Abstract
Background
A great number of studies have investigated changes induced by morphine exposure in gene expression using several experimental models. In this study, we examined gene expression changes during chronic exposure to morphine during maturation and differentiation of zebrafish CNS.
Results
Microarray analysis showed 254 genes whose expression was identified as different by at least 1.3 fold change following chronic morphine exposure as compared to controls. Of these, several novel genes (grb2, copb2, otpb, magi1b, grik-l, bnip4 and sox19b) have been detected for the first time in an experimental animal model treated with morphine. We have also identified a subset of genes (dao.1, wls, bnip4 and camk1γb) differentially expressed by chronic morphine exposure whose expression is related to mu opioid receptor gene expression. Altered expression of copb2, bnip4, sox19b, otpb, dao.1, grik-l and wls is indicative of modified neuronal development, CNS patterning processes, differentiation and dopaminergic neurotransmission, serotonergic signaling pathway, and glutamatergic neurotransmission. The deregulation of camk1γb signaling genes suggests an activation of axonogenesis and dendritogenesis.
Conclusions
Our study identified different functional classes of genes and individual candidates involved in the mechanisms underlying susceptibility to morphine actions related to CNS development. These results open new lines to study the treatment of pain and the molecular mechanisms involved in addiction. We also found a set of zebrafish-specific morphine-induced genes, which may be putative targets in human models for addiction and pain processes.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2164-15-874) contains supplementary material, which is available to authorized users.
Keywords: Morphine, Microarray, Zebrafish, Real-time quantitative PCR, Mu-opioid receptor, Gene expression, Addiction, Morpholino
Background
Morphine, the most widely known opioid agent, has been largely used for pain management. Besides non-analgesic uses of this drug, including experimental depression treatments as a control for opium addiction, has been developed [1, 2]. Despite these common uses, morphine produces disruptive negative secondary effects including sleepiness, drowsiness, blurred vision, constipation, and a decrease in blood pressure and appetite. With continuous use, morphine produces physical tolerance and addiction [1, 2]. At present, it is widely known that there are genetic components in the susceptibility to addiction [3, 4], but the specific genes that are involved in this process are scarcely known.
Opioids modify the expression profile of certain mRNAs in many tissues, including central nervous system (CNS), the main target of drugs of abuse [5]. With the use of microarray analysis, extensive studies have been focused on identifying morphine-induced changes in gene expression [6–19]. Other opioid agonist as oxycodone [20] and other drugs of abuse, such as cocaine, cannabis, methamphetamine, amphetamine, ecstasy, alcohol, heroin and nicotine [14, 21–25], have also been studied. Up to now, all microarray studies reporting gene expression changes in response to morphine have been conducted in rodents and humans. In particular, these studies were performed in whole tissues (mainly brain), distinct components of the nervous system (nucleus accumbens, striatum, hippocampus, frontal cortex, spinal cord), or cell cultures (primary and cell lines).
Zebrafish (Danio rerio) has been used in research related to drugs of abuse, studying the changes of gene expression produced by nicotine [26], amphetamine [27], and alcohol [28]. In addition, this teleost exhibits conditioned place preference (CPP), a technique used as a measure of drug reward or reinforcement [29], as responses to amphetamine and opiates like morphine [30–32]. These research works demonstrate the existence of a conserved drug-responsive ‘reward’ or reinforcement pathway in this teleost (as in all vertebrates), suggesting that this species may show adaptive changes and behavioural correlates of addiction after prolonged exposure to addictive drugs.
Zebrafish has also been used to study human disease-related pathways [33, 34], given this organism displays many benefits in comparison to other vertebrate animal models: small size, low cost, rapid development, transparency of the embryos, permeability to drugs, high fecundity, and transient genetic manipulation [35, 36]. In this sense, zebrafish is an important tool to analyze in vivo the molecular mechanisms related to the neurobiology of drug addiction, withdrawal and reward [31, 37–39] that cannot be fully established in other animals models. In contrast to mammalian embryos that develop in the uterus and are influenced by maternal biochemical processes, zebrafish embryos develop externally, avoiding the maternal effect on the embryos. This is essential when dealing with drug exposure, as the effects observed in mammalian embryos might be due to the susceptibility of the mother and not the embryo per se. The study of the direct effects of morphine in the embryos will provide a better understanding of the molecular mechanisms that underlie the physical and neurobehavioral defects shown in fetuses and offsprings after maternal morphine consumption [40, 41].
Therefore, we used microarray technology to obtain a profile of genes that are regulated by chronic morphine exposure on whole zebrafish embryos at 24 h post-fertilization (hpf), at the end of the segmentation period, during which the CNS is being formed and differentiated [37, 42, 43]. We have shown that zebrafish has endogenous opioids and opioid receptors with high homology with other species [44, 45]. The effects of morphine on embryos are probably mediated by the zebrafish mu opioid receptor (opioid receptor, mu1; oprm1), which exhibits highest affinity toward morphine from the known opioid receptors [46], and is the opioid receptor essential to mediate rewarding properties of most drugs of abuse [47, 48]. Our previous studies indicate that at 24 hpf, the expression of oprm1 is higher than at other stages of development [49]. Therefore, the use of 24 hpf zebrafish embryos treated with morphine can provide information on the implication of the opioid system in the maturation and differentiation of CNS compared to any other stages of development. Our goal in this research was to differentiate functional classes of genes and individual candidates involved in the endogenous systems underlying susceptibility to morphine actions; hence, our efforts were focused on elucidating the functional significance of sets of differentially expressed genes related in some way to neuronal function and/or CNS development. To verify the results obtained by microarray, 12 selected genes were analyzed by quantitative reverse transcription real-time PCR (RT-qPCR). After “silencing” (knocking-down; KD) the oprm1 gene expression by morpholino oligonucleotide injection, we identified a subset of genes that are regulated by morphine and are related to oprm1 expression.
Our results show that morphine produces changes in gene expression in zebrafish embryos as has been obtained in rodents and humans [6–19]. Thus, our data, besides being analyzed independently, was also compared to previous studies, in an effort to determine which alterations in gene expression are species-specific (zebrafish vs. mammalians) and which may be common to all species in relation to addiction and pain studies. These results are important since several morphine-induced genes detected in zebrafish embryos may be putative targets in human models for addiction and pain processes.
Results
Microarrays analysis
To examine the global transcriptional profiles of zebrafish embryos, RNAs generated from six pools of control zebrafish embryos and six pools of zebrafish embryos exposed to morphine were individually hybridized to Affymetrix Zebrafish Genome Arrays. A set of gene products corresponding to a total of approximately 14900 probes on the arrays were confidently detected, based on signal intensity at a fixed value above background levels. Differentially expressed genes were identified through the Significance Analysis of Microarrays (SAM) method. As shown in Figure 1, we identified a total of 1023 gene expression changes in the above mentioned comparison (representing 1076 probe sets; Figure 1A-C). Of these, 955 genes were identified as known genes; 401 genes were significantly up-regulated and 561 down-regulated (Figure 1C-D). Moreover, as shown in Figure 1D, seven genes regulated, after chronic exposure to morphine, are represented by more than one probe set in the zebrafish microarray, and each of the probe set is differently deregulated (up- or down-regulated); for example: acireductone dioxygenase 1 (adi1; Dr.20538.1.A1, +1.15 fold change (FC) and Dr.9210.2.S1, −1.23 FC) and selenoprotein X, 1a (sepx1a; Dr.147.1.A1, +1.43 FC; Dr.24956.1.S1, −1.27 FC; and Dr.16417.1.A1; −1.46 FC). Therefore, distinct gene expression changes obtained for each one of the probe sets of each gene might mean that these genes show alternative splicing and each one of them has at least two isoforms that are differently regulated by exposure to morphine. Finally, we specifically identified 255 genes (representing 265 probe sets) showing differential expression with a FC of at least 1.3: 121 genes (126 probe sets) were up-regulated and 134 genes (140 probe sets) were down-regulated by chronic morphine treatment (Figure 1E; see complete list of differentially expressed genes in Additional file 1).
Figure 1.
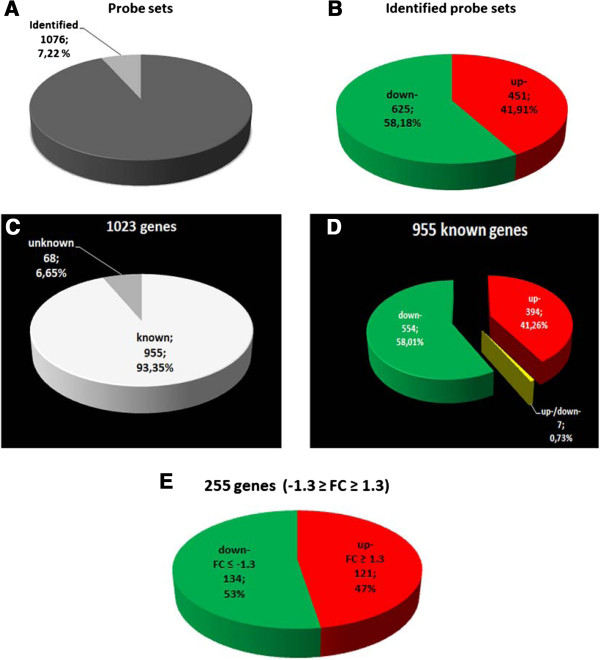
Probe-sets and genes differentially expressed by chronic morphine treatment in zebrafish embryos. Identified probe sets in comparison with total probes on the microarray (A) and detail of identified probe sets up- and down-regulated (B). (C): Number of identified genes (known and unknown). (D) Number of known genes and detail of genes up-, down- and up-/down-regulated. (E): Number of identified genes with a fold change (FC) of at least 1.3. Numbers following the category names indicate the number of probe-set or genes. For the full list of probe-set/genes, see Additional file 1.
Gene ontology analysis
To enhance biological interpretation of the differentially expressed genes from our microarray studies, we sought if any of the biological processes or molecular functions were over-represented by these genes. Therefore we performed a function enrichment analyses for the differentially expressed genes using the functional classification tool DAVID Bioinformatics resources. A representative selection of meaningful categories is illustrated in Figure 2 and Table 1. This analysis shows that some categories are specifically enriched in either up- or down-regulated genes, whereas other categories show both types of regulation. Genes involved in oxidation-reduction processes (Gene Ontology (GO):0055114), proteolysis (GO:0006508), monosaccharide metabolic processes (GO:0005996), macromolecular complex assembly (GO:0065003) and generation of precursor metabolites and energy (GO:0006091) represent an important part of both up- and down-regulated genes. Macromolecule catabolic processes (GO:0009057), DNA metabolic process (GO:0006259), protein catabolic process (GO:0030163), homeostatic process (GO:0042592), protein complex assembly (GO:0006461), proteasome complex (GO:0000502), and in general, post-translational modification, protein turnover, chaperones, mainly include down-regulated genes. There is an over-representation of up-regulated transcripts associated with secondary metabolites biosynthesis, transport, and catabolism (for complete details, see supplementary Additional files 2 and 3).
Figure 2.
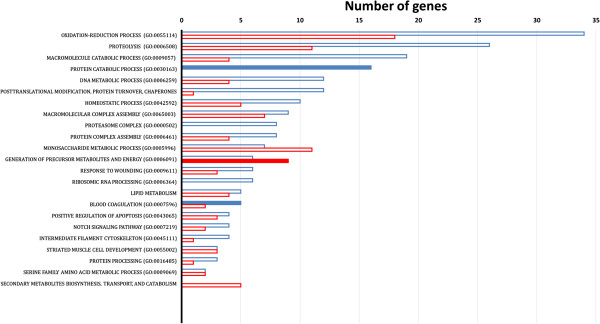
Gene ontology analysis of zebrafish embryos gene collection. Genes were categorized with the Biological Process domain. Significantly enriched GO terms have a probability lower than 0.01 (P value) and include at least three genes. GO terms are shown for both up- (in red) and down-regulated (in blue) groups if they are significantly enriched in at least one of them. Significantly enriched GO terms are indicated as full bars whereas non-significantly enriched terms are displayed as empty bars. Bars represent the number of genes assigned with the corresponding GO term. For the full list of represent genes in each category, see Additional file 3.
Table 1.
Examples of significantly enriched GO annotation for the list of morphine-responsive genes
| Enriched GO annotation | De‐regulation | Up‐regulation | Down‐regulation | |||||
|---|---|---|---|---|---|---|---|---|
| No. genes | P value | No. genes | P value | Genes | No. genes | P value | Genes | |
| Oxidation‐reduction process (GO:0055114) | 52 | 3.4E + 10 | 18 | 0.013 | acox1, cyp11a1, cyp26b1, cyp27a1.4, d2hgdh, gapdhs, | 34 | 3.7E + 10 | adh8b, aldh18a1, cp, cyp2aa4, dao.1, dhfr, dhrs13b, mdh1b, phyhd1, ptgr1, sepx1a |
| Proteolysis (GO:0006508) | 37 | 0.049 | 11 | ube2d2 | 27 | 0.036 | f7, rad23b, spcs2, uchl5, vsg1 | |
| Macromolecule catabolic process (GO:0009057) | 23 | 0.003 | 4 | ube2d2 | 19 | 4.3E + 12 | ctbs. LOC553461, rad23b, uvsg1 | |
| Monosaccharide metabolic process (GO:0005996) | 18 | 6.8E + 08 | 11 | 3.8E + 09 | aldocb,galk2, gapdhs, gpia | 7 | 0.050 | mdh1b, pklr |
| DNA metabolic process (GO:0006259) | 17 | 0.080 | 5 | mlf1, mphosph8 | 12 | 0.054 | orc4, rad23b | |
| Protein catabolic process (GO:0030163) | 16 | 0.002 | 16 | 0.002 | LOC553461, rad23b, uchl5, vsg1 | |||
| Macromolecular complex assembly (GO:0065003) | 16 | 0.003 | 7 | 0.044 | h1fx | 9 | 0.065 | fgg, tubb5 |
| Generation of precursor metabolites and energy (GO:0006091) | 15 | 0.012 | 9 | 0.006 | aldocb, gapdhs, gpia | 6 | mdh1b, pklr | |
| Homeostatic process (GO:0042592) | 15 | 0.025 | 5 | fth1 | 10 | 0.060 | ||
| Posttranslational modification. protein turnover. chaperones | 13 | 0.005 | 1 | 12 | 1.9E + 12 | |||
| Protein complex assembly (GO:0006461) | 12 | 0.005 | 4 | 8 | 0.024 | fgg, tubb5 | ||
| Lipid metabolism | 9 | 0.005 | 4 | 0.087 | acsl1, acsl5 | 5 | 0.072 | |
| Response to wounding (GO:0009611) | 9 | 0.010 | 3 | cxcl12a | 6 | 0.042 | f7, fgg | |
| Proteasome complex (GO:0000502) | 8 | 5.1E + 11 | 8 | 5.1E + 11 | ||||
| Blood coagulation (GO:0007596) | 7 | 7.1E + 11 | 2 | tfpia | 5 | 0.005 | f7,fgg | |
| Positive regulation of apoptosis (GO:0043065) | 7 | 0.010 | 3 | bnip4 | 4 | |||
| Striated muscle cell development (GO:0055002) | 6 | 0.060 | 3 | cxcl12a | 3 | tnnt2a | ||
| Intermediate filament cytoskeleton (GO:0045111) | 5 | 0.043 | 1 | cyp27a1.4 | 4 | 0.043 | krt23 | |
| Notch signaling pathway (GO:0007219) | 5 | 0.072 | 2 | dlc | 3 | vsg1 | ||
| ribosomic RNA processing (GO:0006364) | 5 | 0.086 | 5 | 0.021 | ||||
| Secondary metabolites biosynthesis. transport. and catabolism | 5 | 0.044 | 5 | 0.044 | cyp11a1. cyp26b1. cyp51 | |||
| protein processing (GO:0016485) | 4 | 0.036 | 1 | 3 | spcs2 | |||
| serine family amino acid metabolic process (GO:0009069) | 4 | 0.065 | 2 | gcshb | 2 | dhfr | ||
The complete results of the Gene Ontology (GO) analysis are presented in Additional file 3.
For our study, the different categories within Gene Ontology Biological Process can be classified in other “functional” categories according to databases and consulted publications (see Discussion section). As shown in Figure 3, more than one fourth of differentially expressed genes are related to signal transduction and other biological processes, including energy metabolism, transcription, protein modification and degradation, neuronal function, transport, development, cell structure and organization, apoptosis, amino acid and protein metabolism, replication and stress (see Table 2, and for complete list also see supplementary Additional file 4). In particular, the genes related to neuronal function are shown in a hierarchical cluster in Figure 4 and listed in Table 3. These genes were further investigated in this study resulting in the following categories of GO Biological Process (Additional file 4): regulation of transcription [GO:0045449; as for example, cAMP responsive element binding protein 3-like 3 (creb3l3), developing brain homeobox 1b (dbx1b), distal-less homeobox protein 5a (dlx5a), c-fos FBJ murine osteosarcoma viral oncogene (c-fos), hairy-related 4.2-like (her4.2), jun B proto-oncogene and b (juna and junb), neurogenic differentiation factor 6-B (neurod6b), orthopedia b (otpb), SRY-box containing gene 19b (sox19b), and sox21b], neuron differentiation [GO:0030182; v-erb-b2 erythroblastic leukemia viral oncogene homolog 2 (erbb2), otpb, N-ethylmaleimide-sensitive factor a (nsfa), delta-like protein C Precursor (dlc)], somitogenesis (GO:0001756; dlb, dlc, her4.2) and glutamine biosynthetic process [GO:0006542; solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 (slc1a4) and solute carrier family 1 (glial high affinity glutamate transporter), member 3a (slc1a3a)].
Figure 3.
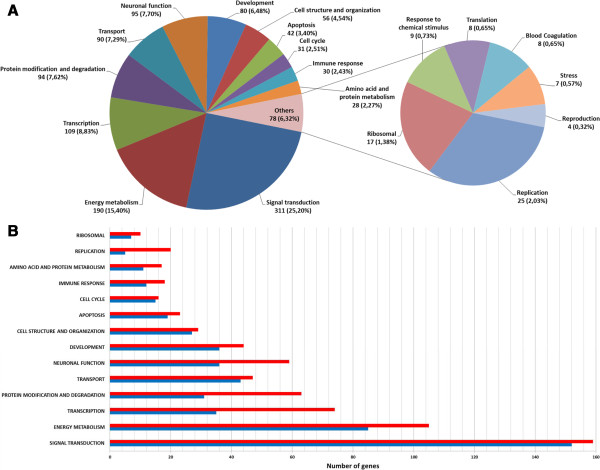
Functional categories and number of genes found to be modified by morphine in each category. (A) Functional categories of genes found to be associated with the exposure to morphine in zebrafish embryos. Numbers following the category names indicate the number of genes in the category, and those in parentheses are the corresponding percentages. (B) Number of genes (up-, in blue, and down-regulated, in red) by functional category. For the full list of genes, see Table 2 and Additional file 4.
Table 2.
Functional categories and number of genes found to be modified by morphine in each category
| Functional categories | N genes | Up‐regulation | Down‐regulation | ||
|---|---|---|---|---|---|
| Signal transduction | 311 | 152 | ppp1r14ab, camk1γb, grb2, sept9a, itih3, ahnak, hrc, sesn3, megf6, btr22, tmem101, magi1b, vldlr*, hspb1*, plg* | 159 | LOC566752, a2ml, sh3bp5la, fam115, txndc9, tctex1d2, rcan2, rnf220b, hspb8, slc1a4*, wls*, grik‐l* |
| Energy metabolism | 190 | 85 | itih3, acox1, soat1, cyp17a2, cyp51, cox4.2, alpp, aldocb, pcmt*, plg* | 105 | aldh8a1, pklr, mpdu1b, mdh1b, phyhd1, hrasls, glb1l, glud1b* |
| Transcription | 109 | 35 | dcps, zbtb16, creb3l3*, junb*, dlx5a*, fos*, dlx4a* | 74 | sox19b, mettl11a, noc2l, dbx1b, junbb, her4.2, otpb*, sox11b*, sox21b*, hdac6*, dnmt3b*, sox19a*, sox11b*, mir429b |
| Protein modification and degradation | 94 | 31 | ube2d2, hspb1*, plg* | 63 | pcmt, rad23b, fbxo25, hspb8 |
| Transport | 90 | 43 | slc31a1, abca12, apoea, abcb5, ctssb.2*, slc3a2*, abt4*, slc1a3a*, slc7a8* | 47 | a2ml, bcap31, aqp12, aqp3a, vamp5, slc1a4*, vamp4*, hiat1b*, apoIV*, brp44*, kctd12.2* |
| Neuronal function | 95 | 36 | bnip4, ppp1r14ab, acox1, serinc5, maptb, magi1b, hspb1*, plg*, aplp*, slc1a3a*, creb3l3*, hsp90a.1*, junba*, dlx5a*, fos*, glula | 59 | pcmt, dao.1, sox19b, a2ml, mettl11a, astn1, noc2l, copb2, dbx1b, junbb, her4.2, slc1a4*, her3*, wls*, grik‐like*, otpb*, cyrano*, sox11b*, sr*, sox21b*, glulb*, glud1b*, dnmt3b*, kctd12.2*, sox11b*, mir429b* |
| Development | 80 | 36 | bnip4, ppp1r14ab, camk1γb, acox1, ahnak, maptb, zbtb16, cyp11a1, magi1b, hspb1*, plg*, dlx5a*, dlx4a* | 44 | sox19b, a2ml, hspb8, noc2l, copb2, dbx1b, her4.2, her3*, wls*, otpb*, cyrano*, sox11b*, sox21b*, hoxb8a*, dnmt3b*, kctd12.2*, sox11b* |
| Cell structure and organization | 56 | 27 | mybpc2b, maptb, col5a1 | 29 | mylz2, krt1‐19d |
| Apoptosis | 42 | 19 | bnip4, hspb1*, bbc3* | 23 | hspb8, noc2l, bcap31, casp9*, mir429b* |
| Cell cycle | 31 | 15 | sept9a, sesn3, ccnt1, hsp90a.1*, ccnd1* | 16 | ccnb3* |
| Immune response | 30 | 12 | ccl‐c11b, cxcl12a, ctssb.2*, plg* | 18 | crfb6 |
| Amino acid and protein metabolism | 28 | 11 | hnmt, eif4a2, eif3hb, eif2c2, slc3a2*, slc7a8* | 17 | fah, dao.1, aldh18a1, iars, eif4g2b*, sr*, eif4g2a*, eif2s1l* |
| Replication | 25 | 5 | h1fx | 20 | rad23b, mettl11a, noc2l, h2afx*, dnmt3b* |
| Ribosomal | 17 | 7 | rpf2 | 10 | nob1, fcf1 |
| Response to chemical stimulus | 9 | 3 | hspb1*, hsp90a.1* | 6 | hspb8 |
| Translation | 8 | 2 | gcn1l1 | 6 | hbs1l* |
| Blood Coagulation | 8 | 3 | serping1 | 5 | fgg |
| Stress | 7 | 3 | sesn3, hspb1*, hsp90a.1* | 4 | hspb8 |
| Reproduction | 4 | 3 | 1 | ||
Some up-regulated genes with FC > 1.0 and down-regulated genes FC < −1.0 are marked with an asterisk (*).
Figure 4.
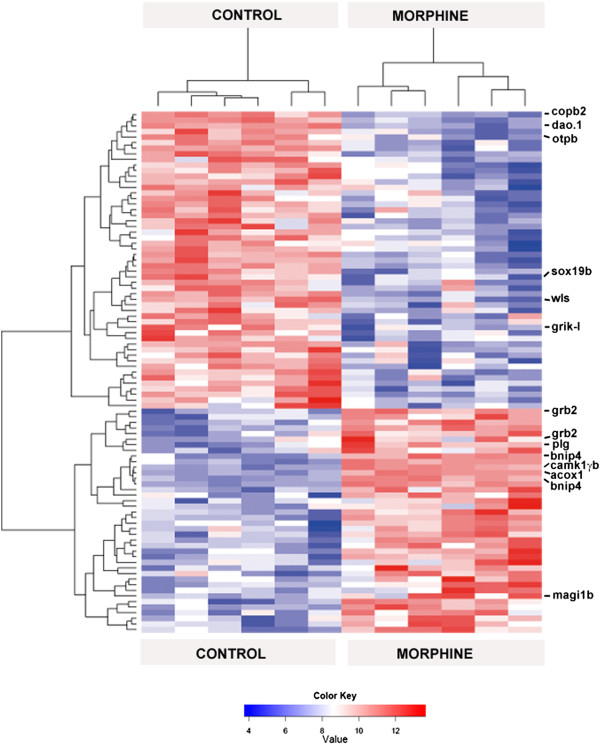
Hierarchical cluster of probe sets regulated in zebrafish embryos following chronic morphine exposure. Raw data from six independent hybridizations are presented for the two experimental groups: control and morphine-treated embryos zebrafish. The probe-set selection was based on a standard statistical analysis with Robust Multi-array Analysis (see Methods for details). Hierarchical cluster analysis was performed using Cluster 3.0 and Treeview software and the tree image represents low (in blue) or high expression (in red) of probe sets. These probe sets represent 87 genes (representing 93 probe sets) involved in Neuronal Function (a complete list is provided in Additional file 4). The twelve genes (fourteen probe sets) chosen for validation of microarray data by RT-qPCR are listed (right hand side of the figure).
Table 3.
Genes involved in neuronal function with −1.30 ≥ R fold ≥ 1.30
| Gene symbol | Gene description | R.fold | Probeset ID |
|---|---|---|---|
| UP‐REGULATION | |||
| bnip4 | BCL2/adenovirus E1B interacting protein 4 | 2,357 | Dr.24320.1.S1_at |
| ppp1r14ab | Protein phosphatase 1, regulatory (inhibitor) subunit 14Ab | 2,239 | Dr.19567.1.S1_at |
| camk1γb | Calcium/calmodulin‐dependent protein kinase Iγb | 2,164 | Dr.9852.1.A1_at |
| acox1 | Acyl‐Coenzyme A oxidase 1, palmitoyl | 1,741 | Dr.3576.1.A1_at |
| serinc5 | Serine incorporator 5 | 1,480 | Dr.7467.1.S1_at |
| maptb | Microtubule‐associated protein tau b | 1,462 | Dr.16118.1.A1_at |
| dlc | Delta‐like protein C Precursor | 1,375 | Dr.16183.1.S1_at |
| dmxl2 | Dmx‐like 2 (rabconnectin‐3) | 1,367 | Dr.12467.1.A1_at |
| lmbr1l | limb region 1 like | 1,346 | Dr.19947.1.S1_at |
| fam120c | Family with sequence similarity 120C (constitutive coactivator of PPAR‐gamma‐like protein 2) | 1,334 | Dr.18209.1.A1_at |
| magi1b | Membrane associated guanylate kinase, WW and PDZ domain containing 1b (BAI1‐associated protein 1) | 1,300 | Dr.4477.1.A1_at |
| DOWN‐REGULATION | |||
| dao.1 | D‐amino‐acid oxidase 1 | −1,943 | Dr.3663.1.A1_at |
| sox19b | SRY‐box containing gene 19b | −1,802 | Dr.25405.1.A1_at |
| a2ml | Alpha‐2‐macroglobulin protein‐like | −1,801 | Dr.3025.1.A1_at |
| astn1 | Astrotactin 1 | −1,484 | Dr.14729.2.A1_at |
| enc1 | Ectodermal‐neural cortex (with BTB‐like domain) | −1,466 | Dr.9565.1.S1_at |
| copb2 | Coatomer protein complex, subunit beta 2 | −1,393 | Dr.14687.1.A1_at |
| mchr1b | Concentrating hormone receptor 1b | −1,387 | Dr.24966.1.S1_at |
| dbx1b | Developing brain homeobox 1b | −1,364 | Dr.8072.1.S1_at |
| junbb | Jun B proto‐oncogene b | −1,356 | Dr.737.1.A1_at |
| her4.2 | Hairy‐related 4.2 | −1,324 | Dr.20386.1.S1_at |
| lrrn1 | Leucine rich repeat neuronal 1 | −1,309 | Dr.24292.3.A1_a_at |
Genes were identified as being differentially regulated (up- and down-regulated) in embryos zebrafish by morphine treatment.
Validation of microarray data by quantitative reverse transcription real-time PCR (RT-qPCR)
RT-qPCR analysis was used to confirm a set of gene expression changes observed in the microarray analysis. Chosen genes for RT-qPCR confirmation were mainly selected based on ontological categories with potential roles in the nervous system.
To choose the most stable genes as internal references for RT-qPCR data normalization, four candidates [β-actin (β-act), ribosomal protein L13a (rpl13a), β-2-microglobulin (β2 mg) and elongation factor-1α (ef1α)] were selected according to their expression levels detected in the microarray studied. The expression of these four genes was also measured by RT-qPCR. The NormFinder software [50] was used to calculate the intra- and inter-group variations in their expression. Our results indicate that β-act is the most stable gene, whereas rpl13a, β2 mg and ef1α are less stable (data not shown). Thus, the mean of threshold cycle (Ct) value and primer efficiency value of β-act was used for normalization.
As shown in Table 4 and Figure 4, we examined a total of 12 regulated genes after chronic morphine exposure on zebrafish embryos (represented by 14 probe sets in the microarray system studied) using RT-qPCR technique. The up-regulated genes include: acyl-Coenzyme A oxidase 1, palmitoyl (acox1), growth factor receptor-bound protein 2 (grb2), and Ca2+/calmodulin-dependent protein kinase Iγb (camk1γb). The group of down-regulated genes is formed by the transcription factor sox19b (also named sox31) and wntless (wls, also known as gpr177).
Table 4.
Comparison between microarray and RT-qPCR data for selected genes
| Transcripts cluster Id | Gene names | Microarray data R.fold | RT‐qPCR data | |
|---|---|---|---|---|
| Fold change | P‐value | |||
| Up‐regulation | ||||
| Dr.24320.1.S1_at Dr.21771.1.S1_at | bnip4 | 2.36/1.23 | 1.90 ± 0.41* | 7.42 E‐05 |
| Dr.9852.1.A1_at | camk1γb | 2.16 | 1.90 ± 0.57* | 1.41 E‐05 |
| Dr.7717.2.A1_at Dr.7717.1.A1_at | grb2 | 2.02/1.58 | 1.27 ± 0.15* | 8.98 E‐04 |
| Dr.3576.1.A1_at | acox1 | 1.74 | 2.03 ± 0.43* | 2.04 E‐05 |
| Dr.4477.1.A1_at | magi1b | 1.30 | 1.55 ± 0.12* | 5.43 E‐05 |
| Dr.3645.1.S1_at | plg | 1.26 | −1.06 ± 0.21 | 0.299 |
| Down‐regulation | ||||
| Dr.3663.1.A1_at | dao.1 | −1.94 | −3.31 ± 0.11* | 3.59 E‐08 |
| Dr.25405.1.A1_at | sox19b | −1.80 | −2.09 ± 0.19* | 0.001 |
| Dr.14687.1.A1_at | copb2 | −1.39 | −1.69 ± 0.19* | 0.004 |
| Dr.3546.1.S1_at | wls | −1.25 | −1.67 ± 0.16* | 0.006 |
| Dr.3211.1.A1_at | grik‐l | −1.23 | −2.94 ± 0.21* | 2.53 E‐04 |
| Dr.8118.1.A1_at | otpb | −1.22 | −1.64 ± 0.23* | 0.004 |
Values of fold change by RT-qPCR are given as mean of fold change ± standard deviation. The study includes samples used for microarray hybridization and samples from independent treatments; a total of n = 8. For better comparison of the results obtained with both techniques, microarray and RT-qPCR, Morphine vs. Control fold changes are indicated with positive and negative values for up- and down-regulations, respectively. A one-way t-test was performed to determine whether fold changes obtained for morphine-regulated genes were different from 1 and significant values are indicated with asterisks.
It is of interest that the probe set that presents the highest gene expression (Dr.26538.1.A1; +4.73 FC), putatively identified by Affymetrix as wls, do not aligned to wls genomic locus. Our in silico analyses demonstrated that when performing a Blastn in the Ensembl site, using as query an unique sequence of 59 nucleotides formed by the overlapping of 16 probes which constitute this probe set, the query sequence is aligned in two unidentified non-coding regions (on chromosomes 6 and 8) that do not belong to wls genomic locus located on chromosome 2. Therefore, Dr.26538.1.A1 probe set cannot be identified as wls gene (see other genes differentially expressed by morphine validation by our in silico studies in Additional file 5). Concerning the wls gene, we have detected that uniquely the Dr.3546.1.S1 probe set is complementary to this gene (Table 4).
We also analyzed four genes regulated by chronic morphine treatment involved specifically in dopaminergic and serotonergic neurotransmission. A gene putatively related to serotonergic neurotransmission is BCL2/adenovirus E1B interacting protein 4 (bnip4). Three genes involved in dopaminergic neurotransmission are the plasminogen (plg), coatomer protein complex subunit beta 2 (copb2), and the transcription factor otpb. Furthermore, we included in our study three genes related to glutamatergic neurotransmission: one up-regulated (membrane associated guanylate kinase, WW and PDZ domain containing 1b, magi1b) and two down-regulated (D-amino-acid oxidase 1, dao.1, and a kainate-like receptor, grik-l).
We showed that two regulated genes, after chronic morphine exposure, are represented by more than one probe set in the zebrafish microarray (bnip4 and grb2; Table 4). Distinct gene expression changes obtained for each one of the probe sets for each gene might mean that these genes show alternative splicing and each one of them has at least two isoforms that are differently regulated by exposure to morphine.
All genes chosen for RT-qPCR confirmation and involved in neuronal functions are also related to other functional categories according to databases and consulted publications, such as development (acox1, camk1γb, sox19b, plg, otpb, copb2, wls, bnip4 and magi1b), signal transduction (camk1γb, grb2, plg, wls, grik-l and magi1b), transcription (sox19b and otpb), apoptosis (bnip4), energy metabolism (acox1 and plg) and, specifically, amino acid and protein metabolism (dao.1) (see details in Table 2). It should be emphasized that among all genes confirmed by RT-qPCR some have previously been identified in distinct studies with mammals, such as Camk1γ, Plg, Dao and Wls [10, 12, 15, 51, 52] (Table 5).
Table 5.
Example of genes differentially expressed by morphine in our study and identified in other species
| Probeset ID | Genename | Description/validation in silico | R.fold | Species | Reference |
|---|---|---|---|---|---|
| Up‐regulation | |||||
| Dr.9852.1.A1_at | camk1γb | Calcium/calmodulin‐dependent protein kinase Igb | 2,16 | Mouse | [12] |
| Dr.23925.1.A1_at | soat1 | Sterol O‐acyltransferase (acyl‐Coenzyme A: cholesterol acyltransferase) 1 | 1,69 | Rat | [16] |
| Dr.19223.1.S2_at | aldocb | Aldolase C, fructose‐bisphosphate, b | 1,49 | Rat | [105] |
| Dr.11457.1.S1_at | zbtb16 | Zinc finger and BTB domain containing 16 | 1,44 | Mouse | [12, 65] |
| Dr.12489.1.S1_at | mlf1 | Myeloid leukemia factor 1 | 1,44 | Mouse | [8] |
| Dr.18505.1.S1_at | ccnt1 | Cyclin‐T1 | 1,44 | Mouse | [6] |
| Dr.822.1.S3_at | cxcl12a | Chemokine (C‐X‐C motif) ligand 12a (stromal cell‐derived factor 1) | 1,38 | Mouse | [8] |
| Dr.23722.1.S1_at | cyp27a1.4 | Cytochrome P450, family 27, subfamily A, polypeptide 1, gene 4 | 1,35 | Mouse | [8] |
| Dr.9423.1.S1_at | ndufb2 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8 kDa | 1,31 | Mouse | [6] |
| Dr.867.1.S1_at | wdr1 | WD repeat domain 1 | 1,28 | Rat | [105] |
| Dr.12378.1.S1_at | hspb1 | Heat shock protein, alpha‐crystallin‐related, 1 | 1,27 | Rat & mouse | [16, 55] |
| Dr.3645.1.S1_at | plg | Plasminogen | 1,26 | Rat & mouse | [12, 15] |
| Down‐regulation | |||||
| Dr.3663.1.A1_at | dao.1 | D‐amino‐acid oxidase 1 | −1,94 | Rat | [51, 52] |
| Dr.9860.1.S1_at | mdh1b | Malate dehydrogenase 1b, NAD (soluble) | −1,79 | Rat | [105] |
| Dr.19735.1.S1_at | phyhd1 | Phytanoyl‐CoA dioxygenase domain containing 1 | −1,56 | Mouse | [55] |
| Dr.5687.1.A1_at | hspb8 | Heat shock protein, alpha‐crystallin‐related, b8 | −1,45 | Mouse | [8, 55] |
| Dr.737.1.A1_at | junbb | Jun B proto‐oncogene b | −1,36 | Rat & mouse | [7] |
| Dr.3546.1.S1_at | wls | wntless homolog (Drosophila) | −1,25 | Mouse | [10] |
For our RT-qPCR analysis, we used the same RNA samples as those used for the microarray hybridization experiment, and additionally, samples from eight independent chronic morphine treatments were also used. Results from all the RT-qPCR experiments are summarized in Table 4 and Figure 5: acox1, bnip4, camk1γb, magi1b, and grb2 appeared as the most up-regulated transcripts with 2.03 ± 0.43, 1.90 ± 0.41, 1.90 ± 0.57, 1.55 ± 0.12, and 1.27 ± 0.15 FC, respectively. We also clearly confirmed down-regulation of sox19b, grik-l, dao.1, copb2, wls and otpb transcripts (−3.31 ± 0.11, −2.94 ± 0.21, −2.09 ± 0.19, −1.69 ± 0.19, −1.67 ± 0.16, −1.64 ± 0.23 FC, respectively). However, the up-regulation of plg was not confirmed by RT-qPCR since the result showed no significant deregulation. Differences between RT-qPCR and microarray experiments occur for several reasons, including the fact that different probes are used for the microarray and RT-qPCR experiments (which can capture differential expression in splice variants), differences in the methods for normalization of expression data and possible false-positive expression changes. In addition, lower correlations between RT-qPCR and microarray results, such as for plg gene (+1.26 FC), were consistently reported for genes exhibiting small degrees of changes, generally less than 1.4 FC [53].
Figure 5.
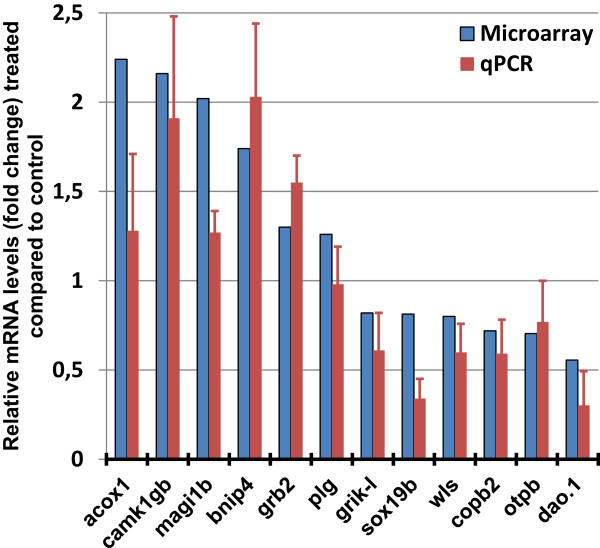
Quantitative real-time PCR (RT-qPCR) was used to validate the microarray data. The genes selected showed similar expression changes when assessed by RT-qPCR as determined by microarray analysis. Data are presented as morphine vs. control fold changes.
Correlation between the microarray and RT-qPCR results for the 12 selected genes was then performed and the statistical significance of the correlation determined. As Morey et al. [53] suggest, prior to performing correlation analysis, the dataset should be tested for normality using the Shapiro-Wilk test; our data points were not normally distributed and therefore Spearman’s rho test was used. This test is the rank-based non-parametric equivalent of the more commonly used Pearson’s correlation calculation [53]. The expression data for this gene set detected by microarray and RT-qPCR are plotted in Figure 6. The RT-qPCR and microarray methods showed excellent qualitative agreement on both up- and down-regulated genes. The correlation between microarray and RT-qPCR data obtained a high statistical significance [r (12) = 0.884, p < 0.001]. As mentioned before, the expression of several morphine-induced genes identified in previous studies (see Table 5), such as camk1γb, dao.1 and wls, was also analyzed to evaluate the data reliability and the sensitivity of microarray analysis. As shown in Figure 5, the induced expression of camk1γb and decreased expression of wls was successfully detected by both RT-qPCR and microarray approaches. These results confirmed the reliability of microarray analysis method in this study.
Figure 6.
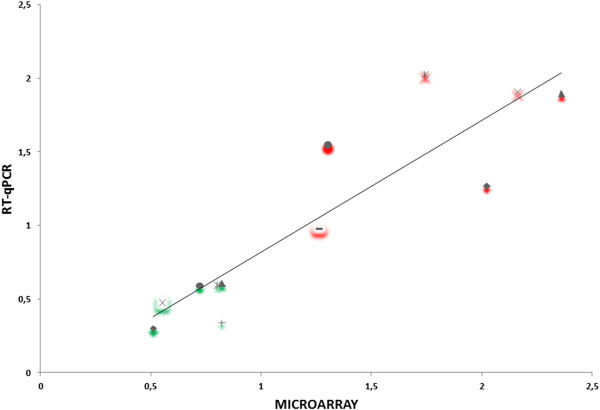
Validation of microarray data by RT-qPCR. Correlation between microarray (x-axis) and RT-qPCR (y-axis) data. This correlation was analyzed by Spearman’s rho test and a high statistical significance [r (12) = 0.884, p < 0.001] was observed. Values used in this graph were taken from results presented in Table 4. Each gene is labeled with a symbol and up- and down-regulated genes are marked in red and green, respectively: ♦, grb2 and dao.1; ●, magi1b and copb2; ▲, bnip4 and otpb; x, camk1gb and sox19b; Ӿ, acox1 and wls; +, grik-l; −, plg. In the case of two regulated genes (bnip4 and grb2) after chronic morphine exposure that are represented by two probe sets each one, we have included in this figure the highest microarray data: Dr. 24320.1, bnip4 + 2.36 FC (▲), and Dr. 7717.2, grb2, +2.02 FC (♦).
The role of oprm1gene in morphine-induced regulation of differentially expressed genes in zebrafish embryos
The effects of morphine in the embryos are probably mediated by oprm1, the opioid receptor that exhibits highest affinity towards morphine [46]. Thus, in order to establish the role of oprm1 in the expression of the identified genes in our study, we microinjected specific morpholino (MO) to knock-down (KD)-oprm1 at the one-to-four-cell stage in the yolk (named oprm1-MO group; Figure 7), embryos were analyzed when the 24 hpf stage was reached.
Figure 7.
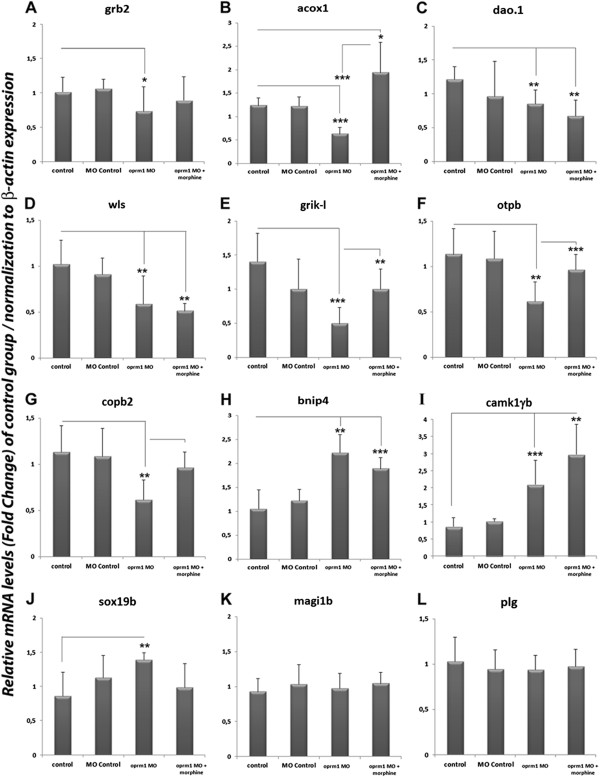
Knockdown of μ opioid receptor ( oprm1 ). Expression levels of grb2, acox1, dao.1, wls, grik-l, otpb, copb2, bnip4, camk1gb, sox19b, magi1b and plg of the Control group, the Morpholine (MO) control, Morpholine oprm1 (oprm1-MO) and oprm1-MO exposure to morphine (oprm1-MO + morphine). The expression levels were quantified using RT-qPCR analysis and were normalized to β-actin expression. Each bar represents the FC ± SD (n = 8). Data were analyzed by one-way ANOVA and using the Tukey post-hoc test. P values of < 0.05 being considered statistically significant (***p < 0.001; **p < 0.01; *p <0.05).
After the MO microinjection of oprm1 we did not detect any morphological alteration in the embryos. We found a decrease of the mRNA expression level approximately 95% (data not shown) in the oprm1 which was similar to that was reported by previous studies of our group [54–56].
To establish that changes observed after MO microinjections were produced by MO action on their target, we also employed two more experimental groups (see Figure 7): embryos injected with standard MO (not targeting any gene in zebrafish, named MO Control group), and embryos with oprm1-MO injection plus exposure to morphine (oprm1-MO + morphine group).
As shown in Figure 7, our results demonstrated that the gene expression of grb2, acox1, dao.1, wls, grik-l, otpb and copb2 in the oprm1 morphants group (oprm1-MO) was decreased with respect to the Control group, indicating that oprm1 is important in the expression of these genes (Figure 7A-G). When the oprm1-MO group was exposed to morphine (oprm1-MO + morphine group), grb2 expression was recovered and similar values were reached with respect to Control groups, including MO Control group (Figure 7A). In contrast, analyzing the gene expression levels of acox1, grik-l, otpb and copb2 in the oprm1-MO + morphine group, we observed an increase of these genes compared to the oprm1-MO group. The comparison of gene expression levels of these four genes in the oprm1-MO + morphine group relative to the two Control groups shows that gene expression levels of grik-l, otpb and copb2 were comparable to the Control groups (Figure 7E-G), but in the case of acox1, it showed a significant increase as compared to Control groups (Figure 7B). Furthermore, dao.1 and wls expression did not undergo any significant change between oprm1-MO + morphine and oprm1-MO groups (Figure 7C-D).
Unlike the previous group of genes, the absence of oprm1-mRNAs (oprm1-MO group) increased the expression of bnip4, camkγ1b and sox19b genes (Figure 7H-J). Thus, silencing gene transcription oprm1 indicates that it appears to be important in some way in the expression of these three genes. When the oprm1-MO group was exposed to morphine (oprm1-MO + morphine group), sox19b expression was recovered and similar values were reached with respect to the Control groups (Figure 7J). In contrast, camkγ1b and bnip4 expression did not undergo any significant change between oprm1-MO + morphine and oprm1-MO groups (Figure 7H-I). Finally, magi1b and plg expression did not present any significant change among all experimental groups (Figure 7K-L).
In conclusion, we found that morphine deregulated (increased or decreased) the expression of grb2, acox1, grik-l, otpb, copb2 and sox19b through an unknown (not related to oprm1) mechanism; however, it required the expression of oprm1 to exert its influence on the expression of dao.1, wls, bnip4 and camkγ1b. In other words, these four genes represent a putative collection of genes whose expression is related to oprm1 expression.
Discussion
Most of the published data addressing addictive drugs have focused on adults or neonates in mammalian models [7–19, 57, 58]; little information has been published concerning embryonic development and addiction (e.g. [59] –alcohol-; [10] –morphine-). Nevertheless, the use of morphine by pregnant women, besides affecting them personally, also affects their fetuses [60]. It is known that exposure to morphine may have additional consequences in the mammalian newborn, affecting immune function, neurodevelopment [61] and long term neurobehavioral effects in combination with stress [10, 62].
In the present study, we used zebrafish embryos, since in this model external development can be observed, which is advantageous for the study of the actions of drugs in a specific embryonic stage (see our recent studies with cocaine: [55, 56, 63]).
We described four groups of genes regulated by chronic morphine treatment in zebrafish embryos. Each group contains both, genes already known to be related to morphine activity, and new genes that have not been related to morphine until now. We have also described some genes whose expression is related to oprm1 expression: (i) Immediate early genes (IEG) and other genes related to transcription; (ii) genes involved in monoaminergic neuromodulation; (iii) genes involved in glutamatergic neurotransmission; and (iv) others genes involved in neuronal function.
Immediate early genes and other genes related to transcription
Genes previously known are IEGs, such as members of the fos and jun families, which have important roles in processes such as brain development, learning and long-term neuronal plasticity [64]. Rapid and transient induction of c-Fos and Jun-B has been described in response to acute morphine administration in different rat brain regions [7]. Our results showed that a duplicate of junb was down-regulated (junbb, −1.36 FC) as well as their ortholog was up-regulated (junba, +1.18 FC). This fact suggests that both genes have different functions, such as reported by Postlethwait et al. [65] indicating a subfunctionalization of a gene can occur after a gene duplication event. Additionally, c-fos was also up-regulated (+1.15 FC). Other known genes deregulated by morphine in zebrafish embryos and related to transcription are the zinc finger and BTB domain containing 16 (zbtb16; +1.44 FC) and the myeloid leukemia factor 1 (mlf1; +1.44 FC). For both genes some similar results have been reported in various studies. Thus, zbtb16 was also induced after acute treatment by morphine in mice [12], and this induction was lower after prolonged administration. Furthermore, this gene was found to be positively related to CPP induced by morphine in mouse [66]. Anghel et al. [8] showed that the Mlf1 was also up-regulated by short-term morphine and conversely down-regulated by long-term morphine in the pituitary gland of mice.
We also detected other genes involved in the regulation of transcription that have not been identified in previous studies after morphine treatment, for example, sox19b (−1.80 FC), sox19a (−1.15 FC), dbx1b (−1.36 FC), and her4.2 (−1.32 FC) that were also deregulated by amphetamine in zebrafish, like other members of these gene families (sox9a, her15 and dlx1a) [27]. With respect to sox19b, a gene that we validated by RT-qPCR, belonging to the family of SoxB1 transcription factors (sox1/2/3/19a/19b), its expression is restricted to the developing CNS, and specifically at 24 h of development in zebrafish, it is expressed in the dorsal spinal cord, in the hindbrain, in the tegmentum of the midbrain and in the telecenphalon [67], acting as an important determinant during such anterioposterior regionalization process of the CNS in zebrafish development [68, 69]. Therefore, we can postulate that the down-regulation of sox19b produced by morphine exposure in zebrafish embryos could intervene in some way by altering the CNS anterioposterior patterning process.
Genes involved in monoaminergic neuromodulation
It is widely known that alterations of biogenic amines, neurotransmitters derived by the enzymatic decarboxylation of naturally occurring amino acids (catecholamines, dopamine and norepinephrine, as well as serotonin and acetycholine), by drugs of abuse play an important role in the CNS [70]. In particular, morphine increases dopaminergic neurotransmission in the nucleus accumbens via the activation of dopamine cells in the ventral tegmental area (VTA), an area that possesses a high density of OPRMs (and/or delta opioid receptors, OPRDs), which may mediate reinforcing effects of morphine [71]. This activation mainly results from the disinhibition of inhibitory GABAergic interneurons in the VTA. In zebrafish, dopaminergic and serotonergic systems share similarities with their mammalian counterparts [72, 73]. Although in mammals, dopaminergic neurons have been observed in the diencephalon, telencephalon and mesencephalon; in zebrafish, these neurons have only been detected in the diencephalon and telencephalon [74, 75], and appear during development between 18–22 hpf in zebrafish embryos [76]. In particular, zebrafish A11-type dopaminergic neurons (homologous to mammals), the major far-projecting dopaminergic neurons in this teleost, are located in the ventral diencephalic (hypothalamus) and posterior tuberculum and express specifically the transcription factor Otp [75, 77]. We hypothesized that the dopaminergic neurotransmission during development may be altered by down-regulation of otpb gene. In zebrafish, two paralogous otpa and otpb genes have been previously reported [78] and the requirement for otpa and otpb function during development of the larval diencephalon is partially redundant. Both genes are essential for the development of specific subsets of diencephalic dopaminergic neurons in zebrafish and mice, of neuroendocrine cells and of specific neurons in the hindbrain in zebrafish [79–83]. Recently, Barreto-Valer et al. [63] showed that cocaine also down-regulates both duplicate otp genes at 24 hpf zebrafish embryos, and these transcription factors, besides LIM homeobox transcription factor 1 and 2-beta (lmx1b.1/2), were important in the expression of tyrosine hydroxylase (th), the enzyme responsible for the biosynthesis of dopamine, during the 24 and 48 hpf zebrafish embryonic stages [79].
Another new identified gene, involved also in dopaminergic neurotransmission and down-regulated by exposure to morphine in zebrafish embryos, is copb2. This gene encodes the subunit (of 7 subunits) of coatomer complex, which is implicated in a variety of functions ranging from signal transduction, vesicular trafficking and transcriptional regulation to cell cycle control and apoptosis [84]. In particular, Copb2 regulates the transport of dopaminergic receptor D1 [85]. Thus, one mechanism by which drugs of abuse can regulate the function of dopaminergic receptors is promoting a decrease in the traffic of dopaminergic receptors from the cytoplasm to the cell membrane or vice versa [15]. The trafficking process could be regulated by changes in gene expression of proteins involved in this process, such as Copb2. On the other hand, Kily et al. [26] detected that copb2 was down-regulated by treatment of both nicotine and ethanol in zebrafish; these authors also demonstrated that copb2 is implied in the development of the notochord.
Concerning genes involved in the serotonergic signaling pathway that in our study are deregulated by chronic morphine treatment, we can mention bnip4 (also named bnip3b), an unknown gene to date in the context of animals treated with morphine. We validated by RT-qPCR the results obtained by microarrays analysis and identified that the expression of this gene is related to oprm1 expression. In zebrafish, three ortholog genes have been described: bnip3 and bnip4, not induced by hypoxia, and bnip3a, which is induced by hypoxia and is much closer to human BNIP3, which is related to cell death/survival [86]. In addition, it is known that rat Bnip3 gene is differentially regulated during development and induces both apoptosis and autophagy. Besides it is also involved in other biological processes, such as mitochondrial dynamics and intracellular calcium regulation [87]. We can hypothesize that the increase of bnip4 expression produced after exposure to morphine may be involved in a possible induction of apoptosis/autophagy.
It has interestingly been suggested that mammalian BNIP3 protein can be a candidate for an intrinsic antidepressive effect-related factor and an antistress reaction factor [88, 89]. Enhanced Bnip3 expression in NG108-15 cells, which possess the serotonin 2C receptor (5-HT2CR) mRNA system, was observed after exposure to Hochu-ekki-to (HET), a (Wakan-yaku) Sino-Japanese traditional drug with antidepressive effects, as well as after exposure to typical antidepressants [88]. In addition, rat brain Bnip3 mRNA expression is enhanced under stress conditions [89]. Thus, both studies speculate that Bnip3 mRNA may be the candidate gene that is controlled by the 5-HT2CR mRNA system, which plays an important role in the action of antidepressants together with serotonin transporters and/or noradrenaline transporters, and also may play a role in the maintenance and/or reformation of synapses in the brain. Therefore, data published up to now [86–89] and our own data suggest that antidepressants in mammals and the treatment with morphine in zebrafish embryos producing an increase in expression of Bnip3-type genes may initially seem to indicate an unfavorable pro-apoptotic state of the neuronal cell. In addition, a second conclusion of functional significance of bnip4 in zebrafish is that the study of this teleost during development may be a good model to confirm that bnip4 mRNA is a candidate for one of the genes that are controlled by the 5-HT(2C)R mRNA system. The relation between morphine and serotonergic neurotransmission is supported by biochemical studies that demonstrated reciprocal functional interactions of the 5-HT(2A) receptor and the OPRM1 following activation by morphine [90].
Genes involved in glutamatergic neurotransmission
In addition to monoaminergic neurons, the glutamatergic system plays a critical role in drug dependence and addiction [91]. In relation to this system, we identified groups of genes that are associated with the modulation of glutamate signaling, producing neuroplasticity, and are regulated by chronic morphine treatment, some of which (as grik-l, slc1a4 and slc1a3a) have not been described before. Plasticity following chronic drug exposure, including morphine, has been described in the extended amygdala (formed by nucleus accumbens and central amygdala) and, in particular, at the interface between glutamatergic and monoaminergic systems [58, 92].
Within the group of genes down-regulated by exposure to morphine in zebrafish embryos, we found dao.1, which we have validated by RT-qPCR and identified as a gene related to oprm1 expression. This gene encodes a peroxisomal flavoprotein (DAO) that catalyzes the oxidative deamination of neutral and polar D-amino acids as D-Ser (a co-agonist of N-methyl-D-aspartate (NMDA) receptor [93]) to hydrogen peroxide, and is expressed in mammals in the kidneys, liver and almost exclusively within astrocytes in the spinal cord [94–96]. Unlike our study, two papers of Yoshikawa et al. [51, 52] showed that acute and chronic treatments with morphine increases the expression of Dao and serine racemase (Sr), which catalyzes the direct formation of D-Ser from L-Ser, in most parts of rat brain, mainly forebrain. In our study, we detected that both genes were down-regulated by morphine exposure in zebrafish embryos (in the case of sr, −1.20 FC). We have found several genes, as for example dao.1, chemokine (C-X-C motif) ligand 12a (cxcl12a), NADH dehydrogenase (ubiquinone) 1 beta subcomplex 2 (ndufb2), phytanoyl-CoA dioxygenase domain containing 1 (phyhd1), and carbonic anhydrase II (ca2) that were regulated in opposite directions in our study in contrast to other studies; such differences may be due to different morphine exposure areas, tissue (brain) region and/or species.
Interestingly, in rodents, spinal DAO contributes to the development of central sensitization-mediated chronic pain and may be a potential target molecule for the treatment of chronic pain and as an efficacious molecule mediating morphine tolerance [96, 97]. In conclusion, the decrease of dao.1 expression by treatment with morphine in zebrafish embryos contrary to that obtained with rodents opens a door hitherto unknown in the use of zebrafish as a model to study chronic pain and the effect of morphine in relation to the dao.1 gene. Future studies will be required to resolve these unknowns, concerning the mechanisms of action of this gene in zebrafish.
Other gene involved in serine metabolism, whose expression was also down-regulated by exposure to morphine in zebrafish embryos, is the transporter of neutral amino acids slc1a4 (named also ACT; −1.27 FC). SLC1A4 seems to be the main uptake system of L-Ser in neurons [98]. Therefore, the presence of at least three genes differentially expressed after exposure to morphine and involved in serine metabolism implies that morphine can alter this type of metabolism influencing multiple processes involved in this amino acid, such as the regulation of NMDA receptors [99]. Other genes related to the primary excitatory amino acid neurotransmitter glutamate, and closely related to signal transduction, whose expression was altered by morphine treatment in zebrafish embryos were: a kainate-like receptor (grik-l); the transporter of glutamate slc1a3a (+1.18 FC); glutamate-ammonia ligases 1a and b (glula: +1.14 FC and glulb: −1.19 FC); and glutamate dehydrogenase 1b (glud1b: −1.18 FC). In the case of grik-l, we identified by in silico studies that Dr.3211.1.A1 probe set target sequence is complementary to deep 3’ untranslation region (UTR) of si:ch211-251b21.1 mRNA, which encodes a protein similar to mammalian glutamate ionotropic receptors, in particular, a kainate-like receptor [99]. Furthermore, Xu et al. [100] demonstrated that si:ch211-251b21.1 as a target gene is regulated by hedgehog signaling during development, and their expression was confined to the dorsal neurons of the spinal cord in wild-type zebrafish embryos at 24 hpf. On the other hand, it is known that morphine causes alterations of gene expression in distinct subunits of postsynaptic glutamatergic receptors, including kainate receptors [101]. Therefore, we suggest that the alteration of gene expression of grik-l by exposure to morphine could modify neural development of the spinal cord. Concerning slc1a3a gene, it is known that alteration of the expression of high-affinity glutamate transporters, like this gene, has been reported in drug dependence and addiction animal models [102]. In zebrafish, two orthologs for slc1a3 were found [103]. slc1a3a was expressed in glia cells of the larval zebrafish brain similar to the expression of mammalian SLC1A3 (also named GLAST or EAAT1) throughout the CNS [97, 103, 104]. SLC1A3 is critical to maintain the extracellular glutamate concentration in a non-neurotoxic range [97]. Interestingly, in mammals, it has been reported that the expression of SLC1A2 mRNA (or GLT1 or EAAT2), but not SLC1A3, is decreased in the striatum/nucleus accumbens and thalamus of morphine-dependent rats [105]. Therefore, we suggest that zebrafish slc1a3a, whose expression was down-regulated in embryos exposed to morphine, could be the key glutamate transporter in the regulation of the glutamate homeostasis in this species, and not slc1a3b or slc1a2. Finally, we also have observed opposite expression for the two orthologs glul genes, suggesting that both genes have different functions, such as reported Postlethwait et al. [65]. glulb expression is diminished in the same way as mice Glul is suppressed by morphine and by other addictive drugs, as methamphetamine, cocaine and alcohol [106].
Others genes involved in neuronal function
Within the group of genes up-regulated by exposure to morphine in zebrafish embryos, we found camk1γb, validated by RT-qPCR and identified as a gene related to oprm1 expression. This gene is involved in Ca2+ signal transduction in neuronal development, such as dendritogenesis and axonogenesis [107, 108]. In particular, CaMKI contributes strongly to Ca2+-mediated transcription in neurons through crosstalk with the Ras/extracellular-signal-regulated kinase (Erk) pathway. In cultured hippocampal neurons or acute slices, NMDA-stimulated activation of Erk is predominantly mediated through Ca2+/calmodulin-dependent protein kinase kinase (CaMKK)/CaMKI. Furthermore, this pathway appears to be important for dendritic arborization where activity-dependent NMDAR activation of the γ isoform of CaMKI results in MEK/Erk-mediated CREB regulated transcription of Wnt-2 and microRNA132 [108]. Therefore, we hipothetize the overexpression of camk1γb produced by morphine exposure in zebrafish embryos may result in pronounced acceleration of axon formation, such as reported by Davare et al. [107].
Another gene down-regulated by exposure to morphine and identified as a gene related to oprm1 expression is the wls, a putative orphan G-protein coupled receptor (GPCR) that encodes a OPRM1 interacting protein, and is conserved from worms to human [108]. Juul et al. [10] reported that Wls was also down-regulated by treatment with morphine in a mouse model of neonatal stress, which suggests that it can promote the inhibition of Wnt secretion. Wls is expressed in various brain regions and peripheral tissues in mammals and during zebrafish embryogenesis suggesting that WLS may play an essential role in regulating secretion of multiple Wnts throughout the body and specifically, it might be critical for neuronal development and morphogenesis among other functions [109–112]. In particular, zebrafish wls persists at 24 hpf in the spinal cord and in different areas of midbrain, hindbrain, midbrain-hindbrain boundary and ventricular zone. Using antisense morpholinos to KD wls mRNA translation in developing zebrafish, Jin et al. [110] suggested that wls expression was required for brain and ear development during zebrafish embryogenesis. WLS may possibly serve as a substrate underlying the alterations in neuronal structure and synaptic organization characteristic of opioid dependence [113]. Regarding these dependence processes, it should be emphasized that WLS and OPRM1 have been co-localized in somata and in dendritic processes in the murine striatum [113, 114], and that proteins that interact directly with the OPRM1, as WLS, influence their biosynthesis, trafficking and signaling [115], suggesting that these proteins could regulate these types of mechanisms, including signaling and trafficking. Reyes et al. [116] hypothesized that when morphine binds with OPRM1, the morphine-enhanced interaction between OPRM1 and WLS causes entrapment of WLS at the cell surface, and WLS is inefficiently internalized. Subsequently, a larger proportion of OPRM1 and WLS are present at the plasma membrane enabling more OPRM1 to be available for activation by morphine. Thus, inhibiting WLS function in mediating Wnt secretion is related to a significant inhibition of Wnt secretion in treatment with morphine. While WLS is inefficiently internalized after morphine treatment, WLS is efficiently internalized in the presence of [D-Ala, N-MePhe, Gly(ol)]-enkephalin (DAMGO) [113, 116] as it is known that happens with morphine. In conclusion, wls down-regulate expression by treatment with morphine suggesting a decrease downstream Wnt signaling, affecting neuronal development (structure and synaptic organization) and ear development in particular [109, 110, 112, 113]. Furthermore, we hypothesize that the decrease in wls expression could be a direct response mediated by the OPRM1. Two reasons suggest this: (i) our demonstration that wls is a gene related to OPRM1 expression, and (ii) a direct interaction at the protein level of WLS and OPRM1. Therefore, both gene expression and protein levels of wls/WLS are regulating actions of morphine through OPRM1.
Conclusions
We present here detailed changes in transcriptome of a critical period of zebrafish development, at 24 hpf, a key stage in the maturation and differentiation of CNS, during chronic treatment of morphine.
Using microarray technology, we identified different functional classes of genes and individual candidates (differentially expressed genes) involved in the mechanisms underlying susceptibility to morphine actions related to CNS development and, in general, the neural function. In particular, we identified 1023 genes whose expression is altered after chronic morphine exposure in zebrafish embryos and, of these, 254 genes had a FC of at least 1.3.
We found morphine-induced changes in gene expression that are specific for the zebrafish and other genes that are similar to mammals. Several morphine-induced genes were exclusively detected in zebrafish embryos, which may be putative targets to analyze, in human models, the problems of addiction and pain. In addition, we demonstrated that some of morphine-induced genes identified in our study might also be related in some way with the opioid system, in particular, the oprm1 expression, which could open new lines for the treatment of pain and the molecular mechanisms involved in addiction.
We suggest that some of the genes differentially expressed after chronic morphine exposure in zebrafish embryos could produce alterations in neuronal development, in particular, in notochord (in the case of copb2), spinal cord (grik-l) and other brain regions (wls and dao.1), CNS patterning processes (sox19b), differentiation and dopaminergic neurotransmission (otpb and copb2). Besides a possible induction of apoptosis and/or autophagy and alteration of serotonergic signaling pathway by the deregulation of bnip4, and activation of processes of axonogenesis and dendritogenesis (camk1γb) could be present in our experimental model. Finally, the down-regulation of dao.1 expression by treatment with morphine in zebrafish embryos contrary to that obtained with rodents opens a door hitherto unknown in the use of zebrafish as a model to study chronic pain and the effect of morphine in relation to this gene.
In conclusion, morphine in distinct species can affect common targets and some of these molecular targets may turn out to be central in understanding pain and addiction processes.
Methods
Animals
Adult zebrafish (wild-type AB strain) were raised in a cycle of 14 h light: 10 h dark at 26°C in a multi-tank system at our Fish Facilities at the Institute of Neuroscience of Castile & Leon, University of Salamanca. Embryos obtained from natural fertilization were selected at 24 hpf using a Discovery V8 stereomicroscope (Carl Zeiss, Germany), after which fish were raised at 28.5°C and maintained in dishes containing sterile E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl, 0.33 mM MgSO4) in distilled water. Embryos staged according to development in hpf according to Kimmel et al. [42].
Ethics statement
All procedures and experimental protocols were carried out in accordance with the guidelines of the European Communities Council directive of 24 November 1986 (86/609/EEC), current Spanish Legislation (RD 1201/2005, BOE 252/34367-91, 2005), and following the Guide for the Care and Use of Laboratory animals as adapted and promulgated by the US National Institute of Health. All efforts were made to minimize the number of embryos used and their possible suffering.
All experiments were performed at the Institute of Neuroscience of University of Salamanca with the approval of the Animal Care and Ethics Committee of this Institution.
Microarray study design and drug treatment
Zebrafish embryos were divided into two experimental groups: control embryos and embryos at 5 hpf (end of blastula) exposed to 10 nM morphine and collected at 24 hpf, in order to study the chronic effects of the exposure to drug. Morphine was administered to the embryos in their water environment, i.e., diluted in E3 embryonic medium. Microarray experiments were performed using six replicates for each condition, which contained the RNA of approximately one hundred embryos to minimize the influence of potential individual differences between the animals and technical variation introduced by tissue preparation. We previously reported that a concentration of 10 nM morphine is the highest concentration that can be used without a toxic effect on the embryos, and close to 5% of the morphine diluted in the E3 medium is detected in the embryo [54]. Morphine was acquired from the Spanish Ministry of Health.
RNA isolation and microarray hybridization
Total RNA was purified using TRIZOL® (Gibco BRL, Gaithersburg, MD, USA) following further RNA purification using an RNeasy Mini Kit for RNA clean-up (Qiagen Sicences, Maryland, USA). RNA quantification and quality was then assessed using Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA), to test the integrity of the 18S and 28S rRNA bands, and samples with an RNA integrity number (RIN) > 8.0 were used.
Microarray analysis was performed in the Cancer Research Center (CIC) of Salamanca according to standard procedures. Labelling and hybridizations were performed according to protocols from Affymetrix. Briefly, 100–300 ng of total RNA were amplified and labeled using the WT Sense Target labelling and control reagents kit (Affymetrix Inc., Santa Clara, CA, USA), and then hybridized to GeneChip® Zebrafish Genome Array (Affymetrix). Washing and scanning were performed using GeneChip System of Affymetrix (GeneChip Hybridization Oven 640, GeneChip Fluidics Station 450 and GeneChip Scanner 7G).
Microarray hybridization data analysis: normalization, differential gene expression and ontological analysis
The RMA (Robust Multi-array Analysis) algorithm [117] was used for background correction and normalization of fluorescent hybridization signals of the microarrays, both at internal (intra-microarrays) and comparative (inter-microarrays) levels. This algorithm was selected over others available (MAS5, (Affymetrix 2001); MBEI, a model-based algorithm) [118] because it was deemed to provide the best precision in signal detection to achieve adequate multiple-chip normalization [119], especially in cases of low-level gene expression [117, 120, 121] by producing efficient quantile normalization of the distribution of probe intensities from each array in the context of a complete set of arrays. We used Bioconductor and R as computational tools (http://www.bioconductor.org), to apply RMA to the data set of 12 microarray hybridizations including six different biological replicas corresponding to each of the different experimental groups under study (Control and Morphine).
After quantitation of expression level of each probe set in all microarrays analyzed, the SAM algorithm [122] was used to identify probe sets displaying significant differential expression when comparing the treat samples to it controls. This algorithm performs statistical discrimination analysis using permutations to check the stability of variables fulfilling the ‘alternative hypothesis’. The method calculates the type I error, or number of expected false positives, using the calculation of the False Discovery Rate (FDR) parameter [123]. In this report, genes with an FDR of 10% or less were considered significant.
Further processing included functional analysis and over representation calculations based on Gene Ontology (GO) Annotation Tool and publication data of Database for Annotation, Visualization, and Integrated Discovery was made with GeneSpring GX 7.3 and DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/) [124].
Data access
The data obtained and discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus [125] and are accessible through GEO Series accession number GSE61062 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE61062).
Quantitative reverse transcription real-time PCR (RT-qPCR)
Total RNA (2 μg), primed with oligo-dT, was reverse-transcribed into cDNA at 37°C for 2 h using the first-strand cDNA synthesis kit (Promega Corporation, Madison, WI, USA) in a 20 μl volume, and stored at −20°C until used, according to manufacturer’s instructions. In all cases, a reverse transcriptase negative control was used for testing genomic DNA contamination.
Quantitative real-time PCR (qPCR) were performed using the SYBR-Green method with a 2× Master Mix (Applied Biosystems). Each reaction contained 10 μL of Master Mix, 0.4 μL of each pair of primers, 3 μL of each cDNA sample in a different serial cDNA quantity for each gene, and MilliQ water up to 20 μl. The amplification reaction took place in an ABI Prism 7000 detection system (Applied Biosystems), with the following conditions: 10 min at 95°C followed by 40 cycles of 15 s at 95°C and 1 min at 60°C depending on each pair of primers. RT-qPCR experiments were performed in replicates of eight and run in triplicate for each gene product examined. The list of used primers is provided in Additional file 6. Zebrafish β-actin was used as housekeeping gene.
The comparative Ct method was used for presenting quantitative data [126]. Following the removal of outliers, raw fluorescence data were used to determine the PCR amplification efficiency (E) according to the formula E = [10 (−1/slope) −1] * 100. All amplifications had an E value of 100 ± 10% the E value close to 100% being an indicator of efficient amplification. The relative gene expression value (FC) for each transcript was calculated according to the equation 2 −(ΔCt “condition 1” − ΔCt “condition 2”), where “condition 1” corresponds to experimental samples (“treatment with morphine”), “condition 2” to samples of control animals and ΔCt of each “condition” is Ct “experimental gene” − Ct “endogenous gene” [126]. A standard error for each relative gene expression value was calculated as a measure of data variation. Significance from qPCR analysis was determined using a one-way t-test for each gene, testing that |FC| > 1 is significant (p < 0.05).
Mu opioid receptor morpholino microinjection
Antisense MO oligonucleotides used were provided by Gene Tools (LLC Philomath, OR, USA). MO injection was performed according to the methodology developed by Nasevicius and Ekker [127], for which a microinjection and micromanipulation system designed especially for the microinjection of zebrafish embryos was used, coupled to a stereoscope (Stereo Microscope Discovery V8, Zeiss, Göttingen, Germany) and a high-resolution camera that allowed videos and photos to be taken. First, the ideal concentration of MO was determined. Accordingly, several concentrations, looking for small lethal effects and maximum embryonic survival, were microinjected. Thus, the highest concentration with minimum mortality was employed; concentrations of 1 M were used for oprm1 (MOR-MO group). Moreover, parameters of pressure and the appropriate time for microinjecting the volume calculated in each pulse (3 nl of solution) were calibrated. Zebrafish embryo yolks microinjected with MOR-MO and the MO controls were used in the single-cell stage of development. Other embryos, in which no solution was microinjected, were used as a control group. The microinjected and the non-microinjected embryos were maintained under the same conditions: 28.5°C and in the E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4 in ddH2O). To inhibit oprm1 mRNA translation we used the following MOR-MO sequence: AATGTTGCCAGTGTTTTCCATCATG. The efficacy and specificity of MO employed was demonstrated by Sanchez-Simon et al. [54].
Electronic supplementary material
Additional file 1: Table listing the probe-sets of genes expressed differentially by chronic morphine treatment in zebrafish embryos. Significance level for each probe-set are presented as d-, p- and q-value. The expression changes between Morphine vs. Control are showed as R fold. List of known genes up-, down-, and up-/down-regulated. (XLSX 199 KB)
Additional file 2: Figure presenting gene ontology analysis of the zebrafish gene collection. Genes were categorized with the Biological Process domain. Significantly enriched GO terms have a probability lower than 0.01 (P value) and include at least three genes. GO terms are shown if they are significantly enriched in at least one of them. Significantly enriched GO terms are indicated as black bars whereas non-significantly enriched terms are displayed as empty bars. Bars represent the number of genes assigned with the corresponding GO term. For more details, see also Additional file 3. (PDF 188 KB)
Additional file 3: Data file providing the detailed description of Gene Ontology analysis presented in Figure 2 and Table 1 . List of probe-sets and gene names classified for each GO category. (XLSX 70 KB)
Additional file 4: Table listing genes differentially expressed by morphine and included in each functional category presented in Figure 3 and Table 2 . The neuronal function category is emphasized in two excel sheet tabs (listing genes and Gene Ontology analysis of genes with neuronal function). (XLSX 98 KB)
Additional file 5: Table listing genes differentially expressed by morphine validation by our in silico analysis. (XLSX 34 KB)
Additional file 6: Table listing the primers used for RT-qPCR. (PDF 35 KB)
Acknowledgements
This work was supported by the Spanish Ministry of Education and Science (Grant SAF2010-18597) and the Regional Government of Castilla y León (Grant B1039/SA25/10).
Abbreviations
- CNS
Central nervous system
- CPP
Conditioned place preference
- Ct
Threshold cycle
- FC
Fold change
- hpf
Hours post-fertilization
- IEG
Immediate early genes
- KD
Knocked down
- OPRM1
Opioid receptor, mu 1
- RT-qPCR
Quantitative reverse transcription real-time PCR
- SAM
Significance analysis of microarrays
- UTR
Untranslated region.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MJHT and RER designed the experiments, discussed the results and wrote the manuscript. RLB prepared the morphine treated experimental groups. MJHT and IRM performed RT-qPCR experiments. MJHT analyzed the data and performed statistical analyses. All authors commented on and revised the manuscript. All authors read and approved the final manuscript.
Contributor Information
M Javier Herrero-Turrión, Email: mjaviht@usal.es.
Iván Rodríguez-Martín, Email: ivan.rodriguez@uem.es.
Roger López-Bellido, Email: rogerlopez@usal.es.
Raquel E Rodríguez, Email: requelmi@usal.es.
References
- 1.Rodriguez RE. Morphine and microRNA Activity: is there a relation with addiction? Front Genet. 2012;3:223. doi: 10.3389/fgene.2012.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stein C. Opioids, sensory systems and chronic pain. Eur J Pharmacol. 2013;716(1–3):179–187. doi: 10.1016/j.ejphar.2013.01.076. [DOI] [PubMed] [Google Scholar]
- 3.Volkow ND, Li TK. Drug addiction: the neurobiology of behaviour gone awry. Nat Rev Neurosci. 2004;5(12):963–970. doi: 10.1038/nrn1539. [DOI] [PubMed] [Google Scholar]
- 4.Everitt BJ, Belin D, Economidou D, Pelloux Y, Dalley JW, Robbins TW. Review. Neural mechanisms underlying the vulnerability to develop compulsive drug-seeking habits and addiction. Philos Trans R Soc Lond B Biol Sci. 2008;363(1507):3125–3135. doi: 10.1098/rstb.2008.0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Przewlocki R. Opioid abuse and brain gene expression. Eur J Pharmacol. 2004;500(1–3):331–349. doi: 10.1016/j.ejphar.2004.07.036. [DOI] [PubMed] [Google Scholar]
- 6.Loguinov AV, Anderson LM, Crosby GJ, Yukhananov RY. Gene expression following acute morphine administration. Physiol Genomics. 2001;6(3):169–181. doi: 10.1152/physiolgenomics.2001.6.3.169. [DOI] [PubMed] [Google Scholar]
- 7.Ammon-Treiber S, Hollt V. Morphine-induced changes of gene expression in the brain. Addict Biol. 2005;10(1):81–89. doi: 10.1080/13556210412331308994. [DOI] [PubMed] [Google Scholar]
- 8.Anghel A, Jamieson CA, Ren X, Young J, Porche R, Ozigbo E, Ghods DE, Lee ML, Liu Y, Lutfy K, Friedman TC. Gene expression profiling following short-term and long-term morphine exposure in mice uncovers genes involved in food intake. Neuroscience. 2010;167(2):554–566. doi: 10.1016/j.neuroscience.2010.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grice DE, Reenila I, Mannisto PT, Brooks AI, Smith GG, Golden GT, Buxbaum JD, Berrettini WH. Transcriptional profiling of C57 and DBA strains of mice in the absence and presence of morphine. BMC Genomics. 2007;8:76. doi: 10.1186/1471-2164-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juul SE, Beyer RP, Bammler TK, Farin FM, Gleason CA. Effects of neonatal stress and morphine on murine hippocampal gene expression. Pediatr Res. 2011;69(4):285–292. doi: 10.1203/PDR.0b013e31820bd165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korostynski M, Kaminska-Chowaniec D, Piechota M, Przewlocki R. Gene expression profiling in the striatum of inbred mouse strains with distinct opioid-related phenotypes. BMC Genomics. 2006;7:146. doi: 10.1186/1471-2164-7-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korostynski M, Piechota M, Kaminska D, Solecki W, Przewlocki R. Morphine effects on striatal transcriptome in mice. Genome Biol. 2007;8(6):R128. doi: 10.1186/gb-2007-8-6-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McClung CA, Nestler EJ, Zachariou V. Regulation of gene expression by chronic morphine and morphine withdrawal in the locus ceruleus and ventral tegmental area. J Neurosci. 2005;25(25):6005–6015. doi: 10.1523/JNEUROSCI.0062-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piechota M, Korostynski M, Solecki W, Gieryk A, Slezak M, Bilecki W, Ziolkowska B, Kostrzewa E, Cymerman I, Swiech L, Jaworski J, Przewlocki R. The dissection of transcriptional modules regulated by various drugs of abuse in the mouse striatum. Genome Biol. 2010;11(5):R48. doi: 10.1186/gb-2010-11-5-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhodes JS, Crabbe JC. Gene expression induced by drugs of abuse. Curr Opin Pharmacol. 2005;5(1):26–33. doi: 10.1016/j.coph.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez Parkitna JM, Bilecki W, Mierzejewski P, Stefanski R, Ligeza A, Bargiela A, Ziolkowska B, Kostowski W, Przewlocki R. Effects of morphine on gene expression in the rat amygdala. J Neurochem. 2004;91(1):38–48. doi: 10.1111/j.1471-4159.2004.02697.x. [DOI] [PubMed] [Google Scholar]
- 17.Tapocik JD, Luu TV, Mayo CL, Wang BD, Doyle E, Lee AD, Lee NH, Elmer GI. Neuroplasticity, axonal guidance and micro-RNA genes are associated with morphine self-administration behavior. Addict Biol. 2013;18(3):480–495. doi: 10.1111/j.1369-1600.2012.00470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tapocik JD, Letwin N, Mayo CL, Frank B, Luu T, Achinike O, House C, Williams R, Elmer GI, Lee NH. Identification of candidate genes and gene networks specifically associated with analgesic tolerance to morphine. J Neurosci. 2009;29(16):5295–5307. doi: 10.1523/JNEUROSCI.4020-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziolkowska B, Korostynski M, Piechota M, Kubik J, Przewlocki R. Effects of morphine on immediate-early gene expression in the striatum of C57BL/6 J and DBA/2 J mice. Pharmacol Rep. 2012;64(5):1091–1104. doi: 10.1016/S1734-1140(12)70906-4. [DOI] [PubMed] [Google Scholar]
- 20.Hassan HE, Myers AL, Lee IJ, Chen H, Coop A, Eddington ND. Regulation of gene expression in brain tissues of rats repeatedly treated by the highly abused opioid agonist, oxycodone: microarray profiling and gene mapping analysis. Drug Metab Dispos. 2010;38(1):157–167. doi: 10.1124/dmd.109.029199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuferov V, Nielsen D, Butelman E, Kreek MJ. Microarray studies of psychostimulant-induced changes in gene expression. Addict Biol. 2005;10(1):101–118. doi: 10.1080/13556210412331308976. [DOI] [PubMed] [Google Scholar]
- 22.Albertson DN, Schmidt CJ, Kapatos G, Bannon MJ. Distinctive profiles of gene expression in the human nucleus accumbens associated with cocaine and heroin abuse. Neuropsychopharmacology. 2006;31(10):2304–2312. doi: 10.1038/sj.npp.1301089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehrmann E, Colantuoni C, Deep-Soboslay A, Becker KG, Lowe R, Huestis MA, Hyde TM, Kleinman JE, Freed WJ. Transcriptional changes common to human cocaine, cannabis and phencyclidine abuse. PLoS One. 2006;1:e114. doi: 10.1371/journal.pone.0000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piechota M, Korostynski M, Sikora M, Golda S, Dzbek J, Przewlocki R. Common transcriptional effects in the mouse striatum following chronic treatment with heroin and methamphetamine. Genes Brain Behav. 2012;11(4):404–414. doi: 10.1111/j.1601-183X.2012.00777.x. [DOI] [PubMed] [Google Scholar]
- 25.Korostynski M, Piechota M, Dzbek J, Mlynarski W, Szklarczyk K, Ziolkowska B, Przewlocki R. Novel drug-regulated transcriptional networks in brain reveal pharmacological properties of psychotropic drugs. BMC Genomics. 2013;14(1):606. doi: 10.1186/1471-2164-14-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kily LJ, Cowe YC, Hussain O, Patel S, McElwaine S, Cotter FE, Brennan CH. Gene expression changes in a zebrafish model of drug dependency suggest conservation of neuro-adaptation pathways. J Exp Biol. 2008;211(Pt 10):1623–1634. doi: 10.1242/jeb.014399. [DOI] [PubMed] [Google Scholar]
- 27.Webb KJ, Norton WH, Trumbach D, Meijer AH, Ninkovic J, Topp S, Heck D, Marr C, Wurst W, Theis FJ, Spaink HP, Bally-Cuif L. Zebrafish reward mutants reveal novel transcripts mediating the behavioral effects of amphetamine. Genome Biol. 2009;10(7):R81. doi: 10.1186/gb-2009-10-7-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan Y, Kaiguo M, Razak Z, Westwood JT, Gerlai R. Chronic alcohol exposure induced gene expression changes in the zebrafish brain. Behav Brain Res. 2011;216(1):66–76. doi: 10.1016/j.bbr.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tzschentke TM. Measuring reward with the conditioned place preference (CPP) paradigm: update of the last decade. Addict Biol. 2007;12(3–4):227–462. doi: 10.1111/j.1369-1600.2007.00070.x. [DOI] [PubMed] [Google Scholar]
- 30.Lau B, Bretaud S, Huang Y, Lin E, Guo S. Dissociation of food and opiate preference by a genetic mutation in zebrafish. Genes Brain Behav. 2006;5(7):497–505. doi: 10.1111/j.1601-183X.2005.00185.x. [DOI] [PubMed] [Google Scholar]
- 31.Ninkovic J, Bally-Cuif L. The zebrafish as a model system for assessing the reinforcing properties of drugs of abuse. Methods. 2006;39(3):262–274. doi: 10.1016/j.ymeth.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Bretaud S, Li Q, Lockwood BL, Kobayashi K, Lin E, Guo S. A choice behavior for morphine reveals experience-dependent drug preference and underlying neural substrates in developing larval zebrafish. Neuroscience. 2007;146(3):1109–1116. doi: 10.1016/j.neuroscience.2006.12.073. [DOI] [PubMed] [Google Scholar]
- 33.Santoriello C, Zon LI. Hooked! Modeling human disease in zebrafish. J Clin Invest. 2012;122(7):2337–2343. doi: 10.1172/JCI60434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lohi O, Parikka M, Ramet M. The zebrafish as a model for paediatric diseases. Acta Paediatr. 2013;102(2):104–110. doi: 10.1111/j.1651-2227.2012.02835.x. [DOI] [PubMed] [Google Scholar]
- 35.Kari G, Rodeck U, Dicker AP. Zebrafish: an emerging model system for human disease and drug discovery. Clin Pharmacol Ther. 2007;82(1):70–80. doi: 10.1038/sj.clpt.6100223. [DOI] [PubMed] [Google Scholar]
- 36.Chakraborty C, Hsu CH, Wen ZH, Lin CS, Agoramoorthy G. Zebrafish: a complete animal model for in vivo drug discovery and development. Curr Drug Metab. 2009;10(2):116–124. doi: 10.2174/138920009787522197. [DOI] [PubMed] [Google Scholar]
- 37.Guo S. Using zebrafish to assess the impact of drugs on neural development and function. Expert Opin Drug Discov. 2009;4(7):715–726. doi: 10.1517/17460440902988464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cachat J, Canavello P, Elegante M, Bartels B, Hart P, Bergner C, Egan R, Duncan A, Tien D, Chung A, Wong K, Goodspeed J, Tan J, Grimes C, Elkhayat S, Suciu C, Rosenberg M, Chung KM, Kadri F, Roy S, Gaikwad S, Stewart A, Zapolsky I, Gilder T, Mohnot S, Beeson E, Amri H, Zukowska Z, Soignier RD, Kalueff AV. Modeling withdrawal syndrome in zebrafish. Behav Brain Res. 2010;208(2):371–376. doi: 10.1016/j.bbr.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 39.Stewart A, Wong K, Cachat J, Gaikwad S, Kyzar E, Wu N, Hart P, Piet V, Utterback E, Elegante M, Tien D, Kalueff AV. Zebrafish models to study drug abuse-related phenotypes. Rev Neurosci. 2011;22(1):95–105. doi: 10.1515/RNS.2011.011. [DOI] [PubMed] [Google Scholar]
- 40.Nasiraei-Moghadam S, Sahraei H, Bahadoran H, Sadooghi M, Salimi SH, Kaka GR, Imani H, Mahdavi-Nasab H, Dashtnavard H. Effects of maternal oral morphine consumption on neural tube development in Wistar rats. Brain Res Dev Brain Res. 2005;159(1):12–17. doi: 10.1016/j.devbrainres.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 41.Nasiraei-Moghadam S, Kazeminezhad B, Dargahi L, Ahmadiani A. Maternal oral consumption of morphine increases Bax/Bcl-2 ratio and caspase 3 activity during early neural system development in rat embryos. J Mol Neurosci. 2010;41(1):156–164. doi: 10.1007/s12031-009-9312-6. [DOI] [PubMed] [Google Scholar]
- 42.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 43.Wilson SW, Brand M, Eisen JS. Patterning the zebrafish central nervous system. Results Probl Cell Differ. 2002;40:181–215. doi: 10.1007/978-3-540-46041-1_10. [DOI] [PubMed] [Google Scholar]
- 44.Gonzalez-Nunez V, Rodriguez RE. The zebrafish: a model to study the endogenous mechanisms of pain. ILAR J. 2009;50(4):373–386. doi: 10.1093/ilar.50.4.373. [DOI] [PubMed] [Google Scholar]
- 45.Herrero-Turrion MJ, Sánchez-Simón FM, Rodríguez RE. Opioids and Opioid Receptors in Fishes. In: Farrell AP, editor. Encyclopedia of Fish Physiology from Genome to Environment. 2011. 2011. pp. 89–97. [Google Scholar]
- 46.de Velasco EM, Law PY, Rodriguez RE. Mu opioid receptor from the zebrafish exhibits functional characteristics as those of mammalian mu opioid receptor. Zebrafish. 2009;6(3):259–268. doi: 10.1089/zeb.2009.0594. [DOI] [PubMed] [Google Scholar]
- 47.Le Merrer J, Becker JA, Befort K, Kieffer BL. Reward processing by the opioid system in the brain. Physiol Rev. 2009;89(4):1379–1412. doi: 10.1152/physrev.00005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Charbogne P, Kieffer BL, Befort K. 15 years of genetic approaches in vivo for addiction research: Opioid receptor and peptide gene knockout in mouse models of drug abuse. Neuropharmacology. 2014;76:204–217. doi: 10.1016/j.neuropharm.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanchez-Simon FM, Rodriguez RE. Developmental expression and distribution of opioid receptors in zebrafish. Neuroscience. 2008;151(1):129–137. doi: 10.1016/j.neuroscience.2007.09.086. [DOI] [PubMed] [Google Scholar]
- 50.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 51.Yoshikawa M, Andoh H, Ito K, Suzuki T, Kawaguchi M, Kobayashi H, Oka T, Hashimoto A. Acute treatment with morphine augments the expression of serine racemase and D-amino acid oxidase mRNAs in rat brain. Eur J Pharmacol. 2005;525(1–3):94–97. doi: 10.1016/j.ejphar.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 52.Yoshikawa M, Shinomiya T, Takayasu N, Tsukamoto H, Kawaguchi M, Kobayashi H, Oka T, Hashimoto A. Long-term treatment with morphine increases the D-serine content in the rat brain by regulating the mRNA and protein expressions of serine racemase and D-amino acid oxidase. J Pharmacol Sci. 2008;107(3):270–276. doi: 10.1254/jphs.08030FP. [DOI] [PubMed] [Google Scholar]
- 53.Morey JS, Ryan JC, Van Dolah FM. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol Proced. 2006;8:175–193. doi: 10.1251/bpo126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanchez-Simon FM, Zhang XX, Loh HH, Law PY, Rodriguez RE. Morphine regulates dopaminergic neuron differentiation via miR-133b. Mol Pharmacol. 2010;78(5):935–942. doi: 10.1124/mol.110.066837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lopez-Bellido R, Barreto-Valer K, Rodriguez RE. Substance P mRNA expression during zebrafish development: influence of mu opioid receptor and cocaine. Neuroscience. 2013;242:53–68. doi: 10.1016/j.neuroscience.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 56.Lopez-Bellido R, Barreto-Valer K, Sanchez-Simon FM, Rodriguez RE. Cocaine modulates the expression of opioid receptors and miR-let-7d in zebrafish embryos. PLoS One. 2012;7(11):e50885. doi: 10.1371/journal.pone.0050885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Befort K, Filliol D, Darcq E, Ghate A, Matifas A, Lardenois A, Muller J, Thibault C, Dembele D, Poch O, Kieffer BL. Gene expression is altered in the lateral hypothalamus upon activation of the mu opioid receptor. Ann N Y Acad Sci. 2008;1129:175–184. doi: 10.1196/annals.1417.028. [DOI] [PubMed] [Google Scholar]
- 58.Befort K, Filliol D, Ghate A, Darcq E, Matifas A, Muller J, Lardenois A, Thibault C, Dembele D, Le Merrer J, Becker JA, Poch O, Kieffer BL. Mu-opioid receptor activation induces transcriptional plasticity in the central extended amygdala. Eur J Neurosci. 2008;27(11):2973–2984. doi: 10.1111/j.1460-9568.2008.06273.x. [DOI] [PubMed] [Google Scholar]
- 59.Downing C, Flink S, Florez-McClure ML, Johnson TE, Tabakoff B, Kechris KJ. Gene expression changes in C57BL/6 J and DBA/2 J mice following prenatal alcohol exposure. Alcohol Clin Exp Res. 2012;36(9):1519–1529. doi: 10.1111/j.1530-0277.2012.01757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones HE, Heil SH, Baewert A, Arria AM, Kaltenbach K, Martin PR, Coyle MG, Selby P, Stine SM, Fischer G. Buprenorphine treatment of opioid-dependent pregnant women: a comprehensive review. Addiction. 2012;107(Suppl 1):5–27. doi: 10.1111/j.1360-0443.2012.04035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saurer TB, Ijames SG, Carrigan KA, Lysle DT. Neuroimmune mechanisms of opioid-mediated conditioned immunomodulation. Brain Behav Immun. 2008;22(1):89–97. doi: 10.1016/j.bbi.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hays SL, McPherson RJ, Juul SE, Wallace G, Schindler AG, Chavkin C, Gleason CA. Long-term effects of neonatal stress on adult conditioned place preference (CPP) and hippocampal neurogenesis. Behav Brain Res. 2012;227(1):7–11. doi: 10.1016/j.bbr.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barreto-Valer K, Lopez-Bellido R, Rodriguez RE. Cocaine modulates the expression of transcription factors related to the dopaminergic system in zebrafish. Neuroscience. 2013;231:258–271. doi: 10.1016/j.neuroscience.2012.11.052. [DOI] [PubMed] [Google Scholar]
- 64.Perez-Cadahia B, Drobic B, Davie JR. Activation and function of immediate-early genes in the nervous system. Biochem Cell Biol. 2011;89(1):61–73. doi: 10.1139/O10-138. [DOI] [PubMed] [Google Scholar]
- 65.Postlethwait JH. The zebrafish genome in context: ohnologs gone missing. J Exp Zool B Mol Dev Evol. 2007;308(5):563–577. doi: 10.1002/jez.b.21137. [DOI] [PubMed] [Google Scholar]
- 66.Orsini C, Bonito-Oliva A, Conversi D, Cabib S. Susceptibility to conditioned place preference induced by addictive drugs in mice of the C57BL/6 and DBA/2 inbred strains. Psychopharmacology (Berl) 2005;181(2):327–336. doi: 10.1007/s00213-005-2259-6. [DOI] [PubMed] [Google Scholar]
- 67.Girard F, Cremazy F, Berta P, Renucci A. Expression pattern of the Sox31 gene during zebrafish embryonic development. Mech Dev. 2001;100(1):71–73. doi: 10.1016/S0925-4773(00)00491-3. [DOI] [PubMed] [Google Scholar]
- 68.Hu SN, Yu H, Zhang YB, Wu ZL, Yan YC, Li YX, Li YY, Li YP. Splice blocking of zygotic sox31 leads to developmental arrest shortly after Mid-Blastula Transition and induces apoptosis in zebrafish. FEBS Lett. 2012;586(3):222–228. doi: 10.1016/j.febslet.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 69.Hu S, Wu Z, Yan Y, Li Y. Sox31 is involved in central nervous system anteroposterior regionalization through regulating the organizer activity in zebrafish. Acta Biochim Biophys Sin (Shanghai) 2011;43(5):387–399. doi: 10.1093/abbs/gmr025. [DOI] [PubMed] [Google Scholar]
- 70.Frederick AL, Stanwood GD. Drugs, biogenic amine targets and the developing brain. Dev Neurosci. 2009;31(1–2):7–22. doi: 10.1159/000207490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gianoulakis C. Endogenous opioids and addiction to alcohol and other drugs of abuse. Curr Top Med Chem. 2009;9(11):999–1015. doi: 10.2174/156802609789630956. [DOI] [PubMed] [Google Scholar]
- 72.Rico EP, Rosemberg DB, Seibt KJ, Capiotti KM, Da Silva RS, Bonan CD. Zebrafish neurotransmitter systems as potential pharmacological and toxicological targets. Neurotoxicol Teratol. 2011;33(6):608–617. doi: 10.1016/j.ntt.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 73.Klee EW, Schneider H, Clark KJ, Cousin MA, Ebbert JO, Hooten WM, Karpyak VM, Warner DO, Ekker SC. Zebrafish: a model for the study of addiction genetics. Hum Genet. 2012;131(6):977–1008. doi: 10.1007/s00439-011-1128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schweitzer J, Driever W. Development of the dopamine systems in zebrafish. Adv Exp Med Biol. 2009;651:1–14. doi: 10.1007/978-1-4419-0322-8_1. [DOI] [PubMed] [Google Scholar]
- 75.Tay TL, Ronneberger O, Ryu S, Nitschke R, Driever W. Comprehensive catecholaminergic projectome analysis reveals single-neuron integration of zebrafish ascending and descending dopaminergic systems. Nat Commun. 2011;2:171. doi: 10.1038/ncomms1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Panula P, Chen YC, Priyadarshini M, Kudo H, Semenova S, Sundvik M, Sallinen V. The comparative neuroanatomy and neurochemistry of zebrafish CNS systems of relevance to human neuropsychiatric diseases. Neurobiol Dis. 2010;40(1):46–57. doi: 10.1016/j.nbd.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 77.Kastenhuber E, Kratochwil CF, Ryu S, Schweitzer J, Driever W. Genetic dissection of dopaminergic and noradrenergic contributions to catecholaminergic tracts in early larval zebrafish. J Comp Neurol. 2010;518(4):439–458. doi: 10.1002/cne.22214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Del Giacco L, Sordino P, Pistocchi A, Andreakis N, Tarallo R, Di Benedetto B, Cotelli F. Differential regulation of the zebrafish orthopedia 1 gene during fate determination of diencephalic neurons. BMC Dev Biol. 2006;6:50. doi: 10.1186/1471-213X-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ryu S, Mahler J, Acampora D, Holzschuh J, Erhardt S, Omodei D, Simeone A, Driever W. Orthopedia homeodomain protein is essential for diencephalic dopaminergic neuron development. Curr Biol. 2007;17(10):873–880. doi: 10.1016/j.cub.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 80.Wolf A, Ryu S. Specification of posterior hypothalamic neurons requires coordinated activities of Fezf2, Otp, Sim1a and Foxb1.2. Development. 2013;140(8):1762–1773. doi: 10.1242/dev.085357. [DOI] [PubMed] [Google Scholar]
- 81.Eaton JL, Holmqvist B, Glasgow E. Ontogeny of vasotocin-expressing cells in zebrafish: selective requirement for the transcriptional regulators orthopedia and single-minded 1 in the preoptic area. Dev Dyn. 2008;237(4):995–1005. doi: 10.1002/dvdy.21503. [DOI] [PubMed] [Google Scholar]
- 82.Eaton JL, Glasgow E. Zebrafish orthopedia (otp) is required for isotocin cell development. Dev Genes Evol. 2007;217(2):149–158. doi: 10.1007/s00427-006-0123-2. [DOI] [PubMed] [Google Scholar]
- 83.Fernandes AM, Beddows E, Filippi A, Driever W. Orthopedia transcription factor otpa and otpb paralogous genes function during dopaminergic and neuroendocrine cell specification in larval zebrafish. PLoS One. 2013;8(9):e75002. doi: 10.1371/journal.pone.0075002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li D, Roberts R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol Life Sci. 2001;58(14):2085–2097. doi: 10.1007/PL00000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang X, Wang W, Bedigian AV, Coughlin ML, Mitchison TJ, Eggert US. Dopamine receptor D3 regulates endocytic sorting by a Prazosin-sensitive interaction with the coatomer COPI. Proc Natl Acad Sci U S A. 2012;109(31):12485–12490. doi: 10.1073/pnas.1207821109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Feng X, Liu X, Zhang W, Xiao W. p53 directly suppresses BNIP3 expression to protect against hypoxia-induced cell death. EMBO J. 2011;30(16):3397–3415. doi: 10.1038/emboj.2011.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cho B, Choi SY, Park OH, Sun W, Geum D. Differential expression of BNIP family members of BH3-only proteins during the development and after axotomy in the rat. Mol Cells. 2012;33(6):605–610. doi: 10.1007/s10059-012-0051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tohda M, Hayashi H, Sukma M, Tanaka K. BNIP-3: a novel candidate for an intrinsic depression-related factor found in NG108-15 cells treated with Hochu-ekki-to, a traditional oriental medicine, or typical antidepressants. Neurosci Res. 2008;62(1):1–8. doi: 10.1016/j.neures.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 89.Tohda M, Mingmalairak S, Murakami Y, Matsumoto K. Enhanced expression of BCL2/adenovirus EIB 19-kDa-interacting protein 3 mRNA, a candidate for intrinsic depression-related factor, and effects of imipramine in the frontal cortex of stressed mice. Biol Pharm Bull. 2010;33(1):53–57. doi: 10.1248/bpb.33.53. [DOI] [PubMed] [Google Scholar]
- 90.Lopez-Gimenez JF, Vilaro MT, Milligan G. Morphine desensitization, internalization, and down-regulation of the mu opioid receptor is facilitated by serotonin 5-hydroxytryptamine2A receptor coactivation. Mol Pharmacol. 2008;74(5):1278–1291. doi: 10.1124/mol.108.048272. [DOI] [PubMed] [Google Scholar]
- 91.Kalivas PW. The glutamate homeostasis hypothesis of addiction. Nat Rev Neurosci. 2009;10(8):561–572. doi: 10.1038/nrn2515. [DOI] [PubMed] [Google Scholar]
- 92.Spijker S, Houtzager SW, De Gunst MC, De Boer WP, Schoffelmeer AN, Smit AB. Morphine exposure and abstinence define specific stages of gene expression in the rat nucleus accumbens. FASEB J. 2004;18(7):848–850. doi: 10.1096/fj.03-0612fje. [DOI] [PubMed] [Google Scholar]
- 93.Shleper M, Kartvelishvily E, Wolosker H. D-serine is the dominant endogenous coagonist for NMDA receptor neurotoxicity in organotypic hippocampal slices. J Neurosci. 2005;25(41):9413–9417. doi: 10.1523/JNEUROSCI.3190-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pollegioni L, Piubelli L, Sacchi S, Pilone MS, Molla G. Physiological functions of D-amino acid oxidases: from yeast to humans. Cell Mol Life Sci. 2007;64(11):1373–1394. doi: 10.1007/s00018-007-6558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang YX, Gong N, Xin YF, Hao B, Zhou XJ, Pang CC. Biological implications of oxidation and unidirectional chiral inversion of D-amino acids. Curr Drug Metab. 2012;13(3):321–331. doi: 10.2174/138920012799320392. [DOI] [PubMed] [Google Scholar]
- 96.Gong N, Li XY, Xiao Q, Wang YX. Identification of a Novel Spinal Dorsal Horn Astroglial D-Amino Acid Oxidase-Hydrogen Peroxide Pathway Involved in Morphine Antinociceptive Tolerance. Anesthesiology. 2014;120(4):962–975. doi: 10.1097/ALN.0b013e3182a66d2a. [DOI] [PubMed] [Google Scholar]
- 97.Zhao WJ, Gao ZY, Wei H, Nie HZ, Zhao Q, Zhou XJ, Wang YX. Spinal D-amino acid oxidase contributes to neuropathic pain in rats. J Pharmacol Exp Ther. 2010;332(1):248–254. doi: 10.1124/jpet.109.158816. [DOI] [PubMed] [Google Scholar]
- 98.Kanai Y, Clemencon B, Simonin A, Leuenberger M, Lochner M, Weisstanner M, Hediger MA. The SLC1 high-affinity glutamate and neutral amino acid transporter family. Mol Aspects Med. 2013;34(2–3):108–120. doi: 10.1016/j.mam.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 99.Sacchi S. D-Serine metabolism: new insights into the modulation of D-amino acid oxidase activity. Biochem Soc Trans. 2013;41(6):1551–1556. doi: 10.1042/BST20130184. [DOI] [PubMed] [Google Scholar]
- 100.Xu J, Srinivas BP, Tay SY, Mak A, Yu X, Lee SG, Yang H, Govindarajan KR, Leong B, Bourque G, Mathavan S, Roy S. Genomewide expression profiling in the zebrafish embryo identifies target genes regulated by Hedgehog signaling during vertebrate development. Genetics. 2006;174(2):735–752. doi: 10.1534/genetics.106.061523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jacobs EH, Wardeh G, Smit AB, Schoffelmeer AN. Morphine causes a delayed increase in glutamate receptor functioning in the nucleus accumbens core. Eur J Pharmacol. 2005;511(1):27–30. doi: 10.1016/j.ejphar.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 102.Nakagawa T, Satoh M. Involvement of glial glutamate transporters in morphine dependence. Ann N Y Acad Sci. 2004;1025:383–388. doi: 10.1196/annals.1307.047. [DOI] [PubMed] [Google Scholar]
- 103.Gesemann M, Lesslauer A, Maurer CM, Schonthaler HB, Neuhauss SC. Phylogenetic analysis of the vertebrate excitatory/neutral amino acid transporter (SLC1/EAAT) family reveals lineage specific subfamilies. BMC Evol Biol. 2010;10:117. doi: 10.1186/1471-2148-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nakagawa T, Kaneko S. SLC1 glutamate transporters and diseases: psychiatric diseases and pathological pain. Curr Mol Pharmacol. 2013;6(2):66–73. doi: 10.2174/18744672113069990033. [DOI] [PubMed] [Google Scholar]
- 105.Ozawa T, Nakagawa T, Shige K, Minami M, Satoh M. Changes in the expression of glial glutamate transporters in the rat brain accompanied with morphine dependence and naloxone-precipitated withdrawal. Brain Res. 2001;905(1–2):254–258. doi: 10.1016/S0006-8993(01)02536-7. [DOI] [PubMed] [Google Scholar]
- 106.Wang J, Yuan W, Li MD. Genes and pathways co-associated with the exposure to multiple drugs of abuse, including alcohol, amphetamine/methamphetamine, cocaine, marijuana, morphine, and/or nicotine: a review of proteomics analyses. Mol Neurobiol. 2011;44(3):269–286. doi: 10.1007/s12035-011-8202-4. [DOI] [PubMed] [Google Scholar]
- 107.Davare MA, Fortin DA, Saneyoshi T, Nygaard S, Kaech S, Banker G, Soderling TR, Wayman GA. Transient receptor potential canonical 5 channels activate Ca2+/calmodulin kinase Igamma to promote axon formation in hippocampal neurons. J Neurosci. 2009;29(31):9794–9808. doi: 10.1523/JNEUROSCI.1544-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wayman GA, Lee YS, Tokumitsu H, Silva AJ, Soderling TR. Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron. 2008;59(6):914–931. doi: 10.1016/j.neuron.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Das S, Yu S, Sakamori R, Stypulkowski E, Gao N. Wntless in Wnt secretion: molecular, cellular and genetic aspects. Front Biol (Beijing) 2012;7(6):587–593. doi: 10.1007/s11515-012-1200-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jin J, Morse M, Frey C, Petko J, Levenson R. Expression of GPR177 (Wntless/Evi/Sprinter), a highly conserved Wnt-transport protein, in rat tissues, zebrafish embryos, and cultured human cells. Dev Dyn. 2010;239(9):2426–2434. doi: 10.1002/dvdy.22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang LT, Wang SJ, Hsu SH. Functional characterization of mammalian Wntless homolog in mammalian system. Kaohsiung J Med Sci. 2012;28(7):355–361. doi: 10.1016/j.kjms.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 112.Zhu X, Zhu H, Zhang L, Huang S, Cao J, Ma G, Feng G, He L, Yang Y, Guo X. Wls-mediated Wnts differentially regulate distal limb patterning and tissue morphogenesis. Dev Biol. 2012;365(2):328–338. doi: 10.1016/j.ydbio.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 113.Jin J, Kittanakom S, Wong V, Reyes BA, Van Bockstaele EJ, Stagljar I, Berrettini W, Levenson R. Interaction of the mu-opioid receptor with GPR177 (Wntless) inhibits Wnt secretion: potential implications for opioid dependence. BMC Neurosci. 2010;11:33. doi: 10.1186/1471-2202-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Reyes AR, Levenson R, Berrettini W, Van Bockstaele EJ. Ultrastructural relationship between the mu opioid receptor and its interacting protein, GPR177, in striatal neurons. Brain Res. 2010;1358:71–80. doi: 10.1016/j.brainres.2010.08.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Milligan G. Opioid receptors and their interacting proteins. Neuromolecular Med. 2005;7(1–2):51–59. doi: 10.1385/NMM:7:1-2:051. [DOI] [PubMed] [Google Scholar]
- 116.Reyes BA, Vakharia K, Ferraro TN, Levenson R, Berrettini WH, Van Bockstaele EJ. Opiate agonist-induced re-distribution of Wntless, a mu-opioid receptor interacting protein, in rat striatal neurons. Exp Neurol. 2012;233(1):205–213. doi: 10.1016/j.expneurol.2011.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 118.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98(1):31–36. doi: 10.1073/pnas.98.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 120.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Barash Y, Dehan E, Krupsky M, Franklin W, Geraci M, Friedman N, Kaminski N. Comparative analysis of algorithms for signal quantitation from oligonucleotide microarrays. Bioinformatics. 2004;20(6):839–846. doi: 10.1093/bioinformatics/btg487. [DOI] [PubMed] [Google Scholar]
- 122.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125(1–2):279–284. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 124.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 125.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 127.Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26(2):216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table listing the probe-sets of genes expressed differentially by chronic morphine treatment in zebrafish embryos. Significance level for each probe-set are presented as d-, p- and q-value. The expression changes between Morphine vs. Control are showed as R fold. List of known genes up-, down-, and up-/down-regulated. (XLSX 199 KB)
Additional file 2: Figure presenting gene ontology analysis of the zebrafish gene collection. Genes were categorized with the Biological Process domain. Significantly enriched GO terms have a probability lower than 0.01 (P value) and include at least three genes. GO terms are shown if they are significantly enriched in at least one of them. Significantly enriched GO terms are indicated as black bars whereas non-significantly enriched terms are displayed as empty bars. Bars represent the number of genes assigned with the corresponding GO term. For more details, see also Additional file 3. (PDF 188 KB)
Additional file 3: Data file providing the detailed description of Gene Ontology analysis presented in Figure 2 and Table 1 . List of probe-sets and gene names classified for each GO category. (XLSX 70 KB)
Additional file 4: Table listing genes differentially expressed by morphine and included in each functional category presented in Figure 3 and Table 2 . The neuronal function category is emphasized in two excel sheet tabs (listing genes and Gene Ontology analysis of genes with neuronal function). (XLSX 98 KB)
Additional file 5: Table listing genes differentially expressed by morphine validation by our in silico analysis. (XLSX 34 KB)
Additional file 6: Table listing the primers used for RT-qPCR. (PDF 35 KB)


