Abstract
Invasive ductal carcinoma (IDC) and invasive lobular carcinoma (ILC) are the two major histological types of breast cancer worldwide. Whereas IDC incidence has remained stable, ILC is the most rapidly increasing breast cancer phenotype in the United States and Western Europe. It is not clear whether IDC and ILC represent molecularly distinct entities and what genes might be involved in the development of these two phenotypes. We conducted comprehensive gene expression profiling studies to address these questions. Total RNA from 21 ILCs, 38 IDCs, two lymph node metastases, and three normal tissues were amplified and hybridized to ∼42,000 clone cDNA microarrays. Data were analyzed using hierarchical clustering algorithms and statistical analyses that identify differentially expressed genes (significance analysis of microarrays) and minimal subsets of genes (prediction analysis for microarrays) that succinctly distinguish ILCs and IDCs. Eleven of 21 (52%) of the ILCs (“typical” ILCs) clustered together and displayed different gene expression profiles from IDCs, whereas the other ILCs (“ductal-like” ILCs) were distributed between different IDC subtypes. Many of the differentially expressed genes between ILCs and IDCs code for proteins involved in cell adhesion/motility, lipid/fatty acid transport and metabolism, immune/defense response, and electron transport. Many genes that distinguish typical and ductal-like ILCs are involved in regulation of cell growth and immune response. Our data strongly suggest that over half the ILCs differ from IDCs not only in histological and clinical features but also in global transcription programs. The remaining ILCs closely resemble IDCs in their transcription patterns. Further studies are needed to explore the differences between ILC molecular subtypes and to determine whether they require different therapeutic strategies.
INTRODUCTION
Invasive ductal carcinoma (IDC) and invasive lobular carcinoma (ILC) are the major histological types of invasive breast cancer among women of different races worldwide, ranging from 47 to 79% and 2 to 15%, respectively (Harris et al., 2000). Although histologically disparate, these tumor types show clinical similarities and differences. Characteristics such as tumor site, size, grade, and stage at presentation are similar for both types (Winchester et al., 1998). ILCs often present with subtler signs on physical examination and mammography due to their characteristic histology and absence of a sclerotic tissue reaction. In contrast to a mammographic mass, asymmetric density or architectural distortion are the predominant mammographic signs in more ILCs than IDCs, whereas malignant calcifications are less frequent in ILCs (Newstead et al., 1992). IDC and ILC are managed similarly, but whether overall survival rates of patients differ is controversial (Yeatman et al., 1995; Toikkanen et al., 1997; Winchester et al., 1998). However, the metastatic patterns of IDC and ILC are clearly different, with gastrointestinal, gynecologic, and peritoneal-retroperitoneal metastases, particularly to endocrine-related sites such as adrenal glands and ovaries, markedly more prevalent in ILCs (Dixon et al., 1991; Borst and Ingold, 1993; Bumpers et al., 1993; Sastre-Garau et al., 1996).
At the molecular level, IDC and ILC seem to show more differences than similarities. They differ in hormone receptor status with 55–72% of IDCs being estrogen receptor (ER) positive compared with 70–92% of ILCs, and 33–70% of IDCs being progesterone receptor (PR) positive compared with 63–67% of ILCs (Harris et al., 2000). A number of proteins have also been found to be differentially expressed in IDC and ILC, including vimentin, cathepsin D, thrombospondin, E-cadherin, vascular endothelial growth factor, cytokeratin 8, and cyclin A (Domagala et al., 1990; Bedner et al., 1995; Serre et al., 1995; Berx et al., 1996; Lee et al., 1998; Lehr et al., 2000; Coradini et al., 2002). In addition, differences in genetic alterations in IDC and ILC have been observed. Genes such as ERBB2 (Rosenthal et al., 2002) and p21 (Rey et al., 1998) show a markedly higher amplification rate in IDC than in ILC. In contrast, loss of chromosome 16q (site of the E-cadherin gene) is observed at a much higher frequency in ILC than in IDC (Serre et al., 1995; Cleton-Jansen, 2002), and particularly more frequent in ILC than poorly differentiated IDC (Buerger et al., 2000). However, IDC and ILC share certain characteristics in gene expression. Well differentiated IDC and ILC show similar expression of some proliferation and cell cycle regulated genes, including cyclin D1, p16, p27, mdm-2, and mib-1 (Geradts and Ingram, 2000; Soslow et al., 2000), and similar bcl-2 and HIF-1α expression (Coradini et al., 2002).
Recent research reported a disproportionate increase of ILCs in the United States and Europe, possibly associated with increased usage of combined hormone replacement therapy (Li et al., 2000a,b, 2003; Daling et al., 2002; Verkooijen et al., 2003). In the United States, ductal carcinoma incidence rates remained essentially constant from 1987 to 1999, whereas lobular carcinoma rates increased steadily, significantly increasing the proportion of breast cancer with a lobular component from 9.5 to 15.6% during that time period. In Switzerland, there has been a mean annual increase in the incidence of IDC of 1.2% compared with a mean annual increase of 14.4% for ILC during the period 1976–1999. Use of combined hormone replacement therapy, but not estrogen replacement therapy alone, seems to increase the risk of developing ILC by 2.7-fold, whereas the increase in IDC risk is only 1.5-fold (Li et al., 2003).
Because ILC is the most rapidly increasing breast cancer phenotype, more difficult to diagnose than IDC, and yet is treated similarly to IDC, it is imperative to determine whether the clinical treatment of ILC should differ from IDC. To individualize breast cancer treatment, a molecular understanding of the mechanisms that underlie the development of these two phenotypes is crucial.
It is not clear whether IDC and ILC represent molecularly distinct entities and what genes might be involved in the development of these two phenotypes. Traditional studies have focused on a small number of genes in a large number of cases, whereas microarray analysis has provided us a powerful tool to explore gene expression on a genome-wide scale. By using this technology, breast cancers have been molecularly classified into a number of subtypes associated with different clinical outcomes (Sorlie et al., 2001, 2003; West et al., 2001; Ahr et al., 2002; van't Veer et al., 2002). Sets of genes have been identified as signatures of each subtype and may be potentially useful in drug design and patient care (Perou et al., 2000; Ahr et al., 2001; van de Vijver et al., 2002). However, IDC has been the dominant histological subtype investigated in most of these studies.
To better understand the biology of the two predominate phenotypes of breast carcinoma at the molecular level, we conducted comprehensive studies on the gene expression profiles of IDC and ILC by using RNA amplification and cDNA microarray technology. A total of 64 breast tissues from American and Norwegian patients, including 21 ILCs, 38 IDCs, two lymph node metastases, and three normal tissues were analyzed on arrays containing >42,000 clones. Hierarchical clustering analysis (Eisen et al., 1998), significance analysis of microarrays (SAM) (Tusher et al., 2001), prediction analysis for microarrays (PAM) (Tibshirani et al., 2002), and Pearson's correlation analysis were used to address whether ILCs and IDCs are molecularly distinct entities, what are the similarities and differences in gene expression profiles between these two phenotypes of breast cancer, and whether there are molecularly distinct subtypes within ILCs.
MATERIALS AND METHODS
Description of Patient Material
We selected a total of 59 primary breast cancer cases of which 28 were from Stanford Hospital (BC samples) and 31 from a series of patients from Ullevål Hospital, Oslo (ULL samples) (Bukholm et al., 1997). Cases had been accrued in accordance with local institutional review board guidelines. Of these, 38 were IDCs and 21 were ILCs. The distribution of cases according to patient source, lymph node status, tumor grade, patient age, the expression of hormone receptors (ER and PR) and a prognostic marker ERBB2/HER2/neu, and the tumor component (the percentage of carcinoma cells on an adjacent frozen or permanent section of the solid tumor) are shown in Table 1. For complete details for each case, see Clinical and Pathology Parameters on the Web supplement at http://genome-www.stanford.edu/breast_cancer/lobular/. The IDCs and ILCs had similar tumor characteristics, except that almost all of the ILCs were grade II tumors and most were from patients >55 years of age at diagnosis. In addition, no HER2/neu-positive tumors were present in the 14 ILCs with known HER2/neu status, whereas 8 of 31 IDCs with known HER2/neu status had positive expression. Two lymph node metastases and three normal breast samples from five IDC patients were also included in the study.
Table 1.
Distribution of clinical and histopathological characteristics of cases included in the study
| IDC (N = 38) | ILC (N = 21) | ||
|---|---|---|---|
| Source | Stanford | 22 (58%) | 6 (29%)a* |
| Norwegian | 16 (42%) | 15 (71%)a | |
| Lymph node | Positive | 18 (53%) | 10 (59%) |
| Negative | 16 (47%) | 7 (41%) | |
| Unknown | 4 | 4 | |
| Grade | I | 5 (13%) | 1 (5%) |
| II | 20 (53%)b | 20 (95%)b | |
| III | 13 (34%)c | 0 (0%)c | |
| Age | ≥55 | 16 (44%)d | 16 (80%)d |
| <55 | 20 (56%)e | 4 (20%)e | |
| Unknown | 2 | 1 | |
| Tumor component | ≥60% | 33 (87%) | 13 (62%) |
| 40–60% | 5 (13%) | 8 (38%) | |
| Estrogen receptor | Positive | 24 (69%) | 14 (78%) |
| Negative | 11 (31%) | 4 (22%) | |
| Unknown | 3 | 3 | |
| Progesterone receptor | Positive | 23 (68%) | 12 (67%) |
| Negative | 11 (32%) | 6 (33%) | |
| Unknown | 4 | 3 | |
| HER2/neu | Positive | 8 (26%)f† | 0 (0%)f |
| Negative | 23 (74%)g | 14 (100%)g | |
| Unknown | 7 | 7 |
Paired labels (a–g) mark statistically significant different values (P value <0.05)
HER2/neu positive, 3+ expression by immunostain; fluorescence in situ hybridization not done on these samples
Tissue Acquisition and Histology Evaluation
Primary breast carcinomas were frozen in either liquid nitrogen or on dry ice within 20 min after devascularization and stored at -80°C. Frozen sections were cut from primary breast carcinoma specimens and stained with hematoxylin and eosin to confirm tumor content. Specimens in which at least 40% of the cells were carcinoma cells were used in this study. Two experienced breast pathologists separately reviewed, classified, and graded all tumor specimens according to the modified Scarff-Bloom-Richardson method (Elston and Ellis, 1993). Cases with discrepancies were reviewed together to obtain consensus. Details of tumor specimen histology are available on the Web at http://genome-www.stanford.edu/breast_cancer/lobular/.
RNA Preparation, Amplification, Labeling, and Hybridization
Total RNA was isolated from primary tumor tissue using by TRIzol solution (Invitrogen, Carlsbad, CA) after homogenization by using a PowerGen model 125 (Fisher Scientific. Pittsburgh, PA). The concentration of total RNA was determined using a GeneSpec I spectrophotometer (Hitachi, Yokohama, Japan), and the integrity of the RNA was assessed using a 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Amplification of total RNA was performed using an optimized protocol described previously (Zhao et al., 2002). Amplified tumor RNA was labeled by Cy5 and amplified RNA from Universal Human Reference total RNA (Stratagene, La Jolla, CA) was labeled by Cy3. The labeling and hybridization of amplified RNA to cDNA microarrays containing >42,000 elements, was performed as described previously (Zhao et al., 2002). Complete experimental protocols can be found at http://genome-www.stanford.edu/breast_cancer/lobular/ or http://www.stanford.edu/group/sjeffreylab/. Multiple batches of arrays were used in this study which did not seem to influence the sample distribution in hierarchical clustering analysis in any significant way. Details of the normalization of the intensity levels can be found at http://genome-www5.stanford.edu/help/results_normalization.shtml.
Imaging and Data Analysis
The arrays with hybridized probes were scanned using an Axon scanner. The scanned images were analyzed first using GenePix Pro3.0 software (Axon Instruments, Foster City, CA), and spots of poor quality determined by visual inspection were removed from further analysis. The resulting data collected from each array was submitted to the Stanford Microarray Database (SMD; http://genome-www5.stanford.edu/microarray/SMD) (Sherlock et al., 2001; Gollub et al., 2003). Only features with a regression correlation (among all pixels within a feature) >0.5 and a signal intensity >50% above background in both Cy5 and Cy3 channels were retrieved from SMD.
Hierarchical Clustering. A hierarchical clustering algorithm (Eisen et al., 1998) was applied to group genes and samples on the basis of their similarities in expression, and the results were visualized using TreeView software (http://rana.lbl.gov/EisenSoftware.htm). The first clustering was performed on all 64 samples by using 4539 clones representing 3341 genes whose expression varied at least threefold from the mean abundance across all samples in at least three samples and was measurable in at least 70% of the samples included in the analysis. The second clustering was performed on 59 IDCs and ILCs by using 78 clones representing 45 named genes identified in PAM analysis. The third clustering was performed on the 59 primary tumors by using 481 genes (represented by 548 clones) of 500 intrinsic genes identified previously (Perou et al., 2000; Sorlie et al., 2001, 2003) (see below) whose expression was measurable in at least 70% of the samples included in the analysis. The criteria for spot quality control and gene filtering before hierarchical clustering are somewhat arbitrary. However, prior work has shown that expression variations selected similarly reliably reflect changes in expression levels measured by other methods (Lossos et al., 2002; Chen et al., 2003).
SAM. Genes with potentially significant changes in expression between ILCs and IDCs were identified using the SAM procedure (Tusher et al., 2001) (http://www-stat.stanford.edu/~tibs/SAM/), which computes a two-sample t-statistic of ILCs and IDCs for the normalized log ratios of gene expression levels for each gene. It thresholds the t-statistics to provide a “significant” gene list and provides an estimate of the false discovery rate (the percentage of genes identified by chance alone) from randomly permuted data. We performed a SAM analysis on 32,345 clones representing 20,375 genes whose expression was measurable in at least 70% of the 59 primary tumors and filtered only by spot quality (regression correlation >0.5, signal intensity >50% above background in both Cy5 and Cy3 channels). We used a selection threshold giving the lowest median estimate of 0.6 false positive genes (false discovery rate of 0.1%).
PAM. PAM does sample classification using the nearest shrunken centroid method with automatic gene selection and cross-validation (Tibshirani et al., 2002). It uses a parameter threshold Δ to select genes for class prediction. The first PAM classification done to compare ILCs with IDCs was performed using the PAM for R package (http://www-stat.stanford.edu/~tibs/PAM/) on the 32,345 clones filtered as described above. We varied Δ to find a value that balanced prediction accuracy with the number of genes in the predictive model. A threshold of 2.9 giving the lowest overall error rate, and the minimum number of predictive genes was selected. Another PAM analysis was performed comparing typical ILCs and IDCs (details available on the Web supplement). A final PAM classification was performed comparing typical ILCs with ductal-like ILCs on 23,914 clones representing 15,281 genes whose expression was measurable in at least 80% of the 21 ILCs and filtered only by spot quality as described above. A threshold of 2.3 was chosen for the same reasons described above.
Pearson's Correlation Analysis Using Centroids. Previously, an “intrinsic” gene list had been selected consisting of 500 genes represented by 561 clones whose expression varied the least in successive samples from the same patient's tumor but which showed the most variation among tumors from different patients (Perou et al., 2000; Sorlie et al., 2001, 2003). Five sets of centroids (i.e., profiles, consisting of the average expression for the 500 intrinsic genes) corresponding to the five subtypes of breast carcinomas were recently published using data from 122 breast samples (Perou et al., 2000; Sorlie et al., 2001, 2003). In total, 455 of the centroid genes (represented by 484 clones) were measurable in at least 70% of the 59 primary tumors in our dataset. We then computed the Pearson's correlation coefficients (r) of each of our samples to each of these five sets of centroids. An r threshold of 0.14 (the 95th percentile) was generated by permutation of the expression values for each gene and computing the maximal correlation of each resulting sample with one of the original five centroids. This was repeated 10 times, and the 95th percentile of these correlations was used as the cutoff to categorize 56 out of 59 primary tumors into the five subtypes.
RESULTS
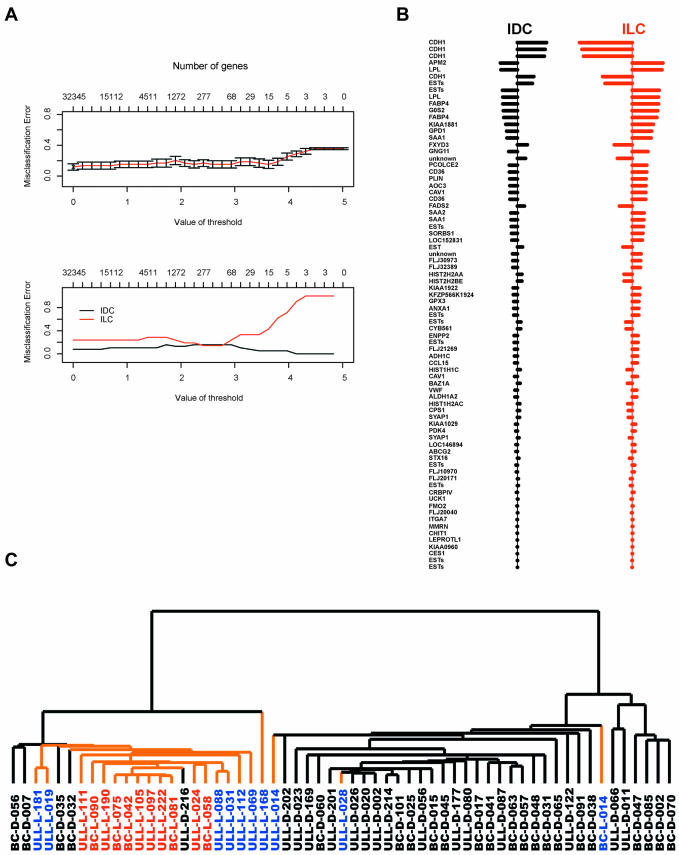
Hierarchical clustering of the 64 samples was performed using the selected 4539 clones representing 3341genes whose expression varied more than threefold from the overall mean abundance in at least three samples (Figure 1). In the dendrogram shown in Figure 1A, four distinct groups of tumors are apparent, suggesting that the tumors can be divided into four types on the basis of the 3341 differentially expressed genes. The association of tumors within this unsupervised cluster is not due to gene filtering criteria because varying data selection criteria still maintains the tumor associations. It also seems that the contents of tumor cells or the adipose and immune components have little influence on this cluster pattern (see Clinical and Pathology Parameters on the Web supplement at http://genome-www.stanford.edu/breast_cancer/lobular/). One striking feature is that 11 of 21 (52%) of the ILCs were found in group IV, which also contains three normal breast samples. This suggests that this group of ILCs is different in gene expression profile from IDCs and has more gene expression similarities with normal breast than IDCs. We refer to this group of ILCs as “typical” ILCs. A fraction of other ILCs share similar gene expression profiles with IDCs and are referred to as “ductal-like” ILCs. The relatedness of typical ILCs to normal samples is not likely due to the composition of the tumors because five of eight ILCs with relatively low percentage of tumor cells (40–60%) clustered elsewhere with IDCs. In addition, genes such as E-cadherin and basal epithelial cell markers (e.g., KRT5, KRT 17, and epidermal growth factor receptor [EGFR]) show significantly different expression levels in typical ILCs and normal samples (Figure 1, D, H, and I). It is also worth noticing that the two lymph node metastases clustered together with the primary tumors they derived from, consistent with our previous findings, suggesting a similar gene expression profile between primary tumor and lymph node metastasis. Each normal sample (derived from the same breast as a corresponding primary tumor but taken from a distant location) exhibited expression profiles similar to other normals (our unpublished data) and different from their corresponding IDC (Figure 1A).
Figure 1.
Unsupervised hierarchical clustering analysis of 64 breast samples. ULL represents the Norwegian samples and BC represents the Stanford samples. (A) Dendrogram representing similarities in the expression patterns between experimental samples. Thirty-eight IDCs are in black, 21 ILCs in orange, and three normal breast samples in green. Two lymph node metastases are marked with pink arrowheads. Three pairs of IDC and normal breast tissue from the same patient are marked with pairs of arrows of the same color. Samples were separated into four groups (group I–IV) by the clustering algorithm. The distributions of lymph node (LN) status and patient age at diagnosis are shown at the bottom. Red indicates positive or at least 55 years old, green is negative or <55 years of age, black is unknown, and gray is not applicable. (B) Overview of the gene expression patterns of 3314 genes whose expression varied more than threefold in at least three samples across the 64 breast samples. Each row represents a single gene, and each column an experimental sample. Colored bars identify the locations of the inserts in C–J. (C) Proliferation/cell cycle regulation cluster. (D) E-cadherin cluster. (E) ERBB2 cluster. (F) ER and its coexpressed gene cluster. (G) Stromal/fibroblast cell cluster. (H) Basal keratin cluster. (I) EGFR cluster. (J) Adipose-enriched cluster.
Group I tumors have high relative expression of ER and its regulated genes (Figure 1F). This group displays low relative expression of basal epithelial cell markers, including basal keratins and EGFR (Figure 1, H and I), adipose (Figure 1J) and stromal tissue markers (Figure 1G). Interestingly, the ER-overexpressing group I tumors differentially express genes involved in proliferation and cell cycle regulation (Figure 1C). Group II IDC tumors exhibit the lowest relative expression of ER and its regulated genes (Figure 1F) and high relative expression of basal epithelial cell markers, EGFR, and proliferation and cell cycle-regulated genes. (Figure 1, C, H, and I). Stromal and adipose tissue markers in group II are present mainly in the ILC samples (Figure 1, G and J). Group III and IV are similar in that they both show relatively low proliferation/cell cycle activities (Figure 1C) but differ in other signatures. Specifically, group III has relatively high expression of ER and its regulated genes (Figure 1F), stromal tissue markers (Figure 1G), and variable expression of basal epithelial cell (although relatively low EGFR) and adipose tissue markers (Figure 1, H, I, and J). Group IV tumors, consisting of the typical ILCs, has mixed expression of ER and its regulated genes (Figure 1F) and stromal tissue markers (Figure 1G), variable expression of basal epithelial markers (with relatively low EGFR expression) (Figure 1, H and I) but very high relative expression of adipose tissue markers (Figure 1J). Two markers, E-cadherin and ERBB2, are almost absent from group IV tumors but present in several tumors in the other three groups (Figure 1, D and E). These results suggest that the typical ILCs are molecularly different from IDCs. It is worth noting that group III mainly consists of patients <55 years of age and most had lymph node metastases. More than one-half the patients in group IV (typical ILCs) also had lymph node metastases but were at least 55 years old at diagnosis.
To identify genes whose expression differs significantly between ILCs and IDCs, we performed SAM analysis (Tusher et al., 2001) (http://www-stat.stanford.edu/~tibs/SAM/). There were 474 clones representing 378 unique genes that were selected at the lowest median number of falsely significant genes, 0.6. Of the 378 clones, 150 have known biological functions, including 75 genes that show high expression in ILCs and low expression in IDCs, and 75 genes vice versa. Most of the 150 genes can be categorized into five biological processes according to Gene Ontology annotations (Ashburner et al., 2000): cell adhesion/motility, lipid/fatty acid metabolism, immune and defense response, electron transport, and nucleosome assembly (Table 2). Many genes involved in signal transduction, regulation of transcription, and small molecule transport and metabolism were also among the genes identified by SAM (see Web supplement for full list).
Table 2.
Genes whose expression significantly differs between ILCs and IDCs identified by SAM
| Cell adhesion/motility | AOC3↑* | Amine oxidase, copper containing 3 (vascular adhesion protein 1) | CD36↑* | CD36 antigen (collagen type 1 receptor, thrombospondin receptor) |
| ANXA1↑* | Annexin A1 | VWF↑* | von Willebrand factor | |
| MMRN↑* | Multimerin | ITGA7↑* | Integrin, alpha 7 | |
| SORBS1↑* | Sorbin and SH3 domain containing 1 | NCAM1↑ | Neural cell adhesion molecule 1 | |
| ENPP2↑* | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) | PTGS2↑ | Prostaglandin-endoparoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | |
| CDH1↓* | Cadherin 1, type 1, E-cadherin (epithelial) | TNF↓ | Tumor necrosis factor (TNF superfamily, member 2) | |
| PAK4↓ | p21(CDKN1A)-activated kinase 4 | PKP3↓ | Plakophilin 3 | |
| F11R↓ | F11 receptor | |||
| Lipid/fatty acid transport and metabolism | CD36↑* | CD36 antigen (collagen type 1 receptor, thrombospondin receptor) | FACL4↑ | Fatty-acid-Coenzyme A ligase, long-chain 4 |
| ANXA1↑* | Annexin A1 | LPL↑* | Lipoprotein lipase | |
| AKR1C2↑ | Aldo-keto reductase family 1, member C2 | PLA2G2A↑ | Phospholipase A2, group IIA (platelets, synovial fluid) | |
| PLIN↑* | Perilipin | CRBPIV↑* | Retinoid binding protein 7 | |
| FABP4↑* | Fatty acid binding protein 4, adipocyte | ALOX15B↑ | Arachidonate 15-lipoxygenase, second type | |
| FADS2↓* | Fatty acid desaturase 2 | SAA2↑* | Serum amyloid A2 | |
| Immune/defense response | AOC3↑* | Amino oxidase, copper containing 3 (vascular adhesion protein 1) | ANXA1↑* | Annexin A1 |
| CCL23↑ | Chemokine (C-C motif) ligand 23 | CHIT1↑* | Chitinase 1 (chitotriosidase) | |
| SAA1↑* | Serum amyloid A1 | SAA2↑* | Serum amyloid A2 | |
| SAA4↑ | Serum amyloid A4 | LY64↑ | Lymphocyte antigen 64 homolog (mouse) | |
| PTGS2↑ | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | TNFSFI2↑ | Tumor necrosis factor (ligand) superfamily, member 12 | |
| CNIH↓ | Cornichon homolog (Drosophila) | F11R↓ | F11 receptor | |
| GIP2↓ | Interferon, alpha-inducible protein (clone IFI-15K) | |||
| Electron transport | ALDH1A1↑ | Aldehyde dehydrogenase 1 family, member A1 | ALDH1A2↑* | Aldehyde dehydrogenase 1 family, member A2 |
| AKR1C1↑ | Aldo-keto reductase family 1, member C1 | AKR1C2↑ | Aldo-keto reductase family 1, member C2 | |
| AOC3↑* | Amine oxidase, copper containing 3 (vascular adhesion protein 1) | ALOX15B↑ | Arachidonate 15-lipoxygenase, second type | |
| COX7A1↑ | Cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) | FMO2↑* | Flavin containing monooxygenase 2 | |
| FTHFD↑ | Formyltetrahydrofolate dehydrogenase | GPX3↑* | Glutathione peroxidase 3 (plasma) | |
| CYB561↓* | Cytochrome b-561 | |||
| Nucleosome assembly | HIST1H1C↓* | Histone 1, H1c | HIST1H2AC↓* | Histone 1, H2ac |
| HIST1H2AL↓ | Histone 1, H2a1 | HIST1H2BL↓ | Histone 1, H2b1 | |
| HIST1H2BD↓ | Histone 1, H2bd | HIST1H3D↓ | Histone 1, H3d | |
| HIST1H2BJ↓ | Histone 1, H2bj | HIST2H2AA↓* | Histone 2, H2aa | |
| HIST1H2BK↓ | Histone 1, H2bk | HIST3H2A↓ | Histone 3, H2a | |
| HIST2H2BE↓* | Histone 2, H2bc |
Genes in bold are involved in multiple biological processes included.
indicates up-regulation in ILCs
indicates down-regulation in ILCs
Genes also identified by PAM analysis performed on the same data as used in the SAM analysis
To explore the question of which genes best discriminate ILCs and IDCs, we performed PAM analysis. This method of nearest shrunken centroids is used in cancer class prediction to find genes that best characterize cancer types. Here, we used PAM to identify a minimal subset of genes that succinctly characterized ILCs and IDCs. By using a threshold of 2.9 (Figure 2A), a set of 78 clones representing 45 named genes were selected (Figure 2B), 44 of which were also present in the list of genes identified by SAM. ILCs and IDCs were separated based on the expression pattern of these genes with an overall error rate of 0.15. Specifically, 18 of 21 ILCs (86%) and 32 of 39 IDCs (82%) were correctly classified. BC-L-014, ULL-L-014, and ULL-L-028 were the exceptions and they all belonged to the ductal-like ILCs. When the 78 clones were used in a hierarchical clustering of all 59 tumor samples, the same three ductal-like ILC samples were placed on a main ductal branch containing most of the IDCs, separate from the lobular branch that contained 18 ILCs (Figure 2C). All typical ILCs clustered together in a core on the lobular branch with ductal-like ILCs positioned at the edges. Two group I IDCs (ULL-D-056 and ULL-D-216) and three group II IDCs (BC-D-007, BC-D-032, and BC-D-035) also are on the lobular branch, although most are on one edge near the ductal-like ILCs. Each of the IDCs on the lobular branch is ER and/or PR positive (see Clinical and Pathology Parameters on the Web supplement).
Figure 2.
Identification of gene expression patterns distinguishing IDCs and ILCs by PAM. (A) Relationships of value of threshold in cross-validation, number of genes identified, and overall misclassification rate or misclassification rate for each tumor type are shown in the top and bottom graph, respectively. (B) Seventy-eight clones were selected at threshold of 2.9 that separated IDCs and ILCs with the lowest overall misclassification rate. Bars to the right of the middle line indicate relative overexpression, and bars to the left indicate relative underexpression. The length of the bar represents the relative degree of variation. (C) Hierarchical clustering analysis of the 59 samples by using the 78 clones identified by PAM. In the dendrogram, IDCs are black and ILCs are orange. Case names in red indicate typical ILCs, those in blue indicate ductal-like ILCs, and those in black indicate IDCs.
The most important discriminator identified by PAM is cadherin 1 (CDH1, E-cadherin). Four different clones representing CDH1 were among the top discriminators (Figure 2B). Their average expression ratio in ILCs was 4.2- fold lower than that in IDCs, consistent with previous immunohistological studies of CDH1 in ILCs and IDCs. It is worth noticing that BC-D-048 has low expression of CDH1 similar to ILCs, which is consistent with invasiveness and unfavorable prognosis (Siitonen et al., 1996; Hunt et al., 1997; Nagae et al., 2002). Seven other genes (SORBS1, VWF, AOC3, MMRN, ITGA7, CD36, and ANXA1) functioning in cell adhesion were also selected as discriminators, suggesting a different cell adhesion feature between ILCs and IDCs. A number of other genes with high ranks among the identified discriminators are involved in lipid/fatty acid transport and metabolism, including FABP4, LPL, PLIN, ANXA1, and CD36, indicating a potential difference in lipid/fatty acid metabolism between ILC and IDC tumor tissue. An interesting electron transport gene overexpressed in ILCs is glutathione peroxidase 3, which catalyzes the reduction of hydrogen peroxide, organic hydroperoxides, and lipid peroxides, protecting cells against oxidative damage. Together, these results demonstrate that the majority of ILCs can be distinguished from IDCs by expression patterns of a small set of genes involved in several biological processes.
When typical ILCs were compared with IDCs by PAM analysis (see Web Supplement), 26 clones representing 14 named genes were identified that best distinguished the two groups with an overall misclassification error rate of 0.102 (0% error rate for the typical ILCs, 13% error rate for the IDCs). Twenty-one of the 26 clones were present among the 78 clones previously identified by PAM that distinguished ILCs and IDCs. Among the five clones not identified, there were two named genes: PDE2A (phosphodiesterase 2A and cGMP-stimulated) and early B-cell factor. These two genes are also present in a PAM analysis that distinguishes typical ILCs from ductal-like ILCs (Figure 4, B and C), discussed below.
Figure 4.
Identification of gene expression patterns that distinguish typical and ductal-like ILCs by using PAM. (A) Relationships of value of threshold in cross validation, number of genes identified and overall misclassification rate or misclassification rate for each tumor type were shown in the top and bottom graph, respectively. (B) Seventy-six clones were selected at threshold of 2.3 that separated typical and ductal-like ILCs with the lowest overall misclassification rate. Bars to the right of the middle line indicate relative overexpression, and bars to the left indicate relative underexpression. The length of the bar represents the relative degree of variation. (C) Hierarchical clustering analysis of the 21 ILC samples by using the 76 clones identified by PAM. Typical ILCs are in red in the dendrogram on top of the image and ductal-like ILCs are in blue.
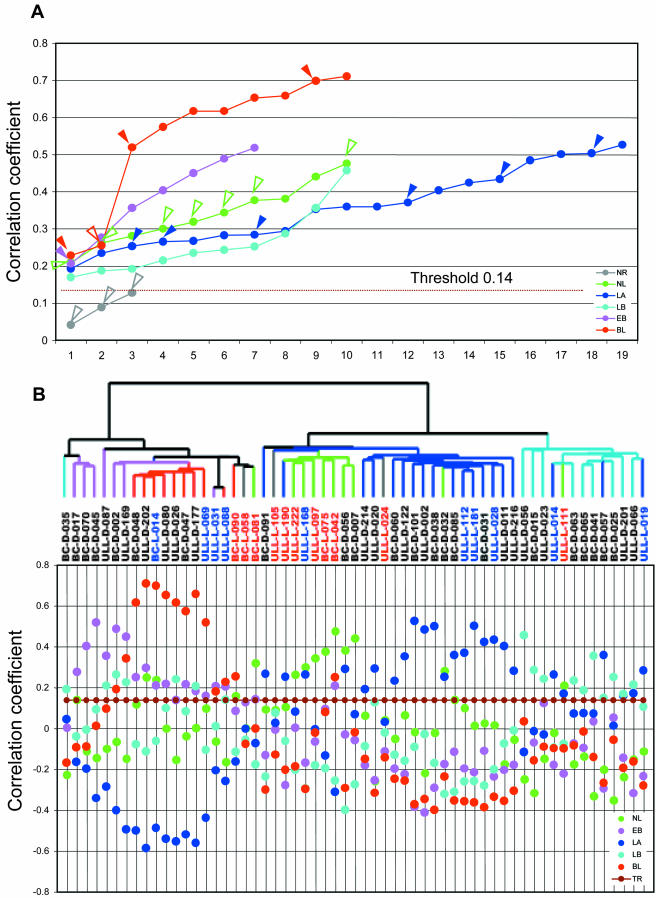
To further assess the degree of differences between gene expression profiles in ILCs and IDCs, and to compare that to the previous classification into five subclasses (luminal A, luminal B, ERBB2, basal, and normal-like), we performed Pearson's correlation by using the five sets of centroids recently defined in Sorlie et al. (2003). These sets of centroids consist of the average expression of the 500 intrinsic genes corresponding to each of the five subtypes. The Pearson's correlation coefficients between the expression ratio of 455 intrinsic genes in our 59 tumor samples, and the five sets of centroids were calculated. Fifty-six of 59 carcinomas were assigned to a subtype by the highest r (Figure 3, A and B), confirming the existence of the five centroids also in this set of tumors. The three tumors that could not be classified using an r threshold of 0.14 (determined by multiple permutations of gene expression values) were all typical ILCs (ULL-L-024, ULL-L-058, and ULL-L-105, colored gray in Figure 3).
Figure 3.
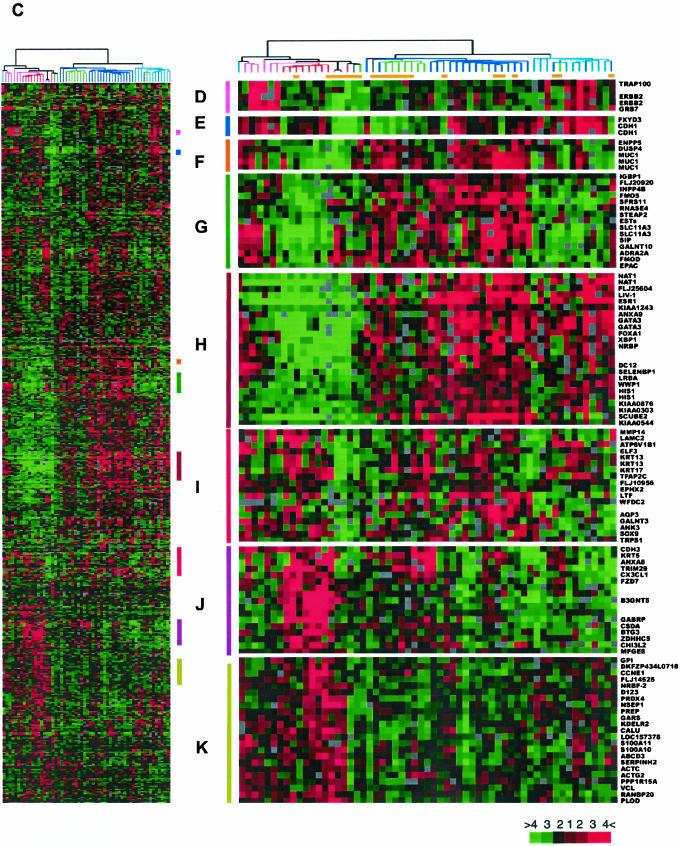
Comparison of gene expression patterns of ILCs and IDCs by using intrinsic genes. (A). The highest Pearson's correlation coefficients between each of the 59 primary tumors and five sets of centroids derived from 122 breast samples published previously were plotted in color corresponding to the subtype to which the samples were assigned. Open arrow heads indicate typical ILCs and filled arrowheads ductal-like ILCs. NR, nonrelated (r < 0.14); NL, normal-like; LA, luminal A; LB, luminal B; EB, ERBB2; and BL, basal-like subtype. (B) Top, dendrogram of hierarchical clustering analysis of the 59 primary tumors by using 481 intrinsic genes after spot quality selection. Fifty-six of 59 samples were categorized into one of the five subtypes of breast carcinomas identified previously based on their Pearson's r. The branches are colored as basal subtype in red, ERBB2 subtype in lavender, normal-like subtype in green, luminal A subtype in blue, and luminal B subtype in teal. Three samples colored in gray showed correlation below threshold. The sample labels are red for typical ILCs, blue for ductal-like ILCs, and black for IDCs. Bottom, Pearson's correlation coefficients between each of the 59 primary tumors and five sets of centroids derived from 122 breast samples published previously (49). Each vertical line corresponds to one sample label on top of the graph. NL, normal-like; LA, luminal A; LB, luminal B; EB, ERBB2; BL, basal-like subtype; and TR, threshold. (C) Overview of the gene expression patterns of 488 genes across the 59 breast samples. Each row represents a single gene, and each column an experimental sample. Colored bars identify the locations of the inserts in D–K. Orange bars under the dendrogram identify positions of ILCs. (D) ERBB2 cluster. (E) E-cadherin cluster. (F) MUC1 cluster. (G) Unknown cluster, expressed in a subset of tumors that overexpress ER and coexpressed genes. (H) ER and its coexpressed gene cluster (luminal epithelial cell cluster). (I) Basal epithelial cell cluster 1. (J) Basal epithelial cell cluster 2. (K) Unknown cluster, inversely correlated with ER and coexpressed genes.
The correlation coefficients between our 59 samples and the centroids of the five subtypes provide additional evidence that typical ILCs are different from ductal-like ILCs and IDCs in their gene expression profile. Seven of the eight typical ILCs that have >0.14 correlation coefficients were assigned to the normal-like subtype (Figure 3A), consistent with hierarchical clustering results shown in Figure 1. Only one typical ILC was assigned to another subtype (BC-L-090, assigned to basal subtype with an r of 0.25 compared with the ductal-like lobular BC-L-014 assigned to basal subtype with an r of 0.7). In contrast, only one of the 10 ductal-like ILCs was present in the normal-like subtype group (ULL-L-168, with an r of 0.3). Five of 10 ductal-like ILCs showed high correlation with the corresponding set of centroids for their subtypes (r > 0.3). Notably, the basal subtype had the highest correlation with the centroids compared with other subtypes, suggesting a highly consistent gene expression pattern associated with basal subtype tumors.
When variation in expression of 481 intrinsic genes was used to order the 59 samples in a hierarchical clustering, two features of the dendrogram were evident (Figure 3B). First, samples tended to cluster based on their correlation to the centroids of the subtypes. For example, seven of 10 basal subtype tumors clustered together, consistent with the high r among basal subtype IDCs observed above. Second, six of the 11 typical ILCs clustered together on the normal-like subtype branch, whereas only one of the 10 ductal-like ILCs clustered with this group, confirming that this group of ILCs has characteristic gene expression patterns different from IDCs and ductal-like ILCs. When we ordered the 38 IDCs only using the intrinsic genes, the dendrogram showed an even clearer separation of the five subtypes (see Web supplement). This is not surprising because the centroids were essentially derived from IDCs and thus have a high power of classification for IDCs.
The expression patterns of the intrinsic genes characterizing the five subtypes are largely in agreement with previous reports. For example, the basal epithelial cell markers, including keratins 5 and 17 were relatively highly expressed in the basal subtype (Figure 3, I and J), whereas ER and most of the other ER coexpressing genes failed to express in this subtype (Figure 3H). Genes representing tumor markers such as ERBB2 and MUC1 also showed relative low expression in the basal subtype (Figure 3, D and F). Interestingly, a cluster of genes with diverse functions is highly expressed in basal and ERBB2 subtypes (Figure 3K) and seem inversely related to ER expression. Another cluster of genes show relative low expression in basal and luminal B subtypes (Figure 3G), with relative overexpression in luminal A and normal-like subtypes.
To identify a minimum set of genes that best discriminate typical ILCs from ductal-like ILCs, PAM was performed on 23,914 clones representing 15,281 genes whose expression was measurable in at least 80% of the 21 ILCs. Seventy-six clones representing 44 genes with known functions were selected at an overall error rate of 9% (Figure 4, A and B). These genes function in a number of biological processes according to Gene Ontology annotations (for details, see Web supplement). Many of these genes are involved in regulation of cell growth (CDKN1C, G0S2, PDGFA, KIT, and F2 relatively overexpressed and MAP3K8 relatively underexpressed in the typical ILCs) and immune response (AOC3, IGJ, F2, F3, and IGLL1 relatively overexpressed and DEFB1, HLA-C relatively underexpressed in the typical ILCs). When the 76 clones were used in hierarchical clustering of the 21 ILCs, typical ILCs and ductal-like ILCs were separated into two groups with 100% accuracy (Figure 4C). The two genes identified in the PAM analysis of typical ILCs compared with IDCs (see Web supplement) but not identified on the original SAM list of clones distinguishing ILCs and IDCs, PDE2A (phosphodiesterase 2A) and EBF (early B-cell factor) are also relatively overexpressed in typical ILCs (Figure 4C). Together, these results strongly suggest the existence of two groups of ILCs differing in gene expression profiles.
DISCUSSION
We have systematically surveyed gene expression of 38 IDCs and 21 ILCs on a genome-wide scale by using RNA amplification and cDNA microarray techniques. Our data strongly suggest that a subgroup of ILCs, that we are calling typical ILCs, differ from IDCs not only in their histological structures and clinical features but also in global transcription programs. Three different statistical methods used to analyze the expression patterns all provided evidence supporting this conclusion. First, hierarchical clustering analyses showed that ILCs separate into two groups: typical ILCs that tend to cluster together and ductal-like ILCs that cluster with different subgroups of IDCs. Second, PAM analysis showed that ILCs could be separated from IDCs at a fairly high success rate on the basis of expression variations of only 78 transcripts and that the typical ILCs were more closely related than the ductal-like ILCs when clustering was performed using these selected genes. Third, Pearson's correlation analysis revealed that the expression pattern of the intrinsic genes in typical ILCs correlates poorly with previously characterized expression patterns of all but one IDC subtype, whereas the correlation between IDCs in this study with previous IDC subtypes is much higher. The differences between ILCs and IDCs we observed are not explained by different cellular composition of the samples, because the overall percentage of malignant epithelial component in ILCs was comparable with the IDCs when assessed by the same pathologists.
It is believed that all breast carcinomas, including both IDC and ILC, start in the terminal ductal lobular unit (TDLU) (Wellings et al., 1975; Wellings, 1980; Russo et al., 1990, 2001; Russo and Russo, 1994). The malignant epithelial cells in IDC or ILC may represent differences in cell of origin within the TDLU (progenitor cell differences) or differences in point when the cancer started during the TDLU lobular maturation process (type 1 lobule for IDC vs. type 2 lobule for ILC). This might explain why we see some lobular carcinomas as a distinct subtype and others with more similar gene expression to ductal carcinoma—there may be a continuum in the occurrence of epithelial carcinomas within the TDLU or from cells derived during the continuum of the TDLU maturation process.
SAM analysis suggests that genes differentially expressed between IDCs and both groups of ILCs are involved in cell adhesion, lipid/fatty acid metabolism, immune/defense/stress responses, electron transport, and nucleosome assembly. How the differences in gene expression between ILCs and IDCs translate to differences in clinical and microscopic properties of the tumors are not clear. However, several hypotheses can be offered based on information from previous studies. First, the differential expression of cell adhesion molecules may account for some of the differences observed in invasion patterns of ILCs and IDCs. The classical invasion pattern of ILCs is characterized by single files or cords of small cohesive cells that diffusely infiltrate the stromal tissues (Harris et al., 2000). In contrast, IDCs are characterized by tubule formation or solid sheets of tumor cells. Different morphological patterns of invasion may be associated with different adhesive properties between the malignant epithelial cells themselves and with surrounding tissues. It is notable that nine of 11 (82%) of the typical ILCs showed classical lobular morphology, seen in only four of 10 (40%) of the ductal-like ILCs. The other two typical ILCs (ULL-L-111 and ULL-L-190) showed classic lobular mixed with trabecular or trabecular/alveolar growth pattern. Importantly, the ductal-like lobular sample that clustered with basal-like IDCs (BC-L-014) grew as a solid variant with <5% ductal edges. In addition to the multiple cell adhesion molecules, two genes involved in cell motility (ANXA1 and ENPP2) are differentially expressed between IDCs and ILCs and may influence differences in migration ability of tumor cells during invasion.
A recent study (Gupta et al., 2003) analyzing IDCs with and without lymphovascular tumor emboli assessed by E-cadherin immunostaining, suggested that, although this cell adhesion molecule is characteristically lost in ILCs and may even show loss in some high grade IDCs, observation of diffuse strong E-cadherin expression in IDCs may play a role in tumor growth as intravascular nests or emboli within lymphatics when lymphovascular invasion exists. In E-cadherin negative tumors that metastasize, individual cells may be able to migrate and travel in the vasculature and lymphatics differently than tumor emboli, which are composed of clusters of cells, potentially explaining the different patterns of distant metastatic spread in ILCs and IDCs.
The differential expression of genes involved in lipid/fatty acid metabolism is complex but may be partially responsible for different proliferation rates of tumor cells in ILCs and IDCs. Breast epithelial cell proliferation and differentiation are controlled by multiple factors, including growth factors, hormones, and fatty acids. Growth factors may cause phospholipid hydrolysis with release of fatty acids and lipoxygenase products that stimulate cell growth. Dietary intake can affect fatty acid metabolism in tissues. Specific polyunsaturated fatty acids found in vegetable oils, such as linoleic acid, may promote tumor growth, whereas other polyunsaturated fatty acids found in fish oil or monounsaturated fats, such as oleic acid in olive oil, are either neutral or inhibitory (Rose et al., 1997; Natarajan and Nadler, 1998; Stoll, 1998; Bartsch et al., 1999). In our series, the adipose-enriched cluster seems to have an inverse relationship with the proliferation/cell cycle cluster. Profiling whole tissue, nonmicrodissected specimens detects gene expression averaged across all cells in the tumor sample. We are just starting studies to define the cell types that contribute to the observed differential expression, but we have not yet identified which cellular components (epithelial or adipose) are expressing the adipose-enriched gene cluster. Differentiated mammary cells are designed to make lipid during lactation and malignant epithelial cells may potentially represent a source of adipose-enriched genes in more highly differentiated tumors or ILCs. Therapy that induces cell differentiation, such as ligands to retinoid X receptors, has been shown to increase expression of adipocyte-related genes that inhibit cellular proliferation and cause tumor regression. The source of the expressed genes seems to be both malignant epithelial cells and preadipocytes (fibroblasts that differentiate into adipocytes) (Agarwal et al., 2000). Preadipocytes, but not mature adipocytes, have also been shown to secrete molecules that inhibit DNA synthesis in murine mammary carcinoma cells (Rahimi et al., 1998), suggesting both paracrine and autocrine effects. Conversely, some ductal-like lobulars and IDCs may elicit a different response caused by inflammatory cells in the extracellular matrix as evidenced by a higher expression of immunoglobulins, chemokines, and collagen. The differential expression of immune and defense response genes may explain the observed favorable prognosis of ILCs found in some studies (Toikkanen et al., 1997). Together, the gene expression profiles of ILCs and IDCs suggest a different interaction between stromal and epithelial cells in these two types of tumors, possibly due to differences in cross talk between stromal and malignant epithelial cells.
Genetic alterations have been proposed to be the basis for tumor initiation and progression. This raises the question of whether the differences in gene expression between ILCs and IDCs are due to differences in genetic makeup of these two types of tumors. Pandis et al. (1996) reported significant differences in karyotypic patterns between ILCs and IDCs in 125 breast carcinomas. In contrast to IDC, ILC were characterized by few, often balanced chromosomal aberrations, yielding a near diploid karyotype. Although no tumor-type specific patterns of aberrations were identified, Flagiello et al. (1998) reported highly recurrent der(1:16)(q10;p10) and other 16q (location of the E-cadherin gene) alterations in ILCs. Recently, Gunther et al. (2001) demonstrated that ILCs have significant losses of 16q, 22, and 17p and q, and gains in 1q and 8q. These chromosomal changes may contribute to type-specific properties of ILCs. Pollack et al. (2002) have compared DNA copy number changes and gene expression in parallel on a genome-wide scale by using the same DNA microarrays in 44 primary breast carcinomas, and revealed a remarkable degree to which variations in DNA copy number changes contribute to gene expression changes and estimated that at least 12% of mRNA variation in breast cancer can be directly attributed to copy number variation. We are currently performing arraybased CGH on our 59 samples for comparison with the gene expression profiles to better understand whether DNA copy number alterations differ between typical ILCs, ductal-like ILCs, and the IDCs that cluster with the ductal-like ILCs to determine whether specific chromosomal abnormalities may play a direct role in type-specific development of these different tumor types.
In conclusion, gene expression profiling has revealed distinct patterns of gene expression among ILCs and IDCs. Differences in a number of biological processes such as cell adhesion and lipid/fatty acid metabolism may contribute to the type-specific properties of IDCs and ILCs. Our data strongly suggest that over half of ILCs (called the typical ILCs) differ from IDCs not only in histological and clinical features but also in global transcription programs. The remaining ILCs (called the ductal-like ILCs) closely resemble IDCs in their transcription patterns. The finding of two subsets of ILCs has important clinical implications about targeted therapies. Further studies would be required to explore whether the ductal-like ILCs should be treated similarly to other IDCs of their particular molecular phenotype (basal-like, luminal A or B, and ERBB2 expressing), and if different and type-specific treatment may be indicated for the typical ILCs. A larger cohort of samples is being analyzed to confirm the existence of different molecular subtypes of ILCs. In addition, further studies on pure populations of epithelial and stromal cells from these two tumor subtypes using microdissection techniques may help us better understand the mechanisms underlying the development and epithelial–stromal interactions of the different tumor phenotypes. Correlative studies using clinical follow up data are in progress.
Acknowledgments
The Stanford Microarray Database group and Stanford Functional Genomics Facilities are acknowledged for supporting the experiments and data analysis. We thank Therese Sørlie for insights in the data analysis, Michelle Ferrari and Maureen Chang for contributions in the management of the breast cancer database, and Susan Overholser for creating the Web supplement and assistance in configuring the database and preparation of this manuscript. This study was supported by Public Health Service grant U01CA85129 from the National Cancer Institute, National Institutes of Health, Department of Health and Human Services, the Norwegian Cancer Society grant 99061, and the Research Council of Norway, grant number 137012/310. Anita Langerød is a fellow of The Norwegian Cancer Society.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-11-0786. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-11-0786.
References
- Agarwal, V.R., Bischoff, E.D., Hermann, T., and Lamph, W.W. (2000). Induction of adipocyte specific gene expression is correlated with mammary tumor regression by the retinoid X receptor-ligand LGD1069 (targretin). Cancer Res. 60, 6033-6038. [PubMed] [Google Scholar]
- Ahr, A., Holtrich, U., Solbach, C., Scharl, A., Strebhardt, K., Karn, T., and Kaufmann, M. (2001). Molecular classification of breast cancer patients by gene expression profiling. J. Pathol. 195, 312-320. [DOI] [PubMed] [Google Scholar]
- Ahr, A., Karn, T., Solbach, C., Seiter, T., Strebhardt, K., Holtrich, U., and Kaufmann, M. (2002). Identification of high risk breast-cancer patients by gene expression profiling. Lancet 359, 131-132. [DOI] [PubMed] [Google Scholar]
- Ashburner, et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch, H., Nair, J., and Owen, R.W. (1999). Dietary polyunsaturated fatty acids and cancers of the breast and colorectum: emerging evidence for their role as risk modifiers. Carcinogenesis 20, 2209-2218. [DOI] [PubMed] [Google Scholar]
- Bedner, E., Harezga, B., Osborn, M., and Domagala, W. (1995). Cathepsin D in invasive ductal NOS, medullary, lobular and mucinous breast carcinoma. An immunohistochemical study. Pol. J. Pathol. 46, 11-15. [PubMed] [Google Scholar]
- Berx, G., Cleton-Jansen, A.M., Strumane, K., de Leeuw, W.J., Nollet, F., van Roy, F., and Cornelisse, C. (1996). E-cadherin is inactivated in a majority of invasive human lobular breast cancers by truncation mutations throughout its extracellular domain. Oncogene 13, 1919-1925. [PubMed] [Google Scholar]
- Borst, M.J., and Ingold, J.A. (1993). Metastatic patterns of invasive lobular versus invasive ductal carcinoma of the breast. Surgery 114, 637-641; discussion 641-632. [PubMed] [Google Scholar]
- Buerger, H., Simon, R., Schafer, K.L., Diallo, R., Littmann, R., Poremba, C., van Diest, P.J., Dockhorn-Dworniczak, B., and Bocker, W. (2000). Genetic relation of lobular carcinoma in situ, ductal carcinoma in situ, and associated invasive carcinoma of the breast. Mol. Pathol. 53, 118-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukholm, I.K., Nesland, J.M., Karesen, R., Jacobsen, U., and Borresen, A.L. (1997). Relationship between abnormal p53 protein and failure to express p21 protein in human breast carcinomas. J. Pathol. 181, 140-145. [DOI] [PubMed] [Google Scholar]
- Bumpers, H.L., Hassett, J.M., Jr., Penetrante, R.B., Hoover, E.L., and Holyoke, E.D. (1993). Endocrine organ metastases in subjects with lobular carcinoma of the breast. Arch. Surg. 128, 1344-1347. [DOI] [PubMed] [Google Scholar]
- Chen, X., et al. (2003). Variation in gene expression patterns in human gastric cancers. Mol. Biol. Cell 14, 3208-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleton-Jansen, A.M. (2002). E-cadherin and loss of heterozygosity at chromosome 16 in breast carcinogenesis: different genetic pathways in ductal and lobular breast cancer? Breast Cancer Res. 4, 5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coradini, D., Pellizzaro, C., Veneroni, S., Ventura, L., and Daidone, M.G. (2002). Infiltrating ductal and lobular breast carcinomas are characterised by different interrelationships among markers related to angiogenesis and hormone dependence. Br. J. Cancer 87, 1105-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daling, J.R., et al. (2002). Relation of regimens of combined hormone replacement therapy to lobular, ductal, and other histologic types of breast carcinoma. Cancer 95, 2455-2464. [DOI] [PubMed] [Google Scholar]
- Dixon, A.R., Ellis, I.O., Elston, C.W., and Blamey, R.W. (1991). A comparison of the clinical metastatic patterns of invasive lobular and ductal carcinomas of the breast. Br. J. Cancer 63, 634-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagala, W., Wozniak, L., Lasota, J., Weber, K., and Osborn, M. (1990). Vimentin is preferentially expressed in high-grade ductal and medullary, but not in lobular breast carcinomas. Am. J. Pathol. 137, 1059-1064. [PMC free article] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston, E.W., and Ellis, I.O. (1993). Method for grading breast cancer. J. Clin. Pathol. 46, 189-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagiello, D., Gerbault-Seureau, M., Sastre-Garau, X., Padoy, E., Vielh, P., and Dutrillaux, B. (1998). Highly recurrent der(1;16)(q10;p10) and other 16q arm alterations in lobular breast cancer. Genes Chromosomes Cancer 23, 300-306. [PubMed] [Google Scholar]
- Geradts, J., and Ingram, C.D. (2000). Abnormal expression of cell cycle regulatory proteins in ductal and lobular carcinomas of the breast. Mod. Pathol. 13, 945-953. [DOI] [PubMed] [Google Scholar]
- Gollub, J., et al. (2003). The Stanford Microarray Database: data access and quality assessment tools. Nucleic Acids Res. 31, 94-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther, K., Merkelbach-Bruse, S., Amo-Takyi, B.K., Handt, S., Schroder, W., and Tietze, L. (2001). Differences in genetic alterations between primary lobular and ductal breast cancers detected by comparative genomic hybridization. J. Pathol. 193, 40-47. [DOI] [PubMed] [Google Scholar]
- Gupta, A., Deshpande, C.G., and Badve, S. (2003). Role of E-cadherins in development of lymphatic tumor emboli. Cancer 97, 2341-2347. [DOI] [PubMed] [Google Scholar]
- Harris, J.R., Lippman, M.E., Morrow, M., and Osborne, C.K., Eds. (2000). Diseases of the Breast 2nd Ed., Philadelphia: Lippincott Willams & Wilkins.
- Hunt, N.C., Douglas-Jones, A.G., Jasani, B., Morgan, J.M., and Pignatelli, M. (1997). Loss of E-cadherin expression associated with lymph node metastases in small breast carcinomas. Virchows Arch. 430, 285-289. [DOI] [PubMed] [Google Scholar]
- Lee, A.H., Dublin, E.A., Bobrow, L.G., and Poulsom, R. (1998). Invasive lobular and invasive ductal carcinoma of the breast show distinct patterns of vascular endothelial growth factor expression and angiogenesis. J. Pathol. 185, 394-401. [DOI] [PubMed] [Google Scholar]
- Lehr, H.A., Folpe, A., Yaziji, H., Kommoss, F., and Gown, A.M. (2000). Cytokeratin 8 immunostaining pattern and E-cadherin expression distinguish lobular from ductal breast carcinoma. Am. J. Clin. Pathol. 114, 190-196. [DOI] [PubMed] [Google Scholar]
- Li, C.I., Anderson, B.O., Porter, P., Holt, S.K., Daling, J.R., and Moe, R.E. (2000a). Changing incidence rate of invasive lobular breast carcinoma among older women. Cancer 88, 2561-2569. [PubMed] [Google Scholar]
- Li, C.I., Weiss, N.S., Stanford, J.L., and Daling, J.R. (2000b). Hormone replacement therapy in relation to risk of lobular and ductal breast carcinoma in middle-aged women. Cancer 88, 2570-2577. [DOI] [PubMed] [Google Scholar]
- Li, C.I., Anderson, B.O., Daling, J.R., and Moe, R.E. (2003). Trends in incidence rates of invasive lobular and ductal breast carcinoma. J. Am. Med. Assoc. 289, 1421-1424. [DOI] [PubMed] [Google Scholar]
- Lossos, I.S., Alizadeh, A.A., Diehn, M., Warnke, R., Thorstenson, Y., Oefner, P.J., Brown, P.O., Botstein, D., and Levy, R. (2002). Transformation of follicular lymphoma to diffuse large-cell lymphoma: alternative patterns with increased or decreased expression of c-myc and its regulated genes. Proc. Natl. Acad. Sci. USA 99, 8886-8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagae, Y., Kameyama, K., Yokoyama, M., Naito, Z., Yamada, N., Maeda, S., Asano, G., Sugisaki, Y., and Tanaka, S. (2002). Expression of E-cadherin catenin and C-erbB-2 gene products in invasive ductal-type breast carcinomas. J. Nippon Med. Sch. 69, 165-171. [DOI] [PubMed] [Google Scholar]
- Natarajan, R., and Nadler, J. (1998). Role of lipoxygenases in breast cancer. Front. Biosci. 3, E81-E88. [DOI] [PubMed] [Google Scholar]
- Newstead, G.M., Baute, P.B., and Toth, H.K. (1992). Invasive lobular and ductal carcinoma: mammographic findings and stage at diagnosis. Radiology 184, 623-627. [DOI] [PubMed] [Google Scholar]
- Pandis, N., et al. (1996). Correlation between karyotypic pattern and clincopathologic features in 125 breast cancer cases. Int. J. Cancer 66, 191-196. [DOI] [PubMed] [Google Scholar]
- Perou, C.M., et al. (2000). Molecular portraits of human breast tumours. Nature 406, 747-752. [DOI] [PubMed] [Google Scholar]
- Pollack, J.R., Sorlie, T., Perou, C.M., Rees, C.A., Jeffrey, S.S., Lonning, P.E., Tibshirani, R., Botstein, D., Borresen-Dale, A.L., and Brown, P.O. (2002). Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc. Natl. Acad. Sci. USA 99, 12963-12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi, N., Tremblay, E., McAdam, L., Roberts, A., and Elliott, B. (1998). Autocrine secretion of TGF-beta 1 and TGF-beta 2 by pre-adipocytes and adipocytes: a potent negative regulator of adipocyte differentiation and proliferation of mammary carcinoma cells. In Vitro Cell Dev. Biol. Anim. 34, 412-420. [DOI] [PubMed] [Google Scholar]
- Rey, M.J., et al., (1998). p21 WAF1/Cip1 is associated with cyclin D1CCND1 expression and tubular differentiation but is independent of p53 overexpression in human breast carcinoma. J. Pathol. 184, 265-271. [DOI] [PubMed] [Google Scholar]
- Rose, D.P., Connolly, J.M., and Liu, X.H. (1997). Fatty acid regulation of breast cancer cell growth and invasion. Adv. Exp. Med. Biol. 422, 47-55. [DOI] [PubMed] [Google Scholar]
- Rosenthal, S.I., Depowski, P.L., Sheehan, C.E., and Ross, J.S. (2002). Comparison of HER-2/neu oncogene amplification detected by fluorescence in situ hybridization in lobular and ductal breast cancer. Appl. Immunohistochem. Mol. Morphol. 10, 40-46. [DOI] [PubMed] [Google Scholar]
- Russo, J., Gusterson, B.A., Rogers, A.E., Russo, I.H., Wellings, S.R., and van Zwieten, M.J. (1990). Comparative study of human and rat mammary tumorigenesis. Lab. Investig. 62, 244-278. [PubMed] [Google Scholar]
- Russo, J., Hu, Y.F., Silva, I.D., and Russo, I.H. (2001). Cancer risk related to mammary gland structure and development. Microsc. Res. Tech. 52, 204-223. [DOI] [PubMed] [Google Scholar]
- Russo, J., and Russo, I.H. (1994). Toward a physiological approach to breast cancer prevention. Cancer Epidemiol. Biomarkers Prev. 3, 353-364. [PubMed] [Google Scholar]
- Sastre-Garau, X., Jouve, M., Asselain, B., Vincent-Salomon, A., Beuzeboc, P., Dorval, T., Durand, J.C., Fourquet, A., and Pouillart, P. (1996). Infiltrating lobular carcinoma of the breast. Clinicopathologic analysis of 975 cases with reference to data on conservative therapy and metastatic patterns. Cancer 77, 113-120. [DOI] [PubMed] [Google Scholar]
- Serre, C.M., Clezardin, P., Frappart, L., Boivin, G., and Delmas, P.D. (1995). Distribution of thrombospondin and integrin alpha V in DCIS, invasive ductal and lobular human breast carcinomas. Analysis by electron microscopy. Virchows Arch. 427, 365-372. [DOI] [PubMed] [Google Scholar]
- Sherlock, G., et al. (2001). The Stanford Microarray Database. Nucleic Acids Res. 29, 152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siitonen, S.M., Kononen, J.T., Helin, H.J., Rantala, I.S., Holli, K.A., and Isola, J.J. (1996). Reduced E-cadherin expression is associated with invasiveness and unfavorable prognosis in breast cancer. Am. J. Clin. Pathol. 105, 394-402. [DOI] [PubMed] [Google Scholar]
- Sorlie, T., et al. (2001). Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 98, 10869-10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie, T., et al. (2003). Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. USA 100, 8418-8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soslow, R.A., Carlson, D.L., Horenstein, M.G., and Osborne, M.P. (2000). A comparison of cell cycle markers in well-differentiated lobular and ductal carcinomas. Breast Cancer Res. Treat. 61, 161-170. [DOI] [PubMed] [Google Scholar]
- Stoll, B.A. (1998). Breast cancer and the western diet: role of fatty acids and antioxidant vitamins. Eur. J. Cancer 34, 1852-1856. [DOI] [PubMed] [Google Scholar]
- Tibshirani, R., Hastie, T., Narasimhan, B., and Chu, G. (2002). Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. USA 99, 6567-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toikkanen, S., Pylkkanen, L., and Joensuu, H. (1997). Invasive lobular carcinoma of the breast has better short- and long-term survival than invasive ductal carcinoma. Br. J. Cancer 76, 1234-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher, V.G., Tibshirani, R., and Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van't Veer, L.J., Dai, H., van de Vijver, M.J., He, Y.D., Hart, A.A., Bernards, R., and Friend, S.H. (2002). Expression profiling predicts outcome in breast cancer. Breast Cancer Res. 5, 57-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vijver, et al. (2002). A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347, 1999-2009. [DOI] [PubMed] [Google Scholar]
- Verkooijen, H.M., Fioretta, G., Vlastos, G., Morabia, A., Schubert, H., Sappino, A.P., Pelte, M.F., Schafer, P., Kurtz, J., and Bouchardy, C. (2003). Important increase of invasive lobular breast cancer incidence in Geneva, Switzerland. Int. J. Cancer 104, 778-781. [DOI] [PubMed] [Google Scholar]
- Wellings, S.R. (1980). Development of human breast cancer. Adv. Cancer Res. 31, 287-314. [DOI] [PubMed] [Google Scholar]
- Wellings, S.R., Jensen, H.M., and Marcum, R.G. (1975). An atlas of subgross pathology of the human breast with special reference to possible precancerous lesions. J. Natl. Cancer Inst. 55, 231-273. [PubMed] [Google Scholar]
- West, M., Blanchette, C., Dressman, H., Huang, E., Ishida, S., Spang, R., Zuzan, H., Olson, J.A., Jr., Marks, J.R., and Nevins, J.R. (2001). Predicting the clinical status of human breast cancer by using gene expression profiles. Proc. Natl. Acad. Sci USA 98, 11462-11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winchester, D.J., Chang, H.R., Graves, T.A., Menck, H.R., Bland, K.I., and Winchester, D.P. (1998). A comparative analysis of lobular and ductal carcinoma of the breast: presentation, treatment, and outcomes. J. Am. Coll. Surg. 186, 416-422. [DOI] [PubMed] [Google Scholar]
- Yeatman, T.J., Cantor, A.B., Smith, T.J., Smith, S.K., Reintgen, D.S., Miller, M.S., Ku, N.N., Baekey, P.A., and Cox, C.E. (1995). Tumor biology of infiltrating lobular carcinoma. Implications for management. Ann. Surg. 222, 549-559; discussion 559-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H., Hastie, T., Whitfield, M.L., Borresen-Dale, A.L., and Jeffrey, S.S. (2002). Optimization and evaluation of T7 based RNA linear amplification protocols for cDNA microarray analysis. BMC Genomics 3, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]