Abstract
Although gemcitabine is the most commonly used drug for treating pancreatic cancers, acquired gemcitabine resistance in a substantial number of patients appears to hinder its effectiveness in successful treatment of this dreadful disease. To understand acquired gemcitabine resistance, we generated a gemcitabine-resistant pancreatic cancer cell line using stepwise selection and found that, in addition to the known mechanisms of upregulated expression of ribonucleotide reductase, 14-3-3σ expression is dramatically upregulated, and that 14-3-3σ overexpression contributes to the acquired resistance to gemcitabine and cross-resistance to cytarabine. We also found that the increased 14-3-3σ expression in the gemcitabine-resistant cells is due to demethylation of the 14-3-3σ gene during gemcitabine selection, which could be partially reversed with removal of the gemcitabine selection pressure. Most importantly, the reversible methylation/demethylation of the 14-3-3σ gene appears to be carried out by DNA methyltransferase 1 under regulation by Uhrf1. These findings suggest that the epigenetic regulation of gene expression may play an important role in gemcitabine resistance, and that epigenetic modification is reversible in response to gemcitabine treatment.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) ranks as the fourth most common cause of human death by cancer in the Western world, with a 5-year survival rate of less than 5% and a median survival of 6 months after diagnosis, thereby exhibiting the poorest prognosis of all solid tumors. Although gemcitabine, a deoxycitidine analog, is currently the standard and most commonly used drug for treating PDAC, almost all PDAC patients eventually develop resistance to gemcitabine, the main cause of relapse and death.
Altered expression of enzymes involved in gemcitabine uptake and metabolism such as hENT1 and ribonucleotide reductase (RRM1 and RRM2) has been shown to contribute to both intrinsic and acquired gemcitabine resistance (Voutsadakis, 2011). Recently, overexpression of 14-3-3σ in PDAC has also been observed and was thought to contribute to intrinsic resistance and poor prognosis (Hustinx et al., 2005; Neupane and Korc, 2008; Li et al., 2010). 14-3-3σ belongs to the human 14-3-3 protein family of seven members (β, ε, θ/τ, ζ, σ, γ, η), which are phosphoserine/phosphothreonine-binding proteins and play important roles in multiple biologic processes (Li et al., 2009). Of these seven 14-3-3 proteins, the σ isoform is particularly intriguing due to its association with poor prognosis, and because its expression is frequently lost in some cancers but increased in other cancers (Li et al., 2009).
Uhrf1 (ubiquitin-like, containing PHD and ring finger domains 1) is a multidomain protein important in epigenetic regulation. Mammalian Uhrf1 also contains a SRA (SET and RING associated) domain, which is responsible for binding to histones and methyl-CpG dinucleotides with a preference for hemimethylated CpG sites. Uhrf1 binds to hemimethylated CpG sites and recruits DNA methyltransferase 1 (DNMT1) to methylate the newly synthesized strands, and thus it plays an important role in facilitating and maintaining DNA methylation (Bostick et al., 2007; Sharif et al., 2007).
In this study, we found that 14-3-3σ expression is dramatically upregulated in a gemcitabine-selected derivative clone of PDAC cell line, MiaPaCa-2, and the overexpression contributes to the acquired resistance to gemcitabine and cross-resistance to cytarabine (Ara-C). We also found that the increased 14-3-3σ expression is due to demethylation of the 14-3-3σ gene during gemcitabine selection, which could be partially reversed with removal of gemcitabine selection. The reversible methylation/demethylation of the 14-3-3σ gene is carried out by DNMT1 under Uhrf1 regulation. Together, we conclude that 14-3-3σ expression can be upregulated in PDAC in response to gemcitabine treatment by reversible gene demethylation, and that the increased 14-3-3σ expression contributes to acquired gemcitabine resistance in PDAC.
Materials and Methods
Metafectene Pro transfection reagent was obtained from Biontex (München, Germany). Small interfering RNAs (siRNAs) targeting 14-3-3σ, Uhrf1, DNMT1, DNMT3a, as well as DNMT3b and antibodies against 14-3-3θ, 14-3-3ζ, DNMT1, and DNMT3a were purchased from Santa Cruz Biotechnology (Dallas, TX). DNMT1 antibody for chromatin immunoprecipitation (ChIP) assay was from Abcam (Cambridge, UK). Antibodies against 14-3-3σ and RRM1, the ChIP Assay kit, and the CpGenome Universal DNA Modification kit were purchased from EMD Millipore (Billerica, MA). Antibodies against Uhrf1 and FASN were from BD Biosciences (San Jose, CA). Antibodies against hENT1, histone H3, and RRM2 were from Epitomics (Burlingame, CA), Cell Signaling Technology (Danvers, MA), and generated in house (Dong et al., 2005), respectively. Lipofectamine, pcDNA3.1(+) plasmid, and G418 were from Invitrogen (Carlsbad, CA). RNeasy Mini kit and Qiagen Blood and Cell Culture DNA Kit were from Qiagen (Germantown, MD). The iScript cDNA synthesis kit and the SYBR Green polymerase chain reaction (PCR) master mix were from Bio-Rad (Hercules, CA) and Applied Biosystems (Grand Island, NY), respectively. Gemcitabine was purchased from Besse Medical (West Chester, OH), whereas Ara-C, 5-fluorouracil (5-FU), Adriamycin (doxorubicin), mitoxantrone, and nocodazole were from Sigma-Aldrich (St. Louis, MO). All other chemicals were purchased from Sigma-Aldrich or Fisher Scientific (Waltham, MA).
Cell Lines, Cultures, and Transfections.
Human pancreatic cancer cell line MiaPaCa-2 (American Type Culture Collection, Manassas, VA) and its derivative lines G3K and G3K/REV were cultured at 37°C, 5% CO2 in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 2.5% horse serum. G3K cells were generated by stepwise selection of MiaPaCa-2 with gradually increasing concentrations of gemcitabine starting at 4 nM. G3K cells were clonal and maintained in the presence of 3 µM gemcitabine. The G3K/REV cell line was generated by culturing the drug-resistant G3K cells in the absence of gemcitabine for 6 months, and it partially lost its gemcitabine resistance phenotype. MCF7 and its derivative lines MCF7/AdVp3000 and MCF7/AdVp3000/Rev were gifts from Dr. Susan E. Bates (National Cancer Institute, Bethesda, MD), and were cultured as previously described (Liu et al., 2006). The cell lines were authenticated by analysis of tandem repeat sequences on September 17, 2013.
For transient knockdown of target genes, cells were plated in a six-well plate at a density of 1.5–3 × 105 cells/well and cultured overnight in complete medium. About 60–120 pmol siRNAs of target genes or control scrambled siRNAs were diluted in serum-free Opti-MEM medium (Invitrogen) and then transiently transfected into cells using Metafectene Pro transfection reagent as previously described (Liu et al., 2008). For stable knockdown, pSilencer-σ containing 14-3-3σ short hairpin RNA (shRNA) engineered in a previous study (Han et al., 2006) was transfected into G3K cells using Lipofectamine, and stable clones were selected using 1 mg/ml G418 as previously described (Han et al., 2006). The stable clones were maintained in complete medium supplemented with 200 μg/ml G418. For stable transfection, the cDNA of 14-3-3σ was engineered into pcDNA3.1(+) and transfected into MiaPaCa-2 cells using Lipofectamine. Stable clones were selected using 1 mg/ml G418 as previously described (Han et al., 2006; Liu et al., 2006). The stable clones were maintained in complete medium supplemented with 200 μg/ml G418.
Cell Lysate Preparation and Western Blot Analysis.
Cultured cells were harvested, washed with phosphate-buffered saline, and lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, 50 mM NaF, 1 mM sodium orthovanadate, 1 mM dithiothreitol, 0.1% SDS, and 2 mM phenyl-methylsulfonyl fluoride) for 30 minutes at 4°C with constant agitation. The cell lysates were then sonicated briefly followed by centrifugation (16,000g at 4°C) for 15 minutes to remove insoluble materials. The protein concentrations of supernatants were measured by Bradford assay.
Cell lysates were separated by SDS-PAGE and transferred to a polyvinylidene fluoride membrane followed by a 2-hour incubation in blocking solution (phosphate-buffered saline containing 5% nonfat dried milk and 0.1% Tween 20) and a 2-hour incubation with primary antibodies. After extensive washing, immunoreactivity was detected with specific secondary antibodies conjugated to horseradish peroxidase. Signals were captured using enhanced chemiluminescence and X-ray film.
Survival Assay.
Survival assay was performed as previously described using MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] colorimetric assay (Yang et al., 2002, 2007). In brief, cells were seeded in a 96-well plate at 2000–3000 cells/well and cultured for 24 hours followed by treatment with a different dose of anticancer drugs and incubated continuously for 3 days followed by addition of MTT (5 mg/ml) to a final concentration of 0.5 mg/ml and incubation of the plates at 37°C for 4 hours. The absorbance at 570 nm was measured using an automated plate reader and analyzed using GraphPad Prism software (GraphPad Software, La Jolla, CA) to generate fitted curve and IC50. Relative resistance factor (RRF) is calculated using the following formula: RRF = IC50(test)/IC50(control).
Quantitative Reverse-Transcription PCR.
Quantitative reverse-transcription PCR (RT-PCR) was performed as described previously (Liu et al., 2007; Dong et al., 2009). In brief, total RNA was extracted using the RNeasy Mini Kit followed by reverse transcription using the iScript cDNA synthesis kit and quantitative PCR using the SYBR Green PCR master mix. The primer pairs used are as follows: 5′- TAGGCGCTGTTCTTGCTCCAA-3′ (forward) and 5′- ACCAGTGGTTAGGTGCGCTCA-3′ (reverse) for 14-3-3σ; 5′- GGCAAGTTCTCCGAGGTCTCTG-3′ (forward) and 5′- TGGTACATGGCTTTTCGATAGGA-3′ (reverse) for DNMT3b, 5′-TCTGGCTTTCTTTGCAGCAA-3′ (forward) and 5′-CAGCGGGCTTCTGTAATCTGA-3′ (reverse) for RRM2, and 5′-AAGGACTCATGACCACAGTCCAT-3′ (forward) and 5′-CCATCACGCCACAGTTTTC-3′ (reverse) for glyceraldehyde-3-phosphate dehydrogenase.
ChIP Assay.
The ChIP assay was performed using the ChIP assay kit following the manufacturer’s instructions. In brief, chromatin DNA was cross-linked by formaldehyde and sheared by sonication in 200 μl of SDS lysis buffer. After centrifugation and dilution, the cross-linked protein-DNA complexes were precipitated by overnight incubation with the primary antibodies against histone H3, Uhrf1, DNMT1, or without any antibody as a negative control. The precipitated DNA was analyzed by PCR using primers 5′-CTGAACAGGCCGAACGGTATGAAGAC-3′ and 5′-GAATCGATGATGCGCTTCTTGTCAC-3′ (for CpG island sequences of 14-3-3σ) and 5′-GCTCTTGGCTAGGTAACTGGACTCTTG -3′ and 5′- AGGGGCTTTCCTCATTCTGCCTGCTAC -3 (for non-CpG island control sequences of 14-3-3σ).
Genomic DNA Isolation, Bisulfite Modification, Methylation-Specific PCR, and Sequencing.
Genomic DNA was isolated from MiaPaCa-2, G3K, and G3K/REV cells using the Qiagen Blood and Cell Culture DNA Kit and modified by sodium bisulfite using the CpGenome universal DNA modification kit according to the supplier's protocol.
Methylation-specific PCR was performed as previously described (Han et al., 2006). In brief, 10 μg of bisulfite-modified genomic DNAs was subjected to PCR analysis using primers 5′-TGGTAGTTTTTATGAAAGGCGTC-3′ and 5′-CCTCTAACCGCCCACCACG-3′ (for methylated sequence) or 5′-ATGGTAGTTTTTATGAAAGGTGTT-3′ and 5′-CCCTCTAACCACCCACCACA-3′ (for unmethylated sequence). The PCR products were then subjected to separation and analysis by agarose gel electrophoresis.
For sodium bisulfite sequencing, 10 μg of bisulfite-modified genomic DNAs was first amplified using primers 5′-GAGAGAGTTAGTTTGATTTAGAAG-3′ and 5′-CTTACTAATATCCATAACCTCC-3′ and subcloned into pGEM-T vectors (Promega, Madison, WI). Six independent clones for MiaPaCa-2 cells and four independent clones for G3K cells were isolated and subjected to DNA sequencing.
Results
A Gemcitabine-Selected PDAC Cell Line Is Cross-Resistant to Ara-C but Not to Other Anticancer Drugs.
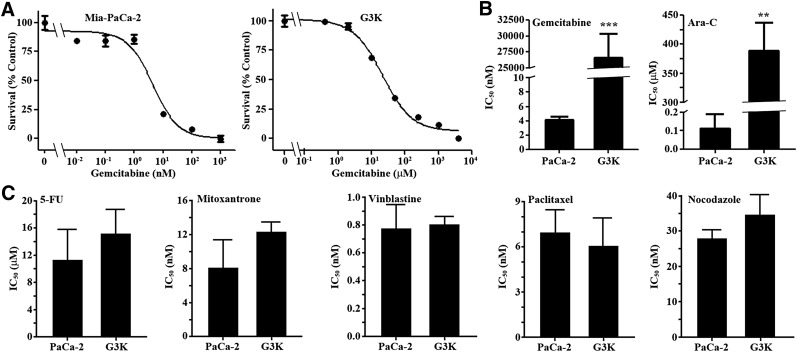
To investigate acquired gemcitabine resistance, we subjected the PDAC cell line MiaPaCa-2 to stepwise selection with escalating concentrations of gemcitabine starting at 4 nM. The final resistant cells were cloned and named G3K, and were viable in the presence of 3000 nM gemcitabine. Authentication using short tandem repeat sequence analysis showed that G3K was derived from the parental MiaPaCa-2 cells (data not shown). The G3K clone has an estimated IC50 of 26.6 ± 3.8 µM, whereas the parental MiaPaCa-2 cells have an IC50 of 4.1 ± 1.3 nM to gemcitabine (Fig. 1, A and B). Thus, G3K cells are ∼6500-fold (RRF) more resistant to gemcitabine than the parental MiaPaCa-2 cells.
Fig. 1.
Drug-response profiles of MiaPaCa-2 and its derivative G3K cells. (A) Dose response of MiaPaCa-2 and G3K cells to gemcitabine treatment. (B and C) IC50 of MiaPaCa-2 and G3K cells to gemcitabine, Ara-C, 5-FU, mitoxantrone, vinblastine, paclitaxel, and nocodazole (n = 4–8). ***P < 0.001; **P < 0.01.
We next examined if G3K cells are cross-resistant to the gemcitabine analog, Ara-C, and other anticancer drugs using MTT assay. As expected, G3K cells are ∼3500-fold more resistant to Ara-C, with an IC50 of 388.4 ± 48.9 µM, than the parental MiaPaCa-2 cells, with an IC50 of 112.8 ± 74.4 nM (Fig. 1B). However, G3K cells did not show any significant resistance to vinblastine, paclitaxel, or nocodazole, although G3K may be slightly more resistant to 5-FU and mitoxantrone than MiaPaCa-2 cells (Fig. 1C).
Ribonucleotide Reductase and 14-3-3σ Are Upregulated in G3K Cells.
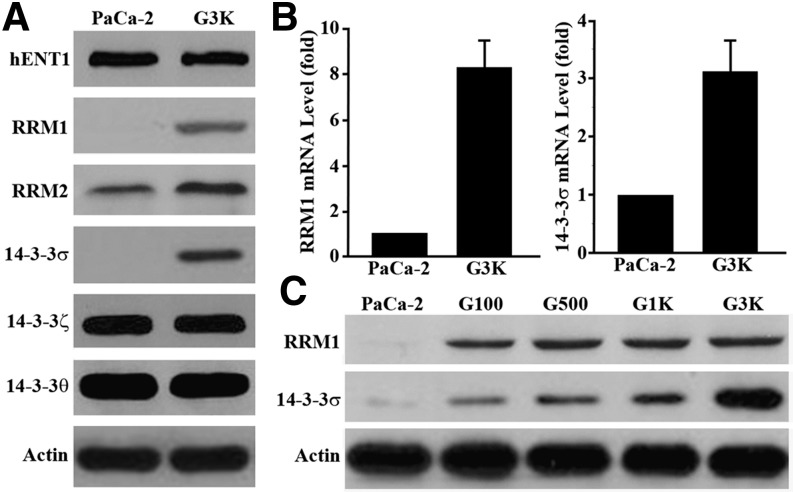
The finding that G3K cells are cross-resistant to Ara-C and lack cross-resistance to multiple other anticancer drugs prompted us to investigate if any known mechanisms of gemcitabine resistance are upregulated in G3K cells. These mechanisms include, but are not limited to, overexpression of hENT1, RRM1, and RRM2 (Voutsadakis, 2011). We also examined 14-3-3σ because it has been shown to contribute to intrinsic resistance to gemcitabine (Li et al., 2010). As shown in Fig. 2A, the protein level of hENT1 in G3K cells remained the same as in MiaPaCa-2 cells, as determined using Western blot analysis. However, the expression of RRM1, RRM2, and 14-3-3σ is drastically upregulated in G3K cells compared with the parental MiaPaCa-2 cells. Interestingly, the other members of the 14-3-3 protein family, 14-3-3θ and 14-3-3ζ, did not increase in expression in the G3K cells.
Fig. 2.
Ribonucleotide reductase and 14-3-3σ are upregulated in G3K cells. (A) Western blot analysis of hENT1, RRM1 and 2, 14-3-3σ, ζ, and θ expression in both MiaPaCa-2 and G3K cells. (B) Real-time RT-PCR analysis of RRM1 and 14-3-3σ mRNA in MiaPaCa-2 and G3K cells (n = 3; P < 0.001). (C) Western blot analysis of RRM1 and 14-3-3σ in the intermediate gemcitabine-resistant cells G100, G500, and G1K. Actin was used as a loading control for Western blot analyses, and glyceraldehyde-3-phosphate dehydrogenase was used as an internal control for PCR analyses.
To further validate the increased expression of RRM and 14-3-3σ, we next performed real-time RT-PCR analysis. As shown in Fig. 2B, the mRNA levels of both RRM1 and 14-3-3σ are significantly increased in G3K cells compared with MiaPaCa-2 cells. We also found that the expression of RRM1 and 14-3-3σ was upregulated early during the selection process by testing the cells with an intermediate level of resistance generated during stepwise selections (Fig. 2C). Although RRM1 remained in the same upregulated level in all intermediate and the final G3K cells, 14-3-3σ appears to be further upregulated in G3K cells compared with the other preceding intermediate cells. This observation suggests that upregulation of both RRM1 and 14-3-3σ may occur as an early event of acquired gemcitabine resistance.
14-3-3σ Overexpression Contributes to the Acquired Gemcitabine Resistance.
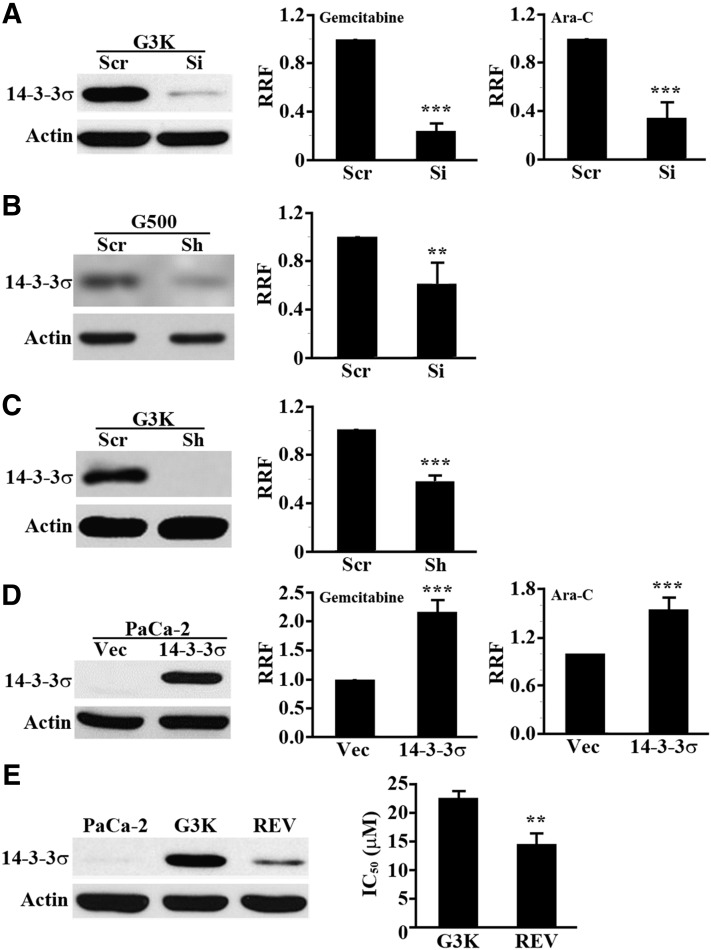
Because RRMs are well known contributors to acquired gemcitabine resistance (Voutsadakis, 2011), we chose to further investigate the potential contribution of 14-3-3σ to the acquired gemcitabine resistance in G3K cells since its potential contribution to acquired gemcitabine resistance has not yet been elucidated. For this purpose, we knocked down 14-3-3σ in G3K cells using siRNA and tested if 14-3-3σ knockdown compromises gemcitabine and Ara-C resistance of G3K cells. As shown in Fig. 3A, 14-3-3σ was successfully knocked down in G3K cells by siRNA, and the reduced 14-3-3σ expression is accompanied by an ∼80% reduction in gemcitabine resistance. Ara-C resistance was also significantly reduced by 14-3-3σ knockdown. Thus, 14-3-3σ overexpression in G3K cells may contribute to both gemcitabine and Ara-C resistance.
Fig. 3.
14-3-3σ upregulation associates with and contributes to gemcitabine and Ara-C resistance. (A) 14-3-3σ knockdown reduces gemcitabine and Ara-C resistance in G3K cells. G3K cells were transiently transfected with 14-3-3σ siRNA (Si) or scrambled control siRNA (Scr) followed by Western blot analysis of 14-3-3σ expression and MTT assay of cellular response to gemcitabine and Ara-C. RRF = IC50(Si or 14-3-3σ)/IC50(Scr or Vec) (n = 3–4; ***P < 0.001). (B) 14-3-3σ knockdown reduces gemcitabine resistance in G500 cells. G500 cells were transiently transfected with 14-3-3σ siRNA or scrambled control siRNA followed by Western blot analysis of 14-3-3σ expression and MTT assay of cellular response to gemcitabine (n = 4; **P < 0.01). (C) 14-3-3σ stable knockdown reduces gemcitabine resistance in G3K cells. G3K cells stably transfected with 14-3-3σ shRNA (Sh) or scrambled control (Scr) were established and subjected to Western blot analysis and MTT assay of cellular response to gemcitabine (n = 3; ***P < 0.001). (D) 14-3-3σ overexpression in MiaPaCa-2 cells causes gemcitabine and Ara-C resistance. Stable MiaPaCa-2 cells with 14-3-3σ overexpression (14-3-3σ) or transfected with vector control (Vec) were established and subjected to Western blot analysis and MTT assay of cellular response to gemcitabine and Ara-C (n = 3–4; ***P < 0.001). (E) Association of 14-3-3σ expression with gemcitabine resistance. Expression of 14-3-3σ and IC50 to gemcitabine in MiaPaCa-2, G3K, and the revertant G3K/REV cells was determined using Western blot analysis and MTT assay, respectively (n = 4; **P < 0.01).
To eliminate any potential issue with the use of the selected final clone G3K in the above studies, we used the pool of the intermediate resistant G500 cells (see Fig. 2C) and transiently knocked down 14-3-3σ expression using siRNA followed by testing gemcitabine resistance. As shown in Fig. 3B, 14-3-3σ was successfully knocked down, and the reduced 14-3-3σ expression resulted in a 40% reduction in gemcitabine resistance. We also tested the effect of 14-3-3σ knockdown on gemcitabine resistance using shRNA of sequences different from that of siRNAs used in the above studies. As shown in Fig. 3C, 14-3-3σ knockdown using shRNA also resulted in a significant reduction in gemcitabine resistance.
To further verify the role of 14-3-3σ in acquired gemcitabine resistance, we established a stable MiaPaCa-2 cell line with overexpression of ectopic 14-3-3σ. Figure 3D shows the stable overexpression of ectopic 14-3-3σ in MiaPaCa-2 cells and the significantly increased resistance of the stable cells to both gemcitabine and Ara-C, confirming that 14-3-3σ overexpression causes gemcitabine and Ara-C resistance.
To validate the role of 14-3-3σ in acquired gemcitabine resistance of G3K cells, we created a partially revertant cell line, G3K/REV, by continuously culturing G3K cells in the absence of gemcitabine selection for 6 months and tested the level of gemcitabine resistance and 14-3-3σ expression. As shown in Fig. 3E, G3K/REV cells have a significantly lower IC50 to gemcitabine than the G3K cells (14.6 ± 1.8 vs. 22.7 ± 1.1 µM). The expression level of 14-3-3σ in the G3K/REV cells is also reduced compared with that of the G3K cells. Taken together, we conclude that the upregulated 14-3-3σ level likely contributes to the acquired gemcitabine resistance in G3K cells, and that both the upregulated 14-3-3σ expression and gemcitabine resistance are partially reversible.
Differential Methylation of 14-3-3σ Gene in MiaPaCa-2 and G3K Cells.
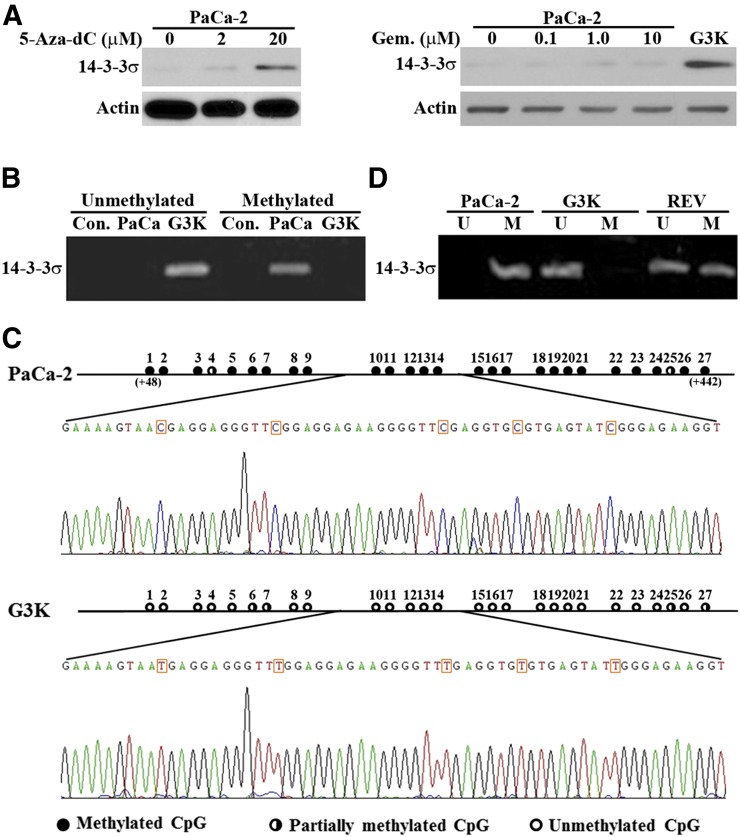
The 14-3-3σ gene was found frequently hypermethylated and, thus, its expression was suppressed in cancer cells (Ferguson et al., 2000). We hypothesize that the 14-3-3σ gene in the parental MiaPaCa-2 cells may be hypermethylated, and that gemcitabine selection may have altered the methylation status of the 14-3-3σ gene, resulting in increased transcription and expression of 14-3-3σ. To test this hypothesis, we first treated the parental MiaPaCa-2 cells with a well known demethylating agent, 5-aza-2'-deoxycytidine (5-Aza-dC, decitabine), that inhibits DNA methyltransferases, and determined 14-3-3σ expression level using Western blot. As shown in Fig. 4A, 5-Aza-dC treatment increased 14-3-3σ expression in a dose-dependent manner. Thus, the 14-3-3σ gene is likely silenced in the parental MiaPaCa-2 cells by methylation and perhaps reactivated in G3K cells.
Fig. 4.
Methylation status of the 14-3-3σ gene in MiaPaCa-2 and G3K cells. (A) Effect of 5-Aza-dC or gemcitabine (Gem.) treatment on 14-3-3σ expression. MiaPaCa-2 cells were treated with increasing concentrations of 5-Aza-dC or gemcitabine followed by Western blot analysis of 14-3-3σ and actin loading control. (B) MSP analysis of MiaPaCa-2 and G3K cells. Con., control MSP without genomic DNA input. (C) Sodium bisulfite sequencing analysis of MiaPaCa-2 and G3K cells. The 27 CpG dinucleotides in the first exon of the 14-3-3σ gene are shown with solid circles for fully methylated, open circles for unmethylated, and half-filled circles for partially methylated CpG dinucleotides. The sequence profile containing CpG dinucleotides 10–14 is shown for both MiaPaCa-2 and G3K cells. The numbers in the parentheses indicate the position of the first and last Cs in this CpG island downstream of AUG start codon. (D) Methylation status of the 14-3-3σ gene in the revertant G3K/REV cells. MSP was used to determine the methylation status of the 14-3-3σ gene in G3K/REV cells with MiaPaCa-2 and G3K cells as controls. M, primers for methylated; U, primers for unmethylated.
Although it has been reported that gemcitabine does not possess the pyrimidine ring modification at position 5, which is responsible for inhibition of DNMTs and, thus, does not inhibit DNA methylation (Goffin and Eisenhauer, 2002), a recent study showed that gemcitabine reactivated several epigenetically silenced genes possibly by inhibiting DNMT1 (Gray et al., 2012). Thus, it is possible that gemcitabine selection reactivated 14-3-3σ gene expression by inhibiting DNMT1. To test this possibility, we treated the parental MiaPaCa-2 cells with different concentrations of gemcitabine followed by Western blot analysis of 14-3-3σ expression. As shown in Fig. 4A, unlike 5-Aza-dC, gemcitabine treatment did not increase 14-3-3σ expression. Thus, reactivation of the 14-3-3σ gene in G3K cells is unlikely due to gemcitabine inhibition of DNMT1.
Next, we compared the methylation status of the 14-3-3σ gene in G3K and the parental MiaPaCa-2 cells using methylation-specific PCR (MSP). As shown in Fig. 4B, the 14-3-3σ gene in the parental MiaPaCa-2 cells was amplified only by primers for methylated sequences, whereas in G3K cells, it was amplified only by primers for unmethylated sequences. Thus, the 14-3-3σ gene in the parental MiaPaCa-2 cells is methylated, whereas it is demethylated in G3K cells. Analysis of the first exon with 27 CpG dinucleotides in the 14-3-3σ gene that are known to be methylated in cancer cells (Ferguson et al., 2000) using sodium bisulfite sequencing shows that 25 of these CpG dinucleotides in the parental MiaPaCa-2 cells are fully methylated, and the remaining 2 are partially methylated (Fig. 4C). However, in G3K cells, 23 of the 27 CpG dinucleotides are unmethylated, and the remaining 4 are partially methylated. Clearly, the methylations of the 14-3-3σ gene in the parental MiaPaCa-2 cells have been removed in G3K cells.
Demethylation of the 14-3-3σ Gene Is Reversible.
As shown earlier, the increased expression of 14-3-3σ in G3K cells is partially reversed in the partially revertant G3K/REV cell line. The partial reversion in 14-3-3σ expression may be due to partial reversal of the methylation status of the 14-3-3σ gene. To test this possibility, we performed MSP of isolated genomic DNAs from G3K/REV cells as described earlier. Figure 4D shows that the 14-3-3σ gene can be amplified by primers for both methylated and unmethylated sequences, indicating that the 14-3-3σ gene in G3K/REV cells is likely partially reversed, and that some cells have regained the methylation of their 14-3-3σ gene while others retain the demethylated 14-3-3σ gene. Thus, methylation and demethylation of the 14-3-3σ gene is likely partially reversible. The incomplete reversal of 14-3-3σ gene methylation following removal of gemcitabine suggests that the increased 14-3-3σ expression in G3K cells is unlikely due to gemcitabine-induced demethylation by inhibiting DNMT1, consistent with the observation shown in Fig. 4A.
Previously, upregulation of 14-3-3σ expression was also observed in an Adriamycin-selected breast cancer cell line, MCF7/AdVp3000, and it was decreased in the revertant MCF7/AdVp3000/Rev cells (Liu et al., 2006) (see also Supplemental Fig. 1A). However, the mechanism of 14-3-3σ regulation in these cell lines is not yet known. To determine if gene methylation is involved in regulating 14-3-3σ expression in these cells, we performed MSP of isolated genomic DNAs from the parental MCF7, Adriamycin-selected MCF7/AdVp3000, and the revertant MCF7/AdVp3000/Rev cells with MiaPaCa-2 cells as a positive control. As shown in Supplemental Fig. 1B, whereas the 14-3-3σ gene in MiaPaCa-2 cells can be amplified only using primers for methylated sequences, in all breast cancer cells it can be amplified only using primers for the unmethylated sequences. Thus, upregulation of 14-3-3σ expression in MCF7/AdVp3000 cells and its reversal in MCF7/AdVp3000/Rev cells is unlikely due to changes in methylation status of the 14-3-3σ gene.
Uhrf1 and DNMT1 Play an Important Role in Regulating 14-3-3σ Expression.
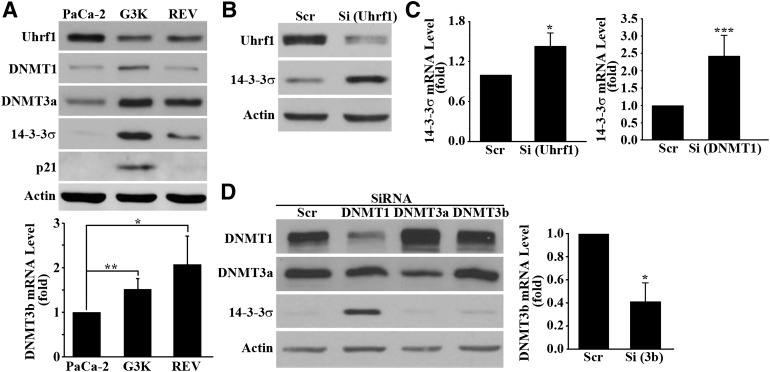
To determine what regulates the reversible methylation of the 14-3-3σ gene in MiaPaCa-2 and G3K cells, we first compared the expression level of enzymes important for DNA methylation between the parental MiaPaCa-2, gemcitabine-resistant G3K, and the partially revertant G3K/REV cells using Western blot or real-time RT-PCR. As shown in Fig. 5A, whereas DNMT3a and DNMT3b were increased in G3K and remained high in G3K/REV cells, DNMT1 is increased in G3K but reduced back to the basal level in G3K/REV cells. The expression pattern of these methyltransferases is peculiar and inconsistent with the expression pattern of 14-3-3σ in MiaPaCa-2, G3K, and G3K/REV cells.
Fig. 5.
Effect of Uhrf1 and DNMT1 on 14-3-3σ expression. (A) Western blot analysis of Uhrf1, DNMT1, DNMT3a, and p21 expression as well as real-time RT-PCR analysis of DNMT3b expression in MiaPaCa-2, G3K, and G3K/REV cells. (B–D) Effect of Uhrf1, DNMT1, DNMT3a, and DNMT3b knockdown on 14-3-3σ expression. MiaPaCa-2 cells were transiently transfected with scrambled control siRNA (Scr) or siRNAs (Si) targeting Uhrf1 (B), DNMT1, DNMT3a, and DNMT3b (D) followed by Western blot analysis of Uhrf1, DNMT3a, and 14-3-3σ or real-time RT-PCR analysis of DNMT3b and 14-3-3σ (B–D) (n = 4–5; *P < 0.05; **P < 0.01; ***P < 0.001). Actin and glyceraldehyde-3-phosphate dehydrogenase were used as a loading control for Western blot and internal control for PCR analyses, respectively.
Another protein of importance in gene methylation, Uhrf1, has been shown to play a major role in recruiting DNMTs (Bostick et al., 2007; Sharif et al., 2007), and loss of Uhrf1 results in a 75% reduction in genomic methylation (Cokus et al., 2008). Thus, we tested the expression pattern of Uhrf1. As shown in Fig. 5A, Uhrf1 protein is reduced in G3K cells and edged back up in the G3K/REV cells. The profile of Uhrf1 expression in these cells is consistent with the partially reversible change in 14-3-3σ expression and gene methylation in these cells (see Discussion). Furthermore, because Uhrf1 has been shown to regulate p21 expression (Kim et al., 2009), we next tested the expression of p21 in MiaPaCa-2, G3K, and G3K/REV cells. As shown in Fig. 5A, the p21 protein level shows an expression profile in these cells similar to that of 14-3-3σ, consistent with the possible regulatory role of Uhrf1 in these cells.
To determine if the reduced Uhrf1 expression is possibly responsible for increased expression of the 14-3-3σ gene in G3K cells, we knocked down Uhrf1 in the parental MiaPaCa-2 cells using siRNA and tested its effect on 14-3-3σ expression. Figure 5B shows that 14-3-3σ protein is dramatically increased by Uhrf1 knockdown. This observation is confirmed by real-time RT-PCR analysis of 14-3-3σ mRNA (Fig. 5C).
To determine if DNMTs also contribute to 14-3-3σ expression regulation, we performed a similar experiment by knocking down DNMT1, DNMT3a, and DNMT3b in MiaPaCa-2 cells. Figure 5D shows successful knockdown of DNMT1, DNMT3a, and DNMT3b as determined using Western blot or real-time RT-PCR. However, only DNMT1 knockdown upregulated the 14-3-3σ protein level. DNMT3a and DNMT3b knockdown did not appear to affect 14-3-3σ expression. The effect of DNMT1 knockdown on 14-3-3σ expression was also confirmed by real-time RT-PCR analysis (Fig. 5C). Taken together, we conclude that both Uhrf1 and DNMT1 likely play important roles in regulating 14-3-3σ expression.
Uhrf1 Binds and Helps Recruit DNMT1 to CpG Islands of 14-3-3σ Gene.
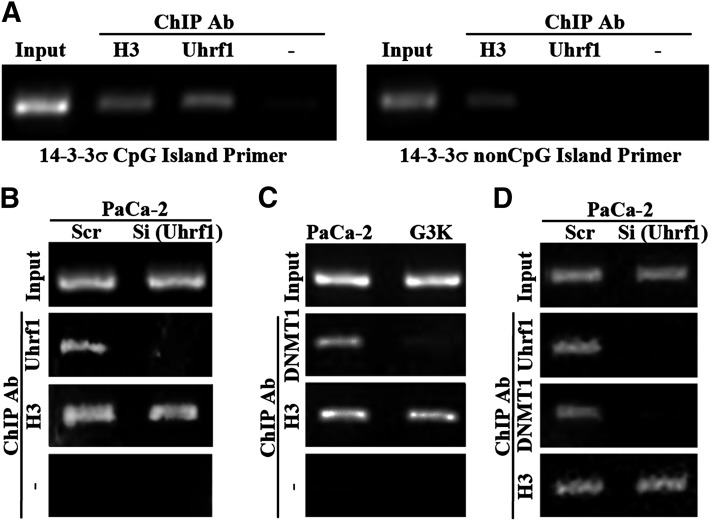
To further determine the role of Uhrf1 and DNMT1 in regulating 14-3-3σ expression and gene methylation, we first determined if Uhrf1 binds to the CpG islands in the first exon of the 14-3-3σ gene in MiaPaCa-2 cells using a ChIP assay. As shown in Fig. 6A, Uhrf1 was bound to the CpG island but not to the CpG-free sequences in the promoter region of the 14-3-3σ gene in MiaPaCa-2 cells. Knocking down Uhrf1 also reduced Uhrf1 binding to the CpG islands of the 14-3-3σ gene (Fig. 6B). Thus, Uhrf1 likely can bind to the CpG island sequence of the 14-3-3σ gene.
Fig. 6.
Binding of Uhrf1 and DNMT1 to the CpG island of the 14-3-3σ gene. (A) ChIP analysis of Uhrf1 binding to the CpG island sequence or non-CpG island sequence of the 14-3-3σ gene. (B and D) Effect of Uhrf1 knockdown on Uhrf1 and DNMT1 binding to the CpG island sequence of the 14-3-3σ gene in MiaPaCa-2 cells. MiaPaCa-2 cells were transiently transfected with scrambled control (Scr) or Uhrf1 siRNAs (Si) followed by ChIP analysis of Uhrf1 and DNMT1 binding. (C) DNMT1 binding to the CpG island of the 14-3-3σ gene in G3K cells is less than that in MiaPaCa-2 cells. ChIPs with histone H3 antibody or without any primary antibody were used as positive and negative controls, respectively. Normal IgG was also used as a negative control, which did not immunoprecipitate any DNA (data not shown). Ab, antibody.
As shown in Fig. 5A, we found that DNMT1 expression was increased in G3K cells, which is peculiar since methylation of the 14-3-3σ gene in G3K cells is reduced. However, because Uhrf1 binding to CpG island sequences helps recruit DNMT1 to maintain the methylation status of the DNA, it is possible that the reduced Uhrf1 expression in G3K cells reduces DNMT1 recruitment despite the higher level of DNMT1 in G3K cells. To test this possibility, we first compared the binding of DNMT1 to the CpG island sequence of the 14-3-3σ gene between MiaPaCa-2 and G3K cells. As shown in Fig. 6C, binding of DNMT1 to the CpG island sequence of the 14-3-3σ gene is indeed much less in G3K than in MiaPaCa-2 cells despite the higher expression level of DNMT1 in G3K cells. Uhrf1 knockdown in MiaPaCa-2 cells also reduced DNMT1 binding to the CpG island sequence of the 14-3-3σ gene (Fig. 6D), indicating that DNMT1 could not be effectively recruited to the CpG island sequence of the 14-3-3σ gene in the absence of Uhrf1. Thus, we conclude that Uhrf1 likely plays a major role in regulating 14-3-3σ expression by binding to its CpG rich sequence, and helps recruit DNMT1 to the site to reversibly methylate the 14-3-3σ gene. The reduced Uhrf1 expression in G3K cells likely decreased DNMT1 recruitment and methylation of the 14-3-3σ gene, whereas the slight increase in Uhrf1 level is responsible for partial reversal of 14-3-3σ gene methylation in G3K/REV cells.
Gemcitabine Treatment Does Not Affect Uhrf1 and DNMT1 Expression.
To investigate if the altered expression of Uhrf1 and DNMT1 in G3K cells is potentially due to gemcitabine treatments during selection, we performed a Western blot analysis of these two proteins in MiaPaCa-2 cells following treatments with gemcitabine at different concentrations. As shown in Supplemental Fig. 2, gemcitabine treatments had no effect on Uhrf1 and DNMT1 expression. Thus, the mechanism of regulation of Uhrf1 and DNMT1 expression in the drug-resistant cells remains to be determined.
Discussion
Stepwise selections with anticancer drugs have been used as a standard method to create model cell lines for laboratory studies and identification of novel mechanisms of acquired resistance. G3K cells selected with gemcitabine in this study are clonal and cross-resistant to Ara-C. Considering the similarity in structure, mechanism of action, and metabolism between gemcitabine and Ara-C, we are not surprised to find the cross-resistance of G3K cells to Ara-C. The observation of increased expression of RRM is also as expected, as these proteins have previously been shown to be upregulated and contribute to acquired gemcitabine resistance in other gemcitabine-selected cells (Goan et al., 1999; Davidson et al., 2004). However, the findings of the upregulated expression of 14-3-3σ via reversible epigenetic regulation during gemcitabine selection and its role in acquired gemcitabine resistance and cross-resistance to Ara-C are unexpected and absolutely novel. These findings will shed light not only on possible targeting of 14-3-3σ for sensitization of acquired gemcitabine resistance but also on further understanding of the molecular mechanisms in epigenetic regulation of 14-3-3σ expression.
14-3-3σ, a homodimeric protein that functions as a chaperone, binds to >100 phosphoserine/phosphothreonine proteins and plays important roles in cell survival (Li et al., 2009). Upregulated expression of 14-3-3σ has been found in PDAC and appears to be associated with poor prognosis of PDAC by causing resistance to gemcitabine (Hustinx et al., 2005; Li et al., 2010). The possible role of 14-3-3σ in resistance to other anticancer drugs such as doxorubicin and cisplatin has also been reported previously (Han et al., 2006, 2009; Liu et al., 2006; Neupane and Korc, 2008). Similar to the finding of this study that 14-3-3σ can be selected and responsible for acquired gemcitabine resistance, it can also be selected and responsible for acquired doxorubicin resistance (Liu et al., 2006). However, in this study, we showed that the increased 14-3-3σ expression for drug resistance in different cell lines is regulated by different mechanisms, which have not been reported in any previous studies. The finding of epigenetic regulation due to methylation changes in the 14-3-3σ gene during gemcitabine selection is novel, and it likely occurs only in the parental cells such as MiaPaCa-2 cells, where the 14-3-3σ gene is epigenetically silenced. It is, however, noteworthy that the methylation change of the 14-3-3σ gene observed here is in the clonal G3K cells. A different G3K clone may not present a reduction in 14-3-3σ gene methylation. It is also not clear if gemcitabine selection of a different cancer cell line with methylation suppression of the 14-3-3σ gene can result in demethylation of the gene. Future studies are warranted to test if gemcitabine selection results in demethylation of the 14-3-3σ gene in different cancer cells.
Although some of the other members of the 14-3-3 protein family, such as 14-3-3ζ and 14-3-3θ/τ, have been reported to also contribute to (Maxwell et al., 2009; Neal et al., 2009) and associate with (Hodgkinson et al., 2012) drug resistance, we did not find upregulation in expression of 14-3-3ζ and 14-3-3θ/τ in the gemcitabine-resistant G3K cells. This observation suggests that 14-3-3ζ and 14-3-3θ/τ may not participate in the acquired gemcitabine resistance in PDAC.
Although the detailed mechanism of 14-3-3σ–mediated drug resistance is unknown, the fact that it binds to various proteins important for different cellular processes suggests that it may work by binding to these proteins. Somatic knockout of 14-3-3σ in colon cancer cells has been shown to cause drug-induced mitotic catastrophe by reducing cellular ability to arrest in the G2/M phase (Chan et al., 1999), and increased 14-3-3σ expression in breast cancer cells makes cancer cells more resistant to drug-induced apoptosis (Liu et al., 2006), possibly due to 14-3-3σ binding and arresting cyclin B1 and CDC2 (Chan et al., 1999; Han et al., 2006) and proapoptosis proteins such as Bax and Bad (Samuel et al., 2001; Subramanian et al., 2001) in cytoplasm. Nevertheless, it remains to be determined if 14-3-3σ–mediated gemcitabine resistance also occurs via mitotic catastrophe and apoptosis.
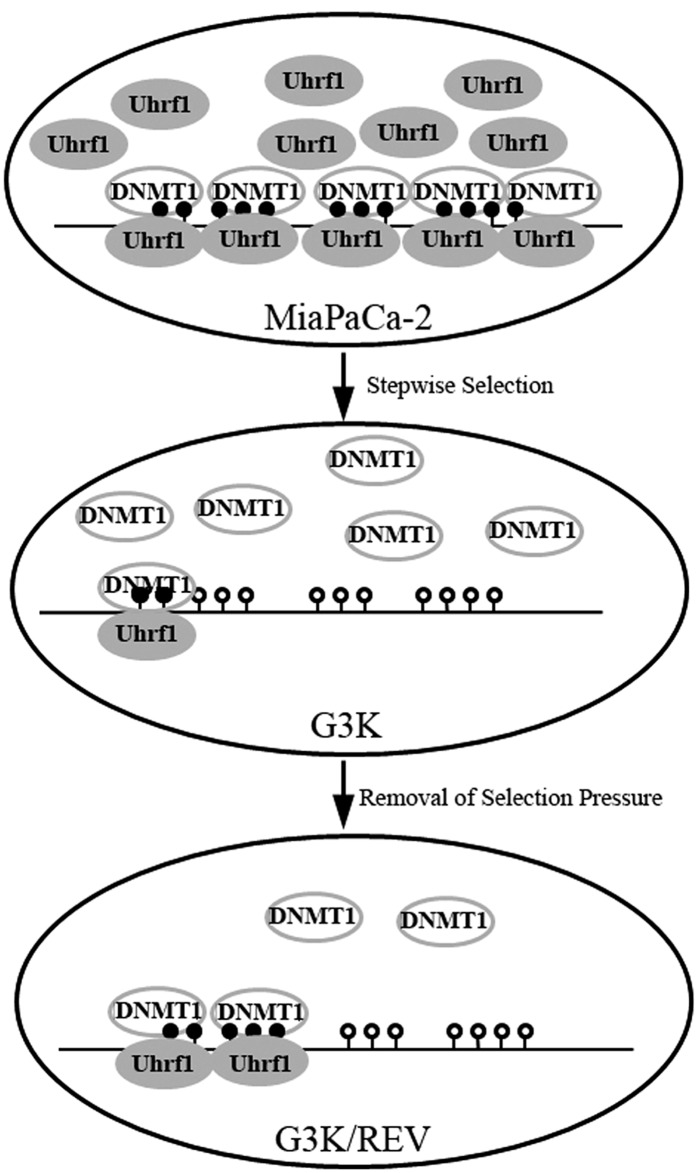
The finding that reversible methylation of the 14-3-3σ gene is regulated by Uhrf1 and DNMT1 is novel and has not been reported, although regulation of 14-3-3σ expression by methylation has been observed in cell line models and clinical samples (Ferguson et al., 2000; Suzuki et al., 2000). The finding that 14-3-3σ expression is progressively upregulated during gemcitabine selection is very intriguing, and suggests that the gene demethylation may be progressive. We are also the first to report that Uhrf1 and DNMT1 expression changes following stepwise gemcitabine selection, which leads to failure of DNMT1 recruitment to the 14-3-3σ gene and subsequent demethylation and increased 14-3-3σ expression (Fig. 7). Although the decreased methylation of the 14-3-3σ gene in the drug-resistant G3K cells is inconsistent with the increased expression of DNMT1, the reduced level of Uhrf1 may be responsible for the decreased methylation.
Fig. 7.
Schematic diagram of regulation of the 14-3-3σ gene by reversible methylation during gemcitabine selection. In MiaPaCa-2 cells, a high level of Uhrf1 binds to and helps recruit DNMT1 to the methylated region of the 14-3-3σ gene to maintain methylation. In G3K cells, Uhrf1 expression is reduced and serves as a limiting factor. Thus, little DNMT1 is recruited to the 14-3-3σ gene to maintain methylation despite the presence of a high level of DNMT1. In G3K/REV cells, the increase in Uhrf1 level helps recruit more DNMT1 protein to the 14-3-3σ gene, resulting in more methylation and partial loss of 14-3-3σ expression.
Uhrf1, a multidomain protein associated with cellular proliferation and epigenetic regulation, binds to histones and methyl-CpG dinucleotides with a preference for hemimethylated CpG sites. The consequence of Uhrf1 binding was recruitment of DNMT1 and histone deacetylase 1, resulting in methylation of nascent DNA strands (Bostick et al., 2007; Sharif et al., 2007). Thus, reduced Uhrf1 expression in G3K cells is likely responsible for reduced methylation of the 14-3-3σ gene by reducing recruitment of DNMT1 protein to the CpG islands, despite the presence of a high level of DNMT1. In MiaPaCa-2 cells, however, the high level of Uhrf1 may efficiently recruit all DNMT1 despite low DNMT1 level for efficient methylation of 14-3-3σ gene. Knocking down either Uhrf1 or DNMT1 in MiaPaCa-2 cells would effectively reduce the level of the recruiter (Uhrf1) or the pool of DNMT1 to be recruited for methylation of the 14-3-3σ gene, and consequently increase 14-3-3σ expression. Furthermore, the slight increase of Uhrf1 expression in the G3K/REV cells may be responsible for the reduced expression and increased methylation of the 14-3-3σ gene.
The findings on altered expression of Uhrf1 and DNMT1 in G3K and G3K/REV cells also suggest that other genes that are under epigenetic regulation, specifically DNA methylation, may also change in their expression, similar to 14-3-3σ. Indeed, we found that p21, a known Uhrf1 downstream target gene, has an expression profile similar to that of 14-3-3σ in the parental MiaPaCa-2, gemcitabine-resistant G3K, and revertant G3K/REV cells. What other genes have similar expression profiles in these cells and whether these genes, such as p21, also contribute to the acquired gemcitabine resistance remain to be investigated. It would also be of interest to determine if use of DNMT inhibitors affects gemcitabine resistance of MiaPaCa-2 cells in xenograft animal models, which may provide guidance on future treatment options for combination therapy. We are currently working toward this direction.
In summary, we identified 14-3-3σ as an important contributor for acquired gemcitabine resistance and for Ara-C cross-resistance in PDAC cells using a stepwise-selected and gemcitabine-resistant cell line, and showed that upregulated 14-3-3σ expression by gemcitabine selection was due to partially reversible demethylation of its gene by reduced Uhrf1 expression in the gemcitabine-resistant derivative cells. Reversible epigenetic regulation may play an important role in gemcitabine response in pancreatic cancer treatment, and targeting epigenetic regulation may provide a new direction for chemosensitization of gemcitabine-resistant human pancreatic cancers.
Supplementary Material
Abbreviations
- Ara-C
cytarabine
- 5-aza-dC
5-aza-2'-deoxycytidine
- ChIP
chromatin immunoprecipitation
- DNMT
DNA methyltransferase
- 5-FU
5-fluorouracil
- MSP
methylation-specific PCR
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PCR
polymerase chain reaction
- PDAC
pancreatic ductal adenocarcinoma
- RRF
relative resistant factor
- RRM
ribonucleotide reductase
- RT-PCR
reverse-transcription PCR
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
Authorship Contributions
Participated in research design: Qin, Dong, Zhang.
Conducted experiments: Qin, Dong.
Performed data analysis: Qin, Dong, Zhang.
Wrote or contributed to the writing of the manuscript: Qin, Dong, Zhang.
Footnotes
This work was supported in part by the National Institutes of Health [Grant R01-CA140582].
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317:1760–1764 [DOI] [PubMed] [Google Scholar]
- Chan TA, Hermeking H, Lengauer C, Kinzler KW, Vogelstein B. (1999) 14-3-3Sigma is required to prevent mitotic catastrophe after DNA damage. Nature 401:616–620 [DOI] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452:215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson JD, Ma L, Flagella M, Geeganage S, Gelbert LM, Slapak CA. (2004) An increase in the expression of ribonucleotide reductase large subunit 1 is associated with gemcitabine resistance in non-small cell lung cancer cell lines. Cancer Res 64:3761–3766 [DOI] [PubMed] [Google Scholar]
- Dong Z, Liu Z, Cui P, Pincheira R, Yang Y, Liu J, Zhang JT. (2009) Role of eIF3a in regulating cell cycle progression. Exp Cell Res 315:1889–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Liu Y, Zhang JT. (2005) Regulation of ribonucleotide reductase M2 expression by the upstream AUGs. Nucleic Acids Res 33:2715–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson AT, Evron E, Umbricht CB, Pandita TK, Chan TA, Hermeking H, Marks JR, Lambers AR, Futreal PA, Stampfer MR, et al. (2000) High frequency of hypermethylation at the 14-3-3 sigma locus leads to gene silencing in breast cancer. Proc Natl Acad Sci USA 97:6049–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goan YG, Zhou B, Hu E, Mi S, Yen Y. (1999) Overexpression of ribonucleotide reductase as a mechanism of resistance to 2,2-difluorodeoxycytidine in the human KB cancer cell line. Cancer Res 59:4204–4207 [PubMed] [Google Scholar]
- Goffin J, Eisenhauer E. (2002) DNA methyltransferase inhibitors-state of the art. Ann Oncol 13:1699–1716 [DOI] [PubMed] [Google Scholar]
- Gray SG, Baird AM, O’Kelly F, Nikolaidis G, Almgren M, Meunier A, Dockry E, Hollywood D, Ekström TJ, Perry AS, et al. (2012) Gemcitabine reactivates epigenetically silenced genes and functions as a DNA methyltransferase inhibitor. Int J Mol Med 30:1505–1511 [DOI] [PubMed] [Google Scholar]
- Han Z, Dimas K, Tian X, Wang Y, Hemmi H, Yamada K, Kato N, Pantazis P, Ramanujam RJ, Anant S, et al. (2009) 14-3-3sigma-dependent resistance to cisplatin. Anticancer Res 29:2009–2014 [PubMed] [Google Scholar]
- Han B, Xie H, Chen Q, Zhang JT. (2006) Sensitizing hormone-refractory prostate cancer cells to drug treatment by targeting 14-3-3sigma. Mol Cancer Ther 5:903–912 [DOI] [PubMed] [Google Scholar]
- Hodgkinson VC, ELFadl D, Agarwal V, Garimella V, Russell C, Long ED, Fox JN, McManus PL, Mahapatra TK, Kneeshaw PJ, et al. (2012) Proteomic identification of predictive biomarkers of resistance to neoadjuvant chemotherapy in luminal breast cancer: a possible role for 14-3-3 theta/tau and tBID? J Proteomics 75:1276–1283 [DOI] [PubMed] [Google Scholar]
- Hustinx SR, Fukushima N, Zahurak ML, Riall TS, Maitra A, Brosens L, Cameron JL, Yeo CJ, Offerhaus GJ, Hruban RH, et al. (2005) Expression and prognostic significance of 14-3-3sigma and ERM family protein expression in periampullary neoplasms. Cancer Biol Ther 4:596–601 [DOI] [PubMed] [Google Scholar]
- Kim JK, Estève PO, Jacobsen SE, Pradhan S. (2009) UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res 37:493–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Dong Z, Myer D, Yip-Schneider M, Liu J, Cui P, Schmidt CM, Zhang JT. (2010) Role of 14-3-3σ in poor prognosis and in radiation and drug resistance of human pancreatic cancers. BMC Cancer 10:598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Liu J-Y, Zhang J-T. (2009) 14-3-3σ, the double-edged sword of human cancers. Am J Transl Res 1:326–340 [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Dong Z, Yang Z, Chen Q, Pan Y, Yang Y, Cui P, Zhang X, Zhang JT. (2007) Role of eIF3a (eIF3 p170) in intestinal cell differentiation and its association with early development. Differentiation 75:652–661 [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu H, Han B, Zhang JT. (2006) Identification of 14-3-3sigma as a contributor to drug resistance in human breast cancer cells using functional proteomic analysis. Cancer Res 66:3248–3255 [DOI] [PubMed] [Google Scholar]
- Liu H, Liu Y, Zhang JT. (2008) A new mechanism of drug resistance in breast cancer cells: fatty acid synthase overexpression-mediated palmitate overproduction. Mol Cancer Ther 7:263–270 [DOI] [PubMed] [Google Scholar]
- Maxwell SA, Li Z, Jaye D, Ballard S, Ferrell J, Fu H. (2009) 14-3-3zeta mediates resistance of diffuse large B cell lymphoma to an anthracycline-based chemotherapeutic regimen. J Biol Chem 284:22379–22389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal CL, Yao J, Yang W, Zhou X, Nguyen NT, Lu J, Danes CG, Guo H, Lan KH, Ensor J, et al. (2009) 14-3-3zeta overexpression defines high risk for breast cancer recurrence and promotes cancer cell survival. Cancer Res 69:3425–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupane D, Korc M. (2008) 14-3-3sigma Modulates pancreatic cancer cell survival and invasiveness. Clin Cancer Res 14:7614–7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel T, Weber HO, Rauch P, Verdoodt B, Eppel JT, McShea A, Hermeking H, Funk JO. (2001) The G2/M regulator 14-3-3sigma prevents apoptosis through sequestration of Bax. J Biol Chem 276:45201–45206 [DOI] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. (2007) The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450:908–912 [DOI] [PubMed] [Google Scholar]
- Subramanian RR, Masters SC, Zhang H, Fu H. (2001) Functional conservation of 14-3-3 isoforms in inhibiting bad-induced apoptosis. Exp Cell Res 271:142–151 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Itoh F, Toyota M, Kikuchi T, Kakiuchi H, Imai K. (2000) Inactivation of the 14-3-3 sigma gene is associated with 5′ CpG island hypermethylation in human cancers. Cancer Res 60:4353–4357 [PubMed] [Google Scholar]
- Voutsadakis IA. (2011) Molecular predictors of gemcitabine response in pancreatic cancer. World J Gastrointest Oncol 3:153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Chen Q, Zhang JT. (2002) Structural and functional consequences of mutating cysteine residues in the amino terminus of human multidrug resistance-associated protein 1. J Biol Chem 277:44268–44277 [DOI] [PubMed] [Google Scholar]
- Yang Y, Liu Y, Dong Z, Xu J, Peng H, Liu Z, Zhang JT. (2007) Regulation of function by dimerization through the amino-terminal membrane-spanning domain of human ABCC1/MRP1. J Biol Chem 282:8821–8830 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.