Abstract
Organelle sequences have a long history of utility in phylogenetic analyses. Chloroplast sequences when combined with nuclear data can help resolve relationships among flowering plant genera, and within genera incongruence can point to reticulate evolution. Plastome sequences are becoming plentiful because they are increasingly easier to obtain. Complete plastome sequences allow us to detect rare rearrangements and test the tempo of sequence evolution. Chloroplast sequences are generally considered a nuisance to be kept to a minimum in bacterial artificial chromosome libraries. Here, we sequenced two bacterial artificial chromosomes per species to generate complete plastome sequences from seven species. The plastome sequences from Glycine syndetika and six other perennial Glycine species are similar in arrangement and gene content to the previously published soybean plastome. Repetitive sequences were detected in high frequencies as in soybean, but further analysis showed that repeat sequence numbers are inflated. Previous chloroplast-based phylogenetic trees for perennial Glycine were incongruent with nuclear gene–based phylogenetic trees. We tested whether the hypothesis of introgression was supported by the complete plastomes. Alignment of complete plastome sequences and Bayesian analysis allowed us to date putative hybridization events supporting the hypothesis of introgression and chloroplast “capture.”
Keywords: incongruence, divergence dates, repetitive sequences, inversions
The flowering plant (angiosperm) chloroplast genome (plastome) has a wide range of uses in plant biology. Much of this is due to its behavior as a single phylogenetic unit of tightly linked genes, typically comprising 125 and 160 kb. In the majority of angiosperms, the plastome is uniparentally (generally maternally) inherited (Corriveau and Coleman 1988). Plastome sequences are useful for species identification (Nock et al. 2011; Kane et al. 2012) and biotechnology applications (Sabir et al. 2014; Ruhlman and Jansen 2014), and have a long history of utility for phylogenetic inference (Palmer et al. 1988b; Jansen et al. 2007; Ruhfel et al. 2014). Plastome sequence data combined with genes from the mitochondrion and nucleus can provide the ability to resolve ancient, higher-order relationships among genera that have remained unresolved until adequate data became available (Soltis et al. 2011). At the genus level and below, uniparentally inherited plastome data can indicate reticulate evolution when incongruence between plastome-derived trees do not agree with other data such as morphology or nuclear loci (Rieseberg and Soltis 1991).
Angiosperm chloroplast sequences generally evolve more slowly than angiosperm nuclear sequences (Palmer 1990). Organelle size, gene content, and gene order have remained quite conserved among land plants as compared with mitochondrial genome sequences. The typical chloroplast genome, comprising approximately 150 kb, is composed of four regions, a large single copy (LSC) region, and a small single copy region (SSC), separated by a pair of large inverted repeats (IR). Rearrangements in chloroplast gene order are generally found in taxa that have one of the following qualities: changes in the size of the IRs or complete loss of one copy of the repeat; a high frequency of small dispersed repeats; biparental chloroplast inheritance; or complete or near-complete absence of photosynthesis (Wicke et al. 2011). Although inversions are not common in angiosperm chloroplasts, there are several families that are known for having a number of chloroplast genome inversions: Geraniaceae (Weng et al. 2014; Guisinger et al. 2011); Onagraceae (Greiner et al. 2008); Campanulaceae (Haberle et al. 2008); Asteraceae (Kim et al. 2005); and Leguminosae (Cai et al. 2008).
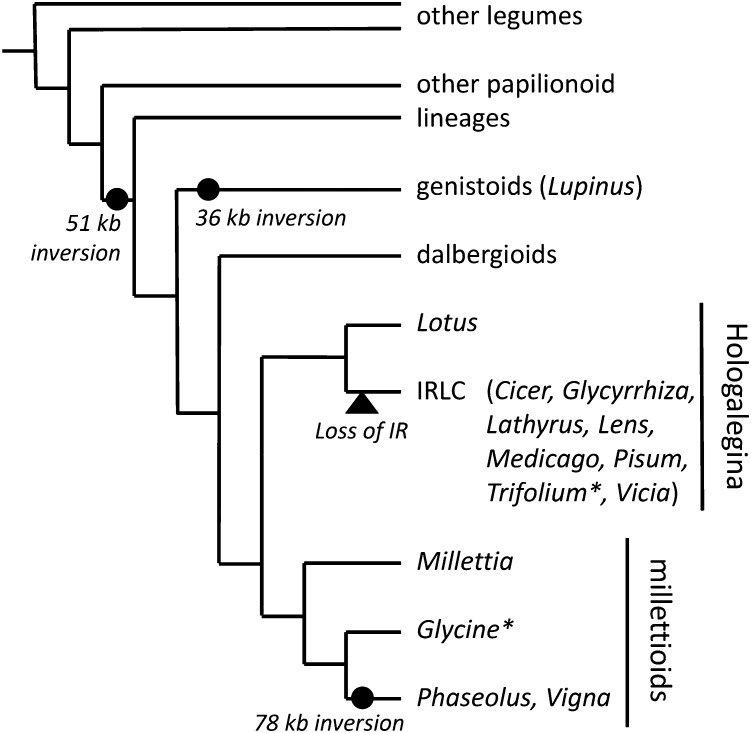
The Leguminosae or legume family is the third largest land plant family. It has been divided into three subfamilies, only two of which, Mimosideae and Papilionoideae, are monophyletic (LPWG: et al. 2013). Research interests centered on the papilionoids are driven by the wide array of grain, oil, and forage legumes in the subfamily, encompassing at least 24 economically important genera from peanuts to beans (Cannon et al. 2009). Prior to the current study, there were 17 complete plastome sequences published from 14 papilionoid legume genera (Figure 1). One of the largest clades of the Papilionoideae is characterized by the loss of one copy of the IR and is thus called the “inverted repeat loss (or lacking) clade” (Wojciechowski et al. 2000). One genistoid chloroplast genome has recently been published, Lupinus luteus, European yellow lupine (Martin et al. 2014). Chloroplast genomes from species representing eight genera in the IRLC have been sequenced: Trifolium aureum; T. grandiflorum; T. repens and T. subterraneum (clover); Medicago truncatula (barrel medic); Cicer arietinum (chickpea); Pisum sativum (pea); Lathyrus sativus (grass pea); Glycyrrhiza glabra (licorice); and Lens culinaris (lentil) and Vicia faba (broad bean) (Sabir et al. 2014; Saski et al. 2005; Cai et al. 2008; Jansen et al. 2008; Magee et al. 2010). The IRLC is sister to the clade that includes the model legume Lotus, which retains both IRs (Kato et al. 2000); together, these clades comprise the Hologalegina (Figure 1). Sister to Hologalegina is the millettioid clade (Wojciechowski et al. 2004), from which complete chloroplast genome sequences have been obtained from Milletia pinnata (= Pongamia pinnata, pongam oiltree), as well as three phaseoloid genera, Vigna radiata (mung bean), Phaseolus vulgaris (common bean), and Glycine max (soybean) (Saski et al. 2005; Tangphatsornruang et al. 2010; Guo et al. 2007; Kazakoff et al. 2012).
Figure 1.
Partial representation of legume phylogeny. Fourteen genera with sequenced plastomes are named. Asterisks indicate genera with multiple species with plastome sequences.
In the legume family, a 51-kb inversion is shared by most members of subfamily Papilionoideae (Doyle et al. 1990b, 1996; Palmer et al. 1988a). This inversion is present in Glycine chloroplast genomes and occurs in the LSC region, changing the gene order between trnK and accD. Three additional inversions have been reported, a newly described 36-kb inversion within the 51-kb inversion is present in Lupinus and other genisotoids (Martin et al. 2014), a 78-kb inversion shared by several closely related genera including Phaseolus and Vigna (Guo et al. 2007; Tangphatsornruang et al. 2010; Bruneau et al. 1990), and a 5.6-kb inversion reported in Milletia (Kazakoff et al. 2012). The 78-kb inversion spans almost the entire LSC between trnH and rps19, returning the genes in the 51-kb inversion to the order found in most land plants (Guo et al. 2007). This change in gene order has also been attributed to the expansion and contraction of the IR, leaving the gene order as described in papilionoids with the 51-kb inversion but changing the genes bordering the IR (Tangphatsornruang et al. 2010; Perry et al. 2002).
Legume chloroplasts also show variation in the presence and absence of genes. All legumes are missing two chloroplast encoded genes, infA and rpl22 (Doyle et al. 1995), and both have nuclear copies targeted to the chloroplast (Gantt et al. 1991; Millen et al. 2001). Loss of rps16 from the chloroplast has been reported in a number of legume lineages excluding Glycine. The mitochondrial copy is dual-targeted to both the mitochondria and chloroplast (Jansen et al. 2008; Sabir et al. 2014). Intron losses in clpP and rps12, found in IRLC lineage (Jansen et al. 2008), are not detected in Glycine (Saski et al. 2005).
The genus Glycine includes at least 28 species divided into two subgenera. The annuals include G. soja and the cultivated soybean, G. max, which are native to eastern Asia, whereas the majority of species are perennials found in Australia. Early investigations grouped Glycine species into “genome groups” (designated by letters A–I) based on the fertility of artificially produced hybrids and the degree to which meiotic chromosomes paired (Singh and Hymowitz 1985). These data, combined with isozyme data and sequences of two nuclear loci [the nuclear ribosomal gene cistron internal transcribed spacer (nrDNA ITS) and the low copy gene histone H3D], have been used to delimit nine genome groups as reviewed by Ratnaparkhe et al. (2011).
Chloroplast data from annual and perennial Glycine species have been used in genetic diversity studies and in phylogenetic studies (Doyle et al. 1990b, c; Sakai et al. 2003; Xu et al. 2000, 2001, 2002) including investigations of neopolyploidy in perennial taxa (Doyle et al. 1990a, 2004a). For the perennial subgenus as a whole, Doyle et al. (1990b) identified three major clades, termed “plastome groups,” which showed varying degrees of agreement with nuclear genome groups. The B-plastomes and C-plastomes were found to characterize species of the B-genome and C-genome groups, respectively, although later work revealed incongruence between nuclear and chloroplast phylogenies within the B-genome group itself (Doyle et al. 1999). The A-plastome group included chloroplast genomes from all of the remaining species in the subgenus. The majority of these species belonged to the histone H3D clade comprising the A-genomes, D-genomes, E-genomes, H-genomes, and I-genomes, a result broadly congruent between chloroplast and nuclear data; however, subsequent studies suggested little agreement between genome groups and groupings within the A-plastome clade (J. T. Rauscher, A. H. D. Brown, and J. J. Doyle, unpublished data). The most obvious disagreement between nuclear and chloroplast phylogenies was the placement of G. falcata, the sole species of the F-genome, which in the histone H3D phylogeny was sister to all other perennials, but in the chloroplast phylogeny was strongly supported as part of the A-plastome clade. A sister relationship between G. falcata and other perennial Glycine agrees with the distinctiveness of this species (Doyle et al. 1996, 2004a).
In this study we describe complete plastome sequences from seven perennial Glycine species and compare these results to G. max (G-genome). We describe the G. syndetika plastome in detail as a representative genome of the perennial Glycine species. Glycine syndetika is currently the perennial species with the most extensive nuclear sequence available (A. Bombarely, J. Schmutz, J. Grimwood, S. A. Jackson, and J. J. Doyle, unpublished data). The perennial Glycine chloroplast genomes are compared with the annual soybean, G. max (Saski et al. 2005) and Phaseolus vulgaris (Guo et al. 2007), as well as other closely related legumes Milletia pinnata and Vigna radiata (Kazakoff et al. 2012; Tangphatsornruang et al. 2010).
Materials and Methods
Five plants of a single accession each from seven perennial Glycine species representing five of the nine genome groups (Table 1) were grown in the greenhouse at Cornell. Plants were placed in the dark for 48 hr prior to collecting leaf tissue. Tissue was frozen and shipped to Arizona Genomics Institute (AGI), where bacterial artificial chromosome (BAC) libraries were prepared from HindIII digests of genomic DNA. BAC ends were sequenced for all BACs. Two BACs per species were selected from the BAC libraries based on BLAST matches of perennial BAC END sequence data to the G. max chloroplast sequence (Supporting Information, Table S1). BAC DNA was sheared to 3 kb to 4 kb using Adaptive Focused Acoustics technology (Covaris, Woburn, MA) cloned into the plasmid vector pIK96, sequenced with Sanger technology to an average depth of 9×, and then assembled (using Phrap Version 0.990319) and finished as previously described (Ferris et al. 2010). Additional BACs were selected as needed to close the chloroplast circles based on BLAST to the finished sequences.
Table 1. Summary statistics from complete Glycine chloroplast sequences.
| Plastome Group | Genome Group | Glycine Species | Total | LSC | SSC | IR | Duplicated ycf1 in IR | rps19 IR | %GC |
|---|---|---|---|---|---|---|---|---|---|
| G | G | G. maxa | 152218 | 83174 | 17896 | 25574 | 478 | 0.34 | |
| A | A | G. syndetika | 152794 | 83844 | 17840 | 25555 | 463 | 61 | 0.353 |
| A | A/D | G. dolichocarpa | 152804 | 83815 | 17807 | 25591 | 490 | 61 | 0.353 |
| A | A | G. canescens | 152533 | 83559 | 17844 | 25565 | 463 | 61 | 0.353 |
| A | D | G. tomentella D3 | 152728 | 83773 | 17829 | 25563 | 463 | 61 | 0.353 |
| A | F | G. falcata | 153023 | 84027 | 17846 | 25575 | 463 | 59 | 0.353 |
| B | B | G. stenophita | 152618 | 83937 | 17817 | 25432 | 463 | 61 | 0.353 |
| C | C | G. cyrtoloba | 152381 | 89368 | 17801 | 25505 | 463 | 61 | 0.353 |
Length in base pairs of the chloroplast and the three major divisions. Extent of border genes within the IR. GC content in each chloroplast genome.
DOGMA (Wyman et al. 2004) was used with default parameters for preliminary annotation of each sequence. The complete chloroplast sequences were arranged so that each sequence started with trnH. The SSC regions were arranged so that the complete ycf1 gene followed IRb, as in the G. max orientation depicted in Saski et al. (2005). Sequences were then aligned using default parameters in Mulan (Ovcharenko et al. 2005), with minor adjustments to the alignment made manually. Glycine max gene features were downloaded from NCBI (DQ317523) and aligned against the eight sequence alignment using Sequencher (Gene Codes). BioEdit (Hall 1999) was used for manual alignment and confirmation of annotations. Analysis of the total chloroplast alignment for GC content and codon bias were performed using DNAsp (Librado and Rozas 2009).
Previously published primers were used for amplification of trnL-trnF (Shaw et al. 2005) from 11 G. falcata accessions. The resulting sequences were aligned by MUSCLE (Edgar 2004) and edited in BioEdit (Hall 1999). This alignment was used to determine pairwise nucleotide diversity (π) between individuals. Levels of nucleotide polymorphism were calculated using DNAsp (Librado and Rozas 2009).
Direct and palindromic repeated sequences from each chloroplast sequence were identified using REPuter (Kurtz et al. 2001). The number of repeats identified was limited by searching for repeats greater than 30 bp in length and with a sequence identity of 90% or better (Hamming distance of 3 as used previously by Saski et al. 2005). To further investigate dispersed repeat sequences, we extracted the sequence of each repeat separated by more than 1000 bp. Dispersed repeat sequences were aligned using SeqMan (version 2.2.0.56; DNA-STAR, Madison WI). In many cases, repeats listed separately in the REPuter output were from the same location, had the same sequence, and the only difference was the length; for example, a 36-bp repeat could also be listed for the same position as a 30-bp repeat. Similarly, repeats were listed as pairs and, if the repeat was found in a third or fourth location, then the original location was listed again with the third location and again with the fourth location; hence, the overall number of repeats was inflated. Repeat analysis was also performed using RepeatScout and Repeat Masker (Price et al. 2005; Smit et al. 1996) using default parameters in the MAKER 2.10 (Cantarel et al. 2008) genome annotation software.
A Bayesian approach was used to generate a phylogenetic tree for the chloroplast genome sequences. The phylogenetic tree was generated with BEAST (Drummond et al. 2012) using a prior assumption that Glycine perennials are a monophyletic group compared with the annual Glycine species (G. max), and that Phaseolus vulgaris is an outgroup to the Glycine genus. The node date of 19.2 million years ago (mya) for P. vulgaris and Glycine in the plastome tree was used for calibration and is based on the mean age estimated from matK sequence divergence in a comprehensive legume phylogeny (Lavin et al. 2005). The substitution model was general time reversible (GTR). The analysis was run for a MCMC length of 10,000,000 iterations, sampling every 1000 iterations. The molecular clock was an uncorrelated relaxed clock with a log normal distribution model (Drummond et al. 2006). FigTree was used to plot the tree (http://tree.bio.ed.ac.uk/software/figtree/).
Percent identity across the chloroplast genomes, excluding the second IR, was visualized using the VISTA tools server (Brudno et al. 2003). The LAGAN shuffle option was used to facilitate inclusion of chloroplast sequences from GenBank for species with inversions not present in Glycine (Phaseolus vulgaris var. Negro Jamapa (DQ886273), Millettia pinnata (JN673818), and Vigna radiata (GQ893027). Pairwise differences in single nucleotides were recorded for each species in an alignment of Glycine species and Phaseolus vulgaris plastomes. Pairwise differences were used to calculate the nucleotide substitution rate by dividing by the length of the alignment and the divergence dates calculated with histone H3D alignments (González-Orozco et al. 2012; Sherman-Broyles et al. 2014).
Results and Discussion
Sequencing, size, gene content, and extent of inverted repeat regions
The two G. falcata BAC sequences used to create a complete chloroplast sequence were polymorphic. No other species had polymorphisms between the two BAC sequences. BAC libraries for all taxa were created by collecting tissue from up to five plants in each accession. The region of overlap between the two BACs was 86,523 bp. There were 72 segregating sites in this region, resulting in pairwise nucleotide diversity or π = 0.00083. This very low level of diversity rules out contamination with chloroplast DNAs from other Glycine species studied here. To determine levels of chloroplast genome polymorphism in G. falcata, we sequenced a noncoding region of the plastome known to be variable in Glycine species, the intergenic spacer between trnL and trnF, in 11 additional accessions of G. falcata (S. Sherman-Broyles, J. A. Doyle, and J. J. Doyle, unpublished data). We found that this sample was an order of magnitude more diverse (π = 0.00225) than the estimate from the two BACs, consistent with the two BACs representing variation among individuals in accession G1718. This could be due to polymorphisms in the chloroplast genome, or to one BAC representing the chloroplast genome and the other being derived from a recent, large nuclear integrant of plastid DNA (NuPT). Similar types of variation were found in the cultivated plant Pelargonium × hortorum (Chumley et al. 2006). Chumley et al. (2006) pointed out that Pelargonium has biparental inheritance of chloroplasts that may contribute to the detection of variation between individuals, but also commented that they found no evidence of heteroplasmy. In Glycine, maternal inheritance of chloroplasts was confirmed in G. max (Hatfield et al. 1985) and in the perennials, determined by Southern blot analysis of two synthetic allopolyploid taxa (Doyle et al. 1990b), so it seems more likely that there are differences between individuals rather than within a single plant. Further analysis of nuclear genome sequences is required to determine if a NuPT was sequenced (Yoshida et al. 2013; Michalovova et al. 2013).
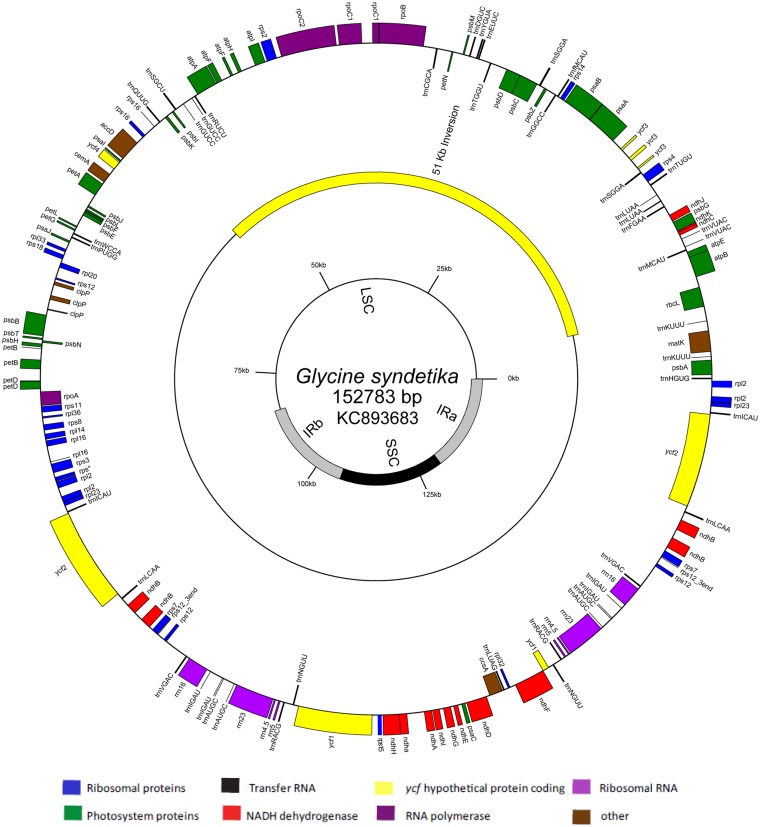
The G. syndetika genome map (Figure 2) is representative of the seven perennial Glycine species’ chloroplast sequences discussed here. No structural rearrangements were detected among the perennial Glycine plastomes. The G. syndetika plastome sequence is 152,794 bp in length, just slightly larger than G. max (152,218 bp) (Saski et al. 2005) (Table 1). The chloroplast genome length in Glycine ranges from G. max (with the smallest genome) to G. falcata at 153,023 bp (with the largest genome) (Table 1).
Figure 2.
Gene map of Glycine syndetika chloroplast genome. Gray bars on inner circle represent the extent of the inverted repeats (IRa and IRb). Yellow bar on middle circle represents 51-kb inversion found in papillionoid legumes. Genes on the outside of the circle are transcribed in a clockwise direction, and genes on the inside of the circle are transcribed in a counter-clockwise direction.
Glycine plastomes contain 111 unique genes, including 77 protein coding genes, 30 tRNA genes, and 4 rRNA genes. Six protein coding genes, seven tRNA genes, and all of the rRNA genes are completely duplicated in the IRs. The perennial Glycine plastomes are similar to the annual soybean plastome, with 19 genes containing introns, of which six are tRNA genes. Two genes have alternative start codons (psbL and ndhD) in all of the Glycine chloroplast sequences as found in other angiosperm plastomes (Sugiura 2013). The GC content in G. max is 34% (Saski et al. 2005), whereas in the perennial taxa the GC content of all species is 35% (Table 1). Codon bias is consistent with the A/T-rich aspect of angiosperm chloroplast genomes, with codons with A/T(U) in the third position being more common.
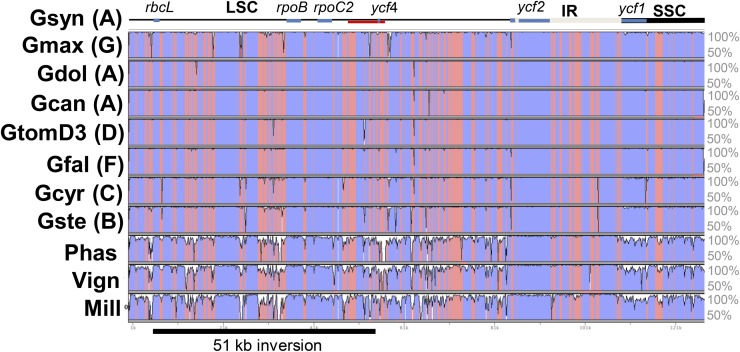
The total number of genes reported above does not include ycf4, which in legumes is divergent from other angiosperm ycf4 sequences (Stefanovic et al. 2009) and was not annotated by Dogma (Wyman et al. 2004) or GenBank for our submissions. Stefanovic et al.(2009) were the first to draw attention to the fact that the gene was present and highly divergent in legumes (Stefanovic et al. 2009) rather than absent, as suggested by DNA hybridization surveys (Doyle et al. 1995) and the G. max annotation (Saski et al. 2005). This gene is thought to be a nonessential Photosystem I assembly factor in higher plants (Krech et al. 2012), and this level of sequence divergence may indicate that if its function is retained in Glycine, it is replaced by a nuclear copy of a plastid gene (NuPT). This region flanks one of several NuPTs found in the soybean genome sequence (A. Bombarely, D. Robinson, S. Sherman-Broyles, and J. J. Doyle, unpublished data). Although ycf4 is divergent from other legume ycf4 sequences, there is no indication that it is a mutational hotspot in Glycine, as it is known to be in the legume genus Lathyrus (Magee et al. 2010).There are high levels of sequence similarity in genes between rps16 and cemA, the hypervariable region in Magee et al. (2010) (Figure 3). Pairwise Ka/Ks values for all Glycine species (data not shown) are misleading because there are so few differences that ratios were based on too few polymorphic sites.
Figure 3.
Sequence similarity plot generated by VISTA tools. Base sequence is G. syndetika. Intergenic regions are pink. Shuffle LAGAN option used to align Phaseolus and Vigna that have 78-kb inversions. Hypervariable region identified by Magee et al. (2010) is demarcated by red line. Gene with low sequence similarity to other angiosperms, ycf4 is indicated, along with five other genes for orientation. Only one inverted repeat is shown.
The genes that mark the beginning and end of the IR are only partially duplicated: 61 bp of rps19 for all species except G. falcata (59 bp), and hypothetical chloroplast RF1 (ycf1), with 478 bp duplicated in G. max, 490 bp in G. dolichocarpa, and 463 bp in the remaining perennial Glycine taxa (Table 1). The full-length ycf1 sequence in G. cyrtoloba, spanning the IRb and SSC region, has a premature stop codon in the single copy region; further analyses are necessary to determine if this is a sequencing error or an edited base, or if ycf1 is a pseudogene in G. cyrtoloba. The function of ycf1 was recently determined to be a translocon protein of the inner chloroplast membrane and in Arabidiposis was renamed Tic214 (Kikuchi et al. 2013). It has been found to have relaxed substitution rates except at its 5′ end, which may coincide with an origin of replication but might also be explained by its duplication in the IR regions, which have lower substitution rates than other chloroplast regions (Wicke et al. 2011).
Repetitive sequences
Repeat sequences are of interest as a possible mechanism for inversions or as remnants of the inversion process, and thus are frequently reported in descriptions of legume plastomes and other taxa that are known to be characterized by one or more inversions.
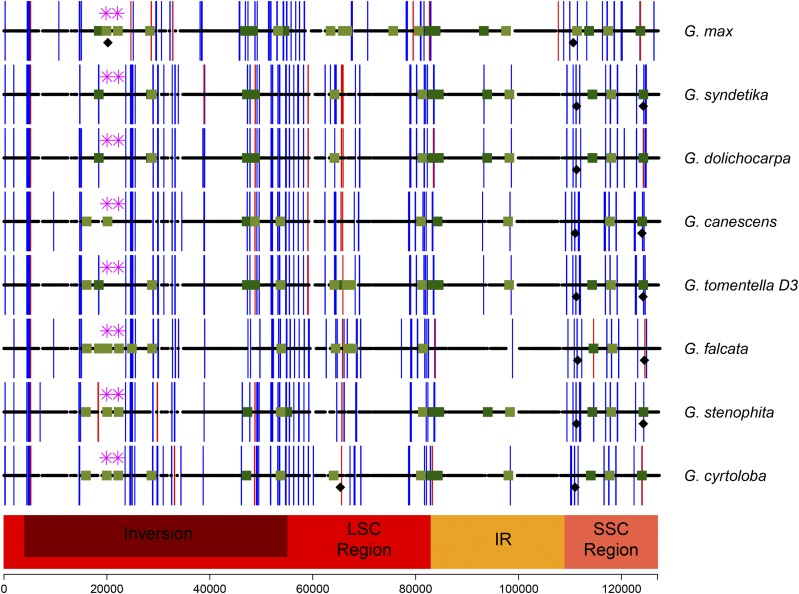
REPuter detected 104 tandem and dispersed repeats of 30 bp in length or longer in G. max (Saski et al. 2005); similarly, in G. syndetika there are 103 repeats. RePuter detected the fewest (85) repeats from the G. falcata plastome (Table 2). The number of repeats detected in Glycine plastomes is much higher than the number of repeats detected previously in Arabidopsis (57 repeats) (Table 2) (Saski et al. 2005). To gain a more thorough understanding of the REPuter output, we investigated the sequence of each repeat identified. If repeat sequences were within 1000 bp of each other, then we considered them tandem repeats. If dispersed repeats occurred only in the IR regions, then we did not consider them further; if they were repeated both within and outside the IR regions, then the repeat sequences were investigated further. An alignment of all the repeats identified by REPuter and separated by more than 1000 bp showed that Glycine dispersed repeats with high levels of sequence similarity are found in multiple locations in the chloroplast genome. This reduced the number of unique sequences to 10 or fewer in each species and allowed us to plot their locations (Figure 4). For example, Repeat sequence 1, TATATATCTATMTATMTATAGATAGATATATAGATAT, is the most common repeat sequence and it was found in up to 11 positions in a single plastome (represented by light green squares in Figure 4). The nine remaining repeat sequences are found less frequently (four or fewer positions, depicted by dark green squares in Figure 4.) Repeats present within a coding region are depicted by an asterisk.
Table 2. Number and types of repeated sequences.
| RePuter | Dispersed Repeats | RepeatMasker/Repeat Finder | Low Complexity | ||||
|---|---|---|---|---|---|---|---|
| Species | Repeat Sequences | Unique Locations | Unique Sequences | Tandem Repeats | Repeat Sequences | Low Complexity | SSR |
| Arabidopsis | 57a | 19 | 9 | 18 | |||
| G. max | 104a | 22 | 10 | 28 | 83 | 56 | 8 |
| G. syndetika | 103 | 20 | 5 | 32 | 95 | 69 | 7 |
| G. dolichocarpa | 104 | 20 | 5 | 29 | 94 | 69 | 7 |
| G. canescens | 86 | 16 | 5 | 29 | 95 | 71 | 5 |
| G. tomentella D3 | 108 | 28 | 8 | 30 | 92 | 69 | 4 |
| G. falcata | 85 | 28 | 7 | 27 | 89 | 64 | 5 |
| G. stenophita | 96 | 19 | 6 | 31 | 82 | 58 | 5 |
| G. cyrtoloba | 101 | 20 | 6 | 34 | 78 | 54 | 6 |
| Phaseolus | 62 | 9 | 3 | 21 | |||
| Vigna | 68 | 10 | 3 | 24 | |||
| Milletia | 46 | 10 | 3 | 21 | |||
Repeated sequences were detected by RePuter, RepeatMasker, and Repeat Finder. Further analysis of dispersed repeats showed that many of the sequences shared similarity, reducing the number of unique repeat sequences and locations.
Figure 4.
Locations of repetitive sequences in Glycine plastomes. Black horizontal lines represent coding regions in each plastome. Light green squares mark the location of dispersed repeat sequence 1 from REPuter. Dark green squares mark the location of 10 additional dispersed sequences. The locations of low complexity repeats are shown by blue lines. SSRs are shown by red lines as determined by RepeatScout. Asterisks represent repeats within coding sequence. Black diamonds represent repeats with sequence similarity to hAT repeat transposons.
Unlike the Geraniaceae (Guisinger et al. 2011) and Oenothera (Onagraceae) (Greiner et al. 2008), where inversions are often flanked by tandem or palindromic repeats, in soybean (Saski et al. 2005) or in the plastomes of any of the other Glycine species reported here, there is not a higher percentage of repeats in the area flanking the 51-kb inversion (Figure 4). The 51-kb inversion shared by most papilionoid species dates back approximately 56 mya (Lavin et al. 2005). The lack of repeats flanking the inversion in modern genomes sequences is not surprising considering the age of the inversions.
The sizes of repeats detected in the Glycine plastomes are mostly less than 50 bp, but within the range necessary for illegitimate recombination. Microhomologies as little as 16 bp in length have been shown to be sufficient for illegitimate recombination while regions of sequence similarity longer than 50 bp are necessary for homologous recombination (Maréchal and Brisson 2010). The higher incidence of repeated sequences in legume plastomes and several other taxa with inversions suggests that there may have been mutations in chloroplast DNA repair genes; this could be tested by examining candidate genes such as genes recently described as responsible for organelle stability through recombination surveillance (Maréchal and Brisson 2010).
RepeatMasker and RepeatScout (Smit et al. 1996; Price et al. 2005) detected 101 repetitive sequences in G. syndetika (Table 2), including short sequences with similarity to transposable elements, low-complexity repeats, and simple sequence repeats (SSRs). The locations of low complexity and SSRs are also shown in Figure 4. Comparison of G. syndetika with G. dolichocarpa indicates that the repetitive sequence profiles are identical except for a 76-bp region in G. syndetika with sequence similarity to hAT DNA transposon (Figure 4). All sequences with similarity to transposable elements are shorter than 506 bp and probably do not represent true transposable element insertions, but rather sequences that are coincidentally similar to transposable elements and, therefore, detected by the software.
Evolution
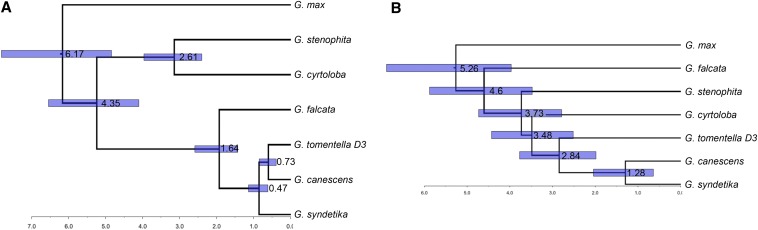
The alignment of the Glycine sequences was deposited in GenBank as Popset 514252878. Distances estimated from this alignment reveal that all species of Glycine outside of the B-genome, C-genome, and G-genome groups have highly similar chloroplast genome sequences, consistent with their previous recognition as the A-plastome group (Doyle et al. 1990b) (Table 1). A Bayesian inference tree (Figure S1), calibrated with Phaseolus vulgaris (Guo et al. 2007) as an outgroup, has a topology that is congruent with previous Glycine chloroplast gene trees (Doyle et al. 1990b; Sakai et al. 2003). All chloroplast-based trees are incongruent with topologies based on nuclear DNA sequences (Doyle et al. 2004b), including histone H3D (Figure 5) (Doyle et al. 1996b; Gonzalez-Orozco et al. 2012), nrDNA ITS (Rauscher et al. 2004), and genome-wide single nucleotide polymorphisms (D. Ilut, P. Cregan, and J. J. Doyle, unpublished data). The most conspicuous incongruence involves the placement of G. falcata as sister to A-genome and D-genome taxa in the chloroplast phylogeny rather than sister to all other perennial taxa as indicated by nuclear data (Figure 5).
Figure 5.
Comparison of Bayesian inference trees based on whole plastome alignment (A) and histone H3D alignment (B). Divergence dates calculated using BEAST. Bars represent the 95% HPD (highest posterior density) interval.
The histone tree lacks a P. vulgaris sequence because, although this locus is useful for nuclear phylogenies, it is mostly intron sequence and the P. vulgaris sequence is too diverged to align properly. The histone tree is instead calibrated using a mean divergence date between annual and perennial Glycine subgenera of 5.25 mya, an average estimated from the divergence dates of several low copy nuclear genes from G. max and G. tomentella D3 (Egan and Doyle 2010; Innes et al. 2008). That date is similar to the 6.17 million years estimated from the BEAST plastome phylogeny and is well within the credible interval for that node (Figure 5A). These estimates of relative divergence times rather than absolute node ages are dependent on calibration using estimates from other studies and not on dates from fossils.
The plastome and nuclear gene trees differ in the depiction of the relationship between B-genome and C-genome groups. The plastome tree shows G. stenophita and G. cyrtoloba as sister to each other, sharing a common ancestor 2.61 mya, whereas the histone tree (Figure 5B) is similar to previously published trees that vary in the placement of the B and C genomes relative to each other depending on taxon sampling and the locus that was used. Regardless of the data set used, the B-genome and C-genome groups are always derived after the F-genome group and before the remaining genome groups. The presence of a B–C clade is consistent with genome-wide SNP data (D. Ilut, P. Cregan, and J. J. Doyle, unpublished data).
The strong similarities among A-plastomes lead to very recent divergence time estimates: less than 500,000 years to the common ancestor of the G. canescens and G. syndetika plastomes, whereas their histone H3D genes are estimated to have diverged 1.28 mya. Similarly, the date for the divergence of the G. tomentella D3 plastome from the plastomes of these A-genome species is less than 1 mya, whereas the histone divergence date is 2.84 mya. As expected, the most dramatic difference involves G. falcata. As sister to the remainder of the perennial subgenus in the histone H3D tree, it is estimated to have last shared a common ancestor with the other species approximately 4.6 mya, yet its chloroplast genome is estimated to have diverged from those of the other A-plastome species only 1.64 mya (Figure 5).
Doyle et al. (1996, 2004b) hypothesized that the plastome from the common ancestor of the entire A-plastome group was introgressed into G. falcata through a hybridization event and subsequent backcrossing to G. falcata. The plastome phylogeny divergence dates suggest that the introgression occurred as recently as 1.6 mya. The transfer of a maternally inherited chloroplast replaced the G. falcata plastome in what is called a chloroplast capture event (Tsitrone et al. 2003; Baack and Rieseberg 2007). The current range of G. falcata overlaps with both A-genome and D-genome species; however, little is known about the historical ranges of the taxa (E. Y. Hwang and P. Cregan, unpublished data). Based on the sample of 11 G. falcata accessions surveyed here for the trnL-trnF spacer region, there is no evidence of polymorphism involving a second, deeply coalescing chloroplast genome in this species.
In the subsequent generations, with backcrossing to G. falcata, the nuclear sequences in G. falcata have eliminated all but a few genes that demarcate the introgression event. Analysis of genome-wide SNP data from transcriptome libraries of the same taxa detected SNPs from only 80 genes out of more than 27,000 genes (< 0.3%), consistent with introgression from the A-genome into G. falcata (D. Ilut, P. Cregan, and J. J. Doyle, unpublished data). Even relatively recent introgression events have been reported to result in only a very low number of retained genes from the nonbackcross parent, perhaps because only genes that are selectively advantageous are retained (McKinnon et al. 2010). To further test the hypothesis of introgression from the common ancestor of A/D-genome ancestors into G. falcata, we are interested in using genotyping by sequencing, an inexpensive method for generating genome-wide markers from many individuals (Twyford and Ennos 2012).
The plastome tree has low posterior probabilities for the placement of G. canescens, G. syndetika, and G. dolichocarpa. The depicted tree shows the plastomes of G. syndetika and G. canescens diverging 0.43 mya, and that of G. dolichocarpa as slightly older at 0.50 mya. The allopolyploid, G. dolichocarpa, is polymorphic for chloroplast sequences from its two diploid progenitors, and this result supports previous work (J. T. Rauscher, A. H. D. Brown, and J. J. Doyle, unpublished data) in showing that the accession sampled here has the G. syndetika plastome. A divergence date of approximately 0.5 million years is consistent with results from nuclear genes sampled from transcriptome data (Bombarely et al. 2014). Hybridization events, resulting in sterile diploid offspring and fertile allopolyploids following whole genome duplication, between taxa that had diverged as many as 3 mya are common in Glycine, a prime example being G. dolichocarpa. The allopolyploid taxa lend credence to the hypothesis that a hybridization event, 1 to 3 mya, led to the capture of an A-plastome within what is probably the most earliest diverging perennial Glycine species, G. falcata.
Most differences between Glycine plastomes involve a handful of insertion/deletion polymorphisms, and A-plastome sequences are virtually identical to one another, with low levels of genetic diversity among taxa (Figure 3). The number of pairwise nucleotide substitutions (Table 3) can be used to estimate the plastome substitution rates in the Glycine taxa, if it is assumed that the annuals and perennials diverged approximately 5.25 mya (Egan and Doyle 2010; Innes et al. 2008) and we use the other divergence dates estimated from the histone H3D phylogeny (Figure 5). In comparisons between G. max and the other taxa (including not only other Glycine species but also Phaseolus vulgaris), the average substitution rate is 2.68 × 10−9 substitutions per site per year (Table 3). These rates are slow relative to other reported chloroplast nucleotide substitution rates (Zhong et al. 2009; Guo et al. 2007). The similarity of the substitution rates between G. max and the other species suggests a clock-like rate of evolution. However, when divergence dates from the nuclear gene are used, substitution rates appear much slower among the A-plastome species, averaging 7.95 × 10−10 substitutions per site per year. Because comparisons of these same taxa with G. max are clock-like, this suggests that the divergence date estimates from the nuclear genome are inappropriately high for estimating substitution rates of chloroplast genomes within this group of taxa. Recent divergence of plastomes within the A-plastome group supports a hypothesis of introgression.
Table 3. Pairwise SNPs and substitution rates.
| Rate/SNPs | G. max | G. syndetika | G. dolichocarpa | G canescens | G. tomentella D3 | G. falcata | G. stenophita | G. cyrtoloba | P. vulgaris |
|---|---|---|---|---|---|---|---|---|---|
| G. max | 2066 | 2044 | 2074 | 2104 | 2094 | 2022 | 2384 | 8234 | |
| G. syndetika | 2.49E−09 | 175 | 210 | 307 | 638 | 1608 | 2026 | 8353 | |
| G. dolichocarpa | 2.47E−09 | 2.22E−09 | 213 | 303 | 635 | 1596 | 2032 | 8341 | |
| G. canescens | 2.50E−09 | 1.04E−09 | 2.70E−09 | 322 | 660 | 1627 | 2054 | 8331 | |
| G. tomentella | 2.54E−09 | 5.60E−10 | 5.53E−10 | 5.87E−10 | 687 | 1640 | 2066 | 8368 | |
| G. falcata | 2.53E−09 | 8.80E−10 | 8.76E−10 | 9.11E−10 | 9.48E−10 | 1640 | 2074 | 8365 | |
| G. stenophita | 2.44E−09 | 2.74E−09 | 2.72E−09 | 2.77E−09 | 2.79E−09 | 2.79E−09 | 1284 | 8351 | |
| G. cyrtoloba | 2.88E−09 | 3.70E−09 | 3.71E−09 | 3.75E−09 | 3.77E−09 | 3.78E−09 | 2.34E−09 | 8616 | |
| P. vulgaris | 2.72E−09 | 2.76E−09 | 2.76E−09 | 2.75E−09 | 2.77E−09 | 2.77E−09 | 2.76E−09 | 2.85E−09 |
The number of SNPs between species appears above the diagonal, and the substitution rate was calculated using histone H3D divergence dates. Substitution rates in bold represent A plastome taxa with substitution rates that appear slower because of inferred chloroplast capture.
Conclusions
Our understanding of the tempo and mode of plastome evolution is enhanced as sequencing technologies have led to the increase in complete plastome sequences, including multiple species within key genera. Rearrangements observed in legumes, when only a few sequences were available, are now being shown to be specific to individual lineages at key times in legume evolution and not indicative of continuous processes leading to changes in gene order. Within Glycine, the chloroplast genome is very stable. Repetitive sequences do not flank the 51-kb inversion shared by the papilionoid legumes and are not as numerous as they might seem without further investigation of their location and sequence. Despite being distributed across the Glycine chloroplast genomes, repetitive sequences have not changed the order of genes among species. Phylogenetic analyses of complete plastome sequences corroborate previous restriction mapping studies (Doyle et al. 1990b) in suggesting an introgression of the A-plastome into G. falcata. Bayesian analysis and dating divergence allows us to hypothesize the introgression occurred 1 to 3 mya.
Supplementary Material
Acknowledgments
This work is part of the SoyMap2 project and was funded by NSF Grant 0822258. We thank Jane Doyle, Dan Ilut and Ashley Egan for technical support and access to unpublished data. NSF Grant 0822258, SoyMap2, Jane Doyle, Dan Ilut, and Ashley Egan for technical support and access to unpublished data.
Footnotes
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.114.012690/-/DC1
GenBank PopSet: 514252878 KC893632-KC893640
Communicating editor: S. A. Jackson
Literature Cited
- Baack E. J., Rieseberg L. H., 2007. A genomic view of introgression and hybrid speciation. Curr. Opin. Genet. Dev. 17: 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely A., Coate J. E., Doyle J. J., 2014. Mining transcriptomic data to study the origins and evolution of a plant allopolyploid complex. PeerJ 2: e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M., Malde S., Poliakov A., Do C. B., Couronne O., et al. , 2003. Glocal alignment: finding rearrangements during alignment. Bioinformatics 19(suppl 1): i54–i62 [DOI] [PubMed] [Google Scholar]
- Bruneau A., Doyle J. J., Palmer J. D., 1990. A chloroplast DNA structural mutation as a subtribal character in the Phaseolae (Leguminosae). Syst. Bot. 14: 378–386 [Google Scholar]
- Cai Z., Guisinger M., Kim H.-G., Ruck E., Blazier J. C., et al. , 2008. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J. Mol. Evol. 67: 696–704 [DOI] [PubMed] [Google Scholar]
- Cannon S. B., May G. D., Jackson S. A., 2009. Three sequenced legume genomes and many crop species: rich opportunities for translational genomics. Plant Physiol. 151: 970–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel B. L., Korf I., Robb S. M., Parra G., Ross E., et al. , 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18: 188–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumley T. W., Palmer J. D., Mower J. P., Fourcade H. M., Calie P. J., et al. , 2006. The complete chloroplast genome sequence of Pelargonium× hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 23: 2175–2190 [DOI] [PubMed] [Google Scholar]
- Corriveau J. L., Coleman A. W., 1988. Rapid Screening Method to Detect Potential Biparental Inheritance of Plastid DNA and Results for Over 200 Angiosperm Species. American Journal of Botany 75 (10): 1443–1458 [Google Scholar]
- Doyle J., Doyle J., Brown A., 1990a Analysis of a polyploid complex in Glycine with chloroplast and nuclear DNA. Aust. Syst. Bot. 3: 125–136 [Google Scholar]
- Doyle J. J., Doyle J. L., Brown A. H. D., 1990b A chloroplast-DNA phylogeny of the wild perennial relatives of soybean (Glycine subgenus Glycine): congruence with morphological and crossing groups. Evolution 44: 371–389 [DOI] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L., Brown A. H. D., 1990c Chloroplast DNA polymorphism and phylogeny in the B genome of Glycine subgenus Glycine (Leguminosae). Am. J. Bot. 77: 772–782 [Google Scholar]
- Doyle J. J., Doyle J. L., Ballenger J., Palmer J., 1996. The distribution and phylogenetic significance of a 50-kb chloroplast DNA inversion in the flowering plant family Leguminosae. Mol. Phylogenet. Evol. 5: 429–438 [DOI] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L., Brown A. H. D., 1999. Incongruence in the diploid B-genome species complex of Glycine (Leguminosae) revisited: histone H3-D alleles vs. chloroplast haplotypes. Mol. Biol. Evol. 16: 354–362 [DOI] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L., Palmer J. D., 1995. Multiple independent losses of two genes and one intron from legume chloroplast genomes. Syst. Bot. 20: 272–294 [Google Scholar]
- Doyle J. J., Doyle J. L., Rauscher J. T., 2004a Evolution of the perennial soybean polyploid complex (Glycine subgenus Glycine): a study of contrasts. Biol. J. Linn. Soc. Lond. 82: 583–597 [Google Scholar]
- Doyle J. J., Doyle J. L., Rauscher J. T., Brown A. H. D., 2004b Diploid and polyploid reticulate evolution throughout the history of the perennial soybeans (Glycine subgenus Glycine). New Phytol. 161: 121–132 [Google Scholar]
- Doyle J. J., Kanazin V., Shoemaker R. C., 1996b Phylogenetic Utility of Histone H3 Intron Sequences in the Perennial Relatives of Soybean (Glycine:Leguminosae). Molecular Phylogenetics and Evolution 6: 438–447 [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Ho S. Y., Phillips M. J., Rambaut A., 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Suchard M. A., Xie D., Rambaut A., 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29: 1969–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan A. N., Doyle J. J., 2010. A comparison of global, gene-specific, and relaxed clock methods in a comparative genomics framework: dating the polyploid history of soybean (Glycine max). Syst. Biol. 59: 534–547 [DOI] [PubMed] [Google Scholar]
- Edgar R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32 (5): 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P., Olson B.J.S.C., De Hoff P.L., Douglass S., Casero D., et al. , 2010. Evolution of an expanded sex-determining locus in Volvox. Science 328 (5976): 351–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantt J. S., Baldauf S. L., Calie P. J., Weeden N. F., Palmer J. D., 1991. Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 10: 3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Orozco C. E., Brown A. H., Knerr N., Miller J. T., Doyle J. J., 2012. Hotspots of diversity of wild Australian soybean relatives and their conservation in situ. Conserv. Genet. 13: 1269–1281 [Google Scholar]
- Greiner S., Wang X., Rauwolf U., Silber M. V., Mayer K., et al. , 2008. The complete nucleotide sequences of the five genetically distinct plastid genomes of Oenothera, subsection Oenothera: I. Sequence evaluation and plastome evolution. Nucleic Acids Res. 36: 2366–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger M. M., Kuehl J. V., Boore J. L., Jansen R. K., 2011. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol. Biol. Evol. 28: 583–600 [DOI] [PubMed] [Google Scholar]
- Guo X., Castillo-Ramírez S., González V., Bustos P., Fernández-Vázquez J. L., et al. , 2007. Rapid evolutionary change of common bean (Phaseolus vulgaris L) plastome, and the genomic diversification of legume chloroplasts. BMC Genomics 8: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle R. C., Fourcade H. M., Boore J. L., Jansen R. K., 2008. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J. Mol. Evol. 66: 350–361 [DOI] [PubMed] [Google Scholar]
- Hall, T., 1999BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Res. 41: 95–98.
- Hatfield P. M., Shoemaker R. C., Palmer R. G., 1985. Maternal inheritance of chloroplast DNA within the genus Glycine, subgenus soja. J. Hered. 76: 373–374 [Google Scholar]
- Innes R. W., Ameline-Torregrosa C., Ashfield T., Cannon E., Cannon S. B., et al. , 2008. Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean. Plant Physiol. 148: 1740–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. K., Cai Z., Raubeson L. A., Daniell H., dePamphilis C. W., et al. , 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 104: 19369–19374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. K., Wojciechowski M. F., Sanniyasi E., Lee S.-B., Daniell H., 2008. Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae). Mol. Phylogenet. Evol. 48: 1204–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane N., Sveinsson S., Dempewolf H., Yang J. Y., Zhang D., et al. , 2012. Ultra-barcoding in cacao (Theobroma spp.; Malvaceae) using whole chloroplast genomes and nuclear ribosomal DNA. Am. J. Bot. 99: 320–329 [DOI] [PubMed] [Google Scholar]
- Kato T., Kaneko T., Sato S., Nakamura Y., Tabata S., 2000. Complete structure of the chloroplast genome of a legume, Lotus japonicus. DNA Res. 7: 323–330 [DOI] [PubMed] [Google Scholar]
- Kazakoff S. H., Imelfort M., Edwards D., Koehorst J., Biswas B., et al. , 2012. Capturing the biofuel wellhead and powerhouse: the chloroplast and mitochondrial genomes of the leguminous feedstock tree Pongamia pinnata. PLoS ONE 7: e51687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S., Bédard J., Hirano M., Hirabayashi Y., Oishi M., et al. , 2013. Uncovering the Protein Translocon at the Chloroplast Inner Envelope Membrane. Science 339: 571–574 [DOI] [PubMed] [Google Scholar]
- Kim K.-J., Choi K.-S., Jansen R. K., 2005. Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae). Mol. Biol. Evol. 22: 1783–1792 [DOI] [PubMed] [Google Scholar]
- Krech K., Ruf S., Masduki F. F., Thiele W., Bednarczyk D., et al. , 2012. The plastid genome-encoded Ycf4 protein functions as a nonessential assembly factor for photosystem I in higher plants. Plant Physiol. 159: 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Choudhuri J. V., Ohlebusch E., Schleiermacher C., Stoye J., et al. , 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29: 4633–4642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M., Herendeen P. S., Wojciechowski M. F., 2005. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Syst. Biol. 54: 575–594 [DOI] [PubMed] [Google Scholar]
- LPWG (Legume Phylogeny Working Group), 2013. Towards a new classification system for legumes: Progress report from the 6th International Legume Conference. South African Journal of Botany 89 (0): 3–9 [Google Scholar]
- Librado P., Rozas J., 2009. DnaSPv5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452 [DOI] [PubMed] [Google Scholar]
- Magee A. M., Aspinall S., Rice D. W., Cusack B. P., Sémon M., et al. , 2010. Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 20: 1700–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A., Brisson N., 2010. Recombination and the maintenance of plant organelle genome stability. New Phytol. 186: 299–317 [DOI] [PubMed] [Google Scholar]
- Martin G. E., Rousseau-Gueutin M., Cordonnier S., Lima O., Michon-Coudouel S., et al. , 2014. The first complete chloroplast genome of the Genistoid legume Lupinus luteus: evidence for a novel major lineage-specific rearrangement and new insights regarding plastome evolution in the legume family. Ann. Bot. (Lond.) 113: 1197–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon G. E., Smith J. J., Potts B. M., 2010. Recurrent nuclear DNA introgression accompanies chloroplast DNA exchange between two eucalypt species. Mol. Ecol. 19: 1367–1380 [DOI] [PubMed] [Google Scholar]
- Michalovova M., Vyskot B., Kejnovsky E., 2013. Analysis of plastid and mitochondrial DNA insertions in the nucleus (NUPTs and NUMTs) of six plant species: size, relative age and chromosomal localization. Heredity 111: 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen R. S., Olmstead R. G., Adams K. L., Palmer J. D., Lao N. T., et al. , 2001. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. The Plant Cell Online 13: 645–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nock C. J., Waters D. L. E., Edwards M. A., Bowen S. G., Rice N., et al. , 2011. Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnol. J. 9: 328–333 [DOI] [PubMed] [Google Scholar]
- Ovcharenko I., Loots G. G., Giardine B. M., Hou M., Ma J., et al. , 2005. Mulan: Multiple-sequence local alignment and visualization for studying function and evolution. Genome Res. 15: 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. D., 1990. Contrasting modes and tempos of genome evolution in land plant organelles. Trends Genet. 6: 115–120 [DOI] [PubMed] [Google Scholar]
- Palmer J., Osorio B., Thompson W., 1988a Evolutionary significance of inversions in legume chloroplast DNAs. Curr. Genet. 14: 65–74 [Google Scholar]
- Palmer J. D., Jansen R. K., Michaels H. J., Chase M. W., Manhart J. R., 1988b Chloroplast DNA variation and plant phylogeny. Ann. Missouri Botanical Garden (USA) 75: 1180–1206 [Google Scholar]
- Perry A. S., Brennan S., Murphy D. J., Kavanagh T. A., Wolfe K. H., 2002. Evolutionary re-organisation of a large operon in adzuki bean chloroplast DNA caused by inverted repeat movement. DNA Res. 9: 157–162 [DOI] [PubMed] [Google Scholar]
- Price A. L., Jones N. C., Pevzner P. A., 2005. De novo identification of repeat families in large genomes. Bioinformatics 21(suppl 1): i351–i358 [DOI] [PubMed] [Google Scholar]
- Ratnaparkhe M. B., Singh R. J., Doyle J. J., 2011. Glycine, pp. 83–116 in Wild Crop Relatives: Genomic and Breeding Resources, edited by Kole C. Springer, Berlin, Heidelberg [Google Scholar]
- Rauscher J. T., Doyle J. J., Brown A. H. D., 2004. Multiple origins and nrDNA internal transcribed spacer homeologue evolution in the Glycine tomentella (Leguminosae) allopolyploid complex. Genetics 166: 987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L. H., Soltis D. E., 1991. Phylogenetic consequences of cytoplasmic gene flow in plants. Evol. Trends Plants 5: 65–84 [Google Scholar]
- Ruhfel B., Gitzendanner M., Soltis P., Soltis D., Burleigh J., 2014. From algae to angiosperms-inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol. Biol. 14: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman T., Jansen R., 2014. The Plastid Genomes of Flowering Plants, pp. 3–38 in Chloroplast Biotechnology, edited by Maliga P. Humana Press, New York, USA [Google Scholar]
- Sabir J., Schwarz E., Ellison N., Zhang J., Baeshen N. A., et al. , 2014. 2014 Evolutionary and biotechnology implications of plastid genome variation in the inverted‐repeat‐lacking clade of legumes. Plant Biotechnol. J. 12: 10.1111/pbi.12179. [DOI] [PubMed] [Google Scholar]
- Sakai M., Kanazawa A., Fujii A., Thseng F., Abe J., et al. , 2003. Phylogenetic relationships of the chloroplast genomes in the genus Glycine inferred from four intergenic spacer sequences. Plant Syst. Evol. 239: 29–54 [Google Scholar]
- Saski C., Lee S.-B., Daniell H., Wood T. C., Tomkins J., et al. , 2005. Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genomes. Plant Mol. Biol. 59: 309–322 [DOI] [PubMed] [Google Scholar]
- Shaw J., Lickey E. B., Beck J. T., Farmer S. B., Liu W., et al. , 2005. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 92: 142–166 [DOI] [PubMed] [Google Scholar]
- Sherman-Broyles, S., A. Bombarely, A.F. Powell, J.L. Doyle, A.N. Egan et al., 2014. The wild side of a major crop: Soybean’s perennial cousins from Down Under. Am. J. Bot. 10.3732/ajb.1400121 DOI [DOI] [PubMed]
- Singh R. J., Hymowitz T., 1985. The genomic relationships among six wild perennial species of the genus Glycine subgenus Glycine Willd. Theor. Appl. Genet. 71: 221–230 [DOI] [PubMed] [Google Scholar]
- Smit, A. F., R. Hubley, and P. Green, 1996 RepeatMasker Open-3.0, http://www.repeatmasker.org
- Soltis D. E., Smith S. A., Cellinese N., Wurdack K. J., Tank D. C., et al. , 2011. Angiosperm phylogeny: 17 genes, 640 taxa. Am. J. Bot. 98: 704–730 [DOI] [PubMed] [Google Scholar]
- Stefanovic S., Pfeil B. E., Palmer J. D., Doyle J. J., 2009. Relationships among phaseoloid legumes based on sequences from eight chloroplast regions. Syst. Bot. 34: 115–128 [Google Scholar]
- Sugiura M., 2013. Plastid mRNA translation. Methods Mol. Biol. 1132: 73–91 [DOI] [PubMed] [Google Scholar]
- Tangphatsornruang S., Sangsrakru D., Chanprasert J., Uthaipaisanwong P., Yoocha T., et al. , 2010. The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: structural organization and phylogenetic relationships. DNA Res. 17: 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitrone A., Kirkpatrick M., Levin D. A., 2003. A model for chloroplast capture. Evolution 57: 1776–1782 [DOI] [PubMed] [Google Scholar]
- Twyford A., Ennos R., 2012. Next-generation hybridization and introgression. Heredity 108: 179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M.-L., Blazier J. C., Govindu M., Jansen R. K., 2014. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol. Biol. Evol. 31: 645–659 [DOI] [PubMed] [Google Scholar]
- Wicke S., Schneeweiss G., dePamphilis C., Müller K., Quandt D., 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76: 273–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski M. F., Lavin M., Sanderson M. J., 2004. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am. J. Bot. 91: 1846–1862 [DOI] [PubMed] [Google Scholar]
- Wojciechowski M. F., Sanderson M. J., Steele K. P., Liston A., 2000. Molecular phylogeny of the “temperate herbaceous tribes” of papilionoid legumes: a supertree approach, pp. 277–298 in Advances in legume systematics, edited by Herendeen P. S. Bruneau A.. Royal Botanic Garden, Kew, UK [Google Scholar]
- Wyman S. K., Jansen R. K., Boore J. L., 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20: 3252–3255 [DOI] [PubMed] [Google Scholar]
- Xu D., Abe J., Gai J., Shimamoto Y., 2002. Diversity of chloroplast DNA SSRs in wild and cultivated soybeans: evidence for multiple origins of cultivated soybean. Theor. Appl. Genet. 105: 645–653 [DOI] [PubMed] [Google Scholar]
- Xu D., Abe J., Kanazawa A., Gai J., Shimamoto Y., 2001. Identification of sequence variations by PCR-RFLP and its application to the evaluation of cpDNA diversity in wild and cultivated soybeans. Theor. Appl. Genet. 102: 683–688 [Google Scholar]
- Xu D., Abe J., Sakai M., Kanazawa A., Shimamoto Y., 2000. Sequence variation of non-coding regions of chloroplast DNA of soybean and related wild species and its implications for the evolution of different chloroplast haplotypes. Theor. Appl. Genet. 101: 724–732 [Google Scholar]
- Yoshida T., Furihata H. Y., Kawabe A., 2014. Patterns of genomic integration of nuclear chloroplast DNA fragments in plant species. DNA Res. 21: 127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B., Yonezawa T., Zhong Y., Hasegawa M., 2009. Episodic evolution and adaptation of chloroplast genomes in ancestral grasses. PLoS ONE 4: e5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.