Abstract
The gap junction channel forming connexins (Cx) Cx31 (Gjb3) and Cx31.1 (Gjb5) are co-expressed in the mouse trophoblast lineage. Inactivation of either gene results in partial embryonic loss at mid gestation (60% and 30%, respectively, between embryonic days E10.5and E13.5) caused by placental phenotypes. Cx31 deficiency results in loss of stem cell potential and enhanced trophoblast giant cell (TGC) differentiation, whereas the molecular role of the co-expressed Cx31.1 remained unclear. It was assumed that both isoforms have overlapping functions and can compete for each loss in placentation as both knockout mice show similar survival rates, reduced placental weights, and growth restricted embryos. Instead, here we show that Cx31.1 has opposed functions in regulating trophoblast differentiation. Cx31.1 deficiency causes a shift in placental subpopulations, reduced area of fetal blood spaces, and a reduced number of secondary TGC in the junctional zone, as shown by stereology at E10.5. Cx31.1 is critical for terminal differentiation of trophoblast cells during placentation resulting in a delayed induction of marker genes Tpbpa, Prl3b1/Pl-2, and Ctsq in Cx31.1-deficient placentas. Derivation and analysis of Cx31.1-deficient trophoblast stem lines clearly indicates a delayed trophoblast differentiation manifested by repression of marker genes for placental subpopulations and continued expression of stem cell marker genes Id2 and Ascl2, which is correlated to enhanced proliferation capacity of differentiating stem cells These findings clarify the disparate actions of Cx31.1 and Cx31 that act in opposition to balance the fate of trophoblast cells during differentiation, with Cx31.1 promoting, and Cx31 delaying terminal differentiation.
Introduction
The placenta is the first fetal organ formed during pregnancy and abnormalities in its development or function can lead to gestational problems such as spontaneous abortion, intra-uterine growth restriction, preeclampsia or preterm labor. Several studies have demonstrated the impact of intercellular communication mediated by gap junction channels in trophoblast development and placental function in mice and humans [1–3]. Gap junctions provide direct cytoplasmatic communication between adjacent cells by exchanging ions, second messengers; small metabolites such as glucose or amino acids, and also small interfering RNAs through the membrane spanning channel pore [4]. The connexin multi-gene family comprises 20 members in the mouse, and regulations in connexin expression levels and pattern can change gap junctional communication, which allows coordination of physiological processes like cell proliferation, growth, homeostasis, differentiation, motility, and signal transduction [5–7].
The mouse placenta shows a tightly regulated spatio-temporal expression pattern of connexins during trophoblast differentiation. Gene-deficient mice demonstrated a critical role for Cx26 (Gjb2), Cx31 (Gjb3), Cx31.1 (Gjb5), and Cx45 (Gjc1) during placental development [1]. The diploid, proliferating cells of the postimplantation trophoblast are characterized by co-expression of the Cx31 and Cx31.1 isoforms. Both genes cluster in distal mouse chromosome 4 and exhibit 65% amino acid sequence identity suggesting a close evolutionary relationship between both isoforms [8]. Inactivation of either the Cx31 or the Cx31.1 gene results in a transient placental phenotype causing death of 60% (Cx31) or 30% (Cx31.1) of the concepti between embryonic days (E) E10.5 and E13.5. Both, Cx31 and Cx31.1-deficient placentas revealed reduced weights over gestation and surviving Cx31 or Cx31.1-deficient embryos were born growth restricted compared to litter mate controls [9–11]. The Cx31-deficient placenta revealed an excessive terminal differentiation into secondary trophoblast giant cells (TGC) in the junctional zone at the expense of the proliferating spongiotrophoblast (SPT) and labyrinthine trophoblast cell populations. The result is a transient phenotype; whereas 60% of the Cx31-deficient placentas cannot support the growing embryo leading to death between E10.5 and E13.5, 40% of placentas are just developed enough to support the conceptus through this critical phase. The surviving placentas and embryos, however, remain growth-restricted throughout pregnancy [9,10]. The analysis of Cx31-deficient trophoblast stem cell (TSC) lines in vitro further showed that the loss of Cx31 changes the temporal differentiation process of trophoblast cells. Upon differentiation, Cx31-deficient TSC show an earlier temporal induction of the Ascl2 (Mash2) gene and subsequently the SPT marker gene Tpbpa, the syncytiotrophoblast marker Cx26 (Gjb2), and the TGC marker Prl3d1 (Pl-1, placental lactogen 1), compared to wild-type controls, which indicates an endogenous, enhanced differentiation of TSC [12]. A recent study analyzing markers for the different TGC populations, showed that Cx31-deficient TSC, in particular, reveal an upregulation of secondary TGC markers Prl2c2/Plf, Prl3b1/Pl-2, and Ctsq [13].
In contrast to the Cx31-deficient strain, placentas of the Cx31.1-deficient mice did not show such dramatic structural alterations. The weight of the Cx31.1-deficient placenta was 15% reduced from E11.5 to E18.5, whereas embryonic weight was significantly reduced about 7%–12% from E16.5 to E18.5, compared to wild-type littermates. In the first evaluation of Cx31.1 placenta only mild effects on placental morphology like a more compact SPT layer could be detected, but based on morphology it was not possible to predict which conceptus will lead to death of the embryos [11]. Due to the co-expression of both genes, sequence identity, comparable survival curves, and reduced placental and embryonic weight in the individual knockout mice it was assumed that both connexins fulfill the same functions. Moreover, it was hypothesized that the loss of either the Cx31 or Cx31.1 gene can be compensated by the remaining isoform [1].
In the present study, we performed a deeper structural analysis of the Cx31.1-deficient placenta using stereology, in situ hybridization, and quantitative polymerase chain reaction (qPCR). In addition, we generated and analyzed Cx31.1-deficiecnt TSC lines. Our results counter the previous compensation hypothesis, by showing that Cx31.1-deficiency delays placental development by blocking terminal differentiation of TSC into placental subpopulations. Thus, Cx31.1 reveals an opposite function to Cx31. These results suggest a new implication for the co-existence of both connexin isoforms in coordination and balance of trophoblast differentiation to ensure proper placental development.
Materials and Methods
Animals
Animal experiments were performed at the Toronto Centre for Phenogenomics, Canada, with ethical approval and guidelines of the Animal Care Committee according to Canadian laws. Mice (Mus musculus) were maintained in a pathogen-free environment with a fixed 12-h/12-h light/dark cycle with food, and water, ad libitum. Mating was performed overnight, and noon of the day of plug detection was considered as E0.5. Cx31.1-deficient mice were obtained from the laboratory of Dr. K. Willecke, University of Bonn, Germany [11] and twice crossed with the in-house C57BL/6 strain.
TSC culture
Isolation of primary mouse embryonic fibroblasts (MEF) and preparation of mitomycin C arrested feeder layers, and MEF-conditioned TS medium (TS-CM), was performed as described before [14]. TSC lines were derived from isolated E3.5 blastocysts as described before [12]. Undifferentiated, feeder-free TSC lines were cultured in 70% TS-CM, 30% TS medium [RPMI-1640 (Life Technologies, Burlington, ON, Canada), 20% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA), 2 mM glutamine (Life Technologies), 1 mM sodium pyruvate (Life Technologies), 100 μM beta-mercaptoethanol (Sigma-Aldrich, Oakville, ON, Canada), and 100 μg/mL penicillin/streptomycin (Lonza, Mississauga, ON, Canada)], supplemented with 25 ng/mL FGF4 (R&D systems, Minneapolis, MI) and 1 U/mL heparin (Sigma). Media were changed every other day and cells were passaged using TrypLE-Express (Life Technologies) dissociation reagent. Differentiation of TSC was induced by culturing cells in TS medium alone without any supplements.
Proliferation of TSC was measured by a colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. TSC were seeded at 40,000 cells in 24 wells and cultured over 6 days in the undifferentiated or differentiating state. To measure relative cell numbers, MTT (Sigma) was added at a concentration of 160 μg/mL to the cultures. After 2 h of incubation at 37°C the medium was removed and cells lysed by adding 0.5 mL of DMSO, releasing the MTT reduction product formazan. The amount of fomazan, representing the relative number of TSC per well, was measured at a wave length of 570 nm using an InfinateM200ORD plate reader (Teacan). Measurements were performed in four technical replicates per day and clone, and data were normalized the first day of measurement (day 0). The linear part of the growth curves was fitted to assess doubling rates.
RNA isolation and qPCR
Total RNA from frozen tissues was isolated using TRIzol reagent (Life Technologies). One hundred microgram of total RNA was incubated with DNase I (Qiagen, Toronto, ON, Canada) for 15 min at room temperature (RT) and further purified using the RNeasy MinElute Cleanup Kit (Qiagen) according to the manufacturer's protocol. Total RNA from TSC was isolated using the RNeasy Plus Mini Kit (Qiagen). For production of cDNA, 1 μg of RNA was reverse transcribed using the TaqMan Reverse Transcription Reagents (ABI, Cobourg, ON, Canada) or the iScript Reverse Transcription Supermix (Bio-Rad, Mississauga, ON, Canada). Five nanogram of cDNA was used in a qPCR reaction in triplicates as described [15] using an Eppendorf (Mississauga, ON, Canada) realplex2-S mastercycler or a CFX384 system (Bio-Rad). The relative expression of genes was determined using the CFX manager 3.0 software (Bio-Rad) based on the delta-delta-Ct method, with beta-actin, hypoxanthine phosphoribosyl-transferase 1 (HPRT-1), and TATA-binding protein (TBP) as reference genes. Specific primers for PCR amplification (see Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/scd) of placental transcripts were generated using the primer designing tool website (NCBI, NIH, USA). For qPCR, primer efficiencies were assessed and specificity of the amplicon was assured by melting curve analysis. Statistical analysis of gestational profiles for specific marker genes was performed using the regression method in SPSS 16.0 to observe significant changes between the wild-type and the Cx31.1 knockout group; and in addition, a Student's t-test was performed to test for significant changes between wild-type and knockout placentas at individual gestational days.
PCR genotyping and cloning
Isolation of genomic DNA from tail biopsies or TSC and subsequent PCR reaction was performed using the REDExtract-N-Amp™ Tissue PCR Kit (Sigma). Specific primers for the Gjb5/Cx31.1 locus were used as described recently [11].
To generate plasmids for RNA probe synthesis, fragments for Tpbpa, Ctsq, and Hand1, were amplified from placental cDNA using specific primers (Supplementary Table S1) and the Taq PCR Master Mix (Fermentas, Thermo Fisher Scientific). Fresh PCR product was ligated into the pCRII-vector and transformed into competent E. coli TOPO10 bacteria using the TA Cloning Kit (Life Technologies). All vectors were verified for sequence and fragment orientation by sequencing. To generate digoxigenin (DIG)-labeled RNA probes, the above constructed pCRII plasmids and plasmids pGEM2-PL-1 [16], pGEM2-PL-2 [17], pGEM1-Plf [18], pBKS-Gcm1, and pBSK-Ascl2/Mash2 [19] were linearized, phenol/chloroform purified, and in vitro transcribed with T3, T7, or SP6 polymerases following the protocol of the DIG RNA labeling Kit (Roche, Mississauga, ON, Canada), or the T7 High Yield Transcription Kit (Thermo Fisher Scientific). Labeling efficiency was assessed by dot blot analysis and probes were purified using the RNeasy Micro Kit (Qiagen), quantified by spectrophotometry and stored at −80°C.
In situ hybridization
Paraffin sections were rehydrated and treated with 0.2 μg/mL proteinase K (Roche) in 20 mM Tris-HCl pH 7.2 and 1 mM EDTA for 30 min at 37°C. After a 5 min wash in phosphate buffered saline (PBS) specimen were postfixed with 4% PFA for 20 min at RT, each twice washed in PBS and 2× saline-sodium citrate (SSC) and incubated in 0.1 M Tris-HCl pH 7.0 and 0.1 M glycine for 30 min at RT. Sections were then overlaid with 100 μL hybridization buffer containing ∼10 ng of DIG-labeled probe (optimal concentration was tested for each probe preparation), 10 mM Tris base, 10 mM sodium phosphate, 200 mM NaCl, 5 mM EDTA, 50% formamide, 10% dextran sulfate, 1× Denhardt's solution, 1 mg/mL yeast tRNA, 10 mM DTT, and 1 mg/mL salmon sperm DNA, pH 7.4. Sections were covered with Parafilm and incubated in a humidified chamber at 60°C overnight. The next day specimen were twice washed for 30 min in 2× SSC, 50% formamide, and 0.1% Tween20 at 65°C, followed by two washes in MABT (100 mM maleic acid, 150 mM NaCl, and 0.1% Tween20, pH 7.4) for 30 min at RT. Samples were equilibrated in NTE buffer (10 mM Tris-HCl pH 7.5 and 5 mM EDTA) for 5 min at 37°C, followed by incubation with 20 μg/mL RNase A in NTE for 30 min at 37°C. After a 5 min wash in NTE at 37°C and a wash in MABT for 5 min at RT specimen were then blocked with 2% blocking reagent (Roche) in MABT, 0.1% Tween20 for 1 h at RT, followed by an overnight incubation with a 1:2,000 dilution of the primary anti-DIG/alkaline phosphatase-conjugated antibody (Roche) at 4°C. The next day, sections were washed four times for 15 min at RT in MABT, equilibrated with NTMT buffer (100 mM Tris-HCl, pH 9.5, 100 mM sodium chloride, 50 mM magnesium chloride, and 0.1% Tween20) and incubated in NTMT plus 2 mM levamisole (Sigma-Aldrich) for 30 min. Color reaction was performed in NTMT, 2 mM levamisole, 50 mg/mL BCIP, and 50 mg/mL NBT (Roche) for 3 h to 2 days at RT. The reaction was stopped by washes in PBS buffer, followed by counterstaining with nuclear fast red.
Histology and immunohistochemistry
Samples were fixed in 4% PFA/PBS overnight, paraffin embedded, sectioned at 7 μm, and stained with hematoxylin and eosin for routine histological analysis as described before [12]. For Ki67 staining slides were rehydrated and boiled for 20 min in 10 mM sodium citrate for antigen retrieval. Endogenous peroxidase was blocked by 3% hydrogen peroxide (Fisher) in methanol for 30 min. After blocking in serum-free protein block solution (DAKO, Mississauga, ON, Canada), a 1:100 dilution of a rabbit anti Ki67 antibody (Thermo Fisher Scientific) in PBS was applied to the sections and incubated overnight at 4°C. For visualization a 1:200 dilution of a biotinylated goat anti-rabbit antibody (Vector Labs, Burlington, ON, Canada) in PBS was applied for 30 min at RT, followed by a 1:50 dilution of the Avidin/Biotin/horseradish-peroxidase complex (Vector Labs) for 30 min. Specific signals were visualized in a colorimetric DAB reaction (DAKO), which was stopped by a short wash in water. Between all steps slides were washed thrice for 5 min in PBS/0.1% Tween20. Sections were counterstained with hematoxylin.
For TUNEL staining sections were rehydrated, and incubated in 10 μg/mL proteinase K in PBS for 10 min at RT. After 3×5 min washes in water, endogenous peroxidase activity was blocked by 3% hydrogen peroxide in methanol for 30 min, followed by three washes in PBS. Slides were then blocked with 2% bovine serum albumin (BSA), 20% newborn bovine serum in PBS for 15 min, and then preincubated in 1× One-Phor-All-Buffer (Trevigen, Gaithersburg, MD), 0.1% Triton X-100 for 10 min at RT. The enzymatic labeling reaction was performed by addition of terminal desoxynucleotidyl transferase (TdT) solution containing 10 μM biotin-16-dUTP (Roche), 1 mM dATP, 4,000 U/mL recombinant TdT enzyme (Roche) in 1× One-Phor-All-Buffer (Trevigen), and 0.1% Triton X-100. Slides were incubated for 2 h at 37°C, then washed in PBS and blocked with 2% BSA and 20% NBT in PBS for 15 min. A 1:50 dilution of an Avidin/Biotin/horseradish-peroxidase complex (Vector Labs) was added to the sections and incubated for 90 min at 37°C. After 2 washes in 0.1% Triton X-100 in PBS, a colorimetric DAB reaction (DAKO) was used to visualize signals. Sections were counterstained with hematoxylin.
Stereology
Slides were scanned with a NanoZoomer digital scanner (Hamamatsu, Middlesex, NJ) and imported to the Visiopharm newCAST™ software program (Horsholm, Denmark). Roughly, 25% of each placental section was randomly counted at 40× magnification with 30 points per visual field, representing 2.7 μm2 per point/count. A total of seven sections per placenta were analyzed. Data were transferred to Excel program and total areas for specific structures were calculated for each section and averaged per placenta. To quantify Ki67 staining, 10% of each placental section was counted for numbers of Ki67-positive nuclei and total nuclei per randomly set counting frames. Data were analyzed using a one-tailed Student's t-test.
Results
Stereological and immunohistological analysis
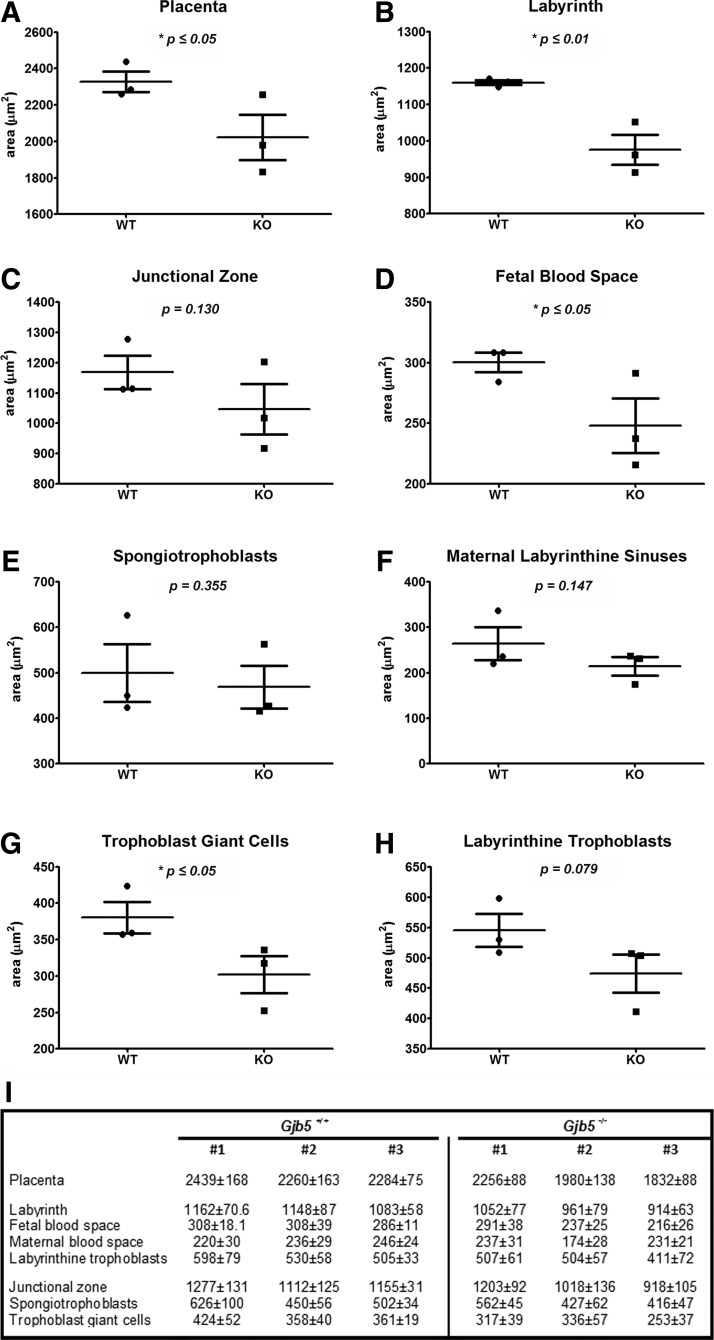
To define the altered placental structural organization of Cx31.1-deficient placentas more precisely, we performed stereological analysis using randomized sampling on the E10.5 placenta. Three wild-type and three Cx31.1-deficient placentas were analyzed, measuring the total area of labyrinth, fetal blood space, maternal blood space, labyrinthine trophoblast, junctional zone, SPT, and parietal TGC (Fig. 1). In the Cx31.1-deficient mice, the total sizes of placenta (Fig. 1A), labyrinth layer (Fig. 1B), fetal blood spaces (Fig. 1D), and parietal TGC were statistical significantly reduced. Interestingly, one of the Cx31.1-deficient placentas (#1, Fig. 1I) takes an intermediate position regarding placental structures between the control group and the two other Cx31.1-deficient littermates (Fig. 1I) showing a gradual effect of Cx31.1-deficiency on placental development. The reduction of individual structures is proportional to the reduction in total size of placentas, though the knockout placentas are affected to a different extent; whereas Cx31.1-deficient placenta #1 shows an average 96% of the structural dimensions of the averaged wild-type controls, the value is 83% for Cx31.1-deficient placenta #2 and 69% for placenta #3.
FIG. 1.
Changed placental structure in the Cx31.1−/− mouse. Stereological analysis on each three Cx31.1+/+ and Cx31.1−/− placentas at E10.5. Areas of placental structures are presented in μm2. Placentas are in average significantly reduced in size (A). The reduction is size is mainly due to a reduced labyrinth layer (B). Fetal blood spaces are reduced in total area (D). There is a trend in reduction of the junctional zone (C) and the area of trophoblast giant cell (TGC) is significantly reduced (G). Areas of spongiotrophoblasts (E), maternal labyrinthine sinuses (F), and labyrinthine trophoblasts (H) were not significantly changed. (I) Tabular result of measurement with standard error of the mean (SEM, n=7).
Altered marker gene expression in the Cx31.1-deficient placenta
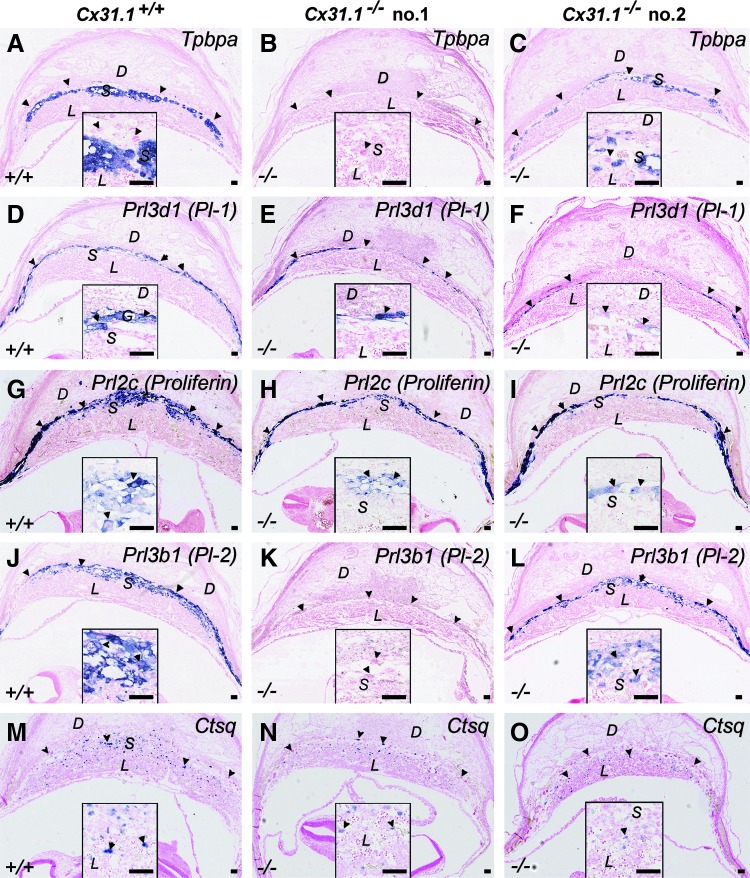
Previous studies demonstrated 30% loss of Cx31.1-deficient embryos between E10.5 and E13.5 [11]. To evaluate whether these structural changes in placentas of Cx31.1-deficient mice are associated to changes in trophoblast lineage differentiation, we performed in situ hybridization analysis for several trophoblast marker genes on sections of placental arrays from littermates at E10.5. The results showed that Cx31.1-deficient placentas exhibited two different marker gene expression phenotypes compared to placentas from wild-type littermate controls (Fig. 2). Specifically, in one group of Cx31.1-deficient placentas, signals for Tpbpa, which marks all SPT cells in the placenta from E10.5 onward (Fig. 2A), was completely absent (Fig. 2B). These placentas showed also absent signals for secondary TGC markers Prl3b1/Pl-2 (Fig. 2J, K), and a reduction in Ctsq-positive TGC in the junctional zone and within the labyrinth layer (Fig. 2M, N). The primary TGC markers Prl3d1/Pl-1 (Fig. 2D, E), and Prl2c/Plf (proliferin) (Fig. 2G, H) were present in these placentas, but the TGC layers appeared thinner compared to wild-type controls. In the second group of Cx31.1-deficient placentas, there were reduced numbers of Tpbpa-positive SPT cell clusters (Fig. 3C) within the junctional zone. These placentas had also Prl3b1/Pl-2-positive TGC (Fig. 2L), though to a lesser extent than the wild-type littermates (Fig. 2J) and reduced numbers of Ctsq-positive TGC (Fig. 2O). These results may indicate a defect in transition of trophoblast cells of the ectoplacental cone (EPC) toward SPT cells and subsequently an altered differentiation of secondary (Prl3b1/Pl-2 and/or Ctsq positive) TGC from the Tpbpa-positive trophoblast cells of this population.
FIG. 2.
In situ hybridization for placental differentiation marker genes. At E10.5 individual Cx31.1−/− placentas show absence of Tpbpa-positive precursors in the spongiotrophoblast (SPT) layer (B), whereas other Cx31.1−/− littermates show Tpbpa-positive cells but a reduced SPT layer (C) compared with Cx31.1+/+ controls (A). The formation of secondary TGC derived from the SPT cells is reduced. Compared with controls there are less Prl3d/Pl-1 and Prl2c-positive TGC in the junctional zone, and the TGC layer appears thinner in the Cx31.1−/− placentas compared to controls (D, E, F, G, H, I). Prl3b1/Pl-2-positive TGC are missing in the Tpbpa-negative placentas (B, K) compared to Tpbpa positive placentas (J, L) and numbers of Ctsq-positive TGC are reduced in Cx31.1−/− placentas compared with controls (M, N, O). Scale bars represent 100 mm. Insets show higher magnification of stained areas. D, decidua; L, labyrinth; S, SPT; arrowheads, TGC.
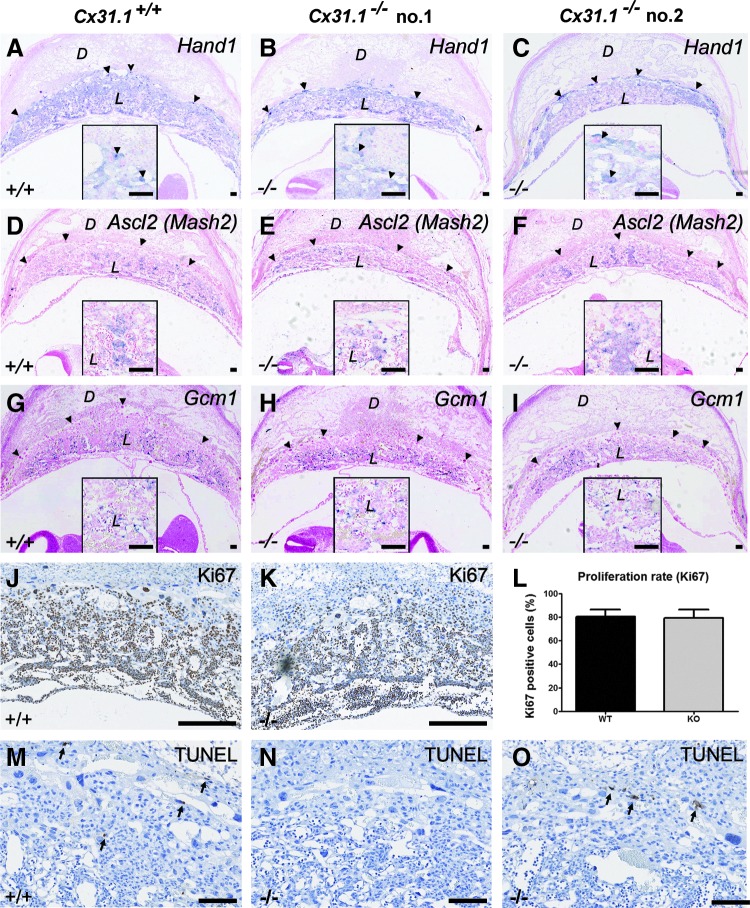
FIG. 3.
In situ hybridization for placental transcription factors. There are no obvious differences in the amount or localization of Hand1 (A–C), Ascl2 (D–F), or Gcm1 (G–I)-positive trophoblast cells in Cx31.1−/− placentas compared to controls. Ki67 staining (J, K) and quantification of Ki67-positive cells (L) reveal no differences in the number of proliferating cells in Cx31.1−/− placentas. TUNEL assay reveals only basal apoptosis levels (arrows) in Cx31.1−/− (N, O), and control placentas (M). Scale bars represent 100 μm in A–I; 200 μm in J–O. Insets show higher magnification of stained areas. D, decidua; L, labyrinth; S, SPT; arrowheads, TGC.
There were no obvious changes in expression or localization of Hand1 (Fig. 3A–C), Ascl2 (Fig. 3D–F), or Gcm1 (Fig. 3I)-positive cells in Cx31.1-deficient placentas. There was no difference in the percentage of Ki67-positive, proliferating cells in the placentas (Fig. 3J–L) and TUNEL assay showed no signs of apoptosis (Fig. 3M–O) in the E10.5 placenta.
Next, we performed hybridizations on placental arrays of surviving concepti at E12.5 and found no obvious changes in spatial distribution of marker genes. In particular, Tpbpa, Prl3b1/Pl-2, and Ctsq-positive cells were present in the Cx31.1-deficient placentas of surviving embryos (Supplementary Fig. S1).
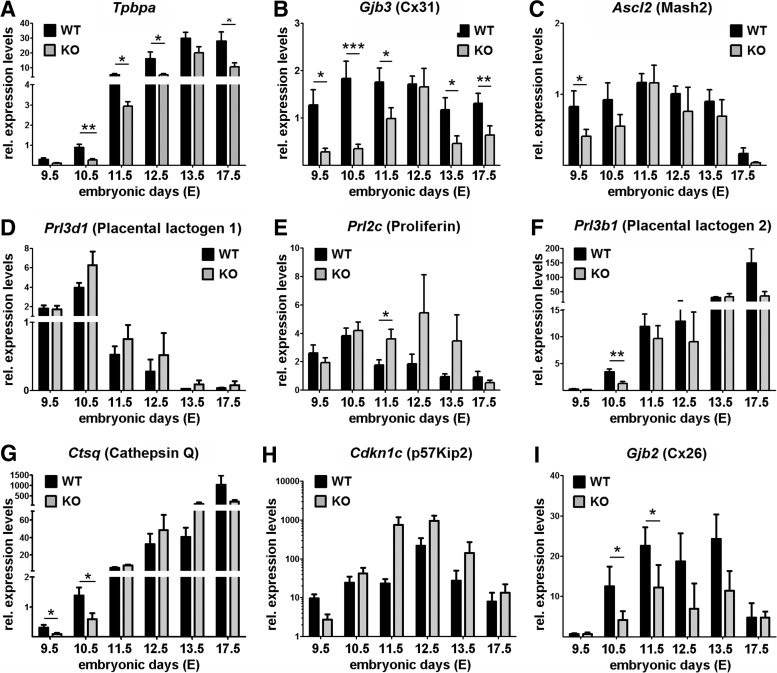
Using qPCR we screened expression profiles for trophoblast marker genes between gestational days E9.5 and E17.5 (Fig. 4), on randomly picked placentas from several Cx31.1+/− crossing. The marker gene Tpbpa was downregulated over gestation. Statistical analysis of the data using regression method reveals a significant influence (P≤0.01) of the genotype on Tpbpa expression over the observed time course (Fig. 4A). As sizes of labyrinth, junctional zone, and SPT layers are reduced proportionally (Fig. 1); the downregulation of markers Tpbpa and Cx31 must be due to a repression/delay on a cellular base rather than on a shift in placental subpopulations. Cx31 that is co-expressed with Tpbpa not only in the SPT, but also in the labyrinthine trophoblast cells, showed decreased expression levels over gestation (statistical regression: P≤0.001). Ascl2 expression was significantly reduced only at E9.5 (Fig. 4C). Interestingly, both Cx31 and Ascl2 appear to recover by E11.5 or E12.5. The secondary TGC marker Prl3b1/Pl-2 was significantly reduced at E10.5 (Fig. 4F) and Ctsq at E9.5 and E10.5 (Fig. 4G), whereas expression levels of both markers recover afterward comparable to the profiles of Cx31 and Ascl2. The marker Gjb2 encoding for gap junction channels protein Cx26, connecting the two syncytiotrophoblast layers, indicates a reduced functional barrier over gestation (Fig. 4I). The parietal TGC markers Prl3d1/Pl-1 (Fig. 4D) and Prl2c/Plf (Fig. 4E) are unchanged in the E9.5 or E10.5 placenta but reveal a trend to upregulation from mid-gestation to the end of pregnancy in surviving Cx31.1−/− placentas. Similarly, the endo-reduplication marker Cdkn1c is persistent at the same time (Fig. 4H), indicating a right shift in expression patterns of TGC markers. Altogether, Cx31.1-deficient placentas show a delayed and/or reduced expression of marker genes Tpbpa, Cx31, Ascl2, Prl3b1/Pl-2, Ctsq, and Gjb2/Cx26 during the early stages of formation of the chorio-allantoic placenta (E9.5 to E11.5).
FIG. 4.
Expression profiles for placental marker genes between E9.5 and E17.5 of gestation. Quantitative PCR was performed on cDNA generated from each 9 (E9.5 to E11.5) or 5 (E12.5 to E17.5) whole placentas. Bars show relative average expression levels (fold change) with SEM. The SPT markers Tpbpa (A) and Cx31 (B) show a strong down regulation in Cx31.1−/− placentas compared with wild-type controls. Ascl2/Mash2 is significantly reduced at E9.5 (C). The TGC markers Prl3b1/Pl-2 (F) and Ctsq (G) are significantly reduced in E9.5 or E10.5 placentas during the phase of embryonic loss. Interestingly, markers catch up until days E12.5 to E13.5. Gjb2/Cx26 (I) a marker for syncytiotrophoblast marker shows downregulation indicating a smaller labyrinth layer in the Cx31−/− placentas. Prl3d1/Pl-1 (D), Prl2c/Plf (E) and Cdkn1c /p57Kip2 (H) reveal a trend to upregulation from mid-gestation. Asterisks indicate statistically significant differences: *P≤0.05; **P≤0.01; ***P≤0.001. PCR, polymerase chain reaction.
Delayed differentiation of Cx31.1-deficient TSC
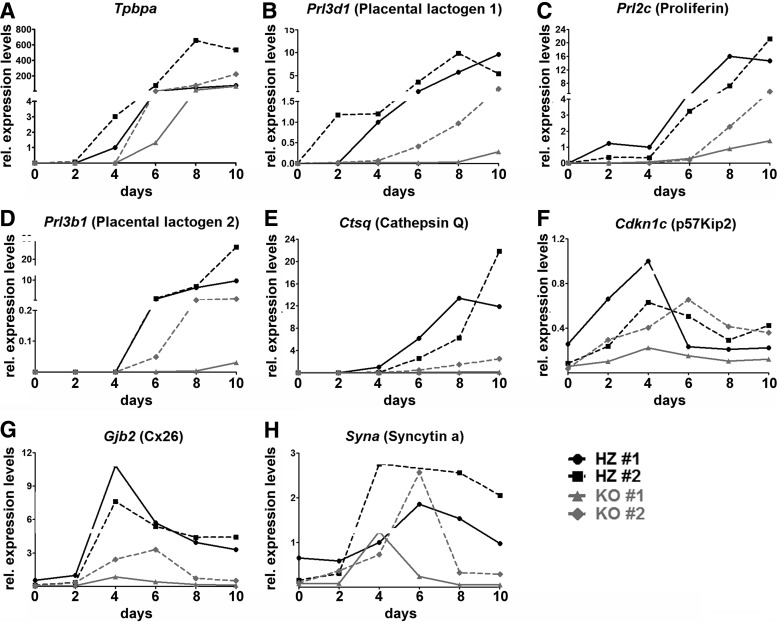
To perform trophoblast cell lineage analysis, we generated TSC lines from blastocysts of the Cx31.1-deficient mouse strain. TSC allow the analysis of trophoblast marker genes without the influence of fetal (ie, mesenchyme, yolk sac) or maternal factors (decidua, immune cells). Two Cx31.1+/− and two Cx31.1−/− TSC lines from the same preparation were analyzed in parallel using identical batches of conditioned medium and growth factors. Quantitative analysis revealed a delayed induction and/or a repression of expression levels of most analyzed marker genes. Tpbpa expression in Cx31.1−/− TSC could first be detected at day 4 of differentiation compared with day 2 in controls (Fig. 5A). Subsequently, induction of the secondary TGC markers Prl3b1/Pl-2 and Ctsq were also delayed and never reached expression levels of Cx31.1+/− TSC (Fig. 5D, E). Interestingly, Prl3d1/Pl-1 and Prl2c/Plf (Fig. 5B, C) also showed a delayed induction time and lower expression levels in Cx31.1−/− TSC compared with controls indicating that primary TGC derived from TSC are also delayed in differentiation. The syncytiotrophoblast marker Gjb2/Cx26 showed lower expression levels in Cx31.1−/− compared to Cx31.1+/− TSC (Fig. 5G) in accordance to results found with qPCR on placentas (Fig. 4I). Cdkn1c showed only a slightly shifted profile in Cx31.1−/− compared to Cx31.1+/− TSC (Fig. 5F). The correlation of low level Cdkn1c and Tpbpa expression with low levels of TGC markers is obvious in both Cx31.1−/− TSC lines. Syna (syncytin a), a regulator for syncytiotrophoblast fusion showed a similar right shift profile (Fig. 5H) compared to Gjb2/Cx26 (Fig. 5G) and Cdkn1c (Fig. 5F) though expression levels were not as strongly repressed. This again indicates a delayed terminal differentiation of Cx31.1−/− TSC into placental trophoblast populations.
FIG. 5.
Expression profile for marker genes during trophoblast stem cell (TSC) differentiation. To analyze trophoblast lineage formation, each two Cx31.1+/− and Cx31.1−/− TSC lines were differentiated for 10 days. Gene expression was analyzed by quantitative PCR and graphed as relative expression levels (fold change) over time. The analyzed marker genes show a block in terminal TGC and syncytiotrophoblast differentiation in the Cx31.1−/− TSC lines. In the Cx31.1−/− TSC lines induction of Tpbpa (A) expression is delayed compared with controls. Subsequently, expression of the secondary TGC markers Prl3b1/Pl-2 (D) and Ctsq (E) is delayed and expression levels remain below those of Cx31.1+/− controls. Also, Cx31.1−/− TSC exhibit a delayed and reduced expression of Prl3d1/Pl-1 (B) and Prl2c/Plf (C) TGC markers, and Cdkn1c (F), mediating cell cycle arrest and endoreduplication, reveals a slight shift in Cx31.1−/− TSC lines. Syncytiotrophoblast marker genes Gjb2/Cx26 (G) and Syna (H) are reduced in Cx31.1−/− TSC, indicating inhibition in of labyrinthine trophoblast differentiation.
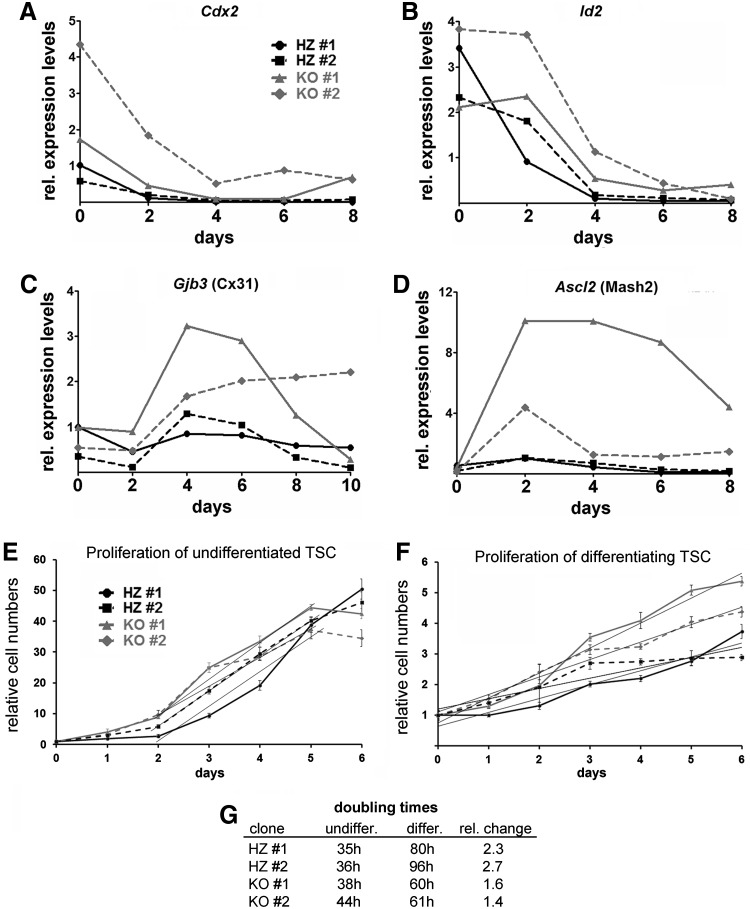
To further define at which point Cx31.1 deficiency affects TSC differentiation, we analyzed expression patterns of stem cell marker genes. The early marker Cdx2 (Fig. 6A) was slightly higher expressed in undifferentiated Cx31.1−/− TSC and showed a slower repression pattern during differentiation. Id2 (Fig. 6B) showed a similar pattern, indicating that the undifferentiated TSC state is only weakly affected by Cx31.1 deficiency. A strong influence of Cx31.1 deficiency was observed on Cx31 and Ascl2 expression patterns (Fig. 6C, D). Both markers showed higher expression levels and slower repression during differentiation of Cx31.1−/− compared with Cx31.1+/− TSC indicating a role of Cx31.1 in transition of the EPC to trophoblast populations in the chorio-allantoic placenta. Proliferation analysis over a 6-day time course indicated that during differentiation Cx31.1−/− TSC remain longer in a proliferative state compared with Cx31.1+/− controls (Fig. 6E, F), as shown by lower doubling times compared with controls during TSC differentiation (Fig. 6G).
FIG. 6.
Cx31.1−/− TSC cultures show prolongation of the intermediate, proliferating state during differentiation. Cx31.1−/− TSC show higher Cdx2 expression levels and, upon differentiation, slower loss of Cdx2 (A) and Id2 (B) expression. In accordance, Cx31.1−/− TSC show a prolonged expression of Ascl2 (C) and Cx31 (D) demonstrating a block in terminal trophoblast differentiation. MTT proliferation assays for undifferentiated (E) and differentiating (F) TSC indicate that the increased expression of Ascl2 and Cx31 is associated with an increased proliferation during TSC differentiation. Differentiating Cx31+/− TSC showed a 2.5-fold increase in doubling times compared with undifferentiated conditions. In contrast, Cx31−/− TSC increased doubling times only 1.5-fold upon differentiation (G). MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
Discussion
In this study, we show that Cx31.1 is critical for terminal differentiation of trophoblast cells during mouse placental development. Cx31.1 deficiency delays TSC differentiation into placental trophoblast subpopulations. As a consequence, formation of the chorio-allantoic placenta is disturbed leading to reduced placental size, and subsequently to fetal death after E10.5. This is the time frame when the placenta's circulation is fully established serving the higher metabolic demand of the conceptus [20]. The more precise stereological analysis revealed that the Cx31.1 phenotype is not a milder phenotype of the Cx31-deficient placenta as hypothesized before [1,11]. Cx31 deficiency enhances terminal differentiation into placental trophoblast subpopulations, whereas Cx31.1 deficiency blocks this process. The delayed expression of Tpbpa and subsequently the delayed onset of Prl3b1/Pl-2 and Ctsq at early stages of placentation (E9.5–E10.5) indicate a delay in the transition of the EPC into the SPT layer combined with a delayed differentiation of secondary TGC from this cell population.
Hu and Cross showed that eliminating Tpbpa-positive precursors, by using an inducible diphtheria toxin A, lead to dramatically reduced numbers of Tpbpa-expressing cells in the SPT, reduced placental sizes, and to embryonic death between E10.5 and E11.5 [21]. The numbers of TGC lining the spiral artery and canal, but not parietal TGC, were reduced. This is in accordance with our results showing that Prl2c/Plf and Prl3d1/Pl-1-positive parietal TGC are always present in Cx31.1−/− placentas, but Prl3b1/Pl-2-positive cells, which differentiate later in gestation, are reduced. Prl3b1/Pl-2 replaces Prl3d1/Pl-1 around mid-gestation and it has been shown that the same TGC express first Prl3d1/Pl-1 followed by Prl3b1/Pl-2 in the junctional zone [22]. However, newer investigations showed that sinusoidal TGC within the labyrinth are also Prl3b1/Pl-2 and Ctsq-positive [23]. As Prl3b1/Pl-2 and Ctsq-positive TGC appear later in Cx31.1−/− placentas it appears that differentiation from the chorionic plate is also inhibited or delayed in the absence of Cx31.1. This is supported by the reduced size of the labyrinth and by the reduced expression of Gjb2/Cx26 and delay of syna in TSC. A developmental delay in all the placental structures might indicate that trophoblast cells in Cx31.1−/− placentas are generally late in differentiation and not yet ready for placental formation. This mechanism fits the expression pattern of Cx31.1, which is expressed last at E8.5 in the extra-embryonic ectoderm and the EPC, but not in the syncytiotrophoblast of the developing labyrinth. In the SPT, it is repressed after E10.5 [11]. Thus, it seems to have a short effect in placental development in promoting the transition of the EPC to SPT and chorionic ectoderm to labyrinthine trophoblast.
The clearest sign of this changed differentiation in Cx31.1 knockouts is the upregulation of Ascl2 in differentiating TSC lines. According to current lineage differentiation models, the delay of Tpbpa induction is caused by an enhanced Ascl2 expression [23]. Ascl2 is absent from undifferentiated TSC and expression peaks around day 3 during differentiation followed by a rapid decline in expression levels [12]. It has been shown that transiently Ascl2 transfected TSC divide longer during differentiation [24] similar to Cx31.1−/− TSC. Cx31−/− TSC instead showed an earlier induction of Ascl2 and faster repression followed by earlier induction of Tpbpa and Prl3d1/Pl-1 [12]. Thus, in both connexin-deficient TSC the induction of TGC markers and Tpbpa are associated to the expression profile of Ascl2. The fact that both Cx31−/− and Cx31.1−/− TSC can be established and that they only revealed their obvious phenotypes under differentiation shows that both connexins, like Ascl2, play a role only in the transition of the EPC to SPT after chorio-allantoic fusion [25]. In fact, the Ascl2−/− mouse dies at E10.5 [19]. However, the placental Ascl2−/− phenotype highly resembles the Cx31−/− mouse with absence of a labyrinth and SPT layers [10]. Cx31.1 had only a mild effect on proliferation associated with increased Cx31 expression in the intermediate Ascl2- positive state. In the placenta itself, no overgrowth could be detected. Thus, the proliferation might be associated with the changed differentiation process rather than a separate effect of gap junctional communication. Gap junctions are somehow influencing the Ascl2-positive state during TSC differentiation. However, in the placenta at E10.5 itself no obvious changes in Ascl2 expression could be observed. This might be due to the fact that TSC cultures represent a 100% enriched population of stem cells, whereas in vivo only individual cells in the extra-embryonic ectoderm are true stem cells. In fact, the markers to identify “true” TSC in vivo are still unknown. The changed differentiation of individual TSC in the trophoblast populations might explain the transient, gradually affected phenotypes of both knockout mice. We also observed a mild effect on downregulation of TSC marker Id2 and Cdx2, indicating that delayed differentiation has already started at the beginning of TSC differentiation and then “arrests” in the Ascl2- positive state.
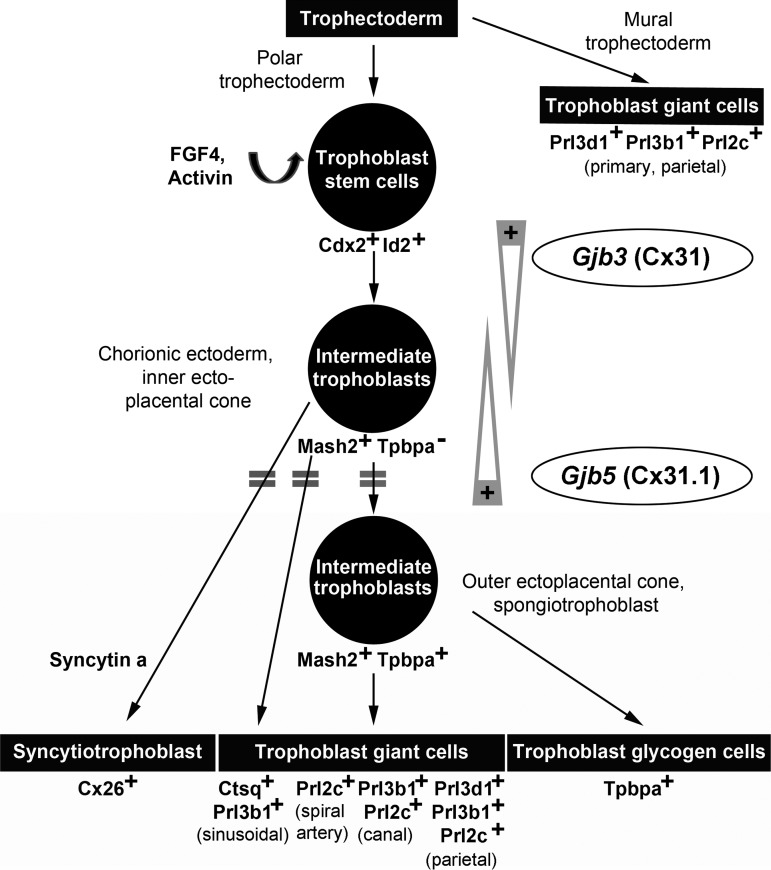
The major finding of our study is that Cx31.1 has a directly opposed function to Cx31 on TSC differentiation and placental development. Cx31 deficiency enhances TSC differentiation into placental subpopulations [10,12,13], whereas the absence of Cx31.1 delays this process. Thus, both connexins are rather collectively balancing TSC fate by having opposite regulatory effects on differentiation processes than serving the same molecular functions by just having redundant properties. In this context, it needs to be considered that the specific phenotypes of the Cx31 or Cx31.1-deficient mouse are not only caused by a “loss of function” of the particular connexin but also by a “gain of function” of the remaining co-expressed isoform (Fig. 7). It is plausible that the downregulation of Cx31 in the Cx31.1−/− placenta is part of a compensatory effect to reduce the “over-function” of Cx31. It would be interesting to see the impact of a Cx31/Cx31.1 double inactivation on trophoblast differentiation, placental development, and physiology of other organs.
FIG. 7.
Model for the role of Cx31 and Cx31.1 in trophoblast lineage differentiation and placental development. The scheme integrates the phenotypes of the Cx31 and Cx31.1 knockout mice into the current spatio-temporal model of murine trophoblast lineage differentiation. Upon differentiation, trophoblast cells pass through a proliferative, Ascl2/Mash2-positive state within the chorionic ectoderm and ectoplacental cone before terminal differentiation into the placental subpopulations. The delayed induction of marker genes (indicated by double bars) in Cx31.1-deficient trophoblasts suggests a coordinated role for Cx31 and Cx31.1 in regulation transition through the Ascl2/Mash2-positive state. Cx31 is required to maintain the stem cells state and preserve the intermediate Ascl2/Mash2-positive state during trophoblast differentiation, whereas Cx31.1 is required to push cells into terminal differentiation, indicating the balancing, opposed function of both connexins (gray arrows) during murine placental development.
Beside the trophoblast lineage, Cx31 and Cx31.1 are also co-expressed in the epidermal layer of the skin, and in humans, mutations in the Cx31 gene cause the skin disease erythrokeratodermia variabilis (EKV) associated with keratinization disorders of the epidermis [26]. Most of these mutations have dominant negative effects, which can also negatively influence properties of the Cx31.1 channels, leading to cell death [27]. Transgenic mice carrying the homozygous human EKV mutation F137L in the Cx31 gene (Cx31F137L/F137L) die in utero after E7.5, whereas mice heterozygous for this mutation (Cx31+/F137L) are viable and fertile [28]. Though the lethal gestational phenotype of this mouse strain has not been further investigated yet it points to an early failure in trophoblast lineage differentiation. Also, the gene-dose effect of this mutation on embryo survival may suggest that the relative amount of functional Cx31 and Cx31.1 protein is responsible for the balance in trophoblast differentiation.
There are a few examples that two connexin isoforms coordinately regulate and balance physiological processes by potentially having opposite physiological functions. For instance, in endothelial cells, Cx37 and Cx40 are collectively essential for vascular development [29]. Recently, it was shown that Cx40 and Cx37 have opposite effects on postischemic limb perfusion, recovery, and tissue survival in the same mouse model [30]. Another example is the expression of Cx31 and Cx43 during preimplantation development. Cx31 and Cx43 are the main isoforms expressed in embryos of several species and it was also thought that these isoforms might compensate for the loss of each other [10,31]. In contrast, the Cx31−/−/Cx43−/− mouse shows both the known placental Cx31−/− and the heart Cx43−/− phenotype, but no new or synergistic effects demonstrating that there are no rescue or compensatory effects of both connexins [9]. This is another example showing that the replacement theory for the co-expression of different connexins has to be viewed carefully. It remains to be shown why the expression of connexins is so strongly regulated in the embryonic and maternal uterine tissues. Gap junctions could mediate positional effects for maintenance of stem cells in the trophoblast lineage by transmitting signals from the embryo and the decidua. This could be addressed that is, by micro-dissecting individual cells of the extra-embryonic or chorionic ectoderm and analyzing its gene expression.
Taken together, our study showed that Cx31.1 is critical for terminal differentiation of TSC into placental trophoblast populations and thus, Cx31 and Cx31.1 have opposite functions in regulating mouse trophoblast differentiation and placental development. This suggests that besides unique and shared functions co-expressed connexins might also have antagonistic effects, and thereby collectively balance physiological processes.
Supplementary Material
Acknowledgments
We would like to thank Dr. K. Willecke (Bonn, Germany) for the Cx31.1-deficient mouse strain, Dr. J. Rossant (Toronto, Canada) for plasmids, and Mr. Biliecki for help with stereology software. This work was supported by grants of the Deutsche Forschungsgemeinschaft (DFG) to M.K. (KI 1278/1-1) and CIHR to S.J.L. (MOP-130550). Material in this publication was presented at the IFPA Meeting 2013 in Whistler, Canada.
Author Disclosure Statement
The authors declare no conflicts of interest.
References
- 1.Kibschull M, Gellhaus A. and Winterhager E. (2008). Analogous and unique functions of connexins in mouse and human placental development. Placenta 29:848–854 [DOI] [PubMed] [Google Scholar]
- 2.Malassine A. and Cronier L. (2005). Involvement of gap junctions in placental functions and development. Biochim Biophys Acta 1719:117–124 [DOI] [PubMed] [Google Scholar]
- 3.Dunk CE, Gellhaus A, Drewlo S, Baczyk D, Potgens AJ, Winterhager E, Kingdom JC. and Lye SJ. (2012). The molecular role of connexin 43 in human trophoblast cell fusion. Biol Reprod 86:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valiunas V, Polosina YY, Miller H, Potapova IA, Valiuniene L, Doronin S, Mathias RT, Robinson RB, Rosen MR, Cohen IS. and Brink PR. (2005). Connexin-specific cell-to-cell transfer of short interfering RNA by gap junctions. J Physiol 568:459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ek-Vitorin JF. and Burt JM. (2013). Structural basis for the selective permeability of channels made of communicating junction proteins. Biochim Biophys Acta 1828:51–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herve JC. and Derangeon M. (2013). Gap-junction-mediated cell-to-cell communication. Cell Tissue Res 352:21–31 [DOI] [PubMed] [Google Scholar]
- 7.Oyamada M, Oyamada Y. and Takamatsu T. (2005). Regulation of connexin expression. Biochim Biophys Acta 1719:6–23 [DOI] [PubMed] [Google Scholar]
- 8.Hennemann H, Dahl E, White JB, Schwarz HJ, Lalley PA, Chang S, Nicholson BJ. and Willecke K. (1992). Two gap junction genes, connexin 31.1 and 30.3, are closely linked on mouse chromosome 4 and preferentially expressed in skin. J Biol Chem 267:17225–17233 [PubMed] [Google Scholar]
- 9.Kibschull M, Magin TM, Traub O. and Winterhager E. (2005). Cx31 and Cx43 double-deficient mice reveal independent functions in murine placental and skin development. Dev Dyn 233:853–863 [DOI] [PubMed] [Google Scholar]
- 10.Plum A, Winterhager E, Pesch J, Lautermann J, Hallas G, Rosentreter B, Traub O, Herberhold C. and Willecke K. (2001). Connexin31-deficiency in mice causes transient placental dysmorphogenesis but does not impair hearing and skin differentiation. Dev Biol 231:334–347 [DOI] [PubMed] [Google Scholar]
- 11.Zheng-Fischhofer Q, Kibschull M, Schnichels M, Kretz M, Petrasch-Parwez E, Strotmann J, Reucher H, Lynn BD, Nagy JI, et al. (2007). Characterization of connexin31.1-deficient mice reveals impaired placental development. Dev Biol 312:258–271 [DOI] [PubMed] [Google Scholar]
- 12.Kibschull M, Nassiry M, Dunk C, Gellhaus A, Quinn JA, Rossant J, Lye SJ. and Winterhager E. (2004). Connexin31-deficient trophoblast stem cells: a model to analyze the role of gap junction communication in mouse placental development. Dev Biol 273:63–75 [DOI] [PubMed] [Google Scholar]
- 13.Koch Y, van Furden B, Kaiser S, Klein D, Kibschull M, Schorle H, Carpinteiro A, Gellhaus A. and Winterhager E. (2012). Connexin 31 (GJB3) deficiency in mouse trophoblast stem cells alters giant cell differentiation and leads to loss of oxygen sensing. Biol Reprod 87:37. [DOI] [PubMed] [Google Scholar]
- 14.Kibschull M. and Winterhager E. (2006). Connexins and trophoblast cell lineage development. Methods Mol Med 121:149–158 [DOI] [PubMed] [Google Scholar]
- 15.Kibschull M, Mileikovsky M, Michael IP, Lye SJ. and Nagy A. (2011). Human embryonic fibroblasts support single cell enzymatic expansion of human embryonic stem cells in xeno-free cultures. Stem Cell Res 6:70–82 [DOI] [PubMed] [Google Scholar]
- 16.Colosi P, Talamantes F. and Linzer DI. (1987). Molecular cloning and expression of mouse placental lactogen I complementary deoxyribonucleic acid. Mol Endocrinol 1:767–776 [DOI] [PubMed] [Google Scholar]
- 17.Jackson LL, Colosi P, Talamantes F. and Linzer DI. (1986). Molecular cloning of mouse placental lactogen cDNA. Proc Natl Acad Sci U S A 83:8496–8500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linzer DI. and Mordacq JC. (1987). Transcriptional regulation of proliferin gene expression in response to serum in transfected mouse cells. EMBO J 6:2281–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guillemot F, Caspary T, Tilghman SM, Copeland NG, Gilbert DJ, Jenkins NA, Anderson DJ, Joyner AL, Rossant J. and Nagy A. (1995). Genomic imprinting of Mash2, a mouse gene required for trophoblast development. Nat Genet 9:235–242 [DOI] [PubMed] [Google Scholar]
- 20.Maltepe E. and Simon MC. (1998). Oxygen, genes, and development: an analysis of the role of hypoxic gene regulation during murine vascular development. J Mol Med (Berl) 76:391–401 [DOI] [PubMed] [Google Scholar]
- 21.Hu D. and Cross JC. (2011). Ablation of Tpbpa-positive trophoblast precursors leads to defects in maternal spiral artery remodeling in the mouse placenta. Dev Biol 358:231–239 [DOI] [PubMed] [Google Scholar]
- 22.Yamaguchi M, Ogren L, Endo H, Thordarson G, Bigsby RM. and Talamantes F. (1992). Production of mouse placental lactogen-I and placental lactogen-II by the same giant cell. Endocrinology 131:1595–1602 [DOI] [PubMed] [Google Scholar]
- 23.Simmons DG, Fortier AL. and Cross JC. (2007). Diverse subtypes and developmental origins of trophoblast giant cells in the mouse placenta. Dev Biol 304:567–578 [DOI] [PubMed] [Google Scholar]
- 24.Hughes M, Dobric N, Scott IC, Su L, Starovic M, St-Pierre B, Egan SE, Kingdom JC. and Cross JC. (2004). The Hand1, Stra13 and Gcm1 transcription factors override FGF signaling to promote terminal differentiation of trophoblast stem cells. Dev Biol 271:26–37 [DOI] [PubMed] [Google Scholar]
- 25.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A. and Rossant J. (1998). Promotion of trophoblast stem cell proliferation by FGF4. Science 282:2072–2075 [DOI] [PubMed] [Google Scholar]
- 26.Richard G, Smith LE, Bailey RA, Itin P, Hohl D, Epstein EH, Jr., DiGiovanna JJ, Compton JG. and Bale SJ. (1998). Mutations in the human connexin gene GJB3 cause erythrokeratodermia variabilis. Nat Genet 20:366–369 [DOI] [PubMed] [Google Scholar]
- 27.Scott CA, Tattersall D, O'Toole EA. and Kelsell DP. (2012). Connexins in epidermal homeostasis and skin disease. Biochim Biophys Acta 1818:1952–1961 [DOI] [PubMed] [Google Scholar]
- 28.Schnichels M, Worsdorfer P, Dobrowolski R, Markopoulos C, Kretz M, Schwarz G, Winterhager E. and Willecke K. (2007). The connexin31 F137L mutant mouse as a model for the human skin disease erythrokeratodermia variabilis (EKV). Hum Mol Genet 16:1216–1224 [DOI] [PubMed] [Google Scholar]
- 29.Simon AM. and McWhorter AR. (2003). Role of connexin37 and connexin40 in vascular development. Cell Commun Adhes 10:379–385 [DOI] [PubMed] [Google Scholar]
- 30.Fang JS, Angelov SN, Simon AM. and Burt JM. (2012). Cx40 is required for, and cx37 limits, postischemic hindlimb perfusion, survival and recovery. J Vasc Res 49:2–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Houghton FD, Barr KJ, Walter G, Gabriel HD, Grummer R, Traub O, Leese HJ, Winterhager E. and Kidder GM. (2002). Functional significance of gap junctional coupling in preimplantation development. Biol Reprod 66:1403–1412 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.