Key Points
Sequencing of Chinese DLBCL reveals novel mutation targets and highlights additional/alternative tumorigenic pathways in these tumors.
DTX1 is frequently mutated in Chinese DLBCL and deleterious mutations in this gene contribute to the activation of the Notch pathway.
Abstract
Next-generation sequencing studies on diffuse large B-cell lymphomas (DLBCLs) have revealed novel targets of genetic aberrations but also high intercohort heterogeneity. Previous studies have suggested that the prevalence of disease subgroups and cytogenetic profiles differ between Western and Asian patients. To characterize the coding genome of Chinese DLBCL, we performed whole-exome sequencing of DNA derived from 31 tumors and respective peripheral blood samples. The mutation prevalence of B2M, CD70, DTX1, LYN, TMSB4X, and UBE2A was investigated in an additional 105 tumor samples. We discovered 11 novel targets of recurrent mutations in DLBCL that included functionally relevant genes such as LYN and TMSB4X. Additional genes were found mutated at high frequency (≥10%) in the Chinese cohort including DTX1, which was the most prevalent mutation target in the Notch pathway. We furthermore demonstrated that mutations in DTX1 impair its function as a negative regulator of Notch. Novel and previous unappreciated targets of somatic mutations in DLBCL identified in this study support the existence of additional/alternative tumorigenic pathways in these tumors. The observed differences with previous reports might be explained by the genetic heterogeneity of DLBCL, the germline genetic makeup of Chinese individuals, and/or exposure to distinct etiological agents.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common and one of the most aggressive types of lymphoma.1 It is a heterogeneous entity with distinct biological and pathological subgroups that may be considered as different diseases.2 At least 2 DLBCL subtypes can be distinguished based on RNA expression profiles3: a germinal center B-cell (GCB) variant that exhibits similarities to normal B cells that populate germinal centers, and a subgroup with a gene expression signature similar to the one present in activated B cells (ABC variant).
A number of genetic alterations are characteristic of GCB and ABC subtypes. The t(14;18) translocation that juxtaposes the BCL2 proto-oncogene and the immunoglobulin (IG) heavy-chain (IGH) locus, thereby dysregulating BCL2 expression, is almost exclusively seen in the GCB group,4 whereas translocations involving the BCL6 locus occur in both subtypes.5 Other genetic lesions, characteristic of the GCB group, include: amplification of the REL oncogene4 and activating mutations in EZH2.6 Constitutive activation of nuclear factor–κB (NF-κB) signaling is a hallmark of the ABC subtype where activating mutations in CARD11,7 CD79B,8 and MYD88 genes,9 or inactivation of the TNFAIP3 gene,10 are often observed. Nevertheless, most of these genes can also be found mutated in a proportion of GCB DLBCL tumors. Genetic lesions in the PRDM1 gene are also common in ABC lymphomas that, thereby, block B-cell differentiation.11 Disruption of the histone-modifying genes CREBBP12 and MLL2,13 inactivation of TP53, and the loss of molecules required for effective tumor immune surveillance such as B2M and CD5814 are additional recurrent genetic aberrations that are found in all DLBCL subtypes.
DLBCLs are postulated to derive from germinal or post-GCB cells.15 At the germinal center, illegitimate somatic hypermutation (SHM) and class switch recombination (CSR) events, resulting from activation-induced cytidine deaminase (AID) activity at non-IG loci, are known to play a major role in the tumorigenesis of DLBCL.16 Proto-oncogenes such as PIM1, BCL6, PAX5, or RHOH are frequently targeted by SHM in these tumors,17 and non-IG SHM targets are also often rearranged in translocations that involve IGH as a partner gene.18 Dysregulation of DNA repair and damage response mechanisms involved in SHM and CSR processes are likely to play an important role in the propagation of genetic instability in DLBCL.19
A number of studies on DLBCL, based on next-generation sequencing (NGS) approaches, have been published during recent years,13,20-23 uncovering novel mutation targets, further underscoring the genetic heterogeneity of this disease. Here, we present the first characterization of the coding genome of DLBCLs derived from Chinese patients.
Methods
Patient materials
DNA was extracted from 31 DLBCL frozen biopsies and the respective paired blood samples were collected at Sun Yat-sen University Cancer Center (Guangzhou, China) to undergo whole-exome sequencing. Eight samples corresponded to relapse DLBCL cases whereas the remaining derived from unrelated primary tumors that had not undergone treatment before biopsy collection. The median age of the DLBCL patients at diagnosis was 61 years and the cohort comprised 13 women and 18 men. Ten DLBCL samples were classified as GCB, whereas 21 DLBCL samples were classified as non-GCB, according to the Hans algorithm.24 The tumor content of the DLBCL tumors was estimated to be at least 80% in all cases after histological evaluation. DNA was also extracted from frozen biopsies from an additional 136 Chinese DLBCL cases for cohort expansion studies. The LYN and DTX1 genes were further screened in 96 and 35 DLBCL samples, respectively, collected at Uppsala’s University Hospital, Sweden. DNA samples that underwent whole-exome sequencing were isolated by QIAGEN’s DNeasy Tissue and Blood kit, according to the manufacturer’s instructions. DNA from subsets of samples composing the expanded cohort was extracted by different methodologies. The institutional review boards at the Sun Yat-sen University Cancer Center and Uppsala University approved this study.
Exome sequencing
Genomic DNA preparation, exome capture, and Illumina sequencing were performed as described previously.19 Sequences were aligned to the genome with the Burrows-Wheeler Aligner software25 and single-nucleotide variants (SNVs) were detected with Varscan26 whereas somatic indels were detected with GATK.27 All reported variants passed visual inspection using the Integrative Genomics Viewer.28
Pathway analysis
Genes affected by nonsilent mutations were assigned to gene sets with the aid of the Molecular Signatures Database,29 where the “canonical pathway” collections from Biocarta and Kyoto Encyclopedia of Genes and Genomes were interrogated. Results were reviewed and only genes with an annotation supported by experimental evidence were considered. Redundant gene sets or pathways from different collections were combined. To compare the mutation loads between different pathways, the number of mutations was normalized against the size of the coding regions of the genes that composed a gene set. Only 1 nonsynonymous mutation per gene was accounted for in each tumor.
Sanger sequencing
Validation of whole-exome sequencing data and screening of the coding regions of CD70, B2M, DTX1, LYN, UBE2A, TMSB4X, TNFAIP3, TP53, and the mutation hotspot regions of NOTCH1 (p.1500-1800 and p.2300-2555) and NOTCH2 (p.2000-2471) genes in a DLBCL cohort, were performed by Sanger sequencing. Specific primers were designed with Primer-BLAST.30 Primer sequences and polymerase chain reaction (PCR) conditions for each gene are available upon request. PCR products were purified and sequenced at Eurofins MWG Operon.
Notch reporter gene assay
The coding region of DTX1 and the Notch intracellular domain (NCID) of NOTCH1 were amplified from human complementary DNA (cDNA) and ligated into a pcDNA 3.1 mammalian expression vector (Life Technologies). DTX1 mutants were generated by site-directed mutagenesis. DTX1 and NCID expression vectors were cotransfected with a CSL transcription factor-dependent firefly luciferase gene reporter (provided by Prof Jon Aster, Harvard Medical School) that has been described previously.31 A Renilla luciferase pRL-TK plasmid was used as an internal control. U2OS cells were cultured and transfected in 24-well plates with 50 ng of Notch1 plasmid, 100 ng of DTX1 plasmid, 500 ng of CSL-dependent firefly luciferase gene reporter, and 10 ng of pRL-TK plasmid, with Turbofect (Fermentas). Gene reporter activities were determined after 48 hours using a dual-luciferase reporter assay (Promega). Luminometric measurements were performed in a Varioskan Flash microplate reader (Thermo Scientific).
Analysis of protein expression of LYN mutants
The coding region of LYN was amplified from human cDNA and ligated into a pcDNA 3.1 mammalian expression vector (Life Technologies). LYN mutants were generated by site-directed mutagenesis. U2OS cells were cultured and transfected in 6-well plates with 1 μg of LYN plasmids, which were cotransfected with 1 μg of a green fluorescent protein (GFP) construct (transfection efficiency control), with Turbofect (Fermentas). The cells were lysed 48 hours after transfection and cell lysates were run on a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). LYN and GFP expression were detected with rabbit polyclonal antibodies (Cell Signaling 2732 and 2956, respectively). β-actin served as a loading control (#4967; Cell Signaling).
Whole-genome sequencing and copy number variation detection
Paired tumor and blood DNA samples from 10 DLBCL patients were fragmented by a Covaris E-210 ultrasonicator. By adjusting shearing parameters, DNA fragments were concentrated in 500-bp peaks with 1 library for each sample. These fragments were purified, end-blunted, “A” tailed, and adaptor-ligated. Ten to 12 cycles of PCR were performed after size selection. Libraries were quantified by a bioanalyzer (Agilent Technologies) and real-time PCR method using ABI StepOne plus real-time PCR system (Life Technologies). Paired-end, 90-bp read-length sequencing was performed in a HiSequation 2000 sequencer according to the manufacturer’s instructions (Illumina). Copy number alterations (CNAs) were detected by means of SegSeq.32 The resulting copy number segment statuses were defined as copy ratio ≥1.25 for gain and ≤0.75 for loss.
Statistical analysis
Statistical tests were performed with GraphPad Prism (version 4.00).
Results
Whole-exome analysis of Chinese DLBCL
Whole-exome capture and sequencing were performed on paired tumor and peripheral blood DNA from 31 Chinese DLBCL patients. A mean sequencing depth higher than 80 times was reached and, on average, 90% of exonic bases were covered by at least 10 sequencing reads (supplemental Table 1, available on the Blood Web site).
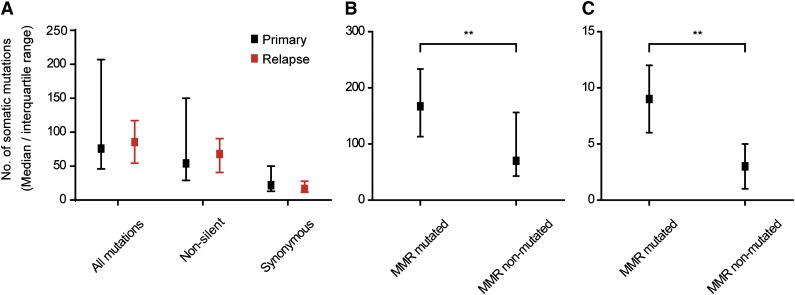
A median of 54 (range, 8-183) somatic, nonsilent mutations per tumor (Figure 1A) was detected, when restricting the analysis to mutations represented by at least 10% of the sequencing reads at a base position. Of note, >75% of these mutations were represented by at least 20% of all sequencing reads at a given nucleotide position (supplemental Figure 1), suggesting that they derived from major tumor clones. Of the 31 sequenced samples, 23 were derived from primary DLBCL tumors whereas the remaining 8 samples consisted of unrelated relapse cases. No differences were observed in the median number of mutations or SNV patterns between primary and relapse DLBCL samples (Figure 1A, supplemental Figure 2).
Figure 1.
Median number of somatic mutations identified by whole-exome sequencing. Comparison of mutation load between primary and relapse DLBCL samples (A), or between tumors with and without mutations in MMR genes (B-C). MMR-mutated tumors presented a significantly higher median of mutations (**P < .01, Mann-Whitney U test) than nonmutated cases. (B) All mutations. (C) InDels.
We previously reported that DLBCL tumors with genetic defects in mismatch repair (MMR) genes carried higher mutation loads, particularly of insertions and deletions (InDels), than tumors without MMR defects.19 We have confirmed these findings in the current cohort, which, partially, overlaps with the previous. Somatic mutations in different MMR genes (MLH1, MSH2, MSH6 [n = 2], and PMS1) were detected in 5 tumor samples and these tumors presented a median number of mutations at least 2 times higher than the one encountered in the remaining DLBCL cases (Figure 1B-C).
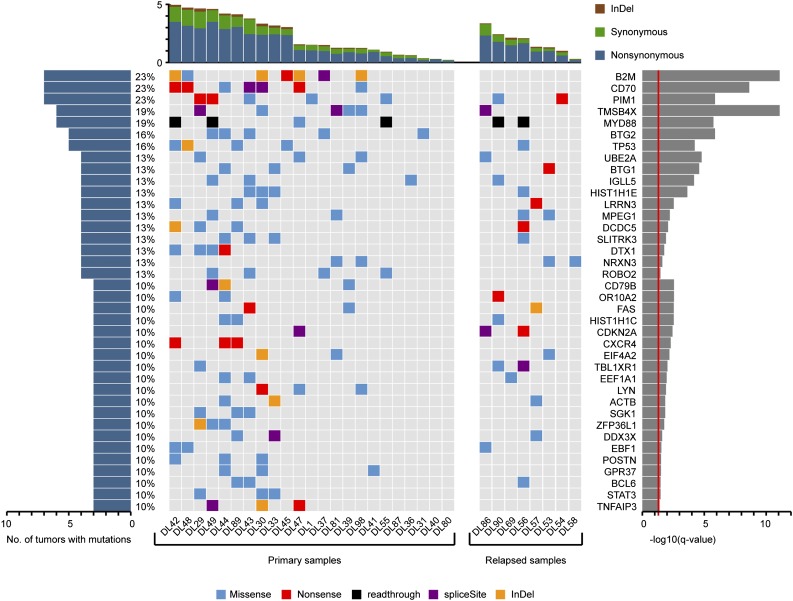
Whole-exome sequencing revealed 72 genes that were affected by nonsilent mutations in at least 3 tumor samples (10% of cases) (supplemental Table 2). Of these, 38 genes (Figure 2) were considered to be significant mutation targets by comparing the prevalence of nonsilent mutations with the background mutation rate as reported previously.33 Twenty-eight genes were targeted by nonsilent mutations in at least 4 (13%) DLBCL samples (Figure 2, supplemental Table 3). These included TMSB4X, DCDC5, IGLL5, and SLITRK3, which had not previously been described as recurrently mutated in DLBCL (Figure 2, supplemental Table 3). Additional notable and novel targets of nonsynonymous mutations included LYN, which was found mutated in 3 tumor samples (10%). Other genes such as CD70 carried alterations at greater frequencies than previously reported (supplemental Tables 2-3).
Figure 2.
Top list of somatic mutation targets in Chinese DLBCL. Genes affected by nonsilent mutations in at least 3 DLBCL samples (10% of cases) and considered to be significantly mutated by cancer gene prediction33 are displayed.
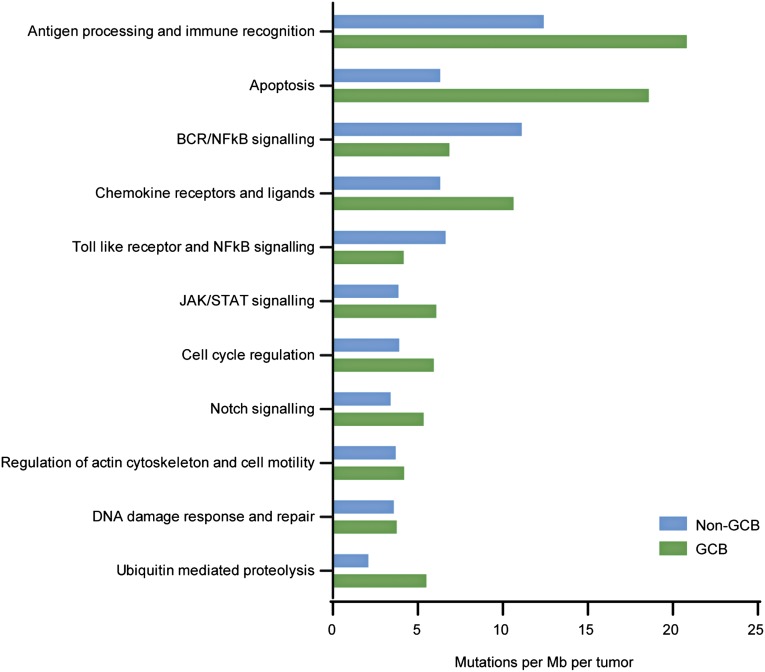
Genes affected by nonsilent mutations were assigned to canonical pathways with the aid of the Molecular Signatures Database.29 Alterations occurred frequently in genes involved in the processing of cellular antigens and immune recognition (eg, B2M, CIITA), apoptotic processes (eg, TP53, FAS), or components of B-cell receptor and NF-κB signaling (eg, MYD88, CD79B, LYN, TNFAIP3), among others (Figure 3). Interestingly, proteins involved in ubiquitination processes were previously unappreciated as mutation targets in DLBCL, but found mutated here in a high proportion of cases. They included the recurrently mutated UBE2A and the CUL4B genes. The Notch pathway was also targeted in a number of DLBCL tumors and, interestingly, DTX1 was the most frequent mutation target within this pathway. No significant differences were observed in the targeting of pathways between GCB and non-GCB DLBCL subtypes due to low sample number, although, as expected, mutations in components of B-cell receptor (BCR) and NF-κB signaling were more frequent in the non-GCB subtype.
Figure 3.
Pathways most frequently affected by somatic mutations in Chinese DLBCL. Mutation rates were calculated by dividing the total number of mutations encountered in a pathway by the size of coding regions of all genes belonging to that gene set and the number of tumors sequenced (n = 31).
Mutation validation and mutation prevalence in an expanded cohort of Chinese DLBCL
The exons of B2M, CD70, DTX1, LYN, UBE2A, and TMSB4X genes were screened by Sanger sequencing in a set of 136 DLBCL cases, including the 31 tumors that underwent whole-exome sequencing, which allowed the validation of the variants discovered by the latter method. All 50 somatic variants identified in those genes by whole-exome sequencing could be validated by Sanger sequencing (supplemental Table 4). In addition, the TNFAIP3 and TP53 genes were sequenced in 91 DLBCL samples, which allowed the validation of 8 mutations identified in these genes by exome sequencing. Finally, we performed an additional validation of the mutation detection pipeline by Sanger sequencing of regions containing 33 randomly selected mutations, of which 30 could be confirmed. Overall, 96.4% of mutations discovered by exome could be validated by Sanger sequencing.
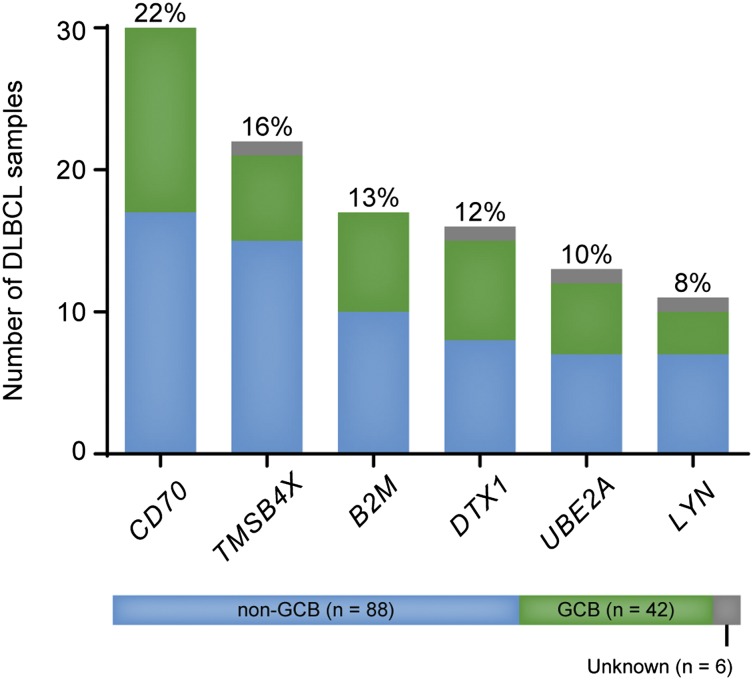
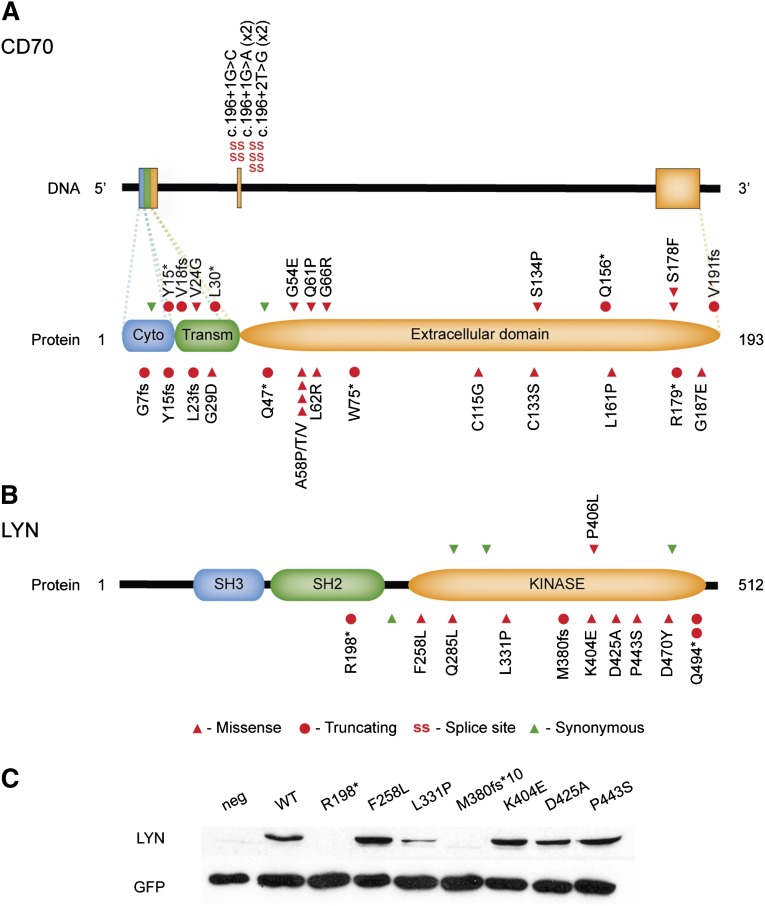
The prevalence of mutations encountered in the expanded cohort were comparable to the one detected by exome sequencing and confirmed CD70 as one of the most frequently mutated genes in Chinese DLBCL (30 of 136 tumors, 22%, Figure 4). In some of the previous NGS studies on DLBCL, CD70 has been found mutated in 7% and 10% of samples analyzed in 2 different studies (supplemental Table 3). Nearly half of mutations identified in CD70 were located at splice sites or led to the production of truncated proteins (supplemental Table 4, Figure 5A), thus indicating a deleterious effect. Furthermore, 10 tumors and their respective controls that underwent whole-exome sequencing have also been analyzed by whole-genome sequencing (L.C. and Q.P.-H., unpublished data) and loss of CD70 copy number was detected in 3 cases, including 1 tumor that carried a splice-site mutation in this gene (DL43, c.196+1G>A, supplemental Table 4). Loss of 1 copy of the CD70 locus was always accompanied by loss of genetic material throughout the whole chromosome. The prevalence of deleterious somatic mutations and loss of genetic material at the CD70 locus are supportive of a tumor suppressor role for this gene.
Figure 4.
Prevalence of nonsilent mutations in CD70, TMSB4X, B2M, DTX1, UBE2A, and LYN in an extended cohort of DLBCL patients (n = 136). CD70 was the most frequent mutation target in the present cohort. TMSB4X and LYN are novel mutation targets in DLBCL.
Figure 5.
Diagrams summarizing the mutations identified in CD70 and LYN. Mutations affecting the CD70 gene were often protein truncating or located at the donor splice site of the exon 2 (A). Mutations in LYN were most often located in the kinase domain of the protein, thus potentially affecting signal transduction through this molecule (B). Wild-type and mutant LYN constructs were transfected into U2OS cells and protein expression detected by immunoblotting. Mutations tested affected LYN’s expression to variable levels (C). neg, negative control (without transfection).
Novel mutation targets in DLBCL TMSB4X and LYN also carried nonsynonymous alterations in a considerable proportion of patients: 22 of 136 (16%) and 11 of 136 (8%), respectively (Figure 4). Of note, TMSB4X was recently described as a target of aberrant SHM in DLBCL.34 However, no details were provided on whether SHM affected the coding regions of the gene. Mutations in LYN have been reported in low frequencies in endometrial, breast, and lung malignancies (http://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=LYN#histo) but, to the best of our knowledge, have not yet been described for DLBCL. Most mutations affected LYN’s kinase domain (Figure 5B), and one-third of nonsilent mutations led to premature STOP codons. The effect of 7 mutants on protein expression was analyzed by transfecting U2OS cells with LYN wild-type and mutant constructs. The transfection efficiency was monitored by cotransfection of the GFP construct as well as measurement of RNA expression of LYN constructs by quantitative PCR (qPCR) (data not shown). The nonsense mutations (p.R198* and M380fs*10) led to abrogated protein expression whereas 1 missense mutation (p. L311P) seemed to cause a reduction on LYN expression (Figure 5C). Of note, LYN mutations were present both in GCB and non-GCB DLBCL cases (Figure 4). A Swedish cohort of 96 DLBCL samples was also screened for mutations in LYN and novel, nonsynonymous mutations were discovered in 2 cases (2%, supplemental Table 4, samples with UL ID).
No significant differences were observed between GCB and non-GCB subsets or between primary and relapse samples in the mutation frequencies of genes screened in this validation cohort, although CD70 and DTX1 mutations were found at a lower frequency in the relapse samples (supplemental Figure 3).
Analysis of DTX1 mutations
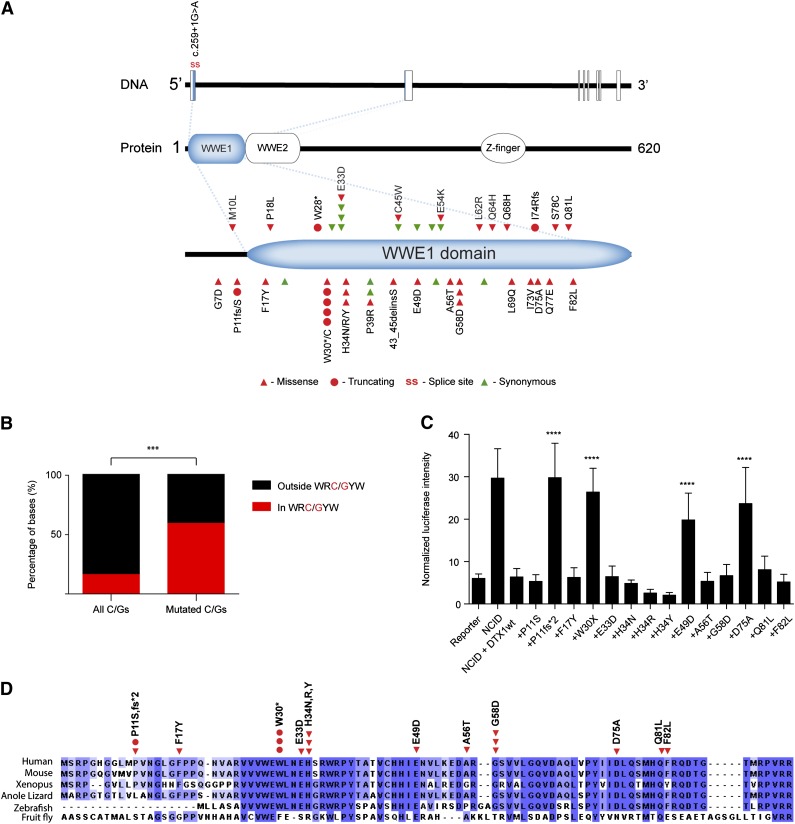
Nonsilent mutations in DTX1 were discovered in 16 of 136 (12%) Chinese DLBCL cases (Figure 4). Remarkably, all mutations identified were located in the first exon of DTX1, which encodes for the most part of an N-terminal protein-interaction domain (WWE1) (Figure 6A). In total, 43 mutations were identified in DTX1 with a nonsilent/silent ratio of 2.9. Several nucleotide positions were hotspots for the accumulation of mutations that preferentially affected cytosines and guanines (84% of all SNVs). These were preferentially located at WRC/GYW motifs (P < .001, Fisher exact test, Figure 6B), thus suggesting that DTX1 is a target for AID activity and aberrant SHM. Furthermore, all recurrently affected cytosines or guanines were located at WRC/GYW motifs. SHM targets are often involved in chromosomal rearrangements but no breakpoints could be observed at the DTX1 locus in 35 DLBCL samples analyzed by fluorescent in situ hybridization (data not shown). To investigate whether mutations in DTX1 could also be identified in an independent cohort from a different ethnic background, this gene was sequenced in 35 DLBCL tumors derived from Swedish patients. Nonsilent mutations were discovered in 3 cases (9%, supplemental Table 4, samples with UL ID).
Figure 6.
Analysis of DTX1 mutations. (A) Diagram displaying mutations identified in DTX1. Mutations were restricted to the first exon of the gene that encodes for most of the WWE1 domain of DTX1. (B) Distribution of DTX1 mutations by AID targeting hotspot. Distribution of all and mutated cytosines and guanines by WRC/GYW domains in the first exon of DTX1. In spite of the majority of cytosines and guanines being located outside WRC/GYW domains, most mutations were encountered at this AID target motif. (C) Functional characterization of DTX1 mutants. Functional characterization of mutations identified in DTX1. Cotransfection of constructs containing the NCID, DTX1 wild-type, or mutant constructs, and a CSL-responsive luciferase reporter, was performed in U2OS cells. A control pRL-TK plasmid was also cotransfected to allow internal normalization of luciferase activity. The capacity to inhibit an NCID-triggered response by DTX1 was diminished in 4 mutants (p.P11fs*2, p.W30*, p.E49D, and p.D75A). Mann-Whitney U tests were performed to compare the firefly luciferase intensity in wild-type and mutant DTX1 transfected cells (***P < .001). The means and standard deviations of 3 independent experiments are displayed, each containing 3 replicates. (D) Multispecies alignment of the N-terminal portion of DTX1, encoded by the first exon of the gene. The mutations tested in the Notch reporter assay are displayed. Multiple sequence alignment was performed with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) and visualized with Jalview.37
DTX1’s N-terminal WWE domains mediate the direct interaction of this protein with the NCID.35 DTX1 was previously shown to inhibit Notch1 activity by impeding the recruitment of transcriptional coactivators by Notch1, thereby repressing the expression of Notch target genes.36 We have characterized a selected set of DTX1 mutations (Figure 6C) by making use of a firefly luciferase gene reporter that is activated by the CSL (also known as RBPJ) transcription factor, a major target of Notch activation. U2OS cells were cotransfected with a CSL-responsive luciferase gene reporter, an intracellular domain of Notch1 (NCID1) construct, and a wild-type or mutant DTX1 constructs. Cotransfection of the wild-type form of DTX1 with NCID completely inhibited the activation of the CSL-responsive reporter (Figure 6C). Four mutants (p.P11fs*2, p.W30*, p.E49D, and p.D75A) displayed significantly decreased ability to repress the NCID-induced transcription of the reporter gene (Figure 6C). Although the p.H34 position is well conserved in vertebrates (Figure 6D37), none of the 3 discovered amino acid substitutions at this position affected the repressor activity of DTX1 in this luciferase assay. Nevertheless, the 4 mutations that displayed a functional consequence were present in 40% of the DLBCL cases that carried nonsilent mutations in DTX1. Furthermore, one would expect that additional truncating mutations (eg, p.W28*) that have not been included in the current assay would compromise DTX1’s function. Comparable RNA expression of DTX1 constructs was monitored by qPCR (data not shown).
In the whole-exome sequencing cohort NOTCH1 and NOTCH2 mutations were present in only 1 and 2 DLBCL cases, respectively. After screening the mutation hotspots of NOTCH1 and NOTCH2, mutations were discovered in 6% and 7% of samples, respectively (supplemental Table 6). DTX1 and NOTCH1/2 mutations were in general mutually exclusive except in one case, DL29, where mutations in both DTX1 and NOTCH2 were observed (supplemental Tables 4 and 6).
Discussion
NGS technologies have allowed a high-throughput, comprehensive characterization of cancer genomes at unprecedented rates. The high prevalence of mutations discovered in the H3K4-methyltransferase gene MLL2 constitutes one of the most significant findings by NGS studies and highlighted the role of epigenetic dysregulation in the progression of DLBCL.13 MEF2B is an additional gene that was found mutated in a considerable proportion of DLBCL tumors by different NGS approaches.13,20 Although reports from different NGS studies on DLBCL display some overlap, significant differences distinguish them in the mutation frequencies reported for particular genes.13,20-23 This observation underscores the heterogeneity of DLBCL as a disease entity and supports the further characterization of these tumors, particularly when derived from different populations.
In our study, we observed mutations in previously known targets in DLBCL such as B2M, CD70, MYD88, PIM1, BTG1, BTG2, TP53, and CD79B but also identified novel or previously unappreciated mutation targets such as TMSB4X, UBE2A, DTX1, CXCR4, and LYN. Of those, 11 genes had not been described as mutated in DLBCL (marked with green shadow, supplemental Table 3). On the other hand, genes such as BCL2, EZH2, MEF2B, and MLL2 were mutated at lower frequencies than previously found (supplemental Table 3). Apart from MEF2B, which presented suboptimal sequencing coverage (median <20 times), the remaining genes were adequately covered by sequencing reads (>40 times) and, thus, their low mutation frequency is unlikely to be caused by technical failures during exome enrichment and sequencing.
CD70 was the most prevalent mutation target in the current cohort of Chinese DLBCL. In normal physiological conditions, CD70 expression is restricted to a subset of activated B, T, and dendritic cells, but its expression has also been described in several cancers, including B-cell lymphomas.38,39 CD70 was previously shown to be able to transduce proliferative signals in malignant B cells, thus promoting tumor development.38 On the other hand, CD70 is believed to function as a costimulatory molecule and a ligand for the CD27 receptor, and its expression is important for the generation of T-cell immunity, including T-cell–mediated antitumor responses.39,40 In light of this, the targeting of CD70 by deleterious mutations and loss of chromosomal material at its locus (this report, Scholtysik et al,41 and Bertrand et al42) could be interpreted as a mechanism of immune escape where malignant B cells circumvent T-cell activation. The observation of CD70 defects in a large proportion of DLBCL cases could support the employment of CD27 agonists to stimulate antitumor immunity in patients,40 although unwanted autoimmune effects must be considered.
One of the most notable novel mutation targets discovered by the present study is LYN. This gene encodes a Src-family tyrosine kinase that is expressed in a variety of cell types, including B cells. LYN plays an important role in the transmission of signals from the BCR through phosphorylation of immunoreceptor tyrosine-based inhibitory/activating motifs (ITIM/ITAM) of proteins that associate in the BCR complex.43 LYN-deficient mice display hyperactivation of BCR signaling and autoimmune phenotypes, suggesting that LYN is required for the induction of B-cell tolerance and supporting its role as a negative feedback regulator of BCR signaling.44,45 LYN has been shown to phosphorylate the ITIM domains of the CD22 inhibitory receptor, among others, leading to the recruitment of phosphatases (eg, SHP-1, SHIP-1) and suppression of BCR signaling.46,47 The accumulation of deleterious mutations in LYN could constitute an additional mechanism by which DLBCLs augment BCR signaling by avoiding negative feedback signals resulting from LYN’s overactivation. The functional elucidation of the mutations discovered here is of utmost importance as an additional subgroup of DLBCL patients could benefit from therapeutic interventions that target the chronically activated BCR signaling.48
DTX1 was affected by nonsilent mutations in 12% of tumors analyzed, both in the discovery and expanded cohorts. To our knowledge, this is the first report describing DTX1 as the most frequently mutated member of the Notch signaling pathway. Previously, mutations in DTX1 were reported at lower frequencies in DLBCL (4%-6%) and splenic marginal zone lymphomas (SMZLs) (2%).21,23,49 Additionally, the DTX1 locus was recently proposed to constitute a target of aberrant SHM in DLBCL, but no protein-coding alterations were reported.34 In addition to being a transcriptional target of Notch,50 DTX1 was also demonstrated to negatively regulate Notch activity at the protein level, thus constituting a potential inhibitory feedback mechanism of the Notch pathway.36 Notch signaling is an evolutionary conserved pathway that exerts distinct effects on cellular development in particular contexts, being able to provide both survival and growth arrest signals to cells.51 Oncogenic activation of the Notch signaling pathway is a hallmark of T-cell acute lymphoblastic leukemia, mainly through the establishment of gain-of-function mutations in the Notch1 receptor.52 This mechanism of Notch activation is also observed in subsets of chronic lymphocytic leukemia, mantle cell lymphomas, and DLBCL.52,53 Notch 2 gain-of-function mutations were recently shown to represent one of the most frequent genetic alterations in SMZL and have also been described in a minority of DLBCL.49,52 In this work, DTX1 truncating mutations that abolished protein function or amino acid substitutions that impaired DTX1’s inhibitory role in Notch signaling were discovered in several tumors. Furthermore, DTX1 mutations that do not show an effect in the Notch1-mediated transcriptional activation of target genes might still be relevant in contexts of noncanonical Notch signaling.54 We propose that the accumulation of deleterious mutations in DTX1, which were in general mutually exclusive with NOTCH1 and NOTCH2 mutations, constitute an additional mechanism by which B cells may acquire oncogenic Notch signaling in DLBCLs. The discovery that additional Notch signaling molecules are mutated in a relevant proportion of DLBCL might support the development of therapeutic strategies that target this pathway in DLBCL patients.
Here we present the first analysis of the coding genome of a Chinese DLBCL cohort. We report novel mutation targets and genes that might be affected at different frequencies than previously reported for Western cohorts. Such differences could not be explained by the inclusion of a few relapse DLBCL cases in the discovery cohort as the overall mutation frequency and mutation spectrum were not significantly different between primary and relapse samples. Furthermore, the mutation frequencies of specific genes of interest in the expanded cohort of 136 DLBCLs were not significantly different between the 2 groups. Our discovery cohort was mainly composed of non-GCB DLBCL cases (68%), which is in line with previous observations that the GCB subtype is less frequent in Asian patients.55,56 Although different prevalence of disease subgroups might partially explain discrepancies between studies, several mutation targets in DLBCL, including the genes studied here, affect GCB and non-GCB subsets at comparable frequencies. A different spectrum of somatic mutations might also be caused by the distinct genetic backgrounds of Asian and Western populations, as germline variations have been shown to influence the somatic alterations that occur in tumors.57 Moreover, exposure to certain etiological agents such as infectious diseases, including oncogenic viruses, or dietary habits could result on the usage of different oncogenic pathways. Indeed, 45% of the Chinese patients screened by exome sequencing presented some types of chronic infection at diagnosis including hepatitis B virus (23%), Epstein-Barr virus (10%), Helicobacter pylori (10%), and tuberculosis (10%), most of which have been shown to be highly prevalent in Chinese populations.58-60 Finally, it is important to note that different technical platforms and analysis pipelines used may lead to variable results and that previously published NGS studies as well as our own have limited power to detect those cancer driver genes that are mutated in <10% to 20% of samples, as the total number of screened DLBCL samples remains low.61 Intertumor heterogeneity carries important implications for the management of DLBCL patients, as targeted therapies are aimed at exploring specific genetic alterations in tumors. Our observations support the need for characterization of a greater number of DLBCLs derived from different populations and the refinement of disease subgroups based on the molecular defects discovered by the NGS studies.
Acknowledgments
The authors are grateful to Profs J. Aster and S. Blacklow (Harvard Medical School) for insightful discussions and assistance in Notch pathway analysis.
This work was supported by the Swedish Cancer Society, the Swedish Research Council, the European Research Council (242551-ImmunoSwitch), and the National Natural Science Foundation of China (81302045).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: N.F.C.C.d.M. collected, analyzed, and interpreted data and wrote the manuscript; L.C. performed bioinformatics analysis; K.G. performed Sanger sequencing and functional studies; C.W. performed Sanger sequencing and was involved in sample preparation; Z.G. supervised bioinformatics analysis; A.Z. was involved in LYN functional studies; S.L. performed cytogenetic analysis; G.E. collected and curated clinical information on DLBCL patients; M.R.T. supervised cytogenetic analysis; Y.Z. was involved in design and supervision of the study; R.P. was involved in sample preparation, collection of clinical information on DLBCL patients, and design of the study; and Q.P.-H. designed and supervised the study, and was involved in analysis and interpretation of data and writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Qiang Pan-Hammarström, Department of Laboratory Medicine, Karolinska Institutet at Karolinska University Hospital Huddinge, SE-14186, Stockholm, Sweden; e-mail: qiang.pan-hammarstrom@ki.se; and Roujun Peng, Department of Medical Oncology, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center of Cancer Medicine, 510060, Guangzhou, Guangdong, China; e-mail: pengrj@sysucc.org.cn.
References
- 1.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC Press; 2008. [Google Scholar]
- 2.Gurbuxani S, Anastasi J, Hyjek E. Diffuse large B-cell lymphoma—more than a diffuse collection of large B cells: an entity in search of a meaningful classification [published correction appears in Arch Pathol Lab Med. 2009;133(8):1186]. Arch Pathol Lab Med. 2009;133(7):1121–1134. doi: 10.5858/133.7.1121. [DOI] [PubMed] [Google Scholar]
- 3.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 4.Rosenwald A, Wright G, Chan WC, et al. Lymphoma/Leukemia Molecular Profiling Project. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(25):1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 5.Iqbal J, Greiner TC, Patel K, et al. Leukemia/Lymphoma Molecular Profiling Project. Distinctive patterns of BCL6 molecular alterations and their functional consequences in different subgroups of diffuse large B-cell lymphoma. Leukemia. 2007;21(11):2332–2343. doi: 10.1038/sj.leu.2404856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lenz G, Davis RE, Ngo VN, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319(5870):1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 8.Davis RE, Ngo VN, Lenz G, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ngo VN, Young RM, Schmitz R, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470(7332):115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Compagno M, Lim WK, Grunn A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459(7247):717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasqualucci L, Compagno M, Houldsworth J, et al. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med. 2006;203(2):311–317. doi: 10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasqualucci L, Dominguez-Sola D, Chiarenza A, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471(7337):189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Challa-Malladi M, Lieu YK, Califano O, et al. Combined genetic inactivation of β2-microglobulin and CD58 reveals frequent escape from immune recognition in diffuse large B cell lymphoma. Cancer Cell. 2011;20(6):728–740. doi: 10.1016/j.ccr.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8(1):22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 16.Pasqualucci L, Bhagat G, Jankovic M, et al. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40(1):108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- 17.Pasqualucci L, Neumeister P, Goossens T, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412(6844):341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 18.Küppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20(40):5580–5594. doi: 10.1038/sj.onc.1204640. [DOI] [PubMed] [Google Scholar]
- 19.de Miranda NF, Peng R, Georgiou K, et al. DNA repair genes are selectively mutated in diffuse large B cell lymphomas. J Exp Med. 2013;210(9):1729–1742. doi: 10.1084/jem.20122842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lohr JG, Stojanov P, Lawrence MS, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA. 2012;109(10):3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morin RD, Mungall K, Pleasance E, et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013;122(7):1256–1265. doi: 10.1182/blood-2013-02-483727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Grubor V, Love CL, et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci USA. 2013;110(4):1398–1403. doi: 10.1073/pnas.1205299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koboldt DC, Chen K, Wylie T, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25(17):2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li R, Li Y, Fang X, et al. SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009;19(6):1124–1132. doi: 10.1101/gr.088013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh JJ, Nofziger DE, Weinmaster G, Hayward SD. Epstein-Barr virus immortalization: Notch2 interacts with CBF1 and blocks differentiation. J Virol. 1997;71(3):1938–1945. doi: 10.1128/jvi.71.3.1938-1945.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiang DY, Getz G, Jaffe DB, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6(1):99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kan Z, Jaiswal BS, Stinson J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466(7308):869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 34.Khodabakhshi AH, Morin RD, Fejes AP, et al. Recurrent targets of aberrant somatic hypermutation in lymphoma. Oncotarget. 2012;3(11):1308–1319. doi: 10.18632/oncotarget.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuno K, Eastman D, Mitsiades T, et al. Human deltex is a conserved regulator of Notch signalling. Nat Genet. 1998;19(1):74–78. doi: 10.1038/ng0598-74. [DOI] [PubMed] [Google Scholar]
- 36.Izon DJ, Aster JC, He Y, et al. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity. 2002;16(2):231–243. doi: 10.1016/s1074-7613(02)00271-6. [DOI] [PubMed] [Google Scholar]
- 37.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lens SM, Drillenburg P, den Drijver BF, et al. Aberrant expression and reverse signalling of CD70 on malignant B cells. Br J Haematol. 1999;106(2):491–503. doi: 10.1046/j.1365-2141.1999.01573.x. [DOI] [PubMed] [Google Scholar]
- 39.Nolte MA, van Olffen RW, van Gisbergen KP, van Lier RA. Timing and tuning of CD27-CD70 interactions: the impact of signal strength in setting the balance between adaptive responses and immunopathology. Immunol Rev. 2009;229(1):216–231. doi: 10.1111/j.1600-065X.2009.00774.x. [DOI] [PubMed] [Google Scholar]
- 40.He LZ, Prostak N, Thomas LJ, et al. Agonist anti-human CD27 monoclonal antibody induces T cell activation and tumor immunity in human CD27-transgenic mice. J Immunol. 2013;191(8):4174–4183. doi: 10.4049/jimmunol.1300409. [DOI] [PubMed] [Google Scholar]
- 41.Scholtysik R, Nagel I, Kreuz M, et al. Recurrent deletions of the TNFSF7 and TNFSF9 genes in 19p13.3 in diffuse large B-cell and Burkitt lymphomas. Int J Cancer. 2012;131(5):E830–E835. doi: 10.1002/ijc.27416. [DOI] [PubMed] [Google Scholar]
- 42.Bertrand P, Maingonnat C, Penther D, et al. The costimulatory molecule CD70 is regulated by distinct molecular mechanisms and is associated with overall survival in diffuse large B-cell lymphoma. Genes Chromosomes Cancer. 2013;52(8):764–774. doi: 10.1002/gcc.22072. [DOI] [PubMed] [Google Scholar]
- 43.Scapini P, Pereira S, Zhang H, Lowell CA. Multiple roles of Lyn kinase in myeloid cell signaling and function. Immunol Rev. 2009;228(1):23–40. doi: 10.1111/j.1600-065X.2008.00758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan VW, Meng F, Soriano P, DeFranco AL, Lowell CA. Characterization of the B lymphocyte populations in Lyn-deficient mice and the role of Lyn in signal initiation and down-regulation. Immunity. 1997;7(1):69–81. doi: 10.1016/s1074-7613(00)80511-7. [DOI] [PubMed] [Google Scholar]
- 45.Harder KW, Parsons LM, Armes J, et al. Gain- and loss-of-function Lyn mutant mice define a critical inhibitory role for Lyn in the myeloid lineage. Immunity. 2001;15(4):603–615. doi: 10.1016/s1074-7613(01)00208-4. [DOI] [PubMed] [Google Scholar]
- 46.Gross AJ, Lyandres JR, Panigrahi AK, Prak ET, DeFranco AL. Developmental acquisition of the Lyn-CD22-SHP-1 inhibitory pathway promotes B cell tolerance. J Immunol. 2009;182(9):5382–5392. doi: 10.4049/jimmunol.0803941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan VW, Lowell CA, DeFranco AL. Defective negative regulation of antigen receptor signaling in Lyn-deficient B lymphocytes. Curr Biol. 1998;8(10):545–553. doi: 10.1016/s0960-9822(98)70223-4. [DOI] [PubMed] [Google Scholar]
- 48.Young RM, Staudt LM. Targeting pathological B cell receptor signalling in lymphoid malignancies. Nat Rev Drug Discov. 2013;12(3):229–243. doi: 10.1038/nrd3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rossi D, Trifonov V, Fangazio M, et al. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med. 2012;209(9):1537–1551. doi: 10.1084/jem.20120904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deftos ML, He YW, Ojala EW, Bevan MJ. Correlating notch signaling with thymocyte maturation. Immunity. 1998;9(6):777–786. doi: 10.1016/s1074-7613(00)80643-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Radtke F, MacDonald HR, Tacchini-Cottier F. Regulation of innate and adaptive immunity by Notch. Nat Rev Immunol. 2013;13(6):427–437. doi: 10.1038/nri3445. [DOI] [PubMed] [Google Scholar]
- 52.South AP, Cho RJ, Aster JC. The double-edged sword of Notch signaling in cancer. Semin Cell Dev Biol. 2012;23(4):458–464. doi: 10.1016/j.semcdb.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fabbri G, Khiabanian H, Holmes AB, et al. Genetic lesions associated with chronic lymphocytic leukemia transformation to Richter syndrome. J Exp Med. 2013;210(11):2273–2288. doi: 10.1084/jem.20131448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huber RM, Rajski M, Sivasankaran B, Moncayo G, Hemmings BA, Merlo A. Deltex-1 activates mitotic signaling and proliferation and increases the clonogenic and invasive potential of U373 and LN18 glioblastoma cells and correlates with patient survival. PLoS ONE. 2013;8(2):e57793. doi: 10.1371/journal.pone.0057793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Y, Dave BJ, Zhu X, et al. Differences in the cytogenetic alteration profiles of diffuse large B-cell lymphoma among Chinese and American patients. Cancer Genet. 2013;206(5):183–190. doi: 10.1016/j.cancergen.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 56.Shiozawa E, Yamochi-Onizuka T, Takimoto M, Ota H. The GCB subtype of diffuse large B-cell lymphoma is less frequent in Asian countries. Leuk Res. 2007;31(11):1579–1583. doi: 10.1016/j.leukres.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 57.Dworkin AM, Ridd K, Bautista D, et al. Germline variation controls the architecture of somatic alterations in tumors. PLoS Genet. 2010;6(9):e1001136. doi: 10.1371/journal.pgen.1001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Custer B, Sullivan SD, Hazlet TK, Iloeje U, Veenstra DL, Kowdley KV. Global epidemiology of hepatitis B virus. J Clin Gastroenterol. 2004;38(10 suppl 3):S158–S168. doi: 10.1097/00004836-200411003-00008. [DOI] [PubMed] [Google Scholar]
- 59.Zhang L, Blot WJ, You WC, et al. Helicobacter pylori antibodies in relation to precancerous gastric lesions in a high-risk Chinese population. Cancer Epidemiol Biomarkers Prev. 1996;5(8):627–630. [PubMed] [Google Scholar]
- 60.Zhao Y, Xu S, Wang L, et al. National survey of drug-resistant tuberculosis in China. N Engl J Med. 2012;366(23):2161–2170. doi: 10.1056/NEJMoa1108789. [DOI] [PubMed] [Google Scholar]
- 61.Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]