Abstract
The KinI kinesin MCAK is a microtubule depolymerase important for governing spindle microtubule dynamics during chromosome segregation. The dynamic nature of spindle assembly and chromosome-microtubule interactions suggest that mechanisms must exist that modulate the activity of MCAK, both spatially and temporally. In Xenopus extracts, MCAK associates with and is stimulated by the inner centromere protein ICIS. The inner centromere kinase Aurora B also interacts with ICIS and MCAK raising the possibility that Aurora B may regulate MCAK activity as well. Herein, we demonstrate that recombinant Aurora B-INCENP inhibits Xenopus MCAK activity in vitro in a phosphorylation-dependent manner. Substituting endogenous MCAK in Xenopus extracts with the alanine mutant XMCAK-4A, which is resistant to inhibition by Aurora B-INCENP, led to assembly of mono-astral and monopolar structures instead of bipolar spindles. The size of these structures and extent of tubulin polymerization in XMCAK-4A extracts indicate that XM-CAK-4A is not defective for microtubule dynamics regulation throughout the cytoplasm. We further demonstrate that the ability of XMCAK-4A to localize to inner centromeres is abolished. Our results show that MCAK regulation of cytoplasmic and spindle-associated microtubules can be differentiated by Aurora B-dependent phosphorylation, and they further demonstrate that this regulation is required for bipolar meiotic spindle assembly.
INTRODUCTION
Assembly of the microtubule-based bipolar spindle, the apparatus that powers chromosome segregation during mitosis and meiosis (M phase), is driven by the inherent dynamic instability of microtubules, as well as by proteins that alter microtubule dynamics (Hyman and Karsenti, 1996; Wittmann et al., 2001). Accordingly, M-phase onset triggers global, cytoplasmic changes in microtubule dynamics that expedite spindle assembly, most notably an increase in the frequency of catastrophes, the transition from microtubule growth to shrinkage (Belmont et al., 1990; Verde et al., 1992; Rusan et al., 2001; Kinoshita et al., 2002). In Xenopus egg extracts, the KinI kinesin MCAK, classified by its internally located motor domain (Wordeman and Mitchison, 1995; Walczak et al., 1996), is essential for governing this change in catastrophe frequency (Walczak et al., 1996; Tournebize et al., 2000). Biochemical studies indicate that MCAK likely accomplishes catastrophe induction directly by catalytically depolymerizing microtubules from their protofilament ends (Desai et al., 1999b; Hunter et al., 2003).
Although spindle assembly demands global regulation of microtubule dynamics, proper spindle function also requires precise control of microtubule dynamics regulation at defined locations within the spindle. Because accurate chromosome segregation requires that each kinetochore of a pair of sister chromatids be attached to microtubules derived from opposing spindle poles (biorientation), it is particularly important to control the interaction of spindle microtubules with kinetochores (Rieder and Salmon, 1998). Proper chromosome biorientation requires the protein kinase Aurora B, and its inhibition increases the frequencies of both mispositioned and malattached chromosomes during preanaphase mitosis (Adams et al., 2001; Oegema et al., 2001; Kallio et al., 2002; Murata-Hori and Wang, 2002; Tanaka et al., 2002; Ditchfield et al., 2003; Hauf et al., 2003). How Aurora B promotes chromosome biorientation is unclear, particularly in vertebrates where the kinase is located at a chromosomal region spatially distinct from kinetochores, the inner centromere (Cleveland et al., 2003). Dynamics regulation of microtubules is a potential mechanism because Aurora B perturbation by either antibody injection or dominant negative approaches cause defects in spindle structure (Kallio et al., 2002; Murata-Hori and Wang, 2002), but Aurora B targets that might regulate microtubule dynamics have not been identified.
Similar to Aurora B, the KinI MCAK exhibits prominent inner centromere localization during mitosis (Wordeman and Mitchison, 1995; Walczak et al., 1996; Ohi et al., 2003) and can also be found at kinetochores that lack tension (Kline-Smith et al., 2004). The complex distribution of MCAK at these chromosomal regions suggests important though mechanistically ill-defined roles for MCAK in regulating the dynamics of microtubules that interact with kinetochores and/or inner centromeres. One possibility is that centromeric MCAK depolymerizes microtubules near inner centromeres to prevent the accumulation of malattached kinetochores (Ohi et al., 2003). Consistent with this notion, depletion of MCAK from inner centromeres in PtK2 cells causes defects in chromosome congression and segregation that are caused by kinetochore-microtubule attachment errors (Kline-Smith et al., 2004). MCAK also has been postulated to provide forces required for chromosome oscillation, powering polewards movement of kinetochores by depolymerizing their attached microtubules (Walczak et al., 1996; Cleveland et al., 2003). Emerging data demonstrating tension-dependent redistribution of MCAK from inner centromeres to kinetochores is consistent with this role (Kline-Smith et al., 2004). MCAK function at kinetochores is also important for polewards sister chromatid movement during anaphase A (Maney et al., 1998; Rogers et al., 2003), when it presumably initiates depolymerization of kinetochore-fiber microtubules from their kinetochore-embedded plus-ends.
A priori, multiple functional populations of MCAK must exist given that this kinesin controls cytoplasmic microtubule dynamics as well as microtubule depolymerization at kinetochores and inner centromeres. One obvious strategy to produce functionally distinct populations of MCAK is to regulate its microtubule depolymerizing activity with spatial and temporal precision. Thus, a major challenge is to identify the factors that control MCAK activity and localization. The biochemical association of MCAK with ICIS, an inner centromere protein that stimulates MCAK activity (Ohi et al., 2003), and Aurora B, raises the possibility that Aurora B may regulate the activity of MCAK by phosphorylation. In this study, we show that Aurora B inhibits Xenopus MCAK activity in vitro in a phosphorylation-dependent manner. We further demonstrate that MCAK resistant to this control still regulates global tubulin polymer levels properly, but it is unable to drive formation of bipolar spindles. Our results identify a mechanism that differentiates the requirement of MCAK in controlling cytoplasmic microtubule polymerization from dynamics regulation of spindle-associated microtubules and show that this control is important for generating the spindle's essential bipolarity.
MATERIALS AND METHODS
Cloning, Mutagenesis, and Baculovirus Construction
Xenopus laevis Aurora B, polymerase chain reaction-amplified from a Xenopus ovary cDNA library (a gift from A. Straight, Stanford School of Medicine. Stanford, CA), was inserted into pFastBac HTa (Invitrogen, Carlsbad, CA) as an EcoRI fragment. pFastBac HTa-XMCAK was constructed by polymerase chain reaction amplifying XMCAK from a full-length expressed sequence tag (GenBank accession no. BE189726; a gift from A. Salic, Harvard Medical School, Boston, MA) and inserting the resulting fragment into pFastBac HTa by using BamHI/XhoI. The XMCAK alanine mutant XMCAK-4A (T95A, S110A, S177A, and S196A) was obtained by site-directed mutagenesis of pFastBac HTa-XMCAK using the QuikChange kit (Stratagene, La Jolla, CA).
Baculoviruses expressing His6Aurora B, wild-type (wt) His6XMCAK, and His6XMCAK-4A were produced according to the Bac-to-Bac system (Invitrogen) using the clones described above. A baculovirus that expresses Xenopus INCENP (a gift from A. Straight, Stanford School of Medicine) also was produced according to the Bac-to-Bac system, by using a plasmid constructed by insertion of its cDNA into pFastBac HTb. Details of plasmid construction and oligonucleotide sequences are available on request.
Antibody Generation
Glutathione S-transferase (GST)-XMCAKNT (Walczak et al., 2002) was expressed and purified as described previously (Ohi et al., 2003) and used to immunize rabbits (Cocalico, Reamstown, PA). Anti-XMCAK antibodies were affinity-purified by passing anti-GST-depleted serum over Affi-Gel 10 covalently coupled to GST-XMCAKNT. Anti-XMCAK antibodies were dialyzed into 10 mM K-HEPES (pH 7.7), 100 mM KCl and frozen in liquid N2 after the addition of sucrose to 150 mM. Alexa 488- and Alexa 594-anti-XMCAK conjugates were prepared as suggested by the manufacturer (Molecular Probes, Eugene, OR).
Expression and Purification of Recombinant Proteins from Sf-9 Insect Cells
Soluble His6Aurora B-His6INCENP were obtained by coinfection of Sf-9 cells with both His6Aurora B and His6INCENP baculoviruses for ∼72 h. During the final 3 h of infection, cells were treated with 0.1 μM okadaic acid to obtain maximally active Aurora B; Aurora B activity is sensitive to its phosphorylation state as well as that of INCENP (Murnion et al., 2001; Bishop and Schumacher, 2002; Bolton et al., 2002; Honda et al., 2003). To purify His6Aurora B-His6INCENP, cells expressing the two proteins were resuspended in lysis buffer (50 mM sodium phosphate, pH 7.8, 500 mM NaCl, 5 mM β-mercaptoethanol, 0.1 mM MgATP, 1 mM sodium fluoride, 20 mM β-glycerophosphate, 10 mM imidazole, 1% IGEPAL CA-630, and protease inhibitors [1 mM phenylmethylsulfonyl fluoride; 1 mM benzamidine; and 10 μg/ml each of leupeptin, pepstatin, and chymostatin]), sonicated, and clarified by centrifugation at 147,505 × g for 1 h. Approximately 1 ml of Ni2+-NTA agarose (QIAGEN, Valencia, CA) was incubated with the supernatant for 2 h at 4°C, and then washed extensively with lysis buffer lacking IGEPAL CA-630 and protease inhibitors. His6Aurora B-His6INCENP were eluted with 50 mM sodium phosphate (pH 7.8), 500 mM NaCl, 5 mM β-mercaptoethanol, 0.1 mM MgATP, 1 mM sodium fluoride, 20 mM β-glycerophosphate, 300 mM imidazole, and the peak fractions desalted into 10 mM K-HEPES (pH 7.7), 300 mM KCl, 1 mM dithiothreitol (DTT), 0.1 mM MgATP, 1 mM sodium fluoride, 20 mM β-glycerophosphate. After the addition of sucrose to 20%, the proteins were aliquoted, frozen in liquid N2, and stored at -80°C.
His6XMCAK and His6XMCAK-4A were separately expressed in Sf-9 cells for ∼72 h. To purify these proteins, cells were lysed in 50 mM sodium phosphate (pH 7.8), 500 mM NaCl, 5 mM β-mercaptoethanol, 0.1 mM MgATP, 10 mM imidazole, 1% IGEPAL CA-630 plus protease inhibitors, and the extracts sonicated and clarified. Approximately 1 ml of Ni2+-NTA agarose (QIAGEN) was incubated with the supernatants for 1 h at 4°C and then washed extensively with lysis buffer lacking IGEPAL CA-630 and protease inhibitors. His6XMCAK and His6XMCAK-4A were eluted with 50 mM sodium phosphate (pH 7.8), 500 mM NaCl, 5 mM β-mercaptoethanol, 0.1 mM MgATP, 300 mM imidazole, and the peak fractions subjected to gel filtration chromatography using either Superose 6 or Superdex 200 columns (Amersham Biosciences, Piscataway, NJ) equilibrated in gel filtration (GF) buffer (10 mM K-HEPES, pH 7.7, 300 mM KCl, 1 mM DTT) containing 0.1 mM MgATP. Powdered sucrose (10%) was added to the peak fractions, and the proteins aliquoted, frozen in liquid N2, and stored at -80°C.
Full-length, untagged XMCAK expressed in Sf-9 cells was purified as described previously (Desai et al., 1999b).
In Vitro Kinase Assays
A 15× kinase stock was prepared by diluting His6Aurora B-His6INCENP to 37.5 μg/ml with kinase dilution buffer (20 mM K-HEPES, pH 7.7, 50 mM KCl, 1 mM DTT), and XMCAK (wt and -4A) concentrations equalized with GF buffer. Kinase reactions (15 μl) contained 2.5 μg/ml His6Aurora B-His6INCENP, and 1 μg of substrate in 1× kinase buffer (20 mM K-HEPES, pH 7.7, 5 mM MgCl2, 1 mM EGTA, 1 mM DTT, 50 μM ATP, 1.5 μCi of [γ-32P] ATP; Amersham Biosciences) and were incubated at 30°C for 30 min. Reactions were terminated by addition of 15 μl of 2× Laemmli sample buffer, resolved by 10% SDS-PAGE, and visualized by autoradiography.
Microtubule Depolymerization Assays
Microtubule depolymerization by XMCAK was monitored by light scattering (Hunter et al., 2003). Stock solutions (100×) of His6Aurora B-His6INCENP, wt His6XMCAK, and His6XMCAK-4A were prepared by diluting the proteins into 20 mM K-HEPES (pH 7.7), 50 mM KCl, 1 mM DTT, 0.2 mg/ml casein immediately before assembling reaction mixtures. Depolymerization reactions (100 μl) with 15 nM kinesin and 0.87 μg/ml His6Aurora B-His6INCENP, as required, were preincubated in 1× BRB80 plus 1 mM MgATP, 1 mM DTT, 0.1 mg/ml casein, 60 mM KCl for 30 min at room temperature and initiated with the addition of 1 μM GMPCPP-stabilized microtubules. When needed, staurosporine (Calbiochem, San Diego, CA) was included at 50 nM. Reactions were mixed, transferred into a quartz cuvette, and the turbidity measured at 100-ms intervals by using a Cary Eclipse fluorescence spectrophotometer (Varian, Palo Alto, CA) with excitation and emission wavelengths set to 350 nm. Absorbance readings over a 15-min period were imported into Excel (Microsoft, Redmond, WA) and converted into percentages assuming a value of 100% for t0. Data are presented using values recorded every 15 s.
MCAK Phosphorylation Site-Mapping by Mass Spectrometry
Coomassie-stained XMCAK protein bands were excised after SDS-PAGE and digested in-gel with trypsin (Shevchenko et al., 1996). Extracted peptides were separated by nanoscale reverse-phase high-performance liquid chromatography. Each peptide was subjected to electrospray ionization after their elution and analyzed by an LCQ-DECA ion trap mass spectrometer (Thermo Finnigan, San Jose, CA). Eluting peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide. Peptide sequences were determined by matching protein databases with the acquired fragmentation pattern using the software program Sequest (Eng et al., 1994).
Preparation of Xenopus egg extracts and In Vitro Spindle Assembly
Cytostatic factor (CSF)-arrested Xenopus egg extracts were prepared as described previously (Murray, 1991) and supplemented with 50 mM sucrose. To immunodeplete MCAK, 20 μg of affinity-purified anti-XMCAK or rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) were incubated with 50 μl of protein A-Dynabeads (Dynal Biotech, Lake Success, NY) overnight at 4°C. Antibody-bound beads were washed once with Tris-buffered saline plus 0.1% Triton X-100, three times with CSF-XB (Murray, 1991), and resuspended in 150 μl of CSF extract. After 90 min on ice, the IgG- and MCAK-depleted extracts were separated from the beads with a magnet and collected.
Before spindle assembly, 50 μl of MCAK-depleted extracts were supplemented with either 1 μl of GF buffer, wt XMCAK, or XMCAK-4A (both at 3.2 μM). IgG-depleted extract received either 1 μl of GF buffer or no solution. X-Rhodamine-labeled tubulin (Hyman et al., 1991) was also included in all extracts at 50 μg/ml to visualize microtubules. Cycled spindles were assembled as described previously (Desai et al., 1999a) by using demembranated Xenopus sperm nuclei (Murray, 1991) at 400/μl. Structures assembled 90 min after the addition of CSF extract were analyzed by squashing 1 μl of extract with 4 μl of fix (Murray, 1991) underneath a coverslip and viewing the sample by fluorescence microscopy.
Immunofluorescence Microscopy
Mitotic structures assembled in Xenopus extracts were processed for immunofluorescence as described previously (Desai et al., 1999a). Coverslips were blocked in AbDil (Tris-buffered saline plus 0.1% Triton X-100 + 2% bovine serum albumin) for 30 min and then incubated with primary antibodies (Alexa 488- and Alexa 594-labeled anti-XMCAK; Alexa 488-labeled anti-XCENP-A, a gift from A. Straight, Stanford School of Medicine) diluted to 1 μg/ml in AbDil for 1 h. After staining DNA with 5 μg/ml Hoechst 33342, coverslips were mounted in 90% glycerol, 0.5% p-phenylenediamine, 20 mM Tris-Cl (pH 8.8) and imaged at the Nikon Imaging Center at Harvard Medical School by using a TE2000 inverted microscope (Nikon, Tokyo, Japan) with a spinning-disk confocal head (PerkinElmer Life and Analytical Sciences, Boston, MA) and a 100×/1.4 numerical aperture objective (Nikon). Images were acquired at 0.25-μm z-steps and used to produce deconvolved reconstructions of the image stacks (Autoquant, Watervliet, NY), which are presented as maximum intensity projections.
Pelleting of Microtubule Structures from Xenopus egg Extracts and Immunoblotting
To pellet microtubule-based structures assembled during metaphase, 20 μl of cycled extract prepared as described above was diluted into 200 μl of 1× BRB80/30% glycerol and centrifuged over 1 ml 1× BRB80/40% glycerol cushions (8500 rpm in a 5415 C Eppendorf microcentrifuge) for 20 min at room temperature. Equivalent amounts of the initial extract, pellet, and supernatant fractions were resolved by 10% SDS-PAGE and analyzed by immunoblotting. Quantification of α-tubulin detected by enhanced chemiluminescence was performed using the public domain software NIH Image.
Typically, immunoblots were blocked with 5% (wt/vol) skim milk in Tris-buffered saline containing 0.2% Tween 20 and then probed with primary antibodies diluted 1:1000 (anti-α-tubulin DM1α; Sigma-Aldrich, St. Louis, MO) or to 1 μg/ml (anti-XMCAK) in blocking solution. Bound antibodies were detected by enhanced chemiluminescence (Amersham Biosciences).
RESULTS
XMCAK Is Inhibited by Aurora B-dependent Phosphorylation In Vitro
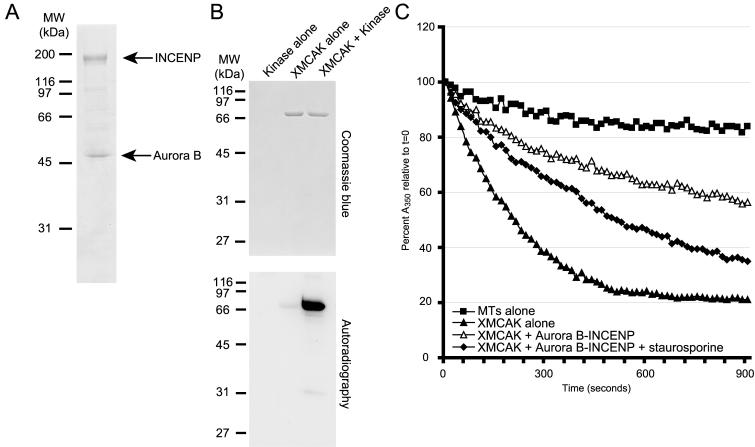
To begin examining whether MCAK is a physiologically relevant substrate of the Aurora B kinase, we first purified recombinant Xenopus Aurora B-INCENP complex after their coexpression in Sf-9 cells using the baculovirus expression system (Figure 1A). The kinase is active as judged by its ability to robustly phosphorylate INCENP, a known Aurora B substrate (Bishop and Schumacher, 2002; Honda et al., 2003), and myelin basic protein (our unpublished data). We tested the ability of Aurora B-INCENP to phosphorylate untagged, full-length, recombinant Xenopus MCAK. As shown in Figure 1B, XMCAK was phosphorylated specifically in the presence of Aurora B-INCENP, indicating that XMCAK is a substrate of Aurora B in vitro.
Figure 1.
Xenopus MCAK is inhibited by Aurora B-dependent phosphorylation. (A) Purification of recombinant His6-tagged Xenopus Aurora B-INCENP. Coomassie Blue staining of 3 μg of Aurora B-INCENP purified after their coexpression in Sf-9 insect cells is shown. (B) Xenopus MCAK is a substrate of Aurora B-INCENP in vitro. Aurora B-INCENP and full-length, untagged XMCAK were mixed in various combinations in the presence of [γ-32P]ATP, and the products of the kinase assays were analyzed by autoradiography and Coomassie Blue staining. (C) XMCAK is inhibited by Aurora B-INCENP in vitro. The stability of GMPCPP-microtubules (1 μM) was measured by light scattering after exposure to XMCAK (15 nM) pretreated without (triangles) or with 0.87 μg/ml Aurora B-INCENP (open triangles), or with Aurora B-INCENP and 50 nM staurosporine (diamonds). Squares, microtubules alone.
To determine whether phosphorylation of XMCAK by Aurora B-INCENP affects the microtubule depolymerizing activity of this KinI kinesin, the ability of XMCAK to disassemble GMPCPP-stabilized microtubules after exposure to Aurora B-INCENP was measured using a light scattering turbidity assay (Hunter et al., 2003). Microtubules (1 μM) alone (Figure 1C, squares) were stable and depolymerized <20% throughout the 15-min time course, whereas inclusion of His6-tagged XMCAK (15 nM) caused catalytic microtubule disassembly (∼80%, triangles). Microtubule depolymerization by XMCAK was ATP dependent and inhibited by 1 mM adenylylimidodiphosphate (our unpublished data). Preincubation of XMCAK with 0.87 μg/ml Aurora B-INCENP led to a reproducible ∼60% inhibition of XMCAK activity such that ∼55% of tubulin polymer remained at the final time point (open triangles). Aurora B-INCENP alone had no effect on the stability of GMPCPP-microtubules (our unpublished data). To verify that the inhibitory effect of Aurora B-INCENP on XMCAK was due to phosphorylation, the broad-spectrum kinase inhibitor staurosporine (Ruegg and Burgess, 1989) was tested for its ability to counteract Aurora B-INCENP activity. Figure 1C (diamonds) shows that ∼65% of tubulin polymer depolymerized in this reaction indicating that XMCAK activity had been restored to 75% of that observed with XMCAK preincubated with no kinase. Collectively, these results indicate that phosphorylation of XMCAK by Aurora B-INCENP in vitro leads to inhibition of its microtubule depolymerizing activity.
Phosphopeptide Mapping of XMCAK
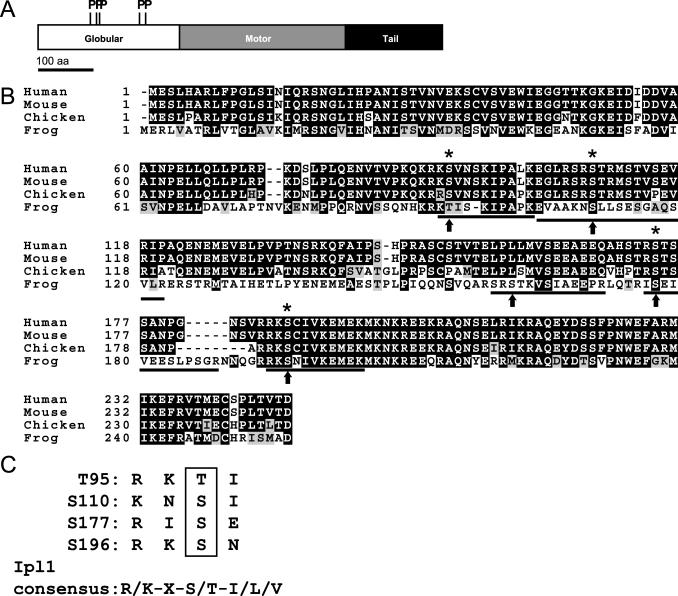
To create a mutant version of XMCAK that is resistant to inhibition by Aurora B-INCENP, residues in XMCAK phosphorylated by Aurora B-INCENP in vitro were first identified by mass spectrometry. By liquid chromatography-tandem mass spectrometry (LC-MS/MS), comparison of tryptic peptides derived from unmodified and phosphorylated XM-CAK revealed that five peptides (Figure 2B; 94-103, 104-122, 159-171, 176-188, and 194-204) specifically within XMCAK exposed to Aurora B contained single phosphate groups, as judged by a +80-Da shift in molecular weight. All five phosphorylation sites (T95, S110, S161, S177, and S196) reside within the N-terminal globular region of XMCAK (Walczak et al., 1996), upstream of the motor domain (Figure 2A). Four of the five phospho-acceptor sites (T95, S110, S177, and S196) conform well to the consensus sequence determined for yeast Ipl1-Sli15 (Figure 2C; Cheeseman et al., 2002). In addition, T95, S110, S177, and S196 are conserved between MCAK homologues from other vertebrates (Figure 2B), providing evidence that these are the relevant XMCAK residues responsible for its regulation by Aurora B-INCENP. Finally, by LC-MS/MS, we have detected phosphorylation of MCAK residues S177 and S196 in protein immunoprecipitated from mitotic XTC tissue cell lysates, indicating that these two residues are indeed phosphorylated in vivo (our unpublished data).
Figure 2.
Identification of Xenopus MCAK residues phosphorylated by Aurora B. (A) Aurora B-INCENP phosphorylates XMCAK at its N terminus. Schematic diagram of XMCAK domains and locations of five phospho-residues generated by in vitro phosphorylation of XMCAK with Aurora B-INCENP as determined by mass spectrometry. (B) Conservation of XMCAK phospho-acceptor sites. Alignment of N termini from frog MCAK and its orthologues from human, mouse, and chicken to depict conservation of T95, S110, S177, and S196 (asterisks). Phosphotryptic peptides identified by LC-MS/MS are underlined, and phosphorylated residues are indicated by arrows. (C) Alignment of XMCAK phosphorylation sites with the Ipl1-Sli15 consensus sequence (Cheeseman et al., 2002).
XMCAK-4A Is Resistant to Inhibition by Aurora B In Vitro
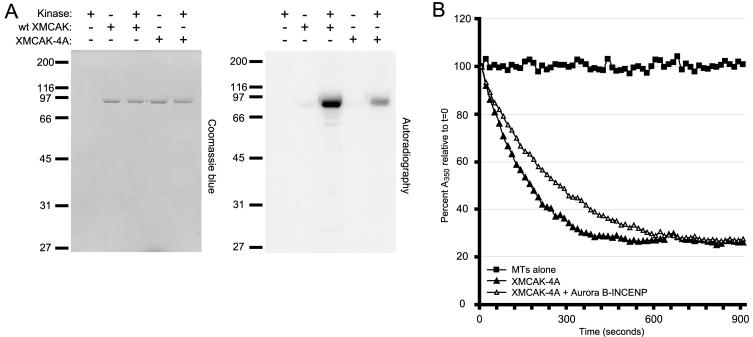
To test whether phosphorylation of XMCAK by Aurora B-INCENP on T95, S110, S177, and S196 were responsible for inhibition of XMCAK activity, these four amino acids were mutated to alanine residues to prevent their posttranslational modification. The full length, His6-tagged mutant protein XMCAK-4A was then expressed in Sf-9 insect cells and purified. When subjected to in vitro kinase assays with Aurora B-INCENP, XMCAK-4A phosphorylation was substantially reduced compared with that of wild-type (wt) XMCAK (Figure 3A), consistent with T95, S110, S177, and S196 being the major XMCAK phosphorylation sites for Aurora B.
Figure 3.
XMCAK-4A is resistant to inhibition by Aurora B-INCENP in vitro. (A) Phosphorylation of XMCAK-4A by Aurora B-INCENP is reduced in comparison with wild-type (wt) XMCAK. Recombinant, full-length wt His6-tagged XMCAK or mutant XMCAK-4A was subjected to kinase assays with Aurora B-INCENP, which were analyzed by autoradiography and Coomassie Blue staining. The absence or presence of Aurora B-INCENP and substrates are indicated with - and +, respectively. (B) XMCAK-4A is resistant to inhibition by Aurora B-INCENP in vitro. XMCAK-4A preincubated with (open triangles) or without (triangles) Aurora B-INCENP was mixed with 1 μM GMPCPP-stabilized microtubules, and the depolymerization were reactions monitored by light scattering. Squares, microtubules alone.
We then tested the ability of XMCAK-4A to depolymerize GMPCPP-stabilized microtubules in the absence and after phosphorylation by Aurora B-INCENP. Importantly, XMCAK-4A alone caused microtubule disassembly (Figure 3B, triangles) with kinetics very similar to that observed for wt XMCAK (Figure 1C), indicating that the alanine substitutions do not grossly alter the enzymatic properties of XMCAK. That XMCAK-4A is properly folded is also suggested by its identical gel filtration profile compared with that of wt recombinant XMCAK (our unpublished data), and its association with ICIS in Xenopus extract as determined by coimmunoprecipitation (Figure S1). In contrast to wt XMCAK however, XMCAK-4A was not inhibited by Aurora B-INCENP. XMCAK-4A depolymerized GMPCPP microtubules with the same efficiency regardless of whether it was preincubated with Aurora B-INCENP (Figure 3B), establishing XMCAK-4A as an Aurora B-INCENP-insensitive XMCAK variant.
XMCAK-4A Is Unable to Support Bipolar Meiotic Spindle Assembly
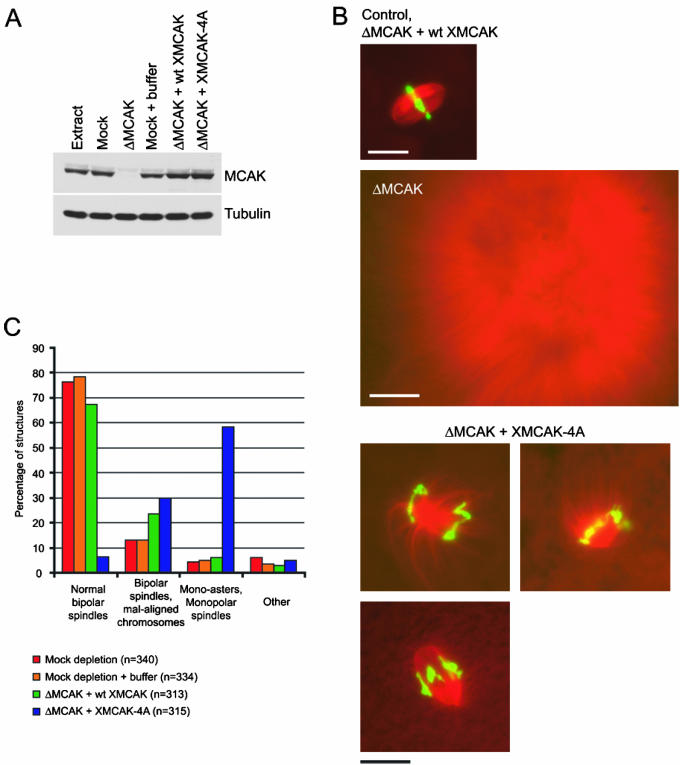
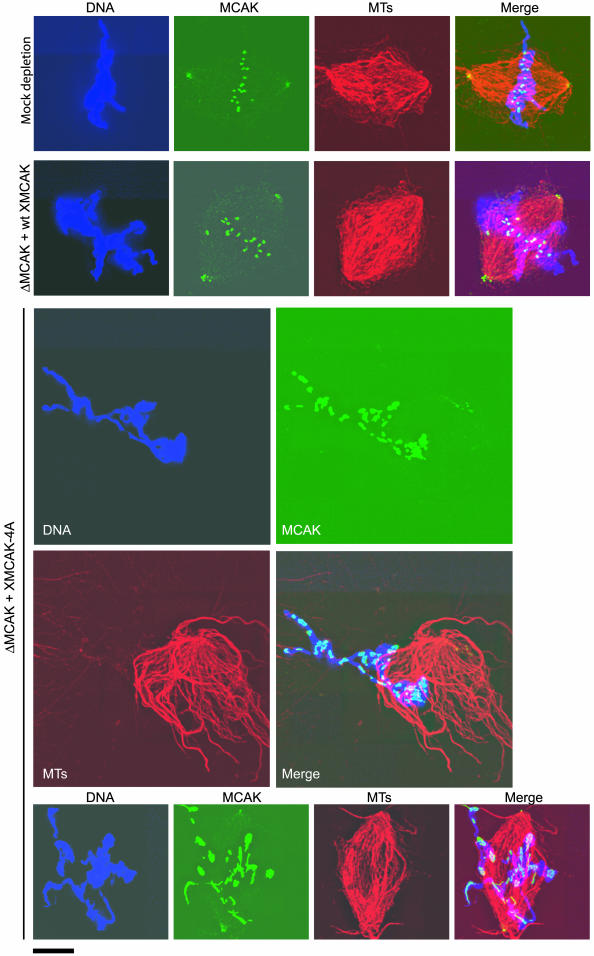
To determine the role(s) of MCAK regulation by Aurora B-INCENP, we assessed the ability of meiotic Xenopus egg extracts lacking endogenous MCAK to assemble spindles when rescued with either recombinant XMCAK or XMCAK-4A. Egg extract was immunodepleted using either control IgG or anti-XMCAK antibodies (Figure 4A). As expected, the majority of assembled structures in the control extract were bipolar spindles (∼90%), whereas MCAK-depleted extracts produced only large sunburst arrays of microtubules (Figure 4B; Walczak et al., 1996).
Figure 4.
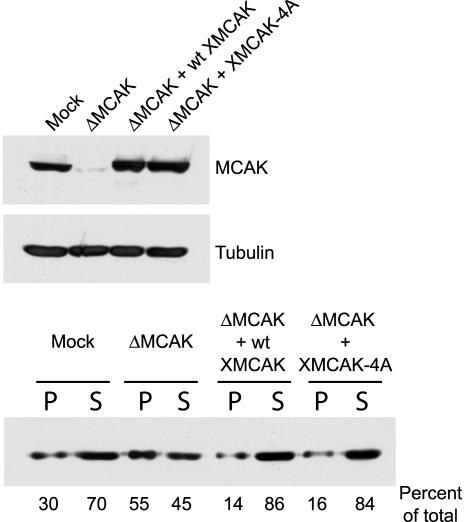
XMCAK-4A is unable to support bipolar spindle assembly in Xenopus egg extracts. (A) MCAK immunodepletion and add-back of wt XMCAK or XMCAK-4A. Xenopus egg extract was immunodepleted using either IgG (Mock) or anti-XMCAK and supplemented with either wt or mutant XMCAK. An immunoblot of 0.5 μl of the extracts indicated was probed with anti-XMCAK and anti-α-tubulin antibodies. (B) XMCAK-4A does not support bipolar spindle assembly efficiently. Representative structures predominantly formed in spindle assembly reactions by using control and XMCAK-depleted extracts, and XMCAK-depleted extracts that were supplemented with either wt XMCAK or XMCAK-4A are shown. Microtubules (red) were visualized with rhodamine-tubulin and DNA (green) with Hoechst 33342. Bars, 20 μM except for the XMCAK-depleted extract, which is 50 μM. (C) Quantitation of structures observed in spindle assembly reactions by using control extracts and MCAK-depleted extracts supplemented with either wt XMCAK or XMCAK-4A. The frequencies of phenotypes observed are averaged from two independent experiments and are expressed as percentages.
To rescue the MCAK-depleted extract, recombinant His6-tagged XMCAK or XMCAK-4A were added to the extract to 10 μg/ml (∼60 nM, dimer concentration), the concentration of endogenous MCAK (Walczak et al., 1996). Figure 4A shows that identical amounts of the wt and mutant kinesins were added to the MCAK-depleted extract and that their levels approximated those of endogenous MCAK.
When induced to assemble mitotic microtubule arrays, the wt XMCAK-supplemented extract generated bipolar spindles to the degree seen in control extracts (91%), although a slight increase (1.7 times) in the percentage of bipolar spindles with misaligned chromosomes was observed (Figure 4C). Similar to wt XMCAK, XMCAK-4A rescued the global microtubule catastrophe defect of MCAK-depleted extract, evident by the lack of long microtubules. However, in contrast to wt XMCAK, XMCAK-4A was severely compromised in its capacity to drive bipolar spindle formation. The majority (59%) of chromatin-containing microtubule structures in this reaction were mono-asters or monopolar spindles (Figure 4, B and C). Of the bipolar spindles that did assemble (37%), ∼80% failed to align their constituent chromosomes at the metaphase plate (Figure 4, B and C). These results indicate that XMCAK-4A has lost an important aspect of its functionality, and further, suggest that its regulation by Aurora B-INCENP-mediated phosphorylation is critical for bipolar spindle assembly.
XMCAK-4A Regulates Tubulin Polymer Levels Correctly
Although the sizes of microtubule-based structures in XMCAK-4A extracts suggest that the mutant protein is not defective for regulating global microtubule dynamics, we sought to measure this more accurately. To this end, demembranated sperm nuclei were replicated in control IgG-depleted, MCAK-depleted, and MCAK-depleted extracts containing either wt XMCAK or XMCAK-4A (Figure 5), and the extracts induced to enter metaphase. Sixty minutes later, microtubule arrays were isolated by centrifugation, and the amount of tubulin present in the pellet and supernatant fractions analyzed by immunoblotting (Figure 5). MCAK depletion led to an approximately twofold increase in the amount of pelleted tubulin polymer compared with that in control extract. This defect was completely rescued by supplementing MCAK-depleted extracts with either wt XMCAK or XMCAK-4A, confirming that bulk microtubule assembly is correctly regulated by the XMCAK phospho-mutant.
Figure 5.
XMCAK-4A controls tubulin polymer levels normally. Control IgG-depleted (mock), MCAK-depleted (ΔMCAK), and MCAK-depleted extracts supplemented with either wt XMCAK or XMCAK-4A were used to assemble metaphase microtubule-based structures. These arrays were isolated by centrifugation and the amount of tubulin in the pellet (P) and supernatant (S) fractions analyzed by immunoblotting. Top, an immunoblot of 0.5 μl of the indicated extracts was probed with anti-XMCAK and anti-α-tubulin antibodies. Bottom, immunoblot of equivalent amounts of pellet and supernatant fractions from the indicated extracts probed with anti-α-tubulin antibodies.
XMCAK-4A Fails to Localize to Inner Centromeres
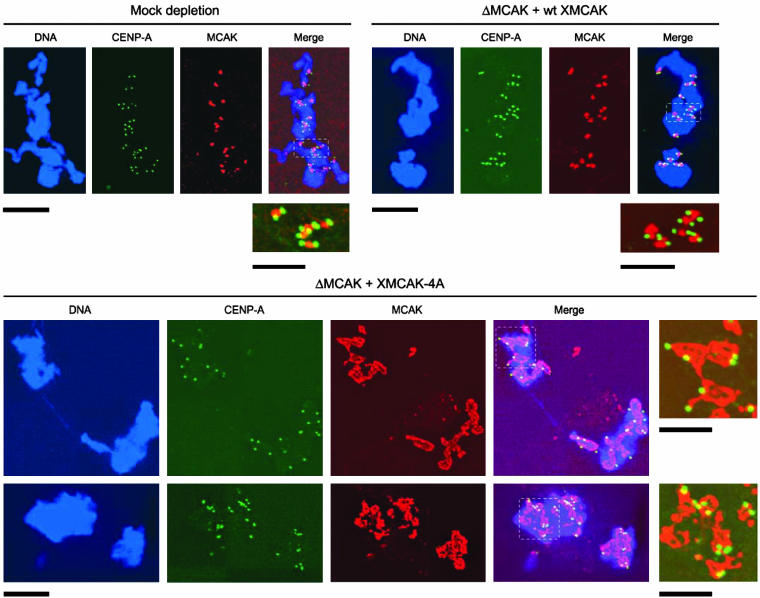
We analyzed the localization of XMCAK on the structures assembled in the immunodepletion/add-back experiment described above. By immunofluorescence with anti-XMCAK antibodies, both endogenous MCAK in the control extract and wt XMCAK added to the MCAK-depleted extract exhibited strong inner centromere labeling and faint staining at spindle poles (Figures 6 and 7). In contrast, XMCAK-4A failed to localize specifically to inner centromeres and instead showed irregular labeling of chromosomes. Chromosome-staining of XMCAK-4A was equally abnormal in both mono-asters and bipolar spindles with misaligned chromosomes (Figure 6), indicating that severity of the spindle assembly phenotype does not correlate with the efficiency of MCAK inner centromere targeting. Notably, the mal-localization of XMCAK-4A is likely not a consequence of a failure to bind ICIS because ICIS coimmunoprecipitates XMCAK-4A with equal efficiency to wt XMCAK from MCAK-depleted extracts supplemented with these recombinant proteins (Figure S1).
Figure 6.
XMCAK-4A fails to localize to inner centromeres. Structures assembled in mock-depleted extract, and MCAK-depleted extracts (ΔMCAK) that were supplemented with either wt XMCAK or XMCAK-4A were fixed and stained with anti-XMCAK antibodies (green). Tubulin (MTs; red) was visualized with rhodamine-tubulin and DNA (blue) with Hoechst 33342. Bar, 10 μM.
Figure 7.
XMCAK-4A does not exhibit preferential localization to centromeres. Structures assembled in mock-depleted extract, and MCAK-depleted extracts (ΔMCAK) supplemented with either wt XMCAK or XMCAK-4A were fixed and double stained with antibodies specific for XMCAK (red) and CENP-A (green). DNA (blue) was visualized with Hoechst 33342. The boxed regions are enlarged and shown without DNA staining to allow more careful comparison of XMCAK and CENP-A immunofluorescence. Bars, 10 μM except for the enlarged images, which are 5 μM.
We analyzed the abnormal localization of XMCAK-4A more carefully by costaining structures assembled in XMCAK-4A extracts with antibodies specific for the kinetochore protein CENP-A (Maddox et al., 2003) and XMCAK. As expected, both endogenous MCAK and wt XMCAK added to MCAK-depleted extracts localized precisely in between sister kinetochores (Figure 7). CENP-A immunostaining on chromosomes assembled in XMCAK-4A extracts was indistinguishable from that observed in control extracts, suggesting that XMCAK-4A does not adversely affect kinetochore assembly. Interestingly, XMCAK-4A immunolabeling did not correlate with the location of kinetochores and inner centromeres, indicating that XMCAK-4A may bind randomly to chromosome arms. Together, these data strongly suggest that phosphorylation of MCAK by Aurora B-INCENP is required for the motor to target inner centromeres.
DISCUSSION
MCAK Is a Substrate of Aurora B
The protein kinase Aurora B has emerged as a central regulator of chromosome segregation whose activity is important for M-phase chromosome assembly (Swedlow and Hirano, 2003) and chromosome biorientation (Tanaka, 2002; Andrews et al., 2003). As such, it is critical to identify substrates of Aurora B relevant to each of its functions. Our knowledge of Aurora B substrates that influence kinetochore-microtubule interactions is derived mainly from studies using budding yeast, where phosphorylation of two kinetochore proteins, Ndc10 and Dam1, by Ipl1 (yeast Aurora) influences the microtubule-affinity of kinetochores (Sassoon et al., 1999; Cheeseman et al., 2002). In vertebrates, it is plausible that nonkinetochore substrates of Aurora B may also contribute to chromosome biorientation given that Aurora B localizes to the inner centromere, a chromosomal region morphologically and spatially distinct from the kinetochore (Cleveland et al., 2003).
In Xenopus extracts, Aurora B and the microtubule depolymerase MCAK, which also localizes to the inner centromere during preanaphase mitosis, associate with a third inner centromere protein, ICIS (Ohi et al., 2003). An intriguing possibility is that ICIS and Aurora B associate with MCAK to regulate the activity of the KinI kinesin, an idea consistent with the MCAK-stimulatory activity possessed by ICIS. Our present demonstration that Aurora B phosphorylates MCAK and thereby inhibits its microtubule depolymerizing activity in vitro further strengthens this notion.
Genetic analysis in Saccharomyces cerevisiae and real-time studies of PtK1 cells treated with the small molecule Aurora B inhibitor Hesperadin indicate that Ipl1/Aurora B are required to destabilize incorrect kinetochore-microtubule attachments (Tanaka et al., 2002; Hauf et al., 2003; Lampson et al., 2004). Aurora B may accomplish this by either reducing microtubule affinity of kinetochores or activating a protein that can destabilize improper linkages. That Aurora B inhibits MCAK in vitro seems inconsistent with the latter possibility and suggests that the biochemistry underlying attachment error correction is complex. To address this issue further, it will be interesting to measure Aurora B activation and MCAK phosphorylation as a function of different types of kinetochore-microtubule attachment errors. Additionally, it will be important to assess the impact of ICIS on MCAK regulation by Aurora B both in vitro and in the context of the inner centromere.
Cytoplasmic Control of Microtubule Polymerization Does Not Require Aurora B-mediated Inhibition of MCAK
MCAK is indispensable for the control of cytoplasmic microtubule dynamics as demonstrated by the long microtubules that organize into large, sunburst arrays upon its removal from Xenopus egg extract (Walczak et al., 1996). Complementation of MCAK-depleted Xenopus extracts with an alanine mutant of MCAK (XMCAK-4A) that cannot be inhibited by Aurora B-dependent phosphorylation in vitro rescued the cytoplasmic microtubule phenotype, evident by the formation of structures that were similar in size and shape to those which transiently form during spindle assembly in normal Xenopus extracts. This indicates that XMCAK-4A fulfills the functional requirement of MCAK in controlling global microtubule polymerization, a notion we confirmed biochemically by measuring tubulin polymer that was incorporated into precipitable microtubule assemblies. It is reasonable that MCAK inhibition by Aurora B is not essential for cytoplasmic microtubule dynamics regulation; indeed, the increase in catastrophe frequency observed during mitosis (Kinoshita et al., 2001; Rusan et al., 2001) and meiosis (Belmont et al., 1990; Verde et al., 1992) is more likely to require activation of MCAK than inhibition. Factors that contribute to cytoplasmic MCAK activation during M-phase onset remain to be identified. The Cdc2 kinase is the master regulator of M-phase microtubule dynamics (Verde et al., 1992), suggesting that MCAK activity may also be modulated by Cdc2-dependent control.
Regulation of MCAK by Aurora B Is Important for Bipolar Meiotic Spindle Assembly
Although XMCAK-4A is competent to prevent abnormal cytoplasmic microtubule polymerization, the Aurora B-resistant MCAK mutant is defective in its ability to drive bipolar spindle assembly. Replacing endogenous MCAK in Xenopus extracts with XMCAK-4A led predominantly to the formation of mono-asters and monopolar spindles. This indicates that regulation of MCAK by Aurora B is normally important to control aspects of spindle microtubule dynamics that are essential for spindle bipolarization. We have also found that MCAK phosphorylation by Aurora B is required for MCAK to target inner centromeres as XMCAK-4A exhibits aberrant chromosome arm localization. It is possible that maltargeting of XMCAK-4A exacerbates the effect of uncoupling MCAK activity from Aurora B, but it cannot be the principle cause for spindle assembly failure because a small, but reproducible percentage of bipolar spindles with mislocalized MCAK does form in XMCAK-4A extracts.
It is difficult to postulate how Aurora B regulation of MCAK tailors microtubule dynamics to drive spindle bipolarization, primarily because little is known about the role of microtubule dynamics in this process. In meiotic Xenopus egg extracts, spindle assembly is triggered by chromatin in a manner involving Ran-GTP (Carazo-Salas et al., 1999; Kalab et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999) and possibly by other factors that control microtubule dynamics such as the chromokinesin XKlp1 (Bringmann et al., 2004). Bipolarity is then established by microtubule minus- and plus-end-directed motors that organize microtubule minus ends at the spindle poles, and slide antiparallel microtubules apart, respectively (Walczak et al., 1998; Sharp et al., 2000). Determining whether MCAK regulation by Aurora B is downstream of the Ran pathway, or whether this regulation comprises a separate mechanism by which chromosomes influence microtubule dynamics is an important goal for the future.
An unresolved issue concerns whether MCAK regulation by Aurora B is required for spindle bipolarization in all cell types. Although defects in spindle structure are observed in somatic cells microinjected with anti-Aurora B antibodies or overexpressing kinase-defective Aurora B (Kallio et al., 2002; Murata-Hori and Wang, 2002), the abnormal spindles are nonetheless bipolar. Inactivation of Aurora B in somatic cells or nematode embryos by using small molecule Aurora B inhibitors or RNAi have similarly revealed no role for the kinase in spindle assembly (Andrews et al., 2003). All of these studies, however, have used systems in which spindle bipolarity is achieved primarily by a microtubule organizing center (MTOC)-driven pathway. In this pathway, the MTOC duplicates, separates before nuclear envelope breakdown, and a bipolar spindle assembles between the paired MTOCs. As discussed above, Xenopus meiotic spindles, as well as those in insect oocytes (Matthies et al., 1996) and mitotic spindles in higher plant cells (Smirnova and Bajer, 1998), form using a second pathway involving chromosome-stimulated microtubule assembly and motor-dependent microtubule sorting. It is possible that this second pathway uses mechanisms to regulate microtubule dynamics that are not used in the MTOC-driven pathway. Indeed, it will be informative to further examine the role of Aurora B during spindle assembly in Xenopus extracts.
Dissection of MCAK Functions during Chromosome Segregation
MCAK is required for numerous aspects of chromosome segregation during M phase. MCAK controls global microtubule dynamics (Walczak et al., 1996), contributes to polewards chromosome movement during anaphase A (Maney et al., 1998; Rogers et al., 2003), and has a preanaphase function(s) at inner centromeres that involves suppression of abnormal kinetochore-microtubule attachments (Kline-Smith and Walczak, 2002; Walczak et al., 2002; Rogers et al., 2003; Kline-Smith et al., 2004). By investigating Aurora B-mediated regulation of MCAK, we have separated the requirement of MCAK for cytoplasmic microtubule dynamics regulation from a fourth, novel function in affecting spindle microtubule dynamics that influence spindle bipolarity.
In addition to being critical for bipolar spindle assembly, Aurora B-mediated regulation of MCAK is expected to influence kinetochore-microtubule attachments given the importance of the kinase in chromosome biorientation. MCAK activity is likely to be regulated by Aurora B in vivo because we have detected phosphorylation of MCAK S177 and S196 in mitotic Xenopus tissue cells (our unpublished data). In addition, recently published studies indicate that MCAK is phosphorylated at S196 (Lan et al., 2004) and S92 (equivalent to T95 in Xenopus MCAK; Andrews et al., 2004) in Xenopus and human somatic cells, respectively. Consistent with a function for MCAK regulation by Aurora B in affecting chromosome-microtubule interactions, these studies also showed that overexpression of MCAK phosphorylation site mutants or blocking MCAK function with phospho-specific antibodies lead to chromosome biorientation, alignment, and segregation defects (Andrews et al., 2004; Lan et al., 2004).
It is clear that further analysis of MCAK during chromosome segregation requires more sophisticated reagents and approaches that will specifically affect one of its several functional pools. One successful method has been to develop dominant negative MCAK fragments that block its recruitment to centromeres (Maney et al., 1998; Walczak et al., 2002; Kline-Smith et al., 2004). We have demonstrated here that an additional strategy is to uncouple MCAK from kinases that modulate its microtubule depolymerizing activity by appropriate mutagenesis. Continued efforts to identify mechanisms that control MCAK activity and localization will undoubtedly aid us in understanding how the microtubule depolymerizing activity of MCAK is harnessed to affect the dynamics of multiple classes of spindle microtubules.
Supplementary Material
Acknowledgments
We thank C. Walczak, J. Swedlow, L. Wordeman, and T. Stukenberg for sharing unpublished information; A. Straight and A. Salic for reagents; and members of the Mitchison laboratory as well as M. Ohi for fruitful discussions. The authors are grateful to Jennifer Waters-Shuler for assistance with deconvolution fluorescence microscopy that was performed at the Nikon Imaging Center at Harvard Medical School. We thank Ross Tomaino at the Taplin Biological Mass Spectrometry Facility at Harvard Medical School for expert analysis of protein phosphorylation. M. Ohi and B. Brieher are appreciated for comments on the manuscript. This work was funded by National Institutes of Health grants GM-20309 (to R.O.), AR-40593 (to J.H.) and GM-39565 (to T.J.M.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-02-0082. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-02-0082.
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Adams, R.R., Maiato, H., Earnshaw, W.C., and Carmena, M. (2001). Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153, 865-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, P.D., Knatko, E., Moore, W.J., and Swedlow, J.R. (2003). Mitotic mechanics: the auroras come into view. Curr. Opin. Cell Biol. 15, 672-683. [DOI] [PubMed] [Google Scholar]
- Andrews, P.D., Ovechkina, Y., Morrice, N., Wagenbach, M., Duncan, K., Wordeman, L., and Swedlow, J.R. (2004). Aurora B regulates MCAK at the mitotic centromere. Dev. Cell 6, 253-268. [DOI] [PubMed] [Google Scholar]
- Belmont, L.D., Hyman, A.A., Sawin, K.E., and Mitchison, T.J. (1990). Real-time visualization of cell cycle-dependent changes in microtubule dynamics in cytoplasmic extracts. Cell 62, 579-589. [DOI] [PubMed] [Google Scholar]
- Bishop, J.D., and Schumacher, J.M. (2002). Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B kinase stimulates Aurora B kinase activity. J. Biol. Chem. 277, 27577-27580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton, M.A., Lan, W., Powers, S.E., McCleland, M.L., Kuang, J., and Stukenberg, P.T. (2002). Aurora B kinase exists in a complex with Survivin and INCENP and its kinase activity is stimulated by Survivin binding and phosphorylation. Mol. Biol. Cell 13, 3064-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann, H., Skiniotis, G., Spilker, A., Kandels-Lewis, S., Vernos, I., and Surrey, T. (2004). A kinesin-like motor inhibits microtubule dynamic instability. Science 303, 1519-1522. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas, R.E., Guarguaglini, G., Gruss, O.J., Segref, A., Karsenti, E., and Mattaj, I.W. (1999). Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400, 178-181. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., Anderson, S., Jwa, M., Green, E.M., Kang, J., Yates, J.R., 3rd, Chan, C.S., Drubin, D.G., and Barnes, G. (2002). Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111, 163-172. [DOI] [PubMed] [Google Scholar]
- Cleveland, D.W., Mao, Y., and Sullivan, K.F. (2003). Centromeres and kinetochores. From epigenetics to mitotic checkpoint signaling. Cell 112, 407-421. [DOI] [PubMed] [Google Scholar]
- Desai, A., Murray, A., Mitchison, T.J., and Walczak, C.E. (1999a). The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61, 385-412. [DOI] [PubMed] [Google Scholar]
- Desai, A., Verma, S., Mitchison, T.J., and Walczak, C.E. (1999b). Kin I kinesins are microtubule-destabilizing enzymes. Cell 96, 69-78. [DOI] [PubMed] [Google Scholar]
- Ditchfield, C., Johnson, V.L., Tighe, A., Ellston, R., Haworth, C., Johnson, T., Mortlock, A., Keen, N., and Taylor, S.S. (2003). Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161, 267-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng, J.K., McCormack, A.L., and Yates, J.R., 3rd. (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass. Spectrom. 5, 976-989. [DOI] [PubMed] [Google Scholar]
- Hauf, S., Cole, R.W., LaTerra, S., Zimmer, C., Schnapp, G., Walter, R., Heckel, A., van Meel, J., Rieder, C.L., and Peters, J.M. (2003). The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161, 281-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, R., Korner, R., and Nigg, E.A. (2003). Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol. Biol. Cell 14, 3325-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, A.W., Caplow, M., Coy, D.L., Hancock, W.O., Diez, S., Wordeman, L., and Howard, J. (2003). The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol. Cell 11, 445-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman, A., Drechsel, D., Kellogg, D., Salser, S., Sawin, K., Steffen, P., Wordeman, L., and Mitchison, T. (1991). Preparation of modified tubulins. Methods Enzymol. 196, 478-485. [DOI] [PubMed] [Google Scholar]
- Hyman, A.A., and Karsenti, E. (1996). Morphogenetic properties of microtubules and mitotic spindle assembly. Cell 84, 401-410. [DOI] [PubMed] [Google Scholar]
- Kalab, P., Pu, R.T., and Dasso, M. (1999). The ran GTPase regulates mitotic spindle assembly. Curr. Biol. 9, 481-484. [DOI] [PubMed] [Google Scholar]
- Kallio, M.J., McCleland, M.L., Stukenberg, P.T., and Gorbsky, G.J. (2002). Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr. Biol. 12, 900-905. [DOI] [PubMed] [Google Scholar]
- Kinoshita, K., Arnal, I., Desai, A., Drechsel, D.N., and Hyman, A.A. (2001). Reconstitution of physiological microtubule dynamics using purified components. Science 294, 1340-1343. [DOI] [PubMed] [Google Scholar]
- Kinoshita, K., Habermann, B., and Hyman, A.A. (2002). XMAP 215, a key component of the dynamic microtubule cytoskeleton. Trends Cell Biol. 12, 267-273. [DOI] [PubMed] [Google Scholar]
- Kline-Smith, S.L., Khodjakov, A., Hergert, P., and Walczak, C.E. (2004). Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol. Cell 15, 1146-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith, S.L., and Walczak, C.E. (2002). The microtubule-destabilizing kinesin XKCM1 regulates microtubule dynamic instability in cells. Mol. Biol. Cell 13, 2718-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson, M.A., Renduchitala, K., Khodjakov, A., and Kapoor, T.M. (2004). Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 6, 232-237. [DOI] [PubMed] [Google Scholar]
- Lan, W., Zhang, X., Kline-Smith, S.L., Rosasco, S.E., Barrett-Wilt, G.A., Shabanowitz, J., Hunt, D.F., Walczak, C.E., and Stukenberg, P.T. (2004). Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14, 273-286. [DOI] [PubMed] [Google Scholar]
- Maddox, P., Straight, A., Coughlin, P., Mitchison, T.J., and Salmon, E.D. (2003). Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J. Cell Biol. 162, 377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney, T., Hunter, A.W., Wagenbach, M., and Wordeman, L. (1998). Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 142, 787-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies, H.J., McDonald, H.B., Goldstein, L.S., and Theurkauf, W.E. (1996). Anastral meiotic spindle morphogenesis: role of the non-claret disjunctional kinesin-like protein. J. Cell Biol. 134, 455-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata-Hori, M., and Wang, Y. (2002). The kinase activity of Aurora B is required for kinetochore-microtubule interactions during mitosis. Curr. Biol. 12, 894-899. [DOI] [PubMed] [Google Scholar]
- Murnion, M.E., Adams, R.R., Callister, D.M., Allis, C.D., Earnshaw, W.C., and Swedlow, J.R. (2001). Chromatin-associated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J. Biol. Chem. 276, 26656-26665. [DOI] [PubMed] [Google Scholar]
- Murray, A.W. (1991). Cell cycle extracts. Methods Cell Biol 36, 581-605. [PubMed] [Google Scholar]
- Oegema, K., Desai, A., Rybina, S., Kirkham, M., and Hyman, A.A. (2001). Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153, 1209-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba, T., Nakamura, M., Nishitani, H., and Nishimoto, T. (1999). Selforganization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science 284, 1356-1358. [DOI] [PubMed] [Google Scholar]
- Ohi, R., Coughlin, M.L., Lane, W.S., and Mitchison, T.J. (2003). An inner centromere protein that stimulates the microtubule depolymerizing activity of a KinI kinesin. Dev. Cell 5, 309-321. [DOI] [PubMed] [Google Scholar]
- Rieder, C.L., and Salmon, E.D. (1998). The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 8, 310-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, G.C., Rogers, S.L., Schwimmer, T.A., Ems-McClung, S.C., Walczak, C.E., Vale, R.D., Scholey, J.M., and Sharp, D.J. (2003). Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature 427, 364-370. [DOI] [PubMed] [Google Scholar]
- Ruegg, U.T., and Burgess, G.M. (1989). Staurosporine, K-252 and UCN-01, potent but nonspecific inhibitors of protein kinases. Trends Pharmacol. Sci. 10, 218-220. [DOI] [PubMed] [Google Scholar]
- Rusan, N.M., Fagerstrom, C.J., Yvon, A.M., and Wadsworth, P. (2001). Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-alpha tubulin. Mol. Biol. Cell 12, 971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassoon, I., Severin, F.F., Andrews, P.D., Taba, M.R., Kaplan, K.B., Ashford, A.J., Stark, M.J., Sorger, P.K., and Hyman, A.A. (1999). Regulation of Saccharomyces cerevisiae kinetochores by the type 1 phosphatase Glc7p. Genes Dev. 13, 545-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D.J., Rogers, G.C., and Scholey, J.M. (2000). Microtubule motors in mitosis. Nature 407, 41-47. [DOI] [PubMed] [Google Scholar]
- Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850-858. [DOI] [PubMed] [Google Scholar]
- Smirnova, E.A., and Bajer, A.S. (1998). Early stages of spindle formation and independence of chromosome and microtubule cycles in Haemanthus endosperm. Cell Motil. Cytoskeleton 40, 22-37. [DOI] [PubMed] [Google Scholar]
- Swedlow, J.R., and Hirano, T. (2003). The making of the mitotic chromosome: modern insights into classical questions. Mol. Cell 11, 557-569. [DOI] [PubMed] [Google Scholar]
- Tanaka, T.U. (2002). Bi-orienting chromosomes on the mitotic spindle. Curr. Opin. Cell Biol. 14, 365-371. [DOI] [PubMed] [Google Scholar]
- Tanaka, T.U., Rachidi, N., Janke, C., Pereira, G., Galova, M., Schiebel, E., Stark, M.J., and Nasmyth, K. (2002). Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108, 317-329. [DOI] [PubMed] [Google Scholar]
- Tournebize, R., Popov, A., Kinoshita, K., Ashford, A.J., Rybina, S., Pozniakovsky, A., Mayer, T.U., Walczak, C.E., Karsenti, E., and Hyman, A.A. (2000). Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol. 2, 13-19. [DOI] [PubMed] [Google Scholar]
- Verde, F., Dogterom, M., Stelzer, E., Karsenti, E., and Leibler, S. (1992). Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. J. Cell Biol. 118, 1097-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak, C.E., Gan, E.C., Desai, A., Mitchison, T.J., and Kline-Smith, S.L. (2002). The microtubule-destabilizing kinesin XKCM1 is required for chromosome positioning during spindle assembly. Curr. Biol. 12, 1885-1889. [DOI] [PubMed] [Google Scholar]
- Walczak, C.E., Mitchison, T.J., and Desai, A. (1996). XKCM 1, a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell 84, 37-47. [DOI] [PubMed] [Google Scholar]
- Walczak, C.E., Vernos, I., Mitchison, T.J., Karsenti, E., and Heald, R. (1998). A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 8, 903-913. [DOI] [PubMed] [Google Scholar]
- Wilde, A., and Zheng, Y. (1999). Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science 284, 1359-1362. [DOI] [PubMed] [Google Scholar]
- Wittmann, T., Hyman, A., and Desai, A. (2001). The spindle: a dynamic assembly of microtubules and motors. Nat. Cell Biol. 3, E28-E34. [DOI] [PubMed] [Google Scholar]
- Wordeman, L., and Mitchison, T.J. (1995). Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol. 128, 95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.