Abstract
Through cDNA microarray analysis of gene expression in human cochlea and vestibule, we detected strong expression of μ-crystallin (CRYM; also known as “NADP-regulated thyroid hormone-binding protein”) only in these inner-ear tissues. In a subsequent search for mutations of CRYM, among 192 patients with nonsyndromic deafness, we identified two mutations at the C-terminus; one was a de novo change (X315Y) in a patient with unaffected parents, and the other was a missense mutation (K314T) that segregated dominantly in the proband’s family. When the mutated proteins were expressed in COS-7 cells, their subcellular localizations were different from that of the normal protein: the X315Y mutant showed vacuolated distribution in the cytoplasm, and the K314T mutant localized in perinuclear areas, whereas normal protein was distributed homogeneously in the cytoplasm. Aberrant intracellular localization of the mutated proteins might cause dysfunction of the CRYM product and result in hearing impairment. In situ hybridization analysis using mouse tissues indicated its expression in the lateral region of the spiral ligament and the fibrocytes of the spiral limbus, implying its possible involvement in the potassium-ion recycling system. Our results strongly implicate CRYM in normal auditory function and identify it as one of the genes that can be responsible for nonsyndromic deafness.
Introduction
Hearing loss that disturbs normal communication is a common sensory disorder worldwide. The incidence of congenital deafness is ∼1 in 1,000 newborns, and half of those cases are thought to result from genetic factors (Marazita et al. 1993). Most congenital or childhood-onset hearing impairments are nonsyndromic. So far, >70 genetic loci linked to nonsyndromic deafness have been described, and 26 genes whose mutations can cause deafness have been cloned (Hereditary Hearing Loss Homepage). Those data indicate that deafness is a highly heterogeneous disorder, and that genes responsible for deafness encode a large diversity of molecules. However, little is known of the molecular basis of inner-ear function, because the tissues in question are too small to be investigated in detail. The classical genetic approach through linkage analysis has limitations because the causes of deafness are so heterogeneous, and because linkage analysis requires DNA from a relatively large number of affected and unaffected members in a single family. Hence, we need to establish an effective alternative approach to searching for as-yet-unidentified genes that may be involved in hearing loss.
Obviously, genes that are expressed specifically in auditory tissues are likely to be good candidates to screen for genetic alterations in patients with deafness. In fact, several genes associated with deafness have been efficiently identified by way of organ-specific approaches involving, for example, subtractive human and mouse cDNA cochlear libraries (Robertson et al. 1994; Yasunaga et al. 1999; Simmler et al. 2000; Verpy et al. 2000). Several databases are now available which contain information about genes expressed in cochlea or in the developing ear (Morton Cochlear EST Database, Table of Gene Expression in the Developing Ear Web site, and Corey Lab Inner Ear Gene Expression Database).
It follows that identification of transcripts specific to the inner ear should also be helpful for studying hearing disorders. Therefore, in the work reported here we applied a genome-wide cDNA microarray analysis to investigate gene-expression profiles in human cochlea and vestibule, and focused on one of the genes that was expressed at high levels in both of those tissues. Mutant alleles of this gene were responsible for nonsyndromic deafness in two individuals among the group of probands we studied.
Family, Material, and Methods
Preparation of Tissues and RNA
Tissues from one cochlea and seven vestibules were obtained with written informed consent from different adult patients undergoing labyrinthectomy; each patient had been diagnosed with a nonlabyrinthine disorder, such as temporal-bone tumor or acoustic tumor.
Total RNA was extracted from each inner-ear sample using Trizol (Life Technologies) according to the manufacturer’s instructions. After treatment with DNase I, T7-based RNA amplification was performed as described elsewhere (Luo et al. 1999), with some modifications. Using an estimated 3 ng of total RNA from the cochlear tissue, we performed three rounds of amplification. For vestibular tissues, we performed two rounds of RNA amplification, using 3 μg of total RNA. We obtained 70–80 μg of each amplified RNA (aRNA) sample. As a control, we mixed PolyA(+) RNAs derived from 29 normal human tissues (bone marrow, brain, heart, kidney, liver, lung, lymph node, mammary gland, pancreas, placenta, prostate, salivary gland, skeletal muscle, small intestine, spinal cord, spleen, stomach, testis, thymus, thyroid, trachea, uterus, fetal brain, fetal kidney, fetal liver, fetal lung [Clontech], colon, ovary [Biochain], and mesenteric adipose tissue).
cDNA Microarray
Microarray slides containing 23,040 cDNA spots selected from the UniGene database of the National Center for Biotechnology Information were utilized for our analysis of gene expression in the human inner ear. As described elsewhere (Ono et al. 2000; Saito-Hisaminato et al. 2002), aRNAs from each tissue sample (2.5–5 μg) were labeled with Cy3-dCTP (Amersham Pharmacia Biotech), and an equal amount of aRNAs from a mixture of 29 human tissues was labeled with Cy5-dCTP (Amersham Pharmacia Biotech). Labeling and hybridization were performed according to protocols noted elsewhere (Ono et al. 2000). The intensity of each hybridized spot was measured and analyzed with the ArrayVision computer program (Amersham Bioscience), and background signals were subtracted. Each Cy3- and Cy5-fluorescence intensity was normalized by use of averaged signals from 52 housekeeping genes. We calculated a cut-off value for each gene’s expression to dissolve background fluctuation, according to parameters established previously (Saito-Hisaminato et al. 2002).
RT-PCR
To confirm our cDNA microarray data we performed semiquantitative RT-PCR experiments using cDNAs derived from the vestibule and from the 29 normal tissues, using single-round aRNAs. GAPDH served as an internal control. The amount of cDNA, as judged by the intensity of the GAPDH signal, was optimized in both samples. Primers used to amplify specific CRYM gene products were F: TCTGGAGATGTCCTGCTGTC and R: GGCTACCTAGCTTTGCTTTC; the PCR proceeded for 25 cycles of 95°C for denaturing, 55°C for annealing, and 72°C for extension. The PCR products were electrophoresed on a 2% agarose gel and were visualized by ultraviolet light.
Family Selection and Mutational Search
We screened DNA from 192 Japanese families in which probands were found to have congenital or childhood-onset bilateral nonsyndromic sensorineural hearing loss and no history of drug toxicity, infections or injury. Of these families, 41 showed dominant inheritance of deafness and 21 showed recessive inheritance; the other 130 were represented by only one affected individual (simplex cases). Each patient was clinically well characterized by a series of auditory examinations (pure-tone audiometry) that indicated sensorineural hearing loss ranging from mild to profound in severity. All participants in the project had provided written informed consent and had already been examined for mutations in the GJB2 gene [MIM 121011] and the mitochondrial gene encoding 12S rRNA [MIM 561000], but no mutations were detected in either of those genes although both had been described as common causes of deafness (Abe et al. 2000; Usami et al. 2000). On the basis of our microarray results, we analyzed genomic DNA isolated from peripheral blood leukocytes of one affected member of each family for mutations in the CRYM gene (MIM 123740).
We determined the genomic structure of the CRYM gene by comparison with a BAC sequence (GenBank accession number AF001550) on chromosome 16. We amplified each of its eight exons and their flanking intronic sequences using the following oligonucleotide primers: F1, AGGCTGGGCTGTGACCAGCA; R1, AGCTGTTAGCAA CGGTTAGG; F2, TGTCTAAGGGAAGGGCAGAG; R2, TGTTGCTGGTATCCAGTCAC; F3, AGGAATCGGATCCAGGTCTGA; R3, TCTGGAGTTCCAGCTATGTC; F4, ATTGCCTGCAAGCTCTTGAG; R4, CCTGACTCTTATCCTCCATC; F5, CGGTCTCATCAAGTTGAAAGG; R5, CTGCACCCAGCCAAATATTG; F6, GGGAATGAGGGGGTATTTTG; R6, GCCCATATTTTTCTGGAATGG; F7, CAGTGTACAAAGGATCTCTC; R7, TGACCTGAATGATGGAGCAC; F8, TAGGCATTGGCAACATGGAC; and R8, GGTAGAACAGAAGAAATGGC.
Each genomic DNA (5–10 ng) was amplified by the PCR, using Ex-Taq polymerase (Takara), for 2 min at 95°C, followed by 37 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 50 s, with a final extension of 5 min. After the products were purified by Multiscreen PCR (MILLIPORE), we performed standard cycle-sequencing reactions in an ABI 3700 autosequencer. DNA samples from 96 unrelated Japanese volunteers with normal hearing were used as controls.
Epitope Tagging of CRYM
To obtain constructs expressing the wild-type CRYM protein, as well as the X315Y and K314T variants, we PCR amplified the corresponding parts of the CRYM cDNA sequence (GenBank accession number NM_001888), using KOD-Plus-DNApolymerase (Toyobo) and the following primer sequences: (1) a W-Tag-F forward primer common to the wild type and both mutants, containing sequence corresponding to HA-tag (5′-CGTGAATTCCAGACCGTGCATCATGTACCCATACGACGTCCCAGACTACGCTAGCCGGGTACCAGCGTTCC-3′); and (2) reverse primers W-R (5′-AAGCTCGAGTTATTTACCAGATGACCAGGAATCA-3′) for the wild type, M(1)-R (5′-AAGCTCGAGTCAAGTTCCTTTGTTATATTTACCAG-3′) for the X315Y mutant, and M(2)-R (5′-AAGCTCGAGTTATGTACCAGATGACCAGGAATC-3′) for the K314T mutant. PCR products were cloned into the pcDNA 3.1(+) vector (Invitogen).
Microscopic Analysis of Immunofluorescence
For transfection of COS-7 cells, we used 1 μg of each construct and 3 μl of FuGENE6 (Roche Molecular Biochemicals) diluted in OPTIMEM medium (Gibco BRL) according to the manufacturer’s instructions. In two-well glass chamber slides, 1 × 104 to 5 × 104 cells were seeded. Cell samples were fixed 24 h, 48 h, 72 h, and 96 h after transfection and were treated as described elsewhere (Tsujikawa et al. 1999). They were first incubated overnight at 4°C with mouse anti-HA tag antibody (1:1,500) (Santa Cruz) and then for 60 min at 25°C with FITC-conjugated goat anti-mouse IgG (1:3,000) (ICN/Cappel). Images were viewed by means of fluorescence microscopy. We examined a total of six independent experiments.
In Situ Hybridization
The mouse cochlea at postnatal days (Pn) P0 was dissected, was fixed by 10% neutral formalin, and was embedded in paraffin. Tissue sections (4 um) were dewaxed and hybridized as described elsewhere (Hoshino et al. 1999), with some modifications. A 701-bp DNA fragment corresponding to the nucleotide positions 260–960 of mouse Crym cDNA (GenBank accession number NM_016669) was subcloned into pBlueScript SK(−) vector (Stratagene) and was used for generation of sense or antisense RNA probes. We carried out hybridization with digoxigenin-labeled RNA probes at 42°C for 18 h. The bound label was detected using NBT-BCIP, an alkaline phosphatase color substrate, and tissue slides were stained with Kernechtrot stain solution.
Results
Verification of cDNA Microarray Data
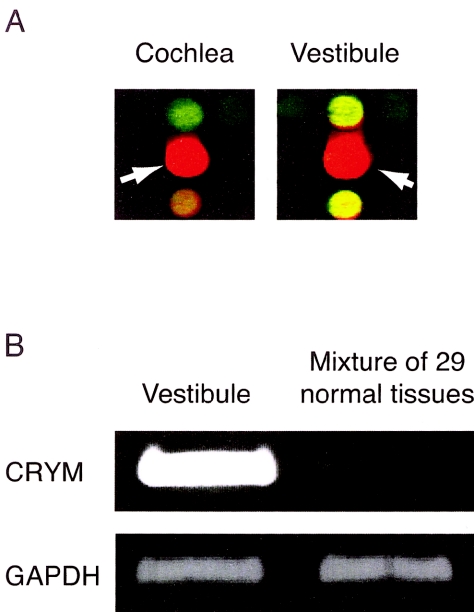
Through analysis of expression profiles of human inner-ear tissues on a cDNA microarray containing 23,040 genes, we found 52 genes whose signal intensities were more than 10-fold higher in cochlea and vestibule than in a mixture of 29 other tissues (table 1). Of the 52 genes expressed preferentially in the inner ear, 14 were located in one of five chromosomal regions known to contain loci linked to human deafness (DFNA4 [MIM 600652], DFNA7 [MIM 601412], DFNB15 [MIM 601869], and DFNB33 [MIM 607239]). Among nine genes known to be responsible for nonsyndromic deafness (COCH [MIM 603196], GJB2, GJB6 [MIM 604418], MYO7A [MIM 276903], USH1C [MIM 605242], MYH9 [MIM 160775], MYO6 [MIM 600970], CLDN14 [MIM 605608], and POU3F4 [MIM 300039]) that were present in our array, we confirmed expression in the inner ear of all except CLDN14 and POU3F. Five of these seven genes showed relatively high levels of expression (ratio >5 compared with the control) in the cochlea and of those, four (COCH, GJB2, GJB6, and USH1C) were considered to be expressed specifically or preferentially (ratio >10) (table 2). Among the genes specifically or preferentially expressed in the inner ear, we focused on CRYM because it represented the second-highest Cy3/Cy5 ratio, after COCH: 119.55 in the cochlea and 59.74 in the vestibule (table 1; fig. 1A). We confirmed predominant expression of CRYM in the inner ear by semiquantitative RT-PCR (fig. 1B) and considered this gene to be a candidate for playing an important role in auditory function.
Table 1.
Genes Preferentially Expressed in the Inner Ear with a Cy3/Cy5 Ratio of >10
| Cy3/Cy5 Ratio in |
|||||
| GenBankAccessionNo. | Cochlea | Vestibule | Description | Locus | Correspondence with Deafness Locus |
| AA669336 | 156.51 | 136.15 | Coagulation factor C homology (cochlin, COCH) | 14q12-q13 | |
| L02950 | 119.55 | 59.74 | μ-crystallin (CRYM) | 16p13.11-p12.3 | |
| M61901 | 114.30 | 70.40 | Prostaglandin D synthase | 9q34.2-34.3 | DFNB33 (9q34.3) |
| NM_020157 | 79.87 | 27.51 | Otoraplin | 20p12.1-p11.23 | Cochlear genea |
| NM_003460 | 77.36 | 185.57 | Zona pellucida glycoprotein 2 (sperm receptor) | 16p12 | |
| M64722 | 69.52 | 104.29 | clusterin (TRPM-2, apolipoprotein J) | 8p21-p12 | |
| J02611 | 68.59 | 108.76 | Apolipoprotein D | 3q26.2-qter | |
| XM_051860 | 56.54 | 22.55 | KIAA1199 protein | 15q24 | |
| W39428 | 55.10 | 67.96 | F-box only protein 2 | 1p36.23 | |
| AA972852 | 43.41 | 45.94 | Retinol-binding protein 1, cellular | 3q23 | DFNB15 (3q21-25/19p13) |
| J05096 | 41.98 | 55.99 | ATPase, Na+/K+ transporting, alpha 2 (+) polypeptide | 1q21-q23 | DFNA7 (1q21-q23) |
| AA252389 | 38.77 | 54.70 | Lipoma HMGIC fusion partner | 13q12 | |
| AA292179 | 37.41 | 16.65 | Ubiquitin A-52 residue ribosomal protein fusion product 1 | 19p13.1-p12 | DFNB15 (3q21-25/19p13) |
| M62402 | 30.46 | 44.97 | Insulin-like growth factor binding protein 6 | 12q13 | |
| AF039699 | 30.22 | 22.61 | USH1C | 11p14.3 | |
| X99920 | 26.20 | 70.30 | S100 calcium-binding protein A13 | 1q21 | DFNA7 (1q21-q23) |
| U59832 | 26.04 | 13.68 | Forkhead box D1 | 5q12-q13 | |
| X53331 | 23.79 | 16.19 | Matrix Gla protein | 12p13.1-p12.3 | |
| J02984 | 21.92 | 10.26 | Ribosomal protein S15 | 19p-tel | |
| X06617 | 20.19 | 17.83 | Ribosomal protein S11 | 19q13.3 | DFNA4 (19q13) |
| X75450 | 18.48 | 19.39 | Melanoma inhibitory activity (SH3 domain+) | 19q13.32-q13.33 | DFNA4 (19q13) |
| AA308743 | 17.91 | 11.85 | Ribosomal protein L35 | 9q34.1 | |
| AA526377 | 17.90 | 10.60 | Ribosomal protein L15 | 19q13.3 | DFNA4 (19q13) |
| AA496786 | 17.64 | 33.33 | Collagen, type IX, alpha 3 | 20q13.3 | |
| X96484 | 16.42 | 11.71 | DiGeorge syndrome critical region gene 6 | 22q11.21 | |
| AA058578 | 16.00 | 46.30 | Homo sapiens cDNA FLJ10158 fis | 3q12.3-21.3 | DFNB15 (3q21-25/19p13) |
| U14970 | 15.76 | 12.25 | Ribosomal protein S5 | 19q13.4 | DFNA4 (19q13) |
| AF052685 | 15.27 | 10.71 | Protocadherin gamma subfamily C, 3 | 5q32 | |
| F22593 | 15.27 | 34.13 | Vesicle-associated membrane protein 5 (myobrevin) | 2p11.2 | |
| W73992 | 15.15 | 11.10 | Serologically defined colon cancer antigen 43 | 9q22.2 | |
| AA633908 | 15.00 | 11.10 | ESTs | 20p12 | |
| X03342 | 14.89 | 15.31 | Ribosomal protein L32 | 3q13.3-q21 | |
| AA205528 | 14.79 | 14.90 | Carbonic anhydrase XIV | 1q21 | DFNA7 (1q21-q23) |
| AI344213 | 14.28 | 21.21 | Copper chaperone for superoxide dismutase | 11q13 | |
| AA057243 | 13.00 | 10.90 | PH domain containing protein in retina 1 | 11q13.5-q14.1 | |
| AA434038 | 12.96 | 11.83 | Gap junction protein, beta 2, 26kD (connexin 26) | 13q11-q12 | |
| AA148265 | 12.60 | 12.00 | Ribosomal protein L21 | 13q12.13 | |
| W84565 | 12.50 | 10.60 | Secreted protein of unknown function | 1q32.3 | |
| AI268685 | 12.33 | 11.38 | HSPC023 protein | 19p13.13 | DFNB15 (3q21-25/19p13) |
| AY043487 | 12.22 | 29.28 | Selenoprotein SelM (SELM) | 22q12 | |
| S72043 | 11.95 | 13.10 | Metallothionein 3 (growth inhibitory factor [neurotrophic]) | 16q13 | |
| AF284751 | 11.90 | 12.00 | Hypothetical protein HT036 | 1q34.1 | |
| X54412 | 11.88 | 35.98 | Collagen, type IX, alpha 1 | 6q12-q14 | |
| T55019 | 11.83 | 17.47 | Ribosomal protein L28 | 19q13.4 | DFNA4 (19q13) |
| AB003184 | 11.60 | 30.36 | Immunoglobulin superfamily containing leucine-rich repeat | 15q23-q24 | |
| X89401 | 11.43 | 20.16 | Ribosomal protein L21 | 13q12.13 | |
| AA625532 | 11.37 | 14.36 | Discoidin domain receptor family, member 2 | 1q12-q23 | DFNA7 (1q21-q23) |
| M68864 | 10.88 | 16.00 | ORF | 11cen-q12.1 | |
| AI240945 | 10.60 | 13.80 | Ubiquitously transcribed tetratricopeptide repeat gene, Y chromosome | Yq11 | |
| AA446913 | 10.51 | 13.71 | Ubiquitin specific protease 11 | Xp21.2-p11.2 | |
| N71750 | 10.46 | 26.54 | Zinc-finger protein 288(BC015587) | 3q13 | |
| X13916 | 10.31 | 17.43 | Low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) | 12q13-q14 | |
Otoraplin (OTOR [MIM 606067]) has been identified from a human fetal cochlear. So far, there is no report for human deafness-causing mutation. Abbreviations: DFNA, autosomal dominant loci; DFNB, autosomal recessive loci.
Table 2.
Nonsyndromic Deafness Gene Expression in the Cochlea and Vestibule by cDNA Microarray Analysis[Note]
| Cy3/Cy5 Ratio in |
|||
| Gene | Cochlea | Vestibule | Association with Human Deafness |
| COCH | 156.51 | 136.15 | DFNA9 |
| GJB2 (Cx26) | 12.96 | 11.83 | DFNB1/DFNA3/deafness with skin disorders |
| GJB6 (Cx30) | 34.71 | 4.55 | DFNB1/DFNA3 |
| USH1C | 30.22 | 22.61 | DFNB18/Usher syndrome type 1C |
| MYO7A | 6.20 | 3.63 | DFNB2/Usher syndrome type 1B |
| MYH9 | 1.50 | 1.00 | DFNA17/Epstein and Fechtner syndrome |
| MYO6 | 1.20 | 1.60 | DFNA22 |
| CLDN14 | No signal | No signal | DFNB29 |
| POU3F4 | No signal | No signal | DFN3 |
Note.— MIM numbers are as follows: DFNA9 (MIM 601369), DFNB1 (MIM 220290), DFNA3 (MIM 601544), deafness with skin disorders (MIM 148350), DFNB18 (MIM 602092), Usher syndrome type 1C (MIM 605242), DFNB2 (MIM 600060), Usher syndrome type 1B (MIM 6276903), DFNA17 (MIM 03622), Epstein and Fechtner syndrome (MIM 153650), DFNA22 (MIM 606344), DFNB29 (MIM 60568), and DFN3 (MIM 304400).
Figure 1.
Analysis and confirmation of cDNA microarray results. A, Signals of CRYM in the microarray (white arrows). aRNA from the human inner ear (cochlear and vestibular tissues) was labeled with Cy3-dCTP (red); mixed aRNA from 29 other tissues yielded green signals from Cy5-dCTP (green). B, Confirmation of microarray data by semiquantitative RT-PCR of CRYM with one-round amplified RNAs.
Mutational Analyses
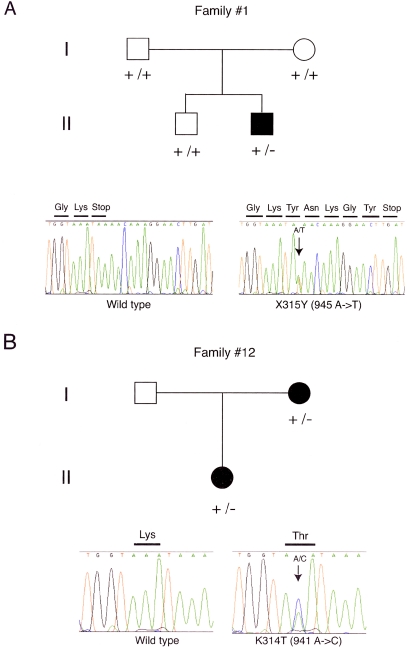
We analyzed the CRYM gene for mutations in 192 patients with nonsyndromic hearing loss. Direct DNA sequencing identified four genetic polymorphisms in this panel: one mutation in exon 8, causing an amino acid substitution, and one mutation affecting the stop codon (table 3). The latter mutation (X315Y; 945A→T) caused addition of five amino acids at the C-terminal end of the protein. This mutation was detected only in the proband, not in either of his unaffected parents or in his brother (family 1; fig. 2A). We confirmed paternity and maternity in family 1, using 12 highly polymorphic microsatellite markers (data not shown), and concluded that this mutation had occurred de novo.
Table 3.
Nucleotide Changes Detected by Mutation Analyses in the CRYM Gene
|
Allele Frequency |
|||||
| Change | Exon/Intron | Amino Acid | Family No. | in Patients | in Control Subjects |
| IVS1-116C→T | Intron1 | 90/364 | Not done | ||
| IVS1-119G→A | Intron 1 | 3/364 | Not done | ||
| IVS4+38C→G | Intron 4 | 84/336 | .12a | ||
| 864G→C | Exon 7 | Thr288Thr | 94/330 | Not done | |
| 941A→C | Exon 8 | Lys314Thr | 12 | 1/384 | 0/192 |
| 945A→T | Exon 8 | X315Tyr → extended protein | 1 | 1/384 | 0/192 |
See “A Database of Japanese Single Nucleotide Polymorphisms” Web site.
Figure 2.
Pedigrees and electrophoregrams showing mutations in the CRYM gene in two families. Corresponding normal sequences are shown in the left-hand panels. A, In family 1, only patient II-2 shows a heterozygous A→T substitution, changing the stop codon to a Tyr residue and bringing about an extension of the protein by five amino acids at the C-terminal. B, In family 12, the patient (II-1) and her affected mother (I-2) are both heterozygous for a Lys314Thr mutation.
The other mutation, K314T (941A→C), was detected in the proband and her affected mother in family 12 (fig. 2B). Although the unaffected father was not available for testing, the mutation appeared to cosegregate with hearing loss. To exclude a possibility that this alteration represented a polymorphism, we examined 192 control chromosomes but did not find the substitution in any of them.
Clinical Analyses
Affected individuals with the X315Y and the K314T mutations have normal stature and intelligence. There is no vestibular, visual, renal, or muscular involvement for all affected individuals with the CRYM mutations. Hearing impairment has been identified in the individual with mutation X315Y at age 19 mo. Auditory brainstem responses (ABR), conditioned orientation reflex audiometry, and pure tone audiometry examinations showed bilateral moderate sensorineural hearing loss (average 50–60 dB) affecting all frequencies by a down-sloping shaped audiogram pattern. Hearing loss progressed from moderate to severe (70 dB), bilaterally at age 13 years, and subsequent Carolic test has shown normal vestibular function. The age at diagnosis of hearing loss in affected members with the other K314T missense mutation has been documented as being 1 year. The K314T case showed bilateral severe (80–90 dB) sensorineural hearing loss affecting all frequencies with no progression. Temporal bone CT scan was performed for all affected individuals carrying a CRYM mutation, but no abnormal findings were observed.
Subcellular Localization of Mutant Proteins
To examine the consequences of CRYM mutations, we expressed HA-tagged wild-type CRYM protein, as well as variants corresponding to each of the two mutations in COS-7 cells. Expression of all three proteins was confirmed by western blotting of protein extracts from transfected cells with mouse anti-HA antibodies. Fluorescent microscopy revealed diffuse cytoplasmic staining in cells transfected with the wild-type expression construct, and, 24 h and 48 h after transfection, the subcellular localizations of both mutant proteins were similar to the distribution of wild-type protein. However, 72 h and 96 h after transfection, we observed different patterns in significant proportions of transfected cells (fig. 3); the wild-type protein still revealed diffuse cytoplasmic distribution, but nearly 20% of the cells expressing the X315Y mutant revealed a vacuolation pattern in the cytoplasm, and approximately one-fourth of those expressing the K314T mutant showed strong staining, predominantly in perinuclear regions. The aberrant staining patterns were observed in almost none of the cells expressing wild-type protein.
Figure 3.
Fluorescent images of COS-7 cells expressing HA-tagged CRYM proteins. Nuclei are counterstained with DAPI (blue). Cells representative of six independent experiments are shown at a magnification of 60×. Subsequently, the numbers of cells were counted in 8–10 randomly selected fields (60×) using fluorescence microscopy (Nikon) at 24 h, 48 h, 72 h, and 96 h, respectively. A, Wild-type (WT) CRYM protein (green) localizes to cytoplasm diffusely throughout the time course of the experiments. B, The X315Y mutant with an extended C-terminal tail leads to vacuolated cytoplasmic distribution. The K314T mutant protein is localized predominantly in the perinuclear area.
Expression Analysis of Crym in the Mouse Cochlea
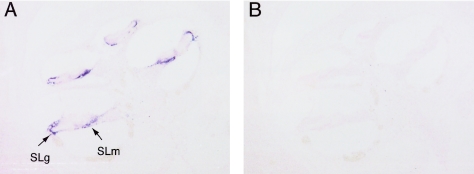
To study a possible biological function of CRYM, we cloned mouse Crym cDNA and performed an in situ hybridization experiment using mouse tissue. As shown in fig. 4A, hybridization signals with the Crym probe were observed in the lateral fibrocytes of the spiral ligaments (SLg) and the fibrocytes of the spiral limbus (SLm), whereas no in situ hybridization signal was detected with the control sense RNA probe (fig. 4B). Crym expression was not detected in neurosensory epithelium such as inner and outer hair cells.
Figure 4.
In situ hybridization analysis of Crym expression. The mouse cochlea reveals distinct labeling by the Crym antisense probe (arrows). Crym mRNA expression in the fibrocytes of spiral ligaments (SLg) and the fibrocytes of spiral limbus (SLm) in all turns of the mouse cochlea. Control hybridization was performed with a sense probe on consecutive tissue sections.
Discussion
To explore molecules essential for auditory function and to isolate genes responsible for deafness, we analyzed expression profiles of human inner-ear tissues on a cDNA microarray. The cochlea (for sound detection) and vestibule (for balance) have the same origin in the developmental process and possess similar properties both structurally and physiologically. We identified a set of 52 genes that were specifically or preferentially expressed in the inner ear and are investigating their possible roles in patients with nonsyndromic deafness. Among nine known nonsyndromic deafness genes that were present on our microarray, seven were confirmed to be expressed in the inner ear by our microarray analysis. Four of them, COCH, GJB2, GJB6, and USH1C, showed high Cy3/Cy5 ratios (>10) as compared with other tissues, indicating specific expression in the inner ear; one additional gene, MYO7A, revealed a relatively high Cy3/Cy5 ratio (6.20) in the cochlea. These results indicated that screening genes that are highly expressed in the inner ear might be an efficient approach to identifying aberrant molecules involved in deafness.
By screening DNA from a panel of patients with nonsyndromic deafness for mutations in CRYM, a gene that was especially abundant in the inner ear, we found possible disease-causing mutations in two cases. In one family, only the proband carried a mutation (X315Y), indicating that this change had arisen de novo in the affected individual, adding five amino acids at the C-terminal end of the CRYM protein. Mutations of a similar type have been reported as causes of some other hereditary conditions (Arlt et al. 1994; Marr et al. 2002). We underscored the deafness-causing potential of the X315Y mutation by demonstrating aberrant subcellular localization of the mutant protein.
The other mutation caused a nonconservative amino acid change of lysine to threonine at codon 314. Since the lysine residue at this position is conserved across mammalian species—including human, mouse, rat, and marsupial—according to alignments of human CRYM sequence with those of other mammals (human μ-crystallin [GenBank accession number Q14894], mouse μ-crystallin [GenBank accession number NP_057878], rat μ-crystallin [GenBank accession number NP_446407] and kangaroo μ-crystallin [GenBank accession number Q28488]), we assume that the K314T mutation would introduce a significant change in protein structure. Aberrant intracellular localization of the corresponding mutant protein supported the importance of this residue in human CRYM. Our findings imply that both mutations at the C-terminal end are likely to have deleterious effects on CRYM function. Although the precise mechanisms of these mutational effects remain to be determined, we consider that both of the heterozygous mutations we detected in the CRYM gene in deaf patients are likely to have affected auditory function, probably in a dominant-negative manner.
CRYM is one of the taxon-specific crystallins and is also called “μ-crystallin.” It was first identified as a major structural protein of the ocular lens in some marsupials (Kim et al. 1992). CRYM mRNA expression in fetal cochlea has been also demonstrated in the Morton Cochlear EST Database (GenBank accession number N73414). Although expression of CRYM is not absolutely specific to the inner ear, our microarray results indicate that its product is extremely abundant in the cochlea and vestibule. Initially, the predicted amino acid sequence showed similarity to the ornithine cyclodeaminase (OCD) and to glutamyl-tRNA reductase (gluTR) of bacteria and plants; both of these proteins represent a diverse superfamily of enzyme. The similarity suggested to some that μ-crystallin might perform an enzymatic rather than a structural role in lens tissue (Kim et al. 1992; Segovia et al. 1997). However, now it is thought to be a cytosolic NADP-regulated thyroid hormone-binding protein (THBP), a member of a group of molecular entities responsible for most of the intracellular high-affinity binding of T3 and T4 (Vie et al. 1997). THBPs are involved in sequestration and release of intracellular thyroid hormones (homeostasis). Whereas the cytosolic binding sites for T3 and T4 are similar to those of thyroid hormone receptors (TRs), the binding activity of THBPs is 100 times greater than that of TRs (Vie et al. 1997). Therefore, we speculate that mutant CRYM could abrogate the affinity of thyroid hormone, an essential agent for development of the auditory system. Cochlear structures are extremely vulnerable to thyroid-hormone deficiency during critical developmental periods; such deficiency results in defective morphological differentiation and maturation in the organ of Corti (Uziel 1986).
In the mouse, Crym was expressed in the cochlea and uticle at P2 and P32 stages (Corey Lab Inner Ear Gene Expression Database). We detected localization of Crym mRNA in two distinct tissues of mouse cochlea; the lateral fibrocytes of the spiral ligaments and the spiral limbus fibrocytes (fig. 4A). The postulated roles for these cells are thought to be K+ circulation/K+ recycling (Spicer and Schulte 1996; Steel and Kros 2001), suggesting that CRYM dysfunction may be interfering with potassium ion recycling, thus disturbing its maintenance of K+ rich endolymph and a positive electrical potential. We hypothesize that dysfunction of μ-crystallin may cause critical morphogenetic abnormalities of the labyrinth induced from thyroid-hormone deficiency and synergic auditory dysfunction involved in ionic homeostasis.
In conclusion, our results demonstrate that mutations of CRYM can be responsible for nonsyndromic deafness and that using the cDNA microarray approach to identify genes expressed specifically in the inner ear may be an efficient means of determining other good candidates for involvement in nonsyndromic deafness.
Acknowledgments
We would like to thank all of the families for their contribution to this study. This work was supported in part by Research for the Future Program Grant #00L01402 from the Japan Society for the Promotion of Science.
Footnotes
Nucleotide sequence data reported herein are available in the DDBJ/EMBL/GenBank databases; for details, see the Electronic-Database Information section of this article.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Corey Lab Inner Ear Gene Expression Database http://www.mgh.harvard.edu/depts/coreylab/index.html [DOI] [PMC free article] [PubMed]
- Database of Japanese Single Nucleotide Polymorphisms, A, http://snp.ims.u-tokyo.ac.jp/
- GenBank, http://www.ncbi.nlm.nih.gov/GenBank/ (for human μ-crystallin [accession number Q14894], mouse μ-crystallin [accession number NP_057878], rat μ-crystallin [accession number NP_446407], kangaroo μ-crystallin [accession number Q28488], human CRYM cDNA [accession number NM_001888], mouse Crym cDNA [accession number NM_016669], BAC sequence [accession number AF001550], and a Morton fetal cochlea EST [accession number N73414])
- Hereditary Hearing Loss Homepage, http://www.uia.ac.be/dnalab/hhh/
- Morton Cochlear EST Database, http://hearing.bwh.harvard.edu/estinfo.htm
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for GJB2 [MIM 121011], 12S rRNA [MIM 561000], CRYM [MIM 123740], DFNA4 [MIM 600652], DFNA7 [MIM 601412], DFNB15 [MIM 601869], DFNB33 [MIM 607239], COCH [MIM 603196], GJB6 [MIM 604418], MYO7A [MIM 276903], USH1C [MIM 605242], MYH9 [MIM 160775], MYO6 [MIM 600970], CLDN14 [MIM 605608], POU3F4 [MIM 300039], OTOR [MIM 606067], DFNA9 [MIM 601369], DFNB1 [MIM 220290], DFNA3 [MIM 601544], deafness with skin disorders [MIM 148350], DFNB18 [MIM 602092], Usher syndrome type 1C [MIM 605242], DFNB2 [MIM 600060], Usher syndrome type 1B [MIM 276903], DFNA17 [MIM 603622], Epstein and Fechtner syndrome [MIM 153650], DFNA22 [MIM 606344], DFNB29 [MIM 60568], and DFN3 [MIM 304400])
- Table of Gene Expression in the Developing Ear, http://www.ihr.mrc.ac.uk/hereditary/genetable/index.shtml
References
- Abe S, Usami S, Shinkawa H, Kelley PM, Kimberling WJ (2000) Prevalent connexin 26 gene (GJB2) mutations in Japanese. J Med Genet 37:41–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt G, Brooks DA, Isbrandt D, Hopwood JJ, Bielicki J, Bradford TM, Bindloss-Petherbridge CA, von Figura K, Peters C (1994) Juvenile form of mucopolysaccharidosis VI (Maroteaux-Lamy syndrome): a C-terminal extension causes instability but increases catalytic efficiency of arylsulfatase. B J Biol Chem 269:9638–9643 [PubMed] [Google Scholar]
- Hoshino M, Sone M, Fukata M, Kuroda S, Kaibuchi K, Nabeshima Y, Hama C (1999) Identification of the stef gene that encodes a novel guanine nucleotide exchange factor specific for Rac1. J Biol Chem 274:17837–17844 [DOI] [PubMed] [Google Scholar]
- Kim RY, Gasser R, Wistow GJ (1992) μ-crystallin is a mammalian homologue of Agrobacterium ornithine cyclodeaminase and is expressed in human retina. Proc Natl Acad Sci USA 89:9292–9296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Salunga RC, Guo H, Bittner A, Joy KC, Galindo JE, Xiao H, Rogers KE, Wan JS, Jackson MR, Erlander MG (1999) Gene expression profiles of laser-captured adjacent neuronal subtype. Nat Med 5:117–122 [DOI] [PubMed] [Google Scholar]
- Marazita ML, Ploughman LM, Rawlings B, Remington E, Arnos KS, Nance WE (1993) Genetic epidemiological studies of early-onset deafness in the U.S. school-age population. Am J Med Genet 46:486–491 [DOI] [PubMed] [Google Scholar]
- Marr N, Bichet DG, Lonergan M, Arthus MF, Jeck N, Seyberth HW, Rosenthal W, van Os CH, Oksche A, Deen PM (2002) Heteroligomerization of an Aquaporin-2 mutant with wild-type Aquaporin-2 and their misrouting to late endosomes/lysosomes explains dominant nephrogenic diabetes insipidus. Hum Mol Genet 11:779–789 [DOI] [PubMed] [Google Scholar]
- Ono K, Tanaka T, Tsunoda T, Kitahara O, Kihara C, Okamoto A, Ochiai K, Takagi T, Nakamura Y (2000) Identification by cDNA microarray of genes involved in ovarian carcinogenesis. Cancer Res 60:5007–5011 [PubMed] [Google Scholar]
- Robertson NG, Khetarpal U, Gutierrez-Espeleta GA, Bieber FR, Morton CC (1994) Isolation of novel and known genes from a human fetal cochlear cDNA library using subtractive hybridization and differential screening. Genomics 23:42–50 [DOI] [PubMed] [Google Scholar]
- Saito-Hisaminato A, Katagiri T, Kakiuchi S, Nakamura T, Tsunoda T, Nakamura Y (2002) Genome-wide profiling of gene expression in 29 normal human tissues with a cDNA microarray. DNA Res 9:35–45 [DOI] [PubMed] [Google Scholar]
- Segovia L, Horwitz J, Gasser R, Wistow G (1997) Two roles for μ-crystallin: a lens structural protein in diurnal marsupials and a possible enzyme in mammalian retinas. Mol Vision 3:9 [PubMed] [Google Scholar]
- Simmler MC, Cohen-Salmon M, El-Amraoui A, Guillaud L, Benichou JC, Petit C, Panthier JJ (2000) Targeted disruption of otog results in deafness and severe imbalance. Nat Genet 24:139–143 [DOI] [PubMed] [Google Scholar]
- Spicer SS, Schulte BA (1996) The fine structure of spiral ligament cells relates to ion return to the stria and varies with place-frequency. Hear Res 100:80–100 [DOI] [PubMed] [Google Scholar]
- Steel KP, Kros CJ (2001) A genetic approach to understanding auditory function. Nat Genet 27:143–149 [DOI] [PubMed] [Google Scholar]
- Tsujikawa M, Kurahashi H, Tanaka T, Nishida K, Shimomura Y, Tano Y, Nakamura Y (1999) Identification of the gene responsible for gelatinous drop-like corneal dystrophy. Nat Genet 21:420–423 [DOI] [PubMed] [Google Scholar]
- Usami S, Abe S, Akita J, Namba A, Shinkawa H, Ishii M, Iwasaki S, Hoshino T, Ito J, Doi K, Kubo T, Nakagawa T, Komiyama S, Tono T, Komune S (2000) Prevalence of mitochondrial gene mutations among hearing impaired patients. J Med Genet 37:38–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel A (1986) Periods of sensitivity to thyroid hormone during the development of the organ of Corti. Acta Otolaryngol Suppl 429:23–27 [DOI] [PubMed] [Google Scholar]
- Verpy E, Leibovici M, Zwaenepoel I, Liu XZ, Gal A, Salem N, Mansour A, Blanchard S, Kobayashi I, Keats BJ, Slim R, Petit C (2000) A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nat Genet 26:51–55 [DOI] [PubMed] [Google Scholar]
- Vie MP, Evrard C, Osty J, Breton-Gilet A, Blanchet P, Pomerance M, Rouget P, Francon J, Blondeau JP (1997) Purification, molecular cloning, and functional expression of the human nicodinamide-adenine dinucleotide phosphate-regulated thyroid hormone-binding protein. Mol Endocrinol 11:1728–1736 [DOI] [PubMed] [Google Scholar]
- Yasunaga S, Grati M, Cohen-Salmon M, El-Amraoui A, Mustapha M, Salem N, El-Zir E, Loiselet J, Petit C (1999) A mutation in OTOF, encoding otoferlin, a FER-1-like protein, causes DFNB9, a nonsyndromic form of deafness. Nat Genet 21:363–369 [DOI] [PubMed] [Google Scholar]
- Zuo J, Rarey KE (1996) Responsiveness of α 1 and β 1 cochlear Na, K-ATPase isoforms to thyroid hormone. Acta Otolaryngol 116:422–428 [DOI] [PubMed] [Google Scholar]