Abstract
Kaposi's sarcoma–associated herpesvirus (KSHV) is an oncogenic virus, the etiological agent of Kaposi's sarcoma (KS) and primary effusion lymphoma (PEL). One of the key viral proteins that contributes to tumorigenesis is vFLIP, a viral homolog of the FLICE inhibitory protein. This KSHV protein interacts with the NFκB pathway to trigger the expression of antiapoptotic and proinflammatory genes and ultimately leads to tumor formation. The expression of vFLIP is regulated at the translational level by an internal ribosomal entry site (IRES) element. However, the precise mechanism by which ribosomes are recruited internally and the exact location of the IRES has remained elusive. Here we show that a 252-nt fragment directly upstream of vFLIP, within a coding region, directs translation. We have established its RNA structure and demonstrate that IRES activity requires the presence of eIF4A and an intact eIF4G. Furthermore, and unusually for an IRES, eIF4E is part of the complex assembled onto the vFLIP IRES to direct translation. These molecular interactions define a new paradigm for IRES-mediated translation.
Keywords: IRES, eIF4E, viruses, RNA structure, translation
INTRODUCTION
Kaposi's sarcoma–associated herpesvirus (KSHV), or human herpes virus 8 (HHV-8), is an oncogenic virus and the etiological agent of Kaposi's sarcoma (KS) (Chang et al. 1994; Mesri et al. 2010; Ballon et al. 2011). KSHV is a lymphotropic herpesvirus, containing a large DNA genome and inducing latent and lytic phases during infection (Renne et al. 1996; Russo et al. 1996). It primarily infects B cells (Ambroziak et al. 1995), but also infects the endothelium and is found in spindle cells and cells lining the vessels of KS lesions (Dupin et al. 1999; Colman and Blackbourn 2008). KS is a multifocal angioproliferative neoplasm and the most common tumor in HIV-infected patients, where it is a leading cause of mortality and morbidity, their immunodeficiency promoting KSHV infection and development of KS (Aversa et al. 2005; Mesri et al. 2010). As a result of the HIV pandemic, KS has become one of the most predominant cancers affecting men and children in subequatorial African countries; it is seen in 50% of HIV-infected individuals and results in a poor prognosis (Parkin 2006). KSHV is also associated with two lymphoproliferative diseases, multicentric Castleman's disease (MCD), a polyclonal lymphoproliferative disorder, and primary effusion lymphoma (PEL), an aggressive subtype of non-Hodgkin's B-cell lymphoma (Cesarman et al. 1995; Soulier et al. 1995; Bouvard et al. 2009).
KSHV infection is mainly latent in KSHV-induced lymphoid tumors (Dupin et al. 1999; Dittmer 2003). During latency, only a limited subset of genes is expressed encoding proliferative, pro-angiogenic, and antiapoptotic signals. These latency-associated genes consist of the latency-associated nuclear antigen (LANA), viral-cyclin (v-cyclin), v-FLICE inhibitory protein (vFLIP) and kaposins A, B, and C (Dittmer et al. 1998; Jenner et al. 2001; Ganem 2010). vFLIP has been proposed to play a role in KSHV pathogenesis by inhibiting FAS-induced apoptosis (Thome et al. 1997). Recent evidence suggests that the key role of vFLIP is to activate the NF-κB pathway by interacting with the IκB kinase γ (IKKγ) (Chaudhary et al. 1999; Field et al. 2003; Bagneris et al. 2008). This in turn induces the expression of antiapoptotic and proinflammatory genes (Guasparri et al. 2004; Matta and Chaudhary 2004; Sun et al. 2006), extending the lifespan of infected cells and explaining the inflammatory phenotype of KS lesions (Efklidou et al. 2008; Ballon et al. 2011).
A cluster of latency-associated genes is transcribed from a common promoter encoding consecutive open reading frames (ORFs) for LANA (ORF73), vCyclin (ORF72), and vFLIP (ORF71), yielding a tricistronic transcript and a bicistronic transcript encoding vCyclin and vFLIP only (Talbot et al. 1999; Bieleski and Talbot 2001; Pearce et al. 2005). Recently, ribosome profiling studies revealed that the transcripts accumulation profiles differ in SLK cells (Arias et al. 2014). During latency only the tricistronic transcript LANA-vCyclin-vFLIP is present while the bicistronic transcript vCyclin-vFLIP could only be detected in lytic SLK cells, confirming that vFLIP is expressed from the bicistronic message but that its expression is mainly regulated at the RNA level during latency (Arias et al. 2014). However, SLK cell lines are contaminated with renal-cell carcinoma and cannot serve as model for KS-derived endothelial tumor cell biology or oncogenesis; nevertheless, they support KSHV replication and are useful for the study of KSHV gene expression in nonendothelial contexts (Sturzl et al. 2013). The absence of a transcript encoding vFLIP alone hinted at a peculiar vFLIP expression mechanism and several studies proposed that vFLIP is expressed via a cap-independent mechanism involving an internal ribosome entry site (IRES) element located within the vCyclin coding region (Bieleski and Talbot 2001; Grundhoff and Ganem 2001; Low et al. 2001).
The majority of cellular mRNAs are translated via a cap-dependent mechanism during which the 5′ cap structure of the mRNA is recognized by eukaryotic initiation factor complex 4F (eIF4F), comprising eIF4E, eIF4A, and eIF4G (Hinnebusch 2014). Then, eIF4F recruits onto the mRNA a 43S preinitiation complex consisting of a 40S ribosomal subunit, the ternary complex eIF2-GTP-MettRNAi, eIF3, eIF1, and eIF1A, thereby priming the canonical scanning mechanism. In contrast, several viral mRNAs and some cellular mRNAs initiate translation via a cap-independent mechanism in which IRES elements within the mRNA internally recruit the ribosome, bypassing the need for many of the eIFs and a 5′ cap recognition event (Jackson et al. 2010). IRES elements have been discovered in the genomes of a number of RNA viruses, mainly within the 5′ untranslated region (UTR) of picornaviruses and flaviviruses, and most studies suggest that structured RNA domains specifically interact with eIFs or even directly with the ribosome to mediate translation (Balvay et al. 2007; Kieft 2008; Plank and Kieft 2012). Unlike most viral IRESs discovered to date, the KSHV vFLIP IRES is unusual in that it is located within a DNA virus genome, and a coding region, not a 5′ UTR. However, some controversy remains concerning the location of the IRES element and little is known about its function. Studies proposed that the IRES resides within the 363 or 232 nt of ORF72 directly upstream of ORF71; while another report detected no IRES activity associated with these fragments and suggested that the IRES lies within an internal 233 nucleotide fragment of ORF72 (Bieleski and Talbot 2001; Grundhoff and Ganem 2001; Low et al. 2001). To address those discrepancies and understand how the vFLIP IRES controls translation we redefined the vFLIP IRES boundaries both in the in vitro rabbit reticulocyte lysate (RRL) model system, and in 293 cells, identifying a 252-nt IRES upstream of the vFLIP ORF. Subsequently, we characterized its functional requirement and demonstrate that vFLIP IRES activity requires the presence eIF4A, an intact eIF4G, but also eIF4E, and that these factors are part of the complex assembled onto the vFLIP IRES. This requirement for the entire eIF4F complex is unprecedented. Furthermore, we characterized the RNA structure of the IRES domain in solution revealing a compact RNA scaffold.
RESULTS
Identification of a minimal IRES domain
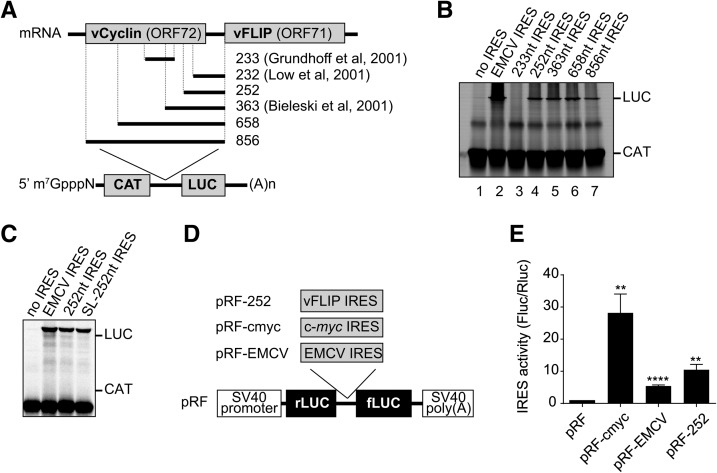
Previous reports suggested the presence of an IRES element within the vCyclin coding region driving vFLIP expression (Fig. 1A). First, using bicistronic luciferase reporter constructs in SLK cells, a minimal IRES was identified within the 232 nt located upstream of ORF71 (Grundhoff and Ganem 2001). However in BCP-1 cells, no IRES activity could be detected for this fragment and bicistronic luciferase reporter constructs suggested instead that the IRES activity lay within an internal 233-nt fragment (Bieleski and Talbot 2001). Moreover in 293 cells and using bicistronic reporter constructs, the IRES activity was assigned to the 363 nucleotides directly upstream of ORF71, for which no IRES activity was found in BCP-1 cells (Low et al. 2001). In order to resolve these discrepancies, we set out to delineate the minimal sequence required for IRES activity in the in vitro RRL model system. First, the following plasmids were assayed in in vitro translation reactions in RRL: the bicistronic reporter plasmid containing the 856 nt comprising of ORF72 and the intergenic region, inserted between the CAT and LUC open reading frames (pGEM-CAT/vFLIP IRES/LUC; lane 7), the negative-control plasmid lacking any IRES sequence (pGEM-CAT/LUC; lane 1), and the bicistronic plasmid, pGEM-CAT/EMCV/LUC (lane 2), containing the EMCV IRES as a positive control (Fig. 1A,B). SDS-PAGE and autoradiography for CAT and LUC expression showed that all plasmids efficiently expressed CAT, as expected (Fig. 1B). Furthermore, the EMCV IRES directed efficient LUC expression; in addition the 856 nucleotides upstream of ORF71 display IRES activity, as indicated by LUC expression (Fig. 1B, lanes 1, 2, and 7). To further define the minimal sequence required for IRES activity we systematically shortened the 856 sequence, to generate 658, 363, 252-nt fragments as indicated in Figure 1A, and assessed the ability of truncated sequences to promote IRES activity. We identified a minimal domain of 252 nt supporting IRES activity (Fig. 1B, lane 4), and shortening this domain further to 189 nt abolished IRES activity (Supplemental Fig. 1A). However, no LUC expression could be detected when we used the previously identified internal 233-nt fragments to drive IRES activity (Fig. 1B, lane 3). Furthermore, adding a stable stem–loop directly upstream of the 252 nt IRES (SL-252) had no impact on internal initiation, supporting its role as an IRES (Fig. 1C). To support these data, we investigated the activity of the vFLIP IRES in 293 cells (Fig. 1D). To this end, the 252-nt vFLIP (pRF-252), EMCV (pRF-EMCV), and c-myc (pRF-cmyc) IRESs were inserted in the intergenic region of a bicistronic pGL3 plasmid between the renilla luciferase (rLUC) and firefly luciferase (fLUC) open reading frames (Stoneley et al. 2000). The resulting plasmids were transfected into 293 cells and the expression from both renilla and firefly luciferase cistrons was assayed (Supplemental Fig. 1B). The integrity of the transcripts produced from bicistronic reporter plasmids was verified at the end of the reaction to ensure that firefly luciferase reflected IRES activity using RT-PCR and Northern blotting as described previously (Van Eden et al. 2004), and only one product was detected (data not shown). Subsequently, the IRES activity was expressed as the ratio of fLUC to rLUC normalized to the empty plasmid (pRF). The positive controls EMCV and c-myc IRES directed efficient internal initiation of translation, although the well-characterized EMCV IRES was less efficient than the cellular c-myc IRES (5.4 versus 28.4 relative IRES activity). In agreement with our in vitro translation results the 252-nt vFLIP IRES was able to support IRES activity (10.4 relative IRES activity, Fig. 1E). Furthermore, we also investigated the IRES activity of the vFLIP IRES in SLK cells, which support KSHV replication, but do not represent a model for KS (Herndier and Ganem 2001; Sturzl et al. 2013). While we could not detect EMCV IRES activity above background in those cells, the positive control pRF-cmyc IRES and the pRF-252 both displayed IRES activity, confirming our previous results (Supplemental Fig. 1C). These results suggest that the optimally effective IRES element lies within the 252 nt directly upstream of ORF71.
FIGURE 1.
Assessment of IRES activity in rabbit reticulocyte lysate and 293 cells. (A) Schematic representation of the bicistronic mRNAs used. The indicated fragments from the vCyclin-vFLIP mRNA were cloned between the CAT and LUC ORFs, and previously identified IRES are highlighted. (B,C) In vitro transcribed capped bicistronic mRNAs were translated in RRL containing 35S Methionine and reactions analyzed by SDS-PAGE and autoradiography. The CAT and LUC products are indicated. The results are representative from three independent experiments. (D) Schematic representation of the bicistronic mRNAs used; the indicated IRESs were inserted between the rLUC and fLUC reporter genes as described in Materials and Methods. (E) Two hundred ninety-three cells were transfected with bicistronic reporter plasmids as indicated. Rluc and Fluc activities were measured as described in Material and Methods. The relative Fluc/Rluc ration was set to 1 for the empty pRF plasmid as relative IRES activity. Values are the mean ± SEM from three independent experiments and were analyzed by one-way ANOVA with Dunnett's multiple comparison test: (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, and (****) P < 0.0001 (GraphPad Prism 6.0).
vFLIP IRES activity requires intact eIF4G and eIF4A
To date the study of vFLIP IRES activity has been limited to delineating the IRES domain and assessing its interaction with a general ITAF, PTB (Bieleski et al. 2004). To evaluate the contribution of specific eukaryotic initiation factors in vFLIP-mediated translation we used the in vitro RRL translation system in combination with chemical and protein inhibitors of translation. First, we investigated the functional role of eIF4F components. While eIF4F, composed of eIF4A, eIF4G, and eIF4E, is strictly required for cap-dependent translation, some IRESs can function either in the absence of eIF4F or in the presence of a subset of the complex (Balvay et al. 2009). For example, IRES elements from picornaviruses function in the presence of only eIF4A and a cleaved fragment of eIF4G (Pause et al. 1994; Pestova et al. 1996; de Breyne et al. 2009; Yu et al. 2011). During infection, the poliovirus (PV) 2A protease and the foot and mouth disease virus (FMDV) L protease each inhibit cap-dependent translation by inducing the cleavage of eIF4G, separating the eIF4E interacting domain (Gingras et al. 1999), while the resulting eIF4G C-terminus domain can still support IRES-mediated translation (Balvay et al. 2009; Jackson et al. 2010). To investigate the role of eIF4G in vFLIP IRES activity, we analyzed the effect of its cleavage on the in vitro RRL translation reactions using the bicistronic pGEM-CAT/LUC reporters used to define the minimal IRES domain. Adding L-protease to the RRL induced a specific eIF4G cleavage as previously described (Supplemental Fig. 2A; Ohlmann et al. 1996). The bicistronic plasmids were assayed in RRL in the presence and absence of FMDV L-protease. As expected the FMDV L-protease severely reduced translation of the first upstream CAT cistron, while the translation of the LUC cistron controlled by the EMCV IRES, which can function in the presence of the cleaved eIF4G fragment, was not affected (Fig. 2A). However, when placed under the control of the vFLIP IRES the translation of the LUC cistron was impaired by the addition of FMDV L-protease (Fig. 2A). These results suggest that an intact eIF4G, including the amino-terminal domain mediating the interaction with eIF4E, is required for vFLIP-dependent IRES activity.
FIGURE 2.
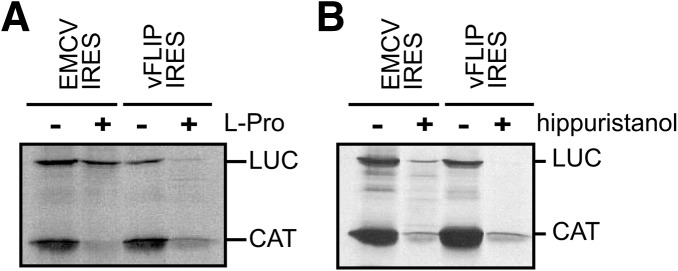
vFLIP IRES activity requires intact eIF4G and eIF4A. Bicistronic-capped mRNAs, in the form of CAT/IRES/LUC were translated in vitro in RRL, pretreated with FMDV L-protease (L-pro) as indicated (A), or in the presence (+) or absence (−) of the eIF4A inhibitor hippuristanol (B). The CAT and LUC products are indicated. The data shown are representative from three independent experiments.
We then investigated whether eIF4A was also involved. To evaluate the requirement for eIF4A, we used hippuristanol a natural compound that binds the carboxy-terminal region of eIF4A and thus inhibits its RNA binding, helicase, and ATPase activities (Bordeleau et al. 2006). Addition of 1 μM hippuristanol inhibits the translation of both the control EMCV IRES-driven luciferase and the vFLIP IRES-driven luciferase suggesting a functional requirement for eIF4A (Fig. 2B). In contrast, the addition of hippuristanol has no effect on IRES activity driven by the HCV IRES which does not require eIF4A activity for translation (Supplemental Fig. 2B).
Inactivation of eIF4E inhibits vFLIP IRES activity
The requirement of the eIF4E interacting domain of eIF4G for vFLIP IRES activity prompted us to investigate whether eIF4E and the eIF4E–eIF4G interaction are required as well. First we assessed the role of the eIF4G:eIF4E interaction using 4EGI-1, a previously described inhibitor of the eIF4E:eIF4G interaction (Moerke et al. 2007). While concentrations of up to 50 μM of the inhibitor did not affect the efficiency of the EMCV or HCV IRES-driven translation, both the cap-dependent translation, as expected, but also the vFLIP IRES activity were impaired (Fig. 3A; Supplemental Fig. 2B). This suggests that the vFLIP IRES activity depends on the eIF4E–eIF4G interaction. Although it is widely used to characterize IRES activity, a recent report proposed that 4EGI-1 could also inhibit the translation mediated by IRES elements that do not require the eIF4E–eIF4G interaction (Redondo et al. 2013). Therefore, to confirm our results, we performed similar experiments using 4E1RCat, another compound identified by high-throughput screening for inhibitors of the eIF4E–eIF4G interaction (Cencic et al. 2011). 4E1RCat blocks cap-dependent translation but does not impair translation mediated by the EMCV IRES, which does not require the eIF4E–eIF4G interaction (Cencic et al. 2011). As shown previously, adding increasing amounts of 4E1RCat inhibited the cap-dependent production of CAT (Fig. 3B). A dose-dependent inhibition of the LUC expression could also be detected for LUC cistron placed under the control of the vFLIP IRES (Fig. 3B). However, the addition of 4E1RCat has no effect on translation driven by the HCV IRES, which does not require eIF4E or eIF4G activity (Supplemental Fig. 2B). Therefore, these results confirmed that the vFLIP IRES activity is sensitive to inhibition of the eIF4G–eIF4E interaction. To further determine whether the vFLIP IRES–eIF4E interaction plays a significant role for IRES activity, translation efficiency was examined in RRL treated with the cap analog m7GpppG to impair eIF4E function (Merrick 2004). As expected, the addition of increasing amounts of cap analog impaired the translation of the CAT upstream cistron, inhibiting cap-dependent translation, while the activity of the EMCV IRES was unaffected (Fig. 3C). However, and surprisingly for an IRES, the activity of the vFLIP IRES displayed a dose-dependent inhibition response to the addition of the cap analog (Fig. 3C). All these results strongly suggest that functional eIF4E and the eIF4E binding domain of eIF4G are required for the internal entry of ribosomes mediated by the vFLIP IRES.
FIGURE 3.
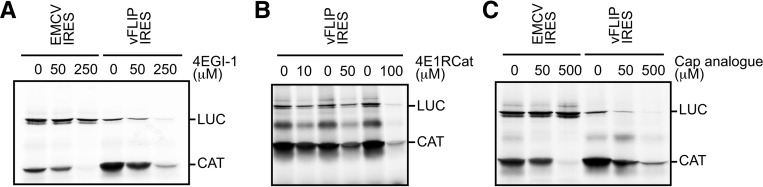
vFLIP IRES activity requires eIF4E. (A) Bicistronic-capped mRNAs, CAT/vFLIP IRES/LUC, or CAT/EMCV IRES/LUC were translated in RRL in vitro in the presence of 0, 50, or 250 μM of the eIF4E inhibitor 4EGI-1 as indicated. (B) Bicistronic-capped mRNAs and CAT/vFLIP IRES/LUC were translated in RRL in vitro in the presence of 0, 10, 50, or 100 μM of the eIF4E inhibitor 4E1RCat. (C) Bicistronic-capped mRNAs, CAT/vFLIP IRES/LUC, or CAT/EMCV IRES/LUC were translated in RRL in vitro in the presence of 0, 50, or 500 μM of the cap analog. The CAT and LUC products are indicated. The data shown are representative from three independent experiments.
eIFs are recruited onto the vFLIP IRES
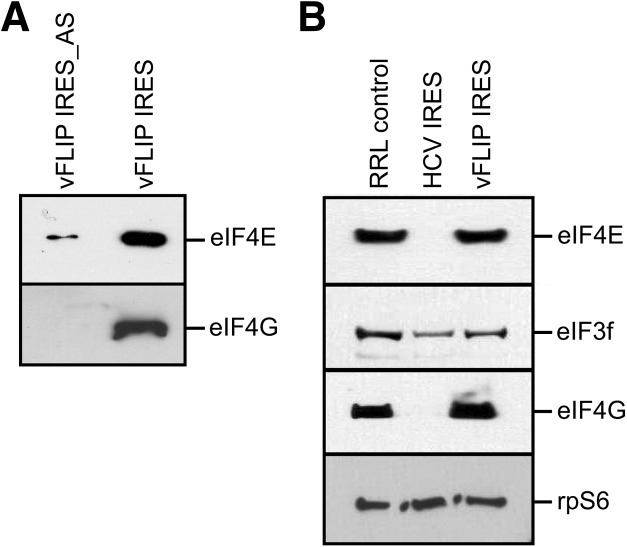
Taken together these results indicate that the vFLIP IRES requires the whole-eIF4F complex for its activity, including eIF4E, and therefore it could interact with the IRES either directly, or indirectly through eIF4G. To further probe the eIF requirements of the vFLIP IRES for the eIF4F complex we investigated whether eIF4E and eIF4G could bind to the vFLIP IRES in vitro. The fragment encoding the 252-nt IRES or an antisense control were inserted into the pSP64 Poly(A) transcription vector. Following linearization of these plasmids, RNA transcripts corresponding to the vFLIP IRES with a 30-nt poly(A) stretch added to its 5′ end, were generated in vitro. These transcripts were then immobilized on oligo(DT) magnetic beads as previously described (Stassinopoulos and Belsham 2001). The presence of the poly(A) stretch at the 5′ extremity prevented nonspecific eIF recruitment via the 5′ end. Then, to monitor the association of eIFs with the vFLIP IRES, RRL was incubated with the beads–RNA complexes. After washing, the bound proteins were separated by PAGE and analyzed by immunoblotting. As shown in Figure 4A both eIF4G and eIF4E form a complex assembled onto the vFLIP IRES but not on the antisense control, although minor eIF4E binding was detected. To ensure that the recruitment of eIF4E could not occur from the 5′end of the RNA reporter, we repeated the experiment with RNA capped with a nonfunctional cap analog to block the 5′end; and we observed similar results (data not shown). To further confirm that eIF4E and eIF4G are part of the complex assembled onto the vFLIP IRES we applied a methodology previously developed to isolate IRES-bound translation complexes (Boehringer et al. 2005; Locker et al. 2006). Briefly, hybrid RNA containing the vFLIP IRES or the HCV IRES, as a control, and the tobramycin aptamer were generated by in vitro transcription. Hybrid RNAs were then incubated in RRL in the presence of GMP-PNP to assemble initiation complexes, which were then isolated by affinity chromatography using tobramycin-coupled sepharose. Following elution of the complexes, the presence of initiation factors was analyzed by Western blotting. As expected and in agreement with previous studies (Locker et al. 2007), complexes assembled onto the HCV IRES contained eIF3 but not eIF4E or eIF4G (Fig. 4B). However, the complexes assembled onto the vFLIP IRES contained eIF3, eIF4E, and eIF4G (Fig. 4B). Therefore, these results confirm our previous results and suggest that both eIF4G and eIF4E are part of the complex assembled onto the vFLIP IRES that mediate IRES activity.
FIGURE 4.
vFLIP IRES forms an RNA–protein complex with eIFs. (A) Immunoblotting analysis of protein binding to the vFLIP IRES or an antisense control (vFLIP IRES_AS). Poly(A) tagged RNAs were immobilized onto oligo(DT) beads and following washes and elution, the proteins were separated by SDS-PAGE and analyzed by immunoblotting against eIF4E and eIF4G. (B) Immunoblotting analysis of complexes assembled onto the vFLIP and HCV IRES. Hybrid RNA, containing the IRES fused to the tobramycin aptamer, were incubated with RRL pretreated with puromycin, in the presence of GMPNP, then immobilized onto tobramycin-linked sepharose. RNA–protein complexes were eluted in the presence of tobramycin and separated by SDS-PAGE, following immunoblotting analysis against rpS6, eIF4E, eIF4G, and eIF3. A control lane of RRL was included as a reference for these factors.
vFLIP IRES directly interact with the ribosomes and eIFs
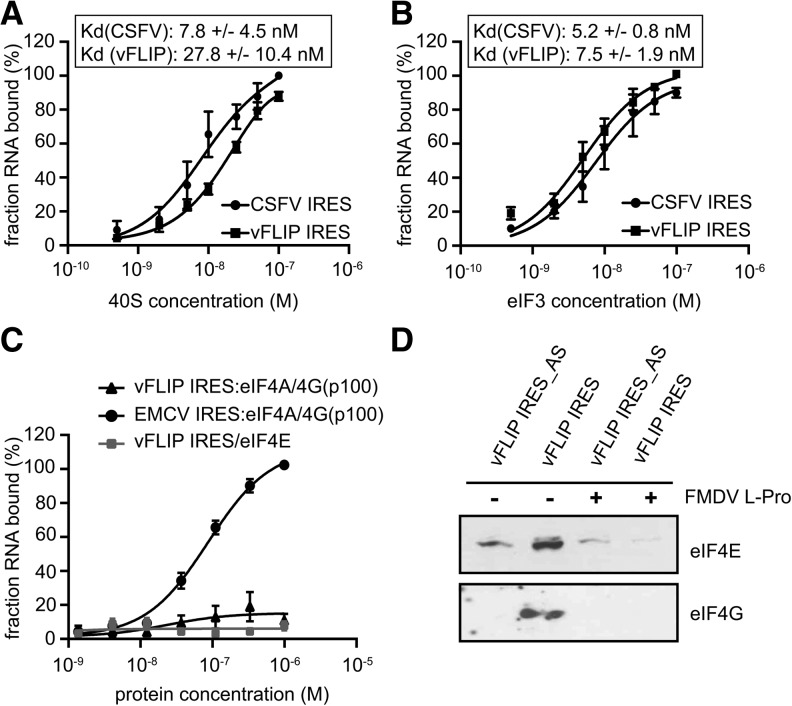
To identify the interactions between the translation machinery and the vFLIP IRES and their contribution to 48S complex formation, we assayed whether individual initiation factors or the 40S ribosomal subunit could directly interact with the IRES. Therefore, eIFs and ribosomal subunits were purified from HeLa cells according to described procedures (Pisarev et al. 2007). Then, individual purified components were incubated with 32P-labeled vFLIP IRES before conducting filter-binding assays to analyze the affinity and specificity of putative interactions (Willcocks et al. 2011). We determined that the vFLIP IRES binds the 40S subunit with an apparent equilibrium dissociation constant (Kd) of 28 nM, while the CSFV IRES (as a control) bound with an affinity of 8 nM (Fig. 5A). This shows that the vFLIP IRES can bind directly to the 40S subunit in the absence of any initiation factor, a property in common with HCV-like and dicistrovirus IRESs, supporting its function as an IRES. Furthermore, the vFLIP IRES also binds to eIF3 with an affinity of 8 nM (Fig. 5B). In contrast, no specific interactions could be detected between the vFLIP IRES and eIF4E (Fig. 5C). While it was not possible to obtain sufficient yield of intact full-length eIF4G or the entire eIF4F complex from HeLa cytoplasmic extract, it was possible to obtain the eIF4A/p100 complex (p100 corresponding to the cleaved fragment of eIF4G that lacks the eIF4E binding site). Again, we could not detect any direct interaction between eIF4A/p100 and vFLIP IRES while it binds the EMCV IRES as previously proposed (Fig. 5C). These results suggest that an intact eIF4G is important for the assembly of the initiation complex onto the vFLIP IRES. To support this we repeated the oligo(DT) pull down experiment in the presence of the FMDV L-protease and showed that eIF4G cleavage abolished eIF4E and eIF4G recruitment to the vFLIP IRES (Fig. 5D). Therefore, we propose that the intact eIF4F complex interacts with the vFLIP IRES rather than its individual components.
FIGURE 5.
vFLIP IRES directly binds to 40S subunits and eIF3. (A) Binding curves of 32P-labeled CSFV and vFLIP IRES RNAs to purified human 40S subunits. Labeled RNAs were incubated with 40S subunits and binding assessed by filter-binding assay. (B) Binding curves of 32P-labeled CSFV and vFLIP IRES RNAs to purified human eIF3. Labeled RNAs were incubated with eIF3 and binding assessed by filter-binding assay. (C) Binding curves of 32P-labeled EMCV and vFLIP IRES RNAs to purified human eIF4E and eIF4A/4G(p100) complex. Labeled RNAs were incubated with eIF4E and eIF4A/4G(p100) complex and binding assessed by filter-binding assay. Reported values are the average from a minimum of three repetitions with standard errors. All calculations were performed with GraphPad Prism 7. (D) Immunoblotting analysis of protein binding to the vFLIP IRES or an antisense control (vFLIP IRES_AS). Poly(A) tagged RNAs were immobilized onto oligo(DT) beads and incubated with RRL treated with or without 2.5 μg of recombinant FMDV L-protease (L-pro) as indicated. Following washes and elution, the proteins were separated by SDS-PAGE and analyzed by immunoblotting against eIF4E and eIF4G as indicated.
Determination of the vFLIP IRES structure
Most viral IRES elements contain structured RNA domains that are crucial for IRES function by interacting with eIFs or the ribosome. For example, within the HCV IRES, domain III stem–loops coordinate small ribosomal subunit recruitment, while we previously demonstrated that a loop structure within domain II mediates the 60S subunit joining event (Locker et al. 2007). Therefore, a deep understanding of the structures accessible to RNA is required to decipher IRES-mediated regulation during translation. To further characterize the vFLIP IRES we analyzed its structure in solution using Selective 2′ Hydroxyl Acylation analysis by Primer Extension (SHAPE), chemical and enzymatic probes. SHAPE analysis interrogates the RNA backbone flexibility at single-nucleotide resolution as flexible nucleotides can sample local conformations increasing the nucleophilic reactivity of 2′-hydroxyl groups toward 1M7 (Mortimer and Weeks 2007). The sites of modification were then mapped as stops by primer extension reaction, followed by capillary electrophoresis analysis, allowing us to assign quantitative SHAPE reactivity to individual nucleotides (Supplemental Fig. 4). The SHAPE studies were repeated three times and the mean of reactivity for each position was used to model the IRES structure in combination with accessibility information yielded by other chemical and enzymatic probes such as RNAse V1 reactivity, to detect double stranded regions, CMCT and DMS reactivity, to detect single stranded regions (Fig. 6). The SHAPE data were then used as constraints for “RNA structure” an RNA secondary structure prediction software implemented with the “shapeknot module” (Bellaousov et al. 2013; Hajdin et al. 2013). The different models obtained were evaluated for consistency with the V1, DMS, and CMCT data. The model that best fits all the data is shown in Figure 6. In only a very few places the model is in conflict with the reactivity data, which may reflect the formation of a tertiary structure, for which we could not identify any constraints. Most V1 hits are in helical region, but a few are found in loops (Id, II, and IV), which could reflect the formation of tertiary structure or local noncanonical base pairs, yet to be identified. Furthermore, “shapeknot” highlighted a potential “kissing complex” by pairing dII 5′-171GCUUGUG177-3′ with dIV 5′-239UACAAGC245-3′. Although we have no further evidence to support such interaction, this would be in agreement with the ambivalent nature of probing reactivities observed in this region, as shown by weak SHAPE reactivity, the CMCT and DMS hits and the few RNAse V1 hits observed. Overall, most DMS and CMCT hits are in single stranded region or at the edges of helices, only the hits in the 3′ region of domain II challenges our model. They may reflect the breathing of a helix that comprises mainly A–U and G–U base pairs. Some positions are reactive to DMS or CMCT, but not to 1M7 (G58 or U180 for example), this is not necessarily a discrepancy since DMS and CMCT probe the involvement of the Watson–Crick position in hydrogen bonds, while 1M7 probes the flexibility of the ribose. Thus, nucleotides constrained by noncanonical interactions not involving Watson–Crick positions are predicted to be unreactive toward 1M7, but may be reactive to DMS or CMCT.
FIGURE 6.
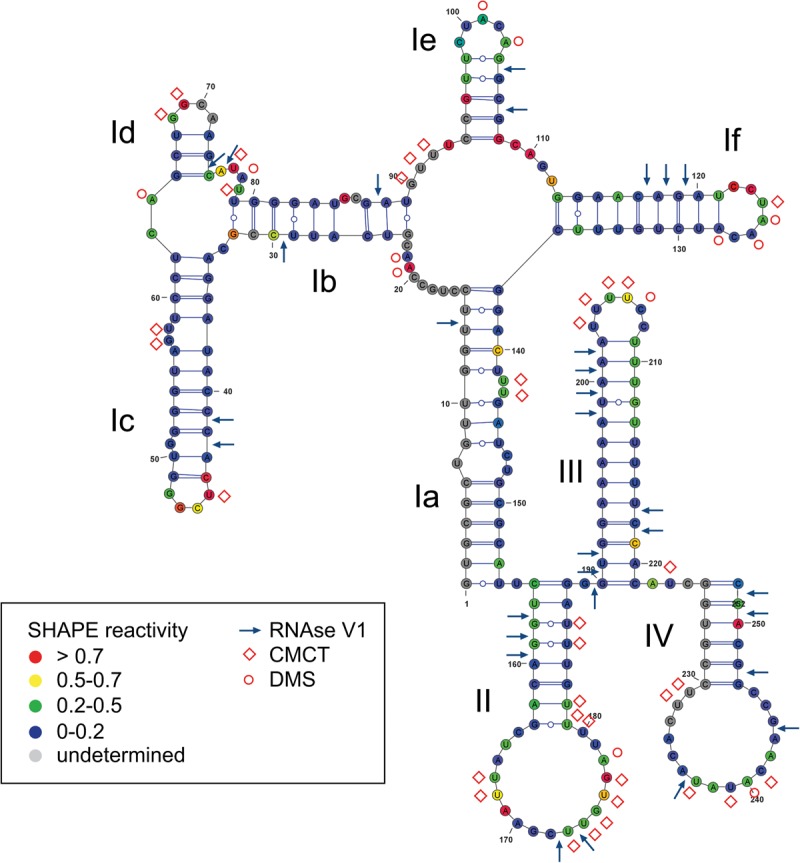
Secondary structure model of the vFLIP IRES. The mean SHAPE reactivity from three independent experiments is indicated for each position on the vFLIP IRES according to the color code boxed. The chemical and enzymatic modifications induced by DMS, CMCT, and RNAse V1 are indicated using red circles, red diamonds, and blue arrows, respectively. The names of the major domains determined are indicated in the model.
The vFLIP IRES adopts a compact structure, which is largely reflected by the overall low SHAPE reactivity. Two main domains can be identified, the first corresponding to nucleotides 1–154, while the second consists of 155–252 nt. The first part of the IRES structure is formed by domain I which consists of subdomains Ia to If organized into two consecutive junctions: a four-helix junction made of Ia, Ib, Ie, and If, followed by a three-helix junction made of Ib, Ic, and Id. Domains Ic and Id are capped by a UCGG and a GGCA tetraloop, respectively; while domain Ie and If are capped by CUACA and CCUAAC loops, respectively. The second part of the IRES structure consists of three stem–loop structures, domains II, III, and IV. This part of the structure provides an attractive platform for protein recruitment as domain II and IV are capped by large loops consisting of 17 and 16 nt, respectively, while domain III is capped by a pyrimidine-rich (U4C2) loop.
DISCUSSION
Previous studies have suggested that expression of vFLIP, a key KSHV tumorigenesis factor during latency, is mediated by an IRES element (Bieleski and Talbot 2001; Grundhoff and Ganem 2001; Low et al. 2001). Adding to the controversy surrounding its exact location with different studies in cells or in vitro defining different domains for internal entry, the vFLIP IRES is atypical in nature, being located in the coding region of a DNA virus, while most viral IRESs characterized so far are located in the 5′ UTR of positive-sense RNA viruses. Therefore, its presence hinted at a molecular mechanism mediating internal entry of ribosomes that is specific to the KHSV IRES.
To reconcile previous studies, and to establish the boundaries of the vFLIP IRES, different plasmid constructs were generated containing a decreasing length of the terminal vCyclin coding region, and included the previously identified 233-nt minimal IRES domain. First, we defined a minimal 252-nt vFLIP IRES in vitro using a translation assay in RRL. Subsequently, the ability of this segment to direct translation was confirmed in 293 and SLK cells—which have been extensively used in the past to characterize KSHV gene expression, but do not represent a KS model system (Herndier and Ganem 2001; Sturzl et al. 2013). Furthermore, we showed that this RNA fragment can directly recruit the ribosome subunit with nanomolar affinity, a property shared with other IRES (Kieft 2008).
To characterize its mode of action, we then dissected which initiation factors are required for IRES activity using a variety of specific inhibitors of translation. As the IRES activity is sensitive to the addition of both hippuristanol and the FMDV L-protease, we concluded that both eIF4A and an intact eIF4G are required for cap-independent translation. While the carboxy-terminal domain of eIF4G interacts with eIF4A and eIF3 to recruit the ribosome, the amino-terminal domain cleaved by the L-protease is responsible for eIF4E interaction and holding the eIF4F complex together. Therefore, we further assessed whether eIF4E and the eIF4E:eIF4G interaction played a role in vFLIP IRES-mediated translation. Using inhibitors of the eIF4E:eIF4G interaction, we established that this interaction is critical to mediate vFLIP IRES activity. These findings were supported by further studies demonstrating that the addition of a cap analog, to inactivate eIF4E, also impaired vFLIP IRES activity. Altogether, these results suggest that not only eIF4G and eIF4A, but also eIF4E are required for vFLIP IRES activity. As the requirement for eIF4E seemed unusual for a mechanism of ribosome internal entry, we further analyzed the association of the core initiation factors eIF4G and eIF4E with either isolated vFLIP IRES or within initiation complexes. We found that eIF4G can associate with the vFLIP IRES but also that eIF4E interacts with the vFLIP IRES during translation initiation, which is unprecedented for an IRES to our knowledge. Those results support a mode of action in which both eIF4E and intact eIF4G are involved in tethering ribosomes to the vFLIP IRES to direct translation. We could not detect any direct interaction with isolated eIF4E and the eIF4A/p100 complex, this leads us to propose that the intact eIF4F is recruited to the vFLIP IRES, and that this is likely to be through the direct interaction between the IRES, the 40S subunit and eIF3. These results also suggest that viral IRES display a much larger functional requirement than those used to define the four groups of IRES elements. Indeed while the functional requirement for eIF4A and the interaction with eIF4G, could be reminiscent of type I and type II IRESs, such as PV or EMCV IRESs respectively, these IRESs are able to function in the presence of a cleaved eIF4G, unlike the vFLIP IRES (Yu et al. 2011; Sweeney et al. 2014). Lentiviral gag IRESs also require eIF4A activity and form initiation complexes that comprises eIF4A and eIF4G, but do not interact directly with eIF4G, nor require the eIF4G–eIF4E interaction (Chamond et al. 2010; Locker et al. 2011). Meanwhile, the ability to interact directly with the 40S subunit and eIF3 is a property shared with HCV-like and primate lentiviruses IRESs (Lukavsky 2009; Locker et al. 2011).
Most viral IRESs adopt complex RNA structures that coordinate the recruitment of the ribosome and initiation factors (Kieft 2008). We therefore established a model of the vFLIP IRES structure using SHAPE analysis combined with traditional modification using chemical and enzymatic probes (Fig. 5). Probing in solution revealed a very compact structure which is the hallmark of many viral IRESs, reflected in the overall poor reactivity toward the single strand specific probes. This finding is also in agreement with the presence of two stretches of nucleotides (82–95 and 223–233) in which we repeatedly observed strong reverse transcription stops reflecting the presence of a structure that the reverse transcriptase is unable to resolve. The vFLIP IRES structure scaffold is separated into two domains, consisting of domain Ia to If and domain II to domain IV, respectively (Fig. 5). The data strongly suggest that elements of the modular structures could be involved in direct interaction with the ribosome, initiation factors or ITAFs, and further studies should aim at characterizing the structure–function relationship of the vFLIP IRES.
IRES elements are by their nature cap-independent, so the requirement for eIF4E here is intriguing. Previously the hepatitis A virus (HAV) IRES also displayed particular eIFs requirement. It was proposed that HAV IRES activity could require an interaction between the eIF4E cap-binding pocket and the IRES or that the eIF4E–eIF4G interaction was necessary for eIF4G to adopt a conformation promoting IRES activity, independently from RNA binding by the cap-binding pocket (Ali et al. 2001; Borman et al. 2001). However, recent findings suggest that the IRES activity is stimulated following eIF4G cleavage by the L-protease in cells and therefore the exact initiation factor requirement for HAV IRES activity remains unclear (Redondo et al. 2012). In addition we cannot rule out a more direct role for eIF4E in cap-independent translation. Several positive-sense RNA plant viruses lack a 5′ cap structure and rely on cap-independent translation elements (CITEs), located in 3′ UTRs, to mediate translation (Nicholson and White 2011). CITEs are highly folded RNA structures that recruit initiation factors to the viral RNA and engage in long-range RNA–RNA base-pairing between the 3′ and 5′ UTR to direct translation (Nicholson and White 2011; Kraft et al. 2013). For example, the pea enation mosaic virus RNA 2 translation element (PTE) contains a pseudoknot that binds to eIF4E with high affinity to recruit eIF4F and act as a translation enhancer (Wang et al. 2009). SHAPE analysis and 3D modeling further showed that the cap-binding pocket of eIF4E clamps the PTE pseudoknot in the place of the canonical cap (Wang et al. 2011). Finally, the ORF of histone H4 mRNA contains a regulatory RNA element that promotes translation by recruiting eIF4E, and eIF4F, internally and independently from the cap (Martin et al. 2011). The direct binding of eIF4E allows the recruitment of eIF4G and eIF4A onto the mRNA while another structural element hinders the 5′ cap. The 5′ cap is subsequently made available to eIF4E through a structural rearrangement mediated by eIF4A (Martin et al. 2011). However, our finding that eIF4E cannot bind the vFLIP IRES in the absence of other factors would indicate another mechanism of translation. Recently, eIF4E was shown to exercise an unexpected second function in translation initiation by stimulating eukaryotic initiation factor 4A helicase activity (Feoktistova et al. 2013). Importantly, it was demonstrated that the eIF4E-mediated eIF4A activity promotes mRNA restructuring independently from its cap-binding function (Feoktistova et al. 2013). This new function of eIF4E could explain how eIF4E can selectively stimulate the translation of mRNAs that possess a structured 5′ UTR, which could apply to IRES elements such as HAV or vFLIP IRESs and would support our results. Therefore eIF4E is involved in different noncanonical initiation events and its role in KSHV IRES-mediated translation emphasizes even further the fact it can act beyond its cap-binding activity to mediate translation. This could reflect a more general mechanism of initiation by which RNA elements, including the vFLIP IRES, can tether eIF4E and eIF4F to mRNA internally to recruit ribosomes and initiate translation.
Much effort has been dedicated to the development of drugs that could impair IRES-mediated translation as a means of blocking viral replication, mainly focusing on the HCV IRES (Dibrov et al. 2012, 2014). Recently it has been proposed that cap-independent expression of vFLIP could provide a mechanism to control the balance between vCyclin and vFLIP levels, thereby allowing vFLIP to suppress autophagy and inhibit senescence during latent infection (Leidal et al. 2012). By unraveling the molecular mechanism by which the vFLIP IRES directs translation and the RNA structure involved, we reveal a new potential target in the design of drugs that could prevent the oncogenic properties of KSHV.
MATERIALS AND METHODS
Cell lines
SLK cells were maintained in Roswell Park Memorial Institute 1640 medium (RPMI 1640) supplemented with 1% penicillin/streptomycin, 1% L-glutamine, and 10% fetal bovine serum (FBS). Two hundred ninety-three cells were maintained in Dulbecco's modified Eagle's MEM supplemented with 1% penicillin/streptomycin, 1% nonessential amino acids and 1% L-glutamine and 10% FBS. The cells were cultured at 37°C in an atmosphere of 5% CO2.
Plasmids
To generate the bicistronic constructs, fragments encoding the different IRES sequences were amplified by PCR from a genomic fragment corresponding to the second exon of the bicistronic ORF72/71 mRNA (Genbank U75698). The c-myc IRES was kindly provided by Anne E. Willis (MRC Toxicology Unit, University of Leicester) and the IRES sequence from encephalomyocarditis (EMCV, nucleotides 406–930) and hepatitis C virus (HCV; nucleotides 1–426) were described previously (Easton et al. 2009). For in vitro translation, the IRES fragments were cloned into the pGEM-CAT/LUC plasmid described previously which encodes chloramphenicol acetyltransferase (CAT) and firefly luciferase (fLUC) under the control of a T7 promoter (Willcocks et al. 2011). The sequence of the stem–loop sequence inserted upstream of the IRES is 5′-CAGATCTACGCGGTTCGCCGCGTAGATCTG-3′. The constructs were confirmed by restriction enzyme digestion, PCR, and sequencing. For transfection into cells and luciferase assays, the IRES fragments were cloned into the pGL3-rLUC/fLUC plasmid described previously which encode Renilla luciferase (rLUC) and firefly luciferase (fLUC) under the control of an SV40 promoter (Stoneley et al. 2000).
Transfections and luciferase assays
Transient DNA transfections of SLK and 293 cells was performed in 35-mm dishes using 4 μL FuGENE HD transfection reagent (Promega) and 2 μg of plasmid DNA according to the manufacturer's instructions. The cells were harvested 28-h post-transfection. The sample cell lysates were frozen and thawed twice before assaying for luciferase activity using the Dual-luciferase Reporter assay system (Promega) and detection in a luminometer (Labtech). To check that the firefly luciferase activity originated from IRES activity rather than aberrant splicing events or cryptic promoter activity, total RNA was extracted from cells and analyzed by RT-PCR as described by Van Eden et al. (2004), and by Northern blotting as described by Bushell et al. (2006).
In vitro transcription and translation
In vitro synthesis and purification of capped RNA was carried out using the mMESSAGE mMACHINE kit (Ambion) according to the manufacturer's instructions. The bicistronic reporter RNAs (0.5 μg) were translated in the rabbit reticulocyte lysate (RRL; Flexi RRL system; Promega; 3.2 mM endogenous Mg2+) in the presence of 20 µM amino acids (lacking methionine), 0.5 mM MgOAc2, 100 mM KCl, 0.8 U/µL of RNasin Plus RNase Inhibitor (Promega), and 0.6 mCi/mL [35S]-methionine. Translation reactions, in a final reaction volume of 12.5 µL, were incubated for 90 min at 30°C. Products were analyzed by SDS-PAGE and autoradiography. For inhibition experiments, RRL lysates were treated with various concentrations of hippuristanol (kind gift of Jerry Pelletier, McGill University), 4E1RCat (idem), L-Protease (kind gift of Simon J. Morley, University of Sussex), 4EGI-1 (Merck), cap analog m7GpppG (Ambion) or 5% DMSO as control.
Luciferase assays
RNA affinity chromatography
The IRES fragments were cloned into the pSP64 poly(A) plasmid (Promega) using standard techniques. Subsequently RNAs were in vitro transcribed and purified using the MEGAscript T7 kit (Ambion) with or without the addition of ApppG (New England Biolabs). The transcripts were then immobilized onto oligo(DT) magnetic beads (Life Technologies) and incubated with RRL (5 μg) as previously described (Stassinopoulos and Belsham 2001). The bound proteins were washed, eluted in 2× SDS sample buffer and separated by SDS-PAGE electrophoresis. Proteins were then transferred onto PVDF membrane and detected using antibodies against eIF4E and eIF4G (kind gift of Simon J. Morley, University of Sussex).
For translation complex analyses, the IRESs were evaluated using tobramycin affinity purification (Hartmuth et al. 2002). Briefly, the IRES fragments were cloned into a pUC18 plasmid between a T7 promoter and the tobramycin aptamer sequence using standard protocols. The RNAs were then transcribed using the MEGAscript T7 kit (Ambion) and purified as described (Sargueil et al. 2000). Affinity purification of translation complexes assembled in RRL in the presence of 5 mM GMP-PNP was then performed as previously described (Boehringer et al. 2005) and the presence of individual eIFs assayed by immunoblotting using antibodies against eIF3f (Santacruz; sc-28856) and rpS6 (Santacruz; sc-4426).
Filter-binding assays
The filter-binding assays were performed as described previously without modification (Willcocks et al. 2011). Control RNA encoding the CSFV IRES (1–427; Paderborn strain) was generated by standard molecular cloning using a bicistronic plasmid containing the CSFV IRES (kind gift of Graham Belsham, National Veterinary Institute). Briefly vFLIP, CSFV or EMCV RNA transcripts were transcribed in vitro in the presence of α-32P-UTP (3000 mCi/mmol). The 40S ribosomal subunits and eIF3 were prepared following previously established procedures from HeLa cells (Pisarev et al. 2007). eIF4E, eIF4A and eIF4G-p100 were expressed as recombinant proteins in Escherichia coli using the following plasmids: pET16-eIF4A (kind gift of Dr Frank Martin, Université de Strasbourg), pGEX-eIF4E (kind gift of Dr Theophile Ohlmann, ENS Lyon), and pET22-p100. Radiolabeled RNA (50 fmol) incubated was with serial dilutions of initiation factors (eIFs) or 40S subunit in binding buffer (20 mM Tris [pH 7.6], 100 mM KCl, 2 mM DTT, 2 mM MgCl2) and incubated at 37°C for 15 min before performing filter-binding assays. Bound RNA was quantified using a Typhoon FLA7000 (GE Healthcare). To determine the apparent dissociation constant (Kd), the data were fitted to a Langmuir isotherm described by the equation θ = P/(P + Kd), where θ is the fraction of RNA bound and P is either the 40S subunit or eIF concentration. Reported values are the average of results from three repetitions with standard errors. All calculations were performed with GraphPad Prism 7.
RNA structure determination
The secondary structure of the KSHV IRES was probed using dimethyl sulfate (DMS), N-cyclohexyl-N′-[N-methylmorpholinoethyl]-carbodiimid-4-toluenesulfonate (CMCT) and RNAse V1 and analyzed by reverse extension as described previously (James and Sargueil 2008; Weill et al. 2010; Willcocks et al. 2011). RNA Selective 2′ Hydroxyl Acylation analysis by Primer Extension (SHAPE) analysis was conducted using 1-methyl-7-nitroisatoic anhydride (1M7) as a modifying agent as previously described (Mortimer and Weeks 2007; Deforges et al. 2012). Modifications were revealed using RNAse H− M-MLV RT (Promega) and primers labeled with WellRed D2, D4 (Sigma-Aldrich), or IR-800 (MWG Eurofins) fluorophores, cDNA fragments were resolved by capillary electrophoresis (Beckman Coulter CEQ 8000). Data were then interpreted and analyzed using the software “QuSHAPE” (Karabiber et al. 2013) (http://bioinfo.unc.edu).
The reverse transcription reactions were performed using the following primers: 5′-ACAGCTATGACCATGATTACGA-3′, 5′-TCCACCCTAAACAAAATCAC-3′, 5′-ACCTCGCTATACTAAGCC-3′, 5′-GTTTCCGTTCTACAGGCGG-3′, 5′-CGGACTTTGATCTGCGCA-3′, and 5′-TTGTGATTTTGTTTAGGGTG-3′.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
Z.O. was supported by a fellowship from the Ministry of Higher Education of Malaysia. M.K.S. was supported by a fellowship from the Tertiary Education Fund Trust and University of Ilorin, Nigeria. B.S. and N.U. are supported by CNRS and Université Paris Descartes.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.045328.114.
REFERENCES
- Ali IK, McKendrick L, Morley SJ, Jackson RJ. 2001. Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J Virol 75: 7854–7863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambroziak JA, Blackbourn DJ, Herndier BG, Glogau RG, Gullett JH, McDonald AR, Lennette ET, Levy JA. 1995. Herpes-like sequences in HIV-infected and uninfected Kaposi's sarcoma patients. Science 268: 582–583 [DOI] [PubMed] [Google Scholar]
- Arias C, Weisburd B, Stern-Ginossar N, Mercier A, Madrid AS, Bellare P, Holdorf M, Weissman JS, Ganem D. 2014. KSHV 2.0: A comprehensive annotation of the Kaposi's sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog 10: e1003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aversa SM, Cattelan AM, Salvagno L, Crivellari G, Banna G, Trevenzoli M, Chiarion-Sileni V, Monfardini S. 2005. Treatments of AIDS-related Kaposi's sarcoma. Crit Rev Oncol Hematol 53: 253–265 [DOI] [PubMed] [Google Scholar]
- Bagneris C, Ageichik AV, Cronin N, Wallace B, Collins M, Boshoff C, Waksman G, Barrett T. 2008. Crystal structure of a vFlip-IKKγ complex: insights into viral activation of the IKK signalosome. Mol Cell 30: 620–631 [DOI] [PubMed] [Google Scholar]
- Ballon G, Chen K, Perez R, Tam W, Cesarman E. 2011. Kaposi sarcoma herpesvirus (KSHV) vFLIP oncoprotein induces B cell transdifferentiation and tumorigenesis in mice. J Clin Invest 121: 1141–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balvay L, Lopez Lastra M, Sargueil B, Darlix JL, Ohlmann T. 2007. Translational control of retroviruses. Nat Rev Microbiol 5: 128–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balvay L, Soto Rifo R, Ricci EP, Decimo D, Ohlmann T. 2009. Structural and functional diversity of viral IRESes. Biochim Biophys Acta 1789: 542–557 [DOI] [PubMed] [Google Scholar]
- Bellaousov S, Reuter JS, Seetin MG, Mathews DH. 2013. RNA structure: web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res 41: W471–W474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieleski L, Talbot SJ. 2001. Kaposi's sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. J Virol 75: 1864–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieleski L, Hindley C, Talbot SJ. 2004. A polypyrimidine tract facilitates the expression of Kaposi's sarcoma-associated herpesvirus vFLIP through an internal ribosome entry site. J Gen Virol 85: 615–620 [DOI] [PubMed] [Google Scholar]
- Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. 2005. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure 13: 1695–1706 [DOI] [PubMed] [Google Scholar]
- Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J. 2006. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol 2: 213–220 [DOI] [PubMed] [Google Scholar]
- Borman AM, Michel YM, Kean KM. 2001. Detailed analysis of the requirements of hepatitis A virus internal ribosome entry segment for the eukaryotic initiation factor complex eIF4F. J Virol 75: 7864–7871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, et al. 2009. A review of human carcinogens—part B: biological agents. Lancet Oncol 10: 321–322 [DOI] [PubMed] [Google Scholar]
- Bushell M, Stoneley M, Kong YW, Hamilton TL, Spriggs KA, Dobbyn HC, Qin X, Sarnow P, Willis AE. 2006. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol Cell 23: 401–412 [DOI] [PubMed] [Google Scholar]
- Cencic R, Hall DR, Robert F, Du Y, Min J, Li L, Qui M, Lewis I, Kurtkaya S, Dingledine R, et al. 2011. Reversing chemoresistance by small molecule inhibition of the translation initiation complex eIF4F. Proc Natl Acad Sci 108: 1046–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med 332: 1186–1191 [DOI] [PubMed] [Google Scholar]
- Chamond N, Locker N, Sargueil B. 2010. The different pathways of HIV genomic RNA translation. Biochem Soc Trans 38: 1548–1552 [DOI] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266: 1865–1869 [DOI] [PubMed] [Google Scholar]
- Chaudhary PM, Jasmin A, Eby MT, Hood L. 1999. Modulation of the NF-κ B pathway by virally encoded death effector domains-containing proteins. Oncogene 18: 5738–5746 [DOI] [PubMed] [Google Scholar]
- Colman R, Blackbourn DJ. 2008. Risk factors in the development of Kaposi's sarcoma. AIDS 22: 1629–1632 [DOI] [PubMed] [Google Scholar]
- de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CU. 2009. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc Natl Acad Sci 106: 9197–9202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deforges J, Chamond N, Sargueil B. 2012. Structural investigation of HIV-1 genomic RNA dimerization process reveals a role for the Major Splice-site Donor stem loop. Biochimie 94: 1481–1489 [DOI] [PubMed] [Google Scholar]
- Dibrov SM, Ding K, Brunn ND, Parker MA, Bergdahl BM, Wyles DL, Hermann T. 2012. Structure of a hepatitis C virus RNA domain in complex with a translation inhibitor reveals a binding mode reminiscent of riboswitches. Proc Natl Acad Sci 109: 5223–5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibrov SM, Parsons J, Carnevali M, Zhou S, Rynearson KD, Ding K, Garcia Sega E, Brunn ND, Boerneke MA, Castaldi MP, et al. 2014. Hepatitis C virus translation inhibitors targeting the internal ribosomal entry site. J Med Chem 57: 1694–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer DP. 2003. Transcription profile of Kaposi's sarcoma-associated herpesvirus in primary Kaposi's sarcoma lesions as determined by real-time PCR arrays. Cancer Res 63: 2010–2015 [PubMed] [Google Scholar]
- Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J Virol 72: 8309–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupin N, Fisher C, Kellam P, Ariad S, Tulliez M, Franck N, van Marck E, Salmon D, Gorin I, Escande JP, et al. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc Natl Acad Sci 96: 4546–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton LE, Locker N, Lukavsky PJ. 2009. Conserved functional domains and a novel tertiary interaction near the pseudoknot drive translational activity of hepatitis C virus and hepatitis C virus-like internal ribosome entry sites. Nucleic Acids Res 37: 5537–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efklidou S, Bailey R, Field N, Noursadeghi M, Collins MK. 2008. vFLIP from KSHV inhibits anoikis of primary endothelial cells. J Cell Sci 121: 450–457 [DOI] [PubMed] [Google Scholar]
- Feoktistova K, Tuvshintogs E, Do A, Fraser CS. 2013. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc Natl Acad Sci 110: 13339–13344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field N, Low W, Daniels M, Howell S, Daviet L, Boshoff C, Collins M. 2003. KSHV vFLIP binds to IKK-γ to activate IKK. J Cell Sci 116: 3721–3728 [DOI] [PubMed] [Google Scholar]
- Ganem D. 2010. KSHV and the pathogenesis of Kaposi sarcoma: listening to human biology and medicine. J Clin Invest 120: 939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68: 913–963 [DOI] [PubMed] [Google Scholar]
- Grundhoff A, Ganem D. 2001. Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus. J Virol 75: 1857–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasparri I, Keller SA, Cesarman E. 2004. KSHV vFLIP is essential for the survival of infected lymphoma cells. J Exp Med 199: 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hajdin CE, Bellaousov S, Huggins W, Leonard CW, Mathews DH, Weeks KM. 2013. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc Natl Acad Sci 110: 5498–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, Luhrmann R. 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc Natl Acad Sci 99: 16719–16724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndier B, Ganem D. 2001. The biology of Kaposi's sarcoma. Cancer Treat Res 104: 89–126 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. 2014. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 83: 779–812 [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James L, Sargueil B. 2008. RNA secondary structure of the feline immunodeficiency virus 5′UTR and Gag coding region. Nucleic Acids Res 36: 4653–4666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner RG, Alba MM, Boshoff C, Kellam P. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J Virol 75: 891–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabiber F, McGinnis JL, Favorov OV, Weeks KM. 2013. QuShape: rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS. 2008. Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci 33: 274–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft JJ, Treder K, Peterson MS, Miller WA. 2013. Cation-dependent folding of 3′ cap-independent translation elements facilitates interaction of a 17-nucleotide conserved sequence with eIF4G. Nucleic Acids Res 41: 3398–3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidal AM, Pringle ES, McCormick C. 2012. Evasion of oncogene-induced senescence by γ-herpesviruses. Curr Opin Virol 2: 748–754 [DOI] [PubMed] [Google Scholar]
- Locker N, Easton LE, Lukavsky PJ. 2006. Affinity purification of eukaryotic 48S initiation complexes. RNA 12: 683–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker N, Easton LE, Lukavsky PJ. 2007. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J 26: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker N, Chamond N, Sargueil B. 2011. A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40S subunit and eIF3. Nucleic Acids Res 39: 2367–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low W, Harries M, Ye H, Du MQ, Boshoff C, Collins M. 2001. Internal ribosome entry site regulates translation of Kaposi's sarcoma-associated herpesvirus FLICE inhibitory protein. J Virol 75: 2938–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky PJ. 2009. Structure and function of HCV IRES domains. Virus Res 139: 166–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Barends S, Jaeger S, Schaeffer L, Prongidi-Fix L, Eriani G. 2011. Cap-assisted internal initiation of translation of histone H4. Mol Cell 41: 197–209 [DOI] [PubMed] [Google Scholar]
- Matta H, Chaudhary PM. 2004. Activation of alternative NF-κ B pathway by human herpes virus 8-encoded Fas-associated death domain-like IL-1 β-converting enzyme inhibitory protein (vFLIP). Proc Natl Acad Sci 101: 9399–9404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick WC. 2004. Cap-dependent and cap-independent translation in eukaryotic systems. Gene 332: 1–11 [DOI] [PubMed] [Google Scholar]
- Mesri EA, Cesarman E, Boshoff C. 2010. Kaposi's sarcoma and its associated herpesvirus. Nat Rev Cancer 10: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, Gross JD, Degterev A, Yuan JY, Chorev M, et al. 2007. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 128: 257–267 [DOI] [PubMed] [Google Scholar]
- Mortimer SA, Weeks KM. 2007. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc 129: 4144–4145 [DOI] [PubMed] [Google Scholar]
- Nicholson BL, White KA. 2011. 3′ Cap-independent translation enhancers of positive-strand RNA plant viruses. Curr Opin Virol 1: 373–380 [DOI] [PubMed] [Google Scholar]
- Ohlmann T, Rau M, Pain VM, Morley SJ. 1996. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J 15: 1371–1382 [PMC free article] [PubMed] [Google Scholar]
- Parkin DM. 2006. The global health burden of infection-associated cancers in the year 2002. Int J Cancer 118: 3030–3044 [DOI] [PubMed] [Google Scholar]
- Pause A, Methot N, Svitkin Y, Merrick WC, Sonenberg N. 1994. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J 13: 1205–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce M, Matsumura S, Wilson AC. 2005. Transcripts encoding K12, v-FLIP, v-cyclin, and the microRNA cluster of Kaposi's sarcoma-associated herpesvirus originate from a common promoter. J Virol 79: 14457–14464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU, Shatsky IN. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol 16: 6859–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Unbehaun A, Hellen CU, Pestova TV. 2007. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol 430: 147–177 [DOI] [PubMed] [Google Scholar]
- Plank TD, Kieft JS. 2012. The structures of nonprotein-coding RNAs that drive internal ribosome entry site function. Wiley Interdiscip Rev RNA 3: 195–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo N, Sanz MA, Steinberger J, Skern T, Kusov Y, Carrasco L. 2012. Translation directed by hepatitis A virus IRES in the absence of active eIF4F complex and eIF2. PLoS One 7: e52065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo N, Garcia-Moreno M, Sanz MA, Carrasco L. 2013. Translation of viral mRNAs that do not require eIF4E is blocked by the inhibitor 4EGI-1. Virology 444: 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renne R, Lagunoff M, Zhong W, Ganem D. 1996. The size and conformation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J Virol 70: 8151–8154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, et al. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc Natl Acad Sci 93: 14862–14867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargueil B, McKenna J, Burke JM. 2000. Analysis of the functional role of a G·A sheared base pair by in vitro genetics. J Biol Chem 275: 32157–32166 [DOI] [PubMed] [Google Scholar]
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay MF, Clauvel JP, Raphael M, Degos L, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86: 1276–1280 [PubMed] [Google Scholar]
- Stassinopoulos IA, Belsham GJ. 2001. A novel protein-RNA binding assay: functional interactions of the foot-and-mouth disease virus internal ribosome entry site with cellular proteins. RNA 7: 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M, Chappell SA, Jopling CL, Dickens M, MacFarlane M, Willis AE. 2000. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol Cell Biol 20: 1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturzl M, Gaus D, Dirks WG, Ganem D, Jochmann R. 2013. Kaposi's sarcoma-derived cell line SLK is not of endothelial origin, but is a contaminant from a known renal carcinoma cell line. Int J Cancer 132: 1954–1958 [DOI] [PubMed] [Google Scholar]
- Sun Q, Matta H, Lu G, Chaudhary PM. 2006. Induction of IL-8 expression by human herpesvirus 8 encoded vFLIP K13 via NF-κB activation. Oncogene 25: 2717–2726 [DOI] [PubMed] [Google Scholar]
- Sweeney TR, Abaeva IS, Pestova TV, Hellen CU. 2014. The mechanism of translation initiation on Type 1 picornavirus IRESs. EMBO J 33: 76–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot SJ, Weiss RA, Kellam P, Boshoff C. 1999. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology 257: 84–94 [DOI] [PubMed] [Google Scholar]
- Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer JL, Schroter M, et al. 1997. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386: 517–521 [DOI] [PubMed] [Google Scholar]
- Van Eden ME, Byrd MP, Sherrill KW, Lloyd RE. 2004. Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA 10: 720–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Treder K, Miller WA. 2009. Structure of a viral cap-independent translation element that functions via high affinity binding to the eIF4E subunit of eIF4F. J Biol Chem 284: 14189–14202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Parisien M, Scheets K, Miller WA. 2011. The cap-binding translation initiation factor, eIF4E, binds a pseudoknot in a viral cap-independent translation element. Structure 19: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill L, James L, Ulryck N, Chamond N, Herbreteau CH, Ohlmann T, Sargueil B. 2010. A new type of IRES within gag coding region recruits three initiation complexes on HIV-2 genomic RNA. Nucleic Acids Res 38: 1367–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcocks MM, Locker N, Gomwalk Z, Royall E, Bakhshesh M, Belsham GJ, Idamakanti N, Burroughs KD, Reddy PS, Hallenbeck PL, et al. 2011. Structural features of the Seneca Valley virus internal ribosome entry site (IRES) element: a picornavirus with a pestivirus-like IRES. J Virol 85: 4452–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Abaeva IS, Marintchev A, Pestova TV, Hellen CU. 2011. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res 39: 4851–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.