Abstract
To define the role that RasC plays in motility and chemotaxis, the behavior of a rasC null mutant, rasC−, in buffer and in response to the individual spatial, temporal, and concentration components of a natural cyclic AMP (cAMP) wave was analyzed by using computer-assisted two-dimensional and three-dimensional motion analysis systems. These quantitative studies revealed that rasC− cells translocate at the same velocity and exhibit chemotaxis up spatial gradients of cAMP with the same efficiency as control cells. However, rasC− cells exhibit defects in maintaining anterior-posterior polarity along the substratum and a single anterior pseudopod when translocating in buffer in the absence of an attractant. rasC− cells also exhibit defects in their responses to both the increasing and decreasing temporal gradients of cAMP in the front and the back of a wave. These defects result in the inability of rasC− cells to exhibit chemotaxis in a natural wave of cAMP. The inability to respond normally to temporal gradients of cAMP results in defects in the organization of the cytoskeleton, most notably in the failure of both F actin and myosin II to exit the cortex in response to the decreasing temporal gradient of cAMP in the back of the wave. While the behavioral defect in the front of the wave is similar to that of the myoA−/myoF− myosin I double mutant, the behavioral and cytoskeletal defects in the back of the wave are similar to those of the S13A myosin II regulatory light-chain phosphorylation mutant. Expression array data support the premise that the behavioral defects exhibited by the rasC− mutant are the immediate result of the absence of RasC function.
The Ras GTPases function as molecular switches in the regulation of a variety of responses to extracellular signals (3, 21, 35, 36). Dictyostelium discoideum, like higher eukaryotes, contains a number of Ras GTPases (6, 21, 22). Because of its attributes, Dictyostelium provides a unique experimental system for exploring the roles played by the Ras GTPases in cell motility and chemotaxis (6, 21, 49). First, because it is haploid, null mutations are readily generated and rescued (16, 18). Second, because the behavior of Dictyostelium amoebae in buffer and in response to the temporal, spatial, and concentration components of the natural chemotactic wave has been characterized in detail by computer-assisted methods (33, 34), a unique contextual framework exists for identifying specific behavioral defects manifested in mutants (29) and for deducing from them the specific role played by mutated genes in motility and/or chemotaxis (4, 8, 10, 42, 45, 50, 51).
In a previous study, it was demonstrated that rasC− cells could not progress through the early stages of development or form aggregates unless they were pulsed with the chemoattractant cyclic AMP (cAMP), indicating that RasC was necessary for signaling (20). rasC− cells artificially pulsed with cAMP were then capable of forming aggregates on filter pads. When mixed with a majority of normal cells, they could also enter aggregates and exhibit chemotaxis towards the source of a spatial gradient of cAMP emitted by a micropipette (20). Although these results suggested that exogenously added cAMP rendered rasC− cells aggregation competent and that RasC was not required for chemotaxis in spatial gradients of the attractant, experiments were not performed to test whether RasC played a direct role in the basic motile behavior of a cell in the absence of a chemotactic signal or in the complex cellular responses to the temporal and concentration components of the natural cAMP wave (29, 46).
Employing a set of experimental protocols developed to analyze the basic motile behavior of amoebae in the absence of attractant and their capacity to respond to the individual spatial, temporal, and concentration components of the chemotactic wave (29), we demonstrate that rasC− cells can efficiently exhibit chemotaxis up a spatial gradient of cAMP, the mechanism regulating orientation at the onset of the front of a natural wave. However, rasC− cells exhibit a number of discrete behavioral defects, including fragmentation and z-axis extension of the anterior pseudopod in buffer, absence of a velocity surge in response to the increasing temporal gradient of cAMP in the front of a wave, and abnormal polarization in response to the decreasing temporal gradient of cAMP in the back of a wave. The behavioral defects in the front and the back of the wave are accompanied by abnormalities in the localization of F actin and myosin in the cortex. Because our experimental protocols allowed us to correlate regulatory pathways with specific phases of the natural wave and associated behavioral responses, we formulated a working model in which RasC plays roles both in the response of cells to the increasing temporal gradient of cAMP in the front of the wave, upstream of MyoA/MyoF, and in the response of cells to the decreasing temporal gradient of cAMP in the back of the wave, upstream of the phosphorylation of the myosin II regulatory light chain.
MATERIALS AND METHODS
Origin and maintenance of control, rasC−, and rescued strains.
The parental Ax2 strain (control), the rasC− null strain, and the rasC− rescued strain, referred to here as rasC−/rasC+, have been described previously in detail (20). Briefly, the rasC− mutant was generated by targeted gene replacement via homologous recombination with a rasC− construct that was disrupted by a blasticidin S resistance-selectable marker. An ectopically encoded RasC expression vector was constructed by directionally ligating the rasC cDNA to the rasC promoter that was cloned by PCR from Ax2 genomic DNA. Transformation of this vector into the rasC− mutant resulted in the rasC−/rasC+ strain. RasC expression in the rescued strain was verified by Western blotting against a RasC-specific rabbit polyclonal antibody. rasC− and rasC−/rasC+ cells were grown in HL-5 medium supplemented with 10 μg of blasticidin S/ml and 10 μg of G418/ml, respectively. Cells were grown on plates since attachment promoted a higher proportion of mononuclear cells. For experimental purposes, cells were harvested from plates at the confluent-monolayer stage.
Obtaining behaviorially equivalent cells.
Cells were harvested from growth cultures, washed with buffered salts solution (BSS; 20 mM KCl, 2.5 mM MgCl2, 20 mM KH2PO4 [pH 6.4]), and distributed on filter pads saturated with BSS at a cell density of 5 × 106 per cm2 (30). Cells were washed from the filter pads at the ripple stage, which represents the onset of aggregation (30) and the time at which cells achieve maximum velocity (40). To obtain rasC− cells that had achieved maximum velocity during development equivalent to that of control cells, a number of protocols were tested and the following one was selected (see Results). rasC− cells were harvested from growth cultures, washed with BSS, resuspended in BSS at a cell density of 5 × 106 per ml and maintained in suspension on a rotary water bath shaker at 22°C. After 1 h, cells were pulsed every 6 min with 50 nM cAMP for 6 h. Cells were then washed in BSS and dispersed on a filter pad as described for control cells. rasC− cells were then harvested from filter pads after 2 h of incubation, at which time cells had achieved maximum velocity (see Results).
Microarray analysis.
The methods used have been described in detail elsewhere (14, 15, 24). At various time points, cells were collected from cAMP-pulsed suspension cultures, pelleted, and dissolved in Trizol reagent (Gibco-BRL, Gaithersburg, Md.) for preparation of RNA. For analysis, Corning slides were microarrayed with 6,345 cDNA and genomic targets by using a Molecular Dynamics GenIII robot in the BioGEM facility, University of California, San Diego, as previously described. Inserts from 5,655 cDNAs were generously provided by the Japanese EST Project. A list of genes is available at http://www.biology.ucsd.edu/loomis-cgi/microarray/rasC-array.html. All genes in this study were sequence verified. Probes were prepared from total RNA collected at 2-h intervals during wild-type and mutant development as well as from time-averaged reference RNA (15). Superscript II DNA polymerase (Invitrogen, Carlsbad, Calif.) was used to incorporate either Cy-5- or Cy-3-conjugated dCTP (Amersham, St. Louis, Mo.) into DNA. Following incubation at 42°C for 3 h, unincorporated dyes were removed using Microcon-30 columns (Millipore, Burlington, Mass.) and three washes with 450 μl of Tris-EDTA buffer, before drying and resuspension in a solution containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.3% sodium dodecyl sulfate, and 25% formamide. Labeled probes from the time points and the time-averaged RNA were mixed and hybridized at 42°C to microarrays for 6 to 12 h. Probed microarrays were analyzed in an Axon Genepix 4000B scanner (Axon Instruments, Foster City, Calif.), and the measurements were processed with the associated software. Total Cy3 signal was normalized to total Cy5 signal after background subtraction to allow independent slides to be compared. The Cy3/Cy5 ratios for individual genes were then calculated at each time point for subsequent analyses. The expression patterns of rasC− cells were compared to those of strain Ax4, which are indistinguishable from those of strain Ax2 (37). The expression of each gene was based on measurements taken at 13 independent time points during the developmental time course. Each analysis was repeated at least twice. The selected cutoff at a threefold increase was based on the reproducibility of the data from 10 independent time course experiments.
Basic motile behavior.
Cells were washed from filter pads and distributed on the glass wall of a Sykes-Moore perfusion chamber (Bellco Glass, Vineland, N.J.) positioned on the stage of an upright microscope with a long-distance working condenser and a ×25 bright-field objective as previously described (8, 48, 50, 51). After 5 min of incubation, the chamber was perfused with BSS at a rate that turned over 1 chamber volume equivalent every 15 s.
Spatial gradient of cAMP.
Cells were dispersed either on the bridge of a Plexiglas chemotaxis chamber (38) modeled after that of Zigmond (52) for bright-field analysis or on the bridge of a quartz chamber developed for differential interference contrast (DIC) microscopy (27, 28). BSS alone was added to one of the two wells bordering the bridge, and BSS containing 10−6 M cAMP was added to the other. Cells were incubated for 5 min to facilitate cell adhesion and allow for the genesis of a steep gradient of chemoattractant across the bridge (27, 28).
Simulated temporal waves of cAMP.
Cell behavior was analyzed in a series of four temporal waves of cAMP generated in the absence of established spatial gradients in a Sykes-Moore perfusion chamber as previously described (8, 32, 33, 39, 41, 50, 51) by using NE-1000 Multi-Phaser programmable syringe pumps (New Era Pump Systems, Wantagh, N.Y.) (11). The shapes of the cAMP waves within the chamber were verified using xylene cyanol FF dye (Molecular Probes, Eugene, Oreg.) (11).
Rapid addition of 10−6 M cAMP.
Cells were distributed on the wall of a Sykes-Moore chamber and perfused with BSS for 10 min as described above. The perfusion solution was then switched to BSS containing 10−6 M cAMP (40). Previous analyses with fluorescent dye demonstrated that the concentration of cAMP increases from 0 to 10−6 M in approximately 12 s (40).
Natural cAMP waves.
To test the ability of rasC− cells to respond to natural waves of cAMP generated by wild-type cells, mixing experiments were performed according to previously described methods (50, 51) with modifications. Growth-phase rasC− cells were incubated overnight in HL-5 medium containing 5 × 10−5 M DiI (Molecular Probes). Labeled cells were then washed in BSS, resuspended to a final concentration of 5 × 106 cells per ml, maintained in suspension for 1 h, and then pulsed every 6 min for 5 h with 50 nM cAMP. rasC− cells were then mixed with unlabeled control cells at a ratio of 1:9. Control cells were prestarved for 3 h before mixing so that the developmental timings of the two strains were comparable (see Results). The mixed population was plated on a 35-mm-diameter petri dish at a density of 2.4 × 106 cells per ml in 2 ml, the dish was placed on the stage of an Axiovert 100STV Zeiss microscope, and the sample was examined with a NORAN laser scanning confocal microscope (LSCM). Transmitted and fluorescent images were simultaneously collected every 20 s, averaged using Intervision software, and saved on a hard drive in Silicon Graphics format (SGI Inc., Mountain View, Calif.). These movies were then converted to Quick Time format, and the motion of labeled rasC− and unlabeled neighboring control cells was analyzed.
2D computer-assisted analysis.
2D-DIAS software was used for computer-assisted two-dimensional (2D) analyses (32, 33). Video images of all preparations were digitized onto the hard drive of a Macintosh computer at a rate of 15 frames per min (4-s intervals) with a frame-grabber board (Data Translation Inc., Marlboro, Mass.). Cell perimeters from the digitized movies were automatically outlined using the grayscale threshold algorithm of 2D-DIAS. Outlines were converted to beta-spline replacement images. Motility parameters were computed from the centroid positions and dynamic morphology parameters were computed from contours of the replacement images according to formulas discussed elsewhere in detail (32, 33). To compute the instantaneous velocity of a cell in frame n, a line was drawn from the centroid in frame n − 1 to the centroid in frame n + 1 and the length of that line was divided by twice the time interval between frames. Directional change was computed as the direction in the interval (n −1, n) minus the direction in the interval (n, n + 1). Directional change values greater than 180° were subtracted from 360°, resulting in a positive value between 0° and 180°. Maximum length was defined as the longest chord between any two points along the cell perimeter. Maximum width was defined as the longest chord perpendicular to the maximum length chord. Roundness was calculated as (4π × area/perimeter squared) × 100. Convexity was defined as the absolute value of the sum of positive turn angles, in degrees, of line segments connecting the vertices of a cell's shape. The chemotactic index (CI) was calculated as the net distance traveled directly towards the source of chemoattractant in a spatial gradient chamber divided by the total distance traveled in that time period. The percent positive chemotaxis was the proportion of cells exhibiting a positive CI in a spatial gradient of cAMP over the period of analysis. Difference images were generated by superimposing the perimeter outline of the cell in frame n onto the perimeter outline in frame n − 1. Expansion zones, color coded green, were demarcated as regions of the cell in frame n not overlapping the outlined cell in frame n − 1. Contraction zones, color coded red, were demarcated as regions of the cell in frame n − 1 not overlapping the outlined cell in frame n.
3D computer-assisted analysis.
The 3D-DIAS software program was used for computer-assisted three-dimensional (3D) reconstruction and motion analysis (13, 32, 33, 34, 43, 47). For 3D reconstructions in buffer and in temporal waves of cAMP, 350 μl of a dilute suspension of cells was inoculated into a Dvorak-Stotler chamber (Lucas-Highland, Inc., Chantilly, Va.). The chamber was placed on the stage of a Zeiss ICM 405 inverted microscope equipped with DIC optics and a Planapo ×63 oil immersion objective. The chamber was either perfused with BSS or treated with four temporal waves of cAMP as described above. For 3D reconstructions in spatial gradients of cAMP, cells were dispersed on the bridge of a quartz chemotaxis chamber (28). To obtain optical sections of a cell, the coarse focus knob of the microscope was connected to a stepper-motor programmed for 60 optical sections in 2 s in the z axis at 0.3-μm increments. This entire process was repeated at 5-s intervals. Images were recorded through an Optronics cooled charge-coupled device camera directly into a Macintosh iMAC computer at 30 frames per s by using iMovie software. After the movie was compressed into the DIAS format, 3D-DIAS software in the newly developed JAVA-based DIAS 4.0 platform (E. Voss and D. R. Soll, unpublished data) automatically outlined the perimeter of the in-focus portion of the image in each optical section by using a pixel complexity algorithm (33). The interface between the particulate cytoplasm of the main cell body and the nonparticulate cytoplasm of the pseudopodial extensions was readily identified in DIC images of optical sections. The distal nonparticulate zones of pseudopodial regions were manually outlined in the in-focus portions of each optical section to generate a faceted 3D reconstruction of pseudopods, which were color coded red and inserted into the faceted cell image.
The construction of a 3D faceted image of the cell surface has been described elsewhere in detail (13, 33, 43, 47). For computation of 3D parameters, the 3D position of the centroid of the cell was computed by averaging the x, y, and z coordinates of all points interior to the 3D faceted cell image. Volume and height were computed as the mathematical volume of the faceted cell and as the maximum height of the faceted image in the z axis, respectively. The overhang parameter represented the percent cell area in the x and y axes not in contact with the substratum.
F-actin and myosin II staining.
Cells were fixed with 4% paraformaldehyde in 10 mM 2(N-morpholino)ethanesulfonic acid (MES; Sigma, St. Louis, Mo.) buffer (pH 6.1) containing 138 mM KCl, 3 mM MgCl2, and 2 mM EGTA. To localize F actin, cells were stained with Oregon green-phalloidin (Molecular Probes, Inc.) according to methods previously described (50, 51). Prior to immunostaining for myosin II, antigen retrieval was performed by processing cells in target retrieval solution (DAKO Corp., Carpinteria, Calif.) in a steamer for 20 min at 90°C according to the manufacturer's recommendations. The solution and coverslips were removed from heat and allowed to cool to room temperature prior to being rinsed in Tris-HCl-buffered saline (50 mM Tris-HCl-150 mM NaCl [pH 7.4]). Nonspecific binding was blocked with 10% normal goat serum (Sigma) in Dulbecco's phosphate-buffered saline (PBS; pH 7.1; Gibco, Grand Island, N.Y.). To localize myosin II, cells were incubated with rabbit anti-myosin II antibody (1/1,000), a generous gift from Arturo De Lozanne (University of Texas, Austin), in PBS for 45 min at 37°C. Following extensive rinsing with PBS, cells were stained with fluorescein isothiocyanate-labeled goat anti-rabbit antibody (1/200; Molecular Probes, Inc.) for 30 min at room temperature. Coverslips containing the F-actin- or myosin II-stained preparations were rinsed and mounted using Gelvatol supplemented with 0.6% N-propylgallate (Sigma). Cells were then scanned with a Bio-Rad Radiance 2100MP LSCM at 0.2-μm increments in the z axis, and the z series was projected as a single image (projection image). To obtain a ratio of the intensity of either F-actin or myosin II staining in the cortex to that in the cytoplasm, a stained Ax2 cell was scanned 1 μm above the substratum and the image was optimized. The optimized LSCM parameters for this reference cell (laser power, iris aperture, gain, and scan rate) were then used to obtain scans 1 μm above the substratum for subsequent control and rasC− cells. Line profiles of intensity through the cortex and cytoplasm were obtained just posterior to the cell nucleus by using Bio-Rad Laser Sharp 2000 5.0 software. Digital images of xy planes, line profiles, and projected z series image stacks were processed using Adobe Photoshop software.
RESULTS
Developmental regulation of motility.
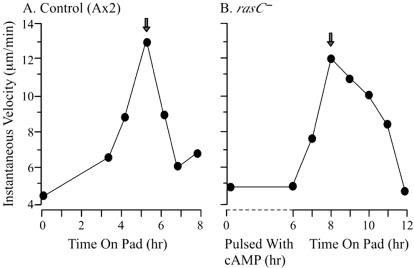
The increase in cell motility associated with the acquisition of chemotactic responsiveness to cAMP and the onset of aggregation (40) provides a developmental landmark for comparison of strains (8, 31, 40, 50, 51). Because rasC− cells fail to aggregate in homogeneous monolayers due to their inability to relay a cAMP signal (20), it was first necessary to identify conditions that produced rasC− cells that were comparable to aggregation-competent control (Ax2) cells. We therefore analyzed motility under a variety of conditions, including starvation on filter pads, starvation in suspension with or without cAMP pulses, and starvation in suspension with cAMP pulses, followed by plating on pads. In the case of the parental control strain (Ax2), single cells achieved maximum velocity between 5 and 6 h of starvation on development pads (Fig. 1A), coincident with the ripple stage, which represents the onset of aggregation (30). Similar results were obtained with the rasC−/rasC+ strain (data not shown). In the case of the rasC− strain, however, cells attained similar peak velocity when they were pulsed with 50 nM cAMP at 6-min intervals during the final 5 h of a 6-h period in shaking cultures and then starved for 2 subsequent h on a filter pad (Fig. 1B).
FIG. 1.
Developmental regulation of motility in rasC− cells. The developmental regulation of cell motility was assessed by plotting single-cell velocity as a function of development time. (A) For control (Ax2) cells, the peak velocity (arrow) occurred 5 to 6 h after cells washed free of growth medium were plated on a development pad saturated with BSS. (B) For rasC− cells, cells washed free of growth medium were suspended in buffer and pulsed with cAMP every 6 min between 1 and 6 h of incubation. Cells were then plated on a development pad saturated with BSS. The peak velocity (arrow) occurred 2 h after rasC− cells were plated on the development pad. The mean instantaneous velocity at each time point was computed from the average instantaneous velocities of 20 amoebae, each analyzed for 10 min.
Developmental regulation of gene expression.
To be sure that the defects in motility and chemotaxis identified in rasC− cells were due directly to the lack of RasC function in regulatory pathways, Lim et al. (20) previously demonstrated by Northern analysis that cAMP-pulsed rasC− cells expressed the developmentally regulated cAMP receptor CAR1 and the coupled heterotrimeric G protein Gα2. To further explore gene expression, microarray analyses were performed with rasC− cells by using slides that carried 6,345 cDNA and genomic targets and the results were compared with those for Ax4 cells developed in the same manner (14, 15, 24). It has previously been demonstrated that Ax2 (the control for rasC−) and Ax4 cells exhibit similar changes in gene expression during development (37). Only mRNAs whose expression increased threefold or more in cAMP-pulsed control cells were compared with those in cAMP-pulsed rasC− cells in order to set a stringent condition for identifying genes that may not be expressed in the latter.
All of the preaggregation genes, including those for CAR1, Gα2, the secreted cAMP-phosphodiesterase and its inhibitor, the adenylyl cyclase ACA, and the internal phosphodiesterase RegA, began accumulating immediately upon induction of development and reached peak levels at the same time in rasC− and Ax4 cells (data available at http://www.biology.ucsd/loomis-cgi/microarray/rasC-array.html). Likewise, mRNAs for the cell-cell adhesion proteins gp80 (CsaA) and gp150 (LagC) accumulated to peak values within 12 h of development in mutant and control cells. All of the postaggregative mRNAs that accumulated at least threefold in wild type, including those encoding the spore coat proteins, accumulated at least threefold in rasC− cells. While genes that did not give clear signals on the microarrays could have been affected in rasC− cells, the transcriptional profiles that characterize developmental stages through aggregation were not significantly perturbed by the loss of RasC as long as the cells were pulsed with cAMP (see http://www.biology.ucsd/loomis-cgi/microarray/rasC-array.html).
Basic motile behavior is defective in rasC− cells.
Cells distributed at low density on the glass wall of a chamber were perfused continuously with BSS to ensure that they did not condition their soluble microenvironment. The mean instantaneous velocity of rasC− cells was statistically indistinguishable from that of control Ax2 and rasC−/rasC+ cells (Tables 1 and 2). However, a histogram of the instantaneous velocities of individual cells suggested a broader distribution for rasC− cells than for control cells at both low and high velocities (data not shown). The broadened range of instantaneous velocities of rasC− cells was accompanied by other behavioral abnormalities. First, perimeter tracks of rasC− cells possessed more side projections, which upon further analysis proved to be due to instability at the anterior ends of cells (data not shown), suggesting either a more dynamic anterior pseudopod or less secure attachment of the anterior end to the substratum. Second, the directional change parameter of rasC− cells was significantly higher than that of control or rasC−/rasC+ cells (Tables 1 and 2), again suggesting less persistent translocation. Third, both the mean roundness and mean convexity parameters of rasC− cells were significantly higher than those of control cells (Table 1). Together, these results indicated abnormalities in rasC− behavior in the absence of attractant.
TABLE 1.
Quantitative 2D-DIAS analysis of the basic motile behavior of rasC− cells in buffer reveals subtle defectsa
| Cells | Mean ± SD
|
|||||
|---|---|---|---|---|---|---|
| Instantaneous velocity (μm/min) | Directional change (°/4 s) | Maximum length (μm) | Maximum width (μm) | Roundness (%) | Convexity (°) | |
| Ax2 (control) | 13.5 ± 4.6 | 28.6 ± 11.0 | 20.0 ± 2.4 | 8.1 ± 1.4 | 42.1 ± 7.3 | 647 ± 71 |
| rasC− | 12.3 ± 7.9 | 35.6 ± 10.4 | 19.8 ± 4.3 | 8.2 ± 1.2 | 55.8 ± 12.8 | 799 ± 52 |
| rasC−/rasC+ | 11.7 ± 3.3 | 27.7 ± 9.9 | 19.8 ± 4.1 | 7.7 ± 1.6 | 47.6 ± 7.8 | 606 ± 69 |
A total of 50 cells of each strain were examined. P values were computed by the Student t test; a P value of >0.05 was considered nonsignificant. P values for all comparisons of Ax2 with rasC− cells and of Ax2 with rasC−/rasC+ cells were nonsignificant, except for P values for comparisons of Ax2 with rasC− cells for directional change (P = 5 × 10−3), roundness (P < 10−5), and convexity (P < 10−5).
TABLE 2.
Quantitative 3D-DIAS analysis of the basic motile behavior of rasC− cells in buffer reveals subtle defectsa
| Cells | Mean ± SD
|
|||||
|---|---|---|---|---|---|---|
| Instantaneous velocity (μm/min) | Directional change (°/4 s) | Vol (μm3) | Surface area (μm2) | Ht (μm) | Overhang (%) | |
| Ax2 (control) | 19.1 ± 3.9 | 30.5 ± 2.6 | 678 ± 164 | 721 ± 133 | 9.3 ± 1.7 | 18.3 ± 4.3 |
| rasC− | 19.8 ± 8.1 | 37.2 ± 5.7 | 732 ± 304 | 808 ± 226 | 12.7 ± 2.2 | 40.9 ± 12.0 |
A total of 10 cells of each strain were examined. P values were computed by the Student t test; a P value of >0.05 was considered nonsignificant. P values for all comparisons of Ax2 with rasC− cells were nonsignificant, except for P values for comparisons of Ax2 with rasC− cells for height (P = 0.020) and overhang (P = 0.004).
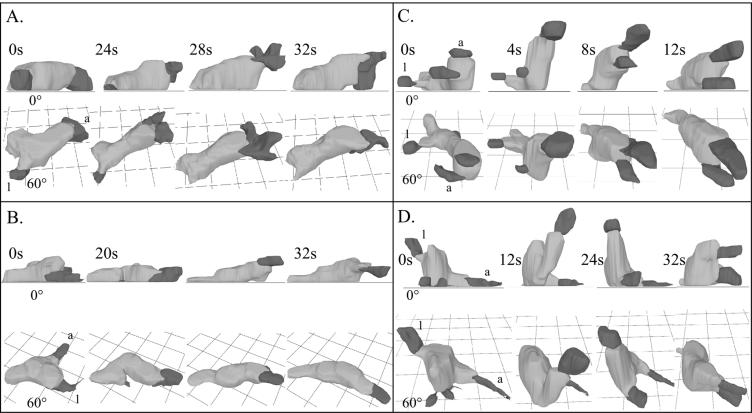
To investigate further the aberrant behavior in buffer, control and rasC− cells were reconstructed in three dimensions at 4-s intervals using 3D-DIAS software (13, 33, 47). The two representative control cells shown in Fig. 2A and B were elongate and extended a single dominant anterior pseudopod (labeled “a”) along the substratum. In both cases, lateral pseudopods (labeled “l”) that had formed prior to reconstruction were retracted back into the cell bodies. Intermittently, the dominant anterior pseudopod lifted off the substratum and then returned to the substratum during the period of analysis (Fig. 2A and B). Eight additional control and 10 rasC−/rasC+ cells reconstructed in three dimensions behaved in a similar fashion (data not shown).
FIG. 2.
3D-DIAS reconstruction of two representative control (Ax2) cells (A and B) and two representative rasC− cells (C and D) reveal rasC− defects in both z-axis extension and the number of anterior pseudopods. Cell bodies are color coded light gray and pseudopods are color coded dark gray. Each cell is viewed at 0° and 60° angles. a, anterior pseudopod; l, lateral pseudopod. Time is presented in seconds.
In contrast to those of control or rasC−/rasC+ cells, the anterior ends of the two representative rasC− cells shown in Fig. 2C and D frequently extended off the substratum in the z axis. When extending their anterior ends in the z axis, rasC− cells remained attached to the substratum at their posterior ends (Fig. 2C and D). The posterior end, or uropod, was identified in those cells by assessing the direction of translocation from earlier frames. Frequently, either the anterior pseudopod of rasC− cells bifurcated or multiple anterior pseudopods extended from the general anterior half of the cell (Fig. 2C and D). In some cases, it was difficult to keep track of the dominant anterior pseudopod, as is evident during z-axis extension of the cell in Fig. 2D. Eight additional rasC− cells reconstructed in three dimensions exhibited similar behavior (data not shown). Qualitative 3D analyses of vegetative rasC− cells migrating on a glass surface revealed defects in the anterior end, including multiple anterior pseudopods, similar to the defects of cAMP-pulsed rasC− cells (data not shown).
Even though the anterior ends of rasC− cells extended off the substratum in the z axis far more frequently than those of control cells and even though rasC− cells at times had problems maintaining a single anterior pseudopod, 3D cell tracks revealed that rasC− cells translocated at instantaneous velocities similar to those of control cells (Table 2). However, height and percent overhang, the latter being a measure of the proportion of the cell body extending over the substratum, were both significantly greater for rasC− cells than for control cells (Table 2), supporting the conclusion that, on average, the anterior ends of rasC− cells migrating in buffer are more often above the substratum than are the anterior ends of control cells.
rasC− cells exhibit chemotaxis efficiently in a spatial gradient of cAMP.
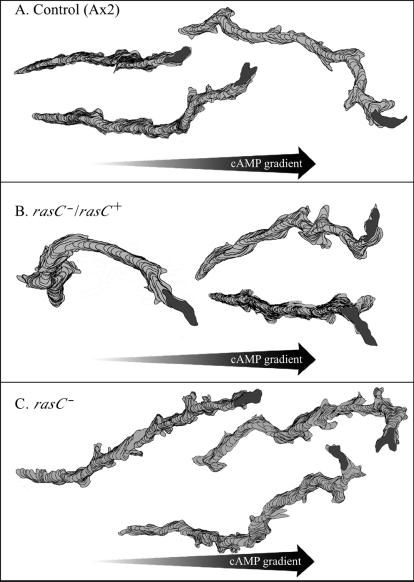
At the onset of the front of a natural, relayed wave of cAMP in an aggregation territory (phase A), Dictyostelium amoebae polarize in the direction of the aggregation center, presumably by assessing the direction of the positive spatial gradient (29). The initial characterization of rasC− cells demonstrated that they were capable of correctly orienting in and exhibiting chemotaxis up a spatial gradient of cAMP released from a micropipette (20). To test whether the efficiency of chemotaxis of rasC− cells was similar to that of control cells, computer-assisted methods were used to quantify single-cell chemotaxis in a spatial gradient of cAMP. The mean CI and percent positive chemotaxis (Table 3) as well as the distributions of CIs (data not shown) of control, rasC−/rasC+, and rasC− cells were statistically indistinguishable. Instantaneous velocity and directional change, as well as the morphology parameters mean maximum length and mean maximum width, were also statistically indistinguishable between control, rasC−/rasC+, and rasC− cells exhibiting chemotaxis in spatial gradients of cAMP (Table 3). However, both the roundness parameter and convexity parameter of rasC− cells differed statistically from those of control cells (Table 3), suggesting that even though chemotaxis and velocity parameters were similar, some aspect of rasC− cell behavior was still abnormal. This was supported by a comparison of perimeter tracks. Although the tracks of rasC− cells (Fig. 3C) were as aligned as those of control cells (Fig. 3A) and rasC−/rasC+ cells (Fig. 3B) in a spatial gradient of cAMP, they were not as smooth. Lateral projections were far more frequent along the tracks of rasC− cells, an aberrant characteristic also observed in perimeter tracks of cells in buffer. An analysis of individual perimeter images along the track revealed that these projections were at the anterior ends of the cells exhibiting chemotaxis, suggesting again a defect in the integrity of the anterior pseudopod. This suggestion was tested with 3D reconstructions of control and rasC− cells exhibiting chemotaxis up spatial gradients of cAMP. The 3D reconstructions revealed that both control (Fig. 4A and B) and rasC− (Fig. 4C and D) cells were highly polarized along the substratum in the direction of increasing cAMP concentration. However, while the anterior pseudopod of control cells remained intact (Fig. 4A and B), the anterior pseudopod of rasC− cells exhibiting chemotaxis frequently bifurcated or fragmented (Fig. 4C and D).
TABLE 3.
2D-DIAS analysis of rasC− cells reveals efficient chemotaxis in a spatial gradient of cAMPa
| Cells | No. of cells | Mean ± SD
|
% Positive chemotaxis | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Instantaneous velocity (μm/min) | Directional change (°/4 s) | Maximum length (μm) | Maximum width (μm) | Roundness (%) | Convexity (°) | Chemotactic index | |||
| Ax2 (control) | 38 | 11.8 ± 5.2 | 26.0 ± 12.5 | 21.3 ± 3.3 | 7.4 ± 1.1 | 50.2 ± 5.5 | 586.3 ± 29.2 | 0.66 ± 0.22 | 92 |
| rasC− | 32 | 16.4 ± 3.7 | 20.6 ± 8.0 | 21.5 ± 4.0 | 8.9 ± 2.3 | 42.4 ± 6.0 | 690.4 ± 113.3 | 0.68 ± 0.17 | 90 |
| rasC−/rasC+ | 20 | 13.9 ± 3.1 | 30.3 ± 13.5 | 19.9 ± 1.8 | 7.6 ± 1.0 | 50.1 ± 5.1 | 616.5 ± 55.3 | 0.57 ± 0.14 | 90 |
P values were computed by the Student t test; a P value of >0.05 was considered nonsignificant. P values for all comparisons of Ax2 with rasC− cells and of Ax2 with rasC−/rasC+ cells were nonsignificant, except for P values for comparisons of Ax2 with rasC− cells for roundness (P = 0.019) and convexity (P = 0.005).
FIG. 3.
Perimeter tracks of cells exhibiting chemotaxis in spatial gradients of cAMP reveal that while chemotactic efficiencies were similar for control (A), rasC−/rasC+ (B), and rasC− (C) cells, the tracks of rasC− cells included more lateral protrusions.
FIG. 4.
3D-DIAS reconstruction of two representative control (Ax2) cells (A and B) and two representative rasC− cells (C and D) reveals that although spatial gradients of cAMP suppress z-axis extension of rasC− cells and lateral pseudopod formation, they do not suppress the bifurcation of anterior pseudopods. The full cells and the pseudopodial zones alone are viewed at 60° and 30° angles, respectively. Cell bodies are color coded light gray, and pseudopodial zones are color coded dark gray. The direction of the cAMP gradient is indicated by an arrow in each panel. Time is presented in seconds.
These results demonstrate that although a spatial gradient of cAMP suppressed the aberrant extension of the anterior end of rasC− cells off the substratum in the z axis, it did not suppress the fragmentation of the anterior pseudopod. Remarkably, this defect did not detract from the efficiency of chemotaxis or cause a decrease in velocity (Table 3).
rasC− cells cannot exhibit chemotaxis in natural cAMP waves.
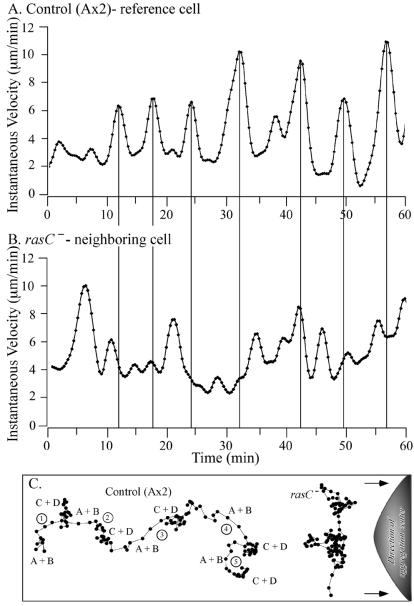
To test whether rasC− cells exhibited chemotaxis normally in natural waves of cAMP, the cells were vitally stained with DiI, mixed with unstained control cells at a ratio of 1:9, and allowed to undergo aggregation on a plastic surface. The behavior of stained rasC− cells was then compared to that of neighboring, unstained control cells no farther than four cell diameters away as waves of cAMP were relayed by the control cells, which represent the majority of cells, across all cells in the natural aggregation territory. The velocity plot of the representative control cell shown in Fig. 5A revealed seven major velocity surges with an average periodicity of 7.3 ± 1.6 min (mean and standard deviation). A second control cell in this neighborhood surged in concert with the reference control cell (data not shown). The rasC− cells in this neighborhood, however, did not surge in concert with reference control cells (Fig. 5B). The time intervals between surges and the extent of the surges were far more variable. Few of the apparently random surges in the rasC− plot (Fig. 5B) lined up with the peaks of the control plot (Fig. 5A). Five additional rasC− cells analyzed in this manner exhibited the same defect.
FIG. 5.
rasC− cells are incapable of exhibiting chemotaxis in natural waves of cAMP relayed through an aggregation territory of control cells. rasC− cells were mixed with control (Ax2) cells at a ratio of 1:9. rasC− cells were pulsed with cAMP in suspension for the last 5 h in a 6-h period, while control cells were starved on filters for 3 h prior to mixing. rasC− cells were stained with DiI before mixing, while control cells were unstained. Transmitted and fluorescent images were simultaneously collected at 4-s intervals. (A) The instantaneous velocity of a control cell over time reveals surges in velocity with a relatively constant periodicity. (B) The instantaneous velocity of a neighboring rasC− cell over time reveals erratic surges that do not correspond to those of the control cell. (C) Centroid plots of control and rasC− cells. The instantaneous velocity plots were smoothed with Tukey windows of 5, 15, 60, 15, and 5. Waves are indicated by 1 to 5, and phases are indicated by A+B for front and C+D for front and back of wave.
Centroid tracks revealed the impact of the rasC− defect on chemotaxis in a natural wave of cAMP. The centroid track of the reference control cell revealed net progress towards the aggregation center (i.e., towards the source of the cAMP waves) in the deduced front (phases A and B) in each of five relayed waves (Fig. 5C). The deduced front of the waves in the control centroid track in Fig. 5C coincided with the peaks in the velocity plot in Fig. 5A. The deduced peak and back (phases C and D) of the waves for the control centroid track in Fig. 5C coincided with the troughs in velocity in Fig. 5A. The centroid tracks of two additional control cells in the neighborhood were similar to that of the reference control cell in Fig. 5C (data not shown).
In marked contrast, the centroid track of the neighboring rasC− cell revealed no net progress towards the aggregation center through the entire period that encompassed the five natural waves (Fig. 5C). Centroid tracks of additional rasC− cells exhibited the same lack of net progress towards the aggregation center (data not shown).
rasC− cells respond abnormally to the temporal gradients of the cAMP wave.
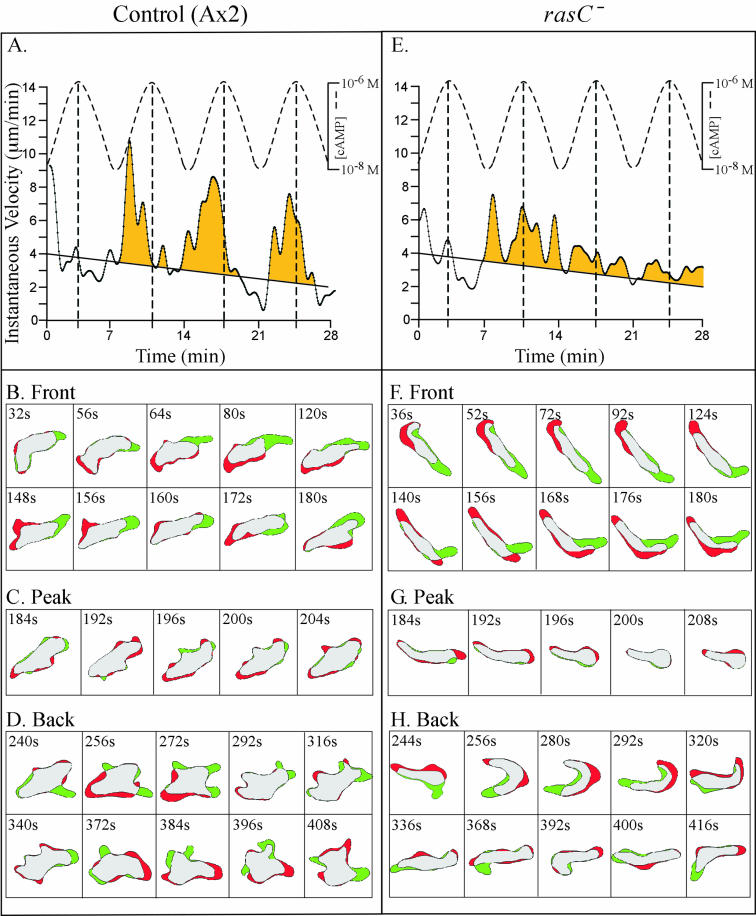
Given that rasC− cells exhibited chemotaxis efficiently in a spatial gradient of cAMP but were incapable of chemotaxis in a natural cAMP wave, we considered the possibility that they were defective in responding to the temporal and concentration components of the natural wave. To test this directly, rasC− cells were treated with a series of four temporal waves of cAMP generated in a round chamber in which spatial gradients were not established. The period between waves was 7 min, the concentrations at the peak and trough of each wave were 10−6 M and 10−8 M, respectively, and the shape of each successive wave was symmetric (i.e., 3.5 min for the increasing phase and 3.5 min for the decreasing phase). As previously described (8, 39, 50, 51), control cells did not surge in the front of the first wave but then transiently surged in the front (phase B) of each of the succeeding three waves (Fig. 6A). Difference images of control cells in the front, peak, and back of the third in a series of four waves revealed associated shape changes and pseudopod dynamics. In the front (phase B) of the wave, control cells were elongate and expansion zones were confined primarily to the front end of the elongate cell (Fig. 6B). At the peak of the wave (phase C), control cells became less elongate and partially retracted their anterior pseudopod (Fig. 6C). In the back of the wave (phase D), control cells once again extended pseudopods, but they did so in a relatively random fashion around the cell perimeter and made no net progress in any one direction (Fig. 6D). The multiple expansion zones formed in the back of the wave were smaller on average than the dominant anterior pseudopod formed in the front of the wave. Similar results were obtained for nine additional control cells analyzed in a similar fashion.
FIG. 6.
rasC− cells behave abnormally in the front (increasing gradient) and in the back (decreasing gradient) of the last three in a series of four simulated temporal waves of cAMP. Instantaneous velocities of a representative control (Ax2) cell (A) and a rasC− cell (E) are plotted as a function of time through four temporal waves of cAMP generated in a perfusion chamber. Instantaneous velocity plots were smoothed with Tukey windows of 5, 15, 60, 15, and 5. Velocity above an arbitrary threshold line drawn from 4 to 2 μm/min over the 28-min period of analysis is color coded yellow to accentuate the surges of the control cell in the front of the last three waves and their absence in rasC− cells. (B to D and F to H) Difference images of a representative control cell and a representative rasC− cell in the front (B and F, respectively), at the peak (C and G, respectively), and in the back (D and H, respectively) of the third in a series of four simulated temporal waves of cAMP. In difference images, contraction zones are color coded red and expansion zones are color coded green.
In marked contrast, rasC− cells did not surge in the front of the last three in a series of four waves (Fig. 6E). rasC− cells were, however, elongate in the front of the wave and extended an anterior pseudopod (Fig. 6F), like control cells (Fig. 6B). At the peak of the wave, rasC− cells became less elongate and partially retracted their anterior pseudopod (Fig. 6G), like control cells (Fig. 6C). In the back of the wave, however, rasC− cells abnormally exhibited an elongate shape and extended an anterior pseudopod (Fig. 6H), in direct contrast to the apolar shape and randomly positioned expansion zones of control cells (Fig. 6D). Similar results were obtained for nine additional rasC− cells analyzed in a similar fashion.
A comparison of the frequencies of lateral pseudopod formation in the front of the wave supported this interpretation. Control cells formed lateral pseudopods at a frequency of 2.1 ± 0.9 (mean and standard deviation) per 3.5 min in buffer and at a decreased frequency of 1.4 ± 0.9 per 3.5 min in the increasing cAMP gradient in the front of a temporal wave. rasC− cells formed lateral pseudopods at a frequency of 3.1 ± 1.0 per 3.5 min in buffer and at a frequency of 0.7 ± 0.9 per 3.5 min in the increasing cAMP gradient in the front of a temporal wave. Therefore, the capacity to suppress lateral pseudopod formation in the front of the wave was intact in rasC− cells. In the decreasing gradient of cAMP in the back of the wave, control cells formed lateral pseudopods at a frequency of 4.5 ± 1.6 per 3.5 min, which is more than three times the frequency in the front of the wave. In the back of the wave, rasC− cells formed lateral pseudopods at a frequency of 1.0 ± 0.9 per 3.5 min, close to the frequency in the front of the wave. Therefore, the suppression of lateral pseudopod formation was normal in rasC− cells in the front of the wave, but suppression of lateral pseudopod formation persisted abnormally in rasC− cells in the back of the wave, in direct contrast to what was seen with control cells, which renewed lateral pseudopod formation.
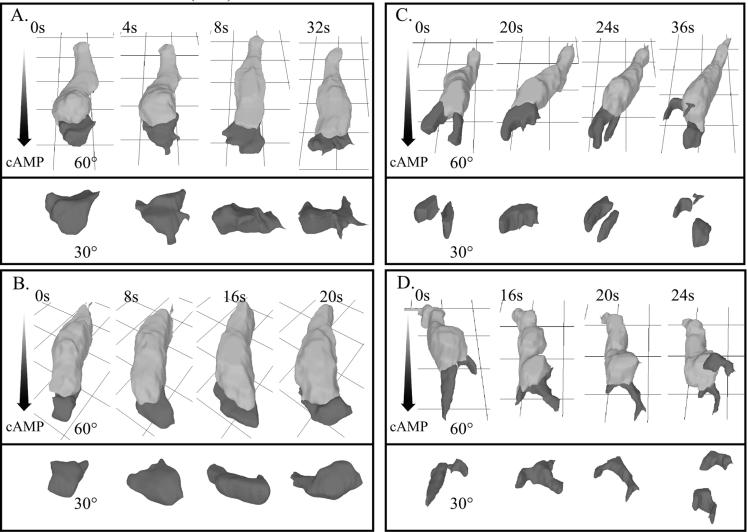
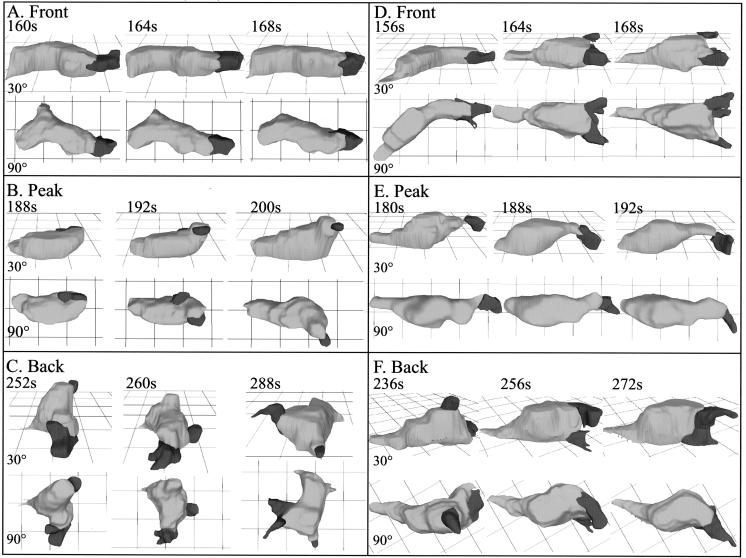
To obtain a more complete description of the aberrant behavior of rasC− cells in temporal waves of cAMP, control and rasC− cells were reconstructed in three dimensions in the front, at the peak, and in the back of the third in a series of four temporal waves of cAMP. 3D reconstructions of control cells translocating in the front of the cAMP wave revealed that they were highly elongate, extending one dominant anterior pseudopod along the substratum (Fig. 7A). At the peak of the wave, they partially retracted their original anterior pseudopod and became less elongate (Fig. 7B). In the back of the wave, they extended multiple pseudopods around their periphery and appeared relatively apolar (Fig. 7C).
FIG. 7.
3D-DIAS reconstruction of a representative control (Ax2) cell and a representative rasC− cell in the front (A and D, respectively), at the peak (B and E, respectively), and in the back (C and F, respectively) of the third in a series of four simulated temporal waves of cAMP reveals a major defect in the response to the back (decreasing temporal gradient) of the third wave. The representative cells are viewed from 30° and 90° angles. The cell bodies are color coded light gray, and the pseudopodial zones are color coded dark gray.
3D reconstructions of rasC− cells revealed that in the front of the wave, they were elongate like control cells, but the anterior pseudopod continued to fragment (Fig. 7D), as it did in buffer (Fig. 2C and D) and in spatial gradients of cAMP (Fig. 4C and D). At the peak of the wave, rasC− cells partially retracted their anterior pseudopod and became less elongate (Fig. 7E), like control cells (Fig. 7B). In the back of the wave, however, the rasC− cells abnormally remained elongate and extended a single dominant pseudopod, which bifurcated (Fig. 7F).
rasC− cells respond normally to the rapid addition of 10−6 M cAMP.
The results of the temporal wave experiments suggested that rasC− cells responded normally to the high concentration of cAMP at the peak of the wave. To examine this point further, cells translocating in buffer were rapidly exposed to the high concentration of cAMP (10−6 M) at the peak of a wave, a procedure that causes a rapid reduction in instantaneous velocity in wild-type cells (48). The rapid addition of 10−6 M cAMP resulted in a decrease in instantaneous velocity in both control cells (9 to 3 μm per min) and rasC− cells (7 to 2 μm per min) within 30 s of the addition of cAMP, supporting the conclusion that rasC− cells respond normally to the peak of a cAMP wave.
Differences in F-actin localization in the back of the wave.
To test whether the aberrant behaviors displayed by rasC− cells were accompanied by abnormalities in the localization of F actin, cells were stained with Oregon green-conjugated phalloidin in buffer and in the front and back of the third in a series of four temporal waves of cAMP. Stained cells were then analyzed by LSCM.
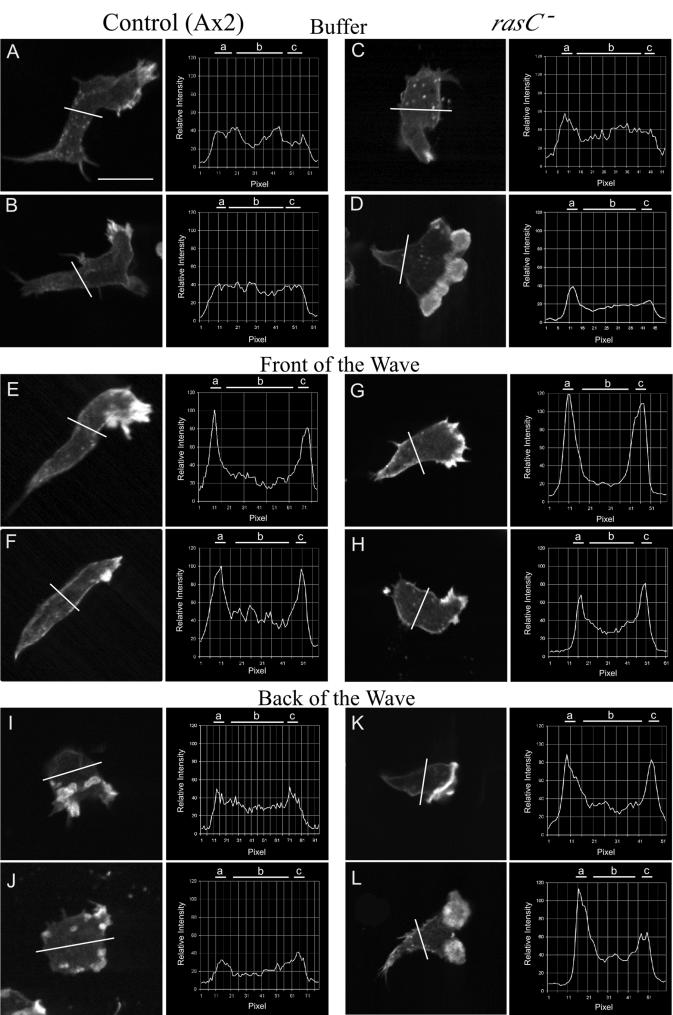
Projection images of control cells, which include the entire stack of LSCM scans obtained at 0.2-μm intervals through a cell, revealed that F actin stained most intensely in the dominant anterior pseudopods in buffer and in the front of the wave and that multiple, small pseudopods randomly located around the cell periphery stained most intensely in the back of the wave (data not shown). These staining patterns were also evident in individual scans 1 μm above the substratum (Fig. 8A and B, E and F, and I and J). These single scans also revealed that while the cytoplasm and cortex of control cells stained diffusely in buffer, there was an increase in relative cortical staining in the front of the wave and a return to the diffuse pattern in the back of the wave. These changes in F-actin distribution were confirmed in line profiles of staining intensity in buffer (Fig. 8A and B), in the front of the wave (Fig. 8E and F), and in the back of the wave (Fig. 8I and J). To quantitate these changes, the ratio of cortical to cytoplasmic staining was computed from line profiles by the formula [(a + c)/2]/b, where a and c are the intensities of cortical staining on the two sides of the cell and b is the intensity of cytoplasmic staining. The ratio of the intensity of staining in the cortex to that in the cytoplasm increased from 1.1 ± 0.2 (mean and standard deviation) in buffer to 2.3 ± 0.5 in the front of the wave and then returned to 1.1 ± 0.2 in the back of the wave (Table 4).
FIG. 8.
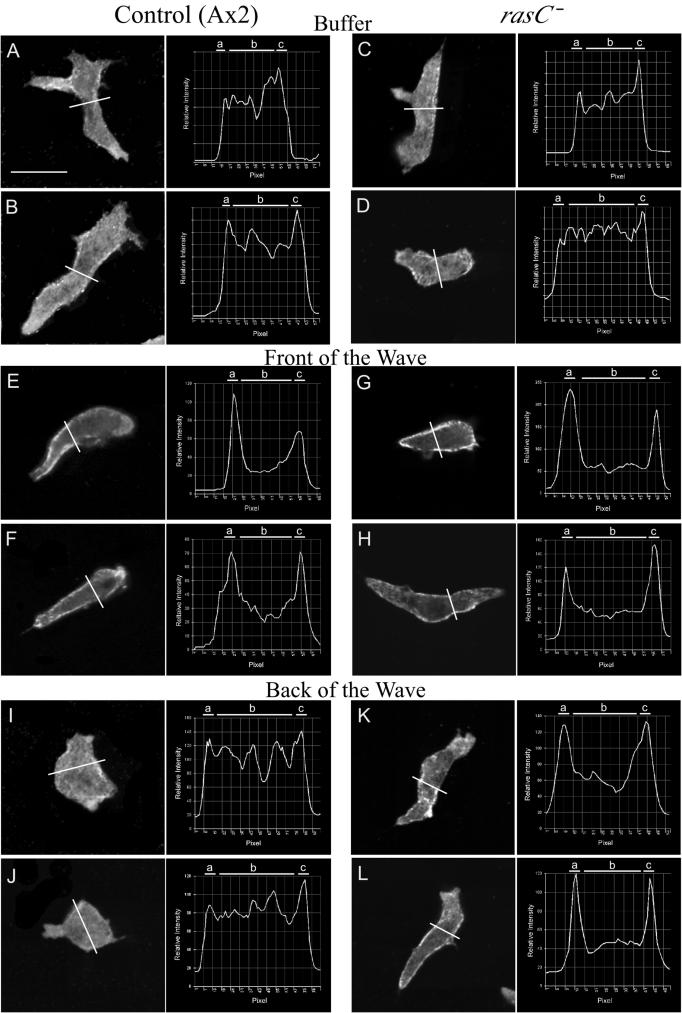
Line profiles of the intensity of F-actin staining in the cortex and cytoplasm reveal subtle differences between control (Ax2) and rasC− cells in buffer and in the front of the wave. In each panel, the position of the line profile across the stained cell image, just posterior to the nucleus, and a plot of the line profile are presented. The analyzed optical section in each case was 1 μm above the substratum. In each line profile, relative intensity is plotted as a function of distance in pixels. Cells were analyzed in the third in a series of four simulated temporal waves of cAMP. The average ratios of the intensity of staining in the cortex to that in the cytoplasm are presented in Table 4. a and c, cortical zones; b, cytoplasmic zone. Scale bar, 5 μm.
TABLE 4.
Ratio of F-actin staining in the cortex and the cytoplasm and ratio of myosin II staining in the cortex and the cytoplasm of representative cells in buffer in relation to the temporal wavea
| Staining | Condition | Ax2 Cells (control)
|
rasC− cells
|
||
|---|---|---|---|---|---|
| No. of cells | Ratio | No. of cells | Ratio | ||
| F actin | Buffer | 10 | 1.1 ± 0.2 | 6 | 1.1 ± 0.1 |
| Wave front | 9 | 2.3 ± 0.5 | 9 | 2.1 ± 1.1 | |
| Wave back | 10 | 1.1 ± 0.2 | 8 | 2.0 ± 0.6 | |
| Myosin II | Buffer | 6 | 1.1 ± 0.1 | 12 | 1.1 ± 0.2 |
| Wave front | 12 | 1.8 ± 0.4 | 14 | 1.7 ± 0.5 | |
| Wave back | 12 | 1.1 ± 0.1 | 15 | 1.6 ± 0.3 | |
The ratios of the staining intensity of cortical regions (a and c) to that of the cytoplasm (b) were computed by the formula [(a + c)/2]/b. The intensities in the cortical regions at the two ends of the scan (approximately six pixels) were averaged. The intensities in the regions between the cortical regions were considered cytoplasmic and were averaged.
Projection images of rasC− cells revealed that F actin stained most intensely in the dominant anterior pseudopods in buffer, in the front of the wave, and in the back of the wave, just as it did in control cells (data not shown). These staining patterns were also evident in individual scans 1 μm above the substratum (Fig. 8C and D, G and H, and K and L). As in the case of control cells, while the cytoplasm and cortex of rasC− cells migrating in buffer stained diffusely, there was an increase in cortical staining in the front of the wave. However, there was no return in the back of the wave to the staining pattern observed in buffer, as was observed in control cells. The ratio of the intensity of staining in the cortex to that in the cytoplasm increased from 1.1 ± 0.1 in buffer to 2.1 ± 1.1 in the front of the wave, approximately the same increase seen in control cells (Table 4). However, in contrast to what was seen with control cells, the ratio did not decrease in the back of the wave (Table 4). The results demonstrate that the increasing temporal gradient of cAMP in the front of a wave stimulates cortical localization of F actin, as in control cells, but the decreasing temporal gradient of cAMP in the back of the wave does not stimulate the normal reduction in F actin in the cortex, as it does in control cells.
Abnormalities in myosin II localization in the back of the wave.
Both projection images (data not shown) and individual scans 1 μm above the substratum (Fig. 9A and B) of control cells migrating in buffer revealed that pseudopods did not stain and that the cortex and cytoplasm stained diffusely for myosin II, as they did for F actin. The ratio of the intensity of staining in the cortex to that in the cytoplasm computed from line profiles of cells in buffer was 1.1 ± 0.1 (Table 4). The intensity of staining increased in the cortex in the front of the wave (Fig. 9E and F). The ratio of the intensity of cortical staining to that of cytoplasmic staining increased from 1.1 ± 0.1 in buffer to 1.8 ± 0.4 in the front of the wave (Table 4). In the back of the wave, however, the staining pattern of myosin II again became diffuse across the cortex and cytoplasm (Fig. 9I and J), as it was in buffer (Fig. 9A and B). The ratio returned from 1.8 ± 0.4 in the front of the wave to 1.1 ± 0.1 in the back of the wave, the latter being the same ratio obtained in buffer (Table 4).
FIG. 9.
Line profiles of the intensity of myosin II staining in the cortex and the cytoplasm reveal a major difference between control (Ax2) and rasC− cells in the back of the wave. In each panel, the position of the line profile across the cell, just posterior to the nucleus, and a plot of the line profile are presented. The analyzed optical section in each case was 1 μm above the substratum. Cells were analyzed in the third of a series of four simulated temporal waves of cAMP. The average ratios of the intensity of staining in the cortex to that in the cytoplasm are presented in Table 4. a and c, cortical zones; b, cytoplasmic zone. Scale bar, 5 μm.
Both projection images (data not shown) and single scans 1 μm above the substratum of rasC− cells in buffer (Fig. 9C and D) and in the front of the wave (Fig. 9G and H) revealed staining patterns similar to those for control cells (Fig. 9A and B and E and F, respectively). The ratio of the intensity of cortical staining to that of cytoplasmic staining for rasC− cells increased from 1.1 ± 0.2 in buffer to 1.7 ± 0.5 in the front of the wave, values close to those for control cells under the respective conditions (Table 4). However, in the back of the wave, myosin II was abnormally retained in the cortex (Fig. 9K and L), in direct contrast to what was seen with control cells (Fig. 9I and J). While the staining intensity ratio of control cells decreased from 1.8 ± 0.4 in the front of the wave to 1.1 ± 0.1 in the back of the wave, the ratios of rasC− cells stayed approximately the same, 1.7 ± 0.5 and 1.6 ± 0.3, respectively, in the front and back of the wave (Table 4). These results demonstrate that while in control cells myosin II exits the cortex in response to the decreasing temporal gradient of cAMP in the back of the wave, myosin II, like F actin, is abnormally retained in the cortex of rasC− cells.
DISCUSSION
Guanine nucleotide exchange factors activate Ras proteins by catalyzing the exchange of GDP with GTP (2). GTPase-activating proteins inactivate Ras proteins by catalyzing GTP hydrolysis (2). Receptor-coupled Ras proteins are responsible for the activation of a variety of signaling pathways upon receptor occupancy (3, 21, 35, 36). In Dictyostelium, RasC, which is expressed throughout the life cycle (6), is involved in the activation of adenylate cyclase and Akt/PKB, the latter possibly resulting from the activation of phosphatidylinositol 3-kinase (PI3K), which catalyzes the phosphorylation of PIP2 to PIP3 (5). Both PI3K1 and PI3K2, potential targets of RasC, have been shown to play roles in chemotaxis (9, 23). The original characterization revealed that while rasC− cells that had been artificially pulsed with cAMP could not signal, they could exhibit chemotaxis up a spatial gradient of cAMP released from a micropipette. Pulsed rasC− cells were also capable of forming aggregates at high density and of entering aggregates formed by control cells (20). Although these experiments suggested that RasC was not involved in the behavioral responses of cells to a chemoattractant, they did not test whether RasC was involved in the basic motile behavior of a cell in the absence of an attractant or in single-cell responses to the temporal and concentration components of a natural wave.
RasC plays a role in basic motile behavior.
Although rasC− cells moved at approximately the same average velocity as did control cells, they had problems maintaining their front end on the substratum and in maintaining an intact anterior pseudopod. The front end of rasC− cells frequently extended off the substratum in a manner similar to that seen with the null mutant for the vasodilator-stimulated phosphoprotein gene (vasp−) (12) and the null mutant for clathrin (chc−) (47). Since rasC− cells exhibited these defects in the absence of chemoattractant, it seems likely that its role in maintaining elongate cells on a substratum and in maintaining an intact anterior pseudopod is independent of cAMP receptor (CAR1) occupancy. Observations of vegetative cell behavior revealed similar abnormal behavior in buffer (data not shown), supporting this conclusion. Wessels et al. (44) have demonstrated that when lateral pseudopods contact the substratum, they are stabilized. Hence, RasC may function immediately downstream of adhesion sites or mechanoreceptors involved in pseudopod stabilization during migration in buffer as well as downstream of the cAMP receptor CAR1 during chemotaxis.
RasC is not necessary for chemotaxis in a spatial gradient of a chemoattractant.
In the original analysis, it was demonstrated that rasC− cells exhibited chemotaxis up a spatial gradient of cAMP released from a micropipette (20). Here, we performed a quantitative analysis of the efficiency of chemotaxis as well as a detailed analysis of the behavior of individual cells during chemotaxis. Our results demonstrate that a spatial gradient of cAMP normalizes the z-axis extension defect in buffer but not the tendency of rasC− pseudopods to bifurcate or fragment. Our results demonstrate, however, that RasC is not essential for cell polarization or efficient chemotaxis in a spatial gradient of cAMP. Therefore, it is unlikely that RasC plays a role in those pathways that regulate cell polarization in response to a chemotactic signal (e.g., the pathway that includes PAKa) (4, 5).
RasC plays an essential role in responding to temporal gradients of cAMP.
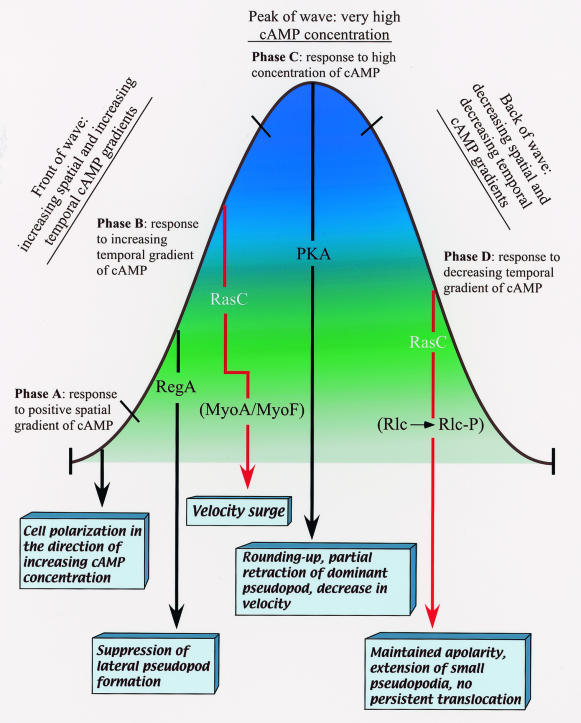
The temporal gradients of cAMP in the front and back of a natural wave play fundamental roles in regulating Dictyostelium chemotaxis in a natural aggregation territory (29). In response to the increasing temporal gradient of cAMP in the front of a simulated temporal wave, rasC− cells suppressed lateral pseudopod formation like control cells but they did not surge (i.e., exhibit a transient increase in velocity). This response was opposite to that of the regA− mutant (45). RegA is an intracellular phosphodiesterase (19). Therefore, regA− cells should have abnormally increased levels of intracellular cAMP, while rasC− cells would be expected to have abnormally reduced levels of intracellular cAMP, a possible explanation for the opposite phenotypes exhibited in the front of the wave. Hence, one might also expect cells with constitutively active protein kinase A activity (pkAR−) (51) to behave in the same manner as regA− cells and opposite that of rasC− cells. The finding that pkAR− cells were constitutively ovoid under all conditions, affecting their behavior in the front of a wave (51), precludes this comparison. However, it is just as likely that RegA, which carries a two-component response regulator domain, and RasC, which regulates pathways other than those involving cAMP, also affect behavior through pathways independent of cAMP. What is clear is that regA and rasC deletion mutants affect different components of the behavioral response to the increasing temporal gradient of cAMP in the front of the wave. Hence, we separated RegA and RasC into two independent pathways emanating from the front of the wave and ending in distinctly different behavioral responses in the working model shown in Fig. 10. Since ERK2 has been postulated to be a negative regulator of RegA (19), an erkB-null mutant should respond abnormally to the front of the wave in the same manner as rasC− cells and opposite that of regA− cells. It would therefore be of considerable interest to analyze the detailed chemotactic behavior of erkB− cells (25) to see whether they exhibit defects in the front of the wave similar to those exhibited by rasC− cells. In contrast to regA−, cells of the myoA−/myoF− double mutant do not surge in response to an increasing temporal gradient of cAMP (8), a response similar to that of rasC−. For that reason, we very tentatively placed MyoA/MyoF downstream of RasC in the RasC-dependent pathway emanating from the front of the wave in the model in Fig. 10.
FIG. 10.
Model of regulation of the sequence of behaviors associated with the four phases of the wave, based on data obtained here for the rasC− mutant and in the following studies of other mutants that used the same contextual framework and experimental protocols (31) for identifying the exact behavioral defects in the different components of the natural wave: regA− (45), myoA−/myoF− (7), pkaR− (47), S13A (48). PKA, protein kinase A; Rlc, regulatory light chain; Rlc-P, phosphorylated regulatory light chain.
Although partially defective in their response to the increasing temporal gradient in the front of the wave, rasC− cells responded normally to the high concentration of cAMP at the peak of the wave. However, rasC− cells then responded abnormally to the decreasing temporal gradient of cAMP in the back of the wave. Rather than becoming apolar and extending small pseudopods in random directions as control cells do, rasC− cells once again extended a dominant anterior pseudopod, usually from the original front end of the cell, and exhibited a relatively elongate, polar morphology along the substratum. The abnormal retention of polarity and the abnormal suppression of lateral pseudopod formation in the back of the wave were the same defects exhibited by the myosin II regulatory light chain phosphorylation mutant S13A (50), suggesting that RasC and the phosphorylation of RLC function along a common independent pathway emanating from the back of the wave, as in the model in Fig. 10. S13A mutant cells respond normally to the front of the wave, supporting the independence of the RasC pathway in the back of the wave.
Previous qualitative results (20) suggested that rasC− cells could aggregate. In chimeric mixes, rasC− cells were observed to be late in entering streams and aggregates of normal cells (20), suggesting a fundamental defect in behavioral responses to relayed cAMP waves. The mixing experiments performed here demonstrate that rasC− cells do not surge or orient in response to a natural wave and therefore do not make net progress towards an aggregation center in an aggregation territory. We conclude that it is the defective responses to temporal gradients of cAMP in the front and back of a wave that preclude chemotaxis in a natural wave of cAMP.
RasC and cytoskeletal reorganization.
Staining experiments revealed a major defect in the localization of F actin and myosin II in the cortex in response to the decreasing temporal gradient of cAMP in the back of a wave in association with abnormal maintenance of an elongate cell morphology. Myosin II has been demonstrated to play a fundamental role in regulating the shape of a Dictyostelium amoeba (26, 42). Our results indicate that the normal loss of anterior-posterior polarity and the random extension of pseudopods in control cells in the back of a wave are dependent on RasC activity and are associated with the reduction of F actin and myosin II in the cortex. Although one might assume that this reduction of F actin and myosin II is the direct result of myosin heavy-chain (MHC) phosphorylation based on previous in vitro (17) and in vivo (7) studies, the retention of myosin II in the cortex of rasC− cells and maintenance of an elongate shape are also characteristic of the S13A mutant, in which the myosin regulatory light chain cannot be phosphorylated (50). This may be explained by the observations of Berlot et al. (1) that the addition of cAMP to aggregation-competent Dictyostelium amoebae suspended in buffer results in an increase in the phosphorylation of both the MHC and regulatory light chain, suggesting coordinate phosphorylation and function. Interestingly, both in vitro (17) and in vivo (7) studies have demonstrated that phosphorylation of the MHC results in the depolymerization of myosin II and its movement from the cortex to cytoplasm. The phenotype of the S13A mutant suggests that the myosin II regulatory light chain must also be phosphorylated for the reduction in F actin and myosin II in the cell cortex in the back of the wave. Therefore, the model in Fig. 10 may eventually be expanded to include both myosin regulatory light chain and MHC phosphorylation as downstream targets of the RasC pathway emanating from the back of the wave. Analyses of the 3XALA mutant, in which MHC was engineered to mimic a permanently unphosphorylated state, and the 3XASP mutant, in which MHC was engineered to mimic a permanently phosphorylated state (7), are now in progress to test this possibility.
An emerging model.
The model that has begun to emerge includes independent pathways emanating from the different phases of the chemotactic wave (Fig. 10). Two parallel pathways, one dependent on RegA (45, 51) and another dependent on RasC, emanate from the front of the wave and are induced by increasing receptor occupancy with time (i.e., a positive temporal gradient of cAMP). The parallel pathway dependent on the activation of PKA is activated by the very high concentration of cAMP at the peak of the wave, presumably the result of full receptor occupancy (51). Finally, a second RasC-dependent pathway emanates from the back of the wave and is induced by decreasing receptor occupancy with time (i.e., a negative temporal gradient of cAMP). Each pathway is responsible for a phase-specific behavior; in sequence, the behaviors represent the complex response to the natural wave (Fig. 10). One interesting, albeit tentative, aspect of this model is that at least two downstream targets of the regulatory pathways so far delineated are myosins, both myosin II and myosin I. In the case of myosin II, localization is effected by phosphorylation-dephosphorylation of both the heavy chain (7; P. Heid, D. Wessels, K. Daniels, H. Zhang, and D. R. Soll, unpublished data) and light chain (50). As more mutants, including both regulatory and cytoskeletal, are analyzed within the contextual framework developed to distinguish parallel pathways emanating from the different phases of the wave (29), we may find that only a few cytoskeletal components, including the myosins, serve as the targets of regulatory cascades for effecting the phase-specific behaviors that in sequence represent the complex chemotactic response to the natural wave. Because of the complexity of each response, it is likely that each cascade may have more than one target.
Acknowledgments
This research was supported by National Institutes of Health grants HD-18577 to D.R.S. and GM60447 and GM62350 to W.F.L. and a grant from the MRC of Canada to G.W.
REFERENCES
- 1.Berlot, C. H., J. A. Spudich, and P. N. Devreotes. 1985. Chemoattractant-elicited increases in myosin phosphorylation in Dictyostelium. Cell 43:307-314. [DOI] [PubMed] [Google Scholar]
- 2.Boguski, M., and F. McCormick. 1993. Proteins regulating Ras and its relatives. Nature 366:643-654. [DOI] [PubMed] [Google Scholar]
- 3.Campbell, S., R. Khosravi-Far, K. L. Rossman, G. J. Clark, and C. J. Der. 1998. Increasing complexity of Ras signaling. Oncogene 17:1395-1413. [DOI] [PubMed] [Google Scholar]
- 4.Chung, C., and R. A. Firtel. 1999. PAKa, a putative PAK family member, is required for cytokinesis and the regulation of the cytoskeleton in Dictyostelium discoideum cells during chemotaxis. J. Cell Biol. 174:559-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung, C. Y., G. Potikyan, and R. A. Firtel. 2001. Control of cell polarity and chemotaxis by Akt/PKB and PI3 kinase through the regulation of PAKa. Mol. Cell 7:937-947. [DOI] [PubMed] [Google Scholar]
- 6.Daniel, J., G. Spiegelman, and G. Weeks. 1995. Dictyostelium ras genes, p. 100-104. In M. Serial and L. Huber (ed.), Guidebook to the small GTPase. Oxford University Press, New York, N.Y.
- 7.Egelhoff, T. T., R. J. Lee, and J. A. Spudich. 1993. Dictyostelium myosin heavy chain phosphorylation sites regulate myosin filament assembly and localization in vivo. Cell 75:363-371. [DOI] [PubMed] [Google Scholar]
- 8.Falk, D. L., D. Wessels, L. Jenkins, T. Pham, S. Kuhl, M. A. Titus, and D. R. Soll. 2003. Shared, unique and redundant functions of three members of the class I myosins (MyoA, MyoB and MyoF) in motility and chemotaxis in Dictyostelium. J. Cell Sci. 116:3985-3999. [DOI] [PubMed] [Google Scholar]
- 9.Funamoto, S., K. Milan, R. Meili, and R. A. Firtel. 2001. Role of phosphatidylinositol 3′ kinase and a downstream pleckstrin homology domain-containing protein in controlling chemotaxis in Dictyostelium. J. Cell Biol. 153:795-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Funamoto, S., R. Meili, S. Lee, L. Parry, and R. Firtel. 2002. Spatial and temporal regulation of 3-phosphoinositides by PI3-kinase and PTEN mediates chemotaxis. Cell 109:611-623. [DOI] [PubMed] [Google Scholar]
- 11.Geiger, J., D. Wessels, and D. R. Soll. 2003. Human PMNs respond to temporal waves of chemoattractant like Dictyostelium. Cell Motil. Cytoskelet. 56:27-44. [DOI] [PubMed] [Google Scholar]
- 12.Han, Y., C. Chung, D. Wessels, S. Stephens, M. A. Titus, D. R. Soll, and R. A. Firtel. 2002. Requirement of VASP for cell adhesion, filopodia formation, and chemotaxis. J. Biol. Chem. 277:49877-49887. [DOI] [PubMed] [Google Scholar]
- 13.Heid, P., E. Voss, and D. R. Soll. 2002. 3D-DIASemb: a computer-assisted system for reconstructing and motion analyzing in 4D every cell and nucleus in a developing embryo. Dev. Biol. 245:329-347. [DOI] [PubMed] [Google Scholar]
- 14.Iranfar, N., D. Fuller, and W. F. Loomis. 2003. Genome-wide expression analyses of gene regulation during early development of Dictyostelium discoideum. Eukaryot. Cell 2:664-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iranfar, N., D. Fuller, R. Sasik, T. Hwa, M. Laub, and W. F. Loomis. 2001. Expression patterns of cell-type-specific genes in Dictyostelium. Mol. Biol. Cell 12:2590-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kessin, R. H. 2001. Dictyostelium: evolution, cell biology and the development of multicellularity. Cambridge University Press, Cambridge, United Kingdom.
- 17.Kuczmarski, E. R., and J. A. Spudich. 1980. Regulation of myosin self-assembly: phosphorylation of heavy chain inhibits formation of thick filaments. Proc. Natl. Acad. Sci. USA 77:7292-7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuspa, A., and W. Loomis. 1994. Transformation of Dicytostelium—gene disruptions, insertional mutagenesis, and promoter traps. Methods Mol. Genet. 3:3-21. [Google Scholar]
- 19.Laub, M. T., and W. F. Loomis. 1998. A molecular network that produces spontaneous oscillations in excitable cells of Dictyostelium. Mol. Biol. Cell 9:3521-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim, C. J., G. B. Spiegelman, and G. Weeks. 2001. RasC is required for optional activation of adenylyl cyclase and Akt/PKB during aggregation. EMBO J. 20:4490-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim, C. J., G. B. Spiegelman, and G. Weeks. 2002. Cytoskeletal regulation by Dictyostelium Ras subfamily proteins. J. Muscle Res. Cell Motil. 23:729-736. [DOI] [PubMed] [Google Scholar]
- 22.Reymond, C. D., R. H. Gomer, M. C. Medhy, and R. A. Firtel. 1984. Developmental regulation of a Dicytostelium gene encoding a protein homologous to mammalian Ras protein. Cell 39:141-148. [DOI] [PubMed] [Google Scholar]
- 23.Rickert, P., O. D,.Weiner, F. Wang, H. R. Bourne, and G. Servant. 2000. Leukocytes navigate by compass: roles of PI3Kgamma and its lipid products. Trends Cell Biol. 10:466-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasik, R., N. Iranfar, T. Hwa, and W. F. Loomis. 2002. Extracting transcriptional events from temporal gene expression patterns during Dictyostelium development. Bioinformatics 18:61-66. [DOI] [PubMed] [Google Scholar]
- 25.Segall, J. E., A. Kuspa, G. Shaulsky, M. Ecke, M. Maeda, C. Gaskins, R. A. Firtel, and W. F. Loomis. 1995. A MAP kinase necessary for receptor-mediated activation of adenylyl cyclase in Dictyostelium. J. Cell Biol. 128:405-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheldon, E., and D. A. Knecht. 1996. Dictyostelium cell shape generation requires myosin II. Cell Motil. Cytoskelet. 35:59-67. [DOI] [PubMed] [Google Scholar]
- 27.Shutt, D., and D. R. Soll. 1999. HIV-induced T-cell syncytia release a two component T-helper cell chemoattractant composed of Nef and Tat. J. Cell Sci. 112:3931-3941. [DOI] [PubMed] [Google Scholar]
- 28.Shutt, D. C., L. M. Jenkins, E. Carolan, J. Stapleton, K. Daniels, R. Kennedy, and D. R. Soll. 1998. T cell syncytia induced by HIV release T cell chemoattractants: demonstration with a newly developed single cell chemotaxis chamber. J. Cell Sci. 111:99-109. [DOI] [PubMed] [Google Scholar]
- 29.Soll, D. R., D. Wessels, P. Heid, and H. Zhang. 2002. A contextual framework for characterizing motility and chemotaxis mutants in Dictyostelium discoideum. J. Muscle Res. Cell Motil. 23:659-672. [DOI] [PubMed] [Google Scholar]
- 30.Soll, D. R. 1979. Timers in developing systems. Science 203:841-849. [DOI] [PubMed] [Google Scholar]
- 31.Soll, D. R. 1987. Methods for manipulating and investigating developmental timing in Dictyostelium discoideum. Methods Cell Biol. 28:413-431. [DOI] [PubMed] [Google Scholar]
- 32.Soll, D. R. 1995. The use of computers in understanding how animal cells crawl. Int. Rev. Cytol. 163:43-104. [PubMed] [Google Scholar]
- 33.Soll, D. R., and E. Voss. 1998. Two and three dimensional computer systems for analyzing how cells crawl, p. 25-52. In D. R. Soll and D. Wessels (ed.), Motion analysis of living cells. John Wiley, Inc., New York, N.Y.
- 34.Soll, D. R., E. Voss, O. Johnson, and D. J. Wessels. 2000. Three-dimensional reconstruction and motion analysis of living crawling cells. Scanning 22:249-257. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki, J., Y. Yamazaki, G. Li, Y. Kaziro, and H. Koide. 2000. Involvement of Ras and Ra1 in chemotactic migration of skeletal myoblasts. Mol. Cell. Biol. 20:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takai, Y., T. Sasaki, and T. Matozaki. 2001. Small GTP-binding proteins. Physiol. Rev. 81:153-208. [DOI] [PubMed] [Google Scholar]
- 37.Van Driessche, N., C. Shaw, M. Katoh, T. Morio, R. Sucgang, M. Ibarra, H. Kuwayama, T. Saito, H. Urushihara, M. Maeda, I. Takeuchi, H. Ochiai, W. Eaton, J. Tollett, J. Halter, A. Uspa, Y. Tanaka, and G. Shaulsky. 2002. A transcriptional profile of multicellular development in Dictyostelium discoideum. Development 129:1543-1552. [DOI] [PubMed] [Google Scholar]
- 38.Varnum, B., and D. R. Soll. 1984. Effect of cAMP on single cell motility in Dictyostelium. J. Cell Biol. 99:1151-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varnum, B., K. Edwards, and D. R. Soll. 1985. Dictyostelium amoebae alter motility differently in response to increasing versus decreasing temporal gradients of cAMP. J. Cell Biol. 101:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varnum, B., K. Edwards, and D. R. Soll. 1986. The developmental regulation of single cell motility in Dictyostelium discoideum. Dev. Biol. 113:218-227. [DOI] [PubMed] [Google Scholar]
- 41.Varnum-Finney, B., K. Edwards, E. Voss, and D. R. Soll. 1987. Amoebae of Dictyostelium discoideum respond to an increasing temporal gradient of the chemoattractant cAMP with a reduced frequency of turning: evidence for a temporal mechanism in amoeboid chemotaxis. Cell Motil. Cytoskelet. 8:7-17. [DOI] [PubMed] [Google Scholar]
- 42.Wessels, D., D. R. Soll, D. Knecht, W. F. Loomis, A. DeLozanne, and J. Spudich. 1988. Cell motility and chemotaxis in Dictyostelium amoebae lacking myosin heavy chain. Dev. Biol. 128:164-177. [DOI] [PubMed] [Google Scholar]
- 43.Wessels, D., E. Voss, N. Von Bergen, R. Burns, J. Stites, and D. R. Soll. 1998. A computer-assisted system for reconstructing and interpreting the dynamic three-dimensional relationships of the outer surface, nucleus and pseudopods of crawling cells. Cell Motil. Cytoskelet. 41:225-246. [DOI] [PubMed] [Google Scholar]
- 44.Wessels, D., H. Vawter-Hugart, J. Murray, and D. R. Soll. 1994. Three dimensional dynamics of pseudopod formation and the regulation of turning during the motility cycle of Dictyostelium. Cell Motil. Cytoskelet. 27:1-12. [DOI] [PubMed] [Google Scholar]
- 45.Wessels, D., H. Zhang, J. Reynolds, K. Daniels, P. Heid, S. Liu, A. Kuspa, G. Shaulsky, W. F. Loomis, and D. R. Soll. 2000. The internal phosphodiesterase RegA is essential for the suppression of lateral pseudopods during Dictyostelium chemotaxis. Mol. Biol. Cell 11:2803-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wessels, D., J. Murray, and D. R. Soll. 1992. Behavior of Dictyostelium amoebae is regulated primarily by the temporal dynamics of the natural cAMP wave. Cell Motil. Cytoskelet. 23:145-156. [DOI] [PubMed] [Google Scholar]
- 47.Wessels, D., J. Reynolds, O. Johnson, E. Voss, R., Burns, K. Daniels, E. Garrard, T. O'Hallaran and D. R. Soll. 2000. Clathrin plays a novel role in the regulation of cell polarity, pseudopod formation, uropod stability and motility in Dictyostelium. J. Cell Sci. 113:26-36. [DOI] [PubMed] [Google Scholar]
- 48.Wessels, D., N. Schroeder, E., Voss, A. Hall, J. Condeelis, and D. R. Soll. 1989. cAMP mediated inhibition of intracellular particle movement and actin reorganization in Dictyostelium. J. Cell Biol. 109:2841-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilkins, A., and R. H. Insall. 2001. Small GTPase in Dictyostelium. Trends Genet. 17:41-48. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, H., D. Wessels, P. Fey, K. Daniels, R. Chisholm, and D. R. Soll. 2002. Phosphorylation of the myosin regulatory light chain plays a role in cell motility and polarity during Dictyostelium chemotaxis. J. Cell Sci. 115:1733-1747. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, H., P. Heid, D. Wessels, K. Daniels, T. Pham, W. F. Loomis, and D. R. Soll. 2003. Constitutively active protein kinase A disrupts motility and chemotaxis in Dictyostelium discoideum. Eukaryot. Cell 2:62-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zigmond, S. H. 1977. Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factor. J. Cell Biol. 75:606-616. [DOI] [PMC free article] [PubMed] [Google Scholar]