Abstract
Escherichia coli, including the closely related genus Shigella, is a highly diverse species in terms of genome structure. Comparative genomic hybridization (CGH) microarray analysis was used to compare the gene content of E. coli K-12 with the gene contents of pathogenic strains. Missing genes in a pathogen were detected on a microarray slide spotted with 4,071 open reading frames (ORFs) of W3110, a commonly used wild-type K-12 strain. For 22 strains subjected to the CGH microarray analyses 1,424 ORFs were found to be absent in at least one strain. The common backbone of the E. coli genome was estimated to contain about 2,800 ORFs. The mosaic distribution of absent regions indicated that the genomes of pathogenic strains were highly diversified becasue of insertions and deletions. Prophages, cell envelope genes, transporter genes, and regulator genes in the K-12 genome often were not present in pathogens. The gene contents of the strains tested were recognized as a matrix for a neighbor-joining analysis. The phylogenic tree obtained was consistent with the results of previous studies. However, unique relationships between enteroinvasive strains and Shigella, uropathogenic, and some enteropathogenic strains were suggested by the results of this study. The data demonstrated that the CGH microarray technique is useful not only for genomic comparisons but also for phylogenic analysis of E. coli at the strain level.
Escherichia coli strains are divided into several disease phenotypes, including nonpathogenic E. coli, enterohemorragic E. coli (EHEC), enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), and urinary tract infectious or uropathogenic E. coli (UPEC) (6). In addition, the strains belonging to the genus Shigella are recognized as E. coil strains because they have many genetic and phenotypic similarities to E. coli (27). Following determination of the complete genome sequence of nonpathogenic E. coli K-12 laboratory strain MG1655 in 1997 (2), the genomes of the two pathogenic strains with the same serotype (O157:H7) were also sequenced (7, 23). A genomic comparison of strain MG1655 and the O157:H7 strains revealed that each genome contained specific genomic regions designated strain-specific islands or loops (7, 23). Recently, the genome sequence of UPEC strain CFT073 was reported to also have unique chromosome regions that are not present in other E. coli genomes that have been sequenced (35). Similar results were obtained in a genomic study of the bacillary dysentery-causing strain Shigella flexneri 2a (12, 34). These results revealed the extensive divergence of the genomes of E. coli strains and indicated that the unique phenotype of each strain should reflect its genomic content. Although whole-genome sequencing is definitely a powerful method for genetics, it is still laborious and expensive. Recently, comparative genomic hybridization (CGH) has been used to facilitate comparisons of unsequenced bacterial genomes in order to look for characteristic genes or chromosomal regions related to unique phenotypes (1, 4, 9, 10, 25, 30).
The first report of a genomic comparison of E. coli strains in which CGH was used was published in 2000; five strains were used in this analysis (19). Later, the genomic contents of pathogens, mainly UPEC strains, and commensal isolates were analyzed (5). In the latter report, a “pathoarray” with 536 pathogenic genes was used in addition to the commercial K-12 DNA array on a nylon membrane. However, most of sequenced strains were not included in the study; the only exception was MG1655, which was used as a reference. Here we describe the results of a genomic comparison of 22 pathogenic E. coli and Shigella strains based on a CGH analysis performed with Stanford-type DNA microarray slides with K-12 genes. A CGH microarray analysis of the O157:H7 Sakai strain (7) and K-12 was performed first, and the results were revised by using the data from an in silico sequence comparison. The qualified threshold determined in this control experiment was used to identify gene deletions of other strains. In the 22 pathogenic E. coli strains 1,424 open reading frames (ORFs) were found to be absent in at least one strain. Detailed characteristics of conserved and nonconserved ORFs are described below.
By using CGH analysis, chromosomal regions that were replaced by pathogenic islands a posteriori (7, 23, 35) and pathoadaptive deletions (16, 17, 32) could be identified as absent on the K-12 DNA arrays. The results of genomic sequencing and comparative genomics analyses suggested that the evolution of pathogenic bacteria is mainly due to large chromosomal alterations, such as horizontal transfers or deletions (7, 12, 23, 35). It was expected that the pattern of deletions detected on the K-12 DNA arrays could mirror the evolution of the genome of a strain, even if it is a pathogen. The gene contents of the strains tested were used as a matrix for the neighbor-joining method for a phylogenic analysis. The dendrogram obtained was consistent with the results of the previous studies in which multilocus enzyme electrophoresis or a sequence comparison of conserved genes was used (6, 26, 27, 28). This finding demonstrated that K-12 microarrays are useful for phylogenic analysis of pathogenic strains of E. coli.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All of the E. coli and Shigella strains used in this study are described in Table 1. Pathogenic strains were obtained from the Research Institute for Microbial Diseases of Osaka University (Osaka, Japan) and the Institute of Medical Science of the University of Tokyo (Tokyo, Japan). Bacterial strains were grown to the stationary phase at 30°C in Luria broth. Genomic DNA was purified from the overnight cultures by using a DNeasy tissue kit (Qiagen K. K., Tokyo, Japan) according to the manufacturer's instructions.
TABLE 1.
Bacterial strains used in this study and numbers of absent W3110 ORFs
| Strain | Abbreviation | Serotype | Sourcea | No. of absent ORFsb |
|---|---|---|---|---|
| E. coli K-12 W3110 | W3110 | NAIST | 0 | |
| E. coli EHEC O157 Sakai | O157 Sakai | O157:H7 | RIMD | 425 |
| E. coli EHEC O26 | EHEC | O26 | RIMD | 293 |
| E. coli EPEC B171 | EPEC-1 | O111:NM | RIMD | 401 |
| E. coli EPEC E2348/69 | EPEC-2 | O127:H6 | RIMD | 457 |
| E. coli EPEC 5513-51 | EPEC-3 | O55:H6 | RIMD | 602 |
| E. coli EPEC 4394-57 | EPEC-4 | O114:NM | RIMD | 218 |
| E. coli EPEC 1929-55 | EPEC-5 | O126:NM | RIMD | 338 |
| E. coli EPEC 1157-54 | EPEC-6 | O119:H6 | RIMD | 578 |
| E. coli EPEC 1181-83 | EPEC-7 | O142:H6 | RIMD | 698 |
| E. coli ETEC H10407 | ETEC-1 | O78:H11 | RIMD | 230 |
| E. coli ETEC 31-10 | ETEC-2 | O25:H− | RIMD | 286 |
| E. coli EIEC 931-78 | EIEC-1 | O124 | RIMD | 382 |
| E. coli EIEC 14185-83HU | EIEC-2 | O28ac | RIMD | 522 |
| E. coli EIEC 127-82FAV | EIEC-3 | O29 | RIMD | 561 |
| E. coli EIEC 282-83FAV | EIEC-4 | O136 | RIMD | 493 |
| E. coli UPEC Z42 | UPEC-1 | O2:H6 | RIMD | 598 |
| E. coli UPEC C72 | UPEC-2 | O46:H52 | RIMD | 595 |
| E. coli UPEC P17 | UPEC-3 | O129:NT | RIMD | 617 |
| E. coli REPEC REPEC-1 | REPEC | O103:K−:H6 | RIMD | 290 |
| S. flexneri 2a YSH6000 | SF | IMS | 716 | |
| S. boydii IID627 | SB | IMS | 533 | |
| S. sonnei phaseI IID969 | SS | IMS | 613 |
NAIST, Nara Institute of Science and Technology, Nara, Japan; RIMD, Research Institute for Microbial Diseases, Osaka University, Osaka, Japan; IMS, Laboratory of Culture Collection, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
Altogether, 1,424 ORFs were absent in at least one strain, and 96 ORFs were commonly absent in 22 strains.
Microarray, labeling, and hybridization.
DNA microarray slides were supplied by Takara Bio Inc. (Ohtsu, Japan). Full-length fragments of 4,071 (92.7%) of the 4,390 annotated ORFs of K-12 strain W3110 (http://ecoli.aist-nara.ac.jp/) were amplified by PCR and spotted on the slides. Each ORF was spotted in duplicate. Genomic DNA was labeled with FluoroLink Cy3- or Cy5-dCTP (Amersham Biosciences Corp., Piscataway, N.J.) by using the method described by Pollack et al. (24) and the components of the BioPrime DNA labeling system (Invitrogen Corp., Carlsbad, Calif.). Two micrograms of genomic DNA was labeled by using 15 μg of random octamers, 40 U of the Klenow fragment, and 3 nmol of Cy3- or Cy5-dCTP at 37°C for 2 h. Unincorporated fluorescent nucleotides were removed by using CentriCep spin columns (Princeton Separations Inc., Adelphia, N.J.), and probes were purified by phenol-chloroform extraction and ethanol precipitation. Precipitated DNA probes were dried and finally resuspended in 8 μl of sterilized distilled water. Hybridization was conducted essentially by the method described by Oshima et al. (21). Twenty-five microliters of the prehybridization buffer (4× SSC, 0.2% sodium dodecyl sulfate [SDS], 5× Denhardt's solution, 100 ng of salmon sperm DNA per μl [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) was added to the microarray under a coverslip, and prehybridization was performed at 65°C for 1 h. Six microliters of labeled probe from each genomic DNA was added to 30 μl (final volume) of hybridization buffer (4× SSC, 0.2% SDS, 5× Denhardt's solution, 100 ng of salmon sperm DNA per μl) and was denatured by heating at 95°C for 2 min. Prehybridized slides were washed with 2× SSC, dried, and subjected to hybridization with the hybridization buffer at 65°C for 16 h. Slides were washed at 60°C with 2× SSC for 5 min and then at 60°C with 0.2× SSC containing 0.1% SDS for 5 min and finally at room temperature with 0.2× SSC for 2 min. The last step was conducted twice. The slides were immediately dried and scanned for fluorescence intensity by using a GenePix 4000B microarray scanner (Axon Instruments, Union City, Calif.), and the results were recorded in 16-bit multi-image TIFF files. Competitive hybridization was done twice for one strain. In the first experiment, the W3110 reference DNA and the sample DNA from a pathogen were labeled with Cy3 and Cy5, respectively. In the second hybridization, the dyes for labeling were swapped.
Data analysis.
The signal intensity of each spot in the microarray was quantified by using the GenePix Pro 4.0 (Axon Instruments) software. Additional data analyses were conducted by using the computer software programs Microsoft Excel and GeneSpring 5.0.2 (Silicon Genetics, Redwood, Calif.). The local background value was subtracted from the intensity of each spot. The mean of the signal intensities of the control spots hybridized with labeled W3110 genome DNA in each experiment was calculated. The human β-actin gene was used for the controls. The spots that showed intensity with labeled W3110 DNA that was lower than the mean value of the control spots were excluded from further analysis. Sample/reference (W3110) ratios of signal intensity were calculated and were transformed to logarithm base 2. The ratios were normalized by using the median of log2 ratios of all spots as zero. To determine the final value for each ORF tested, the median value was calculated from four log2 ratios obtained from two DNA microarray slides used in two dye-swapping experiments. Thirty-seven ORFs that gave invalid results for more than 5 of the 22 strains tested were excluded from further analysis. ORFs were considered absent if the final ratio of signal intensities was less than −1 on the log2 scale. ORFs whose final ratios were greater than 0.8 on the log2 scale were recorded as putatively duplicated in the genomes of pathogenic strains. We used the ERGO database (Integrated Genomics, Chicago, Ill.) to check functions of ORFs of E. coli (22). Final data sets are available at the website http://biowonderland.com/BioWorld/BiseiGenoDB/Image/index01/Genom.pdf and in the supplemental material.
Genomic comparison of the W3110 and O157 Sakai strains in silico.
Each nucleotide sequence of the ORFs assigned in the genome of strain W3110 (http://ecoli.aist-nara.ac.jp/) was used as a query for a homology search with BLASTN against the genome sequence of the O157 Sakai strain (accession number NC_002695) with the default parameters used in the National Center for Biotechnology Information database. W3110 ORFs were designated by using numbers starting from JW0001 for the thrA gene in the clockwise direction. The region with the highest score for each query was retrieved and classified by using the H value, which was calculated as follows: [(length of highest-score region) × (identities of hit shown in BLASTN)]/(length of query sequence). If there was no sequence with a BLASTN E value less than 0.01, the query ORF was judged to be not present in the O157 Sakai genome, and its H value was zero. When artM (JW0845 corresponding to b0861 in reference 2) was used as the query, BLASTN analysis showed that a 650-nucleotide sequence of O157 Sakai was the region with the highest score, and the level of identity was 0.98 (638/650). The query was 669 bp long. The H value of artM should be 0.95 (650 × 0.98/669). In contrast, queries for genes that were probably absent gave low H values. The H value indicated how much the corresponding sequence of O157 Sakai resembled the W3110 query ORF in terms of length and sequence identity.
Phylogenic analysis.
The gene contents of the strains tested were described by using a two-character matrix (0, absent; 1, present [including putative duplication]) in the order based on the clockwise appearance on the W3110 genome. If there were invalid data for a particular gene for one or more strains, all characters representing the presence of the gene were eliminated from the matrix. The matrix was incorporated into the PAUP software program (http://paup.csit.fsu.edu/). The neighbor-joining method was used to construct a phylogenic tree. A bootstrap analysis was conducted to determine the statistical stability of each node, and the number of bootstrap replicates was 1,000. A high-resolution image of the dendrogram was generated by using the TreeView 1.6.6 software (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
RESULTS
CGH microarray analysis and in silico genomic comparison of E. coli O157 Sakai and W3110.
The genomes of two E. coli strains, W3110 (http://ecoli.aist-nara.ac.jp/) and O157 Sakai (7), were sequenced in Japan. W3110 is a commonly used laboratory strain and has a large inversion compared to the original K-12 isolate and MG1655 (11). The genomic contents of W3110 and O157 Sakai were compared by CGH as described in Materials and Methods. The genomic sequences of the two strains were also compared in silico. W3110 ORFs that were not present in O157 Sakai were predicted by a BLASTN homology search, and the possibilities of gene loss were expressed by H values (see Materials and Methods). The results are summarized in Table 2. The majority of W3110 ORFs were detected in the genome of O157 Sakai with H values greater than 0.9 (group I). A second group consisted of 412 ORFs (9.4%) with H values less than 0.1 (group II). The other ORFs were almost equally distributed in classes with H values ranging from 0.9 to 0.1. An H value greater than 0.9 indicated that a group I ORF should have an almost identical homologue in the O157 Sakai genome. On the other hand, the group II ORFs were probably not present in the pathogen. However, it was difficult to evaluate the presence of a counterpart of an ORF that had an H value in the middle range. Gene splitting and a fusion event in O157 Sakai would lower the H value for the query, since split or fused genes could give only a partial sequence of the original gene used as the query in the BLASTN search and could decrease the H value (see Materials and Methods). It was hard to predict how these chimeric genes would hybridize to K-12 gene fragments spotted on the slide. Table 2 shows the average ratios of signal intensities from the results of the CGH analysis for the H-value groups. In previous studies, when the ratio of the signal intensities for a spot was less than −1 on the log2 scale, the corresponding gene was considered absent (1, 9, 10, 30). This threshold definitely divided groups I and II in Table 2, although there was still uncertainty about the presence of split or fused genes with the threshold. We expected that misclassification of genes would be negligible in terms of the whole genome analysis, since the number of genes with H values in the middle range was relatively small (Table 2). The accuracy of classification into groups I and II was checked. In group I, 3 of 3,528 ORFs had a signal ratio less than −1 on the log2 scale. These ORFs were JW0690 (not annotated in MG1655), JW2373 (b2376), and JW3560 (not annotated in MG1655). These three genes were relatively short (420, 276, and 213 bp, respectively), but they were highly homologous to the corresponding ORFs in the O157 Sakai genome. Shorter DNAs were also spotted on the microarray and gave reasonable signal ratios based on the H values of the corresponding genes. The reason why these genes with high H values produced signals similar to absent signals was unclear. In group II, only JW0542 (b0553, nmpC) had a signal ratio greater than −1 on the log2 scale. JW0542 was found to consist of short sequences that were highly homologous to portions of the O157 Sakai genome (data not shown). This fact could explain the unexpected higher ratio of signal intensities to some extent. Although there were genes with unexpected signal ratios in groups I and II, the numbers of such genes were relatively low (3 of 3,528 genes and 1 of 345 genes, respectively). On the other hand, we detected all of the previously reported absent genes (genes for utilization of 2-methylphenylamine, xanthosine, d-galactonate, l-idonate, glycorate, and short-chain fatty acids; genes for the general secretion pathway; genes for the citrate-dependent iron transport system; and genes for restriction-modification) (7). These results suggested that the threshold was sufficient to identify conserved genes (group I) and lost genes (group II) in the O157 Sakai genome. The sequenced genomes of E. coli strains, including S. flexneri, have revealed that they share backbone regions with sequence identity to the K-12 genome (2, 7, 12, 23, 34, 35). Other E. coli strains should also have a similar genome structure with mosaic distribution of the backbone regions. This genome structure results in the bipolar distribution of genes according to their H values, as shown in Table 2. We supposed that the threshold (−1 for the log2 final ratio) could be used to distinguish between groups I and II, as far as the bipolar distribution of H values could be observed in sample genomes. Consequently, a threshold of −1 was used for CGH analysis of other E. coli and Shigella pathogens. In this study, genes were considered absent when the signal ratio was less than −1 on the log2 scale in the CGH analysis.
TABLE 2.
Distribution of ORFs of W3110 on DNA arrays and of different classes of signal ratios by H value
| H value | No. of ORFs
|
No. of ORFs at a signal ratio of:
|
Avg signal ratioa | |||
|---|---|---|---|---|---|---|
| W3110 | DNA array | ≤-1 | >-1 | Invalid | ||
| <0.1 | 412 | 347 | 344 | 1 | 2 | −4.79 |
| ≥0.1 and <0.3 | 46 | 31 | 30 | 1 | 0 | −3.14 |
| ≥0.3 and <0.5 | 42 | 36 | 23 | 12 | 1 | −1.82 |
| ≥0.5 and <0.7 | 51 | 40 | 12 | 28 | 0 | −0.80 |
| ≥0.7 and <0.9 | 75 | 56 | 13 | 42 | 1 | −0.67 |
| ≥0.9 | 3,764 | 3,561 | 3 | 3,525 | 33 | 0.03 |
| Total | 4,390 | 4,071 | 425 | 3,609 | 37 | −0.44 |
Average signal ratio of spots corresponding to all ORFs within each H-value range.
Overview of the microarray analysis.
The genomic contents of 22 pathogenic strains of E. coli and Shigella were analyzed by CGH by using the genome of nonpathogenic K-12 strain W3110 as the reference. The results are shown in Fig. 1 and Table 3. Final data sets for the CGH analysis are available at http://biowonderland.com/BioWorld/BiseiGenoDB/Image/index01/Genom.pdf and in the supplemental material. Of the 4,071 ORFs spotted on the microarray slides, 1,424 ORFs were considered absent in at least one of the pathogenic strains tested. These ORFs accounted for 32.4 and 35.0% of all ORFs annotated and spotted on the slides, respectively. After the ORFs on the microarrays with invalid data were excluded, the remaining 2,586 ORFs were probably conserved in all of the strains used in this study. These conserved ORFs included all 236 essential protein-encoding genes listed in the PEC database (http://www.shigen.nig.ac.jp/ecoli/pec/index.jsp) except three ORFs (firA/lpxD, rhoL, and yieQ) not spotted on the microarrays. The ratio of variable genes to conserved genes was higher than the ratio reported in the previous CGH study (1,165/3,100), in which a DNA macroarray membrane was used for noncompetitive hybridization of pathogenic E. coli genomes (5). The difference is discussed in detail below.
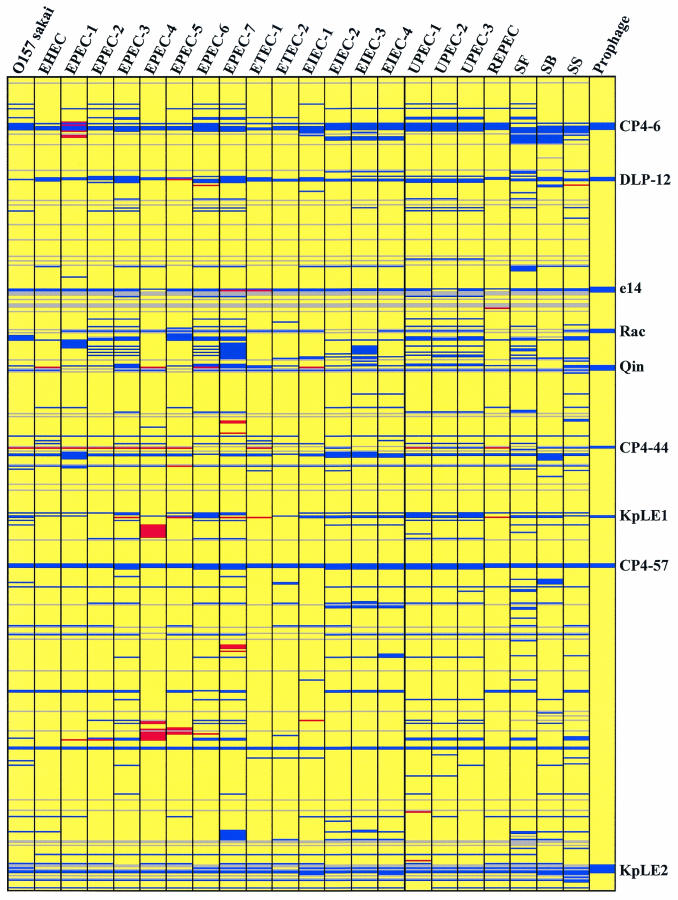
FIG. 1.
Genome composition based on CGH microarray data. Each row corresponds to a specific spot on the array, and the genes are arranged in order of JW number from JW0001 (b0002, thrA) at the top to JW4366 (b4403, lasT) at the bottom. The columns represent the strains analyzed; for an explanation of the abbreviations see Table 1. The colors indicate the status of the ORFs, as follows: blue, absent; yellow, present; red, putatively duplicated; grey, uncertain (not on the DNA array or invalid data). Nine prophage regions in the W3110 genome are indicated by blue bars in the Prophage column; the designations of these regions are indicated on the right.
TABLE 3.
Distribution of W3110 ORFs and identified absent ORFs by functional classes
| Categorya | No. of ORFs | No. of ORFs absent | % of absent ORFs |
|---|---|---|---|
| Amino acid metabolism | 136 | 32 | 23.5 |
| Biosynthesis of cofactors, pros- thetic groups, and carriers | 127 | 11 | 8.7 |
| Cell envelope | 195 | 73 | 37.4 |
| Cellular process | 105 | 27 | 25.7 |
| Central intermediary metabolism | 154 | 57 | 37.0 |
| Energy metabolism | 368 | 130 | 35.3 |
| Fatty acid and phospholipid metabolism | 60 | 7 | 11.7 |
| Nucleotide metabolism | 117 | 16 | 13.7 |
| Regulatory functions | 107 | 29 | 27.1 |
| Replication | 90 | 12 | 13.3 |
| Transport and binding protein | 369 | 131 | 35.5 |
| Translation | 152 | 7 | 4.6 |
| Transcription | 48 | 8 | 16.7 |
| Other categories | 338 | 97 | 28.7 |
| Hypothetical | 2,024 | 787 | 38.9 |
| Total | 4,390 | 1,424 | 32.4 |
Functional categories of W3110 ORFs were based on information from the GanoBase 4.0 database (http://ecoli.aist-nara.ac.jp/gb4/search/function/orffunc.php).
There were differences in the numbers of absent ORFs in different strains. The numbers ranged from 724 in S. flexneri to 221 in EPEC-4 (see Table 1 for strain abbreviations). The mosaic distribution of the absent regions is shown in Fig. 1. These results confirmed the previously reported diversity of the genomes of E. coli strains, including Shigella (2, 7, 12, 19, 23, 35).
There were 96 W3110-specific ORFs that were absent in all 22 other pathogenic strains. At least 10 prophages and phage-like regions were identified in MG1655 (2, 7). W3110 was found to have 9 of the 10 regions corresponding to prophages of MG1655, although several differences at the sequence level and in the gene contents were detected (Fig. 1). As expected, W3110-specific ORFs were frequently found in these prophage regions (75 ORFs). In the CP4-6 region 23 of 26 ORFs spotted were commonly absent. All 26 ORFs of the CP4-57 region were absent in all pathogens tested. These results suggested that these two prophages were specifically acquired only in the K-12 genome.
ORFs of W3110 were functionally categorized in the GenoBase 4.0 database (http://e.coli.aist-nara.ac.jp/). The proportions of the absent W3110 ORFs in the functional category are shown in Table 3. It was relatively hard to find absent genes encoding fundamental cellular functions, replication, translation, and transcription. Genes for fatty acid and phospholipid metabolism, nucleotide metabolism, and biosynthesis of cofactors, prosthetic groups, and carriers were also conserved. In contrast, over 30% of the ORFs in the following five categories were absent: cell envelope, central intermediary metabolism, energy metabolism, transport and binding proteins, and hypothetical proteins. Regulatory genes were also frequently absent (27.1%). These results suggested that W3110 actively acquired accessory genes to respond new environments. It is probable that pathogenic strains also recruited accessory genes for adaptation, including genes for pathogenicity, at the loci considered absent.
Transport and metabolism of carbohydrates.
Variations in the carbohydrate utilization pattern are used for identification of bacteria (3). As mentioned above, genes for transport and binding proteins were frequently absent in pathogenic strains. Loss of transporters should affect utilization of carbon sources and make it possible to identify strains on the basis of their carbohydrate utilization profiles. E. coli is known to utilize various carbohydrates, such as oligosaccharides, sugar alcohols, glycosides, etc. (14). The distribution of 24 gene clusters for carbohydrate transport and metabolism is summarized in Table 4. The substrates of the clusters were diverse and included sugar alcohols (mtl cluster for mannitol), glycosides (mgl cluster for β-d-galactoside), and amino sugars (aga cluster for N-acetylgalactosamine and galactosamine). In general, E. coli utilizes lactose, but Shigella does not utilize this compound (8). The results of CGH analysis indicated that most of the E. coli strains had the lac operon; the only exceptions were EHEC strains, in which only lacA was not present. None of the three Shigella strains contained lacY. This genetic profile for the lac operon contents explained the difference in lactose utilization between E. coli and Shigella well. l-Arabinose, maltose, d-mannitol, d-mannose, and trehalose are good carbon sources for both E. coli and Shigella. Genes for these substrates (ara, mal, mtl, man, tre) were conserved in almost all strains with few exceptions. EIEC-2 was the only strain that did not contain the mal operon (Table 3). EIEC-2 seemed to be deficient for maltose utilization. Also, a few strains did not have genes for glucitol utilization (EPEC-2, EPEC-3, and EPEC-7), d-allose utilization (O157 Sakai, EHEC, EPEC-7, EIEC-2, EIEC-3, EIEC-4, SF, SB, and SS), and melibiose utilization (EPEC-3, EPEC-6, EPEC-7, UPEC-1, UPEC-2, and UPEC-3). These sugars would be useful for distinguishing or tracking the strains.
TABLE 4.
Distribution of carbohydrate utilization gene clusters of W3110 in 22 pathogenic strains
| Gene clustera | Major substrate | O157 Sakai | EHEC | EPEC-1 | EPEC-2 | EPEC-3 | EPEC-4 | EPEC-5 | EPEC-6 | EPEC-7 | ETEC-1 | ETEC-2 | EIEC-1 | EIEC-2 | EIEC-3 | EIEC-4 | UPEC-1 | UPEC-2 | UPEC-3 | REPEC | SF | SB | SS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| araDABC | l-Arabinose | +b | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| lacAYZ1 | Lactose | + | [−] | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | [−] |
| clibGFRACB | N,′N-Diacetyl- chitobiose | [−] | + | + | + | + | + | + | + | + | + | [−] | + | [−] | + | + | [−] | + | + | + | + | + | + |
| manXYZ | d-Mannose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| araHGF | l-Arabinose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| mglCBA, galS | β-d-Galactoside | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | [−] | + |
| srlABD, gutM, srlR | Glucitol | + | + | + | [−] | − | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + |
| ascGFB | Disachharide | + | + | + | − | − | + | + | + | − | + | + | + | + | + | + | + | + | − | + | [−] | [−] | + |
| fucOAPIKUR | l-Fucose | + | + | + | + | + | + | + | + | + | + | + | + | [−] | [−] | [−] | + | + | + | + | + | + | + |
| cmtAB | Uncertain | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + |
| agaRZVWASYBCDI | N-Acetylgalac- tosamine | + | + | + | + | + | + | + | + | + | + | + | + | [−] | [−] | − | + | + | + | + | [−] | + | − |
| xylBAFGHR | d-Xylose | + | + | + | + | + | + | + | + | + | [−] | + | + | + | + | + | + | + | + | + | + | + | [−] |
| imlADR | Mannitol | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| glvGBC | Uncertain | + | + | + | + | − | + | + | − | − | + | + | + | + | + | + | − | − | − | + | + | + | + |
| dgoTAKR | d-Galactonate | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| bglBFG | Disaccharide | − | + | + | + | + | + | + | + | + | + | + | [−] | − | + | + | + | + | + | + | + | − | + |
| frvRXBA | Uncertain | + | + | + | + | [−] | + | + | − | + | + | + | + | − | + | [−] | − | − | − | + | [−] | + | +c |
| rhaDABSRT | l-Rhamnose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| malGFEKLM | Maltose | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + |
| alsKECAB, rpiB | d-Allose | − | − | + | + | + | + | + | + | − | + | + | + | − | − | − | + | + | + | + | − | − | − |
| metRAB | Melibiose | + | + | + | + | [−] | + | + | [−] | − | + | + | + | + | + | + | [−] | [−] | [−] | + | + | + | + |
| sgaTBAHUE | Uncertain | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| treCBR | Trehalose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| sgcREAQCX | Uncertain | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
Genes are arranged in clockwise order on the W3100 genome sequence.
+, all genes were conserved; [−], genes were partially lost; −, all genes were missing.
DNA spots of frvX gave invalid data when labeled S. sonnei genome DNA was used.
Genes for extracellular structures and materials were highly divergent.
Lipopolysaccharides (LPS) and capsular polysaccharides are diverse macromolecules and are known to be involved in expression of pathogenesis (29). CGH analysis revealed that two gene clusters of W3110 for LPS biosynthesis, the rfa and rfb clusters, were frequently absent in pathogenic strains. In particular, 4 of 14 ORFs in the rfa cluster (JW3594 [b3619] to JW3607 [b3632]) and 7 of 10 ORFs in the rfb cluster (JW2017 [b2032] to JW2026 [b2041]) were commonly absent in all pathogenic strains tested. LPS is composed of three domains: lipid A, core, and O antigen. Absent ORFs were involved in core and O-antigen synthesis. It was hypothesized previously that there are alternative genes in the genomes of pathogens (29). Colanic acid is an extracellular polysaccharide that is produced by most E. coli strains (33). Genes related to colanic acid production, JW2027 (b2042) to JW2047 (b2062), are mainly designated wca and are adjacent to the rfb operon in K-12 strains. All genes related to colanic acid production were absent in Shigella boydii. Part of the region (JW2028 to JW2032, including wcaJKLM) was determined to be absent in additional six strains (EPEC-1, EPEC-2, EIEC-2, EIEC-3, EIEC-4, and Shigella sonnei).
Fimbriae and/or pili are present on the surface of E. coli (15). In the W3110 genome, there were 16 regions annotated as fimbria related by the ERGO database (22). The distribution of the gene clusters for fimbriae is summarized in Table 5. In S. flexneri, only five regions were conserved. On the other hand, all K-12 fimbria genes were conserved in EPEC-4. Diversity of fimbria gene contents was also observed among strains with the same pathotype, except that UPEC strains had similar distributions of K-12 fimbria gene clusters. Three fimbria gene clusters (hofCB-ppdD, ppdC-ygdB-ppdBAa, and hofQ-yrfABCD) were conserved in all pathogenic strains tested. These genes were involved in assembly and transport of the type IV fimbriae (36).
TABLE 5.
Distribution of fimbiria-related gene clusters of W3110 in pathogenic strains
| Gene clustera | Fimbrial type (reference[s]) | O157 Sakai | EHEC | EPEC-1 | EPEC-2 | EPEC-3 | EPEC-4 | EPEC-5 | EPEC-6 | EPEC-7 | ETEC-1 | ETEC-2 | EIEC-1 | EIEC-2 | EIEC-3 | EIEC-4 | UPEC-1 | UPEC-2 | UPEC-3 | REPEC | SF | SB | SS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hofCB, ppdD | Putative (22) | +b | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| yadCKLM, htrE, ecpD, yadN | Putative (22) | − | [−] | [−] | [−] | − | + | [−] | − | − | + | + | [−] | [−] | [−] | [−] | − | − | [−] | [−] | − | [−] | [−] |
| yagV[W]XYZ, ykgK | CSI homologue (15, 22) | + | [−] | + | + | + | + | + | + | + | + | + | − | + | − | + | + | + | + | + | − | − | − |
| sfmACDHF, fimZ | Salmonella homo- logue (22) | + | + | + | [−] | − | + | + | − | − | + | + | + | + | [−] | [−] | − | − | − | + | [−] | [−] | [−] |
| ybgPQD | Pap homologue (15, 22) | − | [−] | [−] | − | − | + | [−] | − | − | [−] | [−] | [−] | [−] | [−] | [−] | − | − | − | [−] | [−] | [−] | [−] |
| ycbQRSTUVF | Type 1 homologue (15, 22) | + | [−] | [−] | [−] | − | + | [−] | − | − | + | [−] | [−] | [−] | [−] | [−] | − | − | − | [−] | [−] | [−] | [−] |
| csgGFEDBAC | Curli (15) | + | + | + | + | + | + | + | + | + | + | + | + | + | [−] | [−] | + | + | + | + | − | + | + |
| ydeQRST | Type 1 homologue (15, 22) | + | + | + | + | + | + | + | + | − | [−] | − | − | + | [−] | + | + | + | + | + | [−] | − | [−] |
| yehABCDE | Sef homologue (15, 22) | − | + | [−] | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | [−] | + | + |
| yfcPQRSUV | Pap homologue (22) | − | [−] | [−] | [−] | − | + | [−] | − | − | + | + | [−] | [−] | [−] | [−] | − | − | − | [−] | [−] | [−] | [−] |
| ppdC, ygdB, ppdBA | Putative (22) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| yqiGH | Putative (22) | − | + | + | − | − | + | + | − | − | + | + | [−] | − | + | + | − | − | − | + | − | + | + |
| yraHIJK | Putative (22) | + | + | + | [−] | − | + | + | − | − | + | + | + | [−] | [−] | − | − | − | − | + | + | + | [−] |
| yhcAD | Putative (22) | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| hofQ, yrfABCD | Putative (22) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| fimBEAICFGH | Type 1 (15) | + | + | + | + | + | + | + | + | + | + | + | [−] | + | [−] | [−] | + | + | + | + | + | [−] | − |
Genes are arranged in clockwise order on the W3100 genome sequence. A gene that was not spotted on the DNA array is enclosed in brackets.
+, all genes were conserved; [−], genes were partially lost; −, all genes were missing.
Regulatory genes.
Of the 1,424 genes not conserved, 140 ORFs were annotated with regulatory functions in the ERGO database (22). Most of the absent regulators were the transcriptional regulators for the neighboring operons, but some were global regulators. For example, five two-component regulatory systems (citAB, torRT, atoCS, basRS, and dcuRS) were absent. The atoCS genes were absent in six pathogenic strains, including O157 Sakai, which was shown not to possess these genes on the basis of its genome sequence (7). The other systems were absent as follows: citAB in S. flexneri, torRT in EPEC-3, and both basRS and dcuRS in EPEC-7.
Putative regions of duplication in pathogenic strains.
Some prophage genes were found to be duplicated in the O157 Sakai genome (7). Two of these genes, yeeU and yeeV, were spotted on the DNA microarray slide used in this study. The signal ratios of the two genes in the CGH microarray analysis of O157 Sakai were greater than 0.8 on the log2 scale. This result suggested that duplicated genomic regions in pathogenic strains could be also detected as spots with higher signal ratios in the CGH microarray analysis. To detect absent genes, the threshold was determined by using data sets for hundreds of gene spots that were predicted to be absent by an in silico comparison of O157 Sakai and W3110. Since only two samples (yeeU and yeeV) were used to evaluate the signal intensity of duplication, it was difficult to fix the threshold. ORFs with a signal ratio greater than 0.8 on the log2 scale were putatively considered duplicated. The number of these genes was 369 (Fig. 1). Unexpectedly, duplication of prophage genes was rarely found. Only 36 of the 369 ORFs corresponded to the K-12 prophages. Table 6 shows relatively large regions with concatenated genes that were putatively considered duplicated. Several essential and important genes for fundamental cellular functions were observed in the putative regions of duplication (Table 6).
TABLE 6.
Putative regions of duplication in pathogenic strains detected by CGH microarray analysis
| Position | ORFa | Strain | Ratiob | Characteristic genec | Function |
|---|---|---|---|---|---|
| Start | JW0276 (b0282) | EPEC-1 | 33/36 | eaeH | Attaching and effacing protein homolog |
| End | JW0318 (b0326) | bct genes | Choline/betaine transport and metabolism | ||
| ykgM | Ribosomal protein L31P homologue | ||||
| Start | JW2392 (b2395) | EPEC-4 | 65/73 | gltX | Glutamyl-tRNA synthetase |
| End | JW2465 (b2480) | lig | NAD-dependent DNA ligase | ||
| zipA | Cell division protein ZipA | ||||
| acrD | Acriflavin resistance protein D | ||||
| pts genes | Glucose phosphotransferase system | ||||
| Start | JW3041 (b3070) | EPEC-7 | 41/45 | aer | Aerotaxis receptor |
| End | JW3086 (b3115) | ||||
| Start | JW3451 (b3484) | EPEC-4 | 77/102 | glySQ | Glycyl-tRNA synthetase |
| End | JW3564 (b3591) | aldB | Aldehyde dehydrogenase B | ||
| Start | JW3489 (b3521) | EPEC-5 | 35/35 | cspA | Cold shock protein |
| End | JW3532 (no counterpart) | glySQ | Glycyl-tRNA synthetase | ||
| Start | JW3552 (b3580) | EPEC-2 | 10/10 | aldB | Aldehyde dehydrogenase B |
| End | JW3561 (b3588) |
Regions in which more than 10 ORFs were supposed to be successively duplicated. The corresponding b numbers are indicated in parentheses (2).
Number of putative duplicated ORFs/number of ORFs in the region.
Essential genes that were supposed to be duplicated are indicated by boldface type.
Phylogenic analysis.
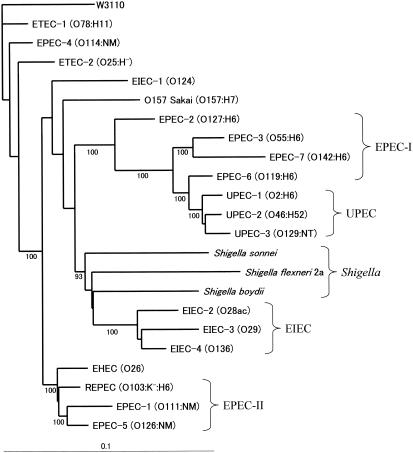
In a previous study (25), the gene contents of Salmonella genomes were analyzed by CGH with a Salmonella enterica serovar Typhimurium LT2 DNA microarray. Genes of the virulence plasmid of serovar Typhimurium were also spotted on the array. The gene content table was subjected to a cluster analysis to construct a phylogenic tree, which revealed a good relationship to the serovar groups of the Salmonella pathogens. In this case, virulence or pathogenic genes seemed to be good markers for a phylogenic analysis of the pathogens. We tried to construct a phylogenic tree from the gene content table consisting only of genes in nonpathogenic strain W3110. The gene contents of the strains tested were recognized as a matrix for the neighbor-joining analysis (see Materials and Methods). The resulting dendrogram is shown in Fig. 2. Three major phylogenic groups (EPEC-I and UPEC, EIEC and Shigella, and EPEC-II) were identified on the basis of bootstrap values greater than 90%. EPEC strains were basically divided into two groups, except for strain EPEC-4. UPEC strains were closely related to each other. Dysentery strains, EIEC strains (18) except strain EIEC-1 (O124), and Shigella were clustered in one phylogenic group. These results showed that the phylogenic tree was consistent with the previous studies in which multilocus enzyme electrophoresis or a sequence comparison of conserved genes was used (6, 26, 27, 28). It should be emphasized that the K-12 microarray was useful for phylogenic analysis of pathogenic strains of E. coli.
FIG. 2.
Unrooted phylogenetic tree for 22 strains. The tree was constructed with the PAUP software by using the neighbor-joining algorithm and 1,000 bootstrap replicates based on the matrix of gene contents of the 22 strains analyzed (see Materials and Methods). Bootstrap confidence values greater than 90% are indicated at the nodes.
The phylogenic distances inside the branch that included the EIEC and Shigella strains were greater than the phylogenic distances for the other two groups. This result suggested that dysentery strains change faster genetically than the strains with other disease phenotypes. UPEC strains were related to the EPEC-I group (Fig. 2). The number of ORFs that were specifically absent in this UPEC-EPEC-I group was 23, and these ORFs included two sets of continuously missing genes. The two regions were JW1289 to JW1295 (b1296 to b1302) and JW2466 to JW2476 (b2481 to b2491). The former region included genes encoding a putative amino acid permease (JW1289, b1296, ycjJ) and 4-aminobutyrate aminotransferase (JW1295, b1302, goaG). The latter consisted of the hyf operon coding for hydrogenase-4 for anaerobic respiration (31), except that in EPEC-2 8 of 14 ORFs in this region were absent. It is possible that strains in the UPEC-EPEC-I group have the specific inserts in these two regions that result in the characteristic phenotype.
DISCUSSION
For CGH analysis with DNA microarray slides, it is important to set an appropriate threshold to detect missing ORFs in sample strains. A signal ratio (sample strain/reference strain) of −1 on the log2 scale was frequently used in previous studies (1, 9, 10, 30). In this study, a threshold value of −1 was found to be appropriate for detection of absent W3110 ORFs in the O157 Sakai strain. In recent studies the workers reported two other thresholds for determining the presence of genes in CGH experiments (13, 25). First, Porwollik et al. used a threshold for the presence of genes based on the median of the microarray hybridization ratios and the standard deviation of data points for each genome (25). We tested this method with our data and calculated an average threshold for 22 pathogenic strains of −2.23, which resulted in a stricter threshold for determining absence. The stricter threshold may result in not considering split or fused genes absent. Chimeric genes are often functionally inactive. A threshold that is too strict would result in considering inactive genes conserved and in a noisy gene content table for functional genomics. The second method for determining a threshold is to use the computer program GACK generated by Kim et al. (13). The GACK program is capable of dynamically generating cutoffs for present/absent gene analysis for each array hybridization based on the shape of the signal ratio distribution. The program was tested in an analysis of the data for each strain. The calculated thresholds were generally greater than −1. The highest threshold was −0.3 for ETEC-2 and resulted in designating an additional 150 ORFs absent; one essential gene, lpxB, was included in this group. In the case of ETEC-1, the threshold calculated by the GACK program was −0.468, and this resulted in designating three essential genes (prfB, ftsX, and valS) as absent. We concluded that the GACK program was not be suitable for processing our data. In the previous study in which the GACK program was used, a short specific region of each gene was selected and spotted on microarrays (4). In our study, full-length DNAs were spotted on microarray slides. The methodological difference for hybridization was thought to be the main reason that the GACK program was not appropriate for analyzing our data.
In a previous E. coli CGH analysis (5), the strains used were different from the strains used in this study. Only one strain, EPEC-2 (E2348/69), was used in both studies. We found that this strain did not contain 457 ORFs of W3110 (Table 1). This number was twofold larger than the number determined in the previous work (209 ORFs). The difference was probably due to the different hybridization methods and thresholds in the two studies. Dobrindt et al. used simple hybridization of sample genomes to the DNA macroarray membranes and used a threshold of 0.3 for the signal-to-noise ratios on a linear scale, which was lower than the value that we used (−1 on the log2 scale). We thought that this threshold of the macroarray CGH for detecting absent genes was rather strict. In the previous study, at least 3,100 ORFs of K-12 were found to be conserved in 28 E. coli genomes (5). This number is larger than our estimate of the number of conserved genes. As described above, we determined that 2,568 genes on the DNA array could be conserved in all strains tested. There are also 319 ORFs of W3110 that were not spotted on our microarray slides. Approximately one-third of these ORFs were insertion element-derived ORFs and were thought to be dispensable. The remaining two-thirds could be considered candidates for conserved genes. Thus, we estimated that the number of common ORFs in the E. coli genome is approximately 2,800 ORFs at most, which is less than the number determined by the macroarray CGH analysis (5). On the other hand, a comparison of the genomic sequences of three E. coli strains, MG1655, O157:H7 EDL933, and UPEC CFT073, showed that these strains shared 2,996 genes (35). Also, 2,881 ORFs were estimated to be shared by a sequence comparison of three strains, S. flexneri 2a strain 2457T, MG1655, and O157:H7 EDL933 (34). Since the number of common ORFs could decrease with increasing numbers of genomic sequences used, our estimate for the number of conserved ORFs seemed to be consistent with the number determined by using three sequenced genomes (34, 35). This consistency suggested that our method for genomic comparison, including the threshold-setting method, could be reasonable and result in better resolution of genome structure than the previous macroarray analysis (5).
In this study 1,424 ORFs were considered absent in at least one of the strains tested. Absent ORFs were frequently detected in a cluster (Fig. 1). This mosaic feature of the E. coli genome was first detected in all of the sequenced strains (7, 12, 23, 35). However, it is still not clear how the genome of E. coli was diversified in terms of mosaic arrangements. Since the phylogenic tree obtained in this study was consistent with the dendrograms constructed by using other indices (6, 26, 27, 28), the absence/presence pattern of the CGH analysis was thought to be a simple interpretation of the mosaic genome and to reflect the lineage of E. coli genomic diversification. Although it was proposed that phage infection and duplication and also recombination between the similar phages could produce lineage-specific regions and deletions (5, 20), the mechanism of genomic rearrangement must still be elucidated. It is expected that CGH data sets combined with phylogenic analysis could be useful for revealing the mechanism and history of E. coli genome diversification.
Missing genes are candidates for pathoadaptive mutations that contribute to the pathogenicity by their absence (16, 17, 32). Although a genomic sequence comparison is expected to be the best method for revealing pathoadaptive mutations, it is hard and less realistic to sequence sufficient numbers of genomes (35). Our results suggested that CGH microarray analysis could be a rapid and powerful method for extracting the candidate regions for pathoadaptive mutations because unsequenced strains can be easily subjected to a genomic comparison analysis.
Even though the CGH microarray method is apparently a powerful method for genomic comparison of related strains, including unsequenced strains, this technique has several limitations. First, it is not possible to detect the genes that are not spotted on the microarray. However, strain-specific and missing regions could suggest strain-specific insertions at the loci. Conserved sequences flanking missing ORFs might serve as good primers for amplifying strain-specific regions. Therefore, our CGH information for pathogenic strains could be useful for rapid identification and isolation of characteristic regions of pathogenic strains, including pathogenicity islands. Actually, comparative analysis of the K-12 strain MG1655 and O157 Sakai genome sequences revealed that there are 296 O157 Sakai-specific regions called S-loops and 325 MG1655-specific regions called K-loops. Indeed, more than the 60% of the loops (203 loops) are in the same interstitial regions of the backbone genome, although the lengths and sequences between the S- and K-loops are different (7).
CGH microarray analyses of pathogenic E. coli strains showed that more than 30% of the K-12 ORFs were variable and that E. coli strains had highly diverse genomic structures (7, 12, 19, 23, 35). Phylogenic analysis based on the CGH microarray data confirmed the authentic relationships among pathogens and revealed specific linkages for EIEC and Shigella and for UPEC and EPEC-I. We concluded that CGH analysis with K-12 DNA microarrays is quite useful for genomic comparison, phylogenetic analysis, and detection of lineage-specific regions of E. coli strains, including Shigella strains.
Supplementary Material
Acknowledgments
This study was carried out as a part of The Project for Development of a Technological Infrastructure for Industrial Bioprocesses on R&D of New Industrial Science and Technology Frontiers by the Ministry of Economy, Trade & Industry (METI) and was supported by the New Energy and Industrial Technology Development Organization (NEDO).
We are grateful to N. Shiraishi and Y. Kawagoe (Tokyo Research Laboratories, Kyowa Hakko Kogyo, Machida, Japan) for their helpful advice about microarray experiments.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Björkholm, B., A. Lundin, A. Sillén, K. Guillemin, N. Salama, C. Rubio, J. I. Gordon, P. Falk, and L. Engstrand. 2001. Comparison of genetic divergence and fitness between two subclones of Helicobacter pylori. Infect. Immun. 69:7832-7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Bochner, B. R. 1989. Sleuthing out bacterial identities. Nature 339:157-158. [DOI] [PubMed] [Google Scholar]
- 4.Chan, K., S. Baker, C. C. Kim, C. S. Detweiler, G. Dougan, and S. Falkow. 2003. Genomic comparison of Salmonella enterica serovars and Salmonella bongori by use of an S. enterica serovar Typhimurium DNA mircroarray. J. Bacteriol. 185:553-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobrindt, U., F. Agerer, K. Michaelis, A. Janka, C. Buchrieser, M. Samuelson, C. Svanborg, G. Gottschalk, H. Karch, and J. Hacker. 2003. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J. Bacteriol. 185:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donnenberg, M. S., and T. S. Whittam. 2001. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J. Clin. Investig. 107:539-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrahgic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 8.Holt, J. G., N. R. Krieg, P. H. A. Sneath, J. T. Staley, and S. T. Williams (ed.). 1994. Bergey's manual of determinative bacteriology, 9th ed. Williams & Wilkins, Baltimore, Md.
- 9.Israel, D. A., N. Salama, C. N. Arnold, S. F. Moss, T. Ando, H. P. Wirth, K. T. Tham, M. Camorlinga, M. J. Blaser, S. Falkow, and R. M. Peek, Jr. 2001. Helicobacter pylori strain-specific differences in genetic content, identified by microarray, influence host inflammatory responses. J. Clin. Investig. 107:611-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Israel, D. A., N. Salama, U. Krishna, U. M. Rieger, J. C. Atherton, S. Falkow, and R. M. Peek, Jr. 2001. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc. Natl. Acad. Sci. USA 98:14625-14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itoh, T., T. Okayama, H. Hashimoto, J. Takeda, R. W. Davis, H. Mori, and T. Gojobor. 1999. A low rate of nucleotide changes in Escherichia coli K-12 estimated from a comparison of the genome sequences between two different substrains. FEBS Lett. 450:72-76. [DOI] [PubMed] [Google Scholar]
- 12.Jin, Q., Z. Yuan, J. Xu, Y. Wang, Y. Shen, W. Lu, J. Wang, H. Liu, J. Yang, F. Yang, X. Zhang, J. Zhang, G. Yang, H. Wu, D. Qu, J. Dong, L. Sun, Y. Xue, A. Zhao, Y. Gao, J. Zhu, B. Kan, K. Ding, S. Chen, H. Cheng, Z. Yao, B. He, R. Chen, D. Ma, B. Qiang, Y. Wen, Y. Hou, and J. Yu. 2002. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 30:4432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, C. C., E. A. Joyce, K. Chan, and S. Falkow. 2002. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 3:research0065.1-0065.17. [Online.] http://genomebiology.com/2002/3/11/research/0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, E. C. C. 1996. Dissimilatory pathways for sugars, polyols, and carboxylates, p. 307-342. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 15.Low, D., B. Braaten, and M. van der Woude. 1996. Fimbriae, p. 146-157. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 16.Maurelli, A. T., R. E. Fernandez, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl Acad. Sci. USA 95:3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakata, N., T. Tobe, I. Fukuda, T. Suzuki, K. Komatsu, M. Yoshikawa, and C. Sasakawa. 1993. The absence of a surface protease, OmpT, determines the intercellular spreading ability of Shigella: the relationship between the ompT and kcpA loci. Mol. Microbiol. 9:459-468. [DOI] [PubMed] [Google Scholar]
- 18.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochman, H., and I. B. Jones. 2000. Evolutionary dynamics of full genome content in Escherichia coli. EMBO J. 19:6637-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohnishi, M., K. Kurokawa, and T. Hayashi. 2001. Diversification of Escherichia coli genomes: are bacteriophages the major contributors? Trends Microbiol. 9:481-485. [DOI] [PubMed] [Google Scholar]
- 21.Oshima, T., C. Wada, Y. Kawagoe, T. Ara, M. Maeda, Y. Masuda, S. Hiraga, and H. Mori. 2002. Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli. Mol. Microbiol. 45:673-695. [DOI] [PubMed] [Google Scholar]
- 22.Overbeek, K., N. Larsen, T. Walunas, M. D'Souza, G. Pusch, E. Selkov, Jr., K. Liolios, V. Joukov, D. Kaznadzey, I. Anderson, A. Bhattacharyya, H. Burd, W. Gardner, P. Hanke, V. Kapatral, N. Mikhailova, O. Vasieva, A. Osterman, V. Vonstein, M. Fonstein, N. Ivanova, and N. Kyrpides. 2003. The ERGOTM genome analysis and discovery system. Nucleic Acids Res. 31:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perna, E. S., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Pósfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. Welch, and F. R. Blattner. 2001. Genomic sequence of enterohaemorrhagic Escherichia coli 0157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 24.Pollack, J. R., C. M. Perou, A. A. Alizadeh, M. B. Eisen, A. Pergamenschikov, C. F. Williams, S. S. Jeffrey, D. Botstein, and P. O. Brown. 1999. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat. Genet. 23:41-46. [DOI] [PubMed] [Google Scholar]
- 25.Porwollik, S. P., R. M.-Y. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:8956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pupo, G. M., D. K. Karaolis, R. Lan, and P. R. Reeves. 1997. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect. Immun. 65:2685-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pupo, G. M., R. Lan, and P. R. Reeves. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. USA 97:10567-10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 29.Rick, P. D., and R. P. Silver. 1996. Enterobacterial common antigen and capsular polysaccharides, p. 104-122. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 30.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skibinski, D. A. G., P. Golby, Y. S. Chang, F. Sargent, R. Hoffman, R. Harper, J. R. Guest, M. M. Attwood, B. C. Berks, and S. C. Andrews. 2002. Regulation of the hydrogenase-4 operon of Escherichia coli by the sigma54-dependent transcriptional activators FhlA and HyfR. J. Bacteriol. 184:6642-6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sokurenko, E. V., D. L. Hasty, and D. E. Dykhuizen. 1999. Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol. 7:191-195. [DOI] [PubMed] [Google Scholar]
- 33.Stevenson, G., K. Andrianopoulos, M. Hobbs, and P. R. Reeves. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei, J., M. B. Goldberg, V. Burland, M. M. Venkatesan, W. Deng, G. Fournier, G. F. Mayhew, G. Plunkett III, D. J. Rose, A. Darling, B. Mau, N. T. Perna, S. M. Payne, L. J. Runyen-Janecky, S. Zhou, D. C. Schwartz, and F. R. Blattner. 2003. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 71:2775-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitchurch, C. B., and J. S. Mattick. 1994. Escherichia coli contains a set of genes homologous to those involved in protein secretion, DNA uptake and the assembly of type-4 fimbriae in other bacteria. Gene 150:9-15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.