Abstract
Serum response factor (SRF) is at the confluence of multiple signaling pathways controlling the transcription of immediate-early response genes and muscle-specific genes. There are active SRF target sequences in more than 50 genes expressed in the three muscle lineages including normal and diseased hearts. However, the role of SRF in heart formation has not been addressed in vivo thus far due to the early requirement of SRF for mesoderm formation. We have generated a conditional mutant of SRF by using Cre-LoxP strategy that will be extremely useful to study the role of SRF in embryonic and postnatal cardiac functions, as well as in other tissues. This report shows that heart-specific deletion of SRF in the embryo by using a new βMHC-Cre transgenic mouse line results in lethal cardiac defects between embryonic day 10.5 (E10.5) and E13.5, as evidenced by abnormally thin myocardium, dilated cardiac chambers, poor trabeculation, and a disorganized interventricular septum. At E9.5, we found a marked reduction in the expression of essential regulators of heart development, including Nkx2.5, GATA4, myocardin, and the SRF target gene c-fos prior to overt maldevelopment. We conclude that SRF is crucial for cardiac differentiation and maturation, acting as a global regulator of multiple developmental genes.
The amount of information on transcription factors that are essential for heart morphogenesis, such as those encoded by homeobox genes (NK family) (16, 21), GATA genes (13, 24, 49), and MADS (for MCM1, agamous, deficiens, serum response factor [SRF]) box genes such as MEF-2 (34, 40) has increased greatly in recent years. The SRF is also a member of the MADS box transcription factor family that has a highly conserved DNA-binding/dimerization domain which binds the core sequence of SRE/CArG boxes (CC[A/T]6GG) as a homodimer (44). The SRF-binding sites are cis-active elements involved in both (i) response of immediate-early genes, e.g., c-fos, egr-1, and the Srf gene itself, to serum and growth factors (45) and (ii) muscle-specific transcriptional activation of a large number of smooth, skeletal, and heart muscle-specific genes (29). Thus, SRF takes part in the control of several cardiac genes, including those encoding α-actin, α-myosin heavy chain (αMHC) and βMHC, and the atrial natriuretic factor (ANF) (8, 19, 30, 31). Consistent with its role in regulating muscle gene expression, the SRF protein is most abundant in embryonic heart, skeletal, and smooth muscle tissues. Strong SRF expression was observed in the myocardium of both the developing chicken and the mouse heart (4, 11). SRF also accumulates during the terminal differentiation of these tissues (5).
The involvement of SRF as an important regulator in both cell growth and muscle differentiation can be explained by its ability to recruit different transcription factors or accessory cofactors. SRF has been shown to associate with the key cardiac Nkx2.5 homeobox transcription factor (8), the myogenic TEF-1 factor (15), and cardiovascular tissue-restricted GATA factors (4, 31) in cardiomyocytes, where it synergistically activates various cardiac promoters. The recent discovery of the cardiac and smooth muscle-specific SRF cofactor, myocardin, and of myocardin-related transcription factors could also explain how SRF selectively activates transcription in cardiomyocytes (17, 46, 47). Several recent findings from transgenic studies and human disorders, indicate the importance of SRF in the myocardium. Heart-specific overexpression of SRF in transgenic mice led to the development of cardiac hypertrophy and cardiomyopathy (50), whereas overexpression of a mutant dominant-negative form of SRF led to a severe dilated cardiomyopathy (51). An increased expression of an alternatively spliced dominant-negative isoform of SRF was also observed in failing human hearts (12). Altogether, these data suggest that SRF could be a critical factor for both cardiomyogenesis and the maintenance of cardiac structure and function in the adult. Gene targeting in mice has been particularly helpful in deciphering the genetic networks underlying cardiac development (41). Classical disruption of both Srf alleles in mice leads to a very early embryonic death, with a defect in mesoderm formation (3), thus precluding any analysis of the role of SRF at later stages. To decipher in more detail the role of SRF in mouse development and in adulthood, we adopted a conditional gene inactivation scheme for SRF by using the Cre/LoxP system and have thus created a transgenic mice line named Srf-flex2neo carrying Srf floxed exon 2 alleles. In order to analyze SRF function during cardiac development, we have also generated and further characterized one transgenic line in which Cre recombinase expression is under the control of the βMHC promoter. Here we show that mice with early depleted cardiac SRF expression die between embryonic day 10.5 (E10.5) and E13.5 of development, display severe cardiac defects, and have impaired expression of critical cardiac transcription factors, thus demonstrating that SRF is one of the essential transcription factors for cardiomyogenesis.
MATERIALS AND METHODS
Construction of the targeting vector and generation of floxed SRF mice.
Genomic fragments encompassing 14 kb of the Srf gene were amplified by PCR from a mouse strain SV129 and completely sequenced. A 10-kb DNA fragment containing the 1,800-bp promoter and exons 1 to 7 was used to construct the targeting vector. A 44-bp fragment containing a 34-bp LoxP site and linker sequence was inserted into the AccI and NsiI sites of the middle part of intron 1. The PGK-Neo expression cassette flanked by LoxP sites in the 5′ and 3′ of the cassette was introduced into the ScaI site of intron 2. A total of 30 μg of the SalI-linearized targeting vector was electroporated into SV129-derived embryonic stem (ES) cells, and ES clones were selected by growing cells in the presence of G418 for 7 days. A total of 900 drug-resistant colonies were selected and analyzed by Southern blotting for homologous recombination by using appropriate 5′ and 3′ probes. The three ES clones that contained the correctly targeted event at the Srf locus were injected into C57BL/6 blastocysts to obtain chimeric mice. Male chimeras were bred with female C57BL/6 mice to obtain heterozygous floxed SRF mice Srf-flex2neo (Sf/+). Crosses between heterozygous animals gave rise to homozygous (Sf/Sf) animals. Mice were genotyped by PCR with primers SF1 (5′-CTGTAAGGGATGGAAGCAGA-3′) and SF2 (5′-TAAGGACAGTGAGGTCCCTA-3′), which give rise to the 448- and 492-bp fragments for wild-type and floxed Srf alleles, respectively. PCR conditions were 94°C for 2 min, followed by 25 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min.
Generation of βMHC-Cre transgenic mice.
An nls-Cre (for nuclear localization signal-Cre) fragment was inserted downstream of a 5.6-kb DNA fragment of the mouse βMHC gene promoter. The linearized βMHC-Cre fragment was injected into (C57BL/6XDBA2) eggs. Injected eggs were implanted in DBA2 foster mothers. The DNA from tail biopsies of F0 mice was genotyped by PCR. The primers used to detect the Cre transgene were CreS (5′-CTTCTGTCCGTTTGCCGGTCGTGG-3′) and CreR (5′-TTTTGCACGTTCACCGGCATCAACG-3′). PCR cycles were as follows: 94°C for 2 min, 25 cycles of 94°C for 1 min, 62°C for 1 min, and 72°C for 1 min. Three βMHC-Cre stable lines were produced and characterized by crosses with CAG-CAT-Z reporter mice (2). The line with the highest recombination efficiency in the heart was selected, further back-crossed on a C57BL/6 background, and used for our experiments. All experiments with animals were conducted in accordance with the European guideline for the care and use of laboratory animals.
Targeting inactivation of the Srf gene in the heart.
βMHC-Cre mice were bred with SRF-floxed homozygous mice (Sf/Sf) to obtain double transgenic mice (βMHC-Cre:Sf/+) in order to inactivate Srf gene in the embryonic heart. Further crosses between βMHC-Cre:Sf/+ mice and Sf/Sf mice were used to obtain βMHC-Cre:Sf/Sf embryos. DNA prepared from the yolk sacs of embryos was used for genotyping by PCR, as described previously. Cre recombinase-mediated excision of the floxed Srf allele was detected by PCR on DNA prepared from pooled hearts from genotyped embryos, by using three primers: SF1, SF2, and SF3. SF1 and SF2 allowed detection of wild-type and floxed Srf alleles. SF1 and SF3 (5′-TTCGGAACTGCCGGGCACTAAA-3′) allowed amplification of a 310-bp DNA fragment when floxed Srf alleles had been recombined by the Cre recombinase.
Reverse transcription-PCR (RT-PCR) assay.
Total RNA was extracted from four pooled hearts previously dissected from genotyped embryos at E9.5 under the microscope, with TRIzol (Invitrogen). RNA extracted from the upper body, which includes the head, cervical somites, upper aortae, pharyngeal arches, and limb buds, was used as a control. The first-strand cDNA synthesis was performed with 1 μg of total RNA in a final volume of 20 μl according to the manufacturer's instructions (Roche). Then, 1 μl of this reaction was used for the PCRs. PCR cycles were as follows: 94°C for 2 min, followed by 25 to 30 cycles of 94°C for 1 min, 62°C for 1 min, and 72°C for 1 min. PCR products were gel electrophoresed on 1.5% agarose gel and stained with ethidium bromide. The following primers were used: skeletal actin, 5′-CTGAGCGCAAGTACTCAGTGTGGA-3′ and 5′TTCCAAAAACAGGCGCCGGCTGCA-3′; cardiac actin, 5′-GAGACTCTCTTCCAGCCCTCTTTC-3′ and 5′-TCAGAAGCACTTGCGGCGGACAAT-3′; αMHC, 5′-ACCGTCTGGACGAGGCAGAGCAGA-3′ and 5′-CGTCGTGCATCTTCTTGGCACCAA-3′; TEF1, 5′-CTGCAATATCCAAGACGACGCCGG-3′/5′-TGTAGATATGGTGCTGTGCTCCGT-3′; GATA4, 5′-TCCCAGGCCTCTTGCAATGCGGAA-3′ and 5′-GCGGTGATTATGTCCCCATGACTG-3′; myocardin, 5′-AGGCCAGATGGCCTTCGGTCACTA-3′/5′-CCACTGCTGTAAGTGGAGATCCAT-3′; ANF, 5′-GCCGCACTTAGCTCCCTCCCCGAG-3′ and 5′-GTACCGGAAGCTGTTGCAGCCTAG-3′; c-fos, 5′-ACCTCCCGCTCTGTGCCAGATGTG-3′ and 5′-TTGCTGCTGCTGCCCTTTCGGTGG-3′; desmin, 5′-GACTCCCTGATGAGGCAGATGAGG-3′ and 5′-CCTCGCTGACAACCTCTCCATCCC-3′; βMHC, 5′-CAGCACCGTCTGGACGAGGCAGAG-3′ and 5′-ATTCAGGCCCTTGGCACCAATGTC-3′; NKX2.5, 5′-GAGCCTACGGTGACCCTGACCCAg-3′ and 5′-TGACCTGCGTGGACGTGAGCTTCA-3′; GAPDH, 5′-AGTCCATGCCATCACTGCCACCCA-3′ and 5′-TCCACCACCCTGTTGCTGTAGCCG-3′; and SRF, 5′-CTCCGCCCCGCTCAGACCCCACCACAGA-3′ and 5′-CAGGTAGTTGGTGATGGGGAAGGA-3′.
X-Gal staining.
Embryos were harvested at different stages and fixed in 1% formaldehyde and 0.2% glutaraldehyde for 30 min at 4°C, rinsed twice in 1× phosphate-buffered saline, and incubated overnight at 37°C in the X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining solution as described previously (28).
Histological analysis.
All embryos were staged, harvested, fixed in 4% paraformaldehyde overnight and embedded in paraffin. Embryos were staged by considering the presence of a vaginal plug as day 0.5 after conception. Slides were stained with hematoxylin and eosin. Cell size was quantified by using the ImageJ software on six sections (n = 6; P < 0.05).
Immunohistochemistry.
Cell proliferation rate was determined by bromodeoxyuridine (BrdU) incorporation. E9.5 and E10.5 embryos were labeled for 2 h with BrdU via intraperitoneal injection of the pregnant female with BrdU labeling solution (100 μg/g [body weight]). Embryos were dissected in phosphate-buffered saline and embedded in paraffin. BrdU-positive cells were detected by immunohistochemistry using anti-BrdU antibody (Roche), followed by horseradish peroxidase (HRP)-conjugated secondary antibody. Sections were counterstained with hematoxylin. The proliferative index of cardiomyocytes was determined as the number of BrdU-positive nuclei reported to the total number of nuclei within a section. Proliferative index of each developmental stage was determined for three embryos per genotype. To determine the mitotic index of cardiomyocytes in E10.5 embryos, 10-μm frozen sections were incubated with an anti-phospho-histone H3 (ser10) polyclonal antibody (Cell Signaling Technologies), followed by TRITC (tetramethyl rhodamine isothiocyanate)-conjugated secondary antibody (Dako). Sections were counterstained with DAPI (4′,6′-diamidino-2-phenylindole). The mitotic index was defined as the number of phosphorylated histone H3-labeled nuclei divided by the total number of nuclei. Apoptotic cells were detected with an anti-cleaved caspase-3 polyclonal antibody (R&D Systems) on frozen sections of E10.5 embryos or paraffin sections of E11.5 embryos, followed by incubation with a TRITC- or HRP-conjugated secondary antibody (Dako).
Southern blot analysis.
A total of 15 μg of DNA from ES cells was digested with EcoRV and NdeI at 37°C overnight. Samples were separated on 0.8% agarose gel overnight and transferred to a HybondN+ membrane (Amersham). The transferred DNA was hybridized at 65°C overnight with either a 5′ or a 3′ 32P-labeled Srf gene fragment. Both probes revealed a 20-kb fragment for the wild-type Srf gene, the 5′ probe revealed an 8-kb fragment for the floxed Srf allele, and the 3′ probe detected a 12-kb fragment.
Western blot analysis.
Tissues from wild-type and homozygous floxed SRF mice were homogenized in 1× Laemmli buffer and sonicated. Equal amount of proteins was loaded on a 12% acrylamide gel, electrophoresed, and transferred to a nitrocellulose membrane. Immunoblotting was performed with the anti-SRF antibody (Santa Cruz) diluted 1/200. Enhanced chemiluminescence revealed two SRF bands by using HRP-conjugated secondary anti-rabbit antibody (Dako).
RESULTS
Homozygous SRF floxed mice are viable and healthy.
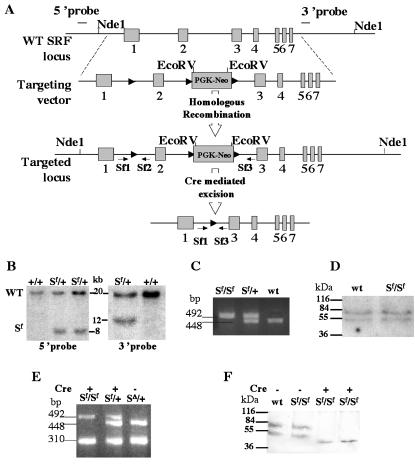
To decipher the biological roles of SRF during mouse development or later in adulthood, we generated mice harboring floxed Srf alleles. A 10-kb DNA fragment of the Srf gene was used to construct the targeting vector. The targeting vector was designed to ensure that the floxed exon 2 of SRF and the floxed PGK-Neo cassette will be inserted into the Srf locus without modifying Srf gene structure and expression after homologous recombination (Fig. 1A and D). This allele was named Srf-flex2neo for floxed exon 2 (abbreviated Sf). The configuration of the LoxP sites was chosen so that the critical exon 2 that encodes most of the MADS box would be excised following Cre delivery. Excision of exon 2 maintains the reading frame of the mRNA between exons 1 and 3. Therefore, an SRF protein truncated in the MADS box domain could theoretically be synthesized from the excised Srf allele. However, deletion of the MADS box in SRF has been shown to result in a protein unable to bind DNA and had no overt negative transdominant effect (4, 36). A total of 900 ES cell clones were analyzed by Southern blot and 3 ES clones, in which the expected homologous recombination event occurred, were selected (Fig. 1B). ES cells heterozygous for the floxed Srf allele (Sf/+) were injected into C57BL/6 blastocysts to generate chimeric males, which transmitted the mutant floxed allele through the germ line. F1 mice heterozygous for the floxed Srf allele (Sf/+) were intercrossed to obtain homozygous floxed mice (Sf/Sf). Homozygous (Sf/Sf) and heterozygous (Sf/+) floxed SRF mice were characterized by PCR with primers SF1 and SF2, allowing the amplification of the first LoxP-containing region (Fig. 1C) and generating, respectively, a 492-bp fragment for the floxed Srf allele and a 448-bp fragment for the wild-type Srf allele. Mice homozygous for the floxed Srf allele (Sf/Sf) were viable, healthy, and fertile. In order to generate mice harboring only a floxed exon 2, without floxed PGK-neomycin cassette, Sf/Sf mice were crossed with the E2a-Cre transgenic line (18). This would enable the generation of random recombination events in the LoxP sites of the gametes. Mice positive for both the Cre transgene and the floxed Srf allele were crossed with wild-type C57BL/6 mice, and the excision events were identified by PCR with SF1, SF2, and SF3 primers in 110 newborns. We obtained no partial excision of exon 2 alone or of the PGK-Neo cassette alone. Only excision of both the PGK-Neo cassette and exon 2, was obtained, leading to a deleted Srf allele (SΔ). This deletion was characterized by a 310-bp amplified PCR fragment (Fig. 1A and E). Consequently, since homozygous floxed mice containing both the floxed exon 2 and the floxed PGK-Neo cassette (Sf/Sf) were found to be perfectly healthy, we decided to use them for all subsequent experiments. Indeed, the expression of SRF between wild-type and Sf/Sf mice appeared the same in Western blot analyses (Fig. 1D), and no abnormal phenotype has been detected in the 400 mice obtained thus far. In agreement with Arsenian et al. (3), intercrossing mice heterozygous for the deleted Srf allele (SΔ/+) resulted in SRF-deficient embryos (SΔ/SΔ) that did not develop to term, and no such embryos was detected after E8.5. Mice heterozygous for both the floxed and the deleted alleles, i.e., SΔ/Sf and SΔ/+, developed normally and were fertile.
FIG. 1.
Generation of floxed SRF transgenic mice by homologous recombination. (A) Targeting strategy. Homologous recombination introduced LoxP sites at the two ends of exon 2 and a floxed neomycin resistance into intron 2. Maps of the wild-type Srf locus, the targeting vector, the recombined allele, and the excised allele are shown. Exons are shown as boxes. The position of the 5′ and 3′ probes used for Southern blot analysis in panel B are indicated. Arrows show primer positions for PCR. Arrowheads indicate the LoxP sites. (B) Southern blot analysis. Genomic DNA from ES cells was digested with NdeI and EcoRV. The 5′ and 3′ probes revealed a 20-kb fragment in the wild-type ES cells (+/+), and two bands (20 and 8 kb for 5′ probe and 20 and 12 kb for 3′ probe) in recombined ES cells (Sf/+). (C) Genotyping of offspring by PCR. Primers SF1 and SF2 amplified a 448-bp fragment for wild-type mice and a 492-bp fragment for homozygous floxed SRF (Sf/Sf) mice due to the presence of the LoxP sequence. Both bands were amplified in heterozygous mice (Sf/+). (D) Western blot analysis. Equal amounts of proteins from the hearts of adult Sf/Sf and wild-type (wt) mice were loaded onto the gel, and two bands corresponding to the SRF protein were revealed with anti-SRF antibody (Santa Cruz Biochemicals). (E) Characterization of exon 2 excision. DNA extracted from E9.5 embryonic hearts was amplified by using SF1, SF2, and SF3 oligonucleotides. A 310-bp fragment was detected after the excision of exon 2 in both homozygous (Sf/Sf) and heterozygous (Sf/+), Cre-expressing embryos. A sample containing DNA extracted from mice containing an allele lacking exon 2 of SRF (SΔ/+) was used as a positive control. (F) Absence of normal SRF protein in E10.5 embryo hearts. Western blot analysis revealed two bands of approximately 67 and 54 kDa in homozygous (Sf/Sf) and heterozygous (Sf/+) control embryos. No bands corresponding to normal SRF was detected in Cre-expressing homozygous (Sf/Sf) hearts. A smaller band of ∼46 kDa corresponding to the exon 2 truncated protein was observed in these samples.
Cardiac tissue-restricted expression of the βMHC-Cre transgenic mice.
In order to investigate the role of SRF in early cardiac development, we created and characterized a cardiac muscle-restricted Cre-expressing transgenic line. The Cre gene was placed under the control of the previously in vivo-characterized 5′ upstream regulatory region of the βMHC gene (22). This gene codes for the beta MHC isoform that is characteristic of the embryonic heart. βMHC gene expression occurs as early as E8 throughout the developing heart tube and later becomes restricted to the ventricular cardiomyocytes (27). Three stable transgenic lines (βMHC-Cre) were obtained and characterized by crossing them with the CAG-CAT-Z reporter transgenic line (2). This line produces β-galactosidase (LacZ) in the cells with Cre recombinase activity. The X-Gal blue staining of double transgenic embryos (βMHC-Cre/CAG-CAT-Z) is illustrated in Fig. 2 for one βMHC-Cre line. This line gave the highest staining level for LacZ in the embryonic heart and was selected for further experiments. A dotted blue staining could be observed in the heart as early as E9.25 (Fig. 2A). The staining is increased considerably at E9.75 (Fig. 2E). At E10.5, X-Gal blue staining remained restricted to the cardiac tissue, mainly in the ventricles (Fig. 2B and F), whereas at E13.5 blue staining extended to the atrial compartment (Fig. 2D, G, and H), as well as to some dorsal, limb, and caudal muscles (Fig. 2C). We conclude that this transgenic line can be used to analyze the steps of cardiac maturation and myocardial differntiation that occur after the linear heart tube has formed.
FIG. 2.
Characterization of the βMHC-Cre transgenic line. βMHC-Cre mice were mated with CAG-CAT-Z reporter lines as indicated in the upper panel. LacZ expression was activated upon Cre-mediated excision of the floxed CAT gene in double transgenic embryos. Embryos at different developmental stages were stained in X-Gal solution. (A, B, and C) Whole-mount blue-stained embryos; (E, F, and G) high magnification of the cardiac region; (D and H) histological sections of E13.5 embryos. A dotted blue staining is observed in E9.25 embryos (A); the staining is increased considerably at E9.75 (E). At E10.5, blue staining remains restricted to the cardiac tissue mainly in the ventricles (B and E), whereas at E13.5 the staining extends to the atrial compartment (H), as well as to the dorsal, limb, and caudal muscles (C). At this stage, the trabeculations were most intensely stained (D and H).
Double transgenic βMHC-Cre:Sf/Sf mice die in utero between E10.5 and E13.5.
We produced mice with impaired SRF expression from early embryonic stages to assess the effect of a lack of functional SRF protein on the developing heart. This was achieved by mating homozygous floxed SRF (Sf/Sf) mice with mice that were heterozygous for the floxed Srf allele and transgenic for the βMHC-Cre (βMHC-Cre:Sf/+). None of the 16 offspring genotyped was both homozygous for the floxed Srf allele and transgenic for the βMHC-Cre transgene (βMHC-Cre:Sf/Sf), suggesting that such a genotype results in embryonic death (Table 1). In order to determine at which stage the βMHC-Cre:Sf/Sf embryos die, we further analyzed both the genotype and the macroscopic aspect of embryos from development stage E9.5 to stage E13.5. Genotyping of the progeny revealed that 20% of Cre-expressing homozygous floxed embryos could be obtained instead of the 25% expected. A total of 41 βMHC-Cre:Sf/Sf were detected among the 209 genotyped embryos from stages E9.5 to E13.5 compared to 58 βMHC-Cre:Sf/+ heterozygous embryos (Table 1). Excision of exon 2 was confirmed by PCR analysis on the DNA extracted from embryonic hearts (Fig. 1E). Further analysis of the embryonic hearts by Western blotting showed the absence of normal SRF protein in the βMHC-Cre:Sf/Sf embryos (Fig. 1F). A weak band corresponding to a truncated SRF protein lacking exon 2 was present in low quantity in the mutant embryonic hearts. To confirm that this protein is inactive, we cloned the corresponding mutant SRF cDNA and found that upon cell transfection this protein was, as expected, unable to bind DNA and could not induce CArG box-dependent luciferase activity (data not shown). Furthermore, the absence of a phenotype in mice heterozygous for the deleted Srf allele (SΔ/+) indicate that this truncated protein had no transdominant-negative effect. Macroscopic analysis showed that the βMHC-Cre:Sf/Sf embryos appeared normal in early developmental stages, with no detectable external cardiac abnormality up to E9.5. Abnormal embryos were found as early as E10.5 and only resorbed or growth-retarded embryos were obtained at E13.5 (Table 1). At E11.5, all unresorbed βMHC-Cre:Sf/Sf, mutant embryos presented severe hemorrhaging in and around the cardiac and the ventral body wall regions (Fig. 3).
TABLE 1.
Numbers of newborns or embryos and their corresponding genotypes and aspects at different stages of development
| Stage | No. of embryos or newborns with genotype:
|
Aspecta | ||||
|---|---|---|---|---|---|---|
| Total | Sf/+ | Sf/+: Cre | Sf/Sf | Sf/Sf: Cre | ||
| E9.5 | 57 | 18 | 16 | 15 | 8 | 8N |
| E10.5 | 63 | 14 | 19 | 18 | 12 | 3H; 2R; 7N |
| E11.5 | 61 | 11 | 14 | 19 | 17 | 10H; 7R |
| E12.5 | 28 | 7 | 9 | 8 | 4 | 4R |
| Newborn | 16 | 6 | 5 | 5 | 0 | |
N, normal; H, hemorrhagic; R, resorbed. Values refer to animals with Sf/Sf: Cre genotype.
FIG. 3.
Analysis of embryonic development in normal and SRF mutant embryos. Control (WT) and mutant (Sf/Sf:βMHC-Cre) embryos at E10.5, E11.5, and E13.5 are shown. Mutant embryos were resorbed at E13.5 and display a hemorrhagic aspect at E10.5 and E11.5.
The SRF mutant embryos show defects both in the cardiac compact layer and in the trabeculations.
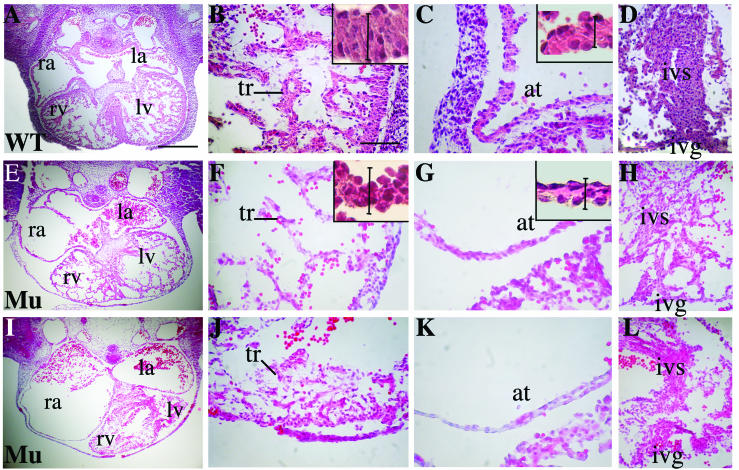
The histology of the βMHC-Cre:Sf/Sf mutant embryos and control littermates was analyzed at various developmental stages. Embryos were embedded in paraffin, and sections were stained with hematoxylin and eosin. At E9.5, mutants showed a normal looping of the linear heart tube and correct cardiac morphology compared to control mice (data not shown). At E10.5, the formation of the cardiac chambers was initiated correctly in mutant embryos that were neither resorbed nor hemorrhagic. However, at E11.5 the histological sections of all mutant embryos revealed striking abnormalities in the myocardium. Two typical examples of SRF mutants are shown in Fig. 4E and I. Severe disorganization of the myocardial wall was observed with variable degrees of ventricular atrophy. The thickness of the compact layer was dramatically reduced in the mutant (Fig. 4E, F, I, and J) compared to the wild type (Fig. 4A and B). The lumens of both the right and the left atria were significantly enlarged (Fig. 4E and I) compared to controls (Fig. 4A). Atrial wall thickness was severely reduced in the mutant (Fig. 4G and K) compared to control (Fig. 4C). Although the control heart at this stage had abundant and thick trabeculae (Fig. 4B), the mutant SRF embryos had thinner and less-abundant trabeculations (Fig. 4F and J). The interventricular septum was also thinner and disorganized in the mutant SRF embryos (Fig. 4H and L) compared to the control (Fig. 4D). At the cellular level, higher magnification of the compact layer in ventricles and atria walls revealed that the reduced wall thickness was mainly due to a reduction of the cytoplasmic volume in the SRF mutant cardiomyocytes (Fig. 4B, C, F, and G [insets]). The cell area was quantified with ImageJ software and showed a reduction of 22% ± 4% in the mutant compared to the wild type.
FIG. 4.
Comparative histological analysis of E11.5 heart structure in control and mutant embryos. Mid-level sections from control (A) and two independent mutant hearts (E and I) are shown. Scale bar, 290 μm. In the remaining panels, the magnifications of the ventricular compact layer and trabeculations (B, F, and J) the atrium (C, G, and K) and the interventricular septum (D, H, and L) are provided. Scale bar, 64 μm. Mutant embryos show an atrial enlargement and ventricular atrophy (I) compared to control (A). Trabeculations in the control heart (B) are denser and thicker than in the mutant hearts (F and J). The ventricular and atrial wall in the mutant embryos are thinner (F, G, J, and K). The reduced thickness of the myocard is mainly due to a reduced cytoplasmic surface (see insets in panels B, F, C, and G). Vertical bars in insets indicate the normal thickness of the ventricular and atrial wall for reference. Vertical bar in panels B and F, 32 μm; vertical bar in panels C and G, 24 μm. The interventricular septum is atrophic and severely disorganized (H and L) compared to the control (D). Abbreviations: ra, right atrium; la, left atrium; rv, right ventricle; lv, left ventricle; at, atrium; tr, trabeculation; ivs, interventricular septum; ivg, interventricular groove.
The hearts of mutant embryos display no modification in cell proliferation.
Given the potential role for SRF in cell homeostasis, we tested whether cell proliferation and/or cell death were affected in the hearts lacking SRF. The proliferation rate was measured in E9.5 and E10.5 embryos by determining the BrdU incorporation in pregnant female mice. BrdU-positive cells were counted on the embryonic heart sections from three mutants and three wild-type embryos at each developmental stage (Fig. 5A). The proliferation rates were 32% ± 4% in wild-type E9.5 embryos and 30% ± 5% in mutant E9.5 embryos. At E10.5, the proliferation rates were 35% ± 7% and 38% ± 5% for wild-type and mutant embryonic hearts, respectively. In addition, we measured the mitotic index at E10.5 by using an anti-phospho-histone H3 (ser10) antibody. The counting performed on six sections from three independent mutants and controls showed no significant differences (Fig. 5B). All together, these data show that no significant change occurs in the proliferation rate of cardiac myocytes after the excision of SRF.
FIG. 5.
Cell proliferation and apoptosis in control and mutant embryonic hearts. (A) Proliferation index of cardiomyocytes was determined as the number of BrdU positive nuclei reported to the total number of nuclei in E9.5 and E10.5 embryos. No significant difference was found between mutants (Mu) and wild-type (Wt) embryos at both developmental stages (n = 12). (B) Mitotic index of cardiomyocytes was defined as the number of phospho-histone H3-positive nuclei reported to the total number of nuclei in E10.5 embryos. No difference was found between mutant (Mu) and wild-type (Wt) hearts (n = 6). (C and D) Typical pictures of immunohistochemistry by anti-cleaved caspase 3 antibody on sections of E11.5 wild-type (Wt) and mutant (Mu) embryonic hearts. At this stage, only a few apoptotic cells were detected in the wild-type heart, whereas 10% of apoptotic cells (indicated by arrows) were observed in mutant embryonic hearts.
Given the important cardiac defects observed in SRF mutant embryos, we also investigated cell death by using the anti-cleaved caspase-3 antibody. At E10.5, only a few cardiac cells that were stained in both mutant (0.53% ± 0.25%) and control (0.71% ± 0.09%) embryos showed the absence of significant cell death by apoptosis at this stage. However, at E11.5, we found a dramatic increase in cells labeled with cleaved caspase-3 antibody in the mutant hearts (Fig. 5D), whereas only a few cells were labeled in the wild-type embryo at this stage (Fig. 5C). At this stage, we also noted that cell apoptosis in the mutant had extended to other regions of the mutant embryos, including the limb buds, epidermis, and vessels (data not shown). This latter observation suggested that apoptosis was not due to lack of SRF per se but rather was linked to global cell death, possibly because of tissue hypoxia due to impaired cardiac function.
Mutant SRF embryos show an impaired expression of critical cardiac transcription factors.
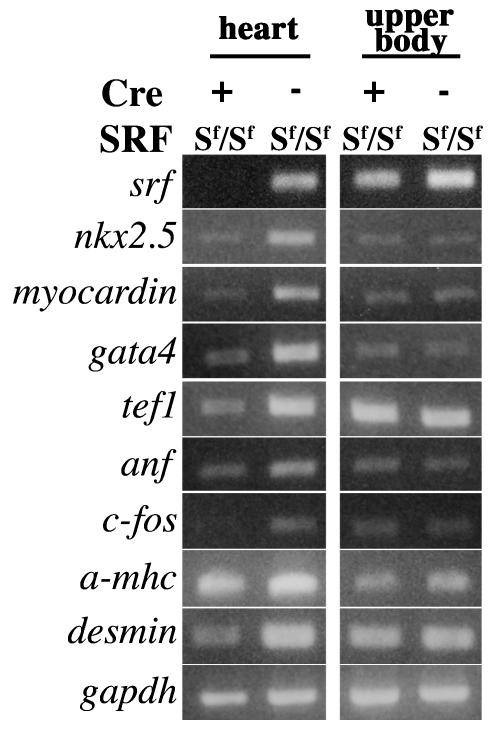
Given the cardiac defects observed in E10.5 and E11.5 mutant embryos at both macroscopic and microscopic levels, we decided to look at 9.5 days of development for potential changes in gene expression pattern. Transcription factors known to be important regulators of cardiac development and to interact with SRF (Nkx2.5, GATA4, myocardin, and TEF-1) or genes whose transcription is controlled by SRF (c-fos, αMHC, and ANF) were analyzed by semiquantitative RT-PCR. Total RNA was extracted from the heart and the upper body of four mutant and four control E9.5 embryos. The RNA extracted from the upper body tissue was used as a control. Evaluation of SRF transcripts in mutant hearts by comparison to controls showed that at this stage an efficient inactivation of SRF expression could already be obtained in the mutant hearts (Fig. 6, compare the first and second lanes). As expected, no alteration in SRF expression could be observed in the upper body of mutant embryos. The loss of SRF expression in the mutant hearts was accompanied by a drastic decrease in the levels of c-fos transcripts. The expression of several cardiac tissue-specific genes was also affected. Indeed, the transcript levels of Nkx2.5, GATA4, and myocardin were markedly affected in mutant hearts. A decrease in transcription of TEF1 was also detected. In contrast, the RT-PCR assay could not detect significant changes in the transcription levels of structural genes such as βMHC, and cardiac actin (data not shown) that are known targets for SRF. At this stage, the expression of desmin, ANF, and αMHC decrease slightly (Fig. 6).
FIG. 6.
Gel electrophoresis of RT-PCR products from E9.5 hearts. Semiquantitative analysis of the gene expression pattern in Cre+ and Cre− homozygous (Sf/Sf) “floxed SRF” embryonic hearts at E9.5 by RT-PCR. The expression patterns of genes involved in cell proliferation (c-fos) and cardiac differentiation (Nkx2.5, GATA4, myocardin, TEF-1, αMHC, and desmin) are presented. RT-PCR on RNA extracted from the upper body of the embryos that does not express Cre activity was also performed. GAPDH amplification was used as a control.
DISCUSSION
There is now considerable evidence that SRF is at the convergence of several regulatory networks controlling cardiac cell growth and differentiation during development (9, 38). In the present study, we used a new βMHC-Cre transgenic line that targets Cre gene expression in cardiomyocytes from E9.25 to produce a cardiac restricted knockout of the Srf gene at early steps of cardiomyogenesis through a Cre recombinase-mediated excision in exon 2 of the Srf gene. Our data show that SRF is required for normal cardiac development and maturation. Mutant embryos die between E10.5 and E13.5 days of development as a result of massive cardiac defects. Poor trabeculations, cardiac dilatation, and a thin myocardium were evident at stage E11.5, leading to embryonic death. At the molecular level, early depletion of SRF results in the downregulation of several essential heart-specific transcription factors, including Nkx2.5, GATA4, and myocardin, which may contribute to subsequent morphological defects.
SRF and heart development.
Heart development is a complex multiple-stage process that requires the coordinated expression of transcription factors such as Nkx2.5, GATA4, SRF, MEF2, etc. At E8.5 of embryonic development the heart is a linear tube that undergoes a rightward looping at stage E9.5. At stage E10.5, the emergence of trabeculations in the luminal layers of ventricles enables the myocardium to increase its mass (37). Subsequent septation leads to the formation of a four-chambered heart.
The role of SRF in postnatal cardiac development was investigated in vivo by perturbing its expression in transgenic mice. Overexpression of SRF or a mutant form of SRF in the heart led to a cardiomyopathy (50, 51). In the present study we investigated the role of SRF in the maturation of the embryonic heart. This maturation process involves cellular outgrowth and movement, leading to the formation of the trabeculations (37) and the interventricular septum (1, 25). For this purpose, targeted depletion of SRF in the developing heart could be obtained from E9.5 days of embryonic development through the expression of Cre recombinase under the control of the βMHC promoter, resulting in excision of the floxed Srf alleles. The onset of Cre activity was assessed by using the CAG-CAT-Z reporter line. It revealed a dotted pattern of blue staining from E9.25, reflecting at this stage a probable heterogeneity in transgene excision within the cardiomyocyte population. Cre activity was significantly increased in E9.75 embryos (Fig. 2). Indeed, this Cre expression level was found to be sufficient in E9.5 mutant embryos to obtain efficient excision of floxed Srf alleles, as reflected by the drastic decrease of Srf transcripts detected by RT-PCR (Fig. 6). However, some variations observed in the phenotype of mutant mice (resorbed embryos versus hemorrhagic) could result from slight differences in the efficiency of floxed Srf alleles excision between individuals.
Mutant embryos lacking SRF died between E10.5 and E13.5 of gestation, a critical period for cardiogenesis. Macroscopic examination of mutant embryos showed no pericardial effusion, a common feature of poor embryonic cardiac function. However, blood retention observed in these E10.5-to-E11.5 embryos suggests impaired cardiac pumping due to the lack of a highly muscularized ventricular myocardium. Indeed, histological sections of these embryos revealed poor trabeculation. At E11.5, severe defects were detected in mutant embryos that displayed abnormally thin compact layers of both ventricles and trabeculations (Fig. 4). At this stage, most of these mutant embryos also had a disorganized interventricular septum that failed to form in some cases. The pronounced hypoplasia of the ventricular myocardium in the mutant embryos is reminiscent of the “thin myocardium syndrome” observed upon inactivation of the genes encoding several transcription or signaling factors involved in cardiogenesis (N-myc, TEF-1, βARK1, WT-1b, MEF2C, and RXRα) (7, 10, 20, 23, 26, 43). Compared to the V-shaped and compact aspect of normal atria, mutant embryos show an enlarged atrial lumen. This could be due to the defective pumping of the ventricles, leading to the accumulation of blood in the atria and subsequent enlargement of the lumen. However, a possible defect in cell adhesion properties could also account for this altered and disorganized structure of the atrial and ventricular myocardium. Indeed, ES cells lacking SRF display impaired cell-cell interactions that are probably linked to the disorganization of the actin cytoskeleton and the downregulation of cell surface proteins (35, 36). Another finding supporting this hypothesis is the phenotype observed in mutant Drosophila for pruned, the SRF homologue in Drosophila, in which the terminal branches in the tracheal (respiratory) system fail to extend cytoplasmic projections (14). This cytoplasmic extension process could be compared to the trabeculation in cardiac development.
SRF seems to be the target of several converging regulatory networks in the developing heart, including activating cofactors such as myocardin (46) and negative modulation by the homeodomain protein HOP, whose expression is dependent on Nkx2.5 (9, 38). Such modulations of SRF activity are thought to ensure a delicate balance between cardiomyocyte proliferation and differentiation during heart development. Most of the observed defects in mutant embryos are consistent with a role of SRF in cardiomyocyte differentiation rather than in proliferation. In support of this is the observation that SRF depletion in E9.5 and E10.5 cardiac cells was found to have no incidence on cell proliferation. Similar results were obtained for SRF−/− ES cells in which the proliferation rate was not altered (36).
The important alterations in the expression of key cardiac transcription factors such as Nkx2.5, GATA4, TEF1, and coactivators such as myocardin, which occur at E9.5 in the mutant embryos, also point to SRF being essential for cardiomyocyte differentiation. The expression of these factors was downregulated to various extents. This downregulation of the cardiac genetic program precedes detectable changes in cardiac morphology. In addition to their crucial role in cardiac differentiation, all of these factors were shown to act in synergy with SRF to activate heart-specific gene expression. Hence, SRF has been shown to interact with Nkx2.5 and GATA4 to activate the ANF and cardiac α-actin promoters and with TEF1, a factor involved in the regulation of several heart-specific genes (8, 15, 31). SRF also acts in synergy with the cofactor myocardin, which interacts with its MADS box (46). The fact that SRF depletion leads to a concomitant reduction in the expression of all of these genes suggests that SRF could act directly or indirectly upstream of these factors in heart development. Alternatively, SRF could play a central role in maintaining their coordinate expression during the maturation stage of heart formation. Indeed, several recent data suggest that SRF could be involved in the control of Nkx2.5. Thus, myocardin that activates transcription through SRF binding sites was shown to strongly transactivate the enhancer of the Nkx2.5 gene (46). Nkx2.5 gene expression was severely reduced in Xenopus embryos in which cardiomyocyte differentiation was impaired by a dominant-negative form of myocardin (47). Our data support the existence of such a control of Nkx2.5 gene expression by SRF. Surprisingly, among the different SRF target genes that were analyzed at E9.5 in mutant embryos, only c-fos transcription was profoundly altered. Expression of desmin, αMHC, and ANF was slightly affected, whereas that of structural genes such as βMHC and cardiac actin does not appear to be affected at this stage. There are several possible explanations for such an observation. First, the need for SRF-mediated activation could vary during development depending upon the analyzed promoters. Some of these SRF targets may be less dependent on SRF activation at this early stage. Accordingly, interactions between Nkx2.5 and SRF in heart-specific gene regulation were shown to produce different transcriptional responses in cardiomyocytes from adults versus those from neonates (33). Second, some mRNAs might be synthesized before inactivation of the Srf gene and be more stable. Although our results clearly identify SRF as crucial for normal cardiac maturation, they also suggest that SRF may act upstream of Nkx2.5 during heart formation. An SRF-related protein was found in the presumptive heart region of the Xenopus early-bud embryo prior to the detection of any marker of cardiac muscle differentiation (6). It is thus tempting to postulate that SRF might also be involved in the very early steps of cardiac determination, in addition to being required for the correct maturation process of the embryonic heart. Our conditional Srf gene inactivation scheme will render it possible to explore whether SRF is also needed for the initiation of the myocardial genetic program with a suitable earlier cardiac Cre-expressing line such as Nkx2.5-Cre transgenic lines (32, 42). In the course of our study, another floxed Srf allele (Srf-flex1neo) was reported by Wiebel et al. (48). This floxed SRF line differs from our own floxed SRF line (Srf-flex2neo). In Srf-flex1neo line, LoxP sequences were placed in the 5′-untranslated region and in intron 1 of the Srf gene. In both transgenic lines the excision of the DNA sequence (exon 1 or exon 2) leads to the inactivation of SRF. It will be interesting in the future to compare the phenotypes resulting from the excision of Srf gene in both SRF floxed mice. In addition, recent transgenic studies (50, 51) and data from human pathology (12) underscore the importance of SRF for the maintenance of postnatal cardiac integrity. Therefore, it will be interesting to examine the function of SRF in the adult heart with the help of an inducible heart-specific Cre-expressing line such as the one developed by Sohal et al. (39).
Acknowledgments
This study was supported by the Association Française contre les Myopathie (AFM) and the Fondation de France (2003005644). A.P. was supported by fellowships from the AFM and Fondation pour la Recherche Médicale. The contribution of the Region Ile de France to Institut Cochin Animal Care Facility equipment is also acknowledged.
We thank P. Vassalli for providing the CAG-CAT-Z line and M. Colbert for providing the βMHC promoter construct. We are grateful to Y. Chéraud and D. Couton for valuable help and to M. Mericskay, G. Butler-Browne, and P. Maire for critical reading of the manuscript.
REFERENCES
- 1.Anderson, R. H., and N. A. Brown. 1996. The anatomy of the heart revisited. Anat. Rec. 246:1-7. [DOI] [PubMed] [Google Scholar]
- 2.Araki, K., M. Araki, J. Miyazaki, and P. Vassalli. 1995. Site-specific recombination of a transgene in fertilized eggs by transient expression of Cre recombinase. Proc. Natl. Acad. Sci. USA 92:160-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arsenian, S., B. Weinhold, M. Oelgeschlager, U. Ruther, and A. Nordheim. 1998. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 17:6289-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belaguli, N. S., J. L. Sepulveda, V. Nigam, F. Charron, M. Nemer, and R. J. Schwartz. 2000. Cardiac tissue enriched factors serum response factor and GATA-4 are mutual coregulators. Mol. Cell. Biol. 20:7550-7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Browning, C. L., D. E. Culberson, I. V. Aragon, R. A. Fillmore, J. D. Croissant, R. J. Schwartz, and W. E. Zimmer. 1998. The developmentally regulated expression of serum response factor plays a key role in the control of smooth muscle-specific genes. Dev. Biol. 194:18-37. [DOI] [PubMed] [Google Scholar]
- 6.Chambers, A. E., M. Logan, S. Kotecha, N. Towers, D. Sparrow, and T. J. Mohun. 1994. The RSRF/MEF2 protein SL1 regulates cardiac muscle-specific transcription of a myosin light-chain gene in Xenopus embryos. Genes Dev. 8:1324-1334. [DOI] [PubMed] [Google Scholar]
- 7.Charron, J., B. A. Malynn, P. Fisher, V. Stewart, L. Jeannotte, S. P. Goff, E. J. Robertson, and F. W. Alt. 1992. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes Dev. 6:2248-2257. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. Y., and R. J. Schwartz. 1996. Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac alpha-actin gene transcription. Mol. Cell. Biol. 16:6372-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, F., H. Kook, R. Milewski, A. D. Gitler, M. M. Lu, J. Li, R. Nazarian, R. Schnepp, K. Jen, C. Biben, G. Runke, J. P. Mackay, J. Novotny, R. J. Schwartz, R. P. Harvey, M. C. Mullins, and J. A. Epstein. 2002. Hop is an unusual homeobox gene that modulates cardiac development. Cell 110:713-723. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Z., G. A. Friedrich, and P. Soriano. 1994. Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev. 8:2293-2301. [DOI] [PubMed]
- 11.Croissant, J. D., J. H. Kim, G. Eichele, L. Goering, J. Lough, R. Prywes, and R. J. Schwartz. 1996. Avian serum response factor expression restricted primarily to muscle cell lineages is required for alpha-actin gene transcription. Dev. Biol. 177:250-264. [DOI] [PubMed] [Google Scholar]
- 12.Davis, F. J., M. Gupta, S. M. Pogwizd, E. Bacha, V. Jeevanandam, and M. P. Gupta. 2002. Increased expression of alternatively spliced dominant-negative isoform of SRF in human failing hearts. Am. J. Physiol. Heart Circ. Physiol. 282:H1521-H1533. [DOI] [PubMed] [Google Scholar]
- 13.Fishman, M. C., and K. R. Chien. 1997. Fashioning the vertebrate heart: earliest embryonic decisions. Development 124:2099-2117. [DOI] [PubMed]
- 14.Guillemin, K., J. Groppe, K. Ducker, R. Treisman, E. Hafen, M. Affolter, and M. A. Krasnow. 1996. The pruned gene encodes the Drosophila serum response factor and regulates cytoplasmic outgrowth during terminal branching of the tracheal system. Development 122:1353-1362. [DOI] [PubMed] [Google Scholar]
- 15.Gupta, M., P. Kogut, F. J. Davis, N. S. Belaguli, R. J. Schwartz, and M. P. Gupta. 2001. Physical interaction between the MADS box of serum response factor and the TEA/ATTS DNA-binding domain of transcription enhancer factor-1. J. Biol. Chem. 276:10413-10422. [DOI] [PubMed] [Google Scholar]
- 16.Harvey, R. P. 1996. NK-2 homeobox genes and heart development. Dev. Biol. 178:203-216. [DOI] [PubMed] [Google Scholar]
- 17.Hauschka, S. D. 2001. Myocardin. a novel potentiator of SRF-mediated transcription in cardiac muscle. Mol. Cell 8:1-2. [DOI] [PubMed] [Google Scholar]
- 18.Holzenberger, M., C. Lenzner, P. Leneuve, R. Zaoui, G. Hamard, S. Vaulont, and Y. L. Bouc. 2000. Cre-mediated germline mosaicism: a method allowing rapid generation of several alleles of a target gene. Nucleic Acids Res. 28:E92. [DOI] [PMC free article] [PubMed]
- 19.Huang, W. Y., J. J. Chen, N. Shih, and C. C. Liew. 1997. Multiple muscle-specific regulatory elements are associated with a DNase I hypersensitive site of the cardiac beta-myosin heavy-chain gene. Biochem. J. 327:507-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaber, M., W. J. Koch, H. Rockman, B. Smith, R. A. Bond, K. K. Sulik, J. Ross, Jr., R. J. Lefkowitz, M. G. Caron, and B. Giros. 1996. Essential role of β-adrenergic receptor kinase 1 in cardiac development and function. Proc. Natl. Acad. Sci. USA 93:12974-12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jamali, M., C. Karamboulas, S. Wilton, and I. S. Skerjanc. 2001. Factors in serum regulate Nkx2.5 and MEF2C function in vitro. Cell. Dev. Biol. Anim. 37:635-637. [DOI] [PubMed] [Google Scholar]
- 22.Knotts, S., A. Sanchez, H. Rindt, and J. Robbins. 1996. Developmental modulation of a beta myosin heavy chain promoter-driven transgene. Dev. Dyn. 206:182-192. [DOI] [PubMed] [Google Scholar]
- 23.Kreidberg, J. A., H. Sariola, J. M. Loring, M. Maeda, J. Pelletier, D. Housman, and R. Jaenisch. 1993. WT-1 is required for early kidney development. Cell 74:679-691. [DOI] [PubMed] [Google Scholar]
- 24.Kuo, C. T., E. E. Morrisey, R. Anandappa, K. Sigrist, M. M. Lu, M. S. Parmacek, C. Soudais, and J. M. Leiden. 1997. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 11:1048-1060. [DOI] [PubMed] [Google Scholar]
- 25.Lamers, W. H., A. Wessels, F. J. Verbeek, A. F. Moorman, S. Viragh, A. C. Wenink, A. C. Gittenberger-de Groot, and R. H. Anderson. 1992. New findings concerning ventricular septation in the human heart: implications for maldevelopment. Circulation 86:1194-1205. [DOI] [PubMed]
- 26.Lin, Q., J. Schwarz, C. Bucana, and E. N. Olson. 1997. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 276:1404-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyons, G. E., S. Schiaffino, D. Sassoon, P. Barton, and M. Buckingham. 1990. Developmental regulation of myosin gene expression in mouse cardiac muscle. J. Cell Biol. 111:2427-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mericskay, M., A. Parlakian, A. Porteu, F. Dandre, J. Bonnet, D. Paulin, and Z. Li. 2000. An overlapping CArG/octamer element is required for regulation of desmin gene transcription in arterial smooth muscle cells. Dev. Biol. 226:192-208. [DOI] [PubMed] [Google Scholar]
- 29.Miano, J. M. 2003. Serum response factor: toggling between disparate programs of gene expression. J. Mol. Cell. Cardiol. 35:577-593. [DOI] [PubMed] [Google Scholar]
- 30.Molkentin, J. D., S. M. Jobe, and B. E. Markham. 1996. Alpha-myosin heavy chain gene regulation: delineation and characterization of the cardiac muscle-specific enhancer and muscle-specific promoter. J. Mol. Cell. Cardiol. 28:1211-1225. [DOI] [PubMed] [Google Scholar]
- 31.Morin, S., P. Paradis, A. Aries, and M. Nemer. 2001. Serum response factor-GATA ternary complex required for nuclear signaling by a G-protein-coupled receptor. Mol. Cell. Biol. 21:1036-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moses, K. A., F. DeMayo, R. M. Braun, J. L. Reecy, and R. J. Schwartz. 2001. Embryonic expression of an Nkx2-5/cre gene using ROSA 26 reporter mice. Genesis. 31:176-180. [DOI] [PubMed] [Google Scholar]
- 33.Muller, J. G., J. T. Thompson, A. M. Edmonson, M. S. Rackley, H. Kasahara, S. Izumo, T. C. McQuinn, D. R. Menick, and T. X. O'Brien. 2002. Differential regulation of the cardiac sodium calcium exchanger promoter in adult and neonatal cardiomyocytes by Nkx2.5 and serum response factor. J. Mol. Cell. Cardiol. 34:807-821. [DOI] [PubMed] [Google Scholar]
- 34.Naya, F. J., B. L. Black, H. Wu, R. Bassel-Duby, J. A. Richardson, J. A. Hill, and E. N. Olson. 2002. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat. Med. 8:1303-1309. [DOI] [PubMed] [Google Scholar]
- 35.Schratt, G., U. Philippar, J. Berger, H. Schwarz, O. Heidenreich, and A. Nordheim. 2002. Serum response factor is crucial for actin cytoskeletal organization and focal adhesion assembly in embryonic stem cells. J. Cell Biol. 156:737-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schratt, G., B. Weinhold, A. S. Lundberg, S. Schuck, J. Berger, H. Schwarz, R. A. Weinberg, U. Ruther, and A. Nordheim. 2001. Serum response factor is required for immediate-early gene activation yet is dispensable for proliferation of embryonic stem cells. Mol. Cell. Biol. 21:2933-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sedmera, D., T. Pexieder, M. Vuillemin, R. P. Thompson, and R. H. Anderson. 2000. Developmental patterning of the myocardium. Anat. Rec. 258:319-337. [DOI] [PubMed] [Google Scholar]
- 38.Shin, C. H., Z. P. Liu, R. Passier, C. L. Zhang, D. Z. Wang, T. M. Harris, H. Yamagishi, J. A. Richardson, G. Childs, and E. N. Olson. 2002. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell 110:725-735. [DOI] [PubMed] [Google Scholar]
- 39.Sohal, D. S., M. Nghiem, M. A. Crackower, S. A. Witt, T. R. Kimball, K. M. Tymitz, J. M. Penninger, and J. D. Molkentin. 2001. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ. Res. 89:20-25. [DOI] [PubMed] [Google Scholar]
- 40.Srivastava, D., P. Cserjesi, and E. N. Olson. 1995. A subclass of bHLH proteins required for cardiac morphogenesis. Science 270:1995-1999. [DOI] [PubMed] [Google Scholar]
- 41.Srivastava, D., and E. N. Olson. 2000. A genetic blueprint for cardiac development. Nature 407:221-226. [DOI] [PubMed] [Google Scholar]
- 42.Stanley E. G., C. Biben, A. Elefanty, L. Barnett, F. Koentgen, L. Robb, and R. P. Harvey. 2002. Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3′UTR-ires-Cre allele of the homeobox gene Nkx2-5. Int. J. Dev. Biol. 46:431-439. [PubMed] [Google Scholar]
- 43.Sucov, H. M., E. Dyson, C. L. Gumeringer, J. Price, K. R. Chien, and R. M. Evans. 1994. RXR alpha mutant mice establish a genetic basis for vitamin A signaling in heart morphogenesis. Genes Dev. 8:1007-1018. [DOI] [PubMed] [Google Scholar]
- 44.Treisman, R. 1987. Identification and purification of a polypeptide that binds to the c-fos serum response element. EMBO J. 6:2711-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Treisman, R., and G. Ammerer. 1992. The SRF and MCM1 transcription factors. Curr. Opin. Genet. Dev. 2:221-226. [DOI] [PubMed] [Google Scholar]
- 46.Wang, D., P. S. Chang, Z. Wang, L. Sutherland, J. A. Richardson, E. Small, P. A. Krieg, and E. N. Olson. 2001. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105:851-862. [DOI] [PubMed] [Google Scholar]
- 47.Wang, D. Z., S. Li, D. Hockemeyer, L. Sutherland, Z. Wang, G. Schratt, J. A. Richardson, A. Nordheim, and E. N. Olson. 2002. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc. Natl. Acad. Sci. USA 99:14855-14860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiebel, F. F., V. Rennekampff, K. Vintersten, and A. Nordheim. 2002. Generation of mice carrying conditional knockout alleles for the transcription factor SRF. Genesis 32:124-126. [DOI] [PubMed] [Google Scholar]
- 49.Xu, R. H., J. Kim, M. Taira, J. J. Lin, C. H. Zhang, D. Sredni, T. Evans, and H. F. Kung. 1997. Differential regulation of neurogenesis by the two Xenopus GATA-1 genes. Mol. Cell. Biol. 17:436-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, X., G. Azhar, J. Chai, P. Sheridan, K. Nagano, T. Brown, J. Yang, K. Khrapko, A. M. Borras, J. Lawitts, R. P. Misra, and J. Y. Wei. 2001. Cardiomyopathy in transgenic mice with cardiac-specific overexpression of serum response factor. Am. J. Physiol. Heart Circ. Physiol. 280:H1782-H1792. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, X., J. Chai, G. Azhar, P. Sheridan, A. M. Borras, M. C. Furr, K. Khrapko, J. Lawitts, R. P. Misra, and J. Y. Wei. 2001. Early postnatal cardiac changes and premature death in transgenic mice overexpressing a mutant form of serum response factor. J. Biol. Chem. 276:40033-40040. [DOI] [PubMed] [Google Scholar]