Abstract
The Saccharomyces cerevisiae RAD6 protein is required for a surprising diversity of cellular processes, including sporulation and replicational damage bypass of DNA lesions. In mammals, two RAD6-related genes, HR6A and HR6B, encode highly homologous proteins. Here, we describe the phenotype of cells and mice deficient for the mHR6A gene. Just like mHR6B knockout mouse embryonic fibroblasts, mHR6A-deficient cells appear to have normal DNA damage resistance properties, but mHR6A knockout male and female mice display a small decrease in body weight. The necessity for at least one functional mHR6A (X-chromosomal) or mHR6B (autosomal) allele in all somatic cell types is supported by the fact that neither animals lacking both proteins nor females with only one intact mHR6A allele are viable. In striking contrast to mHR6B knockout males, which show a severe spermatogenic defect, mHR6A knockout males are normally fertile. However, mHR6A knockout females fail to produce offspring despite a normal ovarian histology and ovulation. The absence of mHR6A in oocytes prevents development beyond the embryonic two-cell stage but does not result in an aberrant methylation pattern of histone H3 at this early stage of mouse embryonic development. These observations support redundant but dose-dependent roles for HR6A and HR6B in somatic cell types and germ line cells in mammals.
Repair of damaged DNA requires a number of complementary, partially overlapping mechanisms, including mismatch repair, nucleotide and base excision repair, and repair pathways that deal with double-strand breaks. S-phase arrest caused by unrepaired DNA damage is prevented by a mechanism known as postreplication repair, which we prefer to rename replicational damage bypass (RDB). This bypass mechanism allows replication across lesions instead of repairing lesions (20, 26).
Based on the mutant phenotype, RAD6 is one of the key players in RDB in yeast. rad6 mutants are extremely sensitive to a wide range of DNA-damaging agents and show an increased spontaneous mutation frequency and a loss of DNA damage-induced mutagenesis (41, 46). In addition, RAD6 is also involved in a number of other cellular processes in yeast, including the degradation of proteins via the N-end rule, retrotransposition, gene silencing, meiosis, and sporulation (14, 15, 28, 52). RAD6 was identified as a ubiquitin-conjugating enzyme (29), and all of the functions of RAD6 in yeast depend on its ability to transfer ubiquitin molecules to a protein substrate (66). The ubiquitin-conjugating pathway requires the consecutive activity of ubiquitin-activating (E1), -conjugating (E2), and -ligating (E3) enzymes and results in mono- or polyubiquitination of substrate proteins. The ubiquitination serves as a signal for modification, activation, or proteolytic degradation via the 26S proteasome. The identification of 11 ubiquitin-conjugating enzymes in yeast and at least 20 genes encoding such proteins in the human genome, as well as the existence of a vast number of E3s, suggests that high substrate specificity can be obtained by combining different E2s and E3s (24). RAD18, UBR1, and BRE1 are E3 proteins that interact with RAD6 and are involved in RDB, degradation of proteins via the N-end rule, and chromatin modification, respectively (7, 15, 77). Ubiquitination of histone H2B at lysine residue 123 by RAD6 is essential for sporulation in yeast (54). Recently, it was shown that ubiquitination of H2B is a prerequisite for the methylation of histone H3 at specific lysine residues, with BRE1 being the putative E3 involved in this process (11, 16, 48, 65). PCNA (proliferating cell nuclear antigen) has only recently been identified as an important target for RAD6 and as having relevance to the process of RDB and resistance to DNA damage. The presence of a functional RAD18 is essential for this (mono)ubiquitination (25). Mammals contain two RAD6-homologous genes, designated HR6A and HR6B, encoding 152-amino-acid proteins that are structurally and functionally highly conserved; the proteins share approximately 70% sequence identity with yeast RAD6 and around 95% identity with each other (33). The mouse homologs mHR6A and mHR6B are 100% identical to the human proteins hHR6A and hHR6B, respectively. Both mammalian homologs are ubiquitously expressed and are able to complement a rad6 mutant strain for repair and mutagenesis functions but not for the sporulation defect which appears to depend on the acidic tail that is present only in yeast RAD6 (31, 53).
Previously, the generation and characterization of mHR6B knockout mice, the first animal model defective in the ubiquitin-conjugating pathway, were described (55). The mHR6B knockout mice show a male-limited sterility phenotype, with spermatogenic derailment which is most pronounced during postmeiotic condensation of chromatin in elongating spermatids (55). This step in spermatogenesis coincides with histone-to-protamine replacement, essential for normal sperm development. As was shown, the level of HR6B exceeds that of HR6A in spermatids in rats and mice, which is in agreement with an important role in postmeiotic cells (6, 32). In addition, a defect in the meiotic prophase in spermatocytes of mHR6B knockout male mice has been identified (6).
In the present report, we describe the effect of using gene targeting in mice to inactivate the other RAD6 homolog, mHR6A. In addition, we have evaluated the viability of mice with different numbers of intact mHR6A and mHR6B alleles. The results obtained are discussed with respect to the specificity and redundancy of the two encoded ubiquitin-conjugating enzymes.
MATERIALS AND METHODS
Isolation of cDNAs and genomic clones.
A 1.7-kb human HR6A cDNA fragment (33) was used to screen a λZAP mouse brain library (Stratagene, La Jolla, Calif.). Out of a number of positive clones, we selected one (designated B4) that contained an insert of approximately 1.7 kb and resembled the major transcript detectable in all tissues (32). This cDNA was sequenced by using T7 polymerase (Amersham Biosciences, Piscataway, N.J.) according to the manufacturer's guidelines. An EcoRI subclone encompassing the first 301 nucleotides was subsequently used to screen an EMBL-3 λ phage-based 129Sv mouse genomic CCE library. Two genomic clones were isolated. These clones were designated X2 and X4, and both contained the complete gene.
Construction of the targeting vector and disruption of mHR6A in ES cells.
A 5.26-kb ScaI fragment containing exons 1 to 3 was selected as the starting point for a targeting construct and cloned in pIC20R (43). The resulting plasmid was digested with SalI and BamHI, and a cassette containing the neomycin resistance gene expressed under the control of the thymidine kinase promoter (69) was inserted as a XhoI/BamHI fragment, thereby replacing the coding part of exon 1 with the dominant selectable marker (Fig. 1A). The resulting plasmid was digested with Asp718I and EcoRV, and the insert was recovered from agarose by electrophoresis. Approximately 10 μg of DNA was electroporated into E14 embryonic stem (ES) cells (a kind gift of A. Berns, NKI, The Netherlands) as described previously (81). Individual neomycin-resistant clones were picked and expanded. Genomic DNA was isolated as described previously (39), digested with EcoRI, and screened for homologous integration by Southern blot analysis.
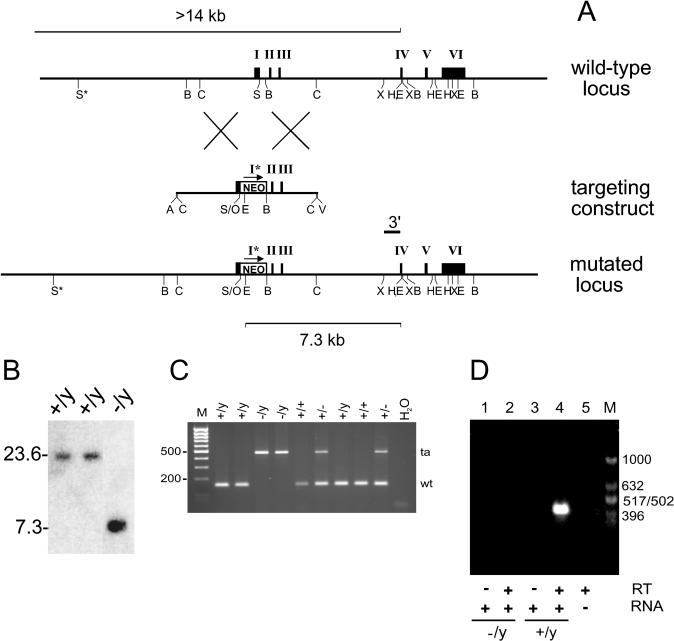
FIG. 1.
Targeted disruption of the mHR6A gene by homologous recombination. (A) Genomic organization and disruption strategy for mHR6A; shown are the gene, the targeting construct, and the targeted mHR6A allele. The neo cassette is inserted between the SalI site of exon 1 and the BamHI site in the first intron, introducing a diagnostic EcoRI site. Note that the insertion of the dominant marker disrupts the gene immediately behind the ATG translation initiation codon and deletes the coding part of exon 1. Shown are the relevant restriction sites (E, EcoRI; S, SalI; B, BamHI; C, ScaI; X, XmnI; H, HinDIII; O, XhoI; A, Asp718I; V, EcoRV). The position of the 3′ probe for Southern blot analysis is indicated above the mutated locus. Lines at the top and bottom indicate the estimated lengths of the fragments detected in Southern blot analysis of EcoRI-digested DNA. Roman numerals mark the exons. The 3′ deletion of exon 1 in the targeted allele is indicated with an asterisk. (B) Southern analysis of EcoRI-digested DNA from three neomycin-resistant ES clones after hybridization with the 3′ probe. The positions of the wild-type allele (23.6 kb) and the targeted allele (7.3 kb) are indicated. Note that the male ES line contains only one X-chromosomal mHR6A gene copy. “−/y” indicates a homologous recombinant ES clone, and “+/y” indicates a wild-type ES clone. The difference in signals between the targeted band and the wild-type band is presumably due to difficulty in the transfer of large fragments. (C) PCR-based genotyping of nine littermates. Primers mHR6A.7B and mHR6A.3B amplify a 152-bp fragment representing the wild-type allele (wt), and primers mHR6A.7B and NEOAS amplify a 473-nucleotide fragment representing the targeted allele (ta). Note that male animals (+/y and −/y) possess only one allele due to the X-chromosomal location of the allele. (D) RT PCR with total mouse testis RNA (lanes 1 to 4) from an mHR6A−/Y animal (lanes 1 and 2) and an mHR6A+/Y male littermate (lanes 3 and 4). Reactions were performed in the absence (lanes 1 and 3) or in the presence (lanes 2, 4, and 5) of RT. Lane 5 contains no RNA. M indicates the marker lane, with fragment lengths indicated on the right.
Generation of mHR6A-deficient mice and fibroblasts.
Individual ES clones with the correct karyotype and a correctly integrated mHR6A-targeting construct were injected into C57BL/6J blastocysts by using standard procedures (9). Male chimeras were bred with wild-type female FVB/n animals, and germ line transmission was recognized on the basis of a gray coat color of the offspring. Tail DNA was isolated as described previously (39), digested, and analyzed by Southern blotting. Starting with the F2 generation, animals were genotyped by multiplex PCR. With primers mHR6A.7B (5′-CTCGGCTCCGGAGCGGCAGT-3′) and mHR6A.3B (5′-CCCTCATGAGGCGGCGCCGGG-3′), a 153-bp fragment representing the wild-type allele was amplified. Primers mHR6A.7B and NEOAS (5′-TAGCCGAATAGCCTCTCCACCC-3′) resulted in the amplification of a 462-nucleotide fragment representing the targeted allele. Primary mouse embryonic fibroblasts (MEFs) were isolated as described elsewhere (71). The UV sensitivity of MEFs was determined by assaying the incorporation of [3H]thymidine at various doses of UV (61).
RNA isolation and RT PCR.
Total RNA was isolated from testes by using LiCl-urea (4), and cDNA was made by using 20 U of AMV-reverse transcriptase (RT) (Promega Benelux, Leiden, The Netherlands) with random hexamer primers (Roche Diagnostics, Almere, The Netherlands) according to the manufacturer's protocol in the presence of 40 U of RNAsin (Promega). mHR6A cDNA was amplified by using forward primer mHR6A.7B in exon 1 and reverse primer mHR6A.10 (5′-TCATAGGTTGGACTCCAACGG-3′) in exon 5.
Gel electrophoresis and immunoblotting.
Two-dimensional gel electrophoresis was done essentially as described by O'Farrell et al. (49). In short, oocytes or ovaria were sonicated in buffer A (0.5% [wt/vol] sodium dodecyl sulfate, 4 M urea, 1 mM dithiothreitol [DTT], 1% [vol/vol] Nonidet P-40, 0.1% [vol/vol] Bio-Lyte [pH 3 to 5; Bio-Rad, Veenendaal, The Netherlands], 0.1% [vol/vol] Bio-Lyte [pH 4 to 6; Bio-Rad]) and spun at 4°C at 13,000 rpm for 10 min, and the supernatant was collected. The protein concentration was determined by using the bichinchoninic acid assay (Pierce, Rockford, Ill.). Protein lysates to be separated in the first dimension were diluted in solution B (9 M urea, 1% [vol/vol] Bio-Lyte [pH 3 to 5], 1% [vol/vol] Bio-Lyte [pH 4 to 6], 1% Nonidet P-40, 100 mM DTT). Samples were boiled for 3 min and cooled on ice. Samples (40 μl) containing approximately 10 μg of protein were loaded on gels consisting of 4% acrylamide-bisacrylamide in solution B without DTT in 12-cm glass tubes. Samples were overlaid with 10 μl of overlay buffer (8 M urea, 1% [vol/vol] Bio-Lyte [pH 3 to 5], 1% [vol/vol] Bio-Lyte [pH 4 to 6]) and run for 16 h at 175 V in a Bio-Rad 175 tube cell (Bio-Rad) with 0.02 M NaOH in the lower chamber (cathode) and 0.01 M H3PO4 in the upper chamber (anode). After the first dimension was run, gels were removed from the glass tubes and either directly used for second-dimension polyacrylamide gel electrophoresis or stored at −20°C. Approximately 2 cm of the gel was cut off at the acidic side, and the rest was equilibrated in sample buffer (10% [vol/vol] glycerol, 2% sodium dodecyl sulfate, 10 mM DTT, 0.01% bromophenol blue, 62.5 mM Tris-HCl [pH 6.8]). Equilibrated gels were placed on top of a 15% phosphonoacetic acid gel with a 4% stacking layer and electrophoresed by using the Mini Protean II system (Bio-Rad, Hercules, Calif.). Subsequently, proteins were blotted on nitrocellulose (0.45 μm; Scleicher & Schull, Dassel, Germany), and mHR6A and mHR6B were detected by using α-HR6A/B antibody as described previously (5).
Superovulation of female mice and isolation of oocytes and embryos.
To isolate unfertilized oocytes for biochemical analysis, follicle growth in female mice was induced by intraperitoneal injection of 5 IU of Folligonan (Intervet, Boxmeer, The Netherlands). Forty-eight hours later, the mice were killed by cervical dislocation and the ovaries were isolated. Isolated ovaries were put in 100-μl drops of IVF medium [98.37 mM NaCl, 4.56 mM KCl, 0.69 mM MgSO4 · 7H2O, 22.54 mM NaHCO3, 0.86 mM NaH2PO4 · 2H2O, 0.085% (wt/vol) glucose, 0.0581% (wt/vol) Ca-l(+)-lactate, and 0.1485% (wt/vol) powdered F-10 supplemented with pasteurized plasma solution (final concentration, 3.5 g/liter)] and extensively punctured with two 30-gauge needles. Oocytes were collected and washed once in fresh IVF medium, pelleted by a quick spin in a table centrifuge, snap frozen, and stored at −80°C pending further use.
To obtain fertilized oocytes and preimplantation embryos, female mice were injected intraperitoneally with 5 IU of Chorulon (Intervet) 48 h after Folligonan injection and housed overnight with male mice. One-cell embryos (fertilized oocytes up to and including the zygote stage), two-cell embryos, and blastocysts were isolated from the oviduct or the uterus at 0.5, 1.5, and 3.5 days postcoitum (dpc) (E0.5, E1.5, and E3.5), respectively, as described previously (27). E1.5-stage embryos were placed in 50-μl droplets of IVF medium under mineral oil in a 5% CO2 incubator at 37°C and cultured for 72 h.
Immunofluorescence.
One-cell-stage embryos were isolated as described above, collected in phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde in PBS for 8 min at room temperature, and subsequently permeabilized in 0.5% Triton X-100 in PBS for 8 min at room temperature. To reduce background fluorescence, embryos were treated with 1% [wt/vol] bovine serum albumin in PBS-0.5% Tween 20 (blocking reagent) for 30 min at room temperature. Primary antibody incubations in blocking reagent were carried out overnight at 4°C, followed by three washes in blocking reagent at room temperature for 10 min. Secondary antibodies were incubated for 2 h at room temperature in blocking reagent, followed by three 10-min washes in blocking reagent at room temperature in the dark. Embryos were mounted on slides in Vectashield (Vector Laboratories, Burlingame, Calif.) containing DAPI (4′,6′-diamidino-2-phenylindole). Images were taken with a fluorescence microscope (Axioplan 2; Carl Zeiss, Jena, Germany) equipped with a digital camera (Coolsnap-Pro; Photometrics, Waterloo, Canada).
Confocal imaging.
Images were made with a confocal scanning laser microscope (Zeiss LSM510NLO; Carl Zeiss). During the observation period, fertilized oocytes were kept in a drop of IVF medium on a round cover glass mounted in a special chamber. Observations were made by using a ×63 oil immersion lens, and fertilized oocytes were imaged in optical planes with a 2-μm distance. DNA was made visible by using the DNA vital stain Syto16 (Molecular Probes Europe BV, Leiden, The Netherlands) added to the IVF medium. A 488-nm argon laser was used for excitation, and the emission was measured with a band-pass filter between 500 and 550 nm. Images were obtained by using the LSM510 image software (Carl Zeiss).
Nucleotide sequence accession number
The B4 HR6A cDNA sequence was submitted to GenBank/EMBL under accession no. AF089812.
RESULTS
Isolation and characterization of mouse mHR6A cDNA.
To isolate the mHR6A cDNA, we screened a mouse brain library with human cDNA as a probe. A mouse cDNA fragment of ∼1.7 kb, probably representing the major transcript (32), was isolated. Sequencing revealed 73% sequence identity with the human transcript overall and 93% sequence identity in the putative open reading frame (ORF). The encoded gene product, 152 amino acid residues long as translated from the submitted mouse sequence (GenBank accession no. AF089812), was 100% identical in sequence to the protein specified by the human cDNA from the public expressed sequence tag and human genome project databases, underlining the importance of this protein. In contrast to the 3′ untranslated regions of mHR6B and hHR6B, no stretch of conserved nucleotides was present in the 3′ untranslated regions of mHR6A and hHR6A, suggesting that this is a specific feature of the HR6B gene (55).
Isolation of genomic sequences and targeting of ES cells.
Two overlapping clones of mouse strain 129Sv DNA, one of which contained the complete gene as outlined in Fig. 1A, were isolated. The gene spans a region of ∼10 kb and is composed of six exons. A 5.36-kb ScaI genomic fragment that was present in both isolated mHR6A clones was selected. This fragment contains the first three exons, including the start codon ATG of the ORF. A dominant-selectable neomycin resistance marker was inserted by using the SalI and the BamHI restriction sites. In this way, the ORF was disrupted immediately after the start codon (Fig. 1A). In addition, amino acids 3 to 14 were deleted, thereby strongly reducing the possibility that an aberrant protein might be formed. Transfection of the targeting construct by electroporation and selection for stable integration of the dominant marker resulted in a frequency of targeted, single integrations of 2.8% (4 out of 146 analyzed), as confirmed by Southern blot analysis. Hybridization with the 3′ external probe visualized a large EcoRI fragment of 23.6 kb in the case of the wild-type allele and a 7.3-kb EcoRI fragment for a targeted allele (Fig. 1B).
Generation of mHR6A-deficient mice.
Two of the targeted ES clones (clones 49 and 102) had a correct karyotype and were used for injection into C57Bl/6 blastocysts to generate chimeras. Four chimeras were obtained, but only one (from clone 49) transmitted the mutated allele as determined by coat color and Southern blot analysis with the 3′ probe (data not shown). Because the mHR6A gene, like the human ortholog, is located on the X chromosome (34, 56), the male chimera transmits the targeted allele only to female pups of the F1 offspring. Indeed, all of the F1 females that had the coat color associated with the 129Sv/FVB hybrid genomic background were heterozygous.
Next, the F1 males and females were bred, resulting in a large number of knockout (mHR6A−/Y) male mice in the F2 offspring. The absence of a functional mHR6A allele in these animals was confirmed by RT PCR with total RNA isolated from testis material of an mHR6A−/Y animal and a wild-type animal. No mHR6A transcripts spanning the (partially) deleted exon 1 could be detected in testis cDNA from the mHR6A−/Y mouse, while a product of the right size (420 nucleotides) was readily amplified from testis cDNA of the wild-type male (Fig. 1D). This finding indicates that targeted integration of the knockout construct resulted in the absence of the original transcripts containing the complete ORF as well as any alternatively spliced mRNAs.
mHR6A-deficient mice and embryonic fibroblasts.
A program of breeding between mHR6A+/− females and mHR6A−/Y males was set up. The offspring showed a Mendelian representation of knockout and heterozygous female animals (91 and 91 animals, respectively) but a significant underrepresentation of knockout male animals compared to wild-type males (71 and 116 animals, respectively) (χ2 test, P = 0.001).
To compare the body weights of the different genotypes, litters that contained animals with both male and/or both female genotypes were included in a paired t test analysis. Mice were weighed at the time of tailing (10 ± 1 days after birth). The average body weight of all animals with the same genotype and sex within one litter was determined and included in the analysis. On average, the body weight of knockout male and female mice at 10 days of age was significantly lower (paired t test, P < 0.01), showing a reduction of approximately 12% compared to that of the respective control littermates (Fig. 2A).
FIG. 2.
Paired t test of the difference between total body weights of mice with a specific genotype and control littermates 10 ± 1 days after birth. Each dot represents the average body weight of all animals with a specific genotype present in one litter. The mean body weights are indicated by horizontal bars. The asterisks indicate statistically significant differences between the indicated groups (P ≤ 0.05). N is the number of litters used in the analysis. (A) Total body weights of mHR6A-deficient animals and control littermates. Included are litters that contain both mHR6A+/Y and mHR6A−/Y male mice and/or both mHR6A+/− and mHR6A−/− female mice. (B) Total body weights of mice with one single functional mHR6B allele and control littermates. Included are only litters that contain both mHR6A+/Y/mHR6B+/+ and mHR6A−/Y/mHR6B+/− male animals and/or both mHR6A+/−/mHR6B+/+ and mHR6A−/−/mHR6B+/− female animals. The genotypes of both genes are indicated under the x axis.
To investigate the possible occurrence of pathological abnormalities, most organs of three adult knockout animals and three control littermates were fixed and hematoxylin and eosin stained, but no aberrations were found (data not shown).
In view of the role of the RAD6 protein in RDB in yeast, mHR6A-deficient MEF cell lines were tested for their growth rates and cellular survival after irradiation with different doses of UV, as measured by [3H]thymidine incorporation. No differences between mHR6A-deficient and mHR6A-proficient cell lines were observed (data not shown). This finding demonstrates that UV resistance in mHR6B-deficient cells (55) does not depend on a functional mHR6A allele but is presumably caused by redundant activities of mHR6A and mHR6B in somatic cell types.
mHR6A−/Y males are normally fertile, but mHR6A−/− females are not able to produce offspring.
In contrast to the phenotype observed in mHR6B knockout mice, whereby females are normally fertile and males are sterile, we observed the reverse for mHR6A-deficient animals; mHR6A−/Y male mice were normally fertile, as shown by their ability to sire litters (Table 1) as well as by a precise analysis of male reproductive parameters (data not shown), but mHR6A−/− female mice appeared infertile. We investigated whether these females were receptive to mating with both mHR6A−/Y and wild-type males. Successful matings, indicated by the presence of copulatory plugs, were observed in mHR6A−/− females regardless of which males were used. The numbers of pups per litter that resulted from the breeding of heterozygous females with wild-type and knockout males were not significantly different (P = 0.81, two-tailed Student's t test). Ovaries were then isolated from knockout and heterozygous littermates, fixed, sectioned, stained, and microscopically analyzed for the presence of the various stages of follicular development. Ovaries from mHR6A−/− females appeared indistinguishable from those of mHR6A+/− littermates, with the presence of a normal number of growing follicles and corpora lutea (data not shown). This finding indicates that the ovaries of mHR6A-deficient females are capable of follicular development and ovulation. To follow up on the initial observation, we superovulated mHR6A−/− and mHR6A+/− females and housed them overnight with FVB/n wild-type males. Fertilized oocytes were recovered from the oviducts and cultured in vitro. The one-cell embryos recovered from mHR6A−/− females appeared morphologically normal and comparable with the embryos isolated from the heterozygous control females. All of the embryos contained two pronuclei (Fig. 3A to D), indicating that they were normally fertilized and did not develop parthenogenetically after fertilization with wild-type sperm. The one-cell embryos recovered from mHR6A+/− and mHR6A−/− females developed up to the two-cell stage in vitro but were not able to overcome a two-cell-stage block. This situation is not unusual for noninbred strains (22) but prevented us from determining the end point of the development of embryos derived from an mHR6A−/− mother. Therefore, embryos were recovered by flushing the oviducts 1.5 dpc, when developing embryos are at the two-cell stage, and subsequently these embryos were cultured for three additional days before reexamination. Total numbers of 37 two-cell embryos and 1 four-cell-stage embryo were recovered from four mHR6A+/− females, and all of these embryos appeared normal (Fig. 3E). All two-cell and four-cell embryos developed into blastocysts, most of which started hatching, and some of these embryos were already completely hatched, leaving behind an empty zona pellucida (Fig. 3F). From four mHR6A−/− females, 13 phenotypically normal two-cell-stage embryos, 2 two-cell-stage embryos with cytoplasmic fragments, 11 fertilized oocytes (of which 5 showed signs of fragmentation), and 1 three-cell embryo were retrieved (Fig. 3G). None of these embryos isolated from the oviducts of the mHR6A−/− females grew out to the blastocyst stage. Eight of them reached or remained in the two-cell stage, and others remained unicellular or showed severe cytoplasmic fragmentation and degeneration (Fig. 3H). This result was not a culture artifact, since embryos isolated at 3.5 dpc from mHR6A−/− female mice were also found to have developed no further than the two-cell stage (data not shown). This finding points to an important role of maternal mHR6A in the very early stages of embryonic development.
TABLE 1.
Average numbers of pups per litter with various parental mHR6A combinations
| Parental mHR6A genotypes | No. of successful matings | Litter size (mean no. of pups ± SD) |
|---|---|---|
| +/+ × +/Y | NDa | |
| +/+ × −/Y | 7 | 5.9 ± 2.6 |
| +/− × +/Y | 35 | 8.7 ± 2.6 |
| +/− × −/Y | 79 | 8.6 ± 2.7 |
| −/− × +/Y | 11 | 0 |
| −/− × −/Y | 13 | 0 |
ND, not determined.
FIG. 3.
Developmental stages of preimplantation embryos obtained from mHR6A+/− (A, B, E, and F) and mHR6A−/− (C, D, G, and H) female mice fertilized by wild-type males. Images obtained with confocal laser scanning microscopy (A to D) show embryos from an mHR6A+/− heterozygous mother (A and B) and an mHR6A−/− knockout mother (C and D) approximately 8 h after fertilization. Two planes are displayed for the fertilized egg, one showing the paternal pronucleus (P in panels A and C) and one showing the maternal pronucleus (M in panels B and D). Phase-contrast microscopy shows examples of embryos obtained from mHR6A+/− (E and F) and mHR6A−/− (G and H) female mice at 1.5 dpc (E and G) and cultured for 3 days (F and H). (E) Embryos at various stages of development, obtained from an mHR6A+/− female. (F) Hatching (left) and hatched (upper right) embryos and an empty zona pellucida (lower right). (G) Embryos obtained from an mHR6A−/− female. (H) One-cell (left), two-cell (upper right), and fragmented (lower right) embryos. Scale bar, 20 μm.
The mHR6A protein level is high compared to that of mHR6B in oocytes.
In a number of tissues and cell types that were analyzed, the levels of mHR6A and mHR6B were comparable (32). In the testis, the postmeiotic cells, the spermatids, showed a relative increase of mHR6B compared to mHR6A (6, 32). This increase might be caused by the inactivation of the X-chromosomal mHR6A gene in spermatogenesis, when the X and Y chromosomes form the heterochromatic sex body during the meiotic prophase (62). In oogenesis, the two X chromosomes are transcriptionally active during the meiotic prophase, which might result in a relatively high level of mHR6A compared to mHR6B (35). To test this possibility, we first prepared protein extracts from ovaries of wild-type and mHR6A heterozygous and knockout mice. The protein contents of these samples were separated by two-dimensional gel electrophoresis and blotted, and mHR6A and mHR6B proteins were visualized by using the α-HR6A/B antibody as described (5). This analysis confirmed the absence of mHR6A in ovaries from mHR6A−/− females (Fig. 4A) but also showed that in the wild-type whole ovary, the levels of mHR6A and mHR6B were about equal. Differential expression of the two proteins in oocytes may go undetected when whole ovary tissue is analyzed. Hence, to determine the relative levels of mHR6A and mHR6B proteins in wild-type oocytes, oocyte maturation was induced in six female mice by injecting the mice with 5 IU of Folligonan. Approximately 240 oocytes were isolated, lysed, and analyzed by two-dimensional gel electrophoresis. It was found that the level of mHR6A in isolated oocytes was relatively high compared to that of mHR6B (Fig. 4B).
FIG. 4.
mHR6A and mHR6B protein expression. Total protein extracts were separated by using two-dimensional gel electrophoresis. (A) Comparison of the amounts of mHR6A and mHR6B proteins (indicated by “A” and “B,” respectively) in ovaries from mHR6A+/+, mHR6A+/−, and mHR6A−/− mice. (B) Expression of mHR6A and mHR6B in oocytes isolated from wild-type mice compared to the amounts of the two proteins in whole ovaries.
Methylation of histone H3 in pronuclei in wild-type and mHR6A-deficient oocytes.
In the very early stages of mammalian development, dramatic epigenetic changes that are involved in spatial and temporal control of gene expression occur (3, 16, 45, 58). These changes include demethylation and de novo methylation of CpG dinucleotide sequences. In addition, much recent evidence shows that epigenetic histone modifications are tightly regulated to assure control of gene expression but also DNA repair and recombination (for a review, see reference 18). Recently, it was shown that Saccharomyces cerevisiae RAD6 plays an important role in the posttranslational modification of histones. Monoubiquitination of histone H2B at lysine 123 is a prerequisite for the methylation of lysine 4 of histone H3 (H3K4) (16, 65), a modification that is associated with active gene expression, cell growth, telomeric and ribosomal DNA silencing, and sporulation (10, 57, 64). In higher eukaryotes, H3K4 methylation is associated with active chromatin domains, whereas methylated H3K9 associates with heterochromatic subdomains and gene silencing (reviewed in reference 38). Using specific antibodies against methylated H3K4 and methylated H3K9, we analyzed the role of mHR6A in controlling the methylation status of histone H3 during fertilization and early embryonic development. Embryos were isolated approximately 12 and 36 h postcoitum from mHR6A−/− and mHR6A+/− females. At 12 h postcoitum, immunofluorescent staining for H3K9 was predominantly associated with the maternal pronucleus (data not shown), a finding that is in full agreement with earlier data (3, 13). Two-cell embryos, isolated 36 h postcoitum, show clear and overall staining of the nuclei of both blastomeres irrespective of the genotype of the mother (data not shown). This finding suggests that H3K9 methylation is not severely affected by the absence of mHR6A during the formation of the first two blastomeres. In a similar experiment, it was observed that methylated H3K4 was also predominantly associated with the maternal pronucleus in one-cell embryos obtained from mHR6A+/− females (Fig. 5A), and no overt difference was observed for one-cell embryos from the mHR6A knockout females (Fig. 5C). After the first cell division, both nuclei fluoresce with similar intensities irrespective of the genotype of the mother (Fig. 5E and G). Immunofluorescent staining of one- and two-cell embryos for histone H3 clearly demonstrates the presence of histones in both pronuclei at this stage (data not shown). These data suggest that there are no marked changes in the methylation status of histone H3 in early embryos that can be attributed to the absence of maternal mHR6A.
FIG. 5.
Immunofluorescent images of one-cell (A to D) and two-cell (E to H) embryos stained with an antibody against methylated H3K4 (A, C, E, and G) and DAPI (B, D, F, and H). The genotype of the mother is indicated on the right. The intense bright spots in panels C to H are caused by the remnants of a polar body. Staining of only one of the two pronuclei with α-methylated H3K4 is observed (A and C), while both nuclei of the two-cell embryo stain equally intensely (E and G).
Mice with losses of mHR6A and mHR6B alleles.
The severe, pleiotropic phenotype of a rad6-null yeast strain (for a review, see reference 40), compared to the relatively mild phenotype that results from either mHR6A or mHR6B deficiency, suggests a strong functional redundancy between these two proteins. To investigate the effect of the complete loss of both RAD6 homologs in mice, a breeding program was set up to obtain double-knockout (DKO) animals. In this breeding program, double heterozygosity has no gross deleterious effect on embryonic development, and mHR6A+/−/mHR6B+/− female mice were mated with mHR6A+/Y/mHR6B+/− males. As shown in Table 2, no DKO animals were found, whereas approximately 11 DKO mice were to be expected (χ2 test, P < 0.001). In addition, female mHR6A+/−/mHR6B−/− animals were also absent. This result is most likely due to random inactivation of the X chromosome early in development, thereby leaving only 50% of the cells in these mHR6B-deficient mice with one functional wild-type X-chromosomal mHR6A allele.
TABLE 2.
Possible genotypes and observed numbers of pups with genotypes for various parental combinations
| Genotype | Observed (expected) no. of pups with genotype for parental combinationa
|
|||
|---|---|---|---|---|
| A+/Y/B+/− × A+/−/B+/− | A−/Y/B+/+ × A+/−/B+/− | A−/Y/B+/− × A+/−/B+/+ | A−/Y/B+/− × A+/−/B+/− | |
| A+/+/B+/+ | 11 (11) | |||
| A+/+/B+/− | 29 (22) | |||
| A+/+/B−/− | 13 (11) | |||
| A+/−/B+/+ | 11 (11) | 75 (59) | 7 (9) | 30 (17) |
| A+/−/B+/− | 20 (22) | 70 (59) | 11 (9) | 56 (34) |
| A+/−/B−/− | 0 (11) | 0 (17) | ||
| A−/−/B+/+ | 61 (59) | 8 (9) | 16 (17) | |
| A−/−/B+/− | 16 (59) | 3 (9) | 20 (34) | |
| A−/−/B−/− | 0 (17) | |||
| A+/Y/B+/+ | 12 (11) | 87 (59) | 11 (9) | 27 (17) |
| A+/Y/B+/− | 37 (22) | 83 (59) | 15 (9) | 54 (34) |
| A+/Y/B−/− | 17 (11) | 29 (17) | ||
| A−/Y/B+/+ | 16 (11) | 60 (59) | 10 (9) | 24 (17) |
| A−/Y/B+/− | 11 (22) | 23 (59) | 6 (9) | 11 (34) |
| A−/Y/B−/− | 0 (11) | 0 (17) | ||
| Total | 177 | 475 | 73 | 267 |
A, mHR6A; B, mHR6B. Boldface numbers are significantly lower than the expected numbers.
With the exception of mHR6A−/Y/mHR6B−/− animals (no functional allele) and the mHR6A+/−/mHR6B−/− mice (only one intact allele, which is subject to X inactivation), all of the other genotypes were represented in the offspring. Mice lacking an intact mHR6A allele and being heterozygous for mHR6B (mHR6A−/Y/mHR6B+/−), however, were clearly underrepresented (χ2 test, P < 0.025) and showed a pronounced decrease in body weight that was already visible at birth. To investigate this result in more detail, mHR6A+/−/mHR6B+/− females were mated with mHR6A−/Y/mHR6B+/+ males and the offspring were genotyped and weighed. Mice that contained only one wild-type mHR6B allele were significantly underrepresented in the offspring (χ2 test, P < 0.001) (Table 2). To quantify the loss of body weight, we compared the body weights of wild-type versus mHR6A−/Y/mHR6B+/− males and those of mHR6A+/−/mHR6B+/+ versus mHR6A−/−/mHR6B+/− females. Only litters that contained pups with both male and/or both female genotypes were included. The average body weights of mHR6A−/Y/mHR6B+/− male and mHR6A−/−/mHR6B+/− female animals at 10 days of age were significantly lower (paired t test, P < 0.01) than those of the control littermates, with a reductions of 37 and 31%, respectively (Fig. 2B). Interestingly, mHR6A−/Y/mHR6B+/− male animals were found to be normally fertile. The average number of pups per breeding sired by these males was not significantly lower then that of pups sired by mHR6A−/Y/mHR6B+/+ males (P = 0.387, two-tailed Student's t test) mated with mHR6A+/−/mHR6B+/− females (7.1 ± 2.7 and 7.6 ± 2.6 pups, respectively). This result indicates that, in the testis, the level of mHR6B protein from one intact and functional allele can compensate for the complete loss of mHR6A and/or that mHR6B has a specific function in spermatogenesis.
DISCUSSION
Over the last few years, a number of mammalian homologs of yeast RDB genes that exhibit a high degree of sequence and functional conservation have been identified, suggesting that RDB mechanisms might be conserved throughout eukaryotic evolution (21, 30, 33, 42, 44, 47, 68, 72, 78-80). However, some remarkable functional differences between yeast and mammalian proteins (8, 17, 73, 76) and the identification of additional Y family DNA polymerases in mammalian species (for a review, see reference 19) forbid simple extrapolation from yeast to mammals. Also, the identification of two mammalian RAD6 homologs provides a complicating factor. Previously, we demonstrated that the inactivation of the mHR6B gene, one of the two mammalian homologs of yeast RAD6, induces only a relatively mild phenotype in mice compared to that induced by a rad6-null allele in yeast. Whereas yeast rad6 mutants display extreme sensitivity to all kinds of genotoxins, low growth rates, and impaired sporulation, the main phenotype of mHR6B−/− mice is a severe derailment of spermatogenesis (55). This observation raised the question whether the second homolog, mHR6A, is responsible for the greater part of the somatic RAD6-like functions in mice or whether there is extensive functional redundancy between the two mammalian proteins.
RAD18 is the E3 that interacts with RAD6 in RDB in yeast. Both human RAD6 homologs can interact with human RAD18, and the overexpression of a dominant-negative human RAD18 gene results in increased UV sensitivity (68, 79). Analysis of mHR6A-deficient MEFs, however, failed to reveal any difference between their response to UV irradiation and that of wild-type cells. This outcome is comparable with results of UV survival assays of mHR6B-deficient MEFs, which also showed no increased UV sensitivity (55). In addition, mHR6A-deficient mice appeared normal, apart from a slight reduction in body weight, and preliminary studies did not reveal an increased cancer predisposition up to 1 year of age.
Besides RAD18, UBR1 is another E3 capable of interacting with RAD6 (15). The human and mouse homologs of UBR1 have been cloned, and their roles in the N-end rule pathway and interaction with mHR6A and mHR6B have been confirmed (36, 37). One of the observed phenotypes in Ubr1−/− mice is a small but significant reduction in body weight (37). Similar reductions in body weight were observed for mHR6A-deficient animals (present data) and for mHR6B-deficient animals (unpublished data). Ubr1−/− mice are, however, normally fertile. This finding indicates that the infertility phenotype of both mHR6B−/− male and mHR6A−/− female mice is not determined by the interaction of mHR6A and/or mHR6B with Ubr1 and does not involve dysregulation of protein degradation via the N-end rule.
In the present breeding program, in addition to the absence of DKO mice, we found that mHR6A−/−/mHR6B+/− females and mHR6A−/Y/mHR6B+/− males were underrepresented and runted and often died around birth. The mHR6A−/Y/mHR6B+/− males that survived were normally fertile. This finding indicates that the level of mHR6B protein obtained from one functional mHR6B allele is sufficient to support spermatogenesis, even in the absence of mHR6A protein. On the one hand, the results obtained indicate that mHR6A and mHR6B have completely redundant activities both in somatic cell types and in germ line cells. On the other hand, the observed female and male infertility phenotypes in mHR6A- and mHR6B-deficient mice, respectively, indicate that germ line cells are subject to a dose-dependent effect which involves spermatogenesis- and oogenesis-specific differences in expression levels of mHR6A and mHR6B. However, the possibility that mHR6A and mHR6B have different functional properties at the molecular level cannot be completely excluded. Several observations that point in this direction have been described. Ubiquitin-protein ligases (E3s) can be found among proteins that contain either HECT or RING motifs, such as RAD18 and UBR1 (reviewed in reference 51), but also among proteins with U-box domains. Hatakeyama et al. (23) showed that the U-box motif-containing cyclophilin CYC4/hCyP-60 needs the E2 enzyme HR6B, but not HR6A, for polyubiquitination. Cyclophilins are thought to have chaperone functions, being involved in the proper folding of protein substrates in vivo. Although ubiquitously expressed, transcript levels of cyclophilins are elevated and differ in size in the testis (74). This finding suggests the possibility that the spermatogenic derailment observed in mHR6B−/− but not in mHR6A−/Y males is not caused by the relatively low level of mHR6A but by the inability of mHR6A to interact with cyclophilin. Additional data on possible specificity come from the identification of mHR6A, and not mHR6B, as an interacting partner of Rfpl4 in a yeast two-hybrid screen (67). Rfpl4 is present in maturing oocytes and remains expressed up to the eight-cell-stage embryo in the mouse. The protein shows the characteristics of a RING domain containing E3 ubiquitin ligase and can specifically interact with mHR6A. This protein could be useful in trying to identify the specificity-determining amino acid residues of mHR6A and mHR6B; mHR6A and mHR6B differ in only seven positions.
The most prominent effect of disrupting mHR6A is the observed complete failure of mHR6A-deficient females to produce offspring, resulting from developmental arrest after the first embryonic cell division. It is unlikely that the phenotype of the female mHR6A−/− mice is due to aberrant levels of gonadotropins, since vaginal smears for mHR6A-deficient females revealed regular 4- to 5-day cycles and histological examination of fixed and stained ovary sections showed the presence of all stages of follicular development. In addition, the animals mated readily, the number of fertilized oocytes was similar to that for mHR6A+/− littermates, and pseudopregnancy was induced. These data indicate a normal hypothalamic-hypophyseal-gonadal axis. Measurable deviations from normal serum gonadotropin levels would have resulted in dysregulated cycles.
The present results identify mHR6A as one of the few mammalian genes that encode a factor essential for the very first steps in embryonic development. Thus far, only Mater and Zar1 have been shown to encode proteins that have their major, if not exclusive, functions in early postfertilization development (12, 70).
The mechanism that is disrupted by the absence of mHR6A is still unclear but could be related to the regulation of transcription and/or chromatin rearrangements. Just recently, it has started to become clear how, and what kind of, core histone tail modifications play a role in chromatin remodeling, higher-order chromatin structure, and transcription (see reference 63 and references therein). These modifications include phosphorylation, acetylation, methylation, and ubiquitination. Robzyk et al. (54) showed that the ubiquitination of histone H2B in yeast is dependent on a functional RAD6 gene. Mutation of lysine residue 123 of H2B, which is essential for ubiquitination, prevents the methylation of H3K4 and induces defects in sporulation and gene silencing. Other RAD6-dependent processes, such as RDB, appear unaffected (54).
During fertilization, the chromatin in the paternal pronucleus is reorganized, with histones of maternal origin replacing protamines. From that point on, acetylated forms of histone H4 are detectable in the paternal pronucleus, associated with the onset of transcription (1, 2). The methylation of histone H3 at lysine residue 9 (H3K9), known to be involved in the formation of stable repressive heterochromatin (50), was found in the maternal pronucleus but could hardly be detected in the paternal pronucleus (3, 13). In addition, H3K4 methylation, a hallmark of active chromatin (64), was also observed predominantly in the maternal pronucleus rather than the paternal pronucleus. The antibody used in the present study recognizes the dimethylated form of H3K4. The presence of trimethylated H3K4 has been shown to be a prerequisite for active genes, and the role of dimethylated H3K4 in mammalian cells is not clear, but the presence of dimethylated H3K4 may correlate with a permissive state of chromatin in which genes are potentially active (59, 60). The observed H3K4 staining in the maternal pronucleus may mark regions that will become active shortly after the pronuclear stage. However, we did not observe a change in the histone H3 methylation pattern in the pronuclei in the mHR6A-deficient oocytes during fertilization.
Both mHR6A and mHR6B cDNAs encode proteins that are able to induce H2B ubiquitination and subsequent methylation of H3K4 in rad6 mutants (65), but there is a clear difference between ubiquitination of H2B in yeast and that in humans. In yeast, ubiquitinated H2B is most abundant and ubiquitinated H2A is undetectable, whereas in human somatic cells about 10% of H2A and only 1 to 1.5% of H2B is ubiquitinated (75). In spermatogenesis, no change in histone ubiquitination patterns has been detected in mHR6B knockout testis (5), but this result does not exclude the possibility of a low, but functionally significant, rate of H2A and/or H2B ubiquitination by mHR6B. Similarly, the possibility that the ubiquitin-conjugating enzyme mHR6A takes part in chromatin remodeling during early embryonic development through low rates of ubiquitination of H2A and/or H2B, which then might affect the methylation of histone H3 in a manner which has not been detected in the present experiments, cannot be excluded. Finally, it is important to note that the ubiquitination of H2B by RAD6 in yeast appears to be unrelated to RDB mechanisms (54) and that the role of mHR6A as a maternal factor for early embryonic development might involve a molecular mechanism of action outside the context of DNA repair.
Acknowledgments
We thank Marja Ooms and Jan Vreeburg for help with and advice on testis parameters and histology and for valuable discussions. Zu-Wen Sun and C. David Allis (University of Virginia, Charlottesville, Va.) are acknowledged for kindly providing us with antibodies against dimethylated forms of H3K4 and H3K9.
This research was supported by the Dutch Science Foundation (NWO) through GB-MW (Medical Sciences) and by the Dutch Cancer Society (grants EUR 92-118 and EUR 99-2003).
REFERENCES
- 1.Adenot, P. G., Y. Mercier, J. P. Renard, and E. M. Thompson. 1997. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development 124:4615-4625. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, F., D. M. Worrad, and R. M. Schultz. 1997. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev. Biol. 181:296-307. [DOI] [PubMed] [Google Scholar]
- 3.Arney, K. L., S. Bao, A. J. Bannister, T. Kouzarides, and M. A. Surani. 2002. Histone methylation defines epigenetic asymmetry in the mouse zygote. Int. J. Dev. Biol. 46:317-320. [PubMed] [Google Scholar]
- 4.Auffray, C., and F. Rougeon. 1980. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur. J. Biochem. 107:303-314. [DOI] [PubMed] [Google Scholar]
- 5.Baarends, W. M., J. W. Hoogerbrugge, H. P. Roest, M. Ooms, J. Vreeburg, J. H. Hoeijmakers, and J. A. Grootegoed. 1999. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev. Biol. 207:322-333. [DOI] [PubMed] [Google Scholar]
- 6.Baarends, W. M., E. Wassenaar, J. W. Hoogerbrugge, G. van Cappellen, H. P. Roest, J. Vreeburg, M. Ooms, J. H. Hoeijmakers, and J. A. Grootegoed. 2003. Loss of HR6B ubiquitin-conjugating activity results in damaged synaptonemal complex structure and increased crossing-over frequency during the male meiotic prophase. Mol. Cell. Biol. 23:1151-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailly, V., J. Lamb, P. Sung, S. Prakash, and L. Prakash. 1994. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 8:811-820. [DOI] [PubMed] [Google Scholar]
- 8.Bemark, M., A. A. Khamlichi, S. L. Davies, and M. S. Neuberger. 2000. Disruption of mouse polymerase zeta (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr. Biol. 10:1213-1216. [DOI] [PubMed] [Google Scholar]
- 9.Bradley, A. 1987. Production and analysis of chimeric mice, p. 113-151. In E. J. Robertson (ed.), Teratocarcinomas and embryonic stem cells: a practical approach. IRL Press, Oxford, United Kingdom.
- 10.Briggs, S. D., M. Bryk, B. D. Strahl, W. L. Cheung, J. K. Davie, S. Y. Dent, F. Winston, and C. D. Allis. 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15:3286-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briggs, S. D., T. Xiao, Z. W. Sun, J. A. Caldwell, J. Shabanowitz, D. F. Hunt, C. D. Allis, and B. D. Strahl. 2002. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418:498. [DOI] [PubMed] [Google Scholar]
- 12.Christians, E., A. A. Davis, S. D. Thomas, and I. J. Benjamin. 2000. Maternal effect of Hsf1 on reproductive success. Nature 407:693-694. [DOI] [PubMed] [Google Scholar]
- 13.Cowell, I. G., R. Aucott, S. K. Mahadevaiah, P. S. Burgoyne, N. Huskisson, S. Bongiorni, G. Prantera, L. Fanti, S. Pimpinelli, R. Wu, D. M. Gilbert, W. Shi, R. Fundele, H. Morrison, P. Jeppesen, and P. B. Singh. 2002. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma 111:22-36. [DOI] [PubMed] [Google Scholar]
- 14.Cox, B. S., and J. M. Parry. 1968. The isolation, genetics and survival characteristics of ultraviolet light-sensitive mutants in yeast. Mutat. Res. 6:37-55. [DOI] [PubMed] [Google Scholar]
- 15.Dohmen, R. J., K. Madura, B. Bartel, and A. Varshavsky. 1991. The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc. Natl. Acad. Sci. USA 88:7351-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dover, J., J. Schneider, M. A. Tawiah-Boateng, A. Wood, K. Dean, M. Johnston, and A. Shilatifard. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277:28368-28371. [DOI] [PubMed] [Google Scholar]
- 17.Esposito, G., I. Godindagger, U. Klein, M. L. Yaspo, A. Cumano, and K. Rajewsky. 2000. Disruption of the Rev3l-encoded catalytic subunit of polymerase zeta in mice results in early embryonic lethality. Curr. Biol. 10:1221-1224. [DOI] [PubMed] [Google Scholar]
- 18.Fischle, W., Y. Wang, and C. D. Allis. 2003. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15:172-183. [DOI] [PubMed] [Google Scholar]
- 19.Friedberg, E. C., R. Wagner, and M. Radman. 2002. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science 296:1627-1630. [DOI] [PubMed] [Google Scholar]
- 20.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, D.C.
- 21.Gibbs, P. E., W. G. McGregor, V. M. Maher, P. Nisson, and C. W. Lawrence. 1998. A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase zeta. Proc. Natl. Acad. Sci. USA 95:6876-6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goddard, M. J., and H. P. Pratt. 1983. Control of events during early cleavage of the mouse embryo: an analysis of the ′2-cell block.' J. Embryol. Exp. Morphol. 73:111-133. [PubMed] [Google Scholar]
- 23.Hatakeyama, S., M. Yada, M. Matsumoto, N. Ishida, and K. I. Nakayama. 2001. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276:33111-33120. [DOI] [PubMed] [Google Scholar]
- 24.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 25.Hoege, C., B. Pfander, G. L. Moldovan, G. Pyrowolakis, and S. Jentsch. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135-141. [DOI] [PubMed] [Google Scholar]
- 26.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366-374. [DOI] [PubMed] [Google Scholar]
- 27.Hogan, B., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo. A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Huang, H., A. Kahana, D. E. Gottschling, L. Prakash, and S. W. Liebman. 1997. The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6693-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jentsch, S., J. P. McGrath, and A. Varshavsky. 1987. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature 329:131-134. [DOI] [PubMed] [Google Scholar]
- 30.Johnson, R. E., C. M. Kondratick, S. Prakash, and L. Prakash. 1999. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285:263-265. [DOI] [PubMed] [Google Scholar]
- 31.Koken, M., P. Reynolds, D. Bootsma, J. Hoeijmakers, S. Prakash, and L. Prakash. 1991. Dhr6, a Drosophila homolog of the yeast DNA-repair gene RAD6. Proc. Natl. Acad. Sci. USA 88:3832-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koken, M. H., J. W. Hoogerbrugge, I. Jasper-Dekker, J. de Wit, R. Willemsen, H. P. Roest, J. A. Grootegoed, and J. H. Hoeijmakers. 1996. Expression of the ubiquitin-conjugating DNA repair enzymes HHR6A and B suggests a role in spermatogenesis and chromatin modification. Dev. Biol. 173:119-132. [DOI] [PubMed] [Google Scholar]
- 33.Koken, M. H., P. Reynolds, I. Jaspers-Dekker, L. Prakash, S. Prakash, D. Bootsma, and J. H. Hoeijmakers. 1991. Structural and functional conservation of two human homologs of the yeast DNA repair gene RAD6. Proc. Natl. Acad. Sci. USA 88:8865-8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koken, M. H., E. M. Smit, I. Jaspers-Dekker, B. A. Oostra, A. Hagemeijer, D. Bootsma, and J. H. Hoeijmakers. 1992. Localization of two human homologs, HHR6A and HHR6B, of the yeast DNA repair gene RAD6 to chromosomes Xq24-q25 and 5q23-q31. Genomics 12:447-453. [DOI] [PubMed] [Google Scholar]
- 35.Kratzer, P. G., and V. M. Chapman. 1981. X chromosome reactivation in oocytes of Mus caroli. Proc. Natl. Acad. Sci. USA 78:3093-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon, Y. T., Y. Reiss, V. A. Fried, A. Hershko, J. K. Yoon, D. K. Gonda, P. Sangan, N. G. Copeland, N. A. Jenkins, and A. Varshavsky. 1998. The mouse and human genes encoding the recognition component of the N-end rule pathway. Proc. Natl. Acad. Sci. USA 95:7898-7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwon, Y. T., Z. Xia, I. V. Davydov, S. H. Lecker, and A. Varshavsky. 2001. Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3alpha) of the N-end rule pathway. Mol. Cell. Biol. 21:8007-8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lachner, M., and T. Jenuwein. 2002. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 14:286-298. [DOI] [PubMed] [Google Scholar]
- 39.Laird, P. W., A. Zijderveld, K. Linders, M. A. Rudnicki, R. Jaenisch, and A. Berns. 1991. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 19:4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawrence, C. 1994. The RAD6 DNA repair pathway in Saccharomyces cerevisiae: what does it do, and how does it do it? Bioessays 16:253-258. [DOI] [PubMed] [Google Scholar]
- 41.Lawrence, C. W., and R. Christensen. 1976. UV mutagenesis in radiation-sensitive strains of yeast. Genetics 82:207-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin, W., H. Xin, Y. Zhang, X. Wu, F. Yuan, and Z. Wang. 1999. The human REV1 gene codes for a DNA template-dependent dCMP transferase. Nucleic Acids Res. 27:4468-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marsh, J. L., M. Erfle, and E. J. Wykes. 1984. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene 32:481-485. [DOI] [PubMed] [Google Scholar]
- 44.Masutani, C., M. Araki, A. Yamada, R. Kusumoto, T. Nogimori, T. Maekawa, S. Iwai, and F. Hanaoka. 1999. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 18:3491-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayer, W., A. Niveleau, J. Walter, R. Fundele, and T. Haaf. 2000. Demethylation of the zygotic paternal genome. Nature 403:501-502. [DOI] [PubMed] [Google Scholar]
- 46.Montelone, B. A., S. Prakash, and L. Prakash. 1981. Recombination and mutagenesis in rad6 mutants of Saccharomyces cerevisiae: evidence for multiple functions of the RAD6 gene. Mol. Gen. Genet. 184:410-415. [DOI] [PubMed] [Google Scholar]
- 47.Murakumo, Y., T. Roth, H. Ishii, D. Rasio, S. Numata, C. M. Croce, and R. Fishel. 2000. A human REV7 homolog that interacts with the polymerase zeta catalytic subunit hREV3 and the spindle assembly checkpoint protein hMAD2. J. Biol. Chem. 275:4391-4397. [DOI] [PubMed] [Google Scholar]
- 48.Ng, H. H., R. M. Xu, Y. Zhang, and K. Struhl. 2002. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277:34655-34657. [DOI] [PubMed] [Google Scholar]
- 49.O'Farrell, P. Z., H. M. Goodman, and P. H. O'Farrell. 1977. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell 12:1133-1141. [DOI] [PubMed] [Google Scholar]
- 50.Peters, A. H., D. O'Carroll, H. Scherthan, K. Mechtler, S. Sauer, C. Schofer, K. Weipoltshammer, M. Pagani, M. Lachner, A. Kohlmaier, S. Opravil, M. Doyle, M. Sibilia, and T. Jenuwein. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323-337. [DOI] [PubMed] [Google Scholar]
- 51.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 52.Picologlou, S., N. Brown, and S. W. Liebman. 1990. Mutations in RAD6, a yeast gene encoding a ubiquitin-conjugating enzyme, stimulate retrotransposition. Mol. Cell. Biol. 10:1017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reynolds, P., M. H. Koken, J. H. Hoeijmakers, S. Prakash, and L. Prakash. 1990. The rhp6+ gene of Schizosaccharomyces pombe: a structural and functional homolog of the RAD6 gene from the distantly related yeast Saccharomyces cerevisiae. EMBO J. 9:1423-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robzyk, K., J. Recht, and M. A. Osley. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287:501-504. [DOI] [PubMed] [Google Scholar]
- 55.Roest, H. P., J. van Klaveren, J. de Wit, C. G. van Gurp, M. H. Koken, M. Vermey, J. H. van Roijen, J. W. Hoogerbrugge, J. T. Vreeburg, W. M. Baarends, D. Bootsma, J. A. Grootegoed, and J. H. Hoeijmakers. 1996. Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell 86:799-810. [DOI] [PubMed] [Google Scholar]
- 56.Roller, M. L., A. C. Lossie, M. H. Koken, E. M. Smit, A. Hagemeijer, and S. A. Camper. 1995. Localization of sequences related to the human RAD6 DNA repair gene on mouse chromosomes 11 and 13. Mamm. Genome 6:305-306. [DOI] [PubMed] [Google Scholar]
- 57.San-Segundo, P. A., and G. S. Roeder. 2000. Role for the silencing protein Dot1 in meiotic checkpoint control. Mol. Biol. Cell 11:3601-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santos, F., B. Hendrich, W. Reik, and W. Dean. 2002. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 241:172-182. [DOI] [PubMed] [Google Scholar]
- 59.Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. Emre, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407-411. [DOI] [PubMed] [Google Scholar]
- 60.Schneider, R., A. J. Bannister, F. A. Myers, A. W. Thorne, C. Crane-Robinson, and T. Kouzarides. 2004. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 6:73-77. [DOI] [PubMed] [Google Scholar]
- 61.Sijbers, A. M., P. J. van der Spek, H. Odijk, J. van den Berg, M. van Duin, A. Westerveld, N. G. Jaspers, D. Bootsma, and J. H. Hoeijmakers. 1996. Mutational analysis of the human nucleotide excision repair gene ERCC1. Nucleic Acids Res. 24:3370-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Solari, A. J. 1974. The behavior of the XY pair in mammals. Int. Rev. Cytol. 38:273-317. [DOI] [PubMed] [Google Scholar]
- 63.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 64.Strahl, B. D., R. Ohba, R. G. Cook, and C. D. Allis. 1999. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. USA 96:14967-14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-108. [DOI] [PubMed] [Google Scholar]
- 66.Sung, P., S. Prakash, and L. Prakash. 1990. Mutation of cysteine-88 in the Saccharomyces cerevisiae RAD6 protein abolishes its ubiquitin-conjugating activity and its various biological functions. Proc. Natl. Acad. Sci. USA 87:2695-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suzumori, N., K. H. Burns, W. Yan, and M. M. Matzuk. 2003. RFPL4 interacts with oocyte proteins of the ubiquitin-proteasome degradation pathway. Proc. Natl. Acad. Sci. USA 100:550-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tateishi, S., Y. Sakuraba, S. Masuyama, H. Inoue, and M. Yamaizumi. 2000. Dysfunction of human Rad18 results in defective postreplication repair and hypersensitivity to multiple mutagens. Proc. Natl. Acad. Sci. USA 97:7927-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thomas, K. R., and M. R. Capecchi. 1987. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51:503-512. [DOI] [PubMed] [Google Scholar]
- 70.Tong, Z. B., L. Gold, K. E. Pfeifer, H. Dorward, E. Lee, C. A. Bondy, J. Dean, and L. M. Nelson. 2000. Mater, a maternal effect gene required for early embryonic development in mice. Nat. Genet. 26:267-268. [DOI] [PubMed] [Google Scholar]
- 71.van der Horst, G. T., H. van Steeg, R. J. Berg, A. J. van Gool, J. de Wit, G. Weeda, H. Morreau, R. B. Beems, C. F. van Kreijl, F. R. de Gruijl, D. Bootsma, and J. H. Hoeijmakers. 1997. Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer predisposition. Cell 89:425-435. [DOI] [PubMed] [Google Scholar]
- 72.van der Laan, R., H. P. Roest, J. W. Hoogerbrugge, E. M. Smit, R. Slater, W. M. Baarends, J. H. Hoeijmakers, and J. A. Grootegoed. 2000. Characterization of mRAD18Sc, a mouse homolog of the yeast postreplication repair gene RAD18. Genomics 69:86-94. [DOI] [PubMed] [Google Scholar]
- 73.Van Sloun, P. P., I. Varlet, E. Sonneveld, J. J. Boei, R. J. Romeijn, J. C. Eeken, and N. De Wind. 2002. Involvement of mouse Rev3 in tolerance of endogenous and exogenous DNA damage. Mol. Cell. Biol. 22:2159-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang, B. B., K. J. Hayenga, D. G. Payan, and J. M. Fisher. 1996. Identification of a nuclear-specific cyclophilin which interacts with the proteinase inhibitor eglin c. Biochem J. 314:313-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.West, M. H., and W. M. Bonner. 1980. Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res. 8:4671-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wittschieben, J., M. K. Shivji, E. Lalani, M. A. Jacobs, F. Marini, P. J. Gearhart, I. Rosewell, G. Stamp, and R. D. Wood. 2000. Disruption of the developmentally regulated Rev3l gene causes embryonic lethality. Curr. Biol. 10:1217-1220. [DOI] [PubMed] [Google Scholar]
- 77.Wood, A., N. J. Krogan, J. Dover, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, A. Golshani, Y. Zhang, J. F. Greenblatt, M. Johnston, and A. Shilatifard. 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11:267-274. [DOI] [PubMed] [Google Scholar]
- 78.Xiao, W., S. L. Lin, S. Broomfield, B. L. Chow, and Y. F. Wei. 1998. The products of the yeast MMS2 and two human homologs (hMMS2 and CROC-1) define a structurally and functionally conserved Ubc-like protein family. Nucleic Acids Res. 26:3908-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xin, H., W. Lin, W. Sumanasekera, Y. Zhang, X. Wu, and Z. Wang. 2000. The human RAD18 gene product interacts with HHR6A and HHR6B. Nucleic Acids Res. 28:2847-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yamaguchi, T., N. S. Kim, S. Sekine, H. Seino, F. Osaka, F. Yamao, and S. Kato. 1996. Cloning and expression of cDNA encoding a human ubiquitin-conjugating enzyme similar to the Drosophila bendless gene product. J. Biochem. (Tokyo) 120:494-497. [DOI] [PubMed] [Google Scholar]
- 81.Zhou, X. Y., H. Morreau, R. Rottier, D. Davis, E. Bonten, N. Gillemans, D. Wenger, F. G. Grosveld, P. Doherty, K. Suzuki, et al. 1995. Mouse model for the lysosomal disorder galactosialidosis and correction of the phenotype with overexpressing erythroid precursor cells. Genes Dev. 9:2623-2634. [DOI] [PubMed] [Google Scholar]