Abstract
The distribution of postmortem damage in mitochondrial DNA retrieved from 37 ancient human DNA samples was analyzed by cloning and was compared with a selection of published animal data. A relative rate of damage (ρv) was calculated for nucleotide positions within the human hypervariable region 1 (HVR1) and cytochrome oxidase subunit III genes. A comparison of damaged sites within and between the regions reveals that damage hotspots exist and that, in the HVR1, these correlate with sites known to have high in vivo mutation rates. Conversely, HVR1 subregions with known structural function, such as MT5, have lower in vivo mutation rates and lower postmortem-damage rates. The postmortem data also identify a possible functional subregion of the HVR1, termed “low-diversity 1,” through the lack of sequence damage. The amount of postmortem damage observed in mitochondrial coding regions was significantly lower than in the HVR1, and, although hotspots were noted, these did not correlate with codon position. Finally, a simple method for the identification of incorrect archaeological haplogroup designations is introduced, on the basis of the observed spectrum of postmortem damage.
Introduction
The extreme susceptibility that research on human ancient DNA (aDNA) has to contamination with modern human DNA has caused skepticism about the authenticity of many results (Richards et al. 1995; Stoneking 1995; Handt et al. 1996; Cooper 1997; Kolman and Tuross 2000; Cooper et al. 2001b). Several studies have shown that, despite rigorous protocols (e.g., see Cooper and Poinar 2000), contaminants can still be identified in amplified products (Richards et al. 1995; Handt et al. 1996; Krings et al. 1997; Kolman and Tuross 2000; Hofreiter et al. 2001). Contaminants are usually identified when repeat amplifications yield differing, or multiple, sequences in cloned products. However, the effect that postmortem damage has on endogenous DNA is often underrated. The majority of postmortem DNA damage occurs as double-strand breaks and oxidative dinucleotide modification, both of which prevent subsequent enzymatic replication (Pääbo 1989; Lindahl 1993; Höss et al. 1996). However, minor sequence modifications, such as hydrolytic deamination and depurination, permit polymerase action and are manifested as limited amounts of base variation among sequenced clones (Krings et al. 1997). Few studies have examined the prevalence of such postmortem damage in detail (Handt et al. 1996; Kolman and Tuross 2000; Hansen et al. 2001; Hofreiter et al. 2001), and it is generally considered that, when the initial template number is >1,000 copies, postmortem-damage rates are unlikely to bias results (Handt et al. 1996; Krings et al. 1997) However, when few DNA templates initiate a PCR, the resulting sequences are likely to contain artifacts.
mtDNA is generally present in archaeological samples in much greater amounts than nuclear DNA is (Greenwood et al. 1999), and, consequently, most human ancient genetic analyses have used the well-characterized hypervariable region 1 (HVR1) (e.g., see Oota et al. 1995, 1999; Ribeiro-dos-Santos et al. 1996; Torroni et al. 1998; Lalueza-Fox et al. 2001). Archaeological HVR1 results can be compared with detailed records of base substitutions within modern populations, and sequences can be located within the commonly used human phylogenetic tree, relative to the Cambridge reference sequence (CRS) (Anderson et al. 1981). Unfortunately, a major limitation of this classification system is that certain haplogroups, especially those of Eurasian origin, are routinely categorized using fewer than five site changes—and often as few as one. If these key sites are commonly modified by postmortem damage, then haplogroup designations may be incorrect. In one of the only studies of this issue, Hofreiter et al. (2001) suggested that postmortem damage appeared to be randomly distributed in the control region of the cave bear (Ursus spelaeus). However, only a small number of samples were studied, and it is also possible that postmortem damage may vary according to environmental or taxonomic factors.
Within modern human populations, certain nucleotide positions within the HVR1, termed “sites,” appear to mutate at significantly higher rates than others, and these sites have been termed “hotspots” (Hasegawa and Horai 1991; Vigilant et al. 1991; Hasegawa et al. 1993; Wakely 1993; Aris-Brisou and Excoffier 1996; Macauley et al. 1997; Excoffier and Yang 1999; Meyer et al. 1999; Stoneking 2000; Finnilä et al. 2001; Heyer et al. 2001). It has been suggested that the variation in rates of damage is related to DNA secondary and tertiary structure (Heyer et al. 2001), and it is possible that the same sites might also be susceptible to postmortem damage. To investigate these issues, we amplified and cloned human HVR1 and cytochrome oxidase subunit III (COIII) mtDNA from a range of archaeological specimens, and we compared the human sequences to aDNA sequences from brown bear (U. arctos), domestic cow (Bos taurus), pig (Sus scrofa), goat (Capra hircus), and sheep (Ovis aries), as well as to published data from other taxa.
Material and Methods
Samples
It is exceedingly difficult to authenticate human aDNA sequences, and it may even be impossible in certain cases (Cooper 1997). Several authors have demonstrated how even stringent controls can fail to prevent or detect contamination (Handt et al. 1996; Kolman and Tuross 2000). Consequently, careful attention was paid to the choice of methods and samples, to provide the best chance of limiting contamination. Forty-three whole teeth were obtained from 34 archaeological human skeletons preserved in a range of temporal, geographical, and environmental situations (table 1). Teeth were used as a DNA source both because the relatively impervious outer enamel layer provides a degree of protection from contaminating DNA sources (M.T.P.G., unpublished data) and because teeth have been shown to yield higher amounts of DNA than bone does in many environments (Kurosaki et al. 1993; Oota et al. 1995). Teeth from cow, pig, goat, and sheep specimens obtained from the Greenland archaeological site were also analyzed for human DNA as a contamination control. Multiple teeth were taken from five specimens, to allow replication of the extraction and amplification procedures. DNA from one of these five specimens was extracted and amplified independently at the Ancient DNA Laboratory (Zoological Institute, Copenhagen) by using similar techniques, to confirm that the spectrum of sequence damage observed was replicable (tooth EA). Two brown bear 12s mtDNA sequences were also analyzed, to allow taxonomic comparisons of sequence damage outside the HVR1. Both bear sequences have previously been replicated at the University of California–Los Angeles and Oxford University (Barnes et al. 2002).
Table 1.
Details of Samples Studied
| HVR1 |
COIIIb |
|||||||||
| Species and Sample | Sitea | Sample Date | Ampsc | Clonesd | UNGe | UNG Clonesf | Ampsc | Clonesd | UNGe | UNG Clonesf |
| Homo sapiens: | ||||||||||
| Tg44 | Greenland | 1000–1200 a.d.g | 1 | 2 | 1 | 8 | ||||
| Tg54 | Greenland | 1000–1200 a.d.g | 1 | 5 | 1 | 8 | ||||
| Tg63h | Britain | Not known | 1 | 32 | 1 | 12 | ||||
| Tg76 | Denmark | 1711 a.d.i | 1 | 5 | 1 | 11 | 1 | 12 | 1 | 12 |
| Tg77 (a)j | Denmark | 1400–1500 a.d.k | 3 | 9 | 1 | 12 | ||||
| Tg80 | Denmark | 1300–1600 a.d.l | 1 | 7 | 1 | 11 | 1 | 10 | 1 | 9 |
| Tg85 | Denmark | 1711 a.d.i | 1 | 5 | 1 | 12 | ||||
| Tg99.1 (e)h,j | Greenland | 1000–1200 a.d.g | 9 | 60 | 1 | 20 | ||||
| Tg99.2 (e)j | Greenland | 1000–1200 a.d.g | 1 | 16 | ||||||
| Tg99.3 (e)j | Greenland | 1000–1200 a.d.g | 1 | 16 | ||||||
| Tg103 | Denmark | 1300–1600 a.d.l | 1 | 4 | 1 | 11 | ||||
| Tg104 | Britain | Not known | 1 | 15 | 1 | 16 | ||||
| Tg105 | Denmark | 400–1000 a.d.l | 1 | 4 | 1 | 6 | ||||
| Tg112 | Greenland | 1000–1200 a.d.g | 1 | 6 | 1 | 7 | ||||
| Tg114 | Denmark | 1711 a.d.i | 1 | 5 | 1 | 12 | 1 | 11 | 1 | 12 |
| Tg116 | Denmark | 700–1000 a.d.l | 2 | 10 | 1 | 12 | ||||
| Tg120 | Denmark | 1711 a.d.i | 1 | 2 | 1 | 8 | ||||
| Tg123 | Denmark | 1400–1500 a.d.k | 1 | 6 | 1 | 8 | ||||
| Tg127 (a)j | Denmark | 1711 a.d.i | 1 | 8 | 1 | 11 | ||||
| Tg128 | Greenland | 1000–1200 a.d.g | 2 | 12 | ||||||
| Tg129 | Greenland | 1000–1200 a.d.g | 2 | 9 | ||||||
| Tg131 | Greenland | 1000–1200 a.d.g | 1 | 7 | 1 | 7 | ||||
| Tg133 | Greenland | 1000–1200 a.d.g | 1 | 2 | 1 | 8 | ||||
| Tg136.1 | Britain | 1172–1536 a.d. | 2 | 17 | 1 | 11 | 1 | 12 | ||
| Tg136.2 | Britain | 1172–1536 a.d. | 2 | 26 | ||||||
| Tg137.1 (c)j | Britain | 675–740 a.d. | 3 | 27 | ||||||
| Tg137.2 (c)j | Britain | 675–740 a.d. | 3 | 38 | ||||||
| Tg138 (b)j | Britain | 873–874 a.d. | 1 | 8 | 1 | 5 | ||||
| Tg141 | Britain | 873–874 a.d. | 1 | 6 | 1 | 6 | ||||
| Tg142 | Britain | 873–874 a.d. | 1 | 7 | 1 | 9 | ||||
| Tg143 | Britain | 874–876 a.d. | 1 | 6 | 1 | 8 | ||||
| Tg145 | Britain | 750–873 a.d. | 1 | 5 | 1 | 7 | ||||
| Tg146 | Britain | 900–1100 a.d. | 1 | 6 | 1 | 8 | ||||
| Tg147h | Britain | 873–874 a.d. | 2 | 9 | ||||||
| Tg148 (c)j | Britain | 675–740 a.d. | 7 | 80 | 1 | 21 | 3 | 33 | 1 | 12 |
| Tg149 (b)j | Britain | 873–874 a.d. | 7 | 57 | 1 | 12 | 2 | 21 | 1 | 10 |
| Tg191h | Britain | Not known | 2 | 8 | ||||||
| Tg192 | Britain | Not known | 2 | 10 | 1 | 11 | 1 | 12 | 1 | 12 |
| Tg196 | Not known | Not known | 2 | 12 | ||||||
| Tg232 (d)j | Britain | Not known | 1 | 8 | 1 | 8 | ||||
| Tg233 (d)j | Britain | Not known | 2 | 14 | ||||||
| Tg200 (e)h,j | Greenland | 1000–1200 a.d.g | 1 | 15 | 2 | 19 | ||||
| EA (e)j | Greenland | 1000–1200 a.d.g | 1 | |||||||
| U. arctos: | ||||||||||
| IB221 | Alaska | Not known | 1 | 12 | ||||||
| IB223 | Alaska | Not known | 1 | 12 | ||||||
| Cow (B. taurus) | Greenland | 1000–1200 a.d.g | 1m | |||||||
| Sheep (O. aries) | Greenland | 1000–1200 a.d.g | 1m | |||||||
| Pig (S. scrofa) | Greenland | 1000–1200 a.d.g | 1m | |||||||
| Goat (C. hircus) | Greenland | 1000–1200 a.d.g | 1m |
|
|
|
|
|
|
|
| Total | 83 | 606 | 31 | 314 | 12 | 134 | 7 | 79 | ||
Greenland samples have been identified as Viking (Lynnerup 1998).
The 12s gene was amplified in bear samples.
Number of untreated (i.e., no UNG) PCRs per sample.
Total number of non-UNG clones sequenced per sample.
Number of PCRs with UNG treatment per sample.
Total number of UNG clones sequenced per sample.
Dated on the basis of radiocarbon and archaeological analysis (Arneborg et al. 1999).
Sample rejected owing to signs of contamination.
Dated on the basis of historical records (Copenhagen City Museum, Copenhagen, Denmark).
Multiple teeth originating from one of five skeletons (labeled, in parentheses, as “a”–“e”).
Dated on the basis of archaeological analysis (Aalborg Museum, Aalborg, Denmark).
Dated on the basis of archaeological analysis (Panum Institute, Copenhagen, Denmark).
No PCR product.
Extraction
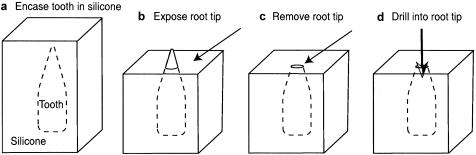
All DNA-preparation and -extraction methods followed strict aDNA-specific requirements (Cooper and Poinar 2000) and were performed in a dedicated aDNA facility, in a building physically isolated from any molecular biology research. Full-body suits, breathing masks, and face shields were used, and gloves were frequently changed. DNA was extracted from teeth by using a new technique that significantly reduces external contamination (fig. 1) (M.T.P.G., unpublished data). Whole teeth were initially washed in 50% bleach for 5 min, followed by exposure to 254-nm-wavelength UV light for 10 min on either side. Teeth were fully encased, upside down, in RTV-11 liquid silicone rubber (Tiranti) and were left overnight while the matrix hardened. The silicone that encases the top 5–10 mm of the root was removed with a horizontal cut, and the root was removed flush with the silicon by using a single-use carborundum cutting disk. Dental pulp was powdered and removed from the pulp chamber by using a dental drill bit and was digested and extracted using a phenol:chloroform DNA-extraction protocol (after Barnes et al. 2002), with 0.2 M PTB (N-phenacylthiazolium bromide) (Poinar et al. 1998). PTB has been shown to increase DNA yield in aDNA extractions, putatively by cleaving cross-linked proteins and DNA formed by condensation reactions (Vasan et al. 1996; Poinar et al. 1998; Bada 1999). The bear DNA extractions were as described elsewhere (Barnes et al. 2002), except with the addition of 0.001 M PTB.
Figure 1.
Powder-extraction method. a, Encasement of tooth, crown down, in liquid silicone, which is left to set. b, Exposure of root tip (5–10 mm) by using horizontal cuts into the silicone. c, Horizontal removal of root tip, flush against silicone surface, with sterile cutting disk. d, Extraction of sterile pulp-cavity powder by drilling into root from cut surface.
PCR Amplification
PCR amplifications used a high-fidelity polymerase (Platinum Taq Hi-Fidelity; Invitrogen) (after Cooper et al. 2001a) to reduce the polymerase-error rate and to increase amplification efficiency (Willerslev et al. 1999; Hansen et al. 2001; Gilbert et al. 2003 [in this issue]). Full details of the mitochondrial regions amplified are given in table 1, and primers and annealing temperatures are given in table 2. All samples were amplified and extensively cloned at least twice. PCR products were purified, by precipitation, using Microclean (Microzone) and were cloned using the Topo TA cloning system (Invitrogen). Colonies were used to initiate PCR reamplifications with vector M13R and T7 primers, were purified as before, and were sequenced on an ABI 377 or 3700 by using the ABI Big Dye 3 PRISM kit. Blank control extractions were performed at a ratio of 1:5 samples, and negative control amplifications were performed at a ratio of 1:1. No positive controls were used at any step of the amplification process.
Table 2.
Details of Primer Used[Note]
| Primer | Sequence | AnnealingTemperature(°C) |
| L09486 | 5′ ttc gca gga ttt ttc tga gc | 56 |
| H09652 | 5′ tgg tga gct cag gtg att ga | 56 |
| 12se | Barnes et al. 2002 | |
| 12sf3 | Barnes et al. 2002 | |
| L16055 | Handt et al. 1996 | |
| H16410 | Handt et al. 1996 | |
| L16209 | Handt et al. 1996 | |
| H16356 | Handt et al. 1996 |
Note.— Conditions for previously reported primers were identical to the references.
Uracil-N-Glycosylase (UNG) Treatment
A 5-μl aliquot of each human DNA extract was treated with 1 U Escherichia coli UNG (Sigma), to excise uracil caused by hydrolytic deamination of cytosine (Dinner et al. 2001). UNG reduces sequence artifacts caused by this common form of postmortem damage, resulting in an apparent C→T/G→A mutation (Pääbo 1989; Hofreiter et al. 2001). After UNG treatment, extracts containing multiple sources of DNA can be identified as those in which cloned sequences still either share no consensus sequence or retain sporadic C→T/G→A base changes that previously may have been attributed to postmortem damage but that now may be recognized as authentic base differences between a contaminant and the sample’s endogenous sequence.
Damage Spectrum in Ancient Specimens
To examine the robustness of the results, we examined samples from a range of sites and time periods (table 1). Previously published cloned aDNA sequences of cave bear (Loreille et al. 2001) and brown bear (Barnes et al. 2002) were obtained (see table 3). Other reported human aDNA clones cannot be included in the present analysis because of an inability to determine the extent of contamination in those studies.
Table 3.
Haplogroups Ranked by Misidentification Potential in Using a Predominantly Haplogroup H and Haplogroup V Data Set[Note]
| Haplogroup and Site | Site Ratea | Lowest Rateb |
| U5: | 15.4 | |
| 270 | 15.4 | |
| B: | 12.5† | |
| 189 | 12.5† | |
| J: | 12.5† | |
| 126 | 12.5† | |
| 69 | 12.5† | |
| JT*: | 12.5† | |
| 126 | 12.5† | |
| D: | 10.3† | |
| 362 | 12.5† | |
| 223 | 10.3† | |
| G: | 10.3 | |
| 223 | 10.3 | |
| L3a*: | 10.3 | |
| 223 | 10.3 | |
| M*: | 10.3 | |
| 223 | 10.3 | |
| N*: | 10.3 | |
| 223 | 10.3 | |
| E: | 10.3 | |
| 223 | 10.3 | |
| A: | 6.4 | |
| 319 | 6.4 | |
| 290 | 7.7 | |
| 223 | 10.3 | |
| C: | 6.4 | |
| 327 | 7.7 | |
| 298 | 6.4 | |
| 223 | 10.3 | |
| Pre-C: | 6.4 | |
| 298 | 6.4 | |
| 223 | 10.3 | |
| T: | 6.4 | |
| 294 | 6.4 | |
| 126 | 12.5† | |
| V: | 6.4 | |
| 298 | 6.4 | |
| X: | 6.4 | |
| 278 | 6.4 | |
| 223 | 10.3 | |
| F: | 3.8 | |
| 304 | 3.8 | |
| M1: | 2.6 | |
| 311 | 6.4 | |
| 249 | 2.6 | |
| 223 | 10.3 | |
| 189 | 12.5† | |
| 129 | 12.5† | |
| U1: | 2.6 | |
| 249 | 2.6 | |
| 189 | 12.5† | |
| W: | 2.6 | |
| 292 | 2.6 | |
| 223 | 10.3 | |
| K: | 2.6 | |
| 311 | 6.4 | |
| 224 | 2.6 | |
| L3a1: | 1.3 | |
| 320 | 1.3 | |
| 223 | 10.3 | |
| I: | .0† | |
| 391 | .0† | |
| 223 | 10.3 | |
| 129 | 12.5† | |
| (continued) | ||
| L3a2: | .0† | |
| 223 | 10.3 | |
| 209 | .0† | |
| M2: | .0 | |
| 343 | .0 | |
| 311 | 6.4 | |
| 265 | 1.3 | |
| 241 | 3.8 | |
| 223 | 10.3 | |
| 148 | .0† | |
| 144 | 37.5† | |
| 129 | 12.5† | |
| N1: | .0 | |
| 223 | 10.3 | |
| 176 | .0† | |
| 145 | .0† | |
| N2: | .0 | |
| 248 | 7.7 | |
| 223 | 10.3 | |
| 147 | .0† | |
| N3: | .0 | |
| 261 | .0 | |
| 257 | 2.6 | |
| 223 | 10.3 | |
| U3: | .0 | |
| 343 | .0 | |
| U4: | .0 | |
| 356 | .0 | |
| U6: | .0† | |
| 219 | 1.3 | |
| 172 | .0† | |
Note.— Rates with a dagger (†) following are estimates on the basis of the OR, and rates without are estimates on the basis of the MR. For full details, see the “Haplogroup Analysis” subsection.
Relative rate per site.
Haplogroup minimum value.
Contamination
Contaminating DNA is a major concern, because it can imitate sequence damage and also permits jumping PCR between endogenous and contaminant strands (Pääbo et al. 1990). Jumping events will increase the apparent number of damaged sites in cloned sequences by introducing positions that differ between the contaminant and authentic DNA. Low-level contamination often is neither observed nor reported, but this insidious problem would be manifested in clone sequences as sporadic, apparently postmortem damage at phylogenetically variable sites. So that this problem may be avoided, we analyzed specimens representing a variety of mitochondrial haplogroups, to detect common contaminants, and used at least two separate PCR amplifications and UNG treatment (which involves a further PCR replication), to confirm that extracts contain only one source of ancient sequence. It could be argued that an old—and therefore damaged—contaminant could generate a solitary consensus sequence from a tooth and that UNG treatment would confirm that the observed sequence heterogeneity had arisen from postmortem damage (as opposed to several contaminating sequences). Although this situation appears to be rare, such results would not affect the analyses, since DNA damage is the only issue under investigation, whether it is from ancient endogenous or contaminant sources.
Determination of Site-Specific Postmortem-Damage Rates
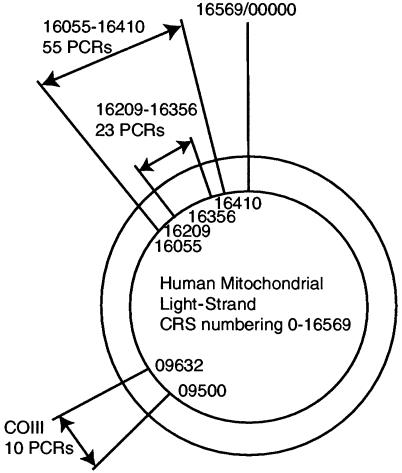
Bases at CRS positions 16209–16356 (here termed the “middle region” [MR]) contain the majority of phylogenetically variable sites and were consequently subject to more amplifications than were the outer regions (OR), at positions 16055–16208 and 16357–16410. Four samples that produced multiple sequences in every cloned product were presumed to be contaminated and were removed from the analysis. Cloned sequences from the remaining samples were aligned manually with the CRS by using the program Sequence Navigator (version 1.0.1; Applied Biosystems). The consensus sequence for each specimen was determined from the sequences shared between all clones, including the UNG-treated amplifications. The remaining intraclone base differences formed the postmortem-damage data set. The few base insertions and deletions observed were excluded from the analysis.
It is difficult to calculate site-specific HVR1 postmortem-damage rates for a number of reasons. A major problem is that damaged sites occurring on several clones within a set derived from one PCR are likely to have arisen from the same ancestral template molecule and, therefore, that the observed number is dependent on the number of clones sequenced. Similarly, the proportion of undamaged starting templates is difficult to assess because of replication advantages over damaged templates during the early amplification cycles. Furthermore, within a set of clones, it is common to see jumping PCR spread damaged sites between daughter amplified strands, generating a few damaged sites at identical nucleotide positions between quite different sequences. Last, the different number of PCR amplifications and clones for each fragment (MR and OR) complicate calculations. Consequently, an approximate relative rate of postmortem damage (ρv) was calculated for each site after the method of Hofreiter et al. (2001), which was modified to account for the dual-fragment amplification of the HVR1 (fig. 2). A modified relative rate of postmortem damage was calculated as ρv=μv/σv, where v is a specific site with reference to the CRS, μv is the number of hits observed at a specific site across all sequences analyzed, and σv is the total number of amplifications for each specific site. This calculation is biased, since it does not take into account the numbers of clones examined at each site (which vary by PCR) or the problems associated with assessing the number of undamaged starting templates. Some previous studies (e.g., see Cooper et al. 2001a) have assumed that the low number of starting templates implies that all undamaged strands effectively arose from one template. The denominator then becomes the number of different clones (as opposed to PCRs). However, our data demonstrate that different genetic regions exhibit heterogeneity in the rates of postmortem damage and, therefore, that an area with larger amounts of background postmortem damage will have a higher denominator in the rate calculation, lowering the apparent relative rates of damage for sites in this region. Consequently, it is difficult to compare postmortem-damage rates between different subregions of the HVR1 or between the HVR1 and COIII, and such calculations should be seen as illustrative, not absolute. It is, however, possible to compare postmortem-damage estimates within a single region of amplification (MR or OR), since all sites have been amplified/cloned the same number of times.
Figure 2.
Human mitochondrial regions amplified
Hotspots
The null hypothesis, H0, that postmortem-damaged sites were randomly distributed across the HVR1 and COIII was tested through comparison with the expected Poisson distribution (Aris-Brisou and Excoffier 1996; Heyer et al. 2001). Heyer et al. (2001) have calculated an expected distribution of substitutions in modern human mitochondrial samples and have tested this against empirical data. It is a simple matter to modify the test for this data set, although, because of the bias described above (see the “Determination of Site-Specific Postmortem-Damage Rates” subsection), the HVR1 analysis was performed separately on the MR and the OR. The probability that sequence sites have exactly X substitutions, P(X), is P(X)=e-λλX/X!, where Poisson parameter λ refers to the observed density of mutations. To estimate the expected count, LP(X), of sequence positions for each category, given the random distribution λ and the length L of the sequence, we multiply the Poisson probability P(X) by the number of sequence sites L, such that LP(X)=L(e-λλX/X!). A χ2 goodness-of-fit test can then be applied to the observed and expected results, to determine whether the H0 that postmortem damage is randomly distributed within each of the regions can be rejected.
Relative-Rate Comparison with Modern Studies
To determine whether there is a correlation between sites with elevated substitution rates in modern populations and sites with high postmortem-damage rates, we obtained estimates of site-specific rates of mutation from recent comprehensive studies (Excoffier and Yang 1999; Meyer et al. 1999). Different rate-calculation methods were used in each study, preventing direct comparison, so the results were standardized onto a quartile scale of 1–4 (with 1 being the lowest and 4 being the highest), excluding invariant sites. Sites with consistently high mutation rates (3 or 4) in both modern studies were contrasted with those in the postmortem data set.
Base Structure
The originally damaged mtDNA strand (light or heavy) was determined according to the method described in the companion article by Gilbert et al. (2003), by using the prevalence of hydrolytic deamination of cytosine to deoxycytidine residues (leading to an observable C→T or G→A change; C→T/G→A), which is by far the most commonly observed form of postmortem damage (Hansen et al. 2001; Hofreiter et al. 2001). The base composition around the most mutable and damaged HVR1 sites (those with a standardized mutation/damage rate of 3 or 4) were examined for patterns by using a window of 3, 5, 7, 9, or 11 bp around the hotspot. Principal-component analysis was performed on the five data sets, and the H0 that the first and second eigenvectors have no correlation with levels of postmortem damage was tested using a general linear model.
Functional Analysis
The rate of postmortem damage and variation in modern data sets was analyzed in three short regions of known function within the OR: 7sDNA, MT5, and TAS (positions 16106–16191, 16194–16208, and 16157–16172, respectively) (Doda et al. 1981; Ohno et al. 1991). An additional area of low variation, observed in the postmortem data set (low-damage region 1 [LDR1]) at positions 16365–16395, was also compared. The H0 that the percentage of variable bases did not differ significantly along the OR was tested using a Student’s unpaired two-sample t test and a Mann-Whitney test. For the COIII postmortem data set, the H0 that postmortem damage (measured as total hits per codon position [hereafter “hits”]) and the number of variable sites at each codon position (hereafter “sites”) were evenly distributed among different codon positions was tested by comparing the observed and expected levels with a χ2 goodness-of-fit test.
Regional Specificity
Postmortem-damage rates in the HVR1 and COIII can be compared if it is assumed that amplifications from the same DNA extract will (a) start from the same template number and combination of postmortem-damaged and undamaged strands and (b) amplify under similar conditions. A comparison of relative rates can be made by using the number of clones screened for HVR1 and COIII and assuming that the number of undamaged sequences results from the same relative number of undamaged templates. The calculation of comparable rate, R, is R=T/LΠ, where T is the total number of hits, L is the total length of sequence, and Π is the number of PCR amplifications. A saturation value, S, was also calculated for each sample, representing the number of different sequences within the clones. An S value of 1 indicates that all sequences are different, and, in such cases, it is probable that further clones would yield different products and, therefore, that postmortem damage for the individual has been underestimated. The H0 that no difference exists in the proportion of sites that show damage between HVR1 and COIII for each sample was tested using a Student’s one-sample t test and a Wilcoxon test.
Haplogroup Analysis
For the analysis of the possible phylogenetic effects of damage-induced sequence artifacts, a Eurasian mitochondrial tree based predominantly on HVR1 sequences (Richards and Macaulay 2000) was modified to show which branches were defined by sites with high postmortem-damage rates. This comparison requires that the strand (light or heavy) on which the postmortem damage occurred be identified, to allow determination of the direction in which the haplogroup assignments will shift on the phylogenetic tree. For the standardization of the postmortem-damage rates between the OR and the MR, the OR rates were multiplied by the ratio of the number of PCRs over the MR and the OR.
Results
Data Authenticity, Postmortem Damage, and UNG
Of the original 43 human teeth samples, 4 (Tg99.1, Tg147, Tg191, and Tg200) appeared to be contaminated, since cloned sequences from a single PCR amplification differed significantly and/or repeat amplifications yielded different products. Tg99.1 contained at least six sequences, whereas Tg147, Tg191, and Tg200 contained two different sequences each. All other samples possessed the same consensus sequence in the separate amplification and cloning experiments. Of the 34 sampled individuals, 7 shared the haplogroup (V) of the researcher (M.T.P.G.) undertaking the experimental analysis (M.T.P.G., unpublished data). However, the sporadic base changes among the clones suggest that these sequences are ancient. All other samples used for the analysis (including the sample sent for independent replication) displayed similar postmortem hydrolytic damage. The four animal teeth examined, from two of the archaeological sites, repeatedly yielded no human DNA, and tests using “spiked” PCR revealed that this did not result from PCR inhibition.
A consistent, single haplotype may easily be obtained when a sample with no endogenous DNA is contaminated by a single modern source or even several sources if one is more concentrated than the others (Cooper 1997). However, cloned sequences of all the postmortem samples show a pattern of a shared consensus sequence with many scattered singleton substitutions, particularly C→T/G→A, characteristic of aDNA (Handt et al. 1996; Krings et al. 1997; Hansen et al. 2001; Gilbert et al. 2003). In samples treated with UNG, no C→T/G→A changes were observed except where shared between all clones (therefore deemed to be part of the original sequence). This strongly suggests that the sequences derive from one ancient source (a damaged contaminant that is either endogenous or old) and are therefore suitable for the present study. The UNG results, together with the very low rate of polymerase misincorporation observed in modern contaminants from both modern DNA and aDNA extractions (Gilbert et al. 2003), indicate that the observed sequence heterogeneity in ancient sequences is not a result of enzyme misincorporation.
Rates of Postmortem Damage
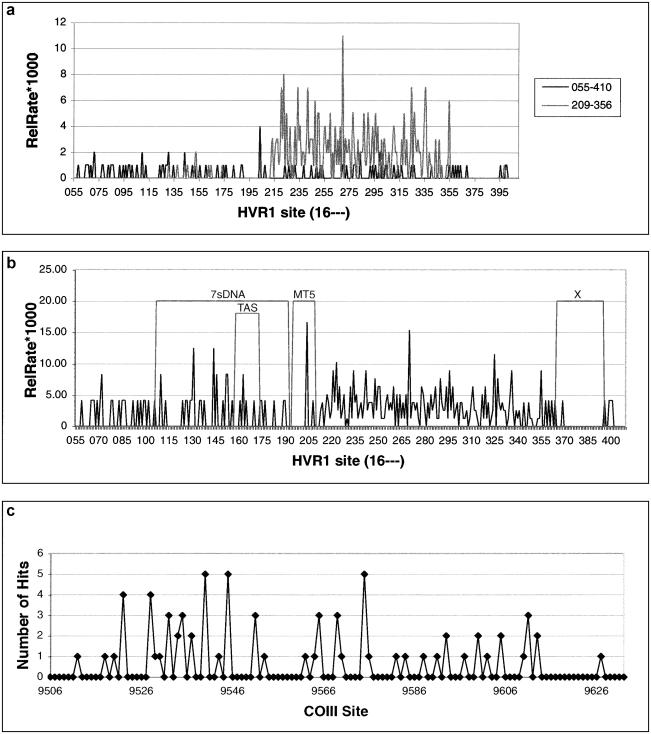
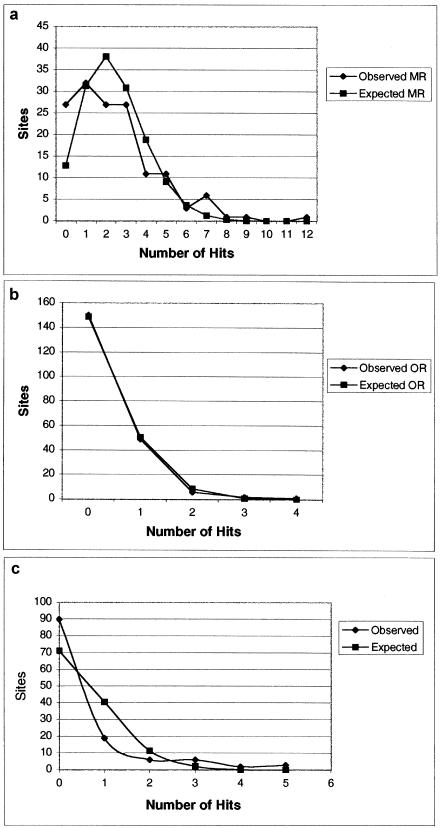
The spatial variation of rates of postmortem damage per site—calculated as absolute number of hits, An, for COIII, and as An in addition to the more comparable relative rate, ρn, of postmortem damage, for the HVR1—can be seen in figure 3. The variation initially appears to be overdispersed, with certain sites being overrepresented, such as sites 16110, 16204, 16223, 16270, 16298, and 16325, in the HVR1, and sites 9540, 9545, and 9570, in the COIII. The H0 that postmortem damage is randomly distributed can be strongly rejected (fig. 4) (MR χ2 P<.00; OR P=.02; COIII P<.00), demonstrating that sites are not being damaged at random in either region.
Figure 3.
Damage variation across HVR1 and COIII. a, Absolute damage measured in hits per site for data sets generated by each PCR primer set across HVR1 (16055–16410). b, Relative-rate variation (×100) across HVR1, calculated as in text. Four areas of interest are marked onto the graph. c, Absolute damage across region of COIII (09500–09632). Numbering is with reference to the CRS (Anderson et al. 1981).
Figure 4.
Observed and expected frequency of absolute damage rates for HVR1 MR (a), HVR1 OR (b), and COIII (c) sites. For full details, see text.
Structural Analysis
The majority (24/30) of sites with a high rate of postmortem damage have also been observed as hypermutable in the modern data sets (table 4). The exceptions (sites 16110, 16131, 16144, 16204, 16242, and 16325) are of special interest, and the surrounding base composition was examined for a possible cause. The first and second eigenvectors of the principal-component analysis explained the majority of the results in all cases, and there was no evidence to reject the H0 that base composition around individual sites plays no role in determining a propensity for postmortem damage (results not shown). However, two of the three HVR1 subregions with known function appear to have a reduced rate of postmortem damage (fig. 4b). A fourth region of low postmortem damage, LDR1, is also apparent at sites 16365–16395 in the HVR1. Two-sample t tests and nonparametric Mann-Whitney tests show that the percentage of postmortem-damaged sites within LDR1 is significantly different from the average rate across the OR, at P<.05, and across MT5, at P<.1 (table 5). Within COIII, there is no evidence to suggest that postmortem damage has a bias toward codon positions (sites χ2 P=.47; hits χ2 P=.14).
Table 4.
Standardized Mutation and Damage Rates[Note]
| HVR1 Base Position | E99 | M99 | TG03 |
| 16072 | 2 | 0 | 3 |
| 16085 | 3 | 0 | 3 |
| 16093 | 4 | 3 | 4 |
| 16110a | 0 | 0 | 4 |
| 16126 | 0 | 4 | 3 |
| 16131a | 0 | 0 | 3 |
| 16144a | 0 | 0 | 3 |
| 16148 | 3 | 3 | 1 |
| 16163 | 4 | 3 | 3 |
| 16172 | 3 | 3 | 1 |
| 16183 | 3 | 3 | 2 |
| 16187 | 4 | 3 | 1 |
| 16189 | 4 | 4 | 1 |
| 16192 | 4 | 3 | 1 |
| 16204a | 0 | 0 | 4 |
| 16219 | 4 | 3 | 1 |
| 16223 | 3 | 4 | 4 |
| 16230 | 3 | 4 | 1 |
| 16242a | 0 | 0 | 3 |
| 16262 | 4 | 0 | 3 |
| 16270 | 3 | 3 | 4 |
| 16278 | 3 | 4 | 2 |
| 16290 | 0 | 2 | 3 |
| 16293 | 3 | 4 | 1 |
| 16294 | 3 | 4 | 2 |
| 16298 | 4 | 2 | 4 |
| 16309 | 3 | 4 | 1 |
| 16319 | 4 | 3 | 2 |
| 16325a | 0 | 0 | 4 |
| 16327 | 4 | 2 | 3 |
Note.— Site-specific in vivo mutation rates taken from two previous studies (Excoffier and Yang 1999 [E99]; Meyer et al. 1999 [M99]) were standardized into quartiles and were compared with the standardized postmortem-damage rates from the present study (TG03).
Seven sites where major disagreement is observed between rates of occurrence of modern mutations and ancient damage.
Table 5.
Statistical Tests on Absolute Number of Sites Changing across the OR and Four Subregions
|
Two-Samplet Test |
Mann-WhitneyNonparametric Test |
||||||
| Region | Bases | Changesb | Averagec | t | P | W | P |
| OR | 208 | 58 | .28 | ||||
| 7sDNA | 86 | 21 | .24 | −.93 | .82 | 12,435 | …d |
| TAS | 16 | 3 | .19 | .27 | .40 | 20,112 | .39 |
| MT5 | 15 | 1 | .07 | 1.62 | .06 | 20,396 | .10 |
| LD1 | 31 | 1 | .03 | 6.07 | .00 | 19,291.5 | .00 |
Size (in bp) of the region.
Number of damaged sites.
Average damage per site.
No P value returned by Minitab.
Regional and Species Specificity
Table 6 presents sample-specific rates of postmortem damage, Π, and saturation, S, for each sample with both HVR1 and COIII data. Six of eight human samples and two of three bear samples demonstrate higher postmortem-damage rates in the HVR1 than in COIII/Cytb, with HVR1 subregions showing 1.1–6.8 times the postmortem damage than corresponding COIII/Cytb regions. Of the three exceptions, two (bear IB221 and human EA) demonstrate HVR1 saturation values of 1, indicating that the postmortem damage has been underestimated. When these two samples are removed, the H0 of an equal amount of postmortem damage in each region can be rejected (one-way t test P=.033; Wilcoxon P=.038).
Table 6.
Region-Specific Damage Rates per Sample
| Species, Sample, and Regiona | DamageRate (Π)b | Saturation (S)c | Sourced |
| H. sapiens: | |||
| 76: | |||
| HVR1 | 1.35 | .40 | |
| COIII | .79 | .50 | |
| 80: | |||
| HVR1 | 2.82 | .86 | |
| COIII | .79 | .20 | |
| 114: | |||
| HVR1 | 4.96 | .80 | |
| COIII | 3.97 | .36 | |
| 136: | |||
| HVR1 | 4.81 | .41 | |
| COIII | .00 | .41 | |
| 148: | |||
| HVR1 | .68 | .18 | |
| COIII | 2.91 | .30 | |
| 149: | |||
| HVR1 | 5.01 | .88 | |
| COIII | 4.76 | .58 | |
| 192: | |||
| HVR1 | .82 | .40 | |
| COIII | .00 | .00 | |
| EA: | |||
| HVR1 | .91 | 1.00 | |
| COIII | 1.59 | .60 | |
| U. spelaeus TAB15: | |||
| CR | 3.4 | 1.0 | Loreille et al. 2001 |
| Cytb | .5 | .9 | Loreille et al. 2001 |
| U. arctos: | |||
| IB221: | |||
| CR | 6.0 | 1.0 | Barnes et al. 2002 |
| 12s | 9.0 | .8 | |
| IB223: | |||
| CR | 5.4 | .7 | Barnes et al. 2002 |
| 12s | 3.3 | .7 |
Regions given are fragments from the following genes/noncoding regions: COIII, control region (CR), apocytochrome b (Cytb), HVR1, and 12s ribosomal RNA (12s).
Sample-specific calculation of damage level (comparable only within regions from one sample).
Proportion of clones that vary from each other within one cloned region.
Where no source is indicated, data are from the present study.
Haplogroup Analysis
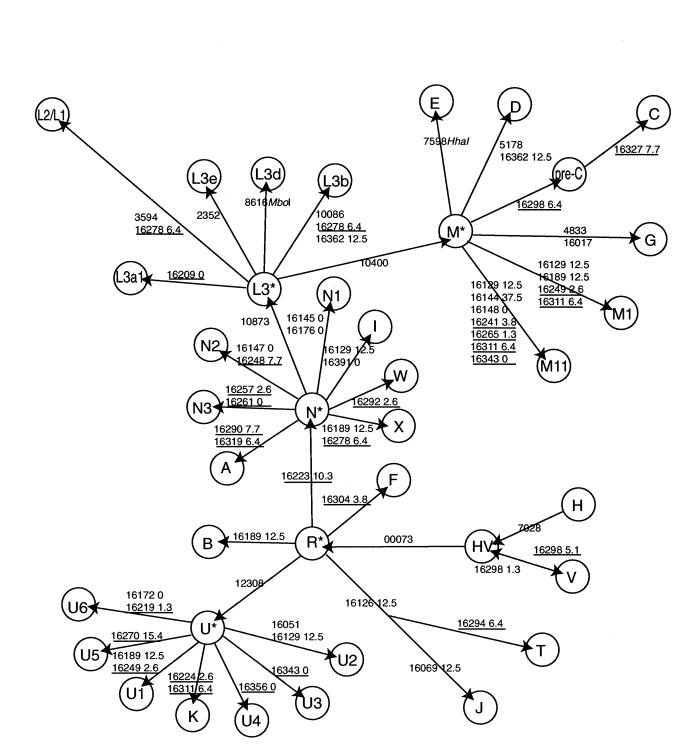
Figure 5 shows the relationship between phylogenetic changes and postmortem-damage rates on each branch of the Eurasian HVR1 mitochondrial tree. The analysis is limited because the majority of the samples that we studied are genetically similar northwestern Europeans (Anglo-Saxons and Scandinavians/Vikings). Contemporary indigenous European populations reach up to 60% haplogroup H and 40% haplogroup V (Torroni et al. 1998), and this is mirrored in our findings (M.T.P.G., unpublished data). Consequently, the postmortem damage in the HVR1 is mostly constrained to non-H and non-V substitutions, and unidirectional changes are almost exclusively observed (e.g., postmortem damage at position 16223 will nearly always be constrained to changes from the haplogroup H or haplogroup V cytosine to a 5-hydroxyuracil).
Figure 5.
Modification of mitochondrial HVR1 haplogroup tree (reprinted, with permission, from Richards and Macaulay 2000) to demonstrate potential for haplogroup misidentification. From a starting sample of haplogroups H or V, arrows indicate the direction along the tree a sample can move because of hydrolytic deamination (or real singleton substitution) of C→T or A→G. Site numbers are with reference to the CRS (Anderson et al. 1981). Relative rates are calculated for the MR (underlined) and OR. For full details, see text.
However, where the opportunity for back mutations exists, such as at position 16298 in haplogroup V (changing back to the CRS state), this is observed in only one clone out of the seven specimens. This scenario is marked on the tree. The data permitted a rough estimation of which haplogroups are most likely to arise through postmortem damage, starting from the CRS (table 3). The limiting factors are naturally the number of sites that are needed to change (two sites are less likely to change than one), as well as the site with lowest rate of postmortem damage in a haplogroup. For example, haplogroup I differs from the CRS at sites 16129, 16223, and 16391, which have relative rates of postmortem damage of 12.5, 10.3, and 0, respectively. Although two of these sites are highly susceptible to postmortem damage, site 16391 effectively prevents the misidentification of haplogroup I.
Discussion
The sequence results of the present study are consistent with derivation from a series of different specimens containing single aDNA sources. The cloning and UNG experiments, along with the repeat extractions and amplifications, suggest that mtDNA heteroplasmy (Gocke et al. 1998), nuclear copies of mitochondrial genes (Mourier et al. 2001), or polymerase misincorporations (Hansen et al. 2001) are not contributing to the observed variation. The results clearly demonstrate that postmortem DNA damage is not distributed randomly across the HVR1 or COIII.
Although the HVR1 contains no functional genes, modern population studies have shown that it does not evolve in a random manner, since sequence variation is concentrated at a few sites. Relative rates of mutations have been calculated using a range of methods (Hasegawa et al. 1993; Wakely 1993; Excoffier and Yang 1999; Meyer et al. 1999; Heyer et al. 2001) that are not directly comparable, although striking similarities are apparent even after standardization has reduced the resolution (table 4). Of the 30 sites that can be compared for postmortem-damage and in vivo mutation rates, 15 show very similar rates, and only 6 (sites 16110, 16144, 16148, 16204, 16242, and 16325) completely disagree (i.e., are not observed to mutate in vivo but experience fast postmortem damage). Of these six, at least three have mutation-rate estimates, in the two modern studies, that also disagree, and this may relate to sampling stochasticity or the standardization approach. If so, further sampling may provide evidence for elevated mutation rates at these sites. Other explanations for different rates include the possibility that some form of in vivo protection for these sites may be removed or degraded after death.
Structural constraints may also explain the correlation between the postmortem-damage and mutation rates of mtDNA in modern studies. For example, if DNA secondary-structural conformation predisposed particular sites to hydrolytic attack, then these could appear both to be hypermutable in modern populations and to be frequently damaged in postmortem samples. Secondary-structural models of the human HVR1 may provide useful insight but are currently insufficiently detailed. Meyer et al. (1999) remark on the correlation between HVR1 site-specific mutation rates and the three known features of structural interest. A short fragment of heavy-strand DNA, 7sDNA, provides the D-loop with its characteristic triple-stranded feature, extending from a trinucleotide stop codon at position 16104 to at least position 00110 (Doda et al. 1981; Meyer et al. 1999). The stop codon itself exhibits neither postmortem damage nor high mutation rates in data from the present study or previous studies (Excoffier and Yang 1999; Meyer et al. 1999), and 7sDNA itself appears to mutate and receive postmortem damage at average rates. The other two structural regions, TAS (the termination-associated sequence) and the putative control element MT5, have low observed in vivo mutation rates, which have previously been suggested to result from functional constraints (Meyer et al. 1999). TAS is located upstream from the trinucleotide stop codon and interacts with sequence-specific binding factors (Doda et al. 1981; Wallace et al. 1995), whereas MT5 is postulated as a protein-binding site (Meyer et al. 1999). Both TAS and MT5 appear to have lower than average postmortem-damage rates, although this cannot be statistically validated at P<.05. Certain proteins have been shown to survive for much longer than DNA (Bada et al. 1999), and it is possible that the low amount of postmortem damage observed in the MT5 region (fig. 3b) is related to the continued binding and protection offered by the putative control element.
The results also identify a previously unrecognized region, LDR1, that possesses significantly reduced levels of postmortem damage. HVR1-mutation-rate studies (Excoffier and Yang 1999; Meyer et al. 1999) do not include all of this region but do characterize low rates of in vivo mutation in the 5′ segment. The absence of postmortem damage over such a large region (and over 600 clones) is unusual and suggests that the DNA sequence may be protected from hydrolytic damage in some way. One possibility would be a protein-DNA association, similar to that postulated for MT5 (Meyer et al. 1999), and, if further studies verify this observation, then LDR1 may be the first structural feature identified by aDNA. The hypothesis that there is a correlation between function and postmortem-damage rate should be tested further by examining ancient sequences of other CR regions with known binding sites, such as the main regulatory elements of transcription and regulation, or the origin of heavy-strand replication (Chang and Wallace 1985; Wallace et al. 1995).
One of the most interesting findings of the present study is the observation in both humans and bear species that postmortem-damage rates are higher in the HVR1 than in coding regions. Three samples did not show this pattern, two of which (one human and one bear) can be excluded because of high “saturation” values of clone sequence diversity within the HVR1, indicating that the true level of postmortem damage is underestimated; the third anomaly, human sample Tg148, cannot be excluded on this basis, and it is possible that varying amounts of template in the original PCRs may be responsible. Finnilä et al. (2001) note that in vivo mutation rates of mitochondrial 3rd codon positions are also lower than those of the HVR1. A plausible explanation is that the secondary-structural conformation of the HVR1 promotes increased rates of both in vivo mutation and postmortem damage, whereas, in the coding region, either this is lacking or there has been some selection to constrain mutation rates. Structural models of the human HVR1 may provide a useful test of this hypothesis.
It is interesting that the postmortem-damage results do not show the selection against 1st- and 2nd-codon-position mutations seen in coding-region sequences of modern populations. The ancient sequences may actually reflect the real mutation rates of COIII sites; these rates are normally masked by the effects of selection. This pattern shows no evidence for codon bias but does shows postmortem-damage hotspots, such as the HVR1 sequences.
Problems with Studies on Human aDNA
Current human aDNA research focuses on the identification of mtDNA haplogroups through either restriction digests (e.g., see Merriweather et al. 1994; Kaestle and Glenn Smith 2001) or sequence analysis (e.g., see Hänni et al. 1994; Krings et al. 1997, 1999; Stone and Stoneking 1998; Adcock et al. 2001) and on sexing individuals through systems such as amelogenin (e.g., see Faerman et al. 1995; Fily et al. 1998; Vernesi et al. 1999). The sexing and restriction-digest studies lack sufficient resolution to allow the assessment of the role of postmortem damage or contamination, so we can only analyze research that uses sequence data to define—or even determine—new haplogroups, such as studies of Neanderthal or archaic human samples (Krings et al. 1997, 1999; Ovchinnikov et al. 2000; Adcock et al. 2001).
The data in the present study have been generated from a small set of northwestern European samples, predominantly of haplogroups H and V. Consequently, the mtDNA tree (fig. 5) has been modified to demonstrate the potential haplogroup alteration caused by damage to these haplogroups only. However, figure 5 demonstrates the ease with which phylogenetic misidentification can occur if postmortem damage is not detected in sequences from any haplogroup. For example, it is not inconceivable that the Amerindian haplogroup A sequence could result from a haplogroup H Viking sample. We suggest that postmortem damage may explain many unusual results obtained from ancient human remains when appropriate techniques were not followed (e.g., the 60,000-year-old “Mungo man” sequences; Adcock et al. 2001). Postmortem damage will also complicate population genetic analyses of ancient humans, and detailed cloning will be needed to avoid overestimation of heterogeneity and population expansion sizes (Lundstrom et al. 1992; Aris-Brisou and Excoffier 1996).
For human aDNA research to progress, it will be necessary to rigorously enforce aDNA guidelines (Cooper and Poinar 2000). As real-time PCR becomes more routinely available, the initial template copy number should be quantified in samples, and detailed cloning experiments should become a requirement. Samples with low template number should be replicated and cloned at least once, since even highly damaged sites, such as 16270, are unlikely to predominate in two independent PCRs. However, the simplest procedure appears to be a comparison of sequences before and after enzymatic treatment with UNG (Pääbo 1989; Hofreiter et al. 2001), EndoIV (Pääbo 1989), AAG (Lau et al. 2000), or T4 with Pol 1 (Pusch et al. 1998; Di Bernardo et al. 2002), to reduce the number of templates with postmortem-damaged bases and the resulting possibility of misidentification. Such treatment is expected to reduce the starting-template copy number, providing a check for authenticity but also limiting the number of samples suitable for study. A variety of other nongenetic factors should also be considered, such as depositional environment (Höss et al. 1996; Nielsen-Marsh 2000) and microbial content (Burger et al. 1999), amino acid racemization (Poinar et al. 1996; Poinar and Stankiewicz 1999) and composition (Bada 1999), and gas chromatography/mass spectroscopy–measured levels of hydantoins (Höss et al. 1996). These issues are now well known, and the heavy burden of proof associated with aDNA research demands that new studies address them.
Acknowledgments
We are indebted to Chris Stringer, Martin Biddle, and Birthe Kjolbe-Biddle, for samples, and Paul Bradshaw and the BBC, who funded initial studies as part of the Blood of the Vikings production. We would also like to thank Ziheng Yang and Arndt von Haesler, for data provided, and Vincent Macaulay, for permission to modify the tree used in figure 5. We are grateful to Eddie Holmes, Paul Johnson, Gil McVean, Svante Pääbo, Mark Stoneking, Ryk Ward, and members of the Henry Wellcome Ancient Biomolecules Centre (Oxford) and the Max Planck Institute for Evolutionary Anthropology (Leipzig), for useful comments. E.W. and A.J.H. are grateful to Kim Aaris-Sorensen and Tina B. Brand, for samples, help, and discussion. M.T.P.G. and A.C. were supported by the Wellcome Trust; E.A. and A.J.H. were supported by the Villumkann Rasmussen Fonden, Denmark; and L.R. was supported by the Danish Research council for the Humanities. Two anonymous reviewers provided helpful comments on the manuscript.
References
- Adcock G, Dennis E, Easteal S, Huttley G, Jermelin L, Peacock W, Thorne A (2001) Mitochondrial DNA sequences in ancient Australians: implications for modern human origins. Proc Natl Acad Sci USA 98:537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Bankier A, Arrell B, de Bruijn M, Coulson A, Drouin J, Eperon I, Nierlich D, Roe B, Sanger F, Schreier P, Smith A, Staden R, Young I (1981) Sequence and organisation of the human mitochondrial genome. Nature 290:457–465 [DOI] [PubMed] [Google Scholar]
- Aris-Brisou S, Excoffier L (1996) The impact of population expansion and mutation rate heterogeneity on DNA sequence polymorphism. Mol Biol Evol 13:494–504 [DOI] [PubMed] [Google Scholar]
- Arneborg J, Heinemeier J, Lynnerup N, Nielsen HL, Rud N, Sveinbjörnsdóttir ÁE (1999) Change of diet of the Greenland Vikings determined from stable carbon isotope analysis and 14C dating of their bones. Radiocarbon 41:157–168 [Google Scholar]
- Bada J, Wang X, Hamilton H (1999) Preservation of key biomolecules in the fossil record: current knowledge and future challenges. Philos Trans R Soc Lond B Biol Sci 354:77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes I, Matheus P, Shapiro B, Jensen D, Cooper A (2002) Dynamics of Pleistocene population extinctions in Beringian brown bears. Science 295:2267–2270 [DOI] [PubMed] [Google Scholar]
- Burger J, Hummel S, Herrmann B, Henke W (1999) DNA preservation: a microsatellite-DNA study on ancient skeletal remains. Electophoresis 20:1722–1728 [DOI] [PubMed] [Google Scholar]
- Chang D, Clayton D (1985) Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc Natl Acad Sci USA 82:351–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A (1997) Reply to Stoneking: ancient DNA—how do you really known when you have it? Am J Hum Genet 60:1001–1002 [PMC free article] [PubMed] [Google Scholar]
- Cooper A, Lalueza-Fox C, Anderson S, Rambaut A, Austin J, Ward R (2001a) Complete mitochondrial genome sequences of two extinct moas clarify ratite evolution. Nature 409:704–707 [DOI] [PubMed] [Google Scholar]
- Cooper A, Poinar H (2000) Ancient DNA: do it right or not at all. Science 289:1139 [DOI] [PubMed] [Google Scholar]
- Cooper A, Rambaut A, Macaulay V, Willerslev E, Hansen AJ, Stringer C (2001b) Human origins and ancient human DNA. Science 292:1655–1656 [DOI] [PubMed] [Google Scholar]
- Di Bernardo G, Del Gaudio S, Cammarota M, Galderisi U, Cascino A, Cipollaro M (2002) Enzymatic repair of selected cross-linked homoduplex molecules enhances nuclear gene rescue from Pompeii and Herculaneum remains. Nucleic Acids Res 30:e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinner A, Blackburn G, Karplus M (2001) Uracil-DNA glycosylase acts by substrate autocatalysis. Nature 413:752–755 [DOI] [PubMed] [Google Scholar]
- Doda ND, Wright CT, Clayton DA (1981) Elongation of displacement-loop strands in human and mouse mitochondrial DNA is arrested near specific template sequences. Proc Natl Acad Sci USA 10:6116–6120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Yang Z (1999) Substitution rate variation among sites in mitochondrial hypervariable region I of humans and chimpanzees. Mol Biol Evol 16:1357–1368 [DOI] [PubMed] [Google Scholar]
- Faerman M, Filon D, Kahila G, Greenblatt C, Smith P, Oppenheim A (1995) Sex identification of archaeological human remains based on amplification of the X and Y amelogenin alleles. Gene 167:327–332 [DOI] [PubMed] [Google Scholar]
- Fily M-L, Crubézy É, Courtaud P, Keyser C, Ébrard D, Ludes B (1998) Analyse paléogénétique des sujets de la grotte sépulcrale d’Elzarreko Karbia (Bronze ancien, Pays Basque). C R Acad Sci III 321:79–85 [DOI] [PubMed] [Google Scholar]
- Finnilä S, Lehtonen M, Majamaa K (2001) Phylogenetic network for European mtDNA. Am J Hum Genet 68:1475–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MTP, Hansen AJ, Willerslev E, Rudbeck L, Barnes I, Lynnerup N, Cooper A (2003) Characterization of genetic miscoding lesions caused by postmortem damage. Am J Hum Genet 72:48–61 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocke C, Benko F, Rogan P (1998) Transmission of mitochondrial DNA heteroplasmy in normal pedigrees. Hum Genet 102:182–186 [DOI] [PubMed] [Google Scholar]
- Greenwood AD, Capelli C, Possner G, Pääbo S (1999) Nuclear DNA sequences from late Pleistocene megafauna. Mol Biol Evol 16:1466–1473 [DOI] [PubMed] [Google Scholar]
- Handt O, Krings M, Ward R, Pääbo S (1996) The retrieval of ancient human DNA sequences. Am J Hum Genet 59:368–376 [PMC free article] [PubMed] [Google Scholar]
- Hänni C, Laudet V, Coll J, Stehelin D (1994) An unusual mitochondrial DNA sequence variant from an Egyptian mummy. Genomics 22:487–489 [DOI] [PubMed] [Google Scholar]
- Hansen A, Willerslev E, Wiuf C, Mourier T, Arctander P (2001) Statistical evidence for miscoding lesions in ancient DNA templates. Mol Biol Evol 18:262–265 [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Di Rienzo A, Kocher T, Wilson A (1993) Toward a more accurate time scale for the human mitochondrial gene tree. J Mol Evol 37:347–354 [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Horai S (1991) Time of the deepest root for polymorphism in human mitochondrial DNA. J Mol Evol 32:37–42 [DOI] [PubMed] [Google Scholar]
- Heyer E, Zietkiewicz E, Rochowski A, Yotova V, Puymirat J, Labuda D (2001) Phylogenetic and familial estimates of mitochondrial substitution rates: study of control region mutations in deep-rooting pedigrees. Am J Hum Genet 69:1113–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreiter M, Jaenicke V, Serre D, von Haeseler A, Pääbo S (2001) DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res 29:4693–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höss M, Jaruga P, Zastawny T, Dizdaroglu M, Pääbo S (1996) DNA damage and DNA sequence retrieval from ancient tissue. Nucleic Acids Res 24:1304–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestle F, Glenn Smith D (2001) Ancient mitochondrial DNA evidence for prehistoric population movement: the numic expansion. Am J Phys Anthropol 115:1–12 [DOI] [PubMed] [Google Scholar]
- Kolman C, Tuross N (2000) Ancient DNA analysis of human populations. Am J Phys Anthropol 111:5–23 [DOI] [PubMed] [Google Scholar]
- Krings M, Geisert H, Schmitz R, Krainitzki H, Pääbo S (1999) DNA sequence of the mitochondrial hypervariable region II from the Neanderthal type specimen. Proc Natl Acad Sci USA 96:5581–5585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings M, Stone A, Schmitz R, Krainitzki H, Stoneking M, Pääbo S (1997) Neanderthal DNA sequences and the origin of modern humans. Cell 90:19–30 [DOI] [PubMed] [Google Scholar]
- Kurosaki G, Matsushita T, Ueda S (1993) Individual DNA identification from ancient human remains. Am J Hum Genet 53:638–643 [PMC free article] [PubMed] [Google Scholar]
- Lalueza-Fox C, Luna Calderón F, Calafell F, Morera B, Bertranpetit J (2001) mtDNA from extinct Tainos and the peopling of the Caribbean. Ann Hum Genet 65:137–151 [DOI] [PubMed] [Google Scholar]
- Lau A, Wyatt M, Glassner B, Samson L, Ellenberger T (2000) Molecular basis for discrimination between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proc Natl Acad Sci USA 97:13575–13578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362:709–715 [DOI] [PubMed] [Google Scholar]
- Loreille O, Orlando L, Patou-Mathis M, Philippe M, Taberlet P, Hänni C (2001) Ancient DNA analysis reveals divergence of the cave bear, Ursus spelaeus, and brown bear, Ursus arctos, lineages. Curr Biol 11:200–203 [DOI] [PubMed] [Google Scholar]
- Lundstrom R, Tavaré S, Ward R (1992) Estimating substitution rates from molecular data using the coalescent. Proc Natl Acad Sci USA 89:5961–5965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynnerup N (1998) The Greenland Norse: a biological-anthropological study. Meddelelser Om Grønland, Man & Society, vol 24. The Commission for Scientific Research in Greenland, Copenhagen [Google Scholar]
- Macauley V, Richards M, Forster P, Bendall K, Watson E, Sykes B, Bandelt H-J (1997) mtDNA mutation rates—no need to panic. Am J Hum Genet 61:983–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merriweather D, Rothhammer F, Ferrel R (1994) Genetic variation in the New World: ancient teeth, bone, and tissue as sources of DNA. Experientia 50:592–601 [DOI] [PubMed] [Google Scholar]
- Meyer S, Weiss G, von Haeseler A (1999) Pattern of nucleotide substitution and rate heterogeneity in the hypervariable regions I and II of human mtDNA. Genetics 152:1103–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourier T, Hansen A, Willerslev E, Arctander P (2001) The Human Genome Project reveals a continuous transfer of large mitochondrial fragments to the nucleus. Mol Biol Evol 18:1833–1837 [DOI] [PubMed] [Google Scholar]
- Nielsen-Marsh C (2000) Patterns of diagenesis in bone. I. Effects of site environments. J Archeol Sci 27:1139–1150 [Google Scholar]
- Ohno K, Tanaka M, Suzuki H, Ohbayashi T, Ikebe S, Ino H, Kumar S, Takahashi A, Ozawa T (1991) Identification of a possible control element, Mt5, in the major noncoding region of mitochondrial DNA by intraspecific nucleotide conservation. Biochem Int 24:263–272 [PubMed] [Google Scholar]
- Oota H, Saitou N, Matsushita T, Ueda S (1995) A genetic study of 2,000-year-old human remains from Japan using mitochondrial DNA sequences. Am J Phys Anthropol 98:133–145 [DOI] [PubMed] [Google Scholar]
- ——— (1999) Molecular genetic analysis of remains of a 2,000-year-old human population in China—and its relevance for the origin of the modern Japanese population. Am J Hum Genet 64:250–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov I, Götherström A, Romanova G, Kharitonov V, Lidén K, Goodwin W (2000) Molecular analysis of Neanderthal DNA from the northern Caucasus. Nature 404:490–493 [DOI] [PubMed] [Google Scholar]
- Pääbo S (1989) Ancient DNA: extraction, characterisation, molecular cloning and enzymatic amplification. Proc Natl Acad Sci USA 86:1939–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pääbo S, Irwin D, Wilson A (1990) DNA damage promotes jumping between templates during enzymatic amplification. J Biol Chem 265:4718–4721 [PubMed] [Google Scholar]
- Poinar H, Hofreiter M, Spaulding G, Martin P, Stankiewicz A, Bland H, Evershed R, Possnert G, Pääbo S (1998) Molecular coproscopy: dung and diet of the extinct ground sloth Nothrotheriops shastensis. Science 281:402–406 [DOI] [PubMed] [Google Scholar]
- Poinar H, Höss M, Bada J, Pääbo S (1996) Amino acid racemization and the preservation of ancient DNA. Science 272:864–866 [DOI] [PubMed] [Google Scholar]
- Poinar H, Stankiewicz B (1999) Protein preservation and DNA retrieval from ancient tissues. Proc Natl Acad Sci USA 96:8426–8431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusch C, Giddings I, Scholz M (1998) Repair of degraded duplex DNA from prehistoric samples using Escherichia coli DNA polymerase I and T4 DNA ligase. Nucleic Acids Res 26:857–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro-dos-Santos A, Santos S, Lucia Machado A, Guapindaia V, Zago M (1996) Heterogeneity of mitochondrial DNA haplotypes in pre-Columbian natives of the Amazon region. Am J Phys Anthropol 101:29–37 [DOI] [PubMed] [Google Scholar]
- Richards M, Macaulay V (2000) Genetic data and the colonization of Europe: genealogies and founders. In: Renfrew C, Boyle K (eds) Archaeogenetics: DNA and the population prehistory of Europe. McDonald Institute for Archaeological Research, Cambridge, UK, pp 139–151 [Google Scholar]
- Richards M, Sykes B, Hedges R (1995) Authenticating DNA extracted from ancient skeletal remains. J Archeol Sci 22:291–299 [Google Scholar]
- Stone A, Stoneking M (1998) mtDNA analysis of a prehistoric Oneota population: implications for the peopling of the New World. Am J Hum Genet 62:1153–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneking M (1995) Ancient DNA: how do you know when you have it and what can you do with it? Am J Hum Genet 57:1259–1262 [PMC free article] [PubMed] [Google Scholar]
- ——— (2000) Hypervariable sites in the mtDNA control region are mutational hotspots. Am J Hum Genet 67:1029–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Bandelt H-J, D’Urbano L, Lahermo P, Moral P, Sellitto D, Rengo C, Forster P, Savontaus M-L, Bonné-Tamir B, Scozzari R (1998) mtDNA analysis reveals a major late Paleolithic population expansion from southwestern to northeastern Europe. Am J Hum Genet 62:1137–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasan S, Zhang X, Zhang X, Kapurniotu A, Bernhagen J, Teichberg S, Basgen J, Wagle D, Shih D, Terlecky I, Bucala R, Cerami A, Egan J, Ulrich P (1996) An agent cleaving glucose-derived protein crosslinks in vitro and in vivo. Nature 382:275–278 [DOI] [PubMed] [Google Scholar]
- Vernesi C, Caramelli D, Carbonell I Sala S, Ubaldi M, Rollo F, Chiarelli B (1999) Application of DNA sex tests to bone specimens from three Etruscan (VII–III century B.C.) archaeological sites. Ancient Biomol 2:295–305 [Google Scholar]
- Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC (1991) African populations and the evolution of human mitochondrial DNA. Science 253:1503–1507 [DOI] [PubMed] [Google Scholar]
- Wakely J (1993) Substitution rate variation among sites in hypervariable region I of human mitochondrial DNA. J Mol Evol 37:613–623 [DOI] [PubMed] [Google Scholar]
- Wallace DC, Lott MT, Brown MD, Huopenen K, Torroni A (1995) Report on the committee on human mitochondrial DNA. In: Cuticchia AJ (ed) Human gene mapping 1995: a compendium. Johns Hopkins University Press, Baltimore, pp 910–954 [Google Scholar]
- Willerslev E, Hansen AJ, Christensen B, Steffensen JP, Arctander P (1999) Diversity of Holocene life forms in fossil glacier ice. Proc Natl Acad Sci 96:8017–8021 [DOI] [PMC free article] [PubMed] [Google Scholar]