Abstract
Kinesins are motor proteins which are classified into 11 different families. We identified 11 kinesin-like proteins in the genome of the filamentous fungus Aspergillus nidulans. Relatedness analyses based on the motor domains grouped them into nine families. In this paper, we characterize KipB as a member of the Kip3 family of microtubule depolymerases. The closest homologues of KipB are Saccharomyces cerevisiae Kip3 and Schizosaccharomyces pombe Klp5 and Klp6, but sequence similarities outside the motor domain are very low. A disruption of kipB demonstrated that it is not essential for vegetative growth. kipB mutant strains were resistant to high concentrations of the microtubule-destabilizing drug benomyl, suggesting that KipB destabilizes microtubules. kipB mutations caused a failure of spindle positioning in the cell, a delay in mitotic progression, an increased number of bent mitotic spindles, and a decrease in the depolymerization of cytoplasmic microtubules during interphase and mitosis. Meiosis and ascospore formation were not affected. Disruption of the kipB gene was synthetically lethal in combination with the temperature-sensitive mitotic kinesin motor mutation bimC4, suggesting an important but redundant role of KipB in mitosis. KipB localized to cytoplasmic, astral, and mitotic microtubules in a discontinuous pattern, and spots of green fluorescent protein moved along microtubules toward the plus ends.
Eukaryotic cells employ a variety of different motors in dynamic processes. Among these are normally one or several dyneins, several myosins, and a large number of kinesins. Kinesins are molecular motors which interact with the microtubule (MT) cytoskeleton. The first kinesin to be identified was isolated from the giant axon of the squid and has since been characterized in many eukaryotes (17, 42). This family of motors has been designated conventional kinesin. Proteins of this family (kinesin heavy chain) convert chemical energy into mechanical force. They are able to move processively, with step sizes of 8 nm, along MTs and generate forces of 4 to 10 pN. They consist of highly conserved globular motor domains, located at the N terminus of the protein, which undergo a small conformational change upon ADP-ATP exchange. The motor domain is attached to a stalk, which consists of extended regions that are predicted to mediate dimerization. This dimerization ensures that the entire motor molecule remains attached once one motor domain detaches and performs a new step. This model was described as the hand-over-hand model (36). Despite our excellent knowledge of biochemical and structural details, little is known about the actual cargoes which are transported within the cell and thus about the biological function of the motors. In filamentous fungi such as Neurospora crassa and Aspergillus nidulans, which rely on long-distance organelle movement along MTs to achieve fast tip growth, conventional kinesin mutants are impaired in hyphal extension, suggesting that conventional kinesin is involved in the secretion of cell wall components (34, 38). Since the first discovery of conventional kinesin, many other kinesins and kinesin-like proteins have been described, and they are currently grouped into 11 different families, 2 of which are the closely related KinI and Kip3 families (37).
The biological function of the KinI family members is less clear than that of conventional kinesin. The KinI family received this nomenclature due to the location of the motor domain and the structure of the protein (33). Kinesins of this family are monomeric proteins and are not able to move along MTs in the conventional sense, but instead catalyze the depolymerization of MTs in vivo and in vitro (6, 18, 21, 29). In mammals a KinI protein, CgMCAK, localizes to the kinetochores in early prophase, and an MCAK deficiency results in chromosome segregation defects. This may be explained through an altered MT depolymerization rate. Overexpression of the gene resulted in the depolymerization of cytoplasmic and spindle MTs, suggesting roles for this kinesin family outside of mitosis (25).
Recently, two kinesins, Klp5 and Klp6, which have catalytic properties similar to those of KinI kinesins, were characterized in the yeast Schizosaccharomyces pombe (45, 46). However, their structure and activity led to the definition of a new family named Kip3, with Saccharomyces cerevisiae Kip3 as the founding member (22, 46). In S. cerevisiae, Kip3 is involved in nuclear migration in preparation for mitosis (5, 7, 27). Interphase nuclei are pushed around in the mother cell through growing and shrinking MTs emanating from the spindle pole body. Prior to mitosis, nuclei move towards the budding neck. This first movement depends on the function of Kip3, whereas the subsequent distribution of the two daughter nuclei is dependent on cytoplasmic dynein, MTs, and cortex-associated proteins (26, 28, 39, 40). MTs emanate from the spindle pole body and grow towards the cortex. Several proteins which are delivered to the cortex and in turn mediate contact between astral MTs and the cortex are associated with the growing plus ends (14, 24). A deletion of Kip3 from S. cerevisiae did not impair its vegetative growth and caused only a slight increase in binucleate mother cells at a low temperature. These effects were much stronger in dynein (dyn1) deletion strains at a low temperature and were largely increased in kip3 dyn1 double mutants at a high temperature. These results suggested that Kip3 is responsible for spindle positioning in the absence of dynein and thus serves overlapping functions with this motor (4, 5). In Schizosaccharomyces pombe, Klp5 and Klp6 are structurally very similar, and the deletion of either one or of both is not lethal (46). In contrast to the case for S. cerevisiae kip3 mutants, nuclear migration is not affected in Δklp5 or Δklp6 strains. However, MTs are stabilized in both fungi, and mitosis and meiosis are impaired in Schizosaccharomyces pombe (45). Klp5 and Klp6 are required for normal chromosome movement in prometaphase, although this function is not essential for successful mitosis. On the other hand, the two kinesins are essential for meiosis. Klp5/6-green fluorescent protein (GFP) fusion proteins localized to spindle and cytoplasmic MTs with no bias to either the plus or the minus end of the filaments (46). The studies of KinI kinesin family members in different organisms demonstrate that despite similar biochemical properties, the cellular processes affected may be different.
Another well-characterized family of mitotic kinesin motors is the BimC family. The founding member of this family was discovered in A. nidulans by a screen for temperature-sensitive lethal mutations (30). Cloning of the bimC (blocked in mitosis) gene revealed that it encodes a kinesin motor protein (8). The bimC4 mutant phenotype at a restrictive temperature is characterized by a block in spindle pole separation and thus the formation of monopolar rather than bipolar spindles. The BimC motor is a homotetramer which cross-bridges antiparallel MTs and produces a force towards the spindle poles. A motor with an opposing force is KlpA, a member of the C-terminal motor domain kinesin family. The deletion of this gene from the genome of A. nidulans causes a suppression of the bimC4 mutation (31).
We studied the Kip3-like motor KipB of A. nidulans to investigate its possible role in nuclear migration and found that this motor is required for spindle positioning within hyphal compartments, for the integrity of the MT cytoskeleton, and for proper chromosome distribution during mitosis.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
Supplemented minimal and complete media for A. nidulans were prepared as described previously, and standard strain construction procedures were used (16). A list of A. nidulans strains used for this study is given in Table 1. Standard laboratory Escherichia coli strains (XL-1 Blue and Top 10 F′) were used. The plasmids and cosmids used are listed in Table 2.
TABLE 1.
A. nidulans strains used for this study
| Strain | Genotype | Source or reference |
|---|---|---|
| SRF200 | pyrG89 ΔargB::trpCΔB pyroA4 veA1 | 20 |
| GR5 | pyrG89 wA3 pyroA4 veA1 | 43 |
| FGSC 26 | biA1 veA1 | FGSC, Kansas |
| GFP-tubA | pyrG89 GFP::tubA::pyr4 pyroA4 wA3 | 15 |
| MO62 | bimC4 argB2 nicA2 | V. Efimov (Piscataway, N.J.) |
| RSM011 | pabaA1 yA2 ΔargB::trpCΔB trpC801 veA1 | 41 |
| RMS012 | Diploid; biA1 ΔargB::trpCΔB methG1 veA1 trpC801/pabaA1 yA2 ΔargB::trpCΔB trpC801 veA1 | 41 |
| SJW02 | GFP-tubA × RMS011 progeny strain; alcAp::gfp::tubA wA3 pyroA4 veA1 ΔargB::trpCΔB | J. Warmbold, Marburg, Germany |
| SPR2 | GR5 transformed with pPR11 and pRG1; alcAp::kipB::sgfp wA3 pyroA4 veA1 | This study |
| SPR13 | SPR1 × RMS011 progeny strain; pabaA1 ΔkipB::argB ΔkipB | This study |
| SPR22 | SPR1 × RMS011 progeny strain; ΔkipB::argB ΔkipB | This study |
| SPR30 | ΔkipB × GFP-tubA progeny strain; GFP::tubA::pyr4 pyroA4 ΔkipB::argB | This study |
| SPR55 | Diploid strain; ΔkipB/kipB; progeny of cross between GR5 and SPR13 | This study |
| SPR60 | Diploid strain; ΔkipB/ΔkipB; progeny of cross between SPR22 and SPR13 | This study |
| SPR88 | bimC4 ΔkipB::argB nicA2; double mutant; progeny of MO62 × SPR13 | This study |
| SPR90 | bimC4 ΔkipB::argB nicA2; double mutant; progeny of MO62 × SPR13 | This study |
| SPR93 | pabaA1 yA2 trpC801 veA1 (RMS011 transformed with pDC1); wild type | This study |
| SPR96 | SRF200 transformed with pPR38, homologous integration; alcAp::sgfp::kipB ΔargB::trpCΔB pyroA4 veA1 | This study |
| SPR98 | SRF200 transformed with pPR38, ectopic integration; alcAp::sgfp::kipB ΔargB::trpCΔB pyroA4 veA1 | This study |
| SPR99 | SJW02 (GFP-tubA) transformed with pPND1 and pDC1, ectopic integration; alcAp::mRFP1::kipB pyroA4 veA1 | This study |
TABLE 2.
Plasmids used for this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pBluescript KS(−) | Cloning vector | Invitrogen (NV Leek, The Netherlands) |
| pCR2.1-TOPO | TA cloning vector for cloning of PCR fragments | Invitrogen (NV Leek, The Netherlands) |
| pCMB17apx | alcAp::GFP pyr4; used for N-terminal fusion of GFP to proteins of interest | V. Efimov (Piscataway, N.J.) |
| pDC1 | A. nidulans argB gene in pIC20R | 2 |
| pHW-arg | KpnI-XhoI-released argB from pDC1 inserted into pBluescript KS(−) | 44 |
| pRG1 | Contains N. crassa pyr4 gene | 43 |
| pPR5 | kipB containing cosmid (BamHI short fragment, 3.3 kb) from pU1 library | This study |
| pGR1 | Subclone BamHI short fragment of kipB from pPR5 into pBluescript KS(−) | This study |
| pPR7 | 3.6-kb SacI-BamHI fragment of kipB cloned into pBluescript KS(−) | This study |
| pPR11 | alcAp::kipB::sgfp (kipB BamHI fragment from pPR7) cloned into pBluescript KS(−) | This study |
| pPR13 | argB gene with BamHI sites cloned into BglII sites of kipB from pPR7 (disruption construct) | This study |
| pPR17 | kipB containing cosmid (entire gene) from pU1 library | This study |
| pPR19 | 4.3-kb kipB SacI subclone from pPR17 cloned into pBluescript KS(−) | This study |
| pPR38 | 1.2-kb kipB fragment from ATG, PCR product with AscI and PacI sites from pPR19 inserted into pCMB17apx | This study |
| pPND1 | GFP replaced with mRFP1 (KpnI-AscI) into pPR38, alcA(p)::mRFP1::kipB, pyr4 | This study |
Molecular techniques.
Standard DNA transformation procedures were used for A. nidulans (48) and E. coli (35). For PCR experiments, standard protocols were applied and a capillary rapid cycler (Idaho Technology, Idaho Falls, Idaho) was used for the reaction cycles. DNA sequencing was done commercially (MWG Biotech, Ebersberg, Germany). Genomic DNA was extracted from the fungus with a DNeasy plant mini kit (Qiagen, Hilden, Germany). RNA was isolated by the use of TRIzol from GibcoBRL (Paisley, Scotland, United Kingdom) according to the manufacturer's protocols. DNA and RNA analyses (Southern and Northern hybridizations) were performed as described previously (35).
Cloning of the ΔkipB disruption construct.
A BamHI-released argB fragment from pHW-arg (44) was cloned into the BglII sites of the kipB gene in order to disrupt the motor domain of the protein. Homologous integration of the construct (pPR13) in A. nidulans would lead to a deletion of 18 bp of the kipB gene. In addition to the deletion, the argB gene disrupts the coding region of kipB. The disruption construct was transformed into the arginine-auxotrophic A. nidulans strain SRF200.
GFP labeling of KipB (pPR11 and pPR38).
For the C-terminal construct of the KipB::GFP fusion protein, a 3.3-kb BamHI fragment derived from pPR5 and including kipB was cloned into pBluescript KS(−). This led to a truncated version of the protein in which the first 690 amino acids were fused to GFP. The alcA promoter was cloned upstream of kipB as a KpnI-XhoI fragment and sgfp was cloned downstream as an XbaI restriction fragment into the polylinker of pBluescript KS(−) (pPR11). For the N-terminal construct, a 1.2-kb (starting from ATG) fragment of kipB from pPR19 was amplified with the primers kipB-Ngfp-fwd (5′-ATGGGGGCCTCAAGCGAC-3′), with an AscI site added at the 5′ end, and kipB-Ngfp-rev (5′-CGCTTTGAGTTCGTTAATTAATGCC-3′) and was cloned into the pCMB17apx plasmid (kindly provided by V. Efimov, Robert Wood Johnson Medical School, Piscataway, N.J.) at AscI-PacI sites (pPR38). Homologous recombination of this construct into the kipB locus led to an N-terminal GFP fusion of the entire KipB protein under the control of the alcA promoter and a truncated 5′ region under the control of the natural promoter.
mRFP1 labeling of KipB (pPND1).
mRFP1 was amplified with the primers KpnI_mRFP1_fwd (5′-CGGTACCATGGTCTCCTCCGAGG-3′) and AscI_mRFP1_rev (5′-CGGCGCGCCGGCGGTGGA-3′) and was cloned into the KpnI-AscI sites of the pPR38 plasmid, replacing the sequence for GFP. This resulted in an N-terminal mRFP1 fusion of 1.2 kb (starting from ATG) of KipB under the control of the alcA promoter.
Fluorescence microscopy and live cell image acquisition and analysis.
Cells were grown in glass-bottomed dishes (World Precision Instruments, Berlin, Germany) in 2 ml of minimal medium containing glycerol, pyridoxine, and/or arginine or minimal medium containing 2% ethanol, pyridoxine, and/or arginine. The cells were incubated at 30°C for 15 h and images were captured at room temperature with an Axiophot microscope (Zeiss, Jena, Germany), a Planapochromatic 63× or 100× oil immersion objective lens, and a 50-W Hg lamp. Images were collected and analyzed with a Hamamatsu Orca ER II camera system and Wasabi software (version 1.2). Time-lapse series were obtained with an automated Wasabi program that acquires series of images with 3, 10, or 20 s of pause time, 0.5 or 0.75 s of exposure time, and about 40 exposures in a sequence.
Nucleotide sequence accession number.
The sequence of A. nidulans kipB is available in the EMBL database under accession number AJ620863.
RESULTS
KipB is a member of the Kip3 family of kinesins.
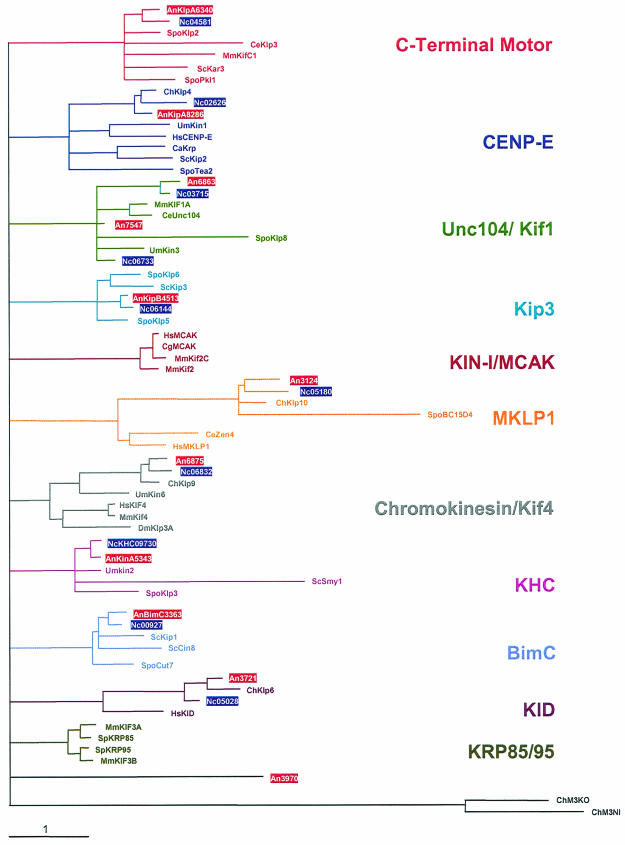
We analyzed the genomic A. nidulans database at Cereon Genomics LLC (Cambridge, Mass.) and at the Whitehead Institute for kinesin-like proteins and used the motor domain of the A. nidulans conventional kinesin for the search (http://www-genome.wi.mit.edu/annotation/fungi/aspergillus/), by which we identified 11 putative kinesin motors. For comparison, in S. cerevisiae there are six kinesins and in Schizosaccharomyces pombe there are nine (37). The motor domains of A. nidulans and other known kinesins were used to unravel their relatedness (Fig. 1). The 11 A. nidulans kinesins grouped into 9 of the 11 families. Two kinesins were found in the Unc104 family and one did not fall into any of the known families. The 10 N. crassa kinesins were closely related to the A. nidulans proteins.
FIG. 1.
Relatedness analysis of the 11 A. nidulans and 10 N. crassa kinesins. A most likely phylogenetic tree for 74 kinesins was built with Treepuzzle (http://www.tree-puzzle.de/), using a maximum-likelihood algorithm. For an evaluation of the statistical significance of the topology, 25,000 replicating puzzle steps were performed. The substitution model Whelan-Goldman 2000 was used because it produced a consensus tree which was in good agreement with the published data (37). For the construction of the tree, we chose fungal kinesin sequences and additional kinesins from other organisms that are characteristic for the different families. Note that the A. nidulans exon-intron borders were only experimentally determined for BimC, KlpA, KinA, KipA, and KipB. In the case of N. crassa, only the conventional kinesin NcKHC has been analyzed experimentally. The other primary structures of the proteins of A. nidulans and N. crassa are based on the predictions in the annotation process at the Whitehead Institute database. The N. crassa Kip3 homologue was annotated manually. The origins of the kinesin sequences were as follows: An, A. nidulans; Nc, N. crassa; Spo, Schizosaccharomyces pombe; Ce, Caenorhabditis elegans; Mm, Mus musculus; Um, Ustilago maydis; Sc, S. cerevisiae; Ch, Cochliobolus heterostrophus; Hs, Homo sapiens; Ca, Candida albicans; Dm, Drosophila melanogaster; Cg, Cricetulus griseus; Sp, Strongylocentrotus purpuratus. The A. nidulans and N. crassa sequences are boxed.
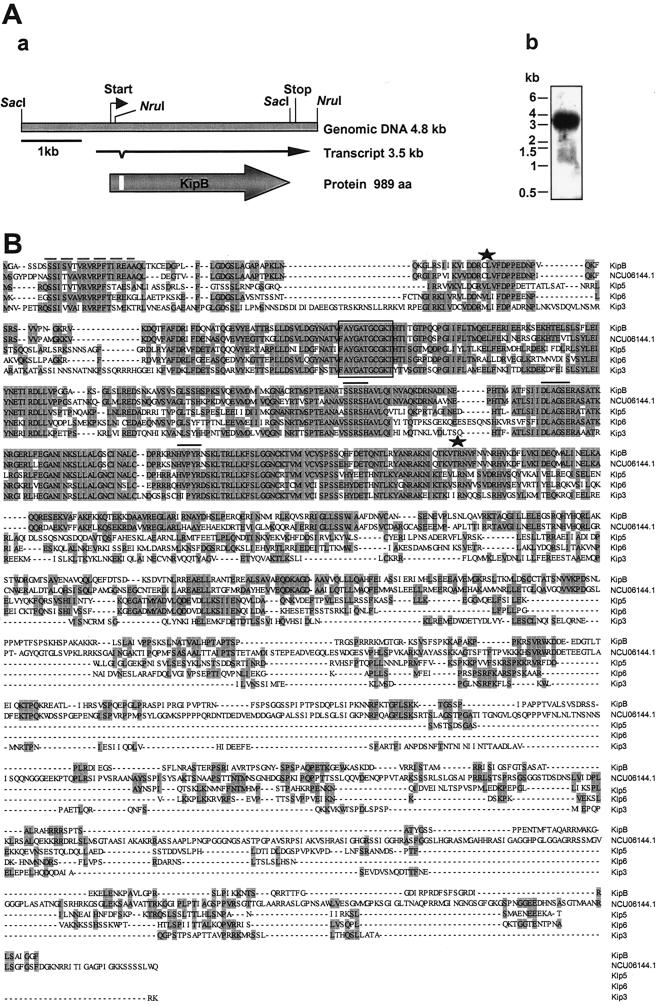
We have characterized KipB as a member of the Kip3 family. The sequence of the kipB gene, localized on chromosome III, was confirmed by sequencing a 4.3-kb SacI restriction fragment obtained from a kipB-carrying cosmid. To determine the intron-exon borders, we amplified a corresponding cDNA by reverse transcription-PCR. A comparison of the genomic DNA and the cDNA sequence revealed one 56-bp intron, located at the N terminus of the predicted protein (Fig. 2A). The position of the intron was conserved compared with those of Schizosaccharomyces pombe klp5 and klp6. A kipB-specific probe hybridized in a Northern blot to a 3.5-kb transcript. The putative KipB protein consists of 989 amino acids, with a predicted molecular mass of 108.7 kDa and a calculated isoelectric point of 9.9. The motor domain starts about 60 amino acids downstream of the initiation codon. A comparison with other Kip3-like proteins revealed 45.7% identity with the N. crassa protein, 33.5 and 35.3% identity with Schizosaccharomyces pombe Klp5 and Klp6, respectively, and 30.4% with S. cerevisiae Kip3. The identities between the proteins were much higher for the motor domains (e.g., 85% with the N. crassa protein), which include ATP- and MT-binding sites (Fig. 2B). In addition, the N-terminal region contains a sequence of 18 amino acids which is conserved among the compared proteins. The significance of the 18-amino-acid motif is yet unknown (46). Outside of the motor domains, the Kip3-like proteins do not share significant sequence similarities. Similar to the proteins from S. cerevisiae and Schizosaccharomyces pombe, we detected short regions with a significant probability of a coiled-coil formation. The positions of these regions are conserved (7, 46).
FIG. 2.
Characterization of A. nidulans kinesin kipB. (A) Panel a shows a schematic of the kipB locus. The open reading frame and the transcript are indicated by arrows, in which the intron position is marked. Panel b shows a Northern blot analysis of RNA isolated from mycelia of strain FGSC26, incubated at 37°C, and harvested after 16 h of vegetative growth. The RNA was isolated from the mycelia and hybridized to a kipB-specific probe. (B) Alignment of A. nidulans KipB with homologous kinesin sequences from N. crassa (NCU06144.1), Schizosaccharomyces pombe (Klp5 and Klp6), and S. cerevisiae (Kip3). The alignment was done with DNAstar by using MEGALIGN (CLUSTAL) with a window size of 5 and a gap length penalty of 10. The beginning and end of the highly conserved motor domains are indicated by asterisks above the sequences. The ATP-binding motif is boxed and the putative MT binding site is indicated by lines above the sequences. The 18-amino-acid motif at the N terminus is highlighted by a dashed line above the sequences.
Disruption of kipB affects MT stability.
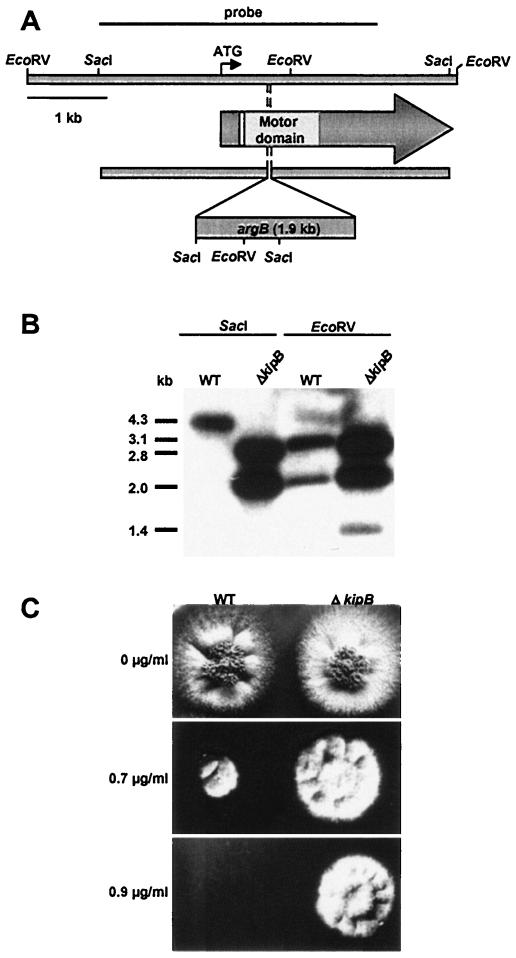
The cellular function of kipB was investigated through the construction of a null mutant by homologous recombination (see Materials and Methods). The motor domain was disrupted by the insertion of the nutritional marker gene argB (Fig. 3A). Colony-purified arginine-prototrophic transformants of strain SRF200 were tested for the integration event at the kipB locus by Southern blot analysis (Fig. 3B). Two of 43 strains harbored the knockout situation. In a cross with an argB deletion mutant, the kipB wild-type strain (RMS011) and the kipB disruption (ΔkipB) cosegregated with the argB marker. Two strains obtained by the cross (SPR13 and SPR22) were chosen for further analysis with respect to polarized growth, mitochondrial movement, and nuclear distribution. We did not detect any differences from the wild type, suggesting that KipB is not involved in those processes or that other kinesin motors are able to substitute for the function of KipB.
FIG. 3.
Disruption of the kipB gene. (A) Schematic of the strategy. The nutritional marker gene argB was inserted into kipB and thereby deleted 18 bp. (B) Southern blot analysis of a ΔkipB disruption strain (SPR13). Genomic DNAs of the wild type (FGSC26) and the kinesin disruptant strain were isolated, digested with SacI (left) and EcoRV (right), separated in a 1% agarose gel, blotted, and hybridized with the probe shown in panel A. (C) Effect of different benomyl concentrations on colony growth of wild type (RMS011) and ΔkipB disruptant (SPR13). Strains were inoculated on agar plates at 37°C and supplemented with the indicated benomyl concentrations dissolved in dimethyl sulfoxide.
As a first step to investigate the role of KipB in the organization of the MT cytoskeleton, we analyzed the sensitivity of cells to the MT-destabilizing drug benomyl. While the wild type failed to grow at concentrations from 0.75 to 0.9 μg/ml, the ΔkipB mutant was still able to grow and was only inhibited at concentrations above 0.95 μg/ml (Fig. 3C). The difference in benomyl sensitivity was independent of the growth temperature. These results suggest that the kinesin KipB destabilizes MTs in A. nidulans.
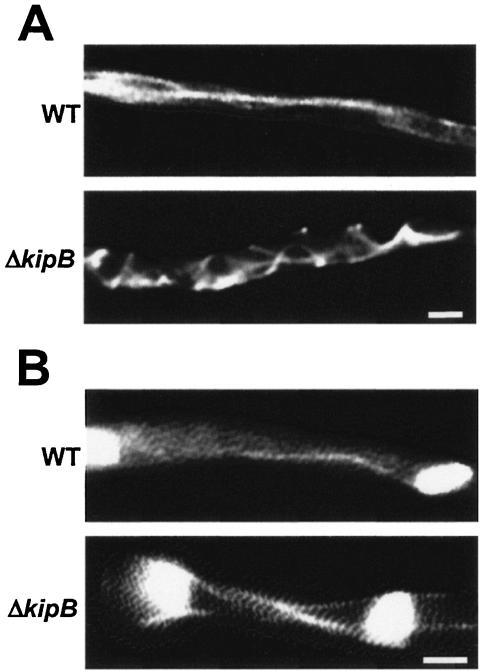
To directly study the effect of the ΔkipB mutation on MT stability, we compared the MT cytoskeletons of the wild type and a ΔkipB mutant strain (SPR30) in which alpha-tubulin was labeled with GFP. In single-cell yeasts such as S. cerevisiae and Schizosaccharomyces pombe, the effects of mutations in genes encoding MT-destabilizing components can be easily documented since the cytoplasmic or astral MTs in those strains elongate even after contacting the cortex. As a result, MTs appear bent along the cortex (3). For filamentous fungi, differences in the cytoplasmic MT network have not yet been detected (34). Likewise, we observed only a slight effect in the ΔkipB mutant in comparison to wild-type interphase cells. The MTs appeared to be more curved, which could have been due to continuous growth after reaching the hyphal tip (Fig. 4A). However, it was difficult to follow a single MT along the entire length through the compartment. The level of alpha-tubulin was not higher in the deletion mutant than in the wild-type strain (data not shown). A striking difference in MT organization became obvious when we looked at mitotic cells (Fig. 4B). During mitosis, cytoplasmic MTs are almost entirely depolymerized to provide tubulin subunits for the assembly of the mitotic spindle and the formation of astral MTs (32). In the ΔkipB mutant strain, however, several cytoplasmic MTs were observed during mitosis. In addition, astral MTs grew into long filaments.
FIG. 4.
Morphology and MT organization in ΔkipB mutant strain and wild type. MTs were observed as α-tubulin-GFP fusions by fluorescence microscopy (see Materials and Methods). Top rows, wild type; bottom rows, ΔkipB strain. (A) Cytoplasmic MTs. In the wild type, they are long and straight, while they display a curved pattern in the ΔkipB mutant. (B) Mitotic spindles. In the wild-type spindle, the cytoplasmic MT remained, while in the ΔkipB strain, three filaments are visible. Bar, 5 μm.
KipB is involved in the positioning of mitotic spindles.
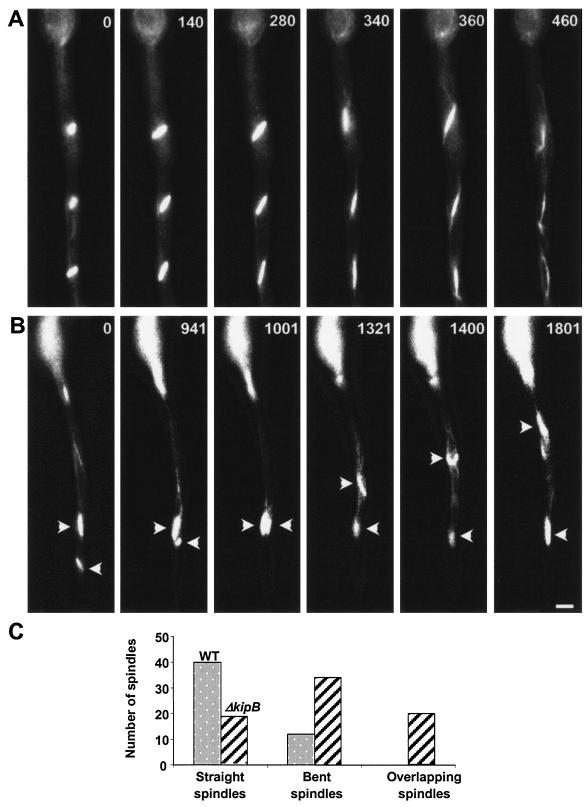
During the course of the analysis of mitotic cells, we noticed that the spindles were not properly distributed in the hyphae (Fig. 5). Mitoses in the wild type are almost synchronous in one hyphal compartment (see Video S1 in the supplemental material), and since nuclei are evenly spaced before entering mitosis, mitotic spindles remain distributed and fixed at their positions. In contrast, mitotic spindles of ΔkipB mutants were highly mobile, overpassed each other, and frequently moved long distances through the cytoplasm (see Video S2 in the supplemental material). However, interphase nuclei were again evenly distributed. During their migration process, 64% of the spindles had a bent shape (53 spindles from two independent strains were analyzed). For the wild type, such a shape was only observed for 23% of the spindles (52 spindles analyzed).
FIG. 5.
Positioning of mitotic spindles in the wild type and the ΔkipB disruptant. Images from a time-lapse series are displayed (times are indicated, in seconds, in the upper right corner of each panel). (A) Wild-type synchronized mitoses, with evenly distributed spindles along the length of the hypha. (B) Mitoses in the ΔkipB mutant strain, with defects in spindle positioning and morphology (sharp angled bow-like structures) and with a high level of spindle mobility through the cytoplasm, which leads to overlapping of the spindles (arrows). Bar, 5 μm. (C) Quantification of different spindle morphologies and spindle behavior in the wild type and the ΔkipB mutant strain (also see Videos S1 and S2 in the supplemental material).
kipB disruption causes a delay in mitotic progression.
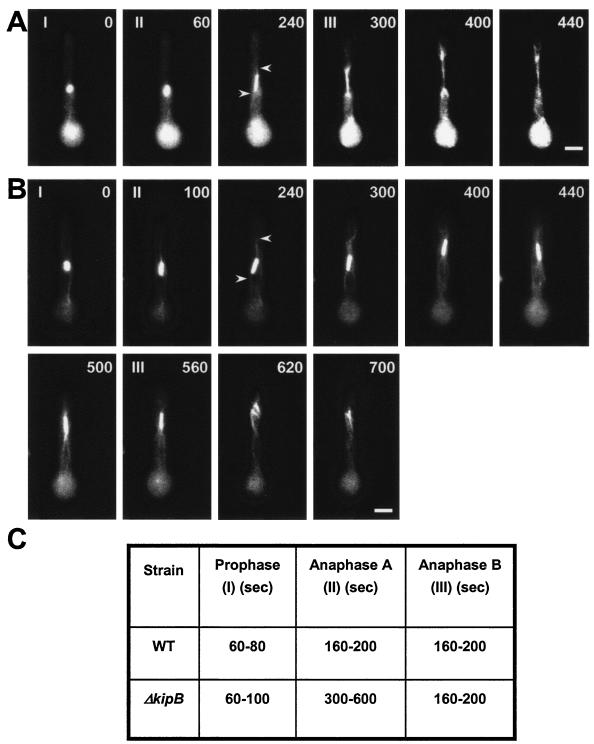
To determine if any stage of mitosis is affected in the ΔkipB mutant, we compared spindle behavior in both the wild type and the mutant by using live cell imaging. In fungi, mitosis starts with an early prophase, during which the spindle pole bodies divide and separate towards opposite sites, generating a half-spindle. During metaphase, the bipolar spindle is completely formed and the chromosomes are attached to the spindle MTs via the kinetochores. The chromosomes are usually scattered over the middle one-third to one-half of the spindle rather than arranged in a metaphase plate. Anaphase occurs in two stages. During anaphase A, the chromatids are separated and migrate asynchronously along the entire length of the spindle to the respective spindle poles, and the astral MTs start to be developed. During anaphase B, the spindle elongates rapidly between the two spindle pole bodies and the astral MTs reach their maximal length (1). The first stage (prophase to metaphase) has a short duration (60 to 80 s). The mutant exhibited a much longer anaphase A, with a duration of 400 to 600 s, than did the wild type (160 to 200 s) (Fig. 6; also see Videos S3 [wild type] and S4 [mutant] in the supplemental material).
FIG. 6.
kipB disruption causes a delay in mitotic progression. The images show time-lapse analyses of mitosis in germlings of the wild type (strain GFP-tubA) (see Video S3 in the supplemental material) (A) and the ΔkipB mutant (SPR30) (see Video S4 in the supplemental material) (B). MTs were labeled with GFP. The stages of mitosis are indicated in the upper left corner of the pictures. I, prophase to metaphase (short spindle); II, anaphase A (spindle elongates very slowly, with the appearance of astral MTs [indicated by arrows]); III, anaphase B (spindle elongates rapidly and doubles or triples in length). The cells were grown overnight at 30°C and were observed at room temperature. Images were taken every 20 s, and a selection of them are displayed here. The time points (in seconds) are indicated in the upper right corner of the pictures. Bar, 5 μm. (C) Summary of the time intervals of different mitotic phases in the wild-type and kipB mutant strains (see Videos S3 [wild type] and S4 [mutant] in the supplemental material).
Genetic interaction of ΔkipB with bimC4.
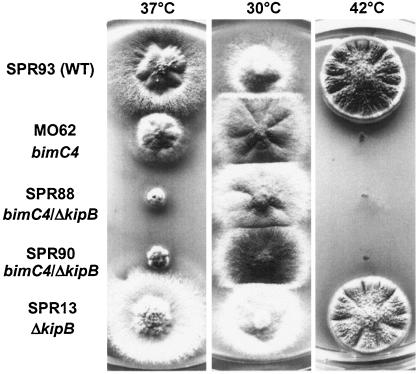
Since KipB appears to play a role in mitosis, we tested the ΔkipB mutation for its genetic interaction with a temperature-sensitive kinesin motor mutation, the bimC4 mutation. We constructed two bimC4 ΔkipB double mutant strains and compared their growth with the growth of strains with corresponding single mutations (Fig. 7). We performed the assay at temperatures that were permissive (30°C), restrictive (42°C), and semipermissive (37°C) for the bimC4 mutant. We found that the double mutant displayed increased temperature sensitivity compared with the two single mutants. At the intermediate temperature, the double mutant had a stronger growth defect than the bimC4 mutant, suggesting that there is a genetic interaction between the two kinesins (Fig. 7).
FIG. 7.
Disruption of kipB is synthetically lethal with bimC4. Comparison of colony growth of SPR93 (wild type), MO62 (bimC4), SPR13 (ΔkipB), and two double mutants (SPR88 and SPR90) at 37 and 42°C for 3 days and 30°C for 5 days.
Gene dosage of kipB determines frequency of chromosome loss in a diploid strain.
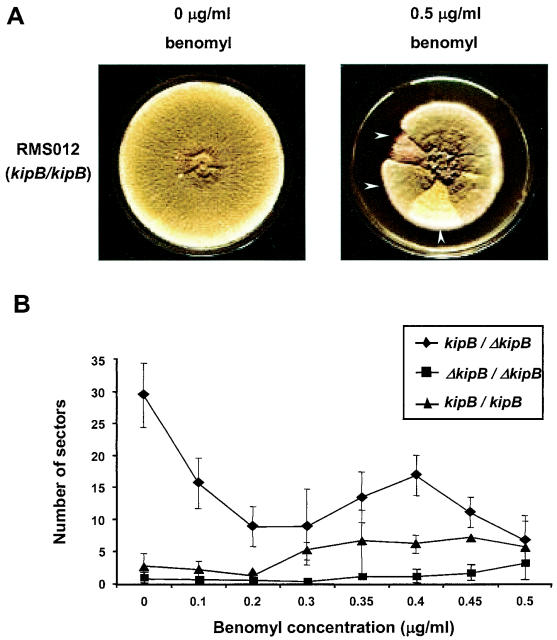
To further investigate the effect of the ΔkipB mutation on nuclear division, we compared the wild type and the ΔkipB mutant for chromosome loss during mitosis. Since conidiospores of A. nidulans are haploid, any loss of one of the eight chromosomes would lead to the death of the spore and thus to a germination defect. However, we found no difference in the viabilities of the conidiospores harvested from the two strains, suggesting that there is a more subtle effect from the mutation. Therefore, we studied putative chromosome loss by using diploid strains (Fig. 8A). These strains can be obtained from conidiospores that are generated in a heterokaryon. Diploid spores are formed at a low frequency and can be stably maintained when a selection pressure is applied. If these spores lose one chromosome during mitosis, they reduce the number of chromosomes to the haploid set, and in the absence of the selection pressure, can grow to mature colonies (19). If strains with green spores are used in combination with strains with yellow spores for the construction of the diploids or if strains with white spores are used in combination with strains with yellow spores, haploidization can be easily visualized via the colors of the colonies and different sectors. Chromosome loss can be stimulated by the application of low concentrations of benomyl. We constructed a heterozygous diploid strain between the ΔkipB mutant (yellow spores; SPR13) and a wild-type strain (white spores; GR5) and a homozygous ΔkipB diploid strain (yellow and green spores for the haploid parents) (SPR13 and SPR22) and compared their frequencies of haploidization with that of a wild-type diploid strain (RMS012). The frequency of sectors with different colors was very low for the wild type and increased with increasing benomyl concentrations. In contrast, diploids that were heterozygous for the ΔkipB mutation (kipB/ΔkipB) (SPR55) formed more haploid sectors than the wild type. Interestingly, this frequency decreased with increasing benomyl concentrations. The ΔkipB homozygous diploid (ΔkipB/ΔkipB) (SPR60) appeared to have an even lower frequency of haploidization than the wild type (Fig. 8B).
FIG. 8.
Haploidization of diploids of kipB/kipB (wild type; RMS012), ΔkipB/kipB (heterozygous; SPR55), and ΔkipB/ΔkipB (homozygous; SPR60) organisms. (A) Colony growth on plates without benomyl (left) and with 0.5 μg of benomyl/ml (right). The arrows point to haploid sectors. (B) The numbers of haploid sectors were counted for the three strains denoted in panel A. For the homozygous ΔkipB strain, we analyzed 10 diploids, for the heterozygous strain we analyzed 6 diploids, and for the wild type, we analyzed 1 diploid. The average numbers of sectors are shown.
In contrast to the effect on mitosis and chromosome segregation in the diploid, we have no evidence for a role of KipB in meiosis. Crosses between different strains did not show any abnormalities, and the number of ascospores per ascus, as well as the viability of the spores, was not altered.
KipB localizes to mitotic, astral, and cytoplasmic MTs.
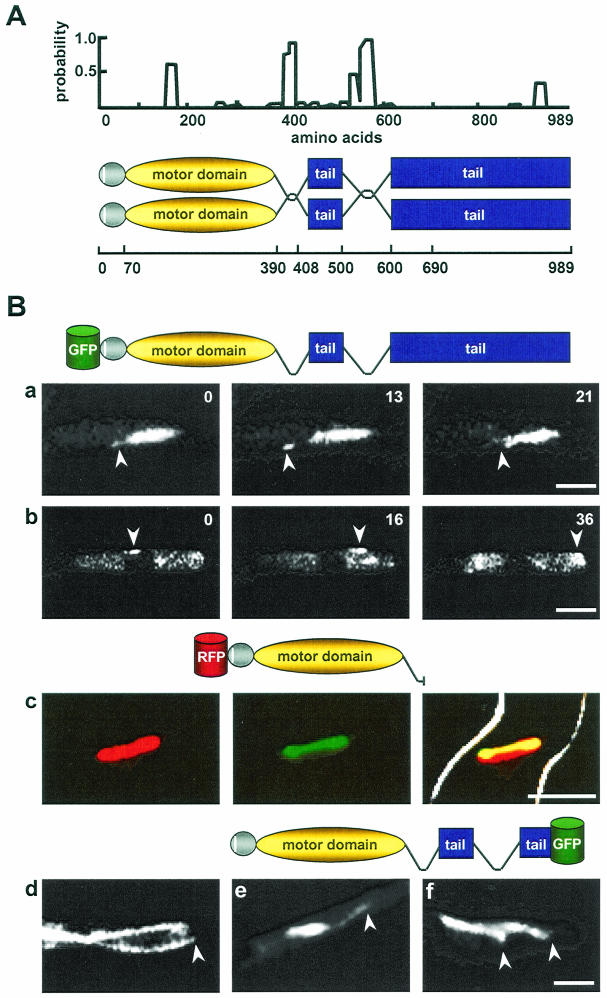
To localize KipB in A. nidulans, we constructed an N-terminal fusion protein with GFP. Approximately 1.2 kb from the 5′ region of the coding sequence was cloned behind GFP. The chimeric protein was expressed under the control of the inducible alcA promoter (pPR38). A. nidulans SRF200 was transformed with the circular plasmid pPR38 and transformants were screened for GFP fluorescence under inducing conditions (with threonine as a carbon source). These strains were analyzed for the integration of the plasmid, and a strain with a single ectopic integration as well as a strain with a single homologous integration was chosen for further analysis (with glycerol and/or ethanol as a carbon source) (Fig. 9). The ectopic copy resulted in an aberrant, nonfunctional fusion protein, whereas homologous integration resulted in a duplication of the region where the plasmid integrated. The full-length KipB protein was N-terminally tagged with GFP. Both strains displayed normal growth on plates. We found that the GFP-KipB protein localized to cytoplasmic MTs in interphase cells and to spindle and astral MTs during mitosis (Fig. 9; also see Videos S5 [spindle] and S6 [cytoplasmic] in the supplemental material). However, the protein displayed a discontinuous pattern along the MTs. The spots disappeared when benomyl was added (results not shown). Patches of GFP-KipB appeared to be aligned in the cell, and time-lapse studies revealed that the spots moved independently along MTs. In the hyphal tip, where MTs are oriented with their plus ends toward the cortex, the spots moved only toward the tip. In the middle and the rear of the hyphal tip compartment, where MT polarity is mixed (unpublished results), GFP-KipB spots moved in both directions (see Video S9 in the supplemental material). To discriminate between the possibilities that GFP-KipB spots are transported together with the growing plus end and that the spots move along MTs independently of the plus ends, we studied the dynamics of plus ends and compared them to the behavior of the GFP-KipB spots. Whereas the MT plus ends grew with a speed of 9 ± 3 μm/min, the GFP-KipB spots showed a larger range of speeds and no pronounced peak. This suggests that GFP-KipB moves toward the MT plus end.
FIG. 9.
Localization of GFP-KipB fusion proteins. (A) Coiled-coil prediction (23) for KipB (window size 14). A significant coiled-coil probability was found for amino acids 390 to 408 and 500 to 600. The schematic drawing shows the domains of the KipB protein, including globular heads (gray), the amino-terminal motif of 18 amino acids (white), the coils (curved lines), and the tail domain (blue). The amino acid positions are represented on the line below. (B) Localization of different GFP-KipB fusion proteins. (a and b) Time-lapse analysis of full-length KipB protein tagged N-terminally with GFP (see the schematic drawings of the different constructs above each series of photos). For strain SPR96, the images show the localization of GFP-KipB onto spindle and astral MTs, with arrows pointing to the plus ends of the astral MTs (see Video S5 in the supplemental material) (a) and spots of GFP-KipB moving onto cytoplasmic MTs (see Video S6 in the supplemental material) (b). (c and d) Colocalization between a truncated version of mRFP1-KipB and the mitotic spindles (see Video S8 in the supplemental material) for strain SPR98. Left, mRFP1-KipB; middle, α-tubulin-GFP; right, merged images of the first two photos. (d) Localization to cytoplasmic MTs. The arrow points to the plus end of the MT in the hyphal tip. (e and f) C-terminal fusion of KipB with GFP (see Video S7 in the supplemental material) in strain SPR2. The images show the localization onto mitotic and astral (arrow) MTs (e) and cytoplasmic MTs (arrows) (f). Bar, 5 μm.
In the case of the truncated GFP-KipB protein, a stronger GFP signal was obtained and the MTs were evenly stained (see Videos S7 and S8 in the supplemental material). We did not observe any abnormality of the MTs which could be caused by a dominant-negative effect of the truncated protein. A similar result was obtained with a construct in which GFP was fused to the C terminus of KipB, replacing about 300 amino acids (Fig. 9).
DISCUSSION
KipB is a member of the Kip3 kinesin family.
In this paper, we identified 11 kinesin motors for A. nidulans. This is in good agreement with the number of kinesins in other fungi, ranging from 6 in S. cerevisiae to 10 in N. crassa and 12 in Cochliobolus heterostrophus (37). Interestingly, two kinesins of A. nidulans and N. crassa were found that belonged to the Unc104 family, whose members are involved in vesicle transportation. This may reflect the importance of the requirement for fast tipward vesicle movement in fast-growing filamentous fungi. Not surprisingly, S. cerevisiae does not have any related kinesin in this family. In addition, S. cerevisiae lacks kinesins from the chromokinesin/Kif4, MKLP1, and KID families, whereas A. nidulans and N. crassa do have representatives in these groups. Since these kinesins are involved in mitotic processes, this may reflect different mechanisms of nuclear division for S. cerevisiae and the two filamentous fungi (37).
In this paper, we characterized a new kinesin, KipB, from A. nidulans. Several lines of evidence showed that it is involved in the turnover of all populations of cellular MTs, including interphase cytoplasmic, mitotic, and astral MTs: (i) ΔkipB mutants were less sensitive to the MT-destabilizing drug benomyl, (ii) the MT cytoskeleton of interphase cells appeared altered, (iii) the spindle positioning was affected, (iv) mitotic progression was delayed, (v) an increased number of cytoplasmic MTs remained intact during mitosis, (vi) the spindle morphology was altered, and (vii) ΔkipB heterozygous strains showed an increased instability of diploid nuclei. Its effect on MT stability and its sequence similarities to other motors in the databases place KipB into the Kip3 family, which thus supports the distinction between the KinI family, consisting of higher eukaryotic kinesin depolymerases, and the fungal Kip3 family.
KipB is involved in spindle positioning and mitosis.
In S. cerevisiae, Kip3 was shown to be involved in nuclear migration to the bud site in preparation for mitosis (7). As a consequence of this migration, the frequency of binucleate mother cells increases. When we looked at the nuclear distribution in germlings of an A. nidulans ΔkipB mutant, we could not find any difference in the distribution pattern. However, when we did time-lapse studies of mitosis using GFP-tagged tubulin strains, we realized that individual mitotic spindles moved within the cytoplasm, although most of the spindles were positioned as in the wild type. In wild-type cells, the spindles are likely to interact with the cortex through astral MTs. Cortical proteins such as ApsA are probably required for the contact (9-11, 47). In the ΔkipB mutant, astral MTs were stabilized and were likely to reach the cortex and subsequently extend further along it. They could then make contact with cortical proteins at a distance from their “own” attachment site, and pulling forces could then cause movement. The spindle movement phenotype was striking and raises the question of why we did not observe an effect on nuclear distribution in interphase cells. This finding suggests that besides nuclear separation through mitosis, other mechanisms exist which further distribute nuclei (47).
Besides the effect of the ΔkipB mutation on spindle positioning, we found a defect in mitosis. The separation of chromosomes requires the coordinated action of motor proteins and MT dynamics. We observed a delay in mitotic progression and an increase in bent mitotic spindles in ΔkipB mutant strains. Both effects may be explained by the effect of the motor deletion on the dynamics of MTs and thus a disruption of the balance of forces exerted on the kinetochores (12, 13, 45). The effect of the mutation on the integrity of the mitotic apparatus is also reflected by the results of the experiment with diploid strains. Heterozygous mutants displayed an increase in chromosome loss. Interestingly, homozygous ΔkipB diploids showed a reduced frequency of chromosome loss in comparison to kipB wild-type diploids. This suggests a gene dosage effect and could mean that haploidization after the initial loss of one chromosome requires a certain amount of the KipB protein to reduce the chromosome number to a haploid set. In the heterozygous situation, the initial chromosome loss might be very frequent, so a further reduction of the chromosome number would occur rapidly. Interestingly, the destabilization of MTs by the addition of increasing amounts of benomyl suppressed this effect. This suggests that the disassembly of MTs in the mitotic spindle and thus chromosome distribution are affected by different concentrations of KipB.
How to reach the end?
We studied the following three GFP constructs: an N-terminal fusion with the full-length KipB protein, an N-terminal fusion with a truncated version of KipB which only consisted of the first 408 amino acids from the N terminus, and a C-terminal GFP fusion, which deleted about 300 amino acids from KipB. Whereas the truncated versions stained the entire length of the filaments, the full-length protein predominantly displayed a discontinuous distribution along MTs, and the GFP spots moved along the filaments. The question was raised regarding how KipB moves along MTs. One accepted model for conventional kinesin movement is the hand-over-hand model, which requires dimerization of the protein. In KipB, as in Klp5, Klp6, and Kip3, only short regions for a potential coiled-coil formation were found. Nevertheless, they might be sufficient for the mediation of an interaction. Indeed, heterodimerization has been shown for Klp5 and Klp6 (13). Therefore, KipB may form homodimers (since we only found one kinesin from this family in the A. nidulans genome) and move along MTs in the conventional sense rather than by diffusion. However, we observed that GFP-KipB formed a discontinuous distribution pattern along MTs. Since the fluorescence signal from single GFP-KipB molecules would be below the detection level, it is likely that we observed aggregates of GFP-KipB or vesicles which contained or were decorated with GFP-KipB. Those vesicle-like structures could be the unit by which KipB is transported toward the MT plus ends, on which the motors would act as a depolymerase. This mechanism would be different from the one described for the transportation of dynein toward the plus ends of MTs in A. nidulans. In that case, conventional kinesin appeared to be the main driving force, but spots along the MTs were not visible. Thus, it is more likely that individual dynein motor proteins are the cargo for kinesin (49). The challenge for future experiments will be to identify motor proteins which may be involved in the transportation of KipB or to prove whether fungal Kip3 kinesins themselves can processively move along MT filaments.
Supplementary Material
Acknowledgments
We thank V. Efimov (Piscataway, N.J.) for sending us the bimC4 and klpA mutant strains and the expression plasmid pCMB17apx and I. Prastio (La Jolla, Calif.) for providing mRFP1. We are grateful to E. Winzenburg for the initial cloning of kipB and thank R. Liese, J. Scheld, and D. Stöhr for their excellent technical assistance. We thank S. Kolb (Marburg, Germany) for help with the phylogenetic analyses and U. Ugalde (San Sebastian, Spain) for stimulating discussions and suggestions on the manuscript.
This work was supported by the Fonds der Chemischen Industrie, the Max-Planck-Institute for Terrestrial Microbiology, and the Deutsche Forschungsgemeinschaft (DFG).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org.
REFERENCES
- 1.Aist, J. R., and N. R. Morris. 1999. Mitosis in filamentous fungi: how we got where we are. Fungal Genet. Biol. 27:1-25. [DOI] [PubMed] [Google Scholar]
- 2.Aramayo, R., T. H. Adams, and W. E. Timberlake. 1989. A large cluster of highly expressed genes is dispensable for growth and development in Aspergillus nidulans. Genetics 122:65-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behrens, R., and P. Nurse. 2002. Roles of fission yeast Tea1p in the localization of polarity factors and in organizing the microtubular cytoskeleton. J. Cell Biol. 157:783-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cottingham, F. R., L. Gheber, D. L. Miller, and M. A. Hoyt. 1999. Novel roles for Saccharomyces cerevisiae mitotic spindle motors. J. Cell Biol. 147:335-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cottingham, F. R., and M. A. Hoyt. 1997. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J. Cell Biol. 138:1041-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desai, A., S. Verma, T. J. Mitchison, and C. E. Walczak. 1999. KinI kinesins are microtubule-destabilizing enzymes. Cell 96:69-78. [DOI] [PubMed] [Google Scholar]
- 7.DeZwaan, T. M., E. Ellingson, D. Pellman, and D. M. Roof. 1997. Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J. Cell Biol. 138:1023-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enos, A. P., and N. R. Morris. 1990. Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in A. nidulans. Cell 60:1019-1027. [DOI] [PubMed] [Google Scholar]
- 9.Farkasovsky, M., and H. Küntzel. 2001. Cortical Num1p interacts with the dynein intermediate chain Pac11p and cytoplasmic microtubules. J. Cell Biol. 152:251-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farkasovsky, M., and H. Küntzel. 1995. Yeast Num1p associates with the mother cell cortex during S/G2 phase and affects microtubular functions. J. Cell Biol. 131:1003-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer, R., and W. E. Timberlake. 1995. Aspergillus nidulans apsA (anucleate primary sterigmata) encodes a coiled-coil protein necessary for nuclear positioning and completion of asexual development. J. Cell Biol. 128:485-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia, M. A., N. Koonrugsa, and T. Toda. 2002. Spindle-kinetochore attachment requires the combined action of KinI-like Klp5/6 and Alp14/Dis1-MAPs in fission yeast. EMBO J. 21:6015-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia, M. A., N. Koonrugsa, and T. Toda. 2002. Two kinesin-like KinI family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr. Biol. 12:610-621. [DOI] [PubMed] [Google Scholar]
- 14.Gundersen, G. G., and A. Bretscher. 2003. Microtubule asymmetry. Science 300:2040-2041. [DOI] [PubMed] [Google Scholar]
- 15.Han, G., B. Liu, J. Zhang, W. Zuo, N. R. Morris, and X. Xiang. 2001. The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr. Biol. 11:19-24. [DOI] [PubMed] [Google Scholar]
- 16.Hill, T. W., and E. Käfer. 2001. Improved protocols for Aspergillus minimal medium: trace element and minimal medium salt stock solutions. F. Genet. Newsl. 48:20-21. [Google Scholar]
- 17.Hirokawa, N. 1998. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 279:519-526. [DOI] [PubMed] [Google Scholar]
- 18.Hunter, A. W., M. Caplow, D. L. Coy, W. O. Hancock, S. Diez, L. Wordeman, and J. Howard. 2003. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP hydrolyzing complex at microtubule ends. Mol. Cell 11:445-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Käfer, E. 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19:33-131. [DOI] [PubMed] [Google Scholar]
- 20.Karos, M., and R. Fischer. 1999. Molecular characterization of HymA, an evolutionarily highly conserved and highly expressed protein of Aspergillus nidulans. Mol. Genet. Genomics 260:510-521. [DOI] [PubMed] [Google Scholar]
- 21.Kinoshita, K., I. Arnal, A. Desai, D. N. Drechsel, and A. A. Hyman. 2001. Reconstitution of physiological microtubule dynamics using purified components. Science 294:1340-1343. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence, C. J., R. L. Malmberg, M. G. Muszynski, and R. K. Dawe. 2002. Maximum likelihood methods reveal conservation of function among closely related kinesin families. J. Mol. Evol. 54:42-53. [DOI] [PubMed] [Google Scholar]
- 23.Lupas, A., M. VanDyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 24.Maekawa, H., T. Usui, M. Knop, and E. Schiebel. 2003. Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J. 22:438-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maney, T., A. W. Hunter, M. Wagenbach, and L. Wordeman. 1998. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 142:787-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, R. K., S. C. Cheng, and M. D. Rose. 2000. Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol. Biol. Chem. 11:2949-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, R. K., K. K. Heller, L. Frisèn, D. L. Wallck, D. Loayza, A. E. Gammie, and M. D. Rose. 1998. The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol. Biol. Chem. 9:2051-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, R. K., and M. D. Rose. 1998. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 140:377-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moores, C. A., M. Yu, J. Guo, C. Beraud, R. Sakowicz, and R. A. Milligan. 2002. A mechanism for microtubule depolymerization by KinI kinesins. Mol. Cell 9:903-909. [DOI] [PubMed] [Google Scholar]
- 30.Morris, N. R. 1976. Mitotic mutants of Aspergillus nidulans. Genet. Res. 26:237-254. [DOI] [PubMed] [Google Scholar]
- 31.O'Connell, M. J., P. B. Meluh, M. D. Rose, and N. R. Morris. 1993. Suppression of the bimC4 mitotic spindle defect by deletion of klpA, a gene encoding a KAR3-related kinesin-like protein in Aspergillus nidulans. J. Cell Biol. 120:153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ovechkina, Y., P. Maddox, C. E. Oakley, X. Xiang, S. A. Osmani, E. D. Salomon, and B. R. Oakley. 2003. Spindle formation in Aspergillus is coupled to tubulin movement into the nucleus. Mol. Biol. Chem. 14:2192-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ovechkina, Y., and L. Wordeman. 2003. Unconventional motoring: an overview of the KinC and KinI kinesins. Traffic 4:367-375. [DOI] [PubMed] [Google Scholar]
- 34.Requena, N., C. Alberti-Segui, E. Winzenburg, C. Horn, M. Schliwa, P. Philippsen, R. Liese, and R. Fischer. 2001. Genetic evidence for a microtubule-destabilizing effect of conventional kinesin and analysis of its consequences for the control of nuclear distribution in Aspergillus nidulans. Mol. Microbiol. 42:121-132. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell. 1999. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schliwa, M., and G. Woehlke. 2003. Molecular motors. Nature 422:759-765. [DOI] [PubMed] [Google Scholar]
- 37.Schoch, C. L., J. R. Aist, O. C. Yoder, and B. G. Turgeon. 2003. A complete inventory of fungal kinesins in representative filamentous ascomycetes. Fungal Genet. Biol. 39:1-15. [DOI] [PubMed] [Google Scholar]
- 38.Seiler, S., F. E. Nargang, G. Steinberg, and M. Schliwa. 1997. Kinesin is essential for cell morphogenesis and polarized secretion in Neurospora crassa. EMBO J. 16:3025-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaw, S. L., P. Maddox, R. V. Skibbens, E. Yeh, E. D. Salmon, and K. Bloom. 1998. Nuclear and spindle dynamics in budding yeast. Mol. Biol. Cell 9:1627-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaw, S. L., E. Yeh, P. Maddox, E. D. Salmon, and K. Bloom. 1997. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J. Cell Biol. 139:985-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stringer, M. A., R. A. Dean, T. C. Sewall, and W. E. Timberlake. 1991. Rodletless, a new Aspergillus developmental mutant induced by directed gene inactivation. Genes Dev. 5:1161-1171. [DOI] [PubMed] [Google Scholar]
- 42.Vale, R. D., T. S. Reese, and M. P. Sheetz. 1985. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 42:39-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waring, R. B., G. S. May, and N. R. Morris. 1989. Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin coding genes. Gene 79:119-130. [DOI] [PubMed] [Google Scholar]
- 44.Wei, H., M. Scherer, A. Singh, R. Liese, and R. Fischer. 2001. Aspergillus nidulans alpha-1,3 glucanase (mutanase), mutA, is expressed during sexual development and mobilises mutan. Fungal Genet. Biol. 34:217-227. [DOI] [PubMed] [Google Scholar]
- 45.West, R. R., T. Malmstrom, and J. R. McIntosh. 2002. Kinesins klp5+ and klp6+ are required for normal chromosome movement in mitosis. J. Cell Sci. 115:931-940. [DOI] [PubMed] [Google Scholar]
- 46.West, R. R., T. Malmstrom, C. L. Troxell, and J. R. McIntosh. 2001. Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast. Mol. Biol. Chem. 12:3919-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiang, X., and R. Fischer. 2004. Nuclear migration and positioning in filamentous fungi. Fungal Genet. Biol. 41:411-419. [DOI] [PubMed] [Google Scholar]
- 48.Yelton, M. M., J. E. Hamer, and W. E. Timberlake. 1984. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc. Natl. Acad. Sci. USA 81:1470-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, J., S. Li, R. Fischer, and X. Xiang. 2003. The accumulation of cytoplasmic dynein and dynactin at microtubule plus-ends is kinesin dependent in Aspergillus nidulans. Mol. Biol. Chem. 14:1479-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.