Abstract
Background
Francisella tularensis, the causative agent of tularemia, is a zoonotic agent that remains across much of the northern hemisphere, where it exists in enzootic cycles. In Ukraine, tularemia has a long history that suggests a need for sustained surveillance in natural foci. To better characterize the host-vector diversity and spatial distribution of tularemia, we analyzed historical data from field collections carried out from 1941 to 2008.
Findings
We analyzed the spatial-temporal distribution of bacterial isolates collected from field samples. Isolates were characterized by source and dominant land cover type. To identify environmental persistence and spatial variation in the source of isolation, we used the space-time permutation and multinomial models in SaTScan. A total of 3,086 positive isolates were taken from 1,084 geographic locations. Isolation of F. tularensis was more frequent among arthropods [n = 2,045 (66.3%)] followed by mammals [n = 619 (20.1%)], water [n = 393 (12.7%)], and farm produce [n = 29 (0.94%)], respectively. Four areas of persistent bacterial isolation were identified. Water and farm produce as sources of bacterial isolation were clustered.
Conclusions
Our findings confirm the presence of long-standing natural foci of F. tularensis in Ukraine. Given the history of tularemia as well as its environmental persistence there exists a possibility of (re)emergence in human populations. Heterogeneity in the distribution of tularemia isolate recovery related to land cover type supports the theory of natural nidality and clusters identify areas to target potential sources of the pathogen and improve surveillance.
Electronic supplementary material
The online version of this article (doi:10.1186/s13071-014-0453-2) contains supplementary material, which is available to authorized users.
Keywords: Tularemia, SaTScan, Spatial clusters, Ukraine, Land cover, Francisella tularensis
Findings
Francisella tularensis, the causative agent of tularemia, is a zoonotic, gram-negative bacterium that is broadly distributed across the Northern Hemisphere, with most human cases caused by either F. tularensis tularensis or F.t. holarctica, with cases from the latter less severe [1]. Generally, F.t. holarctica is most common across Europe, though F.t. tularensis has been recovered in central Europe [2]. Human exposure may occur through various pathways including arthropod bites, ingesting contaminated food products or liquids, inhaling aerosolized bacteria, or handling infected animals [1].
Despite a global decline in reported human cases [3], tularemia has recently (re)emerged in several countries including Sweden, Kosovo, China, Bulgaria [4] and parts of the former Soviet Union (FSU) [5]. Historically, outbreaks in the FSU were linked to small mammals and arthropods (ticks), possibly related to increases in host or vector population abundance [6,7]. More recent waterborne outbreaks of tularemia in Bulgaria [8] and Georgia [9], have reinforced the need for continued surveillance and preventative measures in endemic areas. In Ukraine, the first confirmed cases of tularemia were documented in the 1940's [4,10]. Those cases were associated with occupational exposure in furriers, whereas subsequent outbreaks were linked to rodent populations attracted to sugar factory production [4,10]. Tularemia foci were previously described in the 1960's across a limited geography in the south of Ukraine where several arthropods and small mammals were recognized as competent vectors and hosts [4,11]. However, contemporary characterizations of the spatial distribution and composition of vectors and hosts are incomplete and should be further analyzed.
Active disease surveillance is expensive and labor intensive. The use of historical data can direct such efforts for more efficient allocation of resources. Recent studies have shown that incorporating historical data and spatial analysis can improve vector-borne and zoonotic disease surveillance [12-15]. This study describes the spatial-temporal characteristics of tularemia across Ukraine, as well variation in its hosts and vectors to better understand its distribution, epidemiology, and improve surveillance.
From 1941 to 2008, the Central Sanitary Epidemiological Station (CSES) in Ukraine collected field samples nationally to test for F. tularensis. Confirmation of F. tularensis was performed via traditional bacterial culturing techniques [16]. This isolate collection included positive cultures from mammals, arthropods, and environmental sources, as well as ancillary information on the date and location of sample collections. We mapped F. tularensis positive isolates by year and by source of bacterial recovery. For this study, the historical database did not differentiate subspecies so we refer broadly to isolates as F. tularensis. Land cover (LC) characteristics were derived from the contemporary Globcover LC dataset [17] and assigned to each isolate. GlobCover data were reclassified into five broad categories: rain fed croplands, mosaic croplands, broadleaved forest, grass and shrub lands, and other (water, bare areas, and urban) (Additional file 1: Figure S1). Mapping was performed using ArcGIS v10.1 (ESRI, Redlands, CA).
We tested for space-time clustering of F. tularensis isolates using the retrospective space-time scan statistic in SaTScan™ v9.0 [18] with the space-time permutation model. Significant clusters represented geographic tularemia foci (defined as persistent bacterial isolation over periods >1 year). The space-time permutation model was most appropriate as the isolate database only recorded isolates and not the total field collection effort per isolate. The space-time permutation is described in detail elsewhere [19]. Briefly, the test creates multiple varying sized ‘cylinders’ around each case, where the circular base represents space and the cylinder height represents time. To determine if presence-only data are clustered in space and time, the number of cases in a cylinder is compared to case expectations outside of the cylinder. The model was run using year of isolation with the maximum spatial and temporal windows set to 50% of the population and 50% of the study period. Statistical significance of clusters was evaluated through Monte Carlo simulations, generating 999 random permutations to obtain p-values and selecting clusters with p-values ≤ 0.05, which were overlain with historical foci documented in Pollitzer [20]. A second categorical multinomial model was run in SaTScan, using the aforementioned parameter settings, to test for the presence of space-time clustering among isolate sources grouped as: mammals, arthropods, farm produce, and water. This test identifies geographic areas (here isolate locations) with an increased occurrence of a particular isolate source. To test for differences in LC characteristics, we calculated the proportion of isolates identified as space-time clusters (within foci) and non-clusters within each of the five Globcover-defined LC classes were compared using a chi-square analysis.
From 1941-2008, 3,086 positive isolates of F. tularensis were recovered from samples collected at 1,084 locations. Figure 1 illustrates the spatial distribution of isolates by decade and source of isolation. The fewest number of isolates were obtained during the 1940's (n = 18). The greatest number were recorded during the 1970's (n = 840). Arthropods were the most common source [n = 2,045 (66.3%)] followed by mammals [n = 619 (20.1%)], water [n = 393 (12.7%)], and farm produce [n = 29 (0.94%),] (Additional file 1: Figure S1). The number of isolates per taxa are shown in Figure 2. F. tularensis was most frequently isolated from Dermacentor spp. ticks (29.7%) and Microtus spp. rodents (4.8%), respectively. LC characteristics of the isolates showed a large number were collected within rain-fed croplands [n = 542 (49.1%)], with category 'other' comprising the highest percentage in relation to total available LC (Additional file 2: Figure S2).
Figure 1.
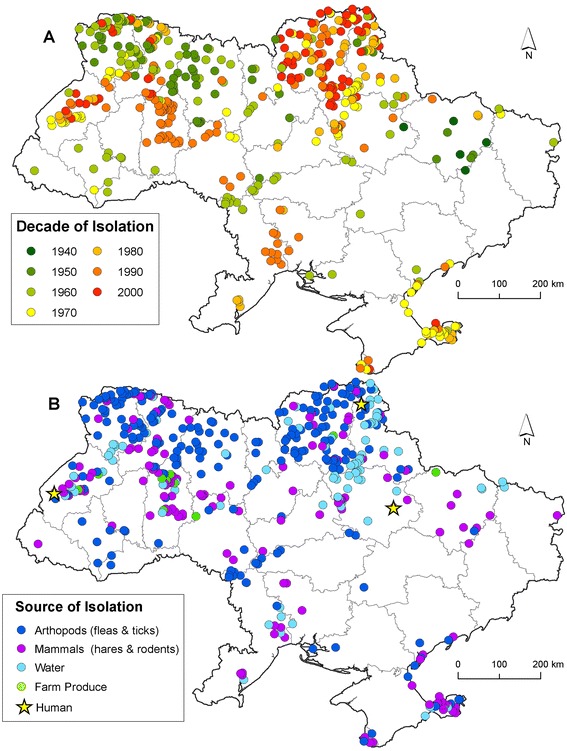
Map inset A shows the distribution of Francisella tularensis isolates collected by decade. Map inset B shows the distribution of isolates by source.
Figure 2.
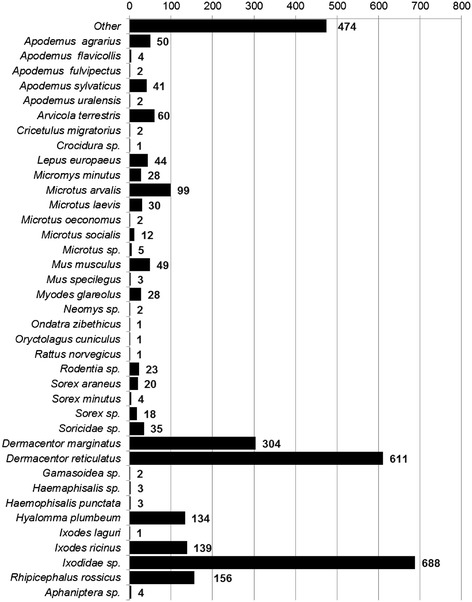
Source of Francisella tularensis isolation by genus and species.
Four tularemia foci (areas of persistent bacterial isolation) were identified by the space-time statistic (Table 1). Foci ranged in duration two years to fourteen years (Figure 3A). The proportion of isolates within the SaTScan defined foci across five LC classes was significantly different from the proportion of isolates within non-foci (χ2 = 48.72, DF = 4, p < 0.001). The Multinomial SatScan model showed that during the period 1971-2000, there was greater occurrence of isolation among the water and farm produce categories, displayed in yellow, whereas during the earlier period 1962-1977 an isolation among arthropods was higher, displayed in light blue (Figure 3B).
Table 1.
Results of the SaTScan space-time permutation analyses identifying foci of Francisella tularensis that persisted > 1 year
| Cluster (Foci) | Isolates | Expected | Test Statistic | p-Value |
|---|---|---|---|---|
| 1 | 217 | 32.69 | 232.12 | 0.001 |
| 2 | 105 | 8.32 | 171.09 | 0.001 |
| 3 | 999 | 571.9 | 168.66 | 0.001 |
| 4 | 26 | 0.45 | 79.82 | 0.001 |
Figure 3.
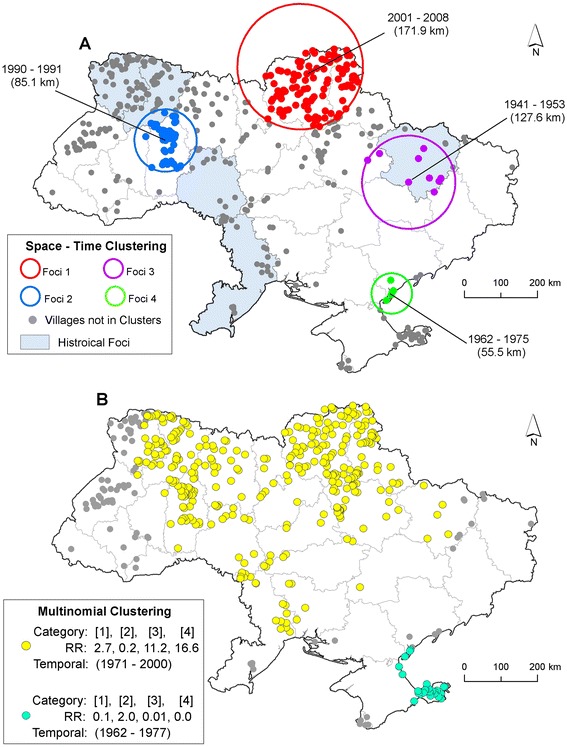
Spatial clusters of Francisella tularensis isolates in Ukraine. Map inset A displays the results of the SaTScan space-time analysis of all Francisella tularensis isolates identifying foci of isolation. Results are overlain with Historical foci documented in Pollitzer 1967 [18]. Circles represent the spatial extent of a given cluster in kilometers (km). Time periods indicate the duration of the cluster in years. Map inset B displays the results of the multinomial cluster model. Categories in brackets refer to the source of isolation: 1 = mammals, 2 = arthropods, 3 = water, and 4 = farm produce. Relative risk (RR) estimates below the brackets indicate whether or not greater than expected number of isolations occurred in a given category. RR >1 represent a greater than expected number of isolations.
Our findings support evidence of long-standing natural foci of F. tularensis in Ukraine. Heterogeneity in the recovery of bacterial isolates by geographic location support the theory of natural nidality [21]. Our findings confirmed observations from the 1960's that documented foci in western Ukraine [20]. We also identified foci in the southeast of the country and elucidated areas of bacterial isolation that persisted through last decade (2000's). The distribution of foci, vectors, hosts, and infectious agents are known to be influenced by ecological characteristics [22]; consistent with this association, our analysis indicated differences in LC among areas identified as foci and non-foci.
Foci in the FSU were historically classified by ecotypes/LC (steppe, forest, foothills) with the most ubiquitous type consisting of flood/marshlands [20,23], associated with areas of sugar beet production and mixed agriculture use [5]. In keeping with previous research we noted that the recovery of F. tularensis in Ukraine occurred primarily in croplands followed by forests, shrub/grasslands (steppe), and water. Interestingly, we found variation in the space-time distribution of the sources of bacterial recovery indicating a greater than expected number of isolates in mammals, water, and farm produce during 1971-2000 (Figure 3B). Farm produce and water can become contaminated by infected mammals, particularly semi-aquatic rodents (Arvicola terrestris), which have been linked to human outbreaks from agriculture and sugar factories [5,20]. This cluster may have indicated an increased abundance in rodents or a more concerted sampling effort brought on by concern over water-borne outbreaks in humans, often the source of epidemics [7-9,24]. Additionally, a cluster of arthropod isolates during 1962-1977 was identified in the southeast where, in some areas, the vector D. marginatus was historically shown to have bacterial recovery rates of 2.5% and played an important role in human transmission and maintaining the enzootic cycle of the pathogen [25]. This cluster is in contrast to the ecotype in western Ukraine, historically classified as steppe [5,20,23] and supports the hypotheses of geographic variation in the epidemiology of human tularemia [5,20,25,26].
Differences in the sampling efforts conducted year to year may partly account for the patterns identified here. Although recovery rates were not available for our data, we incorporated the SaTScan methodology to successfully identify foci and persistence of bacterial isolation due its flexibility in dealing with missing data. Despite the lack of historical human case data, sampling efforts may have been driven by human outbreaks, as were previously documented across Ukraine [25]. High resolution human disease data from the FSU are extremely difficult to obtain. This fact coupled with mass vaccination campaigns and the presence of the less virulent F. tularensis holarcitica likely contributed to an underestimation in reporting [5,27]. While bacterial recovery rates were not available for the entire data set, limited field collections from the 1950's and 60's showed recovery of F. tularensis was 0.2% (265/199,343) from the ticks collected [25]. The contemporary GlobCover database may have misclassified LC characteristics of historical isolates. As a first effort, we calculated the percent change in cropland by decade from 1940 – 2000 using HYDE [28] (Additional file 3; Additional file 4: Figure S3). Overall, there was a slight increase in cropland across the decades.
Disease surveillance is costly and time consuming especially when monitoring is conducted with limited empirical or historical data to identify priority areas. Given the challenges faced by newly independent FSU states, limited public health resources make such monitoring infeasible. This study showed that archived, historical records provide important clues to identify important vectors, hosts, environmental sources, and geographic foci for F. tularensis. The persistence of environmental foci in Ukraine gives rise to the possibility of (re)emergence in human populations. Our findings can be used to inform more efficient targeted surveillance strategies and monitoring of tularemia foci. The recurrent isolation of F. tularensis from several sources (wild rodents, croplands and water) points to the need for continued surveillance. The LC analysis suggests that areas of mixed agriculture (croplands) should be prioritized as areas for future surveillance, with an emphasis on rodent and tick species defined here.
Acknowledgements
This work was funded by the US Defense Threat Reduction Agency through the Cooperative Biological Engagement Program in Ukraine. V. Svyta, Y. Novohatnyi, O. Gubar assisted with the GIS develop and database management. A. Zykov provided a taxonomic review of host and vector species.
Abbreviations
- CSES
Central sanitary epidemiological station
- GIS
Geographic information systems
- FSU
Former soviet union
- LC
Land cover
Additional files
Land cover categories in Ukraine based on the GlobCover dataset and reclassified into five categories: rain-fed croplands, mosaic croplands, broadleaved forests, grass/shrub lands, and other.
The top panel shows the proportion of the Francisella tularensis isolates that fell within each of the five Land cover classes. The bottom panel shows the percentage of the LC in Ukraine by category.
Supplemental information.
Graph illustrates the total area of cropland Land cover (LC) type in Ukraine by decade based on the HYDE historical database. Percent values above bars represent the change in cropland LC for each decade using 2000 as the reference.
Footnotes
Jake Hightower and Ian T Kracalik contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JKB, JH, ITK conceived the study. JKB, NV, and JH developed the database and GIS from CSES records. JH, ITK, JKB, DGG performed the analyses. ITK, JH, GBG, JKB wrote the manuscript. All others reviewed the final draft. All authors read and approved the final manuscript.
Contributor Information
Jake Hightower, Email: jhightower@ufl.edu.
Ian T Kracalik, Email: itk@ufl.edu.
Nataliya Vydayko, Email: vydaykon@ukr.net.
Douglas Goodin, Email: dgoodin@k-state.edu.
Gregory Glass, Email: ggurrigl@jhsph.edu.
Jason K Blackburn, Email: jkblackburn@ufl.edu.
References
- 1.Staples JE, Kubota KA, Chalcraft LG, Mead PS, Petersen JM. Epidemiologic and molecular analysis of human tularemia, United States, 1964–2004. Emerg Infect Dis. 2006;12(7):1113–1118. doi: 10.3201/eid1207.051504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johansson A, Farlow J, Larsson P, Dukerich M, Chambers E, Byström M, Fox J, Chu M, Forsman M, Sjöstedt A. Worldwide genetic relationships among Francisella tularensis isolates determined by multiple-locus variable-number tandem repeat analysis. J Bacteriol. 2004;186(17):5808–5818. doi: 10.1128/JB.186.17.5808-5818.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charette JD. CDC Category explanation (A, B, C) overview. Toxico-Terrorism: emergency response and clinical approach to chemical, biological, and radiological agents. 2007;755:329. [Google Scholar]
- 4.Sjöstedt A. Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann N Y Acad Sci. 2007;1105(1):1–29. doi: 10.1196/annals.1409.009. [DOI] [PubMed] [Google Scholar]
- 5.Pollitzer R: A Review of selected problems of tularemia in the soviet union. Part Ii/A. Vaccines and Vaccination. In New York, NY: Fordham University; 1963.
- 6.Reintjes R, Dedushaj I, Gjini A, Jorgensen TR, Cotter B, Lieftucht A, D’Ancona F, Dennis DT, Kosoy MA, Mulliqi-Osmani G. Tularemia outbreak investigation in Kosovo: case control and environmental studies. Emerg Infect Dis. 2002;8(1):69. doi: 10.3201/eid0801.010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anda P, Del Pozo JS, Garcia JD, Escudero R, Pena FG, Velasco ML, Sellek RE, Chillarón MJ, Serrano LS, Navarro JM. Waterborne outbreak of tularemia associated with crayfish fishing. Emerg Infect Dis. 2001;7(3 Suppl):575. doi: 10.3201/eid0707.017740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kantardjiev T, Ivanov I, Velinov T, Padeshki P, Popov B, Nenova R, Mincheff M. Tularemia outbreak, Bulgaria, 1997–2005. Emerg Infect Dis. 2006;12(4):678. doi: 10.3201/eid1204.050709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chitadze N, Kuchuloria T, Clark D, Tsertsvadze E, Chokheli M, Tsertsvadze N, Trapaidze N, Lane A, Bakanidze L, Tsanava S. Water-borne outbreak of oropharyngeal and glandular tularemia in Georgia: investigation and follow-up. Infection. 2009;37(6):514–521. doi: 10.1007/s15010-009-8193-5. [DOI] [PubMed] [Google Scholar]
- 10.Černý Z. Changes of the epidemiology and the clinical picture of tularemia in Southern Moravia (the Czech Republic) during the period 1936–1999. Eur J Epidemiol. 2001;17(7):637–642. doi: 10.1023/A:1015551213151. [DOI] [PubMed] [Google Scholar]
- 11.Golkovskiĭ G, Mitsevich G, Khaĭtovich A, Alekseev E, Korchevskiĭ P: Natural focus of tularemia on the Kerchen peninsula (Crimea).Zh Mikrobiol Epidemiol Immunobiol 1981, ᅟ(10):99–101. [PubMed]
- 12.Kracalik I, Abdullayev R, Asadov K, Ismayilova R, Baghirova M, Ustun N, Shikhiyev M, Talibzade A, Blackburn JK. Changing patterns of human anthrax in azerbaijan during the post-soviet and preemptive livestock vaccination eras. PLoS Negl Trop Dis. 2014;8(7):e2985. doi: 10.1371/journal.pntd.0002985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kracalik I, Lukhnova L, Aikimbayev A, Pazilov Y, Temiralyeva G, Blackburn JK. Incorporating retrospective clustering into a prospective cusum methodology for anthrax: Evaluating the effects of disease expectation. Spat Spatiotemporal Epidemiol. 2011;2(1):11–21. doi: 10.1016/j.sste.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Kracalik IT, Malania L, Tsertsvadze N, Manvelyan J, Bakanidze L, Imnadze P, Tsanava S, Blackburn JK. Evidence of local persistence of human anthrax in the country of Georgia associated with environmental and anthropogenic factors. PLoS Negl Trop Dis. 2013;7(9):e2388. doi: 10.1371/journal.pntd.0002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris LR, Blackburn JK, Talibzade A, Kracalik I, Ismaylova R, Abdullahyev R. Informing surveillance for the lowland plague focus in Azerbaijan using a historic dataset. Appl Geogr. 2013;45:269–279. doi: 10.1016/j.apgeog.2013.09.014. [DOI] [Google Scholar]
- 16.WHO: World Health Organization Guidelines on Tularemia. In Geneva: World Health Organization; 2007:125.
- 17.Arino O, Bicheron P, Achard F, Latham J, Witt R, Weber JL. GlobCover: the most detailed picture of earth. ESA Bulletin-European Space Agency. 2008;136:24–31. [Google Scholar]
- 18.Kulldorff M: SaTScan user guide for version 9.0. In 2011.
- 19.Kulldorff M, Heffernan R, Hartman J, Assuncao R, Mostashari F. A space–time permutation scan statistic for disease outbreak detection. PLoS Med. 2005;2(3):e59. doi: 10.1371/journal.pmed.0020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollitzer R: History and incidence of tularemia in the Soviet Union: a review. In New York, NY: Institute of Contemporary Russian Studies, Fordham University Bronx; 1967.
- 21.Pavlovsky EN, Plous FK, Jr, Levine ND. Natural nidality of transmissible diseases. Am J Med Sci. 1966;252(5):161. doi: 10.1097/00000441-196611000-00067. [DOI] [Google Scholar]
- 22.Ostfeld RS, Glass GE, Keesing F. Spatial epidemiology: an emerging (or re-emerging) discipline. Trends Ecol Evol. 2005;20(6):328–336. doi: 10.1016/j.tree.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 23.Olsuf'ev N, Rudnev G: Tularemia. In Washington D.C: Joint Publications Research Services; 1962. "Translated into English".
- 24.Akalın H, Helvacı S, Gedikoğlu S. Re-emergence of tularemia in Turkey. Int J Infect Dis. 2009;13(5):547–551. doi: 10.1016/j.ijid.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 25.Stupnitskaya V, Marinov M, Litvinenko E, Slesarenko V, Slesarenko A: Natural tularemia foci on the territory of the Ukrainian SSR. In Fort Detrick, MD: Defense Technical Information Center; 1965.
- 26.Svensson K, Bäck E, Eliasson H, Berglund L, Granberg M, Karlsson L, Larsson P, Forsman M, Johansson A. Landscape epidemiology of tularemia outbreaks in Sweden. Emerg Infect Dis. 2009;15(12):1937–1947. doi: 10.3201/eid1512.090487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark DV, Ismailov A, Seyidova E, Hajiyeva A, Bakhishova S, Hajiyev H, Nuriyev T, Piraliyev S, Bagirov S, Aslanova A. Seroprevalence of tularemia in rural Azerbaijan. Vector Borne Zoonotic Dis. 2012;12(7):558–563. doi: 10.1089/vbz.2010.0081. [DOI] [PubMed] [Google Scholar]
- 28.Goldewijk KK. Estimating global land use change over the past 300 years: the HYDE database. Global Biogeochemical Cy. 2001;15(2):417–433. doi: 10.1029/1999GB001232. [DOI] [Google Scholar]


