Abstract
The heterogeneous environment of the lung of the cystic fibrosis (CF) patient gives rise to Pseudomonas aeruginosa small colony variants (SCVs) with increased antibiotic resistance, autoaggregative growth behavior, and an enhanced ability to form biofilms. In this study, oligonucleotide DNA microarrays were used to perform a genome-wide expression study of autoaggregative and highly adherent P. aeruginosa SCV 20265 isolated from a CF patient's lung in comparison with its clonal wild type and a revertant generated in vitro from the SCV population. Most strikingly, SCV 20265 showed a pronounced upregulation of the type III protein secretion system (TTSS) and the respective effector proteins. This differential expression was shown to be biologically meaningful, as SCV 20265 and other hyperpiliated and autoaggregative SCVs with increased TTSS expression were significantly more cytotoxic for macrophages in vitro and were more virulent in a mouse model of respiratory tract infection than the wild type. The observed cytotoxicity and virulence of SCV 20265 required exsA, an important transcriptional activator of the TTSS. Thus, the prevailing assumption that P. aeruginosa is subject to selection towards reduced cytotoxicity and attenuated virulence during chronic CF lung infection might not apply to all clonal variants.
Pseudomonas aeruginosa is the most dominant bacterial pathogen causing chronic lung infection in cystic fibrosis (CF) patients (16, 30, 44). Although intensive antipseudomonal chemotherapy has greatly improved the prognosis in CF, P. aeruginosa cannot be permanently eradicated from the lung of a CF patient (CF lung) (21). A characteristic microbiological finding is the recovery of different phenotypic variants from the respiratory tracts of CF patients (58) although most patients are colonized with only one or a few P. aeruginosa genotypes (4). The appearance of multiple morphotypes may be facilitated by “hypermutable” strains of P. aeruginosa, which have been isolated at high frequency from CF patients (35). In the context of chronic P. aeruginosa CF lung infection, attention has long focused on the appearance of the most common mucoid P. aeruginosa phenotype (16). Other phenotypes, however, including dwarf, or so-called small colony variants (SCVs), can be isolated from the CF lung. It has recently been shown in our lab that the recovery of SCVs could be correlated to parameters revealing poor lung function and the use of inhalative antibiotics (19). Among the heterogeneous group of clinical SCV isolates from different patients, a subgroup was identified exhibiting hyperpiliation and increased twitching motility as well as increased fitness under stationary growth conditions, better biofilm-forming capabilities, and a high adherence to a pneumocytic cell line in comparison with the clonal fast-growing wild types (20). Similar SCVs from P. aeruginosa that seem to be adapted especially to the formation of biofilms have been described by others (12, 13). As a biofilm mode of growth within the CF lung may be responsible for persistent P. aeruginosa infection (27, 43), we have aimed at elucidating the role of those autoaggregative and highly adherent SCVs in the pathogenicity of CF lung infection. To face this challenge, we used a P. aeruginosa GeneChip (Affymetrix) to compare genome-wide expression patterns of the autoaggregative and highly adherent SCV 20265 with its clonal wild type and a fast-growing revertant that was isolated in vitro from the SCV population. The morphotypes of strain SCV 20265 were already the subject of a recent proteome analysis that revealed significant differences among the profiles of secreted proteins between these clonal variants (50). In this study, we demonstrate that the transcriptional expression-signatures of all three morphotypes are distinct. In comparison to the wild type, SCV 20265 differentially expressed several genes known to be affected in P. aeruginosa living in a biofilm. Moreover, genes involved in the type III secretion system (TTSS) were strongly upregulated in the SCV 20265, matching the results of the recent proteome analysis (50). The increased expression of the TTSS and respective effector proteins in 20265 SCV and other autoaggregative SCVs was shown to be linked to increased in vitro cytotoxicity and enhanced virulence in murine respiratory tract infection.
MATERIALS AND METHODS
Bacterial strains.
P. aeruginosa SCV 20265 and the corresponding wild type were recovered as sequential isolates from a CF patient. SCVs were characterized by their comparatively slower growth on solid medium of 1 to 3 mm in colony diameter after 48 h of incubation. A fast-growing revertant was isolated after serial passages in brain heart infusion medium from the SCV 20265 population (19). Site-directed mutagenesis of exsA in SCV 20265 was carried out as already described (9). Besides SCV 20265, six other autoaggregative SCVs, eight nonautoaggregative SCVs, and their respective clonal wild types were used. All SCVs are isolates from different patients who had been colonized for between 9 and 18 years with P. aeruginosa at the time of SCV isolation. Clonality of different morphotypes from the same patient was shown by identical restriction fragment patterns after SpeI restriction of their genomic DNA and subsequent pulsed-field gel electrophoresis (19, 20). Bacteria were grown in modified Vogel-Bonner (VB) medium (12.5 g of d-gluconic acid, 1.1 g of citric acid · H2O, 4.2 g of K2HPO4 · 3H2O, 2.9 g of NaNH5PO4 · 4H2O, 80-mg/liter MgSO4 · 7H2O, pH 7.4) at 37°C. Growth of bacteria was monitored by optical density at 600 nm (OD600).
RNA isolation.
Total RNA was isolated by a modified hot phenol method. P. aeruginosa was grown in VB medium at 37°C with aeration. The growth rate during the exponential phase was determined for the three 20265 morphotypes in two independent experiments. In the first experiment, the doubling times for the wild type, SCV, and revertant were 78, 86, and 72 min, respectively, and in the second experiment the doubling times were 78, 87, and 73, respectively. Doubling times were also determined based on whole cellular protein by using a Bradford assay. Protein content was tested by incubating bacterial cells for 1 h in 2 M NaOH at 80°C and adding a 40-fold-higher volume of Bradford reagent. Bovine serum albumin (BSA) was used as standard. Despite beginning adherence and clumping of SCV 20265 in the exponential phase, the doubling times diverged only approximately 1, 3, and 5 min for the wild type, SCV, and revertant, respectively, from the OD600-based determination. For convenience, the OD600 measurement was subsequently used as the growth parameter. For the array analysis, bacterial cultures were started at an OD600 of 0.05 and harvested in the exponential phase at an OD600 of 0.8 or in the stationary phase after 27 h of shaking (OD600 of approximately 3.0). For the RNA preparation, two independent precultures were used to inoculate three cultures each and the RNA was prepared from all six of the resulting cultures. The maintenance of the colony morphology after cultivation was checked by plating bacteria from the liquid cultures on Columbia blood agar plates. Total RNA from approximately 1010 bacteria was extracted. Briefly, bacteria were harvested by short centrifugation, resuspended quickly in water, and lysed in the presence of a 65°C hot mixture of 62.5% (wt/vol) phenol (pH 5.5), 2.2 mM sodium dodecyl sulfate, 9 mM Na-acetate, and 0.9 mM EDTA for 10 min with shaking. The aqueous phase was purified by subsequent extraction with phenol-chloroform and chloroform. The nucleic acids were pelleted with ethanol (−20°C overnight), washed with 70% ethanol, and treated with 40 U of DNase I (Roche) and 20 U of SUPERaseIn (Ambion) in DNase I buffer (50 mM Na-acetate, 10 mM MgCl2, 2 mM CaCl2, pH 6.5) for 30 min at 37°C. After purification with RNeasy columns (QIAGEN), the yield of total cellular RNA was determined by UV absorption. RNA with a size below 200 bp (e.g., tRNAs and 5S rRNA) is below the cutoff of the column and therefore could not be recovered.
cDNA generation, fragmentation, and biotinylation.
To reduce variations in the abundance of specific mRNAs due to slight differences in growth conditions and RNA preparation, we pooled equal amounts of RNA from three cultures to get a final amount of 10 μg. The subsequent steps of cDNA generation and biotin-ddUTP terminal labeling were performed as described in the manufacturer's instructions for the P. aeruginosa GeneChip. Ten micrograms of total RNA was mixed with random primers (Invitrogen) and control in vitro transcripts of 10 non-Pseudomonas gene sequences (kindly provided by S. Lory and coworkers) and incubated for 10 min at 70°C followed by 10 min at 25°C. Then cDNA reaction mix was added, which consisted of 5× first strand buffer (10 mM dithiothreitol, 0.5 mM deoxynucleoside triphosphates [dNTPs], 25-U/μl SuperScript II [all from Invitrogen], and 0.5-U/μl SUPERaseIn [Ambion]). This was followed by incubation for 10 min at 25°C, 60 min at 37°C, 60 min at 42°C, and 10 min at 70°C. RNA was removed by adding 1 N NaOH and incubation for 30 min at 65°C; 1 N HCl neutralized the reaction. The cDNA was purified with the QIAquick column (QIAGEN) and quantified by A260. cDNA was fragmented in One Phor-All buffer with 0.5 U of DNase I (Amersham Pharmacia Biotech) per μg of cDNA for 10 min at 37°C and subsequent inactivation for 10 min at 98°C. To check if the majority of cDNA fragments were in a 50- to 200-bp range, 5 μl was loaded on a 2% (wt/vol) agarose gel stained with SYBR Green (Molecular Probes). The fragmentation product was end labeled with the Enzo BioArray terminal labeling kit with biotin-ddUTP (Affymetrix).
Generation of targets from genomic DNA.
For hybridization of genomic DNA of the SCV with the GeneChip, 15 μg of genomic DNA from stationary-phase-grown cells was restricted with 75 U of BstUI (New England Biolabs) at 60°C for 16 h followed by phenol-chloroform-isoamyl alcohol extraction and precipitation with isopropanol. This enzyme cut the majority of the genome to fragments of between 50 and 200 bp, which were suitable for GeneChip hybridization. The DNA pellet was applied to a 2% (wt/vol) agarose gel and labeled as described.
GeneChip hybridization and washing.
MES hybridization buffer (100 mM MES [morpholineethanesulfonic acid], 1 M NaCl, 20 mM EDTA, 0.01% Tween), 50 pM B2 control oligonucleotide (Affymetrix), 0.1-mg/ml herring sperm DNA (Promega), 0.5-mg/ml BSA (Invitrogen), and 7% (wt/vol) dimethyl sulfoxide (DMSO) were added to the labeled cDNA and loaded onto a GeneChip. After incubation for 16 h at 50°C at 60 rpm in an Affymetrix hybridization oven, the GeneChips were put into the Affymetrix fluidics station for washing. First the GeneChips were washed 20 times with nonstringent buffer (6× SSPE [1.08 M NaCl, 60 mM NaH2PO4, 1 mM EDTA; pH 7.7] and 0.01% [vol/vol] Tween 20) at 25°C and then they were washed 60 times with stringent buffer (100 mM MES, 0.1 M NaCl, 0.01% [vol/vol] Tween 20) at 50°C. Then a streptavidin solution mix, consisting of 10-μg/ml streptavidin (Pierce Chemical), 2-mg/ml bovine serum albumin (BSA), 100 mM MES, 0.1 M NaCl, and 0.01% (vol/vol) Tween 20, was applied for 10 min at 25°C to stained bound cDNA. After washing for 40 times with nonstringent buffer at 30°C, the GeneChip was subjected to a secondary stain (5-μg/ml biotin-antistreptavidin antibody [Vector Laboratories], 100-μg/ml normal goat immunoglobulin G, 2-mg/ml BSA, 100 M MES, 0.1 M NaCl, 0.01% [vol/vol] Tween 20) for 10 min at 25°C and to a third stain (10-μg/ml streptavidin-phycoerythrin [Molecular Probes], 2-mg/ml BSA, 100 M MES, 0.1 M NaCl, 0.01% [vol/vol] Tween 20) for 10 min at 25°C. Finally excess label was removed by 60 washings with nonstringent buffer at 30°C. Expression analysis experiments were done in duplicate with cDNA samples derived from two different RNA pools. This minimized variation in the cDNA samples as well as variation in GeneChip performance.
Analysis of P. aeruginosa GeneChip hybridizations.
The P. aeruginosa GeneChip (Affymetrix) contains oligonucleotide probes for 5,549 protein-coding genes, 18 tRNA genes, a representative rRNA cluster, and 199 intergenic regions selected from the annotated genome of P. aeruginosa strain PAO1 (45) as well as 117 genes from P. aeruginosa strains other than PAO1 and 14 genes from other species, which can serve as controls (34). Data analysis was performed using the Affymetrix Microarray Suite software 5.0 with Affymetrix default parameters. As expression analysis was performed in duplicate, a total of two GeneChips per variant and growth stage were scanned at 570 nm with a 3-μm resolution in an Affymetrix GeneChip scanner. The average microarray hybridization signal intensities were scaled to 150. Two different morphotypes from the same growth stage were compared with each other by analyzing the four possible GeneChip pairings (e.g., SCV1 versus WT1, SCV1 versus WT2, SCV2 versus WT1, and SCV2 versus WT2), known as the four-comparison survival method (3, 6). The data were imported into a Microsoft Access database capable of searching for genes, which were found in all four pairings (i) at least twofold regulated and (ii) defined by the Affymetrix Microarray Suite Software as having significant changes in their signal intensities. The arithmetic average and the standard deviation (SD) were calculated. Data were combined with the latest annotation (10 August 2003) from the web site of the P. aeruginosa PAO1 sequence and the community annotation project provided at www.pseudomonas.com.
Proteome analysis.
Preparation of protein samples, two-dimensional gel electrophoresis, and matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) analysis were performed exactly as described earlier (50).
Cytotoxicity assay.
The cytotoxicity of the phenotypic variants was assayed by using the murine macrophage cell line J774.A1. Bacteria were grown in Luria-Bertani (LB) medium (10-g/liter tryptone, 5-g/liter yeast extract, 5-g/liter NaCl, pH 7.2) to the exponential phase (OD600 of 1.2), and macrophages were infected with a multiplicity of infection of 10. Cytotoxicity was determined by measuring the release of the cytosolic enzyme lactate dehydrogenase (LDH) into supernatants by using the cytotoxicity detection kit (Roche) at various time points after infection.
Murine respiratory tract infection.
For intranasal infection, bacteria were grown in LB medium to the exponential phase (OD600 of 1.2). Bacteria were harvested and resuspended in 0.01 M potassium phosphate buffer made isotonic with saline (pH 7.5). BALB/c mice, 8 to 10 weeks old, were anesthetized by a mixture of ketamine hydrochloride and xylazine hydrochloride and subsequently inoculated with ∼108 bacteria in 30 μl into both nostrils. Mortality of animals was monitored daily. Kaplan-Meier survival analysis for time was performed with SPSS version 7.0.
Iron supplementation disk assay.
Bacteria grown overnight on Columbia agar plates (Becton Dickinson) were washed repeatedly with 0.9% (wt/vol) NaCl, and 100 μl containing approximately 2 × 107 to 5 × 107 bacteria was suspended in 3 ml of iron-depleted VB soft agar at 40°C (1.1 or 0.5 mM 2,2-dipyridyl as a chelator for free iron and 0.6% [wt/vol] Difco high-grade agar), mixed well, and poured on iron-depleted VB plates (1.5% [wt/vol] agar). Sterile filter paper disks were placed on the solid soft agar, and disks were spotted with 8 μl of Fe2+ (200 mM ferrous sulfate; Sigma), Fe3+ (100 mM ferric citrate; Sigma), hemin (10 mM; Sigma), or hemoglobin (0.1 mM; Sigma). Plates were incubated at 37°C for 24 h.
H2O2 sensitivity assays.
The H2O2 sensitivity broth assay was adapted from Ma et al. (31). Briefly, all morphotypes were grown aerobically in VB medium until log phase (OD600 = 0.8) or stationary phase for 27 to 48 h (depending on the strain used) at 37°C. After dilution to an OD600 of 0.6 in prewarmed VB medium, the bacteria were incubated with final concentrations of 0.03% (vol/vol) or 0.003% (vol/vol) H2O2 (Merck) for 12 min. The suspensions were then serially diluted in 0.9% (wt/vol) saline containing 60 μg of bovine liver catalase (Sigma) per ml, and aliquots were plated on LB agar. The number of CFU was determined after incubation at 37°C for up to 48 h. The percentage viability was calculated by comparing the CFU with and without exposure to H2O2. All experiments were done in duplicate with three plates each.
The H2O2 sensitivity disk assay was adapted from Hassett et al. (18). All morphotypes were grown aerobically at 37°C in VB medium for 27 to 48 h (stationary phase depended on the strain used). Then, 100 μl of bacteria containing approximately 2 × 107 to 5 × 107 bacteria was suspended in 3 ml of LB soft agar at 40°C (0.6% [wt/vol] agar), mixed well, and poured on LB agar plates (with 1.5% [wt/vol] agar). Sterile filter paper disks were placed on the solid soft agar, and the disks were spotted with 8 μl of 30% (wt/vol) H2O2. Plates were incubated at 37°C for 24 h, and the diameter of zone of growth inhibition around the disks was measured. All experiments were done in triplicate with three plates each.
RESULTS AND DISCUSSION
Genome analysis of P. aeruginosa 20265.
To further characterize SCVs with enhanced biofilm-forming capabilities from P. aeruginosa emerging during CF lung infection, we compared gene expression of the autoaggregative and highly adherent P. aeruginosa SCV 20265 with its isogenic wild type and in vitro-generated revertant (20) showing no enhanced adhesion properties in a genome-wide manner. As genomes from P. aeruginosa strains vary considerably in genome size and content (38), we first assessed whether the P. aeruginosa PAO1 GeneChip is applicable for the approximately 6,644-kb genome of SCV 20265 used in this study, which is 380 kb larger than that of PAO1. The fragmented genome of SCV 20265 hybridized with 5,069 of 5,549 PAO1 genes (91%) (for more details, see Fig. S6 provided in the supplemental material) as well as 46 of 117 (39%) genes from other P. aeruginosa strains. Hence, the P. aeruginosa GeneChip recognizes at least 87% of the SCV 20265 genome.
Transcriptional profiles of P. aeruginosa 20265 morphotypes.
To obtain genome-wide transcriptional profiles of the different clonal morphotypes of strain 20265, bacteria were grown to exponential and to stationary growth phase. A good reproducibility (Pearson correlation of higher than r = 0.97) of transcript levels from duplicates of the same morphotype and growth stage was observed. We identified 309 P. aeruginosa PAO1 genes, 8 genes from other P. aeruginosa strains, and 2 intergenic regions to be differentially expressed (for more details, see Table S3 in the supplemental material). Forty-two percent of these genes encode proteins with unknown functions (confidence class 4; www.pseudomonas.com), which is barely below the proportion of 46% assigned for the genome of P. aeruginosa PAO1 (45).
In our previous study, we showed that in vitro-derived revertants from autoaggregative SCVs exhibit an intermediate phenotype between wild type and SCV with respect to biofilm formation, association with the pneumocytic cell line A549, surface hydrophobicity, and swimming motility. However, revertants showed an even better twitching motility than SCV and the wild type. These data indicate that revertants are not identical to the wild type (20). The comparative gene expression analysis performed here confirmed these results. In the exponential phase, the revertant resembled the wild type more closely than the SCV, indicated by the smallest number of differentially expressed genes (Table 1) and the highest correlation of the transcriptional profiles of all genes determined with Pearson correlation as a distance metric (data not shown). In contrast to the exponential phase, the revertant resembled the SCV more closely during stationary phase, with only 11 genes being differentially expressed (Table 1). As opposed to the pronounced difference in growth rate between SCV 20265, its clonal wild type, and the revertant on solid media (20), only minor differences in growth rate were observed during the exponential phase in liquid media (see Materials and Methods). Although, we cannot completely rule out that some of the observed differences in gene expression might be due to slight differences in growth rate of the different morphotypes, a significant impact is unlikely.
TABLE 1.
Number of differentially expressed genes between P. aeruginosa SCV 20265 and its clonal wild type and revertant during exponential and stationary growth
| Change (fold)a | No. of expressed genesb:
|
|||||
|---|---|---|---|---|---|---|
| Exponential phase
|
Stationary phase
|
|||||
| SCV/WT | SCV/REV | WT/REV | SCV/WT | SCV/REV | WT/REV | |
| Up | ||||||
| ≥10 | 38 | 2 | 16 | 1 | 0 | 11 |
| 5-10 | 13 | 17 | 5 | 10 | 0 | 10 |
| 2-5 | 29 | 51 | 18 | 4 | 0 | 40 |
| Down | ||||||
| ≥10 | 42 | 20 | 18 | 40 | 7 | 26 |
| 5-10 | 18 | 4 | 6 | 20 | 3 | 8 |
| 2-5 | 23 | 1 | 20 | 8 | 1 | 4 |
| Total | 163 | 95 | 83 | 83 | 11 | 99 |
Fold change is the arithmetic average of a change in gene expression that was significant (Affymetrix Microarray Suite Software) and in a magnitude of at least two-fold for all four pairings of each morphotype comparison.
WT, wild type; REV, revertant.
SCV 20265 showed differential expression of genes linked to growth as a biofilm.
It has been shown that type IV pili enable P. aeruginosa to move on surfaces to form confluent films (36) and caps of mushroom-shaped structures in later stages of biofilm development (24). The role of flagella in adherence and biofilm initiation is still controversial and seems to be dependent on the carbon source used as well as the experimental setup (static growth or within a flow chamber) with their various degrees in applied turbulence (25). Still, it was proposed that reduction of flagella may help to stabilize the three-dimensional structure of the mature biofilm (51). The increased ability of the SCV 20265 to form biofilms in a standard biofilm assay (20) might be a consequence of the differential expression of genes involved in the buildup of pili and flagella (Table 2). During the exponential and stationary growth phases, the two most important structural proteins of the bacterial flagellum, the type a flagellin encoded by the gene flaA/fliC (11, 46) and the flagellar capping protein encoded by fliD (2), were downregulated in SCV 20265 in comparison to the wild type. This is in agreement with the reduced swimming motility of SCV 20265 (20). The flaA gene, sequenced from P. aeruginosa PAK, has remarkable sequence differences from the type b flagellin fliC gene of PAO 1 and was therefore also put on the P. aeruginosa GeneChip. The upregulation of pilA in SCV 20265 in comparison to the wild type was confirmed before by Northern hybridizations (20), which correlate with the observed hyperpiliation and increased twitching motility of SCV 20265 (20). Similar to SCV 20265, the fast-growing revertant expressed pilA above the wild-type level but furthermore showed a higher expression of the recently characterized fleP, a gene that is responsible for the stable flagellar attachment to the cellular pole and the control of the length of type IV pili (11). It was shown that a fleP mutant produced elongated pili, fewer and structurally different flagella, and had an impaired twitching motility (11). One might hypothesize that the upregulation of fleP in the revertant of SCV 20265 and its gene product may lead to shorter but still abundant pili through upregulation of pilA and might play a role in the increased twitching and swimming capabilities and the reduced lower autoaggregative and adhesive phenotype of the revertant in comparison to the SCV 20265 (20).
TABLE 2.
Differential expression of selected genes in P. aeruginosa 20265 SCV, the wild type, and revertant during exponential and stationary growth
| Gene identitya | Protein namea | Change as fold arithmetic average (±SD)b
|
|||||
|---|---|---|---|---|---|---|---|
| Exponential phase
|
Stationary phase
|
||||||
| SCV/WT | SCV/REV | WT/REV | SCV/WT | SCV/REV | WT/REV | ||
| Type III secretion | |||||||
| PA0044 (exoT)c | Exoenzyme T | 67.1 (27.0) | 4.2 (1.5) | −15.3 (7.7) | − | − | − |
| PA1692 | Probable translocation protein | 9.6 (1.2) | 3.4 (0.9) | − | − | − | − |
| PA1694 (pscQ) | Translocation protein | 57.1 (46.1) | 3.8 (1.0) | −20.9 (19.2) | − | − | − |
| PA1695 (pscP) | Translocation protein | 7.1 (2.5) | 2.6 (0.3) | −2.7 (0.9) | − | − | − |
| PA1696 (pscO) | Translocation protein | 40.1 (1.4) | 4.2 (0.6) | −10.2 (2.2) | − | − | − |
| PA1697 | ATP synthase | 10.6 (1.0) | 3.4 (0.8) | −3.0 (0.5) | − | − | − |
| PA1698 (popN)c | Outer membrane protein | 29.9 (16.1) | 5.2 (1.0) | −6.0 (2.6) | − | − | − |
| PA1699 | Conserved hypothetical protein | 79.4 (20.4) | 4.7 (0.7) | −16.2 (7.6) | − | − | − |
| PA1700 | Conserved hypothetical protein | 36.9 (13.6) | 5.2 (1.7) | −7.2 (6.0) | − | − | − |
| PA1701 | Conserved hypothetical protein | 34.9 (10.3) | 4.9 (1.6) | −7.9 (3.4) | − | − | − |
| PA1702 | Conserved hypothetical protein | 43.0 (2.8) | 7.0 (1.7) | − | − | − | − |
| PA1703 (pcrD) | Secretory apparatus protein | 7.7 (1.1) | 3.3 (0.6) | − | − | − | − |
| PA1704 (pcrR) | Regulatory protein | 28.1 (4.5) | 4.5 (0.6) | − | − | − | − |
| PA1705 (pcrG) | Regulatory protein | 5.9 (2.5) | − | − | − | − | − |
| PA1706 (pcrV)c | Secretion protein | 268.2 (173.3) | 4.3 (1.0) | −54.7 (55.8) | 26.0 (8.2) | − | −8.8 (3.4) |
| PA1707 (pcrH) | Regulatory protein | 42.7 (7.0) | 4.2 (1.0) | −12.6 (3.0) | 9.6 (5.4) | − | − |
| PA1708 (popB)c | Translocator protein | 137.7 (33.4) | 3.4 (0.8) | −37.9 (10.7) | 7.5 (1.3) | − | −4.2 (0.4) |
| PA1709 (popD)c | Translocator protein | 60.9 (4.0) | 4.6 (1.1) | −12.0 (3.1) | − | − | − |
| PA1710 (exsC) | Exo S synthesis protein C precursor | 13.1 (2.1) | 4.0 (1.5) | −3.3 (0.6) | 8.5 (1.5) | − | −5.6 (1.0) |
| PA1711 | Hypothetical protein | 11.3 (0.6) | 3.2 (0.8) | −3.5 (0.7) | 8.2 (0.9) | −5.0 (0.9) | |
| PA1712 (exsB) | Exo S synthesis protein B | 5.3 (0.5) | 2.8 (0.3) | − | 8.2 (3.4) | − | − |
| PA1713 (exsA) | Transcriptional regulator ExsA | 5.6 (1.0)d | − | − | − | − | − |
| PA1714 | Hypothetical protein | 14.9 (0.0) | 4.9 (1.1) | −3.4 (0.6) | − | − | − |
| PA1715 (pscB) | Export protein | 45.7 (7.3) | 5.6 (1.0) | −13.2 (3.0) | − | − | − |
| PA1716 (pscC) | Secretion protein | 9.0 (1.6) | 3.2 (0.6) | − | − | − | − |
| PA1717 (pscD) | Export protein | 24.2 (12.0) | 5.6 (0.2) | −3.9 (0.9) | − | − | − |
| PA1718 (pscE) | Export protein | 33.2 (2.2) | 4.0 (0.2) | −8.5 (0.9) | − | − | − |
| PA1719 (pscF) | Export protein | 42.3 (27.7) | 3.6 (0.9) | −15.3 (11.0) | − | − | − |
| PA1720 (pscG) | Export protein | 21.1 (6.0) | 6.0 (0.6) | − | − | − | − |
| PA1721 (pscH) | Export protein | 9.9 (2.4) | 4.5 (1.1) | − | − | − | − |
| PA1722 (pscI) | Export protein | 11.5 (3.4) | 3.9 (0.9) | −3.9 (1.0) | − | − | − |
| PA1723 (pscJ) | Export protein | 4.9 (0.7) | 3.8 (0.3) | − | − | − | − |
| PA1724 (pscK) | Export protein | 6.4 (1.4) | 3.5 (0.8) | − | − | − | − |
| PA1725 (pscL) | Export protein | 12.3 (4.3) | 4.0 (0.7) | − | − | − | − |
| PA2191 (exoY)c | Adenylate cyclase | 31.7 (10.6) | 6.8 (2.7) | − | − | − | − |
| PA3841 (exoS)c | Exoenzyme S | 90.6 (33.3) | 3.9 (0.8) | −22.2 (11.2) | 5.5 (1.9) | − | − |
| PA3842 | Probable chaperone | 29.6 (11.6) | 3.2 (0.8) | −8.6 (3.8) | − | − | − |
| PA3843 | Hypothetical protein | 65.4 (6.4) | 4.2 (0.9) | −16.8 (3.5) | − | − | − |
| Motility and attachment | |||||||
| L37109 cds1 (pilA) | Pilin type IV | 7.5 (4.4) | − | −5.6 (2.2) | 5.2 (0.9) | − | −3.2 (0.5) |
| PA1079 (flgD) | Flagellar basal-body rod modification protein | − | − | − | − | − | −2.9 (0.5) |
| PA1080 (flgE) | Flagellar hook protein | − | −5.5 (1.6) | − | − | − | − |
| PA1082 (flgG) | Flagellar basal-body rod protein | − | −4.4 (1.4) | − | − | − | − |
| PA1085 (flgJ) | Flagellar protein | − | −4.2 (0.8) | − | − | − | − |
| AF332547 (orfB) | Putative acyl carrier protein from glycosylation island | −2.4 (0.3) | −3.7 (0.4) | − | − | − | − |
| AF332547 (orfC) | Putative 3-oxoacyl-(acyl-carrier-protein) synthase from glycosylation island | − | −3.0 (0.5) | − | − | − | − |
| AF332547 (orfF) | Large subunit aromatic dioxygenase from glycosylation island | − | −3.2 (0.2) | − | − | − | − |
| M57501 (flaA/fliC) | Flagellin type a | −6.2 (1.0) | −13.6 (2.0) | −2.4 (0.2) | −3.2 (0.6) | −8.2 (1.8) | −2.4 (0.3) |
| L81176 cds3 (fliD) | Flagellar cap protein | −3.0 (0.3) | −4.0 (0.5) | − | − | −2.6 (0.5) | − |
| L81176 cds5 (fliS) | Unknown function | − | −2.8 (0.5) | − | − | − | − |
| L81176 cds6 (fleP) | Type IV pili length control | − | −3.1 (0.6) | − | − | −6.5 (3.8) | − |
| PA1101 (fliF) | Flagellar M-ring protein | − | −5.2 (2.9) | − | − | − | − |
| Transport of small molecules | |||||||
| PA1862 (modB) | Molybdenum transport protein | 9.7 (2.0) | 4.3 (0.7) | − | − | − | − |
| PA1863 (modA) | Molybdate-binding periplasmic protein precursor | 5.5 (0.8) | 6.3 (3.1) | − | − | − | − |
| PA2386 (pvdA) | l-Ornithine N5-oxygenase | − | − | − | −8.4 (5.8) | − | 3.5 (1.1) |
| PA2398 (fpvA) | Ferripyoverdine receptor | − | − | − | −2.4 (0.5) | − | 2.6 (0.3) |
| PA3407 (hasAp) | Heme acquisition protein | − | − | − | −8.2 (4.6) | − | 6.0 (2.0) |
| PA3531 (bfrB) | Bacterioferritin | 4.5 (1.3) | − | −3.5 (0.6) | − | − | − |
| PA3790 (oprC) | Outer membrane protein | 25.8 (11.3) | 9.6 (7.5) | − | − | − | − |
| PA4218 | Probable transporter | 19.0 (15.8) | − | −12.3 (7.2) | − | − | − |
| PA4221 (fptA) | Fe(III)-pyochelin receptor precursor | 15.1 (2.6) | − | −11.0 (1.0) | − | − | −17.7 (15.7) |
| PA4223 (pchH) | Probable ATP-binding component of ABC transporter | 5.6 (0.7) | − | −4.1 (0.8) | − | − | −2.9 (0.5) |
| PA4224 (pchG) | Pyochelin biosynthetic protein | 3.9 (1.0) | − | −3.4 (0.9) | − | − | −4.1 (1.5) |
| PA4225 (pchF) | Pyochelin synthetase | 37.0 (5.5) | − | −36.7 (20.2) | − | − | − |
| PA4226 (pchE) | Dihydroaeruginoic acid synthetase | 37.4 (11.1) | − | −40.5 (16.9) | − | − | − |
| PA4228 (pchD) | Pyochelin biosynthesis protein | 14.2 (13.6) | − | −17.4 (16.7) | − | − | − |
| PA4229 (pchC) | Pyochelin biosynthetic protein | 34.2 (6.5) | − | −40.0 (17.3) | − | − | −4.4 (0.9) |
| PA4230 (pchB) | Salicylate biosynthesis protein | 24.3 (1.9) | − | −28.3 (8.4) | − | − | − |
| PA4231 (pchA) | Isochorismate synthase | 5.8 (0.4) | − | − | − | − | − |
| PA4764 (fur) | Ferric uptake regulation protein | − | 2.7 (0.4) | − | − | − | − |
| Disulfide bond formation and oxidative stress protection | |||||||
| PA0538 (dsbB) | Disulfide bond formation protein | −24.6 (11.1) | − | 6.2 (1.2) | − | − | 7.1 (3.6) |
| PA2475 | Probable cytochrome P450 | −16.3 (0.6) | − | 15.1 (8.5)d | − | − | − |
| PA2476 (dsbG) | Thiol-disulfide interchange protein | −58.9 (5.0) | − | 56.0 (6.3) | −11.7 (1.1) | − | 11.4 (1.8) |
| PA2477 | Probable thiol-disulfide interchange protein | −41.1 (5.6) | − | 132.8 (49.2) | −14.6 (11.7) | − | 37.0 (12.3) |
| PA2478 | Probable thiol-disulfide interchange protein | −111.7 (38.2) | − | 217.0 (56.1) | −49.6 (5.2) | − | 50.6 (22.9) |
| PA2479 | Probable two-component response regulator | −13.9 (4.0) | − | 9.2 (3.3) | −6.9 (1.7) | − | 6.5 (2.0) |
| PA2480 | Probable two-component sensor | −18.8 (4.8) | − | 82.5 (52.3) | −3.8 (0.9) | − | 6.5 (1.1) |
| PA2827 | Conserved hypothetical protein | 2.3 (0.2) | 3.4 (0.4) | − | − | − | − |
| PA3331 | Cytochrome P450 | − | − | − | −3.1 (0.6) | − | 7.6 (1.6) |
| PA5240 (trxA) | Thioredoxin | 2.3 (0.1) | 3.1 (0.7) | − | − | − | − |
| Translation, posttransla- tional modification, degradation | |||||||
| PA3049 (rmf) | Ribosome modulation factor | 4.8 (0.3) | 6.7 (2.6) | − | − | − | − |
Boldface genes were found as well in the proteome analysis of either this study or from Wehmhöner et al. (50). Note that the expression changes from PA2398 (fpvA) and PA3407 (hasAp) diverged from the proteome analysis (see Results and Discussion). PA number, gene, and protein name imposed by the P. aeruginosa Sequencing and Community Annotation Project (http://www.pseudomonas.com). For non-PAO1 genes, the GenBank accession numbers with the number of coding sequences (cds) are used to describe their identity.
For all genes with a significant differential expression (Affymetrix Microarray Suite software) in a magnitude of at least twofold in all four pairings of each morphotype comparison, the arithmetic average and SD were calculated. Upregulated genes are given as positive values, and downregulated genes are given as negative values. −, no significant differential expression. WT, wild type; REV, revertant.
Type III secreted proteins (54).
Expression was significantly different in three out of the four GeneChip pairings.
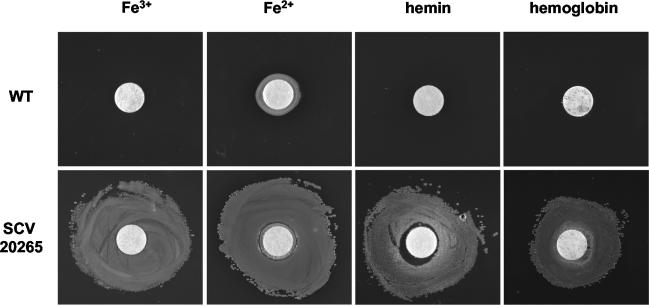
Besides the apparent cluster of motility genes, a second functional group of genes involved in iron supply of P. aeruginosa was found in our analysis to be differentially expressed (Table 2). A sufficient iron supply has recently been demonstrated as essential for the establishment of a P. aeruginosa biofilm (42). During the logarithmic phase of growth in minimal medium with only traces of iron, SCV 20265 and revertant showed a strong upregulation of genes coding for pyochelin-mediated iron uptake (37, 49). In the stationary phase of growth, genes of other systems, such as the Fe(III) siderophore pyoverdin (32) and the gene coding for the hemophore HasAp (33), were downregulated in SCV 20265 and revertant when compared to the wild type (Table 2). This is in contrast to the higher protein abundance of HasAp and the ferripyoverdin receptor protein FpvA found in SCV 20265 during exponential growth, as shown by proteome analysis (50) and an approximately threefold-larger amount of pyoverdin accumulating in the supernatant of SCV 20265 in comparison to the wild type during growth in the stationary phase (data not shown). Interestingly, a posttranscriptional regulation of FpvA and HasAp by quorum sensing has recently been proposed (1). The global iron uptake regulator fur (17) was slightly upregulated in SCV 20265 in comparison to the revertant, but we did not find differential expression between SCV 20265 and the wild type. We thus conclude from our array data that the differential gene expression of fur does not seem to contribute to the described differential gene expression of various iron uptake systems between SCV 20265 and the wild type. The increased expression of bfrB in SCV 20265 encoding the iron storage protein bacterioferritin known to be upregulated under high-iron conditions (42) might indicate that an increased pyochelin, pyoverdin, and HasAp-mediated iron uptake renders the SCV more competitive in iron acquisition during the exponential growth phase than the wild type. To test this hypothesis, growth experiments with SCV 20265 and the wild type on iron-depleted VB plates repleted with Fe3+, Fe2+, hemin, or hemoglobin showed that SCV 20265 can indeed cope with lower concentrations of iron in the medium than the wild type (Fig. 1). Among other autoaggregative SCVs tested, only SCV 52 showed the same growth advantage as the SCV 20265 with Fe3+ and Fe2+ repletion but not with hemin or hemoglobin (data not shown). In future studies, however, it has to be elucidated whether the growth phase-specific usage of iron uptake systems has a functional impact on the abilities of different morphotypes of P. aeruginosa in adapting to the iron-limited environment of the CF lung.
FIG. 1.
Differential response of SCV 20265 and the wild type (WT) to supplementation with different iron sources under iron depletion conditions. Iron-depleted VB agar plates were inoculated with bacteria and made replete by spotting of filter disks laid upon the agar with Fe3+ (100 mM), Fe2+ (200 mM), hemin (10 mM), or hemoglobin (0.1 mM). SCV 20265 shows significant growth around the disks in contrast to the wild type. The same results were obtained in more than three independent experiments.
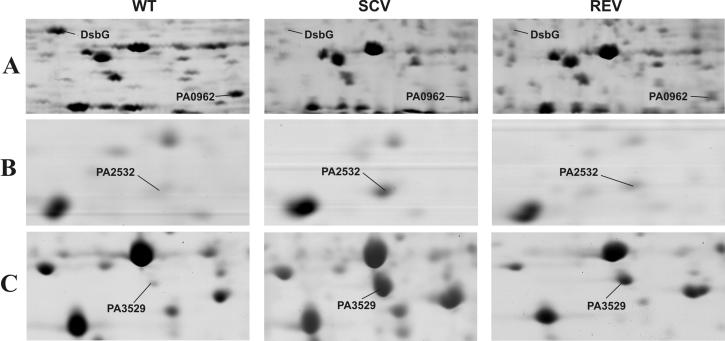
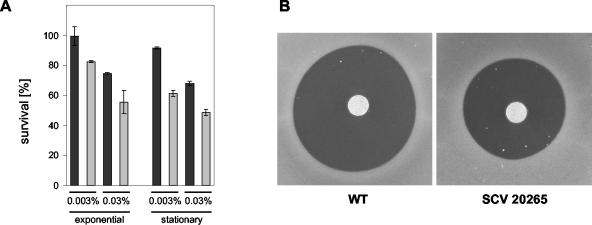
A third functional group of genes that could be linked to the biofilm mode of growth contained genes involved in the maintenance of the cellular redox status. The SCV 20265 in comparison to the fast-growing morphotypes wild type and revertant showed an upregulation of trxA encoding thioredoxin and PA2827 encoding a conserved hypothetical protein with a domain often found in peptide methionine sulfoxide reductases (Pfam assignment PF01641) (Table 2). Thioredoxin and peptide methionine sulfoxide reductases are known to reverse the inactivation of many proteins due to oxidation of thiol-containing residues (28). Genes involved in the periplasmic formation of disulfide bonds (dsbB, dsbG, PA2477, and PA2478), required for the stabilization of exported proteins (7), were reduced in expression in SCV 20265 and the revertant compared with the wild type. Referring to their expression profile, dsbG (PA2476), PA2477, and PA2478 most likely form an operon with PA2475 encoding a probable cytochrome P450. Cytochrome P450 proteins are involved in degradative processes and known to produce superoxide anion radicals as by-products (15). The gene expression data suggest that the operon PA2475-PA2478 is coregulated with a two-component system, PA2479-PA2480, encoded on the opposite strand (Table 2). This may indicate a regulative role of this two-component system for the operon PA2475-PA2478. Besides PA2475, another cytochrome P450 (PA3331) was found during transcriptional analysis. PA3331 was shown to be downregulated in stationary-phase-grown SCV 20265 and the revertant in comparison to the wild type (Table 2). The above-described differential regulation of redox active enzymes may be part of a complex adaptation of SCV 20265 to an altered redox status in the CF lung. This proposal was supported by proteome analysis. Here, the decreased gene expression of dsbG in SCV 20265 and the revertant in both growth phases was shown to result in a negligible amount of protein (Fig. 2A). Furthermore, proteome analysis revealed additional proteins involved in oxidative stress to be differentially expressed. SCV 20265 exhibited an overproduction of the chaperone GroEL in comparison to the wild type (not shown), whereas a probable DNA-binding stress protein (PA0962) was much less expressed in SCV 20265 (Fig. 2A). Moreover in stationary-phase cultures, a thiol peroxidase (PA2532) (Fig. 2B) and a probable peroxidase (PA3529) (Fig. 2C) were shown to be overexpressed in SCV 20265. To test the hypothesis that the differential regulation of these redox active genes might have an impact on the resistance of SCV 20265 against oxidative stress, we tested whether the autoaggregative SCV 20265 is more resistant against H2O2 than its corresponding wild type (Fig. 3). Exposure of exponential- and stationary-phase-grown SCV 20265 to H2O2 in liquid medium (Fig. 3A) as well as on solid agar (Fig. 3B) revealed a higher level of resistance than the respective wild type. However, under these experimental conditions, no other autoaggregative SCV tested revealed a higher H2O2 resistance compared to their respective wild types (data not shown).
FIG. 2.
Differential expression of proteins related to oxidative stress. Cells were grown to the stationary phase. Cellular extracts (A) or supernatants (B and C) were subjected to two-dimensional gel electrophoresis and subsequent MALDI-TOF MS analysis for protein identification. WT, wild type; REV, revertant.
FIG. 3.
Sensitivity of autoaggregative SCV 20265 and its clonal wild type (WT) to H2O2. (A) Bacteria were grown in VB medium to either the exponential or stationary phase and incubated with 0.03% (vol/vol) H2O2 or 0.003% (vol/vol) H2O2 followed by catalase treatment and determination of CFU. Percent survival is given compared to that of nontreated cells. SCV 20265 (black bars) and its respective wild type (gray bars) were tested in two independent experiments with triplicate values each. SCV 20265 showed a higher resistance against H2O2 than the wild type in both growth phases. (B) For filter disk assays, bacteria were grown to the stationary phase in VB medium, inoculated in LB soft agar, and poured on LB agar plates (1.5% [wt/vol]). Sterile filter disks were laid on top, and the disks were spotted with 30% (vol/vol) H2O2. In a representative experiment, SCV 20265 showed a higher resistance against H2O2 with a growth inhibition diameter of 19.7 (0.3) mm while the wild type did not grow within 23.6 (0.2) mm around the disk. Similar results were obtained in more than three independent experiments. Single plates for SCV 20265 and its wild type are shown (B).
The peroxidases as well as the thioredoxin protein TrxA mentioned above have been found to be induced in P. aeruginosa biofilms, especially when the biofilm was raised under anaerobic conditions (57). Other genes known to be upregulated in anaerobic environments were shown to be upregulated in SCV 20265, like modA and modB necessary for molybdate uptake and hence nitrate respiration and denitrification (41) as well as oprC, an outer membrane protein involved in copper uptake (56) and recently shown to be produced in larger amounts in SCV 20265 (51) (Table 2). Copper is a cofactor of the protein azurin probably involved in detoxification of oxidative NO produced during the process of denitrification (48). Another response to stress may be the upregulation of the rmf gene. It encodes a protein necessary for survival of the cell when growth rate decreases (55) and shows increased expression in SCV 20265 during the exponential growth phase (Table 2).
In conclusion, the transcriptional profile of SCV 20265 obtained in oxygenated planctonic culture indicates that this clonal variant might be especially adapted to oxidative stress, low free iron, and anaerobic conditions as well as a biofilm mode of growth. All of these conditions are likely to be encountered within the habitat of the CF lung (26, 53).
SCV 20265 exhibits a constitutive type III secretion-dependent cytotoxicity.
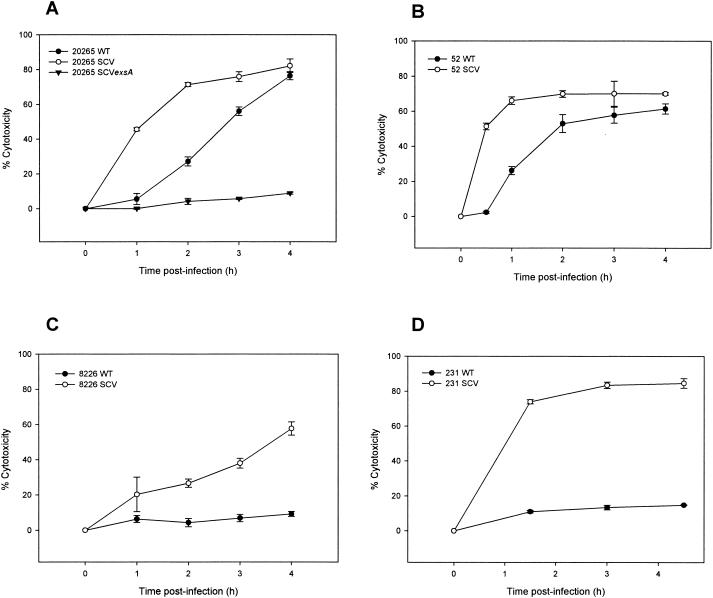
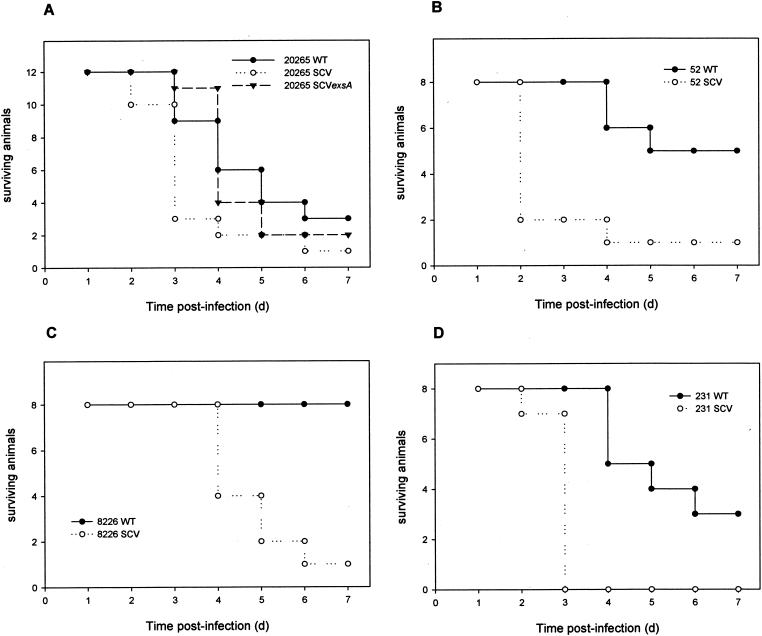
The TTSS and its secreted products are considered to be a major virulence factor of P. aeruginosa and other gram-negative bacterial pathogens (23). After attachment to eukaryotic host cells, the TTSS enables these bacteria to inject effector proteins directly into the cytosol of the host cells (8, 23). TTSS effector proteins of P. aeruginosa are the ADP-ribosyltransferases ExoT and ExoS, the adenylate cyclase ExoY, and the lipase cytotoxin ExoU (PepA) (40). In the previous proteome analysis, we found four TTSS proteins (PopN, PcrV, PopB, and PopD) as well as ExoT and ExoS overproduced in the SCV 20265 in comparison to the wild type (50). With the microarray approach, we found constitutively higher expression of 33 of 36 genes of the TTSS gene cluster (PA1690 to PA1725) in the SCV 20265 and the revertant during exponential growth (Table 2). Most strikingly, expression of genes coding for the effector proteins ExoT, ExoS, and ExoY located outside of the TTSS cluster on the P. aeruginosa chromosome showed massive upregulation of more than 30-fold in the SCV 20265 compared to the corresponding wild type. The exsA gene, encoding a global TTSS regulator (22) was also upregulated in the SCV 20265 compared to the wild type. This upregulation may account for the increased TTSS expression in SCV 20265. To test if the high transcript level of the TTSS and effector proteins in the SCV 20265 is biologically meaningful, we determined the in vitro cytotoxicity by using the murine macrophage cell line J774.A1. Figure 4 shows that SCV 20265 was significantly more cytotoxic than the wild type. The increased cytotoxicity of SCV 20265 was proven to be TTSS dependent, as a chromosomal knockout of exsA in SCV 20265 led to a significantly reduced cytotoxicity (Fig. 4A). The cytotoxicity of SCV 20265 was ExoU independent, as the strain lacks the exoU gene, which was determined by genome analysis with the GeneChips. This is a trait frequently observed for CF isolates (14). It is likely that the cytotoxicity of SCV 20265 is mediated through the introduction of small pores into the macrophage membrane as has been shown for strain CHA (10). The genes encoding PopB, PopD, and PcrV, essential for the formation of these pores (10), were found to be upregulated more than 60-fold in SCV 20265 compared to the wild type. Testing of altogether seven clinical autoaggregative SCV isolates revealed four strains (SCV 20265, SCV 52, SCV 8226, and SCV 231) with increased cytotoxicity (Fig. 4A to D), whereas within a group of eight nonautoaggregative clinical SCVs, none showed this characteristic. A highly significant dependency between the autoaggregative SCV phenotype and an increased cytotoxicity was revealed by a chi-square test (P < 0.025). The analysis of proteins secreted into the culture supernatants showed a significantly higher expression of TTSS effector proteins by the already mentioned two-dimensional gel electrophoresis and subsequent MALDI-TOF MS analysis for SCV 20265 (50) and by one-dimensional sodium dodecyl sulfate gel electrophoresis for ExoT, ExoS, PcrV, and PopB as well as immunoblotting for ExoS and PcrV in strains SCV 20265, SCV 52, SCV 8226, and SCV 231 (data not shown). This indicates that the more pronounced cytotoxicity of these SCVs was also most likely due to type III secretion. Respiratory tract infection of BALB/c mice showed a significantly higher virulence of SCV 20265 than that of the wild type (P = 0.01) and the exsA mutant of SCV 20265 (P < 0.01) at day 3 postinfection (Fig. 5A) and of SCV 52 (day 3; P < 0.01), SCV 8226 (day 5; P < 0.01), and SCV 231 (day 3; P < 0.01) in comparison to their corresponding wild types, respectively (Fig. 5B to D). Within the first 48 h, SCV 20265-infected mice were physically inactive and suffered from more severe dyspnea than the wild-type- and 20265 SCV exsA mutant-infected mice, resulting in 75% dead animals at day 3 compared to 25% for the wild type and 8% for the exsA mutant. However, at later time points, these differences were less pronounced, indicating that TTSS-mediated virulence was important for the acute toxicity observed in SCV 20265 during the early phase of infection. Beyond day 3, the difference in mortality between SCV and the wild type in strains 52, 8226, and 231 was more pronounced than that in strain 20265 (Fig. 5B to D).
FIG. 4.
Cytotoxicity of P. aeruginosa wild types (WT) and autoaggregative SCVs towards J774.A1 macrophages. P. aeruginosa 20265 SCV, 20265 SCV exsA, or wild type (A), 52 SCV or wild type (B), 8226 SCV or wild type (C), and 231 SCV or wild type (D) were used. The percentage of cytotoxicity was calculated according to the release of LDH at various time points. Values are the means ± SD of triplicate determinations. Representative experiments are shown. Note the higher cytotoxicity of SCVs in comparison to wild types and the loss of cytotoxicity of the 20265 SCV exsA mutant (A).
FIG. 5.
Survival of BALB/c mice after intranasal inoculation with either P. aeruginosa 20265 SCV, 20265 SCV exsA, or wild type (WT) (A), 52 SCV or wild type (B), 8226 SCV or wild type (C), and 231 SCV or wild type (D). Pooled data of two independent infection experiments for each strain are shown. At day 3 postinfection, there is a significant difference in mortality between either wild-type 20265 (P = 0.01) or SCV 20265 exsA (P < 0.01) compared to SCV 20265. Mortality is also significantly different between the wild types and the SCVs of strains 52 (day 3; P < 0.01), 8226 (day 5; P < 0.01), and 231 (day 3; P < 0.01).
In this study, we demonstrated that the differential gene expression of a clinical autoaggregative SCV in comparison with its wild type and revertant correlates with functional properties such as motility, iron uptake, resistance against oxidative stress, as well as cytotoxicity. As it is very likely that mutations or small insertions or deletions (which cannot be resolved by SpeI restriction) unrelated to the SCV morphotype are responsible for some of the differential gene expression observed between the 20265 morphotypes, it is not surprising that SCV 20265 also exhibits functional properties, which are not shared by other SCVs. Therefore, one has to be cautious in drawing general conclusions with respect to other autoaggregative SCVs from the expression signature of SCV 20265. However, there is obviously a link between the autoaggregative hyperpiliated SCV morphotype and increased TTSS-mediated cytotoxicity. In this context, upregulation of type IV pili in SCV 20265 not only may enhance adhesion and autoaggregation but also may contribute to increased TTSS-mediated cytotoxicity through increased adhesion to host cells (52). Recently, Wolfgang et al. observed that expression of TTSS genes are not only influenced by the ExsA protein regulator but as well by high levels of cyclic AMP (cAMP) and the cAMP binding protein gene vfr (52). Interestingly, high cAMP levels result along with a higher TTSS expression in higher expression of genes coding for type IV pili as well as in lower expression of genes from the flagellar apparatus of P. aeruginosa (52). Although higher expression of the cAMP-producing adenylate cyclases cyaA and cyaB could not be found in our transcriptome analysis, it might be that an increased cAMP level is involved in the differential expression of flagella, type IV pili, and the TTSS in SCV 20265.
It is generally accepted that the habitat of a chronically infected CF lung selects for less virulent P. aeruginosa strains (5, 29, 39). Isolates from CF patients were reported to produce lower levels of protease, elastase, and exotoxin A (5) and lower levels of type III toxins, such as ExoT, ExoS, and ExoU, as well as less PcrV (39). The results of this study, which show that some P. aeruginosa SCV morphotypes exhibit an increased type III secretion-mediated virulence, contradict this general observation. As the risk of mortality after P. aeruginosa infection was shown to be increased considerably when TTSS effector proteins are expressed (39), SCVs are likely to have an impact on the prognosis of CF patients and diagnostic laboratories should therefore take precautions in detecting this otherwise easily missed morphotype (13, 19).
Supplementary Material
Acknowledgments
This work was supported by a grant from the Cystic Fibrosis Foundation (United States) and the Mukoviszidose e.V. (Germany) to I.S. D.J. S.S.S. and A.S. are supported by the European Graduate College “Pseudomonas: Biotechnology and Pathogenicity” (Deutsche Forschungsgemeinschaft).
We are most grateful for the interest and strong support of Dieter Bitter-Suermann and for the excellent technical assistance of Tanja Toepfer, Jessika Garlisch, Birgit Brenneke, and Jaqueline Chabert.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org.
REFERENCES
- 1.Arevalo-Ferro, C., M. Hentzer, G. Reil, A. Gorg, S. Kjelleberg, M. Givskov, K. Riedel, and L. Eberl. 2003. Identification of quorum-sensing regulated proteins in the opportunistic pathogen Pseudomonas aeruginosa by proteomics. Environ. Microbiol. 5:1350-1369. [DOI] [PubMed] [Google Scholar]
- 2.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1998. The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect. Immun. 66:1000-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakay, M., Y. W. Chen, R. Borup, P. Zhao, K. Nagaraju, and E. P. Hoffman. 31 January 2002, posting date. Sources of variability and effect of experimental approach on expression profiling data interpretation. BMC Bioinformatics 3:4. [Online.] http://www.biomedcentral.com. [DOI] [PMC free article] [PubMed]
- 4.Breitenstein, S., S. Walter, J. Bosshammer, U. Römling, and B. Tümmler. 1997. Direct sputum analysis of Pseudomonas aeruginosa macrorestriction fragment genotypes in patients with cystic fibrosis. Med. Microbiol. Immunol. 186:93-99. [DOI] [PubMed] [Google Scholar]
- 5.Burke, V., J. O. Robinson, C. J. Richardson, and C. S. Bundell. 1991. Longitudinal studies of virulence factors of Pseudomonas aeruginosa in cystic fibrosis. Pathology 23:145-148. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Y. W., P. Zhao, R. Borup, and E. P. Hoffman. 2000. Expression profiling in the muscular dystrophies: identification of novel aspects of molecular pathophysiology. J. Cell Biol. 151:1321-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collet, J. F., and J. C. Bardwell. 2002. Oxidative protein folding in bacteria. Mol. Microbiol. 44:1-8. [DOI] [PubMed] [Google Scholar]
- 8.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 9.Dacheux, D., I. Attree, C. Schneider, and B. Toussaint. 1999. Cell death of human polymorphonuclear neutrophils induced by a Pseudomonas aeruginosa cystic fibrosis isolate requires a functional type III secretion system. Infect. Immun. 67:6164-6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dacheux, D., J. Goure, J. Chabert, Y. Usson, and I. Attree. 2001. Pore-forming activity of type III system-secreted proteins leads to oncosis of Pseudomonas aeruginosa-infected macrophages. Mol. Microbiol. 40:76-85. [DOI] [PubMed] [Google Scholar]
- 11.Dasgupta, N., M. C. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50:809-824. [DOI] [PubMed] [Google Scholar]
- 12.Déziel, E., Y. Comeau, and R. Villemur. 2001. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 183:1195-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drenkard, E., and F. M. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740-743. [DOI] [PubMed] [Google Scholar]
- 14.Feltman, H., G. Schulert, S. Khan, M. Jain, L. Peterson, and A. R. Hauser. 2001. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147:2659-2669. [DOI] [PubMed] [Google Scholar]
- 15.Goeptar, A. R., H. Scheerens, and N. P. Vermeulen. 1995. Oxygen and xenobiotic reductase activities of cytochrome P450. Crit. Rev. Toxicol. 25:25-65. [DOI] [PubMed] [Google Scholar]
- 16.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 18.Hassett, D. J., J. F. Ma, J. G. Elkins, T. R. McDermott, U. A. Ochsner, S. E. West, C. T. Huang, J. Fredericks, S. Burnett, P. S. Stewart, G. McFeters, L. Passador, and B. H. Iglewski. 1999. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 34:1082-1093. [DOI] [PubMed] [Google Scholar]
- 19.Häussler, S., B. Tümmler, H. Weissbrodt, M. Rohde, and I. Steinmetz. 1999. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin. Infect. Dis. 29:621-625. [DOI] [PubMed] [Google Scholar]
- 20.Häussler, S., I. Ziegler, A. Lottel, F. von Götz, M. Rohde, D. Wehmhohner, S. Saravanamuthu, B. Tümmler, and I. Steinmetz. 2003. Highly adherent small-colony variants of Pseudomonas aeruginosa in cystic fibrosis lung infection. J. Med. Microbiol. 52:295-301. [DOI] [PubMed] [Google Scholar]
- 21.Hoiby, N. 1993. Antibiotic therapy for chronic infection of Pseudomonas in the lung. Annu. Rev. Med. 44:1-10. [DOI] [PubMed] [Google Scholar]
- 22.Hovey, A. K., and D. W. Frank. 1995. Analyses of the DNA-binding and transcriptional activation properties of ExsA, the transcriptional activator of the Pseudomonas aeruginosa exoenzyme S regulon. J. Bacteriol. 177:4427-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klausen, M., A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 50:61-68. [DOI] [PubMed] [Google Scholar]
- 25.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 26.Knowles, M. R., and R. C. Boucher. 2002. Mucus clearance as a primary innate defense mechanism for mammalian airways. J. Clin. Investig. 109:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam, J., R. Chan, K. Lam, and J. W. Costerton. 1980. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect. Immun. 28:546-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowther, W. T., N. Brot, H. Weissbach, J. F. Honek, and B. W. Matthews. 2000. Thiol-disulfide exchange is involved in the catalytic mechanism of peptide methionine sulfoxide reductase. Proc. Natl. Acad. Sci. USA 97:6463-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luzar, M. A., and T. C. Montie. 1985. Avirulence and altered physiological properties of cystic fibrosis strains of Pseudomonas aeruginosa. Infect. Immun. 50:572-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma, J.-F., U. A. Ochsner, M. G. Klotz, V. K. Nanayakkara, M. L. Howell, Z. Johnson, J. E. Posey, M. L. Vasil, J. J. Monaco, and D. J. Hassett. 1999. Bacterioferritin A modulates catalase A (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa. J. Bacteriol. 181:3730-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer, J.-M., A. Neely, A. Stintzi, C. Georges, and I. A. Holder. 1996. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 64:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochsner, U. A., Z. Johnson, and M. L. Vasil. 2000. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 146:185-198. [DOI] [PubMed] [Google Scholar]
- 34.Ochsner, U. A., P. J. Wilderman, A. I. Vasil, and M. L. Vasil. 2002. GeneChip(R) expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45:1277-1287. [DOI] [PubMed] [Google Scholar]
- 35.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 36.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 37.Reimmann, C., L. Serino, M. Beyeler, and D. Haas. 1998. Dihydroaeruginoic acid synthetase and pyochelin synthetase, products of the pchEF genes, are induced by extracellular pyochelin in Pseudomonas aeruginosa. Microbiology 144:3135-3148. [DOI] [PubMed] [Google Scholar]
- 38.Römling, U., K. D. Schmidt, and B. Tümmler. 1997. Large genome rearrangements discovered by the detailed analysis of 21 Pseudomonas aeruginosa clone C isolates found in environment and disease habitats. J. Mol. Biol. 271:386-404. [DOI] [PubMed] [Google Scholar]
- 39.Roy-Burman, A., R. H. Savel, S. Racine, B. L. Swanson, N. S. Revadigar, J. Fujimoto, T. Sawa, D. W. Frank, and J. P. Wiener-Kronish. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183:1767-1774. [DOI] [PubMed] [Google Scholar]
- 40.Sato, H., D. W. Frank, C. J. Hillard, J. B. Feix, R. R. Pankhaniya, K. Moriyama, V. Finck-Barbancon, A. Buchaklian, M. Lei, R. M. Long, J. Wiener-Kronish, and T. Sawa. 2003. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 22:2959-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Self, W. T., A. M. Grunden, A. Hasona, and K. T. Shanmugam. 1999. Transcriptional regulation of molybdoenzyme synthesis in Escherichia coli in response to molybdenum: ModE-molybdate, a repressor of the modABCD (molybdate transport) operon is a secondary transcriptional activator for the hyc and nar operons. Microbiology 145:41-55. [DOI] [PubMed] [Google Scholar]
- 42.Singh, P. K., M. R. Parsek, E. P. Greenberg, and M. J. Welsh. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417:552-555. [DOI] [PubMed] [Google Scholar]
- 43.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 44.Smith, J. J., S. M. Travis, E. P. Greenberg, and M. J. Welsh. 1996. Cystic fibrosis airway epithelia fail to kill bacteria because of abnormal airway surface fluid. Cell 85:229-236. [DOI] [PubMed] [Google Scholar]
- 45.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 46.Totten, P. A., and S. Lory. 1990. Characterization of the type a flagellin gene from Pseudomonas aeruginosa PAK. J. Bacteriol. 172:7188-7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vasil, M. L., and U. A. Ochsner. 1999. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol. Microbiol. 34:399-413. [DOI] [PubMed] [Google Scholar]
- 48.Vijgenboom, E., J. E. Busch, and G. W. Canters. 1997. In vivo studies disprove an obligatory role of azurin in denitrification in Pseudomonas aeruginosa and show that azu expression is under control of rpoS and ANR. Microbiology 143:2853-2863. [DOI] [PubMed] [Google Scholar]
- 49.Wang, J., S. Lory, R. Ramphal, and S. Jin. 1996. Isolation and characterization of Pseudomonas aeruginosa genes inducible by respiratory mucus derived from cystic fibrosis patients. Mol. Microbiol. 22:1005-1012. [DOI] [PubMed] [Google Scholar]
- 50.Wehmhöner, D., S. Häussler, B. Tümmler, L. Jansch, F. Bredenbruch, J. Wehland, and I. Steinmetz. 2003. Inter- and intraclonal diversity of the Pseudomonas aeruginosa proteome manifests within the secretome. J. Bacteriol. 185:5807-5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 52.Wolfgang, M. C., V. T. Lee, M. E. Gilmore, and S. Lory. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4:253-263. [DOI] [PubMed] [Google Scholar]
- 53.Worlitzsch, D., R. Tarran, M. Ulrich, U. Schwab, A. Cekici, K. C. Meyer, P. Birrer, G. Bellon, J. Berger, T. Weiss, K. Botzenhart, J. R. Yankaskas, S. Randell, R. C. Boucher, and G. Döring. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Investig. 109:317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yahr, T. L., L. M. Mende-Mueller, M. B. Friese, and D. W. Frank. 1997. Identification of type III secreted products of the Pseudomonas aeruginosa exoenzyme S regulon. J. Bacteriol. 179:7165-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamagishi, M., H. Matsushima, A. Wada, M. Sakagami, N. Fujita, and A. Ishihama. 1993. Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: growth phase and growth rate-dependent control. EMBO J. 12:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoneyama, H., and T. Nakae. 1996. Protein C (OprC) of the outer membrane of Pseudomonas aeruginosa is a copper-regulated channel protein. Microbiology 142:2137-2144. [DOI] [PubMed] [Google Scholar]
- 57.Yoon, S. S., R. F. Hennigan, G. M. Hilliard, U. A. Ochsner, K. Parvatiyar, M. C. Kamani, H. L. Allen, T. R. DeKievit, P. R. Gardner, U. Schwab, J. J. Rowe, B. H. Iglewski, T. R. McDermott, R. P. Mason, D. J. Wozniak, R. E. Hancock, M. R. Parsek, T. L. Noah, R. C. Boucher, and D. J. Hassett. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3:593-603. [DOI] [PubMed] [Google Scholar]
- 58.Zierdt, C. H., and P. J. Schmidt. 1964. Dissociation in Pseudomonas aeruginosa. J. Bacteriol. 87:1003-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.