2-kb locus and mouse recombination hot spots with the nuclear matrix.
Matrix and loop DNA were prepared from mouse total testis. The testis were decapsulated and treated with 0.04% collagenase in 1× phosphate-buffered saline containing 0.2% bovine serum albumin and 0.1% sucrose at 30°C for 20 min to make a cell suspension which was pelleted at 500 × g at 4°C and washed thrice in 1× phosphate-buffered saline. Isolation of nuclei and subsequent preparation of MAR and loop DNA were done according to the low-salt lithium diiodosalicylate (LIS) method of Mirkovitch et al. (31). Briefly, the nuclear pellet was obtained after cell lysis with 20 mM KCl and 0.1% digitonin, and the integrity of the nuclei was confirmed by DAPI (4′,6′-diamidino-2-phenylindole) staining. Nuclei were incubated at 37°C for 20 min, followed by the addition of extraction buffer containing 25 mM LIS. The resulting nucleoid structure was pelleted at 2,400 × g for 20 min at room temperature and washed with digestion buffer to remove LIS. Restriction enzyme digestion was performed using EcoRI and NcoI, each at 500 U/ml in combination at 37°C for 6 h. The S/MAR fraction was pelleted from the digested loop DNA by centrifugation at 2,400 × g for 10 min at 4°C, and both fractions were purified using standard DNA extraction techniques. The authenticity of the S/MAR pellet was also checked by analyzing the acid (0.25 N HCl)-extracted proteins on a sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis. Total genomic DNA was isolated from the nuclei by standard procedures. Total input DNA was estimated by solubilizing the nuclei in 2 M NaCl and 5 M urea and measuring the absorbance at 260 nm. Accurate estimates of the MAR, loop, and total DNA concentrations for PCR were obtained using a spectrofluorimeter with the dye Hoechst 33258 (26). In addition to analyzing the partitioning of different regions of the 17.2-kb locus between the matrix and the loop, we have also analyzed the predicted MAR regions of the mouse recombination hot spots. For this purpose, primers 20 and 21 (for Ea), 22 and 23 (for Psmb9), 24 and 25 (for Pb), and 26 and 27 were designed based on published sequence data for amplifying the mapped crossover segments for the Ea, Psmb9, and Pb hot spots and the experimentally known immunoglobulin (Ig) kappa light chain MAR sequence as a positive control (6). The domain C (13,500 to 14,000 bp; also see Fig. 10) corresponding to the crossover region of the 17.2-kb fragment was amplified using primers 28 and 29. A locus upstream and adjacent to the transcribed noncoding RNA which is flanked by EcoRI and NcoI sites with a predicted low MAR potential (domain E, 360 to 800 bp; also see Fig. 10) was amplified using primers 30 and 31 and used as a negative control. PCR was performed on 150 ng of MAR, loop, and total DNA for 25 cycles, where the amplification was seen to be in the linear range. Subsequently, aliquots of the PCR products were electrophoresed on a 1% agarose gel, stained with ethidium bromide, and quantified densitometrically in a Bio-Rad gel documentation system. The enrichment factor of a particular sequence in the MAR fraction was calculated as the band intensity in the matrix divided by the sum of intensities of matrix plus loop fractions (9). Equal efficiency of PCR amplification in the MAR, loop, and total genomic DNA was judged by the ratio between the sum of intensities of MAR plus loop fractions and band intensity of the PCR product with total genomic DNA as an internal control.
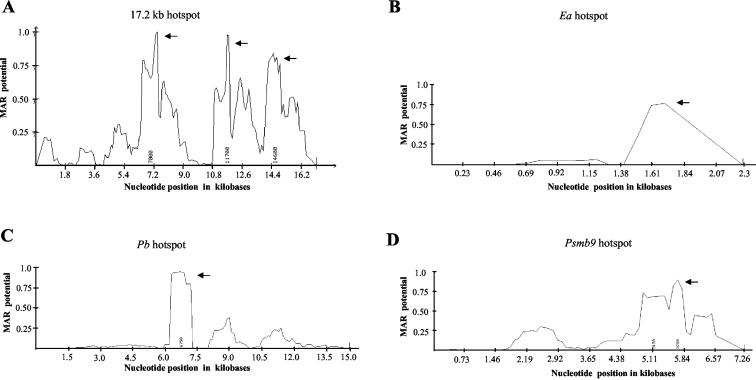
FIG. 10.
S/MAR profile of the Ea, Psmb9, Pb, and the 17.2-kb mouse recombination hot spots. (A) MAR profile of the 17.2-kb fragment. The crossover mapped to domain C is from 13,500 to 14,000 bp. (B) Ea hot spot (2.2 kb), accession number K00971. Crossovers mapped to the fourth intron from 1,163 to 1,780 nucleotides. (C) Pb hot spot (15 kb), accession number AF100956. The crossovers are mapped to a 2.4-kb interval (from 4.6 to 7 kb). (D) Psmb9 hot spot (7.2 kb), accession number D43620. The crossovers are mapped to the 2 kb from the poly(A) signal (5,166 bp) of the Psmb9 gene. Arrows indicate regions which can function as S/MARs in all four panels.
Nucleotide sequence accession numbers.
The nucleotide sequence of the 17.2-kb fragment has been submitted to GenBank under the accession number AF 393505. The sequence of the 1.3-kb fragment from the hamster genome was submitted under accession number AY187064.
RESULTS
A 17.2-kb mouse genomic fragment flanking the mouse 1.3-kb meiotic repair site maps to chromosome 8 C-D.
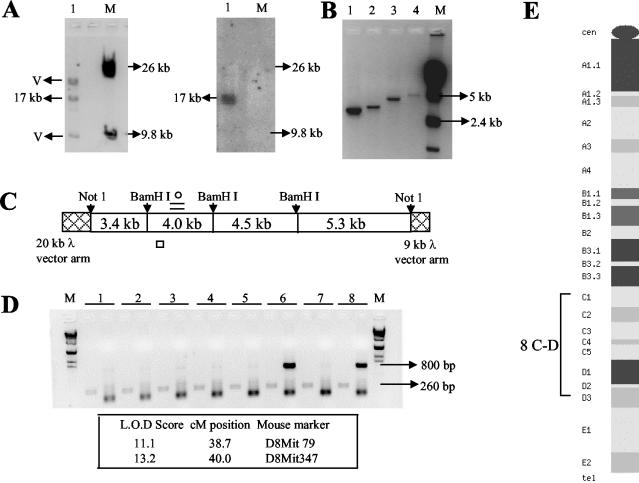
As an initial step towards further understanding the meiotic DNA repair locus (1.3-kb fragment), we embarked on isolating a larger fragment from the mouse genome containing this repair fragment so that we could analyze in greater detail various molecular features associated with this locus. For this purpose, we first amplified the 1.3-kb fragment from the mouse genomic DNA by using primer pairs based on the rat sequence and confirmed its sequence identity. We identified a 564-bp sequence stretch (118 to 682 bp) within this 1.3-kb fragment as being devoid of any repetitive elements, and hence it was used as a radioactive probe to screen a mouse genomic library in λ FIX II vector. This analysis yielded a single positive signal having an insert of 17.2 kb (Fig. 1A), which was completely sequenced after subcloning as four smaller contigs of 3.4, 4, 4.5, and 5.3 kb (Fig. 1B and C). The mouse 1.3-kb sequence was present from 4,560 to 5,800 bp in the 4-kb contig (Fig. 1C), and thus we had around 5 and 11 kb of additional sequence information on either side of the mouse 1.3-kb fragment to study the sequence context in which it functions as a meiotic repair site. The 17.2-kb fragment was mapped to mouse chromosome 8 C-D by PCR analysis of the T31 mouse-hamster radiation hybrid panel, which included 96 independent hybrid cell lines. A representative gel pattern of the PCR products is shown in Fig. 1D. Out of the 96 hybrid cell lines, 29 were positive for the presence of the 800-bp mouse-specific product, while all of them yielded the 260-bp product which is common for both mouse and hamster genomes. Analysis of these data by the Jackson Laboratory RH database server gave a logarithm of odds score of 13.2 and 11.1 with respect to the microsatellite markers D8Mit347 and D8Mit79, both of which are located on chromosome 8 at 38.7 and 40 cM, respectively, corresponding to a cytological position of 8 C-D (Fig. 1E).
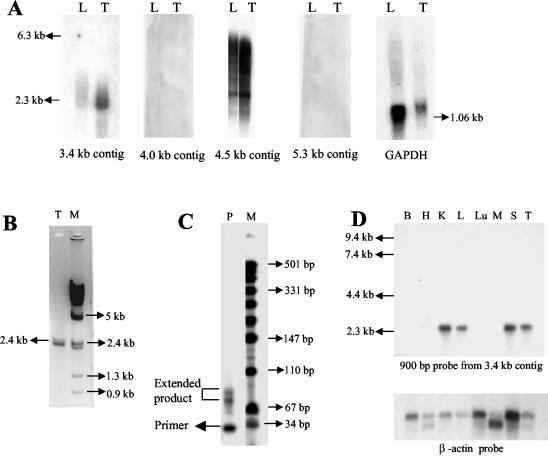
FIG. 1.
Cloning and chromosomal localization of a 17.2-kb mouse genomic fragment flanking the mouse homolog of the 1.3-kb meiotic repair site. (A) Release of the 17.2-kb insert with NotI digestion from the mouse genomic clone and subsequent Southern hybridization with the 564-bp probe (118 to 682 bp within the 1.3-kb repair fragment). M, λ/HindIII marker. (B) Generation of four smaller contigs following a BamHI and NotI double digestion. (C) Position of each of the four contigs and the location of the 1.3-kb fragment (=), the 260-bp fragment (○), and the 800-bp fragment (□) used for screening the mouse-hamster radiation hybrid panel. (D) Representative gel of the PCR analysis of the mouse-hamster radiation hybrid panel. Lanes 6 and 8 show amplification of the 800-bp product while all lanes show amplification of the control 260-bp product. M, λ/HindIII marker. The highest two logarithm of odds (L.O.D.) score values of the mouse 800-bp fragment with known mouse markers are also shown. (E) Cytogenetic map of mouse chromosome 8.
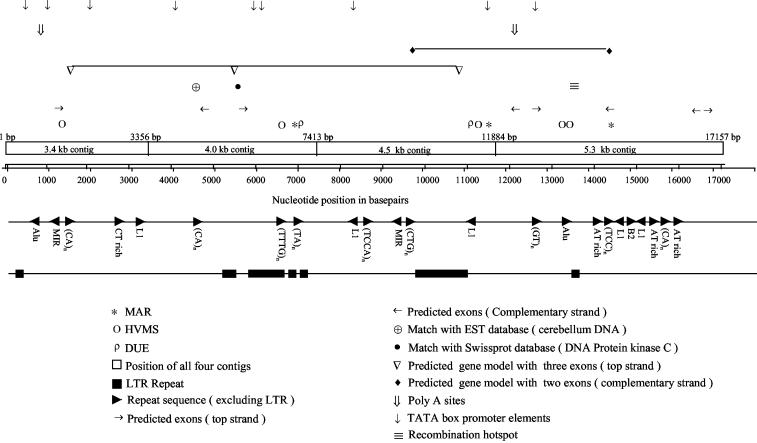
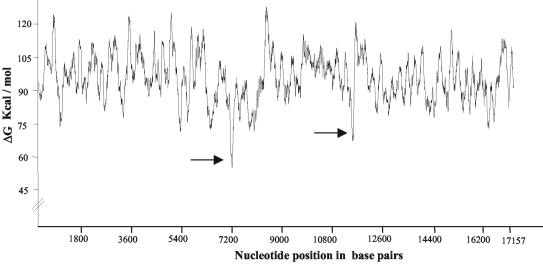
In silico analysis of the 17.2-kb fragment showed it to be 38% GC rich and to have a repeat sequence content of 39%. The major class of repeats observed were the LTRs, which constituted 22% of the sequence and 56% of the total repeat content. This was surprising considering that LTRs normally comprise around 1% of the genome (28). The other repetitive elements were the long interspersed nuclear elements, short interspersed nuclear elements (including the Alu and mammalian interspersed repeats), and microsatellite repeats, all of which are shown in Fig. 2. In silico analysis further revealed two DUEs at 7 and 11.2 kb (Fig. 3) where the helical stability of the duplex, as measured by the ΔG value, is at least 25 kcal/mol less than the average value for the whole sequence (95.9 kcal/mol). DUEs are known to be associated with eukaryotic replication origins, as its intrinsic helical instability facilitates replication initiation (8). DUEs are also a feature of S/MARs due to their property of stress-induced duplex destabilization, which is essential for S/MAR function (3). The presence of sequence features corresponding to the transcribed regions analyzed using the NIX suite of programs resulted in the prediction of several exons, two complete gene models, several promoter elements, and poly(A) sites, as shown in Fig. 2. Interestingly, no CpG islands were detected with either the NIX program or the CpG island searcher program. Two matches to the existing Swissprot and EST databases were also seen. The entire 17.2-kb sequence of the mouse locus was not found in the mouse genome database (http://www2.igh.cnrs.fr/Mouse-Genome_DBS.html); however, complete matches to the high-throughput genomic unfinished mouse draft sequence were seen for many regions of the 17.2-kb fragment.
FIG. 2.
Sequence features of the mouse 17.2-kb fragment. Non-LTRs include long interspersed nuclear elements, short interspersed nuclear elements (including the Alu and MIR [mammalian interspersed repeats]), and microsatellite repeats. In addition to individual exons, two complete gene models have been predicted, one on the top strand with three exons (1,643 to 1,673 bp, 5,668 to 5,767 bp, and 10,922 to 11,045 bp) and one on the complementary strand with two exons (14,354 to 14,552 bp and 9,562 to 9,582 bp). Predicted poly(A) and TATA box promoter sites are indicated, as well as matches to existing EST and Swissprot databases. The position of the recombination hot spot deduced from LD analysis is also shown.
FIG. 3.
Output of the THERMODYN program. Regions of the 17.2-kb fragment which can function as DUEs are indicated by arrows.
A transcribed region encoding a novel non-protein-coding RNA flanks the 1.3-kb meiotic repair site within the 17.2-kb fragment.
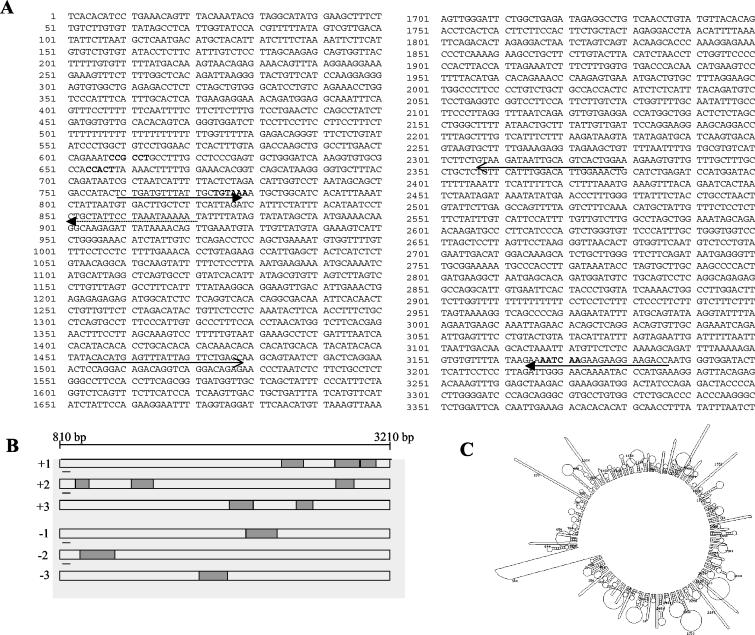
Since there were several exons predicted by different programs, we analyzed whether transcripts were indeed present in the liver and testis total RNA by carrying out RT-PCR analysis using primers across these predicted individual exons in the repeat-free regions of the 17.2-kb sequence. However, no transcripts corresponding to the predicted exons were detected. Northern analysis of poly(A)+ RNA from mouse liver and testis using the four different contigs as probes revealed that two of the contigs (3.4 and 4.5 kb) gave hybridization signals, while the other two contigs (4 and 5.3 kb) did not show any signal (Fig. 4A). Among the two positive contigs, hybridization with the 4.5-kb contig gave a smear showing that the signal might be due to hybridization of several repeat elements (highest in this contig) showing weak homology with several transcribed sequences. This is not unusual, since many classes of repeat elements have been shown to be transcribed into RNA intermediates (28, 43, 45). On the other hand, hybridization with the 3.4-kb contig revealed a single transcript of approximately 2.4 kb in size. This was further confirmed by RT-PCR analysis using primer pair 11 and 12 from within the 3.4-kb contig (Fig. 4B). Artifactual amplification due to contamination with genomic DNA is ruled out, since no other transcripts were detected with other primer pairs. Primer extension analysis (Fig. 4C) positioned the transcript start site to approximately 90 nucleotides (longer extension product) from the 3′ extension primer 13 (Fig. 5A). Expression profiling of this transcript across eight different mouse tissues with a 900-bp (1.5 to 2.4 kb within the 17.2-kb fragment) probe, derived from the transcribed region amplified using primers 9 and 10, revealed that the transcript was expressed in kidney, liver, spleen, and testis but not in brain, heart, lung, and muscle (Fig. 4D). Hybridization of the same multiple-tissue RNA blot using the 4.5-kb contig gave smears similar to Fig. 4A (data not shown). The complete nucleotide sequence encoding this transcript within the 3.4-kb contig is shown in Fig. 5A. Sequence analysis of the RT-PCR product also revealed that the sequence is identical to that present in the 3.4-kb contig. A strict correlation between the genomic sequence and the sequence of the RT-PCR product indicates that it is an unspliced transcript. We also observed that a TATA box and a CAAT box are positioned at 775 and 655 bp, which are −35 and −155 bp away from the transcription start point which has been approximately mapped to 810 bp (Fig. 5A). A GC box in reverse orientation was seen at 610 bp, which is −200 bp away from the transcription start site, and a poly(A) signal sequence with a single nucleotide variation was present at 3,170 bp. Although this transcript has all the known features of polymerase II-transcribed genes, we were surprised to find that no significant open reading frames (ORFs) were seen in the 2.4-kb transcript (Fig. 5B), and none of the short ORFs shown were preceded by a Kozak consensus sequence. It is now becoming increasingly clear that the mouse genome encodes many nontranslatable transcripts (52). The functional significance of these transcripts is not clearly known. Some of them have been known to possess extensive secondary foldback structures. In this context, we analyzed the 2.4-kb RNA transcript for potential secondary structures by using the mfold program (temperature, 37°C; ionic strength, 100 mM NaCl), which revealed that the transcript has considerable propensity to posses secondary structure (Fig. 5C).
FIG. 4.
Transcriptional analysis of the mouse 17.2-kb fragment. (A) Northern analysis carried out on 2 μg of poly(A)+ RNA isolated from mouse liver (L) and testis (T) with each of the four contigs and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as probes. (B) Amplification of the 2.4-kb RT-PCR product amplified from mouse testis (T) total RNA. M, λ/HindIII marker. (C) Primer extension carried out on mouse testis poly(A)+ RNA (P) using primer M. M, pUC 19/MspI digest. The primer position is below the 34-bp marker. (D) Northern analysis carried out on a mouse tissue blot having 2 μg of poly(A)+ RNA from brain (B), heart (H), kidney (K), liver (L), lung (Lu), skeletal muscle (M), spleen (S), and testis (T). A 900-bp fragment (from 1.5 to 2.4 kb) from the 3.8-kb contig and β-actin cDNA were used as probes.
FIG. 5.
The 2.4-kb transcript is a non-protein-coding RNA. (A) Sequence of the 3.4-kb contig which contains the transcribed region with the GC box (610 bp), the CAAT box (655 bp), the TATA box (780 bp), and the poly(A) signal sequence (3,170 bp) in bold. The 2.4-kb transcribed region is shown by two arrows from 810 to 3,210 bp. The dotted arrow shows the position of the 3′ primer M used for primer extension. The open arrows show the 900-bp fragment used as the probe in Northern analysis. (B) Position of predicted ORFs in the transcript in all of the six reading frames. (C) Predicted secondary structure of the transcribed RNA.
Breakdown of LD and gene conversion in a 1-kb hot spot locus within the 17.2-kb fragment.
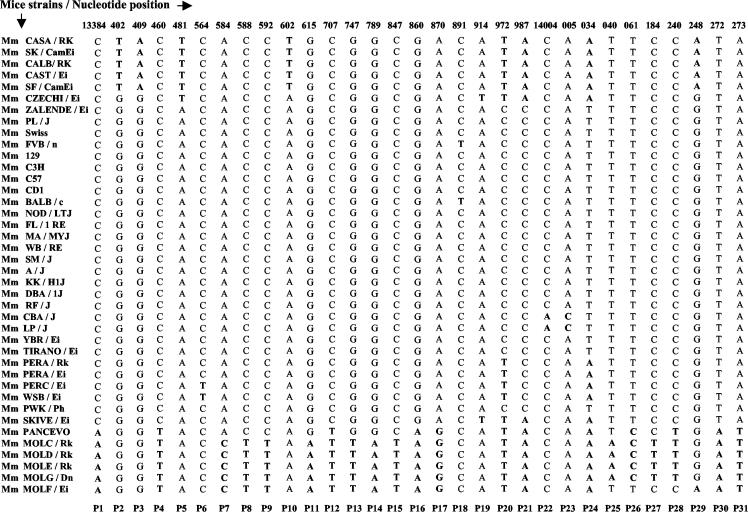
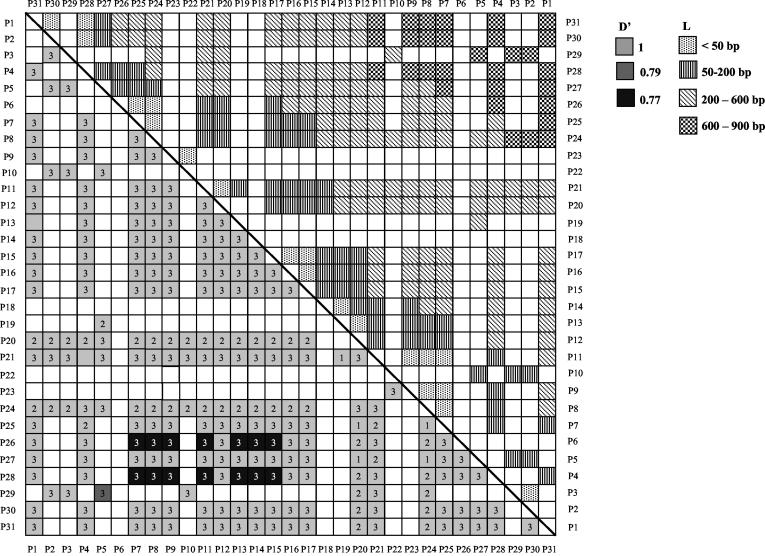
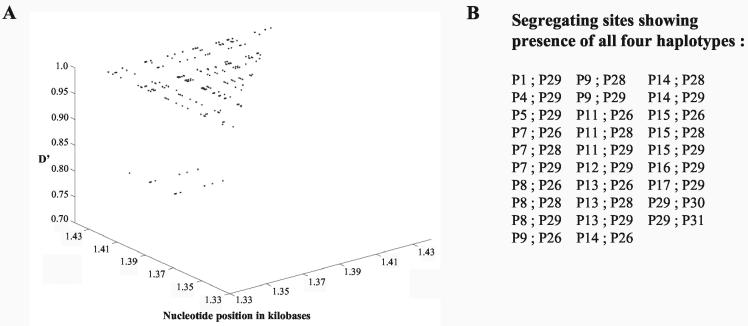
As mentioned earlier, the 1.3-kb EcoRI fragment was initially identified as a meiotic repair fragment in the rat genome. We wanted to further examine whether the 17.2-kb locus harboring the 1.3-kb repair fragment does indeed represent a recombination locus. For this purpose, we have resorted to haplotype diversity analysis in various strains of mice. DNA sequence analysis of the 17.2-kb fragment from a different strain of mouse (M. musculus BALB/c) revealed sequence variation between 9.5 and 14.5 kb. The sequence diversity seen was more pronounced between 13.3 and 14.3 kb, and so this 1-kb region was selected for haplotype diversity analysis across 40 different strains of mice from within the M. musculus species which interbreed freely. This 1-kb region was PCR amplified using the primer pair 14 and 15 from the DNA of all the 40 strains and was sequenced. The region showed considerable sequence polymorphism (Table 2) with 79 polymorphic or segregating sites (excluding insertion-deletion polymorphisms), 31 of which were parsimony informative polymorphic sites with two variants, referred to as P1 to P31 (Fig. 6). Haplotype diversity was further examined by LD analysis of all the pairwise combinations of the 31 parsimony informative polymorphic sites using Lewontin's coefficient D′ (27), with D′ values of <1 being indicative of the presence of recombinants. A total of 465 pairwise analyses were made, of which 239 were statistically significant by Fisher's exact test and the chi-square test (see the supplemental material). These have been plotted as a matrix (Fig. 7). Pairs of markers spaced at very short intervals are expected to exist in complete LD with D′ values of 1. However, D′ values are seen to drop to 0.77 to 0.79 for 15 of these 239 pairwise associations, showing that there are polymorphic loci in linkage equilibrium within the 1-kb interval showing presence of recombinational activity (Fig. 7). These correspond to sites P7 to P15 (13,584 to 13,847 bp), excluding P12, which are in equilibrium with P26 (14,061 bp) and P28 (14,240 bp). This is even more clearly illustrated in the plot of pairwise LD values as a function of the physical map corresponding to this 1-kb interval (Fig. 8A). The position corresponding to the decay of LD in the matrix thereby positions the region of LD breakdown between 13,584 to 13,847 bp on the physical map. However, intermediate sites are seen to be in tight disequilibrium (see D′ values of 1 in Fig. 7 and 8A) with each other, which means that these polymorphisms are relatively recent and could be an indication of continued genetic variability being generated in this region by recent recombination events. Another way of inferring that between two sites at least one recombination event has taken place in the history of the sample is to use the four gamete test. This test looks for the presence of all four haplotypes for any two sites which can arise only if at least one recombination event were to have occurred in the history of the sample between the two sites under the infinite site model. Pairwise analysis of all the 31 parsimony informative diallelic sites by the four gamete test (17) resulted in the finding of 29 pairs of sites which had all four haplotypes (Fig. 8B). The minimum number of recombination events (RM) which is the maximum number of such nonoverlapping pairs is two, P15:P26 and P29:P30. These correspond to nucleotide positions 13,847 to 14,061 bp and 14,248 to 14,272 bp. The actual number of recombination events would be much higher than this, as the four gamete test significantly underestimates the actual number of recombination events (17). The Tajima's D test of neutrality (50) did not detect a significant departure (D = −1.746 at 0.1 > P > 0.05) from neutral expectations (D = 0) in a stationary population. The test was negative, which is an indication of the observed excess of low-frequency variants. Since the region is not under selection pressure, the correlation between decay of LD and presence of crossover sites in this region holds true.
TABLE 2.
Summary of the polymorphisms seen among the 40 strains of mice in the 1-kb sequenced region
| No. of sequences | 40 |
| No. of sites | 1,075 |
| No. of sites (without alignment gaps or missing data) | 866 |
| Avg nucleotide distance between the most distant sites | 972.33 |
| Nucleotide diversity (π) | 0.01158 |
| No. of monomorphic sites | 787 |
| No. of polymorphic or segregating sites | 79 |
| No. of singleton polymorphic sites | 47 |
| No. of parsimony informative polymorphic sites (two or | 32 |
| three variants) |
FIG. 6.
Haplotype diversity analysis of the 17.2-kb mouse fragment. The 31 parsimony informative polymorphic sites (with two variants) marked from P1 to P31 are shown for the 40 strains of mice. Their corresponding nucleotide positions with respect to the 17.2-kb fragment are indicated on top.
FIG. 7.
Pairwise LD values for statistically significant combinations of the parsimony informative polymorphic sites along with nucleotide distance between the sites. The lower triangular panel shows LD values while the upper triangular panel shows intermarker nucleotide distances. Open squares indicate pairwise combinations that were not statistically significant. The label 1 indicates 0.01 < P < 0.05, 2 denotes 0.001 < P < 0.01, and 3 denotes P < 0.001 (chi-square test).
FIG. 8.
Pairwise LD values as a function of the physical map and four gamete test in the 1-kb hot spot locus. (A) D′ measure of LD for all statistically significant pairwise combinations of the parsimony informative polymorphic sites as a function of the physical map from 13,300 to 14,300 bp. (B) Numbers of segregating sites showing presence of all four haplotypes.
While for markers that are several kilobases apart the contribution of gene conversion to the overall level of genetic exchange is negligible, for closely linked markers LD may also be broken down by gene conversion, and its role while interpreting short range LD data is significant (1). The program GENECONV was used to detect conversion events in our sample of DNA sequences from 40 strains of M. musculus. Twenty such significant (P < 0.05) fragment pairs showing gene conversion were detected between different strains, with the size of the conversion tract ranging from 255 to 627 bp (Table 3). This shows that gene conversion has played an important role in the pattern of LD seen at this locus. Most of the conversion events were between 13,595 and 13,971 bp, which corresponds to the region of LD breakdown (13,584 to 13,847 bp; Fig. 8A) and is indicated in Fig. 7. The average size of the conversion tract was around 371 bp, which is comparable to the average conversion tract length of 480 bp reported earlier for the Psmb9 locus by Guillon and de Massy (14).
TABLE 3.
Gene conversion tracts
| M. musculus strain | Sequence | Length (bp) | P value | |
|---|---|---|---|---|
| CALB/RK; CBA/J | 13595 (AAAAAGTCA | TTTGAGTAG) 13971 | 390 | 0.0003 |
| CASA/RK; CBA/J | 13595 (AAAAAGTCA | TTTGAGTAG) 13971 | 390 | 0.0011 |
| SF/CamEi; CBA/J | 13595 (AAAAAGTCA | TTTGAGTAG) 13971 | 390 | 0.0011 |
| CALB/RK; LP/J | 13595 (AAAAAGTCA | TTTGAGTAG) 13971 | 390 | 0.0011 |
| CALB/RK; SM/J | 13595 (AAAAAGTCA | TTTGAGTAG) 13971 | 390 | 0.0011 |
| CALB/RK; YBR/Ei | 13595 (AAAAAGTCA | TTTGAGTAG) 13971 | 390 | 0.0023 |
| CASA/RK; LP/J | 13595 (AAAAAGTCA | TTTGAGTAG) 13971 | 390 | 0.0053 |
| CASA/RK; SM/J | 13595 (AAAAAGTCA | TTTGAGTAG) 13971 | 390 | 0.0053 |
| SF/CamEI; LP/J | 13595 (AAAAAGTCA | TTTGAGTAG) 13971 | 390 | 0.0053 |
| SF/CamEi; SM/J | 13595 (AAAAAGTCA | TTTGAGTAG) 13971 | 390 | 0.0053 |
| CAST/Ei; CBA/J | 13735 (CTAAGGAAG | TTTGAGTAG) 13971 | 255 | 0.0057 |
| CASA/RK; YBR/Ei | 13595 (AAAAAGTCA | TTTGAGTAG) 13971 | 390 | 0.0125 |
| SF/CamEi; YBR/Ei | 13595 (AAAAAGTCA | TTTGAGTAG) 13971 | 390 | 0.0125 |
| CBA/J; SKIVE/Ei | 13607 (GTCAGCTGT | GCTTGCTAC) 13883 | 292 | 0.0218 |
| CAST/Ei; LP/J | 13735 (CTAAGGAAG | TTTGAGTAG) 13971 | 255 | 0.0246 |
| CAST/Ei; SM/J | 13735 (CTAAGGAAG | TTTGAGTAG) 13971 | 255 | 0.0246 |
| Swiss; C57 | 13375 (TTAGTTCTC | TCCTTGCTG) 13705 | 350 | 0.0350 |
| CBA/J; PERA/Rk | 13375 (TTAGTTCTC | TTTGAGTAG) 13971 | 627 | 0.0351 |
| CBA/J; CZECHI/Ei | 13481 (TCTTTAATC | CTCCTGTTA) 13905 | 453 | 0.0389 |
| CAST/Ei; YBR/Ei | 13735 (CTAAGGAAG | TTTGAGTAG) 13971 | 255 | 0.0475 |
Determination of the recombination parameter (R) and recombination intensity value for the 1-kb hot spot locus.
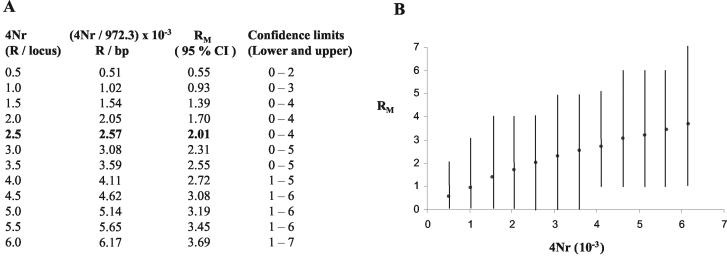
Two parameters that play an important role in DNA sequence variation under the neutral-infinite site model are the mutation parameter (θ = 4Nμ) and the recombination parameter (R = 4Nr). The value of the mutation parameter (θ) has been estimated to be 19.2/gene or 0.02145/bp (total number of sites, 866) from the number of segregating sites. Estimating R is a little more difficult since it is not directly observable in the sample. However, an observable quantity that is related to R is RM, and one can estimate R by using the coalescent approach to simulate the history of the sample for different values of 4Nr that give the observed RM of 2 (17). Such simulations performed 1,000 times for different values of 4Nr (from 0.5 to 6 in increments of 0.5) gave an R value of 0.00257/bp (based on an average nucleotide distance between the most distant sites of 972.33) (Fig. 9 A and B) for this locus. The error bounds on this estimate are wide, which is a known drawback of this method (16, 17). We have therefore also estimated the recombination parameter (R = 4Nr) based on the variance of the average number of nucleotide differences between pairs of sequences (Sk2 in equations 1 and 4 in reference 16). This gave a similar value of 2.50 per locus (1,000 bp) or 0.0026/bp (average nucleotide distance between the most distant sites being 972.33). Given the large data set and the R/θ value of 0.1, which is much less than 4 (17), this would probably represent a reliable estimate of R for this locus. We could not get estimates of the effective population size (N) for the mouse population from the literature, and estimating it from this locus (using θ = 4Nμ) would have a significant upper bias because of the contribution of recombination to the nucleotide diversity at this locus. If one were to assume a value similar to the human population of 104 (most eukaryotic populations have N values from 103 to 104) and the R value of 0.0026, we would get an r value of 6.5 × 10−7, which is 10-fold higher than the average value of 0.6 × 10−8 (4). With an N of 103, we would get an r value of 6.5 × 10−7, which is 100-fold higher than the average value of the mouse recombination rate. Hence, our estimate of the recombination intensity value for this locus is around 10- to 100-fold higher than the genome average.
FIG. 9.
Determination of the recombination parameter (R) and recombination intensity for the 1-kb hot spot. (A) Estimation of R from RM by coalescent simulations of the history of the sample for different values of 4Nr. For each value of 4Nr, the RM value obtained is shown. (B) Graphical representation of the expected number of obligate recombination events (RM) as a function of the population recombination parameter 4Nr. Each point is an average of over 1,000 simulated replicates of the data at 95% confidence interval (CI). Vertical bars show the values at lower and upper limits of the confidence interval.
Association of meiotic crossover DNA segments with the nuclear matrix.
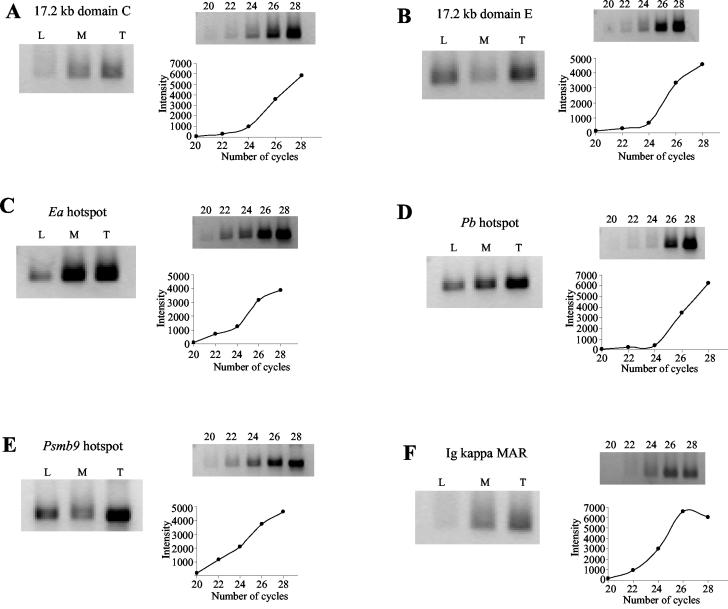
DNA transaction processes like DNA replication and transcription in eukaryotes are known to occur in association with the nuclear matrix (31). When we examined the 17.2-kb sequence with the MARFINDER program, we observed three regions at 7, 11.7, and 14.6 kb as having MAR potential based on the presence of sequence features like AT richness, TG richness, Topo II sites, MAR consensus sequences, and kinked DNA patterns that are known to be associated with S/MARs (Fig. 10A). Interestingly, two of these regions at 7 and 11.7 kb also coincided with the DUEs which have been predicted at 7.2 and 11.2 kb (Fig. 3). Since association of meiotic recombination hot spots with the S/MARs is not well documented in the literature, we have in addition used the known mouse recombination hot spots in the MHC locus for investigating MAR association. For this, we analyzed the DNA sequences of the four mouse MHC hot spots Ea, Pb, Psmb9, and Eb for potential MAR regions by using the MARFINDER program. The three hotspots Ea, Pb, and Psmb9, whose crossover regions have been mapped to an interval of a few kilobases, revealed a strong MAR potential region exactly with their respective crossover regions (Fig. 10B, C, and D). However, such a correlation was not observed with the fourth recombination hot spot locus Eb (data not shown). In order to experimentally verify such an association between crossover sites and the nuclear matrix, we determined the relative enrichment of these sites in S/MAR DNA compared to loop DNA by a PCR-based assay (25). We used a low-salt extraction procedure using LIS, as it has been reported to preserve the molecular interactions at chromatin loop bases much better than that isolated using the high-salt (2 M NaCl) extraction procedure which can cause sliding or rearrangement of the DNA at the matrix attachment site and can also induce precipitation of transcription complexes onto the matrix, leading to an artifactual transcription-dependent enrichment of active genes in the S/MAR fraction (31). Restriction endonucleases EcoRI and NcoI, which do not cleave inside the sequence intervals (10.4 to 15.8 kb for the 17.2-kb locus and the crossover segments for the MHC hot spot loci) being tested for matrix attachment, were used instead of a general endonuclease like DNase I, since we were testing theoretical predictions of the positions of matrix attachment points. The sodium dodecyl sulfate-polyacrylamide gel electrophoresis pattern of the proteins associated with the final 25 mM LIS matrix showed the presence of the cytoskeletal proteins around 60 kDa, which exactly matched with the pattern reported in the literature (47) (data not shown), confirming the authenticity of the matrix preparation being analyzed in the present investigation. In the analysis, we also included a known MAR, Ig kappa MAR, as a positive control. The percentage of matrix (4.04% ± 0.03%) and loop (95.25% ± 0.125%) DNA fractions isolated added up to the amount of total input DNA, showing there were no losses during the DNA isolation procedure. Figure 11A to F shows the amplification pattern seen in the loop, matrix, and total genomic DNA after 25 PCR cycles with primer pairs corresponding to the 17.2-kb domain C (13,500 to 14,000 bp), 17.2-kb fragment domain E (360 to 800 bp), crossover regions for the Ea, Pb, and Psmb9 hot spots, and Ig kappa MAR, respectively. Linearity of the PCR product formation using the same primer pairs is shown graphically with respect to the PCR cycle number. All PCR experiments were carried out in triplicate. The ratio of the sum of the PCR product intensities for the matrix plus loop fractions with respect to the intensity from total genomic DNA for all the primer pairs used shown in Table 4 is around 1 ± 0.03, showing equal efficiency of PCR amplification in matrix and loop fractions and total genomic DNA. R values were set at greater than 0.7 for strong S/MARs, between 0.7 to 0.3 for weak S/MARs, and less than 0.3 for non-S/MARs (9). The R values for the DNA fragments tested are shown in Table 4 and indicate the Ea and 17.2-kb crossover regions and Ig kappa MAR to be strong S/MARs while Pb and Psmb9 crossover regions are weak MARs. The locus upstream and adjacent to the noncoding RNA (domain E) is loop associated. The matrix association potential for these loci is in the following order: Ea > 17.2-kb domain C = Ig kappa MAR > Pb > Psmb9 > 17.2-kb domain E.
FIG. 11.
Experimental detection of predicted MAR regions in matrix DNA preparations from mouse testis. Panels A to F show amplification of PCR fragments corresponding to the predicted MAR regions in the loop (L), matrix (M), and total genomic (T) DNA for (A) 17.2-kb locus domain C, (B) 17.2-kb locus domain E, (C) the Ea hot spot, (D) the Pb hot spot, (E) the Psmb9 hot spot, and (F) Ig kappa MAR. The amount of PCR product formed as a function of the number of PCR cycles (20 to 28), by using 150 ng of mouse testis genomic DNA as template, is also shown for each of the panels.
TABLE 4.
MAR characteristics of DNA fragments tested by PCR-based assay
| Sequence tested | Amplification efficiency | R value | MAR characteristic |
|---|---|---|---|
| 17.2-kb domain C | 0.97 ± 0.000003 | 0.77 ± 0.00023 | Strong MAR |
| Ea hot spot | 0.99 ± 0.0019 | 0.78 ± 0.00093 | Strong MAR |
| Pb hot spot | 0.97 ± 0.00063 | 0.64 ± 0.00090 | Weak MAR |
| Lmp hot spot | 0.97 ± 0.00023 | 0.45 ± 0.00023 | Weak MAR |
| Ig kappa MAR | 1.04 ± 0.0022 | 0.77 ± 0.0021 | Strong MAR |
| 17.2-kb domain E | 0.98 ± 0.0016 | 0.29 ± 0.0013 | Non-MAR |
DISCUSSION
The present study has defined a new recombination hot spot outside the mouse MHC, in a 17.2-kb mouse genomic fragment encompassing the mouse homolog of a 1.3-kb pachytene repair site isolated from rat spermatocytes (38). A summary of all the features of this locus is depicted in Fig. 12. Four distinct domains can clearly be seen to be organized in a sequential manner in the 17.2-kb fragment: a transcribed domain, followed by a pachytene repair site domain which continues to a domain having the crossover breakpoint that overlaps with the MAR domain. Since recombination hot spots quickly destroy LD across small physical regions, knowledge about the location and strength of such hot spots could be valuable for genetic association studies for analyzing complex traits. This is quite important considering that mice offer a large number of advantages, like the availability of mouse models for most common complex human traits, inbred lines that maximize the range of LD, and the ability to increase the power of such analysis by maintaining them in controlled environments that minimize confounding factors (33). In addition, the availability of recombinant inbred strains helps in the dissection of multigenic traits into a series of single traits that can be analyzed separately, and outbred populations of mice mimic isolated human populations with a strong founder effect (51) which reduces genetic heterogeneity, thereby significantly increasing the genotypic relative risk. Identifying meiotic breakpoints in the mouse genome is thus important and unlike in the human genome, where many recombination hot spots in diverse regions of the genome have been identified, only very few have been identified in mice, all of which are in the mouse MHC complex.
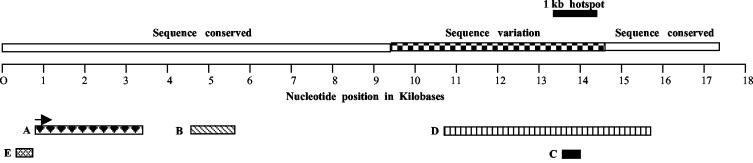
FIG. 12.
Summary of the important domains in the 17.2-kb fragment. The solid box shows the position of the 1-kb region sequenced from all seven strains of mice of the species M. musculus. A larger region from 9.5 to 14.5 kb, within which most of the sequence variation is localized, is shown by a checkered box. A, B, and C indicate the positions of the transcribed region (arrow shows orientation of the transcript), the 1.3-kb meiotic repair site, and the region showing a recombination breakpoint, respectively. D indicates the experimentally determined S/MAR domain flanked by EcoRI and NcoI sites. E indicates the loop-associated region upstream and adjacent to the noncoding RNA.
All of the four (Ea, Eb, Psmb9, and Pb) well-characterized mouse MHC hot spots have been identified using genetic crosses, with sperm typing being used to provide more insights into the molecular processes operating inside them for the Psmb9 and Eb hot spot (14, 56). The presence of meiotic recombination activity in these hot spots is dependent on the MHC haplotype (46). The Ea hot spot is observed in genetic crosses involving the p haplotype, while the Eb hotspot is active in the b, d, and k haplotypes (22). Similarly, the Psmb9 and Pb hot spots show maximal recombination activity in the cas3 and wm7 and the cas4 and wm7 haplotypes, respectively (18). We have used a different population genetics-based approach to show the presence of a recombination hot spot close to the previously determined meiotic repair site. Our estimate of the recombination intensity for this locus is 10- to 100-fold higher than the genome average, and this indicates the locus to be a strong recombination hot spot comparable to the Eb hot spot, which has a recombination intensity 40-fold over the genome average (7). By following a population genetics-based approach, we have also been able to estimate the recombination parameter (R) for this locus to be 0.0026/bp or 2.5/kb. While estimates of R are known for some of the human hot spots (1, 55), it has not been determined for any of the mouse recombination hot spots. We note from the present study that the recombination-gene conversion site is around 10 kb downstream of the 1.3-kb region which we had identified earlier as a meiotic repair site (38). We believe that the 1.3-kb sequence which is rich in recombination potentiating sequence motifs (38) probably acts to stimulate recombination activity in the surrounding genomic regions.
Very little is known about gene conversion in mammals, despite being an important mechanism in breaking down allelic associations. There are few current experimental estimates of conversion tract lengths in organisms other than yeast. In Drosophila spp., for example, estimates are almost exclusively restricted to the rosy locus, where it was estimated to be around 352 bp by crossing strains with very close markers (15). Studies on the human recombination hot spot DNA2 by sperm PCR have provided evidence for a localized width of recombination hot spots of 1 to 2 kb with crossover asymmetry accompanied by biased gene conversion (21), and similar studies by sperm PCR for the mouse Psmb9 hot spot have shown conversion tracts ranging from 315 to 1,018 bp, with an average length of 480 bp (14). We did not observe tract lengths longer than 627 bp, most likely because recombination would quickly reshuffle the polymorphisms, except for those that are very close together. This is particularly true while inferring conversion tract lengths from population-based DNA sequence data, and previous studies based on DNA sequence data have shown short tract lengths of 10 and 16 bp for the Est-5β and Est-5 C genes (23) and an average length of 51 bp for the rp49 locus (40) in Drosophila spp., because of conversion events getting broken up by subsequent recombination events. Hence, calculations of conversion tract length from population-based sequence data are likely to be underestimates for regions with a high density of crossover or conversion events. Interestingly, two HVMS sequences positioned at 13,466 and 13,576 bp and an LTR sequence at 13,748 bp are in close proximity to the location of the conversion tracts in the 1-kb hot spot locus between 13,595 and 13,971 bp. HVMS and LTR sequences are known to be potentiators of meiotic recombination in in vitro assays (11, 46, 54).
A comparison of this recombination hot spot with the cluster of four recombination hot spots in the mouse MHC complex reveals several parallel features. All of the four recombination hot spots in the MHC region contain an LTR element, a middle repetitive element of the mouse transcript family, and tandem repeats of a tetrameric sequence resembling HVMS. The concentration of the crossover points is either in the 3′ end of the genes as seen in the Psmb9 hot spot or in the intron of the genes as seen in the Eb, Ea, and Pb hot spots, with none of the breakpoints located at the 5′ region of the genes as in S. cerevisiae. Our localization of crossover activity follows a similar paradigm, with the crossover regions being present close to LTR and HVMS sequences. Further, they lie at the 3′ end of the transcribed region. Unlike the Pb and Psmb9 hot spots, where transcriptional activity is not seen in meiotic cells (18), we have seen that the 2.4-kb noncoding RNA is present in the testis and flanks the MAR domain in the 17.2-kb fragment.
The noncoding RNAs have been implicated in several functions such as transcriptional regulation, chromosomal replication, and protein translocation (10, 48). The mouse genome has also been reported to have an abundance of noncoding RNAs, most of which are unspliced, single-exon RNA polymerase II-mediated transcripts (52). At this stage we do not know what function this RNA serves, but the presence of transcriptional activity close to a recombination hot spot region is in agreement with a role of open chromatin domains in recombinational activity and is suggestive of an α hot spot. There is also a striking correlation between the presence of DUEs at 11.2 kb, a S/MAR domain between 10.4 and 15.8 kb, and recombinational activity nearby. The presence of such an intrinsically open chromatin structure which would be prone to double-strand breaks shows that it can also function as a β hot spot. The recombination hot spot in the 17.2-kb locus thus has properties of both α and β hot spots. However, the moderate percentage (38.57%) of G+C content rules out a γ hot spot-like nature. While yeast and mammalian recombination hot spots exhibit a global relationship of GC-rich domains and elevated recombination rates (36), the lack of such correlation seen for the 17.2-kb locus is probably due to a high resolution analysis.
Another important new feature of mouse recombination hot spots that has emerged from the present study is the association seen between S/MAR elements and the crossover sites. Both in silico analysis and experimental studies using LIS-extracted matrix DNA have provided evidence for such an in vivo association. LTR elements are also known to integrate preferentially at S/MARs and are the most abundant class of repeats in the 17.2-kb fragment. Thus, the crossover site which has been fine mapped to between 13.3 and 14.3 kb is contained in a S/MAR domain which is characterized by the ease of DNA unwinding, which could be required for different aspects of the meiotic pairing process. The association of meiotic activity with S/MAR elements was also found to be true for the Ea, Pb, and Psmb9 mouse MHC hot spots. These observations suggest that a subset of S/MARs associated with the meiotic chromosome cores are converted into crossover regions, for at least some recombination hot spots, which is a distinct possibility considering that they are capable of undergoing stress-induced duplex destabilization. In conclusion, our studies define a new recombination hot spot in proximity to a novel noncoding RNA transcript. These studies also provide the first estimate for the R value for any hot spot locus in the mouse and also the probable length of gene conversion tracts for a hot spot outside the MHC cluster.
Supplementary Material
Acknowledgments
K. T. Nishant is supported by a fellowship from the Council of Scientific and Industrial Research, New Delhi, India.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org.
REFERENCES
- 1.Ardlie, K., S. N. Liu-Cordero, M. A. Eberle, M. Daly, J. Barrett, E. Winchester, E. S. Lander, and L. Kruglyak. 2001. Lower than expected linkage disequilibrium between tightly linked markers in humans suggests a role for gene conversion. Am. J. Hum. Genet. 69:582-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnheim, N., P. Calabrese, and M. Nordborg. 2003. Hot and cold spots of recombination in the human genome: the reason we should find them and how this can be achieved. Am. J. Hum. Genet. 73:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benham, C., T. Kohwi-Shigematsu, and J. Bode. 1997. Stress-induced duplex DNA destabilization in scaffold/matrix attachment regions. J. Mol. Biol. 274:181-196. [DOI] [PubMed] [Google Scholar]
- 4.Blake, J. A., J. T. Eppig, J. E. Richardson, M. T. Davisson, et al. 2000. The Mouse Genome Database (MGD): expanding genetic and genomic resources for the laboratory mouse. Nucleic Acids Res. 28:108-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bode, J., C. Benham, E. Ernst, A. Knopp, R. Marshaled, R. Strick, and P. Strissel. 2000. Fatal connections: when DNA ends meet on the nuclear matrix. J. Cell. Biochem. 35:3-22. [DOI] [PubMed] [Google Scholar]
- 6.Cockerill, P. N., and W. T. Garrard. 1986. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell 44:273-282. [DOI] [PubMed] [Google Scholar]
- 7.de Massy, B. 2003. Distribution of meiotic recombination sites. Trends Genet. 19:514-522. [DOI] [PubMed] [Google Scholar]
- 8.Dobbs, D. L., W. L. Shaiu, and R. M. Benbow. 1994. Modular sequence elements associated with origin regions in eukaryotic chromosomal DNA. Nucleic Acids Res. 22:2479-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donev, R. M. 2000. The type of DNA attachment sites recovered from nuclear matrix depends on isolation procedure used. Mol. Cell. Biochem. 214:103-110. [DOI] [PubMed] [Google Scholar]
- 10.Eddy, S. R. 2002. Computational genomics of noncoding RNA genes. Cell 109:137-140. [DOI] [PubMed] [Google Scholar]
- 11.Edelmann, W., B. Kroger, M. Goller, and I. Horak. 1989. A recombination hotspot in the LTR of a mouse retrotransposon identified in an in vitro system. Cell 57:937-946. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Diaz, M., O. Dominguez, L. A. Lopez-Fernandez, L. T. de Lera, M. L. Saniger, J. F. Ruiz, M. Parraga, M. J. Garcia-Ortiz, T. Kirchhoff, J. del Mazo, A. Bernad, and L. Blanco. 2000. DNA polymerase lambda (Pol lambda), a novel eukaryotic DNA polymerase with a potential role in meiosis. J. Mol. Biol. 301:851-867. [DOI] [PubMed] [Google Scholar]
- 13.Gerton, J. L., J. DeRisi, R. Shroff, M. Lichten, P. O. Brown, and T. D. Petes. 2000. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:11383-11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guillon, H., and B. de Massy. 2002. An initiation site for meiotic crossing-over and gene conversion in the mouse. Nat. Genet. 32:296-299. [DOI] [PubMed] [Google Scholar]
- 15.Hilliker, A. J., G. Harauz, A. G. Reaume, M. Gray, S. H. Clark, and A. Chovnick. 1994. Meiotic gene conversion tract length distribution within the rosy locus of Drosophila melanogaster. Genetics 137:1019-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hudson, R. R. 1987. Estimating the recombination parameter of a finite population model without selection. Genet. Res. 50:245-250. [DOI] [PubMed] [Google Scholar]
- 17.Hudson, R. R., and N. L. Kaplan. 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111:147-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isobe, T., M. Yoshino, K. Mizuno, K. F. Lindahl, T. Koide, S. Gaudieri, T. Gojobori, and T. Shiroishi. 2002. Molecular characterization of the Pb recombination hotspot in the mouse major histocompatibility complex class II region. Genomics 80:229-235. [DOI] [PubMed] [Google Scholar]
- 19.Jeffreys, A. J., A. Ritchie, and R. Neumann. 2000. High resolution analysis of haplotype diversity and meiotic crossover in the human TAP2 recombination hotspot. Hum. Mol. Genet. 9:725-733. [DOI] [PubMed] [Google Scholar]
- 20.Jeffreys, A. J., J. Murray, and R. Neumann. 1998. High resolution mapping of crossovers in human sperm defines a minisatellite associated hotspot. Mol. Cell 2:267-273. [DOI] [PubMed] [Google Scholar]
- 21.Jeffreys, A. J., L. Kauppi, and R. Neumann. 2001. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat. Genet. 29:217-222. [DOI] [PubMed] [Google Scholar]
- 22.Khambata, S., J. Mody, A. Modzelewski, D. Heine, and H. C. Passmore. 1996. Ea recombination hotspot in the mouse major histocompatibility complex maps to the fourth intron of the Ea gene. Genome Res. 6:195-201. [DOI] [PubMed] [Google Scholar]
- 23.King, L. M. 1998. The role of gene conversion in determining sequence variation and divergence in the Est-5 gene family in Drosophila pseudoobscura. Genetics 148:305-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleckner, N. 1996. Meiosis: how could it work? Proc. Natl. Acad. Sci. USA 93:8167-8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kramer, J. A., and S. A. Krawetz. 1997. PCR-based assay to determine nuclear matrix association. BioTechniques 22:826-828. [DOI] [PubMed] [Google Scholar]
- 26.Labarca, C., and K. Paigen. 1980. A simple, rapid, and sensitive DNA assay procedure. Anal. Biochem. 102:344-352. [DOI] [PubMed] [Google Scholar]
- 27.Lewontin, R. C. 1964. The interaction of selection and linkage. I. General considerations; heterotic models. Genetics 49:49-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lower, R., J. Lower, and R. Kurth. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. USA 93:5177-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majewski, J., and J. Ott. 2000. GT repeats are associated with recombination on human chromosome 22. Genome Res. 10:1108-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maya-Mendoza, A., and A. Aranda-Anzaldo. 2003. Positional mapping of specific DNA sequences relative to the nuclear substructure by direct polymerase chain reaction on nuclear matrix-bound templates. Anal. Biochem. 313:196-207. [DOI] [PubMed] [Google Scholar]
- 31.Mirkovitch, J., M. E. Mirault, and U. K. Laemmli. 1984. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell 39:223-232. [DOI] [PubMed] [Google Scholar]
- 32.Mizuno, K., T. Koide, T. Sagai, K. Moriwaki, and T. Shiroishi. 1996. Molecular analysis of a recombination hotspot adjacent to Lmp2 gene in the mouse MHC: fine location and chromatin structure. Mamm. Genome 7:490-496. [DOI] [PubMed] [Google Scholar]
- 33.Moore, K. J., and D. L. Nagle. 2000. Complex trait analysis in the mouse: the strengths, the limitations and the promise yet to come. Annu. Rev. Genet. 34:653-686. [DOI] [PubMed] [Google Scholar]
- 34.Nicolas, A. 1998. Relationship between transcription and initiation of meiotic recombination: toward chromatin accessibility. Proc. Natl. Acad. Sci. USA 95:87-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orlando, P., R. Geremia, C. Frusciante, B. Tedeschi, and P. Grippo. 1988. DNA repair synthesis in mouse spermatogenesis involves DNA polymerase beta activity. Cell Differ. 23:221-230. [DOI] [PubMed] [Google Scholar]
- 36.Petes, T. D. 2001. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2:360-369. [DOI] [PubMed] [Google Scholar]
- 37.Plug, A. W., C. A. Clairmont, E. Sapi, T. Ashley, and J. B. Sweasy. 1997. Evidence for a role for DNA polymerase beta in mammalian meiosis. Proc. Natl. Acad. Sci. USA 94:1327-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramachandra, L., and M. R. Rao. 1994. Identification and sequence characterization of a 1.3 kb EcoRI repeat fragment that harbors a DNA repair site of rat pachytene spermatocytes. Chromosoma 103:486-501. [DOI] [PubMed] [Google Scholar]
- 39.Reiter, L. T., T. Murakami, T. Koeuth, L. Pentao, D. M. Munzy, R. A. Gibbs, and J. R. Lupski. 1996. A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat. Genet. 12:288-297. [DOI] [PubMed] [Google Scholar]
- 40.Rozas, J., and M. Aguade. 1994. Gene conversion is involved in the transfer of genetic information between naturally occurring inversions of Drosophila. Proc. Natl. Acad. Sci. USA 91:11517-11521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozas, J., and R. Rozas. 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174-175. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., and D. W. Russel. 2000. Bacteriophage λ vectors, p. 6.4-6.11. In Molecular cloning, vol 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Sassaman, D. M., B. A. Dombroski, J. V. Moran, M. L. Kimberland, T. P. Naas, R. J. DeBerardinis, A. Gabriel, G. D. Swergold, and H. H. Kazazian, Jr. 1997. Many human L1 elements are capable of retrotransposition. Nat. Genet. 16:37-43. [DOI] [PubMed] [Google Scholar]
- 44.Sawyer, S. A. 1999. GENECONV: a computer package for the statistical detection of gene conversion. Department of Mathematics, Washington University, St. Louis, Mo.
- 45.Schmid, C. W. 1998. Does SINE evolution preclude Alu function? Nucleic Acids Res. 26:4541-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shiroishi, T., T. Sagai, and K. Moriwaki. 1993. Hotspots of meiotic recombination in the mouse major histocompatibility complex. Genetica 88:187-196. [DOI] [PubMed] [Google Scholar]
- 47.Smith, H. C., R. L. Ochs, D. Lin, and A. C. Chinault. 1987. Ultrastructural and biochemical comparisons of nuclear matrices prepared by high salt or LIS extraction. Mol. Cell. Biochem. 77:49-61. [DOI] [PubMed] [Google Scholar]
- 48.Storz, G. 2002. An expanding universe of noncoding RNAs. Science 296:1260-1263. [DOI] [PubMed] [Google Scholar]
- 49.Stumpf, M. P., and G. A. McVean. 2003. Estimating recombination rates from population genetic data. Nat. Rev. Genet. 4:959-968. [DOI] [PubMed] [Google Scholar]
- 50.Tajima, F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Talbot, C. J., A. Nicod, S. S. Cherny, D. W. Fulker, A. C. Collins, and J. Flint. 1999. High-resolution mapping of quantitative trait loci in outbred mice. Nat. Genet. 21:305-308. [DOI] [PubMed] [Google Scholar]
- 52.The Fantom Consortium and the RIKEN Genome Exploration Research Group. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full length cDNAs. Nature 420:563-573. [DOI] [PubMed] [Google Scholar]
- 53.Tracy, R. B., J. K. Baumohl, and S. C. Kowalczykowski. 1997. The preference for GT-rich DNA by the yeast Rad51 protein defines a set of universal pairing sequences. Genes Dev. 11:3423-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wahls, W. P., L. J. Wallace, and P. D. Moore. 1990. Hypervariable minisatellite DNA is a hotspot for homologous recombination in human cells. Cell 60:95-103. [DOI] [PubMed] [Google Scholar]
- 55.Wall, J. D., L. A. Frisse, R. R. Hudson, and A. Di Rienzo. 2003. Comparative linkage-disequilibrium analysis of the beta-globin hotspot in primates. Am. J. Hum. Genet. 73:1330-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yauk, C. L., P. R. Bois, and A. J. Jeffreys. 2003. High-resolution sperm typing of meiotic recombination in the mouse MHC Eβ gene. EMBO J. 22:1389-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zimmerer, E. J., and H. C. Passmore. 1991. Structural and genetic properties of the Eb recombination hotspot in the mouse. Immunogenetics 33:132-140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.