Abstract
Many spider mites belonging to the genus Tetranychus are of agronomical importance. With limited morphological characters, Tetranychus mites are usually identified by a combination of morphological characteristics and molecular diagnostics. To clarify their molecular evolution and phylogeny, the mitochondrial genomes of the green and red forms of Tetranychus urticae as well as T. kanzawai, T. ludeni, T. malaysiensis, T. phaselus, T. pueraricola were sequenced and compared. The seven mitochondrial genomes are typical circular molecules of about 13,000 bp encoding and they are composed of the complete set of 37 genes that are usually found in metazoans. The order of the mitochondrial (mt) genes is the same as that in the mt genomes of Panonychus citri and P. ulmi, but very different from that in other Acari. The J-strands of the mitochondrial genomes have high (∼84%) A+T contents, negative GC-skews and positive AT-skews. The nucleotide sequence of the cox1 gene, which is commonly used as a taxon barcode and molecular marker, is more highly conserved than the nucleotide sequences of other mitochondrial genes in these seven species. Most tRNA genes in the seven genomes lose the D-arm and/or the T-arm. The functions of these tRNAs need to be evaluated. The mitochondrial genome of T. malaysiensis differs from the other six genomes in having a slightly smaller genome size, a slight difference in codon usage, and a variable loop in place of the T-arm of some tRNAs by a variable loop. A phylogenic analysis shows that T. malaysiensis first split from other Tetranychus species and that the clade of the family Tetranychoidea occupies a basal position in the Trombidiformes. The mt genomes of the green and red forms of T. urticae have limited divergence and short evolutionary distance.
Background
The spider mite genus Tetranychus includes 149 species [1], some of which are of cosmopolitan agronomical pests, such as Tetranychus urticae. With expanded gene families for ABC genes [2], detoxification and digestion [3] in the genome, T. urticae represents one of the most polyphagous arthropod herbivores [1]. The genes of detoxification families have also been reported to transcriptionally respond to the defense chemistry of plants after T. urticae adapts to a challenging host (tomato) [4]. T. urticae and other tetranychid species feed on apples, citrus, cottons, cucumbers, cucurbits, eggplants, grapes, maize, papayas, peppers, soy, strawberries and tomatoes both in the field and in greenhouses. However, it is difficult to identify the tetranychid species by morphological characters because the potential diagnostic morphological characters are limited and often exhibit great phenotypic flexibility. Classification of the green and red forms of T. urticae is also a source of debate. These problems demonstrate the need for combining morphological and molecular approaches to identifying the species [5].
The second internal transcribed spacer (ITS2) of ribosomal DNA can be used as a barcode for distinguishing tetranychid species. For example, the ITS2 sequences of T. kanzawai and T. hydrangea indicate that they are synonymous species, which was confirmed by cross-breeding experiments [6]. One molecular diagnostic tool that has been developed to overcome the difficulty of morphological identification by the ITS2 sequence is restriction fragment length polymorphism (RFLP) [7], [8]. The PCR-RFLP approach has been used to identify tens of Tetranychus species and has the ability to distinguish more species. The 5′ end of the mitochondrial COI gene is extensively used as a barcode to identify Tetranychus species and to analyze their phylogenetic evolution. Its higher divergence makes COI suitable for investigating intraspecific variation, but its usefulness for resolving phylogenetic species relationships remains limited [9], [10]. The lack and occasional unreliability of sequences in public databases restricts their use as molecular diagnostic tools [5].
Thus, to clarify the molecular phylogeny of Tetranychus species, as well as to provide new DNA barcodes, we decided to compare the whole mitochondrial genomes of the green and red forms of T. urticae as well as five other major spider mite pests in China. Most metazoan mitochondrial genomes are circular, have a length of approximately 16 kb and encode 37 genes including 13 protein-coding genes (PCGs), two rRNA genes (rRNAs), and 22 tRNA genes (tRNAs) [11]. The databases presently include the mitochondrial genomes of 37 acarids, including 12 of the superorder Acariformes and 25 of the superorder Parasitiformes (Table S1). Several aspects of the mt-genomes of Acari have been examined, including gene rearrangement [12]–[16], tRNA gene loss [17] and atypical short tRNA [13], [14].
Sequencing these genomes will have other benefits. For example it should provide insights into the molecular evolution of acaricide-resistance genes. The rapid development of acaricide resistance of spider mites is a long-standing problem [18], [19]. Several acaricides have been identified as mitochondrial respiration inhibitors [19]–[21]. Resistance to the acaricide bifenazate has been correlated with mutations in the mitochondrial cytochrome b (cob) gene [22], [23]. The genomes will also provide information on gene rearrangements [24]–[30], evolutionary pattern and structure of the control region [31], [32], strand asymmetry in nucleotide composition [33] and RNA secondary structure [34].
Materials and Methods
Sample origin and identification
Ethics Statement: No specific permits were required for the collection of spider mites because the spider mite is a pest in agriculture and the location is not privately-owned in any way. The field study did not involve endangered or protected species.
Strains of T. kanzawai, T. ludeni, T. malaysiensis, T. phaselus, T. pueraricola and the green and red forms of T. urticae were collected in the field, separately (Table S2). Mites were reared on a leaf of the common bean (Phaseolus vulgaris L.) at 25±1°C, 60% r.h. and under a 16/8 h (light/dark) photoperiod. All the species were classified by morphological characteristics [35] and RFLP analyses [7], [8]. For the RFLP analyses, the ITS2 fragment was amplified by PCR using the Tetranychus universal primers, rD02 and HC2 (Table S3) and genomic DNA as template, and digested by five restriction endonucleases (DraI, RsaI, MboII, DdeI and HinfI) (Fig. S1).
DNA processing
DNA was extracted from individual mites with a Wizard Genomic DNA Purification Kit (Promega) according to the manufacturer's protocol. A fragment of the COI gene was amplified by standard PCR using the Tetranychus universal primers, T-CO1-F and T-CO2-R (Table S3). PCR fragments were ligated into pEASY-T3 cloning vector (Beijing TransGen Biotech) and the resulting plasmid DNAs were transformed into competent Escherichia coli Trans1-T1 cells provided in the cloning kit. The inserted fragments were sequenced with M13f and M13r primers. Long PCR primers for each species were designed according to the COI fragment sequences (Table S3). The mitochondrial genome was amplified by long PCR in one single fragment according to the manufacturer's rapid PCR protocol. The reaction mixture contained 2 µl PrimeSTAR GXL DNA Polymerase (Takara), 10 µl buffer, 4 µl dNTP mixture (2.5 mM each), 1 µl of each primer (10 mM), 1 µl of DNA and water added to total volume of 50 µl. The cycling conditions were 30 cycles of 98°C for 10 s and 68°C for 5 min. Sequencing libraries for the long PCR fragments were prepared by using a TruSeq DNA Sample Prep Kit (Illumina) following the manufacturer's instructions. Each individual library was tagged for a different multiplexing identifier (MID). The libraries were purified with Certified Low Range Ultra Agarose (Bio-Rad), quantified with a TBS380 fluorometer (Invitrogen), pooled and sequenced using a Miseq V2 Reagent kit to generate pair-end reads (read length: 250 bp). The reads for each sample were sorted by tag sequences and assembled into one contig after trimming the tag adapter sequences. The contigs were assembled with COI sequences respectively to obtain integral genome sequences (NCBI GenBank accession numbers: KJ729017 – KJ729023).
Annotation and analysis
Protein-coding genes (PCGs) were identified by ORF Finder implemented at the NCBI website using the invertebrate mitochondrial genetic code. The sequences of PCGs that matched T. urticae mitochondrial genes submitted previously to NCBI [22] were accepted. The tRNA genes were identified using ARWEN with default parameters [36] and the tRNAscan-SE [37] with a cove cutoff score of 0.1, the tRNA-model set to “EufindtRNA-Cove” and source set to “Mixed”. Other tRNA genes and two rRNA genes (rrnL and rrnS) were determined by sequence similarity to genes in Panonychus citri [13], [23], P. ulmi and T. urticae. The secondary structure models for tRNA and rRNA genes were constructed by comparison with the published secondary structures for Dermatophagoides pteronyssinus [16], Leptotrombidium pallidum [38], Panonychus citri [13], Steganacarus magnus [17] and Palmaria palmata [39]. The Map, GC content and GC skew of the mitochondrial genome were drawn with the CGView [40]. The base composition, codon usage, Relative Synonymous Codon Usage (RSCU) values and nucleotide substitution were analyzed with Mega ver. 6 [41]. The secondary structure of the A+T-rich regions (putative control region) were constructed with Mfold Server [42] and RnaViz ver. 2 software [43]. The evolutionary pairwise divergence was estimated with Mega ver. 6 with Kimura 2-parameter model [44]. Multiple alignments of 13 PCGs' amino acid sequences were performed with ClustalW [45] as implemented in Mega ver. 6 and then corrected by eye to ensure that alignment was in agreement with protein coding genes and to minimize the number of uninformative gaps (Appendix S1). The order of the PCGs is atp6, atp8, cox1-3, cob, nad1-4, nad4l and nad5–6. The length of the alignment was 2974 amino acids in the final dataset. ProtTest (http://darwin.uvigo.es/software/prottest2_server.html) [46] was used to select the best model of protein evolution. It selected mtREV+G+I+F as the most appropriate model for the combined amino acid dataset under the Bayesian Information Criterion (BIC) [47]. Maximum-likelihood (ML) analysis of the amino-acid dataset was performed with PhyML ver. 3.1 [48] under the mtREV+G+I+F model, and Maximum-parsimony (MP) analysis was performed with Mega ver. 6. Bootstrap percentages (BPs) were calculated with 1000 replications. MrBayes ver. 3.2.2 was used for Bayesian analysis under the mtREV model of amino acid substitution with the nst = 6 rates = invgamma command. Two runs were performed simultaneously, each with four Markov chains (one cold, three heated). The analyses were run for 1,000,000 generations and the trees were sampled every 100 generations. The Markov chain stationarity and run parameter convergence were evaluated with TRACER ver. 1.6. The first 25 percent of the trees were discarded as burn-in with the relburnin = yes burninfrac = 0.25 command. Multiple alignments of whole genomic DNA sequences of the Tetranychoidea were performed with ClustalW as implemented in Mega ver. 6 (Appendix S1). jModelTest Ver. 2.1.4 [49] selected the GTR+I+G model as the best model for the nucleotide sequences of the mt genome. The ML analysis was performed and bootstrapped with 1000 replications with PhyML v. 3.1 with the GTR+I+G model, and MP analysis was performed by using Mega ver. 6. The Markov chains were run for 10,000,000 generations with sampling every 1000 generations for the Bayesian analysis. The first 25 percent of the trees were discarded and the remaining trees were used to calculate Bayesian posterior probabilities.
Results and Discussion
General features of mitochondrial genome organization
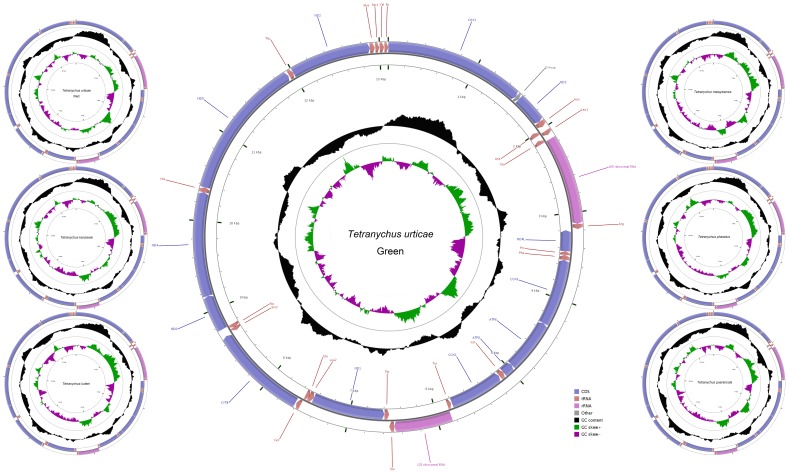
The mitochondrial genomes of the seven common tetranychid mites in China are typical circular DNAs (Fig. 1) with lengths of about 13,000 bp (Table S2). To our knowledge, the mitochondrial genome of T. malaysiensis is the smallest within all Acari genomes accessible in the GenBank (status March 17, 2014). The mitochondrial genome sizes of the red and green forms of T. urticae differ by 4 bp. Thirty-seven genes (13 PCGs, two rRNA genes, and 22 tRNA genes) were identified in each genome (Table S4), which is typical of presently available in metazoan mitochondrial genomes [11]. Twenty genes are encoded on the majority strand (J-strand), whereas the others are encoded on the minority strand (N-strand). The gene order is the same in all seven genomes and the same as that in Panonychus citri and P. ulmi which are in the same family as Tetranychus. However, this gene order is very different from that in other Acari and chelicerates as Yuan's [13] and Van Leeuwen's [23] reports. This suggests that the mitochondrial genome rearrangement event occurred before the divergence of Panonychus and Tetranychus.
Figure 1. Mitochondrial genome maps of T. urticae (green and red forms), T. kanzawai, T. ludeni, T. malaysiensis, T. phaselus and T. pueraricola.
From outer to inner, the 1st circle shows the gene map and tRNA genes are abbreviated by triple letter, with Leu1 = CUN, Leu2 = UUR, Ser1 = AGN and Ser2 = UCN. The 2nd circle shows the GC content and the 3rd shows GC skew calculated as (G−C)/(G+C). GC content and GC skew are plotted as the deviation from the average value of the entire sequence.
Base content
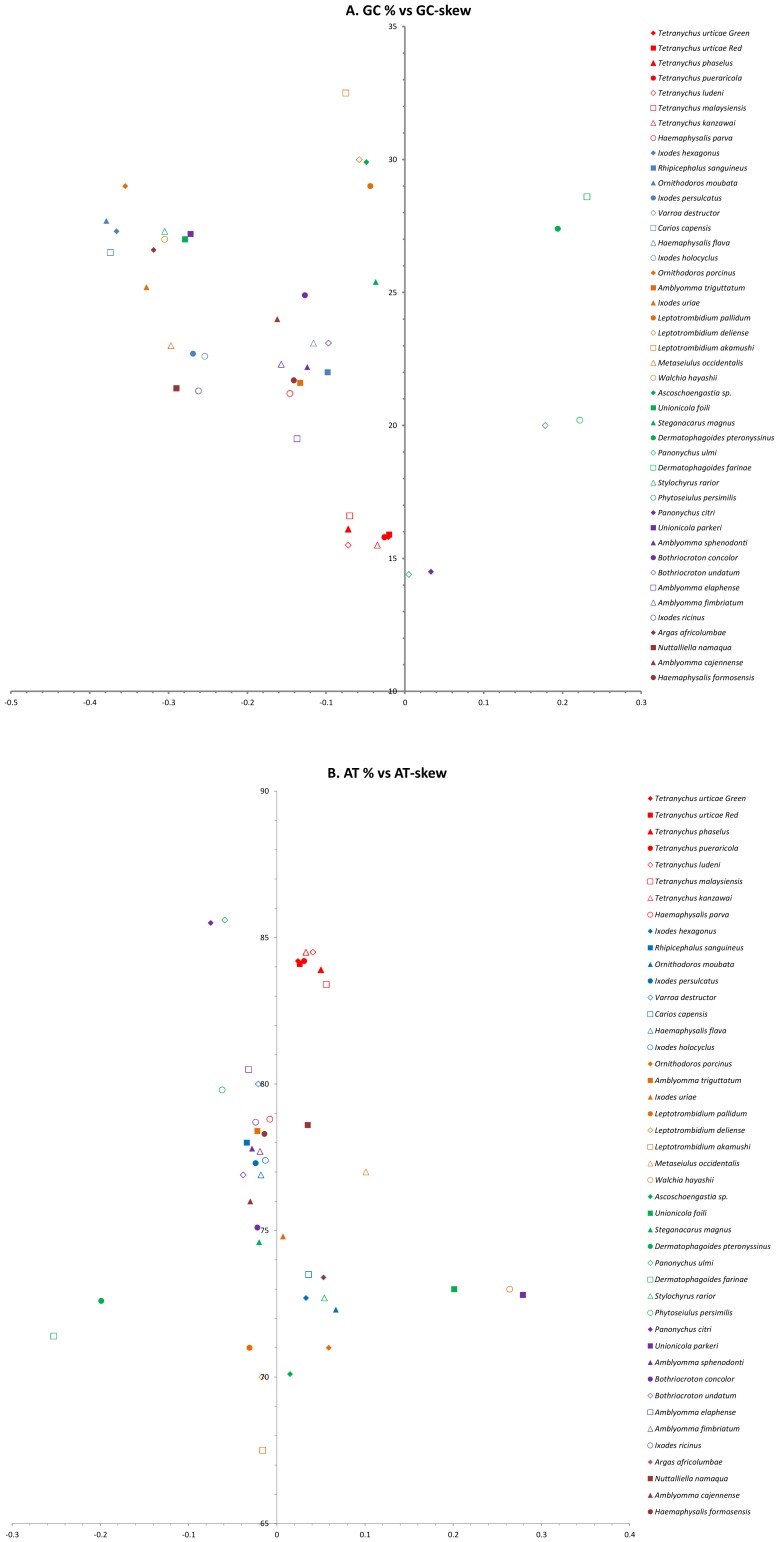
The J-strands of the seven mitochondrial DNAs have high A+T contents (83.4–84.5%) which are higher than those for Acariformes (about 74% without the Tetranychidae family) and Parasitiformes (about 77%) (Fig. 2). In Acari, the AT-skew of the mitochondrial genome (average 0.009±0.013) ranges from 0.279 in Unionicola parkeri to −0.253 in Dermatophagoides farina. Within the genus Tetranychus, the species with the highest AT skews in the mitochondrial DNAs are T. malaysiensis (0.056), T. phaselus (0.050) and T. ludeni (0.041). The average GC-skew of Acari mitochondrial genomes is −0.130±0.024, ranging from −0.379 in Ornithodoros moubata to 0.231 in Dermatophagoides farina. Most Acari have negative mitochondrial genome GC-skews. The exceptions are Varroa destructor (0.178), Dermatophagoides pteronyssinus (0.194), D. farinae (0.231), Phytoseiulus persimilis (0.222), P. citri (0.033) and P. ulmi (0.005). The Tetranychus species with the lowest mitochondrial DNA skews are T. ludeni (−0.072), T. phaselus (−0.072) and T. malaysiensis (−0.070). The AT- and GC-skews are quite similar in the mitochondrial genomes of the two T. urticae forms. Most metazoan species present a clear strand asymmetry, in which the J-strand is biased in favor of A and C and the N-strand is biased in favor of T and G [50]. The J-strands of the seven mitochondrial DNAs exhibit typical GC-skews, but two completed mitochondrial genomes from the genus Panonychus have positive GC-skews. It has been suggested that such reversals are caused by inversions of the A+T-rich regions and replication origin [33].
Figure 2. GC% vs GC-skew and AT% vs AT-skew in the 44 Acari mitochondrial genomes.
Values are calculated for mitochondrial genomes of J-strands. The X-axis provides the nucleotide skew values and the Y-axis shows the nucleotide percentages.
Putative control region
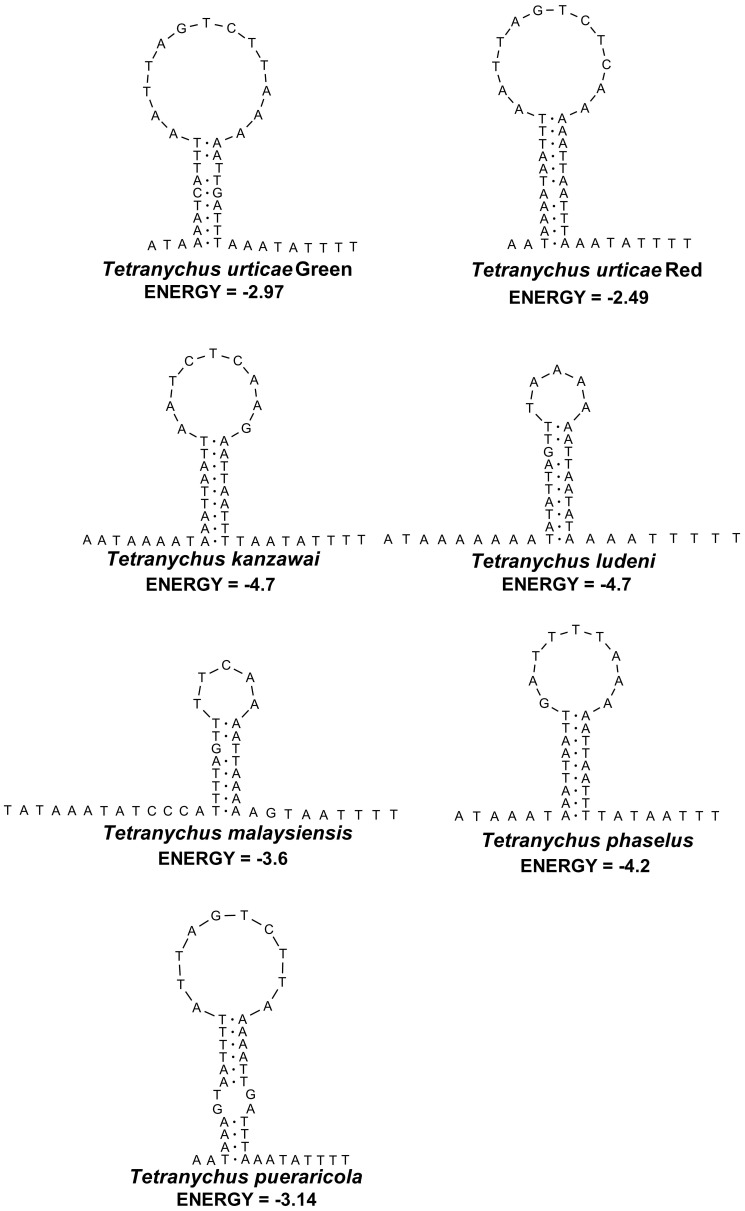
The longest non-coding region, which presumably functions as the mitochondrial control region, is 41–44 bp in length and is flanked by the cox1 and nad3 genes. Among Acari, the genus Tetranychus has the smallest mitochondrial control region. The A+T-rich region is believed to be characterized by a poly-T stretch at the 5′ end, a poly[TA(A)]n stretch close to the poly-T stretch, a stem and loop structure flanked by a TATA motif and a G (A)n T motif [31], [32]. The A+T-rich region in the seven Tetranychus genomes can be folded into one stem-loop secondary structure and an apparent poly [TA(A)]n stretch located near the 5′ end of A+T-rich region (Fig. 3). However,none of the hairpin structures is associated with a poly-T stretch or flanked by a TATA or G (A)n T motif. The loops in the secondary structures of T. ludeni and T. malaysiensis are smaller than they are in other Tetranychus species, and the stem-loop secondary structure in T. urticae and T. pueraricola are located closer to the 5′ end of the A+T-rich region. Although similar poly [TA(A)]n stretches are present near the 5′ end of A+T-rich regions in T. urticae (green form: TAAAA; red form: TAAAA) and T. pueraricola (TAAA), they were folded as stems in the hairpin structures. The A+T-rich regions of P. citri and P. ulmi are not only longer (57 and 55 bp, respectively), but they also can be folded into two hairpin structures. Short poly-T stretches were identified in the A+T-rich regions of P. citri and P. ulmi, but the poly [TA(A)]n stretch, TATA or G (A)n T motif are not present.
Figure 3. Putative stem-loop secondary structure of A+T-rich regions.
The structures were constructed by using Mfold and Watson-Crick bonds are illustrated by black dots. The free energy values (kcal/mol) and the species names are shown below each structure.
Protein-coding genes and codon usage patterns
The total length of the 13 PCGs in T. malaysiensis (10210 bp) is shorter than it is in the other Tetranychus mitochondrial genomes (10224–10227 bp). All the PCGs start with typical ATN codons (where N is any nucleotide), as is the case in other metazoan mitochondria (Table S4). Only three of the mitochondrial genes in T. malaysiensis (cox1, nad6 and nad4) and only one gene in T. phaselus (nad4l) have ATC start codons. In the other genes, ATA, ATT and ATG start codons are more common. In most cases, a given gene has different start codons in the different mitochondrial genomes. The exceptions are cox3, atp6 and cox2, which start with ATG in each genome and nad2, which starts with ATA in each genome. Although most PCGs have TAA or TAG stop codons, many of them have incomplete stop codons, such as T or TA. The two forms of T. urticae have different start codons for nad3 and different stop codons for cox1. In the codons of the 13 PCGs of the seven genomes, the three codon positions have different nucleotide biases (Fig. S2). The A+T content of the third codon position (about 91%) is higher than the A+T contents of the first and second codon positions (about 82% and 78%, respectively). But the third codon position in the T. malaysiensis genome has a lower thymine content (47%) and a higher cytosine content (7%) than do the third codon positions in the others six genomes (in which the average T and C contents are 49% and 5%, respectively).
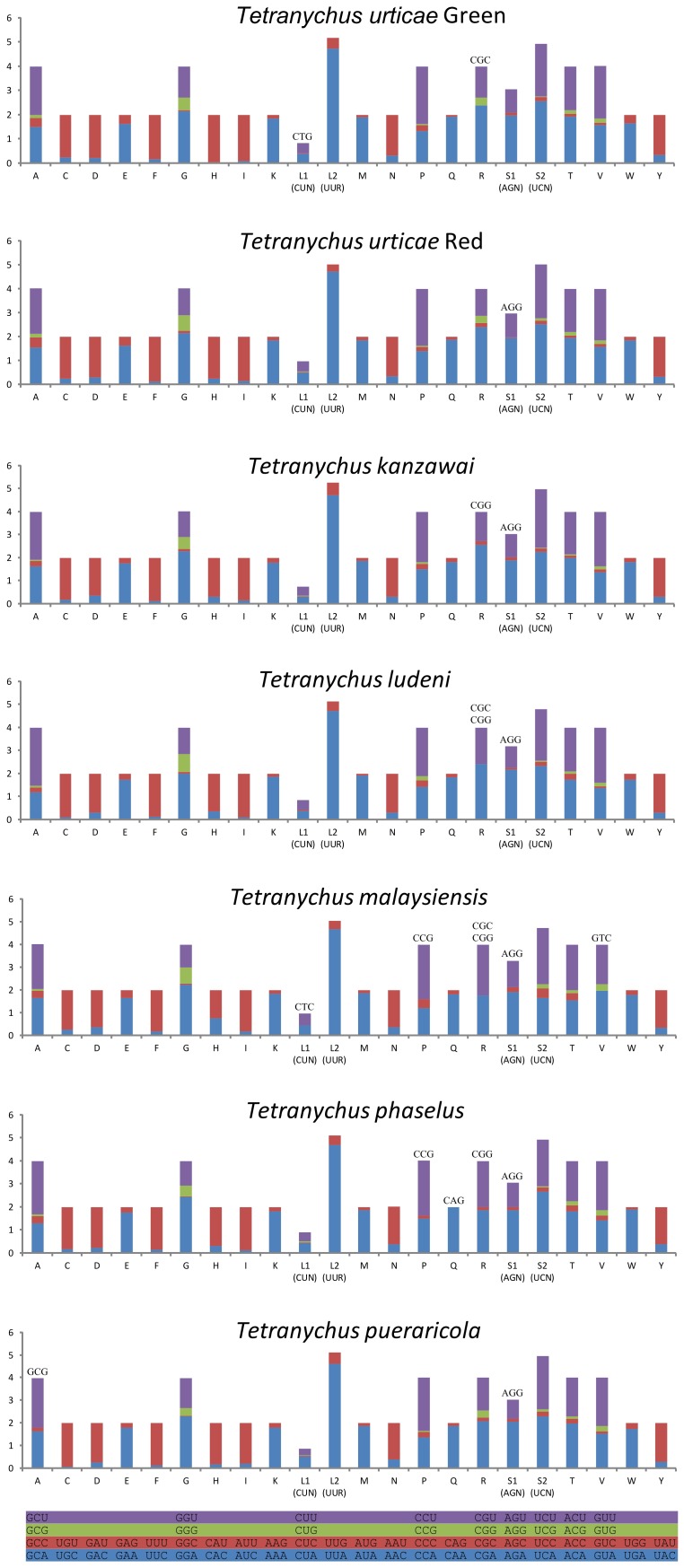
The amino acid frequencies without start and stop codon are similar between the different Tetranychus mitochondrial genomes (Fig. S3). The utmost frequently used amino acids are Phe (14.61–15.26%), Leu (13.14–13.85%), Met (11.25–12.83%), Ile (10.38–11.57%), Ser (9.06–9.65%) and Asn (8.25–8.59%). The mitochondrial genome of T. malaysiensis has a slight variety in the amino acid proportion of Met (12.83%, average: 11.77%) and Ile (10.38%, average: 11.29%). The seven AT-rich codons TTT-Phe (13.31–14.35%), ATT-Ile (9.52–11.02%), TTA-Leu (10.30–10.87%), ATA-Met (10.63–12.00%), AAT-Asn (6.71–7.23%), AAA-Lys (4.52–4.84%), and TAT-Tyr (3.48–3.78%) are the most frequently used codons in the Tetranychus PCGs. The slightly low ATT codon (9.52%, average: 10.54%) and high ATA codon (12.00%, average: 11.03%) usage in the T. malaysiensis mitochondria leads to the variety in the amino acid proportion. A considerable transversion likely occurred between the codon ATT and ATA in the evolution of Tetranychus mitochondria. The Relative Synonymous Codon Usage (RSCU) in Tetranychus PCGs exhibits a similar pattern and an over-usage of A and T at the third codon positions (Fig. 4). Six codons could not be identified in the T. malaysiensis mitochondrial PCGs whereas other Tetranychus PCGs abandon only 1–4 codons. Two codons (CTG and CGC) could not be identified in the PCGs of green form of T. urticae, while only one codon (AGG) is not present in the PCGs of red form of T. urticae. Several arthropods have been reported to translate the codon AGG as lysine instead of serine in mitochondrial genetic codon [51]. However, only one AGG codon was identified in the PSGs of T. urticae green form. The other mitochondrial genomes of spider mites including P. citri and P. ulmi [13] do not use AGG codon. And it is still uncertain which amino acid does the AGG codon in spider mite's mitochondrion translate.
Figure 4. Relative synonymous codon usage (RSCU) for the mitochondrial proteins.
The X-axis shows the codon families, while the Y-axis shows RSCU values. Absent codons are shown at the top of the columns.
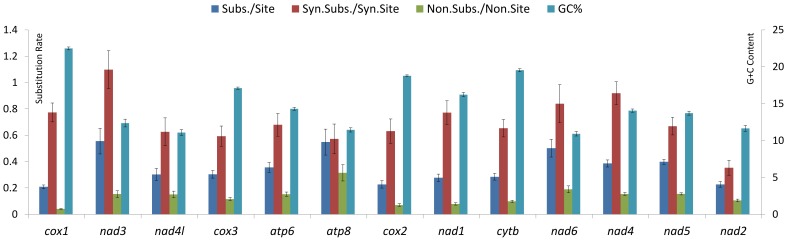
The evolutionary patterns of the PCGs are different (Fig. 5). The cox1 gene is commonly used as a taxon barcode because of its high rates of interspecific sequence change and constraints on intraspecific divergence [52], [53]. But this gene exhibits the lowest nucleotide substitution rate per site (0.208±0.014) and lowest value of nonsynonymous substitutions per nonsynonymous site (Ka) (0.04±0.004) compared to the other genes. The nucleotide substitution rate per site (0.555±0.097) and the number of synonymous substitutions per synonymous site (Ks) (1.098±0.143) of nad3 gene are highest, while the Ka number of atp8 is highest (0.315±0.062). In addition, the GC content was found to be negatively correlated with both the nucleotide substitution rate per site (R = −0.637, P = 0.019) and Ka (R = −0.715, P = 0.006), while the nucleotide substitution rate per site was found to be positively correlated with Ka (R = 0.825, P = 0.001). Ks was not found to be associated with Ka, GC content or nucleotide substitution rate per site.
Figure 5. Different evolutionary patterns among protein coding genes of T. urticae green form, T. kanzawai, T. ludeni, T. malaysiensis, T. phaselus and T. pueraricola.
Sub./Site, the number of nucleotide substitutions per site from averaging over all sequence pairs, was calculated with JC model. Syn.Subs./Syn.Site, the number of synonymous substitutions per synonymous site (Ks); Non.Subs./Non.Site, the number of nonsynonymous substitutions per nonsynonymous site (Ka); Ks-Ka, the value of Ks minus Ka; the analysis were estimated with Kumar model [68]. The rate variation among sites was modelled with a gamma distribution calculated by Mega ver. 6 and the standard error estimations were obtained by bootstrapping 1000 replicates. Substitution rate is shown on the left Y-axis and G+C content is shown on the right Y-axis.
Ribosomal and transfer RNAs
Among the seven Tetranychus mitochondrial genomes, the large subunits of rRNA (rrnL) are from 982 bp (T. ludeni) to 998 bp (T. urticae and T. kanzawai) in length. The first nucleotide downstream of trnE was annotated as the 5′ end of rrnL and the first nucleotide upstream of trnR was annotated as the 3′ end of rrnL. The genes for the small subunit of the rRNA (rrnS) are 629 bp (T. ludeni and T. phaselus) to 645 bp (T. pueraricola) in length and are located between trnY and trnG. Both ribosomal subunits are encoded on the J-strand as is the case in Metaseiulus occidentalis [54], Dermatophagoides farinae [14], D. pteronyssinus [16], Leptotrombidium pallidum [55], P. citri [13] and P. ulmi, whereas the rRNA genes in most species of Acari, arthropods and chelicerates are located on the N-strand [12], [14], [15], [17], [56], [57].
We predicted the rrnL and rrnS secondary structures of T. urticae (Fig S4 and S5). Nucleotides that are identical among the seven Tetranychus mitochondrial DNAs are shown in bold in Figures S4 and S5. In contrast, most of the helices found in Acari rrnL and rrnS genes are present in T. urticae. However, four helices (D10, D14, G9 and H3) of rrnL and one helix (33) of rrnS found in P. citri and L. pallidum are not present, while T. urticae has one more helix (D8) in rrnL and three more helices (4, 18 and 22) in rrnS. Most ribosomal subunit sequences, especially the 3′-end of both rrnL and rrnS, are conserved among the seven mitochondrial genomes. The helices D2, D4, D7, D9, and D17 could be identified in all the mitochondrial rrnL genes with slight variations, whereas their sequences are not conserved. The sequences of helices 24/25/26 as depicted in rrnS are weakly conserved among the genus Tetranychus, and the stem-loops in the deduced secondary structures vary in size.
Eleven of the 22 tRNA genes are encoded on the J-strand and the other 11 are encoded on the N-strand. The secondary structures for all tRNAs of the seven mitochondrial genomes were predicted (Table S5). The aminoacyl acceptor stem (AA-arm) and anticodon arm (AC-arm) in the tRNAs are highly conserved among the seven genomes. With a few exceptions among the seven genomes, only seven of the tRNAs (trnH, trnK, trnL2, trnM, trnN, trnR and trnW) can be potentially folded into a classical cloverleaf structure, whereas four tRNAs (trnD, trnE, trnS1 and trnS2) lose the DHU stem (D-arm), eight (trnA, trnC, trnF, trnG, trnL1, trnT, trnV and trnY) lack the TψC stem (T-arm) and 3 (trnI, trnP and trnQ) appear to lack both the D- and the T-arm. T. ludeni differs from the other species in that the D-arm of trnA was substituted by a D-replacement loop, and T. malaysiensis differs from the others in that the T-arms of trnM, trnR and trnS1 were replaced by a variable loop. trnP of T. malaysiensis and trnQ of T. kanzawai still have T-arms while the trnPs and trnQs in the other species have lost them. The trnD has a only 4 bp well-paired anticodon stem. And the trnP of T. ludeni, trnQ of T. kanzawai and T. phaselus and trnV of T. malaysiensis also have one mismatch. Commonly, the 3 bp anticodon of the tRNAs was flanked by 2 bp up-stream and 2 bp down-stream, but trnI of Tetranychus mitochondria has 3 bp down-stream. These aberrant anticodon loops have been reported for the same tRNA of P. citri [13] and P. ulmi, which implies that this anticodon loop pattern is universal among trnI genes of the Tetranychidae mitochondria. Noncanonical anticodon loop structures are also present in trnL2 (3 bp down-stream) of D. pteronyssinus [16], trnS2 (3 bp up-stream) of Camelus bactrianus ferus [58] and trnH (3 bp up-stream) and trnN (6 bp down-stream) of Mesobuthus gibbosus [59].
Phylogenetic analysis
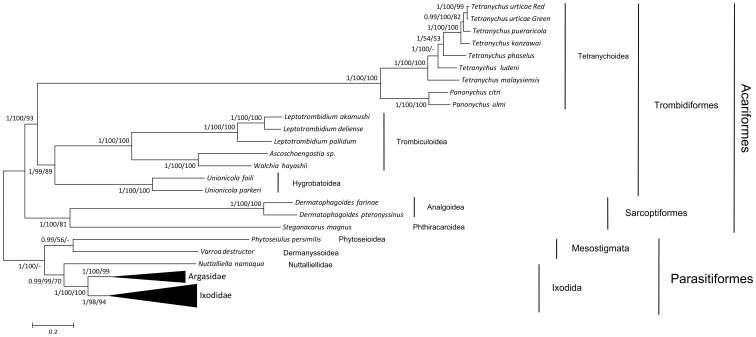
Maximum-parsimony (MP), Maximum-Likelihood (ML) and Bayesian inference (BI) phylogenetic trees were constructed based on the concatenated amino acid sequences of the 13 PCGs of the seven Tetranychus species and other Acariformes. The three trees had identical topologies (Fig. 6). Almost all nodes were well supported, whereas the split between T. phaselus and the other Tetranychus species was not supported by the MP tree. The three species of Sarcoptiformes were monophyletic and clustered as a sister group of Trombidiformes, in agreement with a previous analysis [16]. The superfamily Tetranychoidea, which is considered as a member of Trombidiformes, split as a single clade and formed a sister group to other species of Trombidiformes. Another study [16] obtained a similar topology, except that the bootstrap values and BI posterior probabilities for the Tetranychoidea were low. Mitochondrial genomes evolve at high rates. It has been suggested that the three most important factors are rearrangements in the mitochondrial genome, a parasitic lifestyle and small body size [60]. Most species of Acariformes exhibit all three factors and the great variability in branch lengths suggest a high heterogeneous substitution rate among the mt genomes. These problems led to markedly long branches, and the superabundant phylogenetic signals probably led to the distant phylogenetic position of the Tetranychoidea. Identical topologies were constructed by the three approaches with high bootstrap values and high Bayesian posterior probabilities for the position of Tetranychoidea. However, additional mt genome data or trees based on more conserved sequences are needed to improve the phylogeny of Acariformes.
Figure 6. Phylogenetic tree of Acariformes relationships.
The tree was inferred from amino acid datasets and rooted with Parasitiformes taxa. Numbers at nodes are percentages from Bayesian posterior probabilities (left), ML bootstrapping (middle) and MP bootstrap support values (right). The nodes that did not support Bayesian inference or had low bootstrap support values were marked as midlines.
The seven Tetranychus species clustered monophyleticly. T. malaysiensis was allocated to the basal position of the genus Tetranychus. The position of T. phaselus was supported by the BI and ML trees, whereas the bootstrapping for MP questioned this node. A phylogenetic tree was constructed based on the mitochondrial genomic sequences of Tetranychoidea (Fig. S6). The topologies of the trees constructed with the three approaches were consistent. Furthermore, a phylogenetic analysis based on nuclear gene sequences of rRNA and ITS supported this analysis (data not shown). However, the position of T. phaselus was not supported well by three trees. Some other mitochondrial genomic sequences are needed to improve the phylogenetic analysis for the genus Tetranychus. Especially the species has close relationship with T. phaselus.
The green and red forms of T. urticae
The green and red forms of T. urticae clustered together in the phylogenetic trees and the evolutionary divergence between their mitochondrial genomes was low (0.027±0.002) (Table S6). Pairwise distances among the Tetranychus mt genomes except for the value between the two forms of T. urticae were 0.089 to 0.211 which is much higher than the distance between the two forms of T. urticae. Some slight divergences between two forms are found in the length of 4 bp difference, an AGG codon found in the PCGs of green form, a different start codon in nad3 gene and a different stop codon in cox1 gene. However, these limited differences can not be classified as interspecies divergences. Consequently, the close evolutionary distance and limited mitochondrial genome divergence do not support the red forms of T. urticae as a new species or subspecies. Between the two forms of T. urticae, partial hybrid infertility was discovered [61] and hybrid affinity strongly restricted the gene flow [62]. However, the harboring of Wolbachia and Cardinium by the two forms [63]–[65] confounds the origin of reproductive abnormality in the hybrid analyses. Wolbachia is considered to induce cytoplasmic incompatibility (CI) in T. urticae, whereas the levels of CI in different populations varied greatly [66], [67]. Although Cardinium did not appear to distort reproduction in T. urticae [63], it is still necessary to investigate the CI level between Wolbachia and Cardinium in T. urticae. In conclusion, intracellular bacteria with the ability to manipulate reproduction complicate investigations of the ability of the two forms to interbreed. Further studies are needed to analyze the hybridization in T. urticae without the influence of Wolbachia, Cardinium and other bacteria that can manipulate reproduction.
Supporting Information
Multiple alignments of Acari 13 PCGs' amino acid sequences (13PCGs.meg). The order of the PCGs is atp6, atp8, cox1-3, cob, nad1-4, nad4l and nad5–6. The alignments were performed with ClustalW as implemented in Mega and require the Mega software version 6 to examine it (http://www.megasoftware.net/).
(MEG)
Multiple alignments of Tetranychidae mitochondrial genomic sequence (Tetranychidae.meg). The alignments were performed with ClustalW as implemented in Mega and require the Mega software version 6 to examine it (http://www.megasoftware.net/).
(MEG)
Species identification by PCR-restriction fragment-length polymorphism. PCR products were digested by 5 restriction endonucleases (MboII, HinfI, RsaI, DraI, and DdeI). The white arrowheads indicate interspecific variation. M, 100-bp ladder DNA size marker.
(TIF)
Base composition at each codon position of the 13 PCGs. Y-axis shows the percentage of each nucleotide.
(TIF)
Codon usage pattern of each mitochondrial genome. Numbers to the left refer to the percentage of each codon. Codon families are shown on the X-axis.
(TIF)
Putative secondary structure of the large-subunit ribosomal RNA of T. urticae . Inferred Watson-Crick bonds are illustrated by black dots, whereas GU bonds are illustrated by grey dots. The nucleotides with bold text show 100% identity among the seven mitochondrial genomes. The numbering of stem-loops is after [69].
(TIF)
Putative secondary structure of the small-subunit ribosomal RNA of T. urticae . Inferred Watson-Crick bonds are illustrated by black dots, whereas GU bonds are illustrated by grey dots. The nucleotides with bold text show 100% identity among the seven mitochondrial genomes. The numbering of stem-loops is after [70].
(TIF)
Phylogenetic tree of Tetranychoidea relationships. The tree was inferred from mitochondrial genomic sequences. Numbers at nodes are percentages from Bayesian posterior probabilities (left), ML bootstrapping (middle) and MP bootstrap support values (right).
(TIF)
GenBank accession numbers of mitochondrial genomes for other Acari.
(DOC)
Date and location of mite collections and GenBank accession numbers of mitochondrial genomes.
(DOC)
Initial primers and their sequences for PCR amplifications in this study.
(DOC)
Summary of mitochondrial genome organization of T. urticae (green and red forms), T. kanzawai , T. ludeni , T. malaysiensis , T. phaselus and T. pueraricola .
(DOC)
Comparison of inferred secondary structures of mitochondrial tRNA genes.
(DOC)
Pairwise distance between mitochondrial genomes.
(DOC)
Acknowledgments
We thank Hao-Sen Li for technical assistance for phylogenetic analysis in the lab. Chao Guo and Xiu-Ting Zhao provided important DNA samples of red form of T. urticae.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported in part by a grant-in-aid from the Science and Technology Program of the National Public Welfare Professional Fund (no. 201103020) from the Ministry of Agriculture of China (www.moa.gov.cn), and a grant-in-aid for Scientific Research (nos. 31172131 and 31371944) from the National Natural Science Foundation of China (www.nsfc.gov.cn). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Migeon A, Nouguier E, Dorkeld F (2010) Spider Mites Web: a comprehensive database for the Tetranychidae. Trends in Acarology: 557–560 http://www.montpellier.inra.fr/CBGP/spmweb/.
- 2. Dermauw W, Osborne EJ, Clark RM, Grbic M, Tirry L, et al. (2013) A burst of ABC genes in the genome of the polyphagous spider mite Tetranychus urticae . BMC Genomics 14: 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grbic M, Van Leeuwen T, Clark RM, Rombauts S, Rouze P, et al. (2011) The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479: 487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dermauw W, Wybouw N, Rombauts S, Menten B, Vontas J, et al. (2013) A link between host plant adaptation and pesticide resistance in the polyphagous spider mite Tetranychus urticae . Proc Natl Acad Sci USA 110: E113–E122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Mendonça RS, Navia D, Diniz IR, Auger P, Navajas M (2011) A critical review on some closely related species of Tetranychus sensu stricto (Acari: Tetranychidae) in the public DNA sequences databases. Exp Appl Acarol 55: 1–23. [DOI] [PubMed] [Google Scholar]
- 7. Arimoto M, Satoh M, Uesugi R, Osakabe M (2013) PCR-RFLP analysis for identification of Tetranychus spider mite species (Acari: Tetranychidae). J Econ Entomol 106: 661–668. [DOI] [PubMed] [Google Scholar]
- 8. Osakabe M, Kotsubo Y, Tajima R, Hinomoto N (2008) Restriction fragment length polymorphism catalog for molecular identification of Japanese Tetranychus spider mites (Acari: Tetranychidae). J Econ Entomol 101: 1167–1175. [DOI] [PubMed] [Google Scholar]
- 10. Ros VI, Breeuwer JA (2007) Spider mite (Acari: Tetranychidae) mitochondrial COI phylogeny reviewed: host plant relationships, phylogeography, reproductive parasites and barcoding. Exp Appl Acarol 42: 239–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27: 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edwards DD, Jackson LE, Johnson AJ, Ernsting BR (2011) Mitochondrial genome sequence of Unionicola parkeri (Acari: Trombidiformes: Unionicolidae): molecular synapomorphies between closely-related Unionicola gill mites. Exp Appl Acarol 54: 105–117. [DOI] [PubMed] [Google Scholar]
- 13. Yuan ML, Wei DD, Wang BJ, Dou W, Wang JJ (2010) The complete mitochondrial genome of the citrus red mite Panonychus citri (Acari: Tetranychidae): high genome rearrangement and extremely truncated tRNAs. BMC Genomics 11: 597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Klimov PB, Oconnor BM (2009) Improved tRNA prediction in the American house dust mite reveals widespread occurrence of extremely short minimal tRNAs in acariform mites. BMC Genomics 10: 598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ernsting BR, Edwards DD, Aldred KJ, Fites JS, Neff CR (2009) Mitochondrial genome sequence of Unionicola foili (Acari: Unionicolidae): a unique gene order with implications for phylogenetic inference. Exp Appl Acarol 49: 305–316. [DOI] [PubMed] [Google Scholar]
- 16. Dermauw W, Van Leeuwen T, Vanholme B, Tirry L (2009) The complete mitochondrial genome of the house dust mite Dermatophagoides pteronyssinus (Trouessart): a novel gene arrangement among arthropods. BMC Genomics 10: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Domes K, Maraun M, Scheu S, Cameron SL (2008) The complete mitochondrial genome of the sexual oribatid mite Steganacarus magnus: genome rearrangements and loss of tRNAs. BMC Genomics 9: 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Osakabe M, Uesugi R, Goka K (2010) Evolutionary aspects of acaricide-resistance development in spider mites. Psyche 2009: 1–11. [Google Scholar]
- 19. Van Leeuwen T, Vontas J, Tsagkarakou A, Dermauw W, Tirry L (2010) Acaricide resistance mechanisms in the two-spotted spider mite Tetranychus urticae and other important Acari: a review. Insect Biochem Mol Biol 40: 563–572. [DOI] [PubMed] [Google Scholar]
- 20. Hayashi N, Sasama Y, Takahashi N, Ikemi N (2013) Cyflumetofen, a novel acaricide - its mode of action and selectivity. Pest Manag Sci 69: 1080–1084. [DOI] [PubMed] [Google Scholar]
- 21. Dekeyser MA (2005) Acaricide mode of action. Pest Manag Sci 61: 103–110. [DOI] [PubMed] [Google Scholar]
- 22. Van Leeuwen T, Vanholme B, Van Pottelberge S, Van Nieuwenhuyse P, Nauen R, et al. (2008) Mitochondrial heteroplasmy and the evolution of insecticide resistance: Non-Mendelian inheritance in action. Proc Natl Acad Sci USA 105: 5980–5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van Leeuwen T, Van Nieuwenhuyse P, Vanholme B, Dermauw W, Nauen R, et al. (2011) Parallel evolution of cytochrome b mediated bifenazate resistance in the citrus red mite Panonychus citri . Insect Mol Biol 20: 135–140. [DOI] [PubMed] [Google Scholar]
- 24. Boore JL, Fuerstenberg SI (2008) Beyond linear sequence comparisons: the use of genome-level characters for phylogenetic reconstruction. Philos T R Soc B 363: 1445–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Boore JL (2006) The use of genome-level characters for phylogenetic reconstruction. Trends Ecol Evol 21: 439–446. [DOI] [PubMed] [Google Scholar]
- 26. Dowton M, Castro LR, Austin AD (2002) Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: the examination of genome ‘morphology’. Invertebr Syst 16: 345–356. [Google Scholar]
- 27. Dowton M, Austin AD (1999) Evolutionary dynamics of a mitochondrial rearrangement “hot spot” in the Hymenoptera. Mol Biol Evol 16: 298–309. [DOI] [PubMed] [Google Scholar]
- 28. Boore JL, Lavrov DV, Brown WM (1998) Gene translocation links insects and crustaceans. Nature 392: 667–668. [DOI] [PubMed] [Google Scholar]
- 29. Boore JL, Brown WM (1998) Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr Opin Genet Dev 8: 668–674. [DOI] [PubMed] [Google Scholar]
- 30. Dermauw W, Vanholme B, Tirry L, Van Leeuwen T (2010) Mitochondrial genome analysis of the predatory mite Phytoseiulus persimilis and a revisit of the Metaseiulus occidentalis mitochondrial genome. Genome 53: 285–301. [DOI] [PubMed] [Google Scholar]
- 31. Zhang DX, Hewitt GM (1997) Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary studies. Biochem Syst Ecol 25: 99–120. [Google Scholar]
- 32. Zhang DX, Szymura JM, Hewitt GM (1995) Evolution and structural conservation of the control region of insect mitochondrial DNA. J Mol Evol 40: 382–391. [DOI] [PubMed] [Google Scholar]
- 33. Wei SJ, Shi M, Chen XX, Sharkey MJ, van Achterberg C, et al. (2010) New views on strand asymmetry in insect mitochondrial genomes. PLoS One 5: e12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Masta SE (2010) Mitochondrial rRNA secondary structures and genome arrangements distinguish chelicerates: comparisons with a harvestman (Arachnida: Opiliones: Phalangium opilio). Gene 449: 9–21. [DOI] [PubMed] [Google Scholar]
- 35. Ehara S (1999) Revision of the spider mite family Tetranychidae of Japan (Acari, Prostigmata). Species diversity 4: 63–141. [Google Scholar]
- 36. Laslett D, Canback B (2008) ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 24: 172–175. [DOI] [PubMed] [Google Scholar]
- 37. Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shao R, Barker SC, Mitani H, Takahashi M, Fukunaga M (2006) Molecular mechanisms for the variation of mitochondrial gene content and gene arrangement among chigger mites of the genus Leptotrombidium (Acari: Acariformes). J Mol Evol 63: 251–261. [DOI] [PubMed] [Google Scholar]
- 39. Wuyts J, De Rijk P, Van de Peer Y, Pison G, Rousseeuw P, et al. (2000) Comparative analysis of more than 3000 sequences reveals the existence of two pseudoknots in area V4 of eukaryotic small subunit ribosomal RNA. Nucleic Acids Res 28: 4698–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grant JR, Stothard P (2008) The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36: W181–W184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. De Rijk P, Wuyts J, De Wachter R (2003) RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics 19: 299–300. [DOI] [PubMed] [Google Scholar]
- 44. Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120. [DOI] [PubMed] [Google Scholar]
- 45. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105. [DOI] [PubMed] [Google Scholar]
- 47. Adachi J, Hasegawa M (1996) Model of amino acid substitution in proteins encoded by mitochondrial DNA. J Mol Evol 42: 459–468. [DOI] [PubMed] [Google Scholar]
- 48. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- 49. Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hassanin A, Leger N, Deutsch J (2005) Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of metazoa, and consequences for phylogenetic inferences. Syst Biol 54: 277–298. [DOI] [PubMed] [Google Scholar]
- 51. Abascal F, Posada D, Knight RD, Zardoya R (2006) Parallel evolution of the genetic code in arthropod mitochondrial genomes. PLoS Biol 4: e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hebert PD, Ratnasingham S, de WaardJR (2003) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc B 270: S96–S99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hebert PD, Cywinska A, Ball SL (2003) Biological identifications through DNA barcodes. Proc R Soc B 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jeyaprakash A, Hoy MA (2007) The mitochondrial genome of the predatory mite Metaseiulus occidentalis (Arthropoda: Chelicerata: Acari: Phytoseiidae) is unexpectedly large and contains several novel features. Gene 391: 264–274. [DOI] [PubMed] [Google Scholar]
- 55. Shao R, Mitani H, Barker SC, Takahashi M, Fukunaga M (2005) Novel mitochondrial gene content and gene arrangement indicate illegitimate inter-mtDNA recombination in the chigger mite, Leptotrombidium pallidum . J Mol Evol 60: 764–773. [DOI] [PubMed] [Google Scholar]
- 56. Black W, Roehrdanz RL (1998) Mitochondrial gene order is not conserved in arthropods: Prostriate and metastriate tick mitochondrial genomes. Mol Biol Evol 15: 1772–1785. [DOI] [PubMed] [Google Scholar]
- 57. Mans BJ, de Klerk D, Pienaar R, de Castro MH, Latif AA (2012) The mitochondrial genomes of Nuttalliella namaqua (Ixodoidea: Nuttalliellidae) and Argas africolumbae (Ixodoidae: Argasidae): estimation of divergence dates for the major tick lineages and reconstruction of ancestral blood-feeding characters. PLoS One 7: e49461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cui P, Ji R, Ding F, Qi D, Gao H, et al. (2007) A complete mitochondrial genome sequence of the wild two-humped camel (Camelus bactrianus ferus): an evolutionary history of camelidae. BMC Genomics 8: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Davila S, Pinero D, Bustos P, Cevallos MA, Davila G (2005) The mitochondrial genome sequence of the scorpion Centruroides limpidus (Karsch 1879) (Chelicerata; Arachnida). Gene 360: 92–102. [DOI] [PubMed] [Google Scholar]
- 60. Hassanin A (2006) Phylogeny of Arthropoda inferred from mitochondrial sequences: strategies for limiting the misleading effects of multiple changes in pattern and rates of substitution. Mol Phylogenet Evol 38: 100–116. [DOI] [PubMed] [Google Scholar]
- 61. De Boer R (1982) Partial hybrid sterility between strains of the arrhenotokous spider mite, Tetranychus urticae complex (Acari, Tetranychidae). Genetica 58: 23–33. [Google Scholar]
- 62. Sugasawa J, Kitashima Y, Gotoh T (2002) Hybrid affinities between the green and the red forms of the two-spotted spider mite Tetranychus urticae (Acari: Tetranychidae) under laboratory and semi-natural conditions. Appl Entomol Zool 37: 127–139. [Google Scholar]
- 63. Gotoh T, Noda H, Ito S (2007) Cardinium symbionts cause cytoplasmic incompatibility in spider mites. Heredity (Edinb) 98: 13–20. [DOI] [PubMed] [Google Scholar]
- 64. Vala F, Breeuwer JA, Sabelis MW (2000) Wolbachia-induced ‘hybrid breakdown’ in the two-spotted spider mite Tetranychus urticae Koch. Proc Biol Sci 267: 1931–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Breeuwer JAJ (1997) Wolbachia and cytoplasmic incompatibility in the spider mites Tetranychus urticae and T. turkestani . Heredity 79: 41–47. [Google Scholar]
- 66. Xie RR, Chen XL, Hong XY (2011) Variable fitness and reproductive effects of Wolbachia infection in populations of the two-spotted spider mite Tetranychus urticae Koch in China. Appl Entomol Zool 46: 95–102. [Google Scholar]
- 67. Gotoh T, Sugasawa J, Noda H, Kitashima Y (2007) Wolbachia-induced cytoplasmic incompatibility in Japanese populations of Tetranychus urticae (Acari: Tetranychidae). Exp Appl Acarol 42: 1–16. [DOI] [PubMed] [Google Scholar]
- 68.Nei M, Kumar S (2000) Molecular evolution and phylogenetics. New York: Oxford University Press.
- 69. De Rijk P, Robbrecht E, de Hoog S, Caers A, Van de Peer Y, et al. (1999) Database on the structure of large subunit ribosomal RNA. Nucleic Acids Res 27: 174–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Van de Peer Y, Robbrecht E, de Hoog S, Caers A, De Rijk P, et al. (1999) Database on the structure of small subunit ribosomal RNA. Nucleic Acids Res 27: 179–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple alignments of Acari 13 PCGs' amino acid sequences (13PCGs.meg). The order of the PCGs is atp6, atp8, cox1-3, cob, nad1-4, nad4l and nad5–6. The alignments were performed with ClustalW as implemented in Mega and require the Mega software version 6 to examine it (http://www.megasoftware.net/).
(MEG)
Multiple alignments of Tetranychidae mitochondrial genomic sequence (Tetranychidae.meg). The alignments were performed with ClustalW as implemented in Mega and require the Mega software version 6 to examine it (http://www.megasoftware.net/).
(MEG)
Species identification by PCR-restriction fragment-length polymorphism. PCR products were digested by 5 restriction endonucleases (MboII, HinfI, RsaI, DraI, and DdeI). The white arrowheads indicate interspecific variation. M, 100-bp ladder DNA size marker.
(TIF)
Base composition at each codon position of the 13 PCGs. Y-axis shows the percentage of each nucleotide.
(TIF)
Codon usage pattern of each mitochondrial genome. Numbers to the left refer to the percentage of each codon. Codon families are shown on the X-axis.
(TIF)
Putative secondary structure of the large-subunit ribosomal RNA of T. urticae . Inferred Watson-Crick bonds are illustrated by black dots, whereas GU bonds are illustrated by grey dots. The nucleotides with bold text show 100% identity among the seven mitochondrial genomes. The numbering of stem-loops is after [69].
(TIF)
Putative secondary structure of the small-subunit ribosomal RNA of T. urticae . Inferred Watson-Crick bonds are illustrated by black dots, whereas GU bonds are illustrated by grey dots. The nucleotides with bold text show 100% identity among the seven mitochondrial genomes. The numbering of stem-loops is after [70].
(TIF)
Phylogenetic tree of Tetranychoidea relationships. The tree was inferred from mitochondrial genomic sequences. Numbers at nodes are percentages from Bayesian posterior probabilities (left), ML bootstrapping (middle) and MP bootstrap support values (right).
(TIF)
GenBank accession numbers of mitochondrial genomes for other Acari.
(DOC)
Date and location of mite collections and GenBank accession numbers of mitochondrial genomes.
(DOC)
Initial primers and their sequences for PCR amplifications in this study.
(DOC)
Summary of mitochondrial genome organization of T. urticae (green and red forms), T. kanzawai , T. ludeni , T. malaysiensis , T. phaselus and T. pueraricola .
(DOC)
Comparison of inferred secondary structures of mitochondrial tRNA genes.
(DOC)
Pairwise distance between mitochondrial genomes.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.