Abstract
Recent advances in nuclear medicine have allowed for positron emission tomography (PET) to track transgenes in cell-based therapies using PET reporter gene/probe pairs. A promising example for such reporter gene/probe pairs are engineered nucleoside kinases that effectively phosphorylate isotopically labeled nucleoside analogues. Upon expression in target cells, the kinase facilitates the intracellular accumulation of radionuclide monophosphate, which can be detected by PET imaging. We have employed computational design for the semi-rational engineering of human 2′-deoxycytidine kinase to create a reporter gene with selectivity for l-nucleosides including l-thymidine and 1-(2′-fluoro-5-methyl-β-l-arabinofuranosyl) uracil. Our design strategy relied on a combination of preexisting data from kinetic and structural studies of native kinases, as well as two small, focused libraries of kinase variants to generate an in silico model for assessing the effects of single amino acid changes on favorable activation of l-nucleosides over their corresponding d-enantiomers. The approach identified multiple amino acid positions distal to the active site that conferred desired l-enantioselectivity. Recombination of individual amino acid substitutions yielded orthogonal kinase variants with significantly improved catalytic performance for unnatural l-nucleosides but reduced activity for natural d-nucleosides.
Molecular imaging by positron emission tomography (PET) offers a powerful and versatile method for noninvasive visualization of biological processes in living subjects.1 In preclinical and clinical studies, PET imaging is routinely used to monitor disease development, progression, and treatment. The technique is based on a two-component system, including an isotopically labeled small-molecule reporter and a reporter gene.2−4 The role of the reporter gene is to co-localize with target cells, as well as to interact and trap the radionuclide reporter whose accumulation can be detected via PET. Among the leading reporter systems are nucleoside kinases used in combination with [18F]- or [125I]-labeled nucleoside analogue (NA) reporters.
The phosphorylation of NAs for PET imaging was first implemented with thymidine kinase from Herpes Simplex virus (HSV-tk).5 The enzyme’s high substrate promiscuity enables effective activation of the small-molecule reporter and has found widespread application in cell-culture studies, as well as in translational work with animals and humans.6−12 These studies are typically conducted with wild-type Herpes enzyme or an engineered variant, HSV-sr39TK,13 using various NAs including 1-(2′-fluoro-5-methyl-β-l-arabinofuranosyl) uracil (l-FMAU; Figure 1).14 Ideally, these NAs are not phosphorylated by the endogenous human nucleoside kinases yet are readily activated by the Herpes enzymes. However, the experiments with HSV-tk have revealed significant limitations as this PET reporter gene suffers from problems with activity, specificity, and risk of adverse immune reactions. First, HSV-tk is promiscuous, which is important for NA activation, yet it is still primarily a thymidine kinase. The enzyme’s high thymidine kinase activity, together with elevated expression levels can result in deregulation of the host cell’s tightly controlled dNTP pool, which among other things causes declining DNA replication fidelity.15,16 Second, the catalytic activity of HSV-tk for NAs is far from optimal as reflected in inferior kinetic parameters compared to natural nucleosides.17 Third, the clinical use of viral kinases raises concerns over potential immunogenicity, especially upon its repetitive and long-term application.18
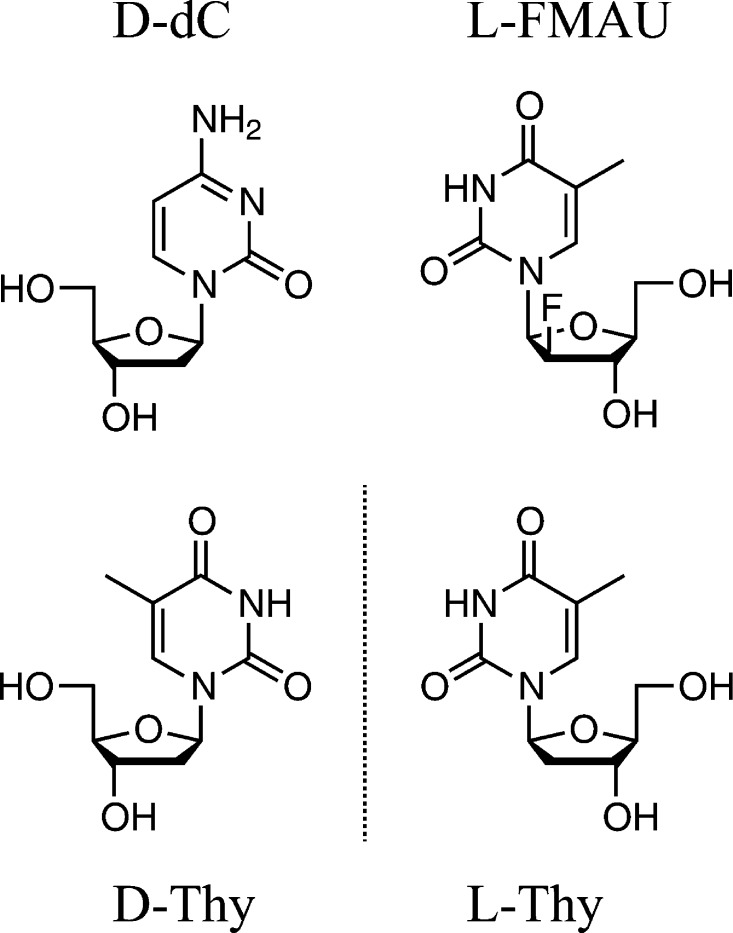
Figure 1.
Structures of native 2′-deoxy-d-ribosyl nucleosides (d-dC and d-Thy), as well as the l-stereoisomeric form of Thy and the nucleoside analogue 2′-fluoro-2′-deoxy-1-β-l-arabinofuranosyl-5-methyluracil (l-FMAU).
Over the past decade, advances in protein engineering have supported scientists in tailoring kinases to address some of these limitations. Recent efforts have concentrated on the exploration of human nucleoside kinases as reporter genes. The application of human enzymes minimizes the risk of an immune response in clinical applications. A majority of these studies have focused on engineering human deoxycytidine kinase (dCK) and human thymidine kinase 2 (TK2) with the goal to broaden and change the substrate specificities of these enzymes.19−25 Improvements in their kinetic parameters compared to the native enzymes have been significant, yet these efforts have tended to identify generalists with broad substrate specificity for native nucleosides and NAs. More recently, a study by Lavie and co-workers specifically focused on tailoring human TK2 for selective l-nucleoside activation.25 Guided by structural information, amino acid substitutions in two positions (N93D and L109F) resulted in a 2-fold decline in activity for d-Thy. The TK2 variants also showed increased turnover for l-FMAU by the same magnitude, resulting in enhanced catalytic activity. However, the enzyme’s specificity constants for these two substrates remained largely unchanged as raised KM values compensate for the activity gains.
In accordance with clinicians’ demands for “ideal” orthogonal kinases as next-generation reporters for high-contrast, low-impact molecular PET imaging,26 we herein report a semi-rational design approach for the generation of l-selective nucleoside kinases based on dCK. More specifically, two previously engineered dCK variants (ssTK1A and ssTK3)22 were chosen as parental kinases due to the enzymes’ broad substrate specificity including phosphorylation of numerous NA prodrugs and l-nucleosides.22,27 Kinetic data for these dCK variants and structural information from crystallographic studies of the wild type enzyme were used to generate a computational model for scoring the impact of amino acid changes on d- or l-Thy bound in the phosphoryl acceptor site. The predictive framework was then tested and refined through two rounds of mutagenesis, using small, focused libraries of dCK variants with single amino acid changes. Combination of individual beneficial mutations were mostly additive and resulted in functional gains, yielding two l-selective candidates with superior kinetic performance for the PET reporter l-FMAU over the natural substrates.
Results and Discussion
For our initial efforts to explore the contributing factors to enantioselectivity in dCK, we assembled a computational model for protein–ligand interactions. The predictive framework was based on the crystal structures of wild type dCK bound with 5-methyl dC as phosphoryl acceptor and ADP (PDB code: 3KFX(28)), as well as the dCK variant R104M/D133A in complex with l-Thy and ADP (PDB code: 3HP1(23)) (Figure 2). Besides the two amino acid changes at positions 104 and 133, the atomic composition of each protein structure was identical. The only significant difference between the modeled structures was the inverted ribose conformation of the two substrates to maintain the correct geometry of their 5′-OH groups for phosphoryl transfer (Figure 2 insert). The impact of amino acid substitutions on the enantioselectivity of dCK was then modeled using the Rosetta macromolecular modeling suite.29 Identical amino acid changes were made to both the d- and l-Thy bound dCK structures via fixed backbone side chain replacement followed by independent energy minimizations to generate a structure ensemble. The ensemble average scores were used to identify substitutions in individual amino acid positions favoring bound l-ribosyl nucleosides. The initial data suggested that the standard Rosetta score function did not provide sufficient resolution to capture experimental findings previous reported for dCK variants.22 For this reason, we re-parameterized Rosetta’s score function using 10 dCK crystal structures (PDB codes: 2NO1, 2NO7, 2NOA, 2NO6, 3KFX, 1P5Z, 2NO9, 1P62, 3HP1, and 2ZI4), along with their corresponding experimental data on catalytic activity.19,23,28,30,31 The new score function significantly improved the correlation with experimental data and hence was used for designing a small test library (Library A).
Figure 2.
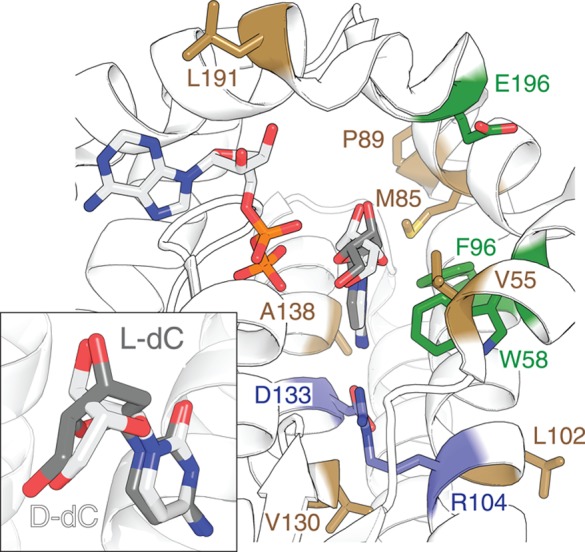
Summary of amino acid substitution in human dCK with bound ADP in the phosphoryl donor site, as well as d- and l-dC in the phosphoryl acceptor binding pocket (PDB access codes: 2NO1 and 2NO7(30)). Variant positions R104M and D133N in ssTK3 are marked in violet. The three positions probed in Library A are highlighted in green, while the seven residues varied in Library B are colored in brown. Insert: Overlay of the d- and l-dC bound in the active site shows the highly similar positioning of the pyrimidine moiety, as well as the 3′- and 5′-hydroxyl groups.
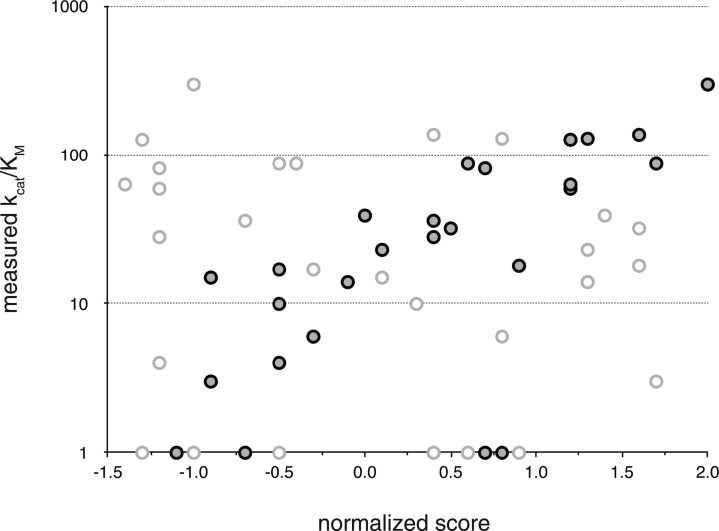
Library A consisted of eight members. The previously described dCK variant ssTK1A (A100V/R104M/D133S) was selected as scaffold (variant A0) due to its exquisitely high activity for d-dC and d-Thy.22 Building on ssTK1A, the top three predicted variants (ssTK1A with F96D, W58E, or E196L) were prepared. In addition, we included three variants with slightly more conservative amino acid changes in the same three positions (ssTK1A with F96Y, W58V, or E196A). Finally, ssTK3 (R104M/D133N) was added to Library A as a control. This variant was identified in the same study as ssTK1A and distinguishes itself by its broad substrate specificity for pyrimidine and purine 2′-deoxynucleosides. Following site-directed mutagenesis, cloning, expression, and purification, all eight Library A variants had their kinetic parameters determined for d- and l-Thy (Table 1). The catalytic properties of the parental ssTK1A and control ssTK3 for the two enantiomers show some interesting differences. Both dCK variants exhibit similar KM values for d- and l-Thy, while turnover rates differ by 2-fold. Overall, both enzymes are effective phosphoryl-transfer catalysts, either showing neutral enantioselectivity (ssTK3) or exhibiting a moderate preference for the d-isomer (ssTK1A) . Among the six new variants of our survey library, the three lead candidates all resulted in inactive enzymes. The rather dramatic amino acid changes in the selected positions are likely responsible for the failure to detect catalytic activity for these variants. In contrast, the three alternatives carrying more conservative amino acid substitutions yielded dCK variants with detectable phosphoryl-transfer activity. Although the catalytic properties of all three enzymes declined relative to the parental ssTK1A, the desired change in enantioselectivity could be observed in two of the three variants. Variants A5 and A6 both showed approximately 3-fold selectivity improvement in favor of the l-enantiomer. While the effect in A5 is associated with worsening KM and kcat values for either substrate, the change in A6 is largely driven by differences in KM values. The experimental data from Library A was used to update and refine our score function in an effort to obtain more accurate predictions on the effects of amino acid changes. Instead of using a traditional linear score function, we investigated the use of non-parametric functions, commonly used in machine learning/data mining applications (Figure 3). After trying various regression methods,32−35 we found the k-nearest neighbor algorithm best captured the results from the initial library and was used to design a second-generation library (Library B).
Table 1. Summary of Kinetic Data for dCK Library A.
|
l-Thy |
d-Thy |
l-Thy/d-Thy | ||||||
|---|---|---|---|---|---|---|---|---|
| variant | mutations | KM (μM) | kcat (s–1) | kcat/KM (s–1 M–1 × 10–3) | KM (μM) | kcat (s–1) | kcat/KM (s–1 M–1 × 10–3) | kcat/KM fold-change |
| A0 | A100V, R104M, D133S | 10.2 ± 1.1 | 1.41 ± 0.03 | 137 | 11.7 ± 1 | 3.49 ± 0.1 | 298 | 0.5 |
| A1 | A0 + F96D | na | na | na | na | |||
| A2 | A0 + W58E | na | na | na | na | |||
| A3 | A0 + E196L | na | na | na | na | |||
| A4 | A0 + F96Y | 489 ± 40 | 0.62 ± 0.03 | 1.2 | 1121 ± 146 | 1.70 ± 0.14 | 1.5 | 0.8 |
| A5 | A0 + W58V | 598 ± 89 | 0.55 ± 0.06 | 0.9 | 1254 ± 347 | 0.84 ± 0.19 | 0.7 | 1.3 |
| A6 | A0 + E196A | 34.4 ± 4.5 | 3.01 ± 0.13 | 87 | 53.4 ± 7.5 | 3.14 ± 0.16 | 59 | 1.5 |
| B0 | R104M, D133N | 18.5 ± 4.3 | 1.61 ± 0.11 | 87 | 18.5 ± 3.1 | 1.49 ± 0.06 | 81 | 1.1 |
Figure 3.
Correlation of linear and non-parametric model scores with experimental data. Data obtained with the standard Rosetta score function are shown as open circles (R2 = 0.02, Pearson’s R = 0.08) while results after re-parametrization are marked by gray circles (R2 = 0.62, Pearson’s R = 0.82).
Similar in size to the previous set of variants, Library B was made up of nine members. Notably, the new library used ssTK3 instead of ssTK1A as parental sequence. The switch in template was based on selectivity data obtained from Library A that indicated neutrality in regard to enantioselectivity for ssTK3 but showed an undesirable 2-fold preference for the d- over l-isomer for ssTK1A. The new template in combination with the refined machine-learned score function was used to identify amino acid substitutions that either destabilized interactions with d-Thy or resulted in structural changes favoring bound l-Thy in the phosphoryl-acceptor site. Besides ssTK3 (B0), eight variants (B1–B8) carrying single amino acid changes in seven positions (V55E/F, M85Y, P89F, L102Y, V130T, A138I, L191A) were prepared. None of these amino acid residues were in immediate contact with the substrates (primary shell residues) but instead were located in second and third shell positions (Figure 2). The steady-state kinetic parameters of the eight variants for d- and l-Thy clearly showed functional improvements over Library A (Table 2). Only two candidates had no detectable activity. The remaining six variants (B3–B8) exhibited either neutral enantioselectivity (B3–B5) or favored the l-isomer by up to 2.3-fold (B6–B8).
Table 2. Summary of Kinetic Data for dCK Library B.
|
l-Thy |
d-Thy |
l-Thy/d-Thy | ||||||
|---|---|---|---|---|---|---|---|---|
| variant | mutations | KM (μM) | kcat (s–1) | Kcat/KM (s–1 M–1 × 10–3) | KM (μM) | kcat (s–1) | Kcat/KM (s–1 M–1 × 10–3) | Kcat/KM fold-change |
| B0 | R104M, D133N | 18.5 ± 4.3 | 1.61 ± 0.11 | 87 | 18.5 ± 3.1 | 1.49 ± 0.06 | 81 | 1.1 |
| B1 | B0 + P89F | na | na | na | na | |||
| B2 | B0 + A138I | na | na | na | na | |||
| B3 | B0 + L102Y | 20.9 ± 2.2 | 2.69 ± 0.11 | 129 | 20.4 ± 2.6 | 2.59 ± 0.12 | 127 | 1 |
| B4 | B0 + M85Y | 53.9 ± 5.3 | 1.73 ± 0.05 | 32 | 47.8 ± 22.2 | 1.34 ± 0.41 | 28 | 1.1 |
| B5 | B0 + V55E | 1145 ± 153 | 1.29 ± 0.12 | 1.1 | 655 ± 96 | 0.67 ± 0.04 | 1 | 1.1 |
| B6 | B0 + L191A | 48.9 ± 6.4 | 1.92 ± 0.08 | 39 | 30.3 ± 5.2 | 0.69 ± 0.04 | 23 | 1.7 |
| B7 | B0 + V130T | 36.5 ± 6 | 1.30 ± 0.08 | 36 | 55.8 ± 15.3 | 0.95 ± 0.11 | 17 | 2.1 |
| B8 | B0 + V55F | 188 ± 13 | 2.65 ± 0.06 | 14 | 391 ± 21 | 2.56 ± 0.06 | 6 | 2.3 |
| B6-II | B0 + V130T, L191A | 99.3 ± 4.9 | 6.21 ± 0.09 | 63 | 158 ± 13 | 2.82 ± 0.09 | 18 | 3.5 |
| B8-II | B0 + V55F, V130T | 91.6 ± 12.5 | 1.34 ± 0.07 | 15 | 210 ± 31 | 0.91 ± 0.07 | 4 | 3.8 |
| B6-III | B0 + V55F, V130T, L191A | 252 ± 34 | 2.40 ± 0.10 | 9.5 | 632 ± 31 | 2.10 ± 0.10 | 3 | 3.2 |
A closer review of the kinetic parameters for active variants in Library B highlights the complexity and subtlety of the enzyme’s functional changes in response to these targeted amino acid substitutions. In variants B3–B5, the functional changes balance out each other, hence conserving the parental d/l-selectivity. The mutation at position 102 in variant B3 does not affect the apparent binding affinity for d- and l-Thy yet causes roughly a 1.7-fold increase in catalytic activity for both substrates. In contrast, the substitution at position 85 in variant B4 results in approximately 2.5-fold higher KM values for the two substrates yet does not significantly affect catalytic rates. Finally, replacing Val with Glu at position 55 (B5) is overall equally detrimental to both substrates, resulting in 80-fold drops in specific activity. However, the particular functional decline for d-Thy originates from a combined 2.5-fold drop in kcat and 30-fold increase in KM, while for l-Thy it comes from a slightly (1.2-fold) lower kcat but a 60-fold increase in KM. Interestingly, substitution of a Phe in the same position (B8) is also detrimental to KM yet reverses the previously observed trend by causing a 20-fold and 10-fold drop in the apparent binding constant for d-Thy and l-Thy, respectively. Furthermore, the V55F substitution boosts catalytic activity for both substrates by 1.5-fold, creating a variant that exhibits a 2.3-fold net gain in enantioselectivity for the l-nucleoside. Similar improvements in enantioselectivity could also be detected upon amino acid changes at positions 191 (variant B6) and 130 (variant B7). Substitutions in these positions resulted in distinct but less dramatic functional changes, causing 2- to 3-fold increases in the KM values for both substrates. However, the unfavorable binding effect for the l-isomer in B6 is more than compensated for by a raise in catalytic activity for l-Thy relative to d-Thy. In variant B7, the desired l-selectivity arises from a more favorable Michaelis–Menten constant for the l-isomer compared to its d-form analogue. In summary, analysis of Library B identified three positions (residues 55, 130, and 191) that individually resulted in a 1.7- to 2.3-fold shift in enantioselectivity in favor of the unnatural nucleoside l-Thy. In comparison to the previously reported l-selective engineered human TK2,25 the kinetic parameters for our three variants (B6–B8) already match the catalytic performance for l-Thy while showing superior discrimination of d-Thy.
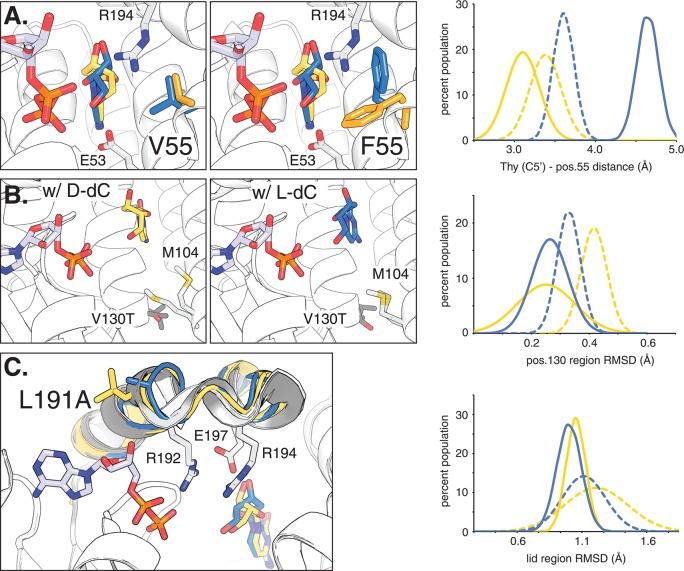
To rationalize the observed functional contributions of amino acid changes in these three locations (positions 55, 130, and 191), we performed molecular dynamics simulations to sample conformational differences in protein structure in the presence of d- or l-Thy. The V55F substitution appears to influence enantioselectivity through sterics as the model predicts the bulky Phe side chain to adopt different rotamer conformations in the presence of the two substrates (Figure 4A). Upon binding of the l-isomer to parent ssTK3 and variant B8, the F55 side chain orients itself parallel to the substrate, moving it away from the substrate by approximately 1 Å relative to V55. Although too far for direct interactions with substrate, the conformational change seems to affect the position of two neighboring catalytic residues, R194 that interacts with the triphosphate and E53 that serves as general base for nucleophilic activation of the substrate’s 5′-hydroxyl group. In contrast, the model in the presence of the d-nucleoside indicates for F88 to assume an alternate orientation, rotating the side chain by ∼120°. The extended conformation protrudes into the substrate binding pocket, causing steric clashes with bound d-Thy and interfering with substrate binding as observed in our experiments. In contrast, the substitution of V130 with the more hydrophilic Thr in variant B7 results in only minor conformational differences (Figure 4B). The analysis of the MD trajectories suggests a stabilizing effect, reflected in a smaller regional RMSD that indicates a decline in protein backbone flexibility of residues surrounding T130. In addition to these changes in dynamics, a notable structural change is observed for the adjacent side chain of M104. This position was originally mutated to enable phosphorylation of thymine nucleosides by dCK.22 Upon substitution of V130 with Thr, energy minimization favors an alternate M104 rotamer, repositioning the side chain closer to the substrate, which could in part explain the measured effects on catalysis. Finally, the mutation at position 191 in variant B6 is located in the kinase’s lid, an extended loop region that undergoes a conformational change upon substrate binding to establish multiple critical binding interactions with phosphoryl donor and acceptor involving residues R192, R194, and E197 (Figure 4C). While the location of the residue on the protein surface is unlikely to directly influence enzyme performance, MD simulations indicate a change in the conformational flexibility of the region upon introduction of the Ala substitution, eliminating side chain torsion constrains. The RMSD of the loop region drops from a broad distribution averaging 1.1–1.2 Å for ssTK3 to a more narrow, less flexible conformational state in B6. Given the increased catalytic activity of B6, these results are consistent with a more defined structural arrangement of the loop in favor of a catalytically competent conformation.
Figure 4.
Active site models based on molecular dynamics simulation of functional amino acid substitutions in dCK variants. For reference, ADP is bound in the phosphoryl donor site. The overlaid structures of d- and l-dC in the phosphoryl acceptor binding pocket are highlighted in yellow and blue, respectively. (A) Comparison of parental ssTK3 (V55) and variant B8 (F55) with their dominant side chain orientations at position 55 highlighted in yellow (for d-isomer) and blue (for l-isomer). Neighboring residues E53 and R194 are marked. The graph shows the overall distribution of preferred side chain conformations as a measure of the distance between the substrate C5′ position and nearest side chain atom for ssTK3 (dotted lines) and B8 (solid lines). (B) Effect of V130T substitution on M104 side chain formation in the presence of d-dC and l-dC, respectively. The graph shows the lower conformational flexibility of residues near T130 (variant B7) versus V130 (B0) via the region’s root square mean deviation (RMSD). (C) Impact of L191A substitution on the enzyme’s lid region (highlighted in gray) and its three catalytic residues (R192, R194, and E197). The reduced conformational flexibility of variant B6 (A191) compared to parent ssTK3 (L191) is reflected in the lower, more narrow RMSD distribution.
The promising results from our single-site mutagenesis of ssTK3 also raised the question whether combination of these individual amino acid changes would result in additive or even synergistic functional effects. We therefore built two double mutants, combining V130T with either L191A (B6-II) or V55F (B8-II), as well as the triple mutant (B6-III). The subsequent kinetic analysis indicated improvement in enantioselectivity for all three variants. While the effects in B6-II and B8-II were roughly additive at 3.5- to 3.8-fold l-selectivity, the preference for the unnatural stereoisomer in B6-III declined to ∼3.2-fold. As seen for variants B3–B8, the overall gains resulted from a combination of KM and kcat effects. In B6-II, the increase in KM values appeared to be additive, while kcat values increased quite dramatically. These findings were contrasted by the kinetic parameters for B8-II, which suggests compensatory effects in KM, averaging the double mutant’s apparent binding affinity for both substrates relative to the single-site variants. At the same time, the catalytic rates in B8-II largely reflect the lower activity seen in variants B7 and B8. For our triple mutant (B6-III), the change in KM values appeared to be mostly additive, raising the apparent binding constant for d- and l-Thy to 632 and 252 μM, respectively. In regards to kcat, the three substitutions were compensatory. Overall, B6-III showed the desired preference for the l-isomer, yet the high KM values make it an unlikely candidate for practical applications.
With B6-II and B8-II as lead candidates, we expanded our evaluation of kinetic properties to two additional substrates; l-FMAU as reference PET reporter and d-dC as the preferred substrate of our original dCK templates. The nucleoside analogue l-FMAU, while a moderately good substrate for ssTK3, has become an excellent substrate for B6-II and B8-II (Table 3). In fact, l-FMAU is preferred over d-Thy and d-dC by a significant margin based on specificity constants (kcat/KM) and apparent binding constants (KM). Compared to d-dC and d-Thy, B6-II has a 2- and 9-fold lower KM value for l-FMAU. In respect to catalytic performance, the nucleoside analogue is turned over faster than d-dC and d-Thy by 1.7- and 12-fold, respectively. Similarly, B8-II favors binding of l-FMAU by 2.5- and 4-fold relative to d-dC and d-Thy, respectively, and shows a preference in specificity constant of 2.6- and 14-fold for the nucleoside analogue over the two natural substrates. The actual kinetic parameters of the two candidates for l-FMAU are equally promising. At KM values of 18 and 50 μM and specific activities that match activity levels of native enzymes with their natural substrates, B6-II and B8-II are effective activators for the PET reporter under physiologically relevant conditions. Finally, the catalytic performance of our two leads surpasses previously reported PET reporter kinases for l-FMAU.13,25 On the basis of in vitro experiments, B6-II and B8-II outperform engineered variants of HSV-tk1 and human TK2 with respect to specific activity and selectivity.
Table 3. Kinetic Parameters for dCK Variants with Native Substrates and l-FMAU.
| substrate | B0 | B6-II | B8-II | |
|---|---|---|---|---|
| d-Thy | KM (μM) | 18.5 ± 3.1 | 158 + 13 | 210 ± 31 |
| kcat (s–1) | 1.49 ± 0.06 | 2.82 ± 0.09 | 0.91 ± 0.07 | |
| kcat/KM (s–1 M–1 × 10–3) | 81 | 18 | 4 | |
| d-dC | KM (μM) | 10.3 ± 1.9 | 33.9 ± 2.7 | 128 ± 4.7 |
| kcat (s–1) | 3.03 ± 0.2 | 4.49 ± 0.09 | 2.75 ± 0.04 | |
| kcat/KM (s–1 M–1 × 10–3) | 294 | 132 | 21 | |
| l-FMAU | KM (μM) | 18.6 ± 1.7 | 18.0 ± 1.0 | 49.9 ± 4.2 |
| kcat (s–1) | 0.46 ± 0.01 | 3.96 ± 0.04 | 2.75 ± 0.05 | |
| kcat/KM (s–1 M–1 × 10–3) | 25 | 220 | 55 | |
| l-FMAU/d-Thy (fold-change) | 0.3 | 12 | 14 | |
| l-FMAU/d-dC (fold-change) | 0.1 | 1.7 | 2.6 | |
In summary, two rounds of semi-rational protein engineering of promiscuous dCK variants have yielded two effective orthogonal kinases with potential use in bioimaging systems in combination with l-FMAU as PET reporter. While benefiting from an extensive collection of structural data for nucleoside kinases, the creation of a predictive framework for enantioselectivity in combination with small libraries to probe the model and refine its accuracy through machine-learning algorithms has enable us to identify lead enzyme variants in a very time- and cost-effective manner. Next, the application of these novel reporter kinases in vivo will need to be tested for validating the predictive power of our bench experiments. At the same time, our findings support a broader, more systematic search for additional amino acid substitutions that might further enhance the functional performance of these kinases as potential PET reporter enzymes.
Methods
Materials
Reagents and chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless indicated otherwise. Restriction enzymes were obtained from New England Biolabs (Ipswich, MA). Oligonucleotides were ordered from Integrated DNA Technologies (Coralville, IA). Pfu DNA polymerase (Stratagene, La Jolla, CA) was used for the PCR. Plasmid DNA was isolated using the QIAprep Spin Miniprep Kit, and PCR products were purified with QIAquick PCR Purification Kit (Qiagen, Valencia, CA).
Computer Models
Structural models for dCK bound to d- and l-Thy were based on PDB codes 3KFX and 3HP1, respectively.23,28 Ensembles of mutant structures were assembled in the Rosetta macromolecular modeling program, using side chain replacement and subsequent energy minimization.29 The predicted structures were used to calculate a feature set of statistically correlated (Rank Sum test) components for machine learning composed of six terms highlighted in Figure S2 (Supporting Information).36−39 Our initial efforts utilized a standard linear score function to predict mutant efficiency, while our subsequent attempts used a non-parametric score function. Several non-parametric functions were evaluated, the k-nearest neighbor algorithm showed the best agreement to known data at various stages of design using leave-one-out validation as the evaluation criteria. Implementation of machine learning to identify potential mutation was performed in MATLAB (MathWorks, Natick, MA).
MD simulations were performed to approximate binding affinities for both d- and l-nucleosides, as well as nucleoside analogues bound to selected dCK variants. Atomic models were assembled using side chain replacement and molecular superposition based on the previously mentioned crystallographic data.23 The structures were evaluated using the CHARMM22 all atom force field in reference to an implicit solvation model (SASA) using the CHARMM package.40 The structures were minimized using cycles of steepest decent and conjugant gradient, heated to 300 K, equilibrated for 200 ps, and trajectories were recording for a 1 ns simulation time.
Site Directed Mutagenesis
Mutations in dck (NCBI code: BT019942) were created by primer overlap extension. The resulting PCR products were cloned into pET-14b vector (Novagen) via NdeI and SpeI endonuclease restriction sites. Individual plasmid constructs were transformed into electrocompetent E. coli strain DH5α and grown on LB-agar in the presence of ampicillin (100 μg/mL). Correct gene constructs were confirmed by DNA sequence analysis.
Protein Expression and Purification
Individual plasmids were transformed into E. coli strain BL21(DE3)pLysS and cultured in 250 mL 2-YT media containing ampicillin (100 μg/mL) and chloramphenicol (34 μg/mL). Cell cultures were grown to an OD600 of ∼0.6 at 37 °C, followed by induction with 0.3 mM IPTG for 2 h at 30 °C. Next, cell cultures were centrifuged (4000g, 4 °C, 30 min), and pellets were resuspended in 10 mL of lysis buffer (50 mM Tris-HCl (pH 8), 300 mM NaCl, 10 mM imidazole), supplemented with 50 μL of protease inhibitor cocktail (Sigma), 5 μL of benzonase (Novagen), and 0.5 mg of lysozyme (Sigma). After incubation on an orbital shaker at 4 °C for 20 min, cells were sonicated (8 × 10 s pulses with 20 s pauses).
Cellular debris was separated via centrifugation (16,000g, 4 °C, 30 min), and the supernatant was equilibrated with 1 mL of Ni-NTA agarose resin (Qiagen) for 90 min at 4 °C. The resin was loaded on a prep-column (BioRad) and washed with 10 mL of lysis buffer, followed by 10 mL of wash buffer (50 mM Tris-HCl (pH 8), 300 mM NaCl, 50 mM imidizole). Finally, target protein was eluted with 2 mL of elution buffer (50 mM Tris-HCl (pH 8), 300 mM NaCl, 250 mM imidizole). The protein was exchanged into storage buffer (50 mM Tris-HCl (pH 8), 500 mM NaCl, 5 mM MgCl2, 2 mM DTT) and concentrated using Amicon ultracentrifugation tubes (MWCO 10 kDa, Millipore). Aliquots were flash frozen in liquid nitrogen and stored at −80 °C. Typical yields for purified protein were 10 mg/L with >95% purity based on SDS-PAGE analysis. Individual kinase variants were evaluated by thermodenaturation experiments in the CD spectrophotometer and showed no significant changes in stability compared to the parental enzymes (data not shown).
Enzyme Kinetics
The catalytic parameters of individual kinase variants were measured using a spectrophotometric coupled-enzyme assay.20 Variants were tested with two substrates; d- or l-Thy was added to reaction stock solution (50 mM Tris-HCl (pH 8), 0.1 M KCl, 5 mM MgCl2, 1 mM DTT, 1 mM ATP, 0.21 mM phosphoenolpyruvate, 0.18 mM NADH, and 2 units/mL pyruvate kinase and 2 units/mL lactate dehydrogenase). Assays were performed in triplicate at 37 °C, and the absorbance change at 340 nm was measured in the presence of 10–100 nM enzyme per reaction with 1–1000 μM substrate. Steady-state kinetic parameters were calculated using nonlinear least regression analysis to the Michaelis–Menten equation in MATLAB (MathWorks, MA).
Acknowledgments
We would like to thank the members of the Lutz lab for helpful comments and suggestions on the manuscript. Financial support in part by the National Institutes of Health [AI028731] is gratefully acknowledged.
Glossary
Abbreviations
- PET
positron emission tomography
- dCK
human 2′-deoxycytidine kinase
- HSV-tk
herpes simplex virus thymidine kinase
- TK2
human thymidine kinase 2
- l-FMAU
1-(2′-fluoro-5-methyl-β-l-arabinofuranosyl)uracil
- d-Thy
d-thymidine
- l-Thy
l-thymidine
- d-dC
d-2′-deoxycytidine
Supporting Information Available
Additional information, tables, and figures on computational design. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Brader P.; Serganova I.; Blasberg R. G. (2013) Noninvasive molecular imaging using reporter genes. J. Nucl. Med. 54, 167–172. [DOI] [PubMed] [Google Scholar]
- Serganova I.; Ponomarev V.; Blasberg R. (2007) Human reporter genes: potential use in clinical studies. Nucl. Med. Biol. 34, 791–807. [DOI] [PubMed] [Google Scholar]
- Yamamoto M.; Curiel D. T. (2010) Current issues and future directions of oncolytic adenoviruses. Mol. Ther. 18, 243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaghoubi S. S.; Campbell D. O.; Radu C. G.; Czernin J. (2012) Positron emission tomography reporter genes and reporter probes: gene and cell therapy applications. Theranostics 2, 374–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjuvajev J. G.; Stockhammer G.; Desai R.; Uehara H.; Watanabe K.; Gansbacher B.; Blasberg R. G. (1995) Imaging the expression of transfected genes in vivo. Cancer Res. 55, 6126–6132. [PubMed] [Google Scholar]
- Jacobs A.; Tjuvajev J. G.; Dubrovin M.; Akhurst T.; Balatoni J.; Beattie B.; Joshi R.; Finn R.; Larson S. M.; Herrlinger U.; Pechan P. A.; Chiocca E. A.; Breakefield X. O.; Blasberg R. G. (2001) Positron emission tomography-based imaging of transgene expression mediated by replication-conditional, oncolytic herpes simplex virus type 1 mutant vectors in vivo. Cancer Res. 61, 2983–2995. [PubMed] [Google Scholar]
- Jacobs A.; Voges J.; Reszka R.; Lercher M.; Gossmann A.; Kracht L.; Kaestle C.; Wagner R.; Wienhard K.; Heiss W. D. (2001) Positron-emission tomography of vector-mediated gene expression in gene therapy for gliomas. Lancet 358, 727–729. [DOI] [PubMed] [Google Scholar]
- Dempsey M. F.; Wyper D.; Owens J.; Pimlott S.; Papanastassiou V.; Patterson J.; Hadley D. M.; Nicol A.; Rampling R.; Brown S. M. (2006) Assessment of 123I-FIAU imaging of herpes simplex viral gene expression in the treatment of glioma. Nucl. Med. Commun. 27, 611–617. [DOI] [PubMed] [Google Scholar]
- Penuelas I.; Mazzolini G.; Boan J. F.; Sangro B.; Marti-Climent J.; Ruiz M.; Ruiz J.; Satyamurthy N.; Qian C.; Barrio J. R.; Phelps M. E.; Richter J. A.; Gambhir S. S.; Prieto J. (2005) Positron emission tomography imaging of adenoviral-mediated transgene expression in liver cancer patients. Gastroenterology 128, 1787–1795. [DOI] [PubMed] [Google Scholar]
- Yaghoubi S. S.; Jensen M. C.; Satyamurthy N.; Budhiraja S.; Paik D.; Czernin J.; Gambhir S. S. (2009) Noninvasive detection of therapeutic cytolytic T cells with 18F-FHBG PET in a patient with glioma. Oncology 6, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton K. N.; Stricker H.; Brown S. L.; Elshaikh M.; Aref I.; Lu M.; Pegg J.; Zhang Y.; Karvelis K. C.; Siddiqui F.; Kim J. H.; Freytag S. O.; Movsas B. (2008) Phase I study of noninvasive imaging of adenovirus-mediated gene expression in the human prostate. Mol. Ther. 16, 1761–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken M. N.; Gschweng E. H.; Nair-Gill E.; McLaughlin J.; Cooper A. R.; Riedinger M.; Cheng D.; Nosala C.; Kohn D. B.; Witte O. N. (2013) Long-term in vivo monitoring of mouse and human hematopoietic stem cell engraftment with a human positron emission tomography reporter gene. Proc. Nat. Acad. Sci. U.S.A. 110, 1857–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambhir S. S.; Bauer E.; Black M. E.; Liang Q.; Kokoris M. S.; Barrio J. R.; Iyer M.; Namavari M.; Phelps M. E.; Herschman H. R. (2000) A mutant herpes simplex virus type 1 thymidine kinase reporter gene shows improved sensitivity for imaging reporter gene expression with positron emission tomography. Proc. Nat. Acad. Sci. U.S.A. 97, 2785–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Likar Y.; Dobrenkov K.; Olszewska M.; Shenker L.; Cai S.; Hricak H.; Ponomarev V. (2009) PET imaging of HSV1-tk mutants with acquired specificity toward pyrimidine- and acycloguanosine-based radiotracers. Eur. J. Nucl. Med. Mol. Imaging 36, 1273–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.; Pursell Z. F.; Copeland W. C.; Longley M. J.; Kunkel T. A.; Mathews C. K. (2005) DNA precursor asymmetries in mammalian tissue mitochondria and possible contribution to mutagenesis through reduced replication fidelity, P. Proc. Nat. Acad. Sci. U.S.A. 102, 4990–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gon S.; Napolitano R.; Rocha W.; Coulon S.; Fuchs R. P. (2011) Increase in dNTP pool size during the DNA damage response plays a key role in spontaneous and induced-mutagenesis in Escherichia coli. Proc. Nat. Acad. Sci. U.S.A. 108, 19311–19316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu C.; Bao H.; Tolstykh T.; Micolochick Steuer H. M.; Murakami E.; Korba B.; Furman P. A. (2010) Evaluation of the in vitro anti-HBV activity of clevudine in combination with other nucleoside/nucleotide inhibitors. Antiviral Ther. 15, 401–412. [DOI] [PubMed] [Google Scholar]
- Riddell S. R.; Elliott M.; Lewinsohn D. A.; Gilbert M. J.; Wilson L.; Manley S. A.; Lupton S. D.; Overell R. W.; Reynolds T. C.; Corey L.; Greenberg P. D. (1996) T-cell mediated rejection of gene-modified HIV-specific cytotoxic T lymphocytes in HIV-infected patients. Nat. Med. 2, 216–223. [DOI] [PubMed] [Google Scholar]
- Sabini E.; Ort S.; Monnerjahn C.; Konrad M.; Lavie A. (2003) Structure of human dCK suggests strategies to improve anticancer and antiviral therapy. Nat. Struct. Biol. 10, 513–519. [DOI] [PubMed] [Google Scholar]
- Gerth M. L.; Lutz S. (2007) Mutagenesis of non-conserved active site residues improves the activity and narrows the specificity of human thymidine kinase 2. Biochem. Biophys. Res. Commun. 354, 802–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev V.; Doubrovin M.; Shavrin A.; Serganova I.; Beresten T.; Ageyeva L.; Cai C.; Balatoni J.; Alauddin M.; Gelovani J. (2007) A human-derived reporter gene for noninvasive imaging in humans: mitochondrial thymidine kinase type 2. J. Nucl. Med. 48, 819–826. [DOI] [PubMed] [Google Scholar]
- Iyidogan P.; Lutz S. (2008) Systematic exploration of active site mutations on human deoxycytidine kinase substrate specificity. Biochemistry 47, 4711–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra S.; Sabini E.; Ort S.; Konrad M.; Lavie A. (2009) Extending thymidine kinase activity to the catalytic repertoire of human deoxycytidine kinase. Biochemistry 48, 1256–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.; Murphy P.; Baker D.; Lutz S. (2010) Computational design of orthogonal nucleoside kinases. Chem. Commun. 46, 8803–8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D. O.; Yaghoubi S. S.; Su Y.; Lee J. T.; Auerbach M. S.; Herschman H.; Satyamurthy N.; Czernin J.; Lavie A.; Radu C. G. (2012) Structure-guided engineering of human thymidine kinase 2 as a positron emission tomography reporter gene for enhanced phosphorylation of non-natural thymidine analog reporter probe. J. Biol. Chem. 287, 446–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J. S.; Machado H. B.; Campbell D. O.; McCracken M.; Radu C.; Witte O. N.; Herschman H. R. (2013) Application of a rapid, simple, and accurate adenovirus-based method to compare PET reporter gene/PET reporter probe systems. Mol. Imaging Biol. 15, 273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner E. S.; Eriksson S. (1995) Mammalian deoxyribonucleoside kinases. Pharmacol. Ther. 67, 155–186. [DOI] [PubMed] [Google Scholar]
- Hazra S.; Ort S.; Konrad M.; Lavie A. (2010) Structural and kinetic characterization of human deoxycytidine kinase variants able to phosphorylate 5-substituted deoxycytidine and thymidine analogues. Biochemistry 49, 6784–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A.; Tyka M.; Lewis S. M.; Lange O. F.; Thompson J.; Jacak R.; Kaufman K.; Renfrew P. D.; Smith C. A.; Sheffler W.; Davis I. W.; Cooper S.; Treuille A.; Mandell D. J.; Richter F.; Ban Y. E.; Fleishman S. J.; Corn J. E.; Kim D. E.; Lyskov S.; Berrondo M.; Mentzer S.; Popovic Z.; Havranek J. J.; Karanicolas J.; Das R.; Meiler J.; Kortemme T.; Gray J. J.; Kuhlman B.; Baker D.; Bradley P. (2011) ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 487, 545–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabini E.; Hazra S.; Konrad M.; Lavie A. (2007) Nonenantioselectivity property of human deoxycytidine kinase explained by structures of the enzyme in complex with l- and d-nucleosides. J. Med. Chem. 50, 3004–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabini E.; Hazra S.; Konrad M.; Burley S. K.; Lavie A. (2007) Structural basis for activation of the therapeutic l-nucleoside analogs 3TC and troxacitabine by human deoxycytidine kinase. Nucleic Acids Res. 35, 186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J. H.; Bentley J. L.; Finkel R. A. (1977) An algorithm for finding best matches in logarithmic expected time. ACM Trans. Math. Software 3, 209–226. [Google Scholar]
- Breiman L., Friedman J. H., Stone C. J., and Olshen R. A. (1984) Classification and Regression trees, CRC Press, New York. [Google Scholar]
- Amit Y.; Geman D. (1997) Shape quantization and recognition with randomized trees. Neural Comput. 9, 1545–1588. [Google Scholar]
- Scholkopf B.; Platt J. C.; Shawe-Taylor J.; Smola A. J.; Williamson R. C. (2001) Estimating the support of a high-dimensional distribution. Neural Comput. 13, 1443–1471. [DOI] [PubMed] [Google Scholar]
- Kortemme T.; Morozov A. V.; Baker D. (2003) An orientation-dependent hydrogen bonding potential improves prediction of specificity and structure for proteins and protein-protein complexes. J. Mol. Biol. 326, 1239–1259. [DOI] [PubMed] [Google Scholar]
- Dunbrack R. L. Jr.; Cohen F. E. (1997) Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci. 6, 1661–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis T.; Karplus M. (1999) Effective energy function for proteins in solution. Proteins 35, 133–152. [DOI] [PubMed] [Google Scholar]
- Kuhlman B.; Baker D. (2000) Native protein sequences are close to optimal for their structures. Proc. Natl. Acad. Sci. U.S.A. 97, 10383–10388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B. R.; Brooks C. L. 3rd; Mackerell A. D. Jr.; Nilsson L.; Petrella R. J.; Roux B.; Won Y.; Archontis G.; Bartels C.; Boresch S.; Caflisch A.; Caves L.; Cui Q.; Dinner A. R.; Feig M.; Fischer S.; Gao J.; Hodoscek M.; Im W.; Kuczera K.; Lazaridis T.; Ma J.; Ovchinnikov V.; Paci E.; Pastor R. W.; Post C. B.; Pu J. Z.; Schaefer M.; Tidor B.; Venable R. M.; Woodcock H. L.; Wu X.; Yang W.; York D. M.; Karplus M. (2009) CHARMM: the biomolecular simulation program. J. Comput. Chem. 30, 1545–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.