Summary
This study represents the first characterization of a Triticeae stigma proteome and highlights numerous proteins important in stigma development, pollen–stigma interactions, and protection against biotic and abiotic stresses.
Key words: Electrophoresis, mass spectrometry, proteomics, stigma, triticale, Triticeae.
Abstract
To our knowledge, this study represents the first high-throughput characterization of a stigma proteome in the Triticeae. A total of 2184 triticale mature stigma proteins were identified using three different gel-based approaches combined with mass spectrometry. The great majority of these proteins are described in a Triticeae stigma for the first time. These results revealed many proteins likely to play important roles in stigma development and pollen–stigma interactions, as well as protection against biotic and abiotic stresses. Quantitative comparison of the triticale stigma transcriptome and proteome showed poor correlation, highlighting the importance of having both types of analysis. This work makes a significant contribution towards the elucidation of the Triticeae stigma proteome and provides novel insights into its role in stigma development and function.
Introduction
Triticale, a cross between wheat (Triticum sp.) and rye (Secale sp.), is the first man-made cereal crop. It is more productive than other cereals under poor soil and environmental conditions, less susceptible to many diseases affecting wheat, and competitive with other cereals in terms of grain yield (Chapman et al., 2005). Triticale is also considered a reduced-input crop as it requires less fertilizer and water, and fewer pesticides. Moreover, triticale has a number of potential advantages as a feedstock for bioethanol production (Pejin et al., 2012). Lastly, triticale represents an interesting opportunity to study the implications of allopolyploidization (Ma and Gustafson, 2008).
In flowering plants, pollination is a critical step in sexual reproduction leading to seed production. In triticale, as in other members of the Triticeae, the process begins with adhesion of the pollen grain to the stigma (the receptive portion of the pistil). The pollen grain then rehydrates and germinates, and the newly formed pollen tube penetrates the stigma, grows down the transmitting tissue of the style, and ultimately reaches the ovule, allowing fertilization to take place (Lausser and Dresselhaus, 2010). The stigma plays a critical role in the capture, recognition, and discrimination of the pollen grain, and in supporting pollen germination and tube growth. Generally, it is a relatively short-lived plant structure and has been broadly divided into two major types, wet and dry stigmas (Heslop-Harrison and Shivanna, 1977). In species with a wet stigma, such as in the Solanaceae and Liliaceae, the epidermis is covered with a viscous surface secretion containing protein, lipid, polysaccharides, and pigments, and the stigma has three distinct zones: an epidermis with papillae, a subepidermal secretory zone, and a zone of parenchyma ground tissue (Kandasamy and Kristen, 1987). In species with a dry stigma, such as in the Brassicaceae and Triticeae, the stigma is composed of specialized papillar cells covered with a waxy cuticle overlaid with a distinct proteinaceous pellicle layer, which interacts directly with the pollen (Zinkl et al., 1999; Ciampolini et al., 2001). Compared with the wet stigma, pollen capture, adhesion, and hydration on the dry stigma is a more highly regulated process (Wolters-Arts et al., 1998). The developmental stages of the triticale plumose dry stigma have been described previously (Tran et al., 2013).
In spite of its crucial role, little is known regarding the proteome underlying stigma development and function. High-throughput transcriptome profiling devoted specifically to the stigma has been reported for a few Brassicaceae species (Swanson et al., 2005; Osaka et al., 2013; Sankaranarayanan et al., 2013), rice (Li et al., 2007; Fujita et al., 2010), maize (Davidson et al., 2011; Xu et al., 2012, 2013), and triticale (Tran et al., 2013). Only a few studies have investigated the stigma proteome and these have usually focused on a subset. In a comparison between species with wet or dry stigmas using two-dimensional electrophoresis/mass spectrometry, stigma-specific or -preferential proteins were identified from maize silk (67 proteins) and tobacco stigmas (47 proteins) (Sang et al., 2012). These authors also identified 132 proteins present within the tobacco stigma exudate, whereas a separate study revealed 51 and 57 exudate proteins from lily and olive (Rejón et al., 2013). In an analysis of Brassica stigmas, differential in-gel electrophoresis/mass spectromtery was used to identify 19 proteins downregulated following self-incompatible pollination, thus bringing new insights into this process (Samuel et al., 2011). Lastly, isobaric tags for relative and absolute quantitation, comparing two developmental stages of Solanum pennellii stigma/styles, identified 2534 proteins and highlighted proteome changes associated with stigma/style maturation and the onset of reproductive barriers (Chalivendra et al., 2013).
The current study represents the largest contribution towards a comprehensive survey of the stigma proteome of any Triticeae species. The possible role of the identified triticale proteins in stigma development and pollen–stigma interactions, as well as stress tolerance, is discussed.
Materials and methods
Plant material
The hexaploid triticale spring cultivar (×Triticosecale AC Alta) was grown as described previously (Tran et al., 2013). In this study, the stigma refers to both the stigma and the style, as these structures are merged in triticale. Mature triticale stigmas were harvested just prior to anthesis (at this stage, the stigma is most receptive to pollen and there is no pollen contamination), frozen immediately in liquid nitrogen, and kept at –80 °C. Bread wheat cultivar Thatcher (Triticum aestivum L.) was grown similarly until the first two true leaves emerged.
Protein extraction
Different procedures were tested to extract triticale stigma proteins, and the most comprehensive and reproducible results were obtained using either SDS-PAGE or isoelectric focusing (IEF) loading buffer directly (results not shown). Frozen stigmas were ground to a fine powder on dry ice and the frozen powder was immediately suspended in 1× SDS-PAGE loading buffer (10% glycerol, 2% SDS, 100mM dithiothreitol (DTT), 1% bromophenol blue and 60mM Tris/HCl, pH 6.8) and sonicated three times for 10 s each using an Ultrasonic Homogenizer 4710 series (Cole-Parmer, Montréal, Canada). After centrifugation at 27 200g for 10min at 18 °C, the supernatant was transferred to another tube and heated for 5min at 95 °C prior to one-dimensional (1D) SDS-PAGE. For the IEF loading buffer extraction, the frozen powder was immediately suspended in sample buffer (8M urea, 2M thiourea, 2% CHAPS, 0.1% ampholytes (pI range 3–10), 50mM DTT, 0.0005% bromophenol blue) and incubated at room temperature on a rotator overnight. The sample was then sonicated three times for 10 s each and centrifuged at 48 400g for 30min at 4 °C. Total protein concentrations were determined with a Bio-Rad protein assay kit (Bio-Rad, Mississauga, Canada) according to the manufacturer’s instructions.
Wheat leaf proteins were extracted by grinding a 2g aliquot in liquid nitrogen, mixing with 25ml of 50mM Tris/HCl (pH 8.3), 2% (v/v) 2-mercaptoethanol, 20mM MgCl2, 1mM PMSF, and grinding further. The sample was centrifuged at 10 000g for 10min at 4 °C and the supernatant frozen overnight at –80 °C. To deplete ribulose bisphosphate carboxylase (RbcL and -S), the sample was thawed at room temperature for 2h and then heated to 42 °C with gentle rotation for 15min. This was followed by centrifugation as above at room temperature. Proteins in the supernatant were precipitated with 9 vols of ice-cold acetone containing 0.7% (w/v) DTT and the pellet was dissolved in 1M urea, 50mM Tris/HCl (pH 8.3), 0.7% (w/v) DTT, sonicated, and clarified by centrifugation as above.
1D liquid chromatography/tandem mass spectrometry (LC-MS/MS)
Stigma proteins (45 μg) extracted with SDS-PAGE loading buffer were submitted to the McGill University Genome Québec Innovation Centre (http://gqinnovationcenter.com/index.aspx) where the protein sample was loaded on a 2.4cm 1D SDS-PAGE gel with a 7–15% acrylamide gradient. The gel was stained with Coomassie Brilliant Blue G (Supplementary Fig. S1 at JXB online) and the entire lane was excised into 15 bands, which were further subdivided into six to seven pieces using a Protein Picking Workstation ProXCISION (PerkinElmer, Shelton, USA). Gel pieces were subjected to reduction, cysteine alkylation, and in-gel tryptic digestion in an automated MassPREP Workstation (Micromass, Manchester, UK) as described previously (Wasiak et al., 2002). A 20 µl volume of tryptic digest solution was injected into a Zorbax 300SB-C18 column (Agilent Technologies, Mississauga, Canada) at 15 µl min–1, and the sample was washed for 5min and then analysed with a Q-Tof microTM MS (Waters Corporation, Milford, MA, USA). The MS survey scan was set to 1 s recording from 350 to 1600 m/z with all doubly and triply charged ions with an intensity higher than 25 counts considered candidates to undergo MS/MS fragmentation, and from these the strongest one was selected. The MS/MS scan was acquired twice, with the second round excluding the most intense precursor of the first round. The precursor was excluded within ±1900 mDa of the entries on the exclusion list.
Two-dimensional (2D) LC-MS/MS
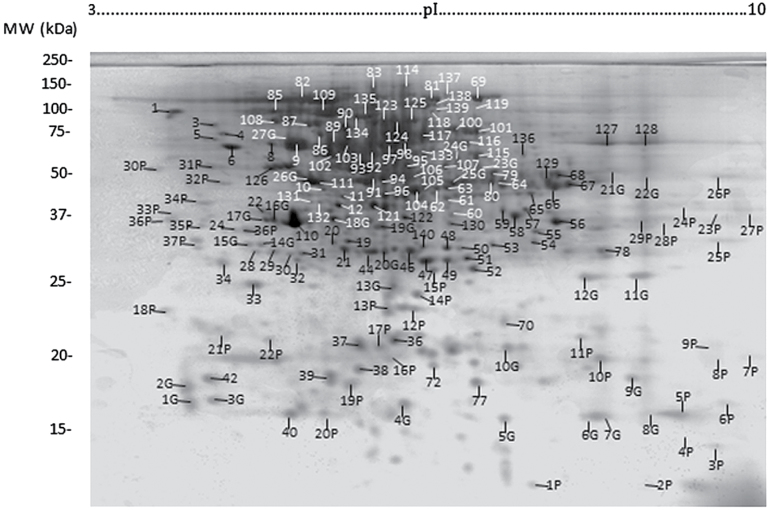
For 2D gels, stigma proteins extracted with IEF loading buffer (200 µg) were first separated on a 17cm pH 3–10 gradient using a PROTEAN IEF system (Bio-Rad) according to the manufacturer’s instructions. After IEF, the strips were first equilibrated for 20min in 6M urea, 2% DTT, 375mM Tris/HCl (pH 8.8), and then for another 20min in the same solution containing 135mM iodoacetamide instead of 2% DTT. SDS-PAGE was performed in a 12% acrylamide gel using a PROTEAN II XI Cell system (Bio-Rad) at a constant voltage of 77V. The gels were stained with Coomassie Brilliant Blue R-250 and gel images were captured with a scanner Astra 2400 SLT (UMAX, Dallas, USA) or Gel Doc XR+ (Bio-Rad). Three 2D gel replicates gave consistent results (Fig. 1). For future reference, the identity of the major triticale stigma proteins was investigated and a total of 170 individual protein spots were excised manually from Coomassie Brilliant Blue stained gels and submitted to the Québec Genomics Center (http://proteomique.crchul.ulaval.ca/en/index.html) for MS. Briefly, the excised proteins were automatically destained, dehydrated, reduced with 10mM DTT, alkylated with 55mM iodoacetamide, and digested with 105mM modified porcine trypsin at 58 °C for 1h using a MassPrep liquid handling robot (Waters Corporation). Digestion products were extracted using 1% formic acid, 2% acetonitrile, followed by 1% formic acid, 50% acetonitrile, and the recovered extracts were pooled, lyophilized, and then resuspended in 7 µl of 0.1% formic acid. A peptide sample volume of 2 µl was separated by online reversed-phase high-pressure liquid chromatography (RP-HPLC) using a Jupiter 300 C18 column (Phenomenex, Torrance, USA) and analysed by electrospray ionization MS/MS. The experiments were performed with a Thermo Surveyor MS pump connected to a LTQ Linear Ion Trap Mass Spectrometer equipped with a nanoelectrospray ion source (ThermoFisher Scientific, San Jose, USA). Mass spectra were acquired using a data-dependent acquisition mode using the Xcalibur software version 2.0. Each full scan mass spectrum (400–2000 m/z) was followed by collision-induced dissociation of the seven most intense ions. The dynamic exclusion (30 s exclusion) function was enabled and the relative collisional fragmentation energy was set at 35%. As many spots contained more than one protein, a quantitative analysis of relative spot intensity was not performed.
Fig. 1.
2D gel electrophoresis of triticale mature stigma proteins also indicating the most highly stained proteins observed in a separate experiment investigating post-translational modifications. P, Pro-Q Diamond phosphoprotein stain; G, Pro-Q Emerald glycoprotein stain (Life Technologies, Burlington, Canada).
OFFGEL electrophoresis (OGE) LC-MS/MS
Triticale stigma proteins (90 μg) extracted with IEF loading buffer were sent to Bioproximity Proteomics Services (http://www.bioproximity.com). Briefly, the protein sample was processed using a filter-assisted sample preparation method (Wiśniewski et al., 2009), digested at a 1:40 trypsin:substrate ratio and incubated overnight at 37 °C. Digested peptides were desalted using C18 stop-and-go extraction tips (Rappsilber et al., 2007) and eluted with 60% acetonitrile, 0.5% acetic acid, and lyophilized. Peptides were then fractionated using an Agilent OFFGEL fractionator (Agilent Technologies) with a 24cm pH 3–10 IPG strip in conjunction with a 24-well tray according to the manufacturer’s instructions. Each fraction was desalted as above and analysed by LC-MS/MS. LC was performed using an Easy Nano-LC II HPLC (ThermoFisher Scientific) using a Zorbax 300SB-C18 column (Agilent Technologies). The LC was interfaced to a LTQ Velos Dual-Pressure Linear Ion Trap mass spectrometer (ThermoFisher Scientific) via nano-electrospray ionization. The mass spectrometer was programmed to acquire, by data-dependent acquisition, tandem mass spectra from the top 15 ions in the full scan from 400 to 1400 m/z. Dynamic exclusion was set to 30 s.
LC-MS/MS of wheat leaf proteins
To prepare the sample for off-line separation by RP-HPLC, each leaf peptide sample replicate was first desalted using a 5cm RP column (MOS1-Hypersil: ThermoFisher Scientific) connected to an analytical HPLC pump (Ultimate 3000; Dionex, Bremen, Germany). The eluent was monitored at 220nm, and a single desalted fraction was collected. The desalted peptides were separated into 60 fractions using a high-pH acetonitrile gradient as described elsewhere (Dwivedi et al., 2008). Fractions were pooled by interlacing fractions 1+21+41; 2+22+42, etc., resulting in 20 fractions analysed by LC-MS/MS on a linear ion trap mass spectrometer (LTQ XL; ThermoFisher Scientific) as described previously (Rampitsch et al., 2013).
Data analysis
All MS/MS spectra were analysed using Mascot version 2. 3. 0 (http://www.matrixscience.com) and X! Tandem version 2007.01.01.1 (http://www.thegpm.org/tandem/) to search the Universal Protein Resource (UniProt) Viridiplantae database. The search was restricted to a maximum of one missed (trypsin) cleavage, fixed carbamidomethyl alkylation of cysteine, and variable oxidation of methionine, as well as a 0.5Da mass unit tolerance for fragment ions and 2.0Da for parent ions. The maximum false discovery rate was set to 0.3% for proteins and 5.3% for peptides. Scaffold version 4.0.4 (Proteome Software, Portland, OR, USA) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at a greater than 95% probability as specified by the peptide Prophet algorithm (Keller et al., 2002). Protein identifications were accepted if they could be established at a greater than 95% probability and contained at least two identified peptides. Scaffold grouped proteins displaying shared peptides to satisfy the laws of parsimony. Proteins with a single peptide match at a greater than 95% probability (Supplementary Table S1 at JXB online) were validated manually based on the following criteria: the peptide had to be identified at least by two search engines (Mascot with significant ion score of 25 and X! Tandem), contain a minimum of 80% coverage of theoretical y or b ions (at least three amino acids in a row in either y or b ions series), and show an absence of prominent unassigned peaks greater than 5% of the maximum intensity (Feng et al., 2009; Prenni et al., 2012). SignalP4.1 (http://www.cbs.dtu.dk/services/SignalP) was utilized to predict the presence of signal peptides. Detailed information about specific proteins (e.g. Mascot scores, sequence of peptides identified) can be provided upon request.
To compare triticale mature stigma protein and mRNA data, the 542 protein sequences identified in the 1D SDS-PAGE shotgun analysis were submitted to a tBLASTn search against an in-house triticale reference transcriptome assembly and the 239 contigs whose derived protein sequences showed ≥95% identity were selected for further analysis. These contigs were then aligned to the sequences used to generate the different probe sets of the 55K Affymetrix GeneChip® wheat genome array and the 180 contigs showing ≥95% identity were selected for expression comparisons with the matching proteins. The mature triticale stigma gene expression levels for these 180 contigs were derived from the microarray study (Tran et al., 2013), whereas the normalization of the corresponding spectral counts was performed using Scaffold.
Results and discussion
Comparison of 1D LC-MS/MS, 2D LC-MS/MS, and OGE LC-MS/MS
In this study, the proteome of the mature triticale stigma was analysed using three different electrophoretic approaches. A total of 2555 proteins were identified comprising 542 proteins using 1D LC-MS/MS, 303 proteins by OGE LC-MS/MS, and 1710 proteins with 2D LC-MS/MS, which represented a total of 2184 unique proteins (Supplementary Table S1). We examined the overlap in the proteins identified using the three different proteomics techniques and, surprisingly, only 66 proteins of the 2184 proteins identified in this study were identified by all three approaches and an additional 237 proteins were detected by two of the three methods (Fig. 2). Half of the proteins found with OGE LC-MS/MS were also identified by 2D LC-MS/MS and this may reflect the fact that they share the same protein extraction buffer, although in the 2D LC-MS/MS it was the proteins that were separated by IEF, whereas it was tryptic peptides with OGE LC-MS/MS. These results clearly indicate that different proteomic approaches can yield distinct proteins from the same tissue, and combining different techniques increases the number of identified proteins.
Fig. 2.
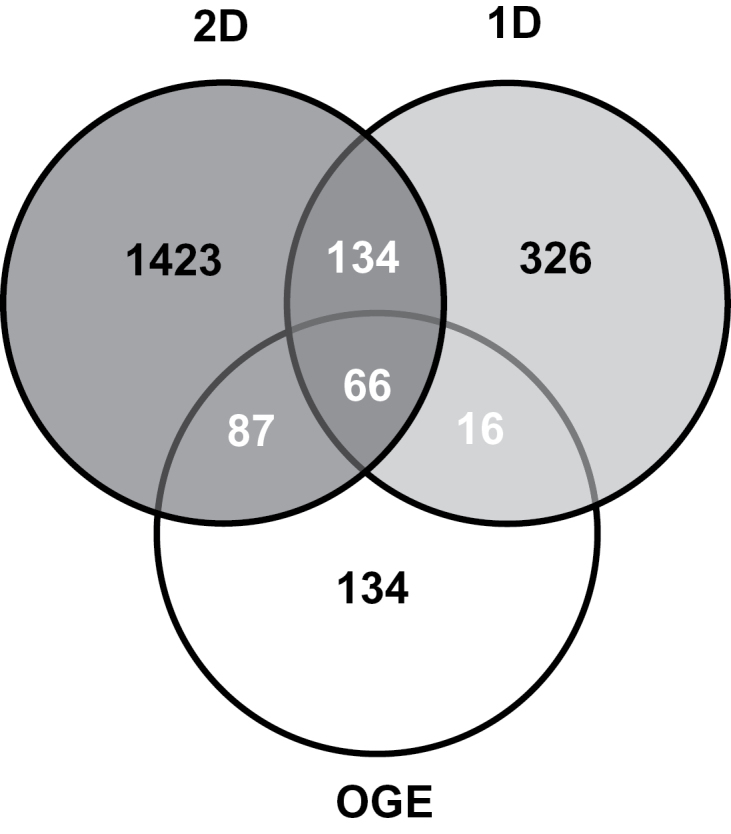
Overlap among the proteins identified by the different methods of analysis. 2D, 2D LC-MS/MS; 1D, 1D LC-MS/MS; OGE, OGE LC-MS/MS.
Evidently, the ‘shotgun’ approaches of 1D LC-MS/MS and OGE LC-MS/MS did not uncover as many stigma proteins as 2D LC-MS/MS, and this was probably the result of a lower number of more highly complex samples. Although labour intensive, 2D LC-MS/MS does provide considerable information pertaining to the proteins detected, as well as a protein profile (Fig. 1) that can be used in comparative analyses such as differential in-gel electrophoresis. For example, almost one-third of the 1710 identified proteins were found in multiple spots with different pIs and molecular masses (Supplementary Table S1), pointing to the existence of isoforms possibly resulting from post-translational modifications, multimeric forms of the protein, proteolytic processing, and alternative transcripts. This is a higher proportion than reported for Arabidopsis floral buds (Feng et al., 2009) and may indicate a higher degree of post-translational modification within the stigma. The 2D gels were also analysed for the presence of phosphorylated or glycosylated proteins using commercially available stains, but MS analysis of intensely fluorescent spots (labelled P and G in Fig. 1) could not confirm the presence of these post-translational modifications (results not shown).
Functional classification of triticale mature stigma proteins
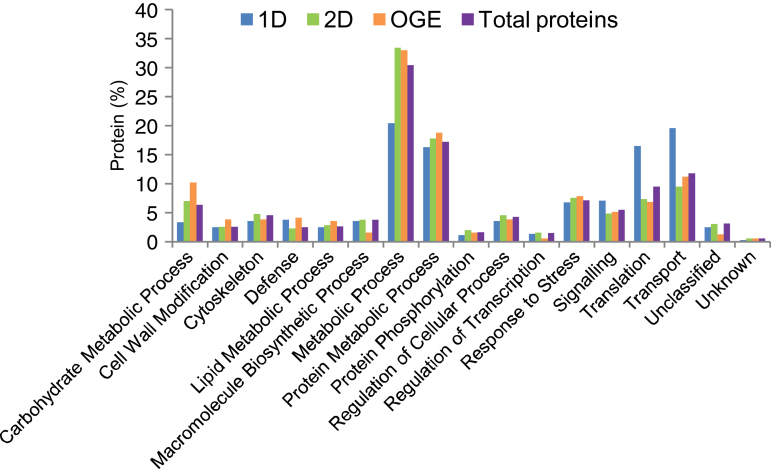
Generally, the proteins identified by the different electrophoretic procedures displayed similar overall functional distribution patterns (Fig. 3), although, for reasons that are not evident, 1D LC-MS/MS differed considerably in the categories Metabolic Process, Translation, and Transport. More than 75% of the 2184 triticale mature stigma proteins identified in this study were found in only five categories: Metabolic Process (30.5%), Protein Metabolic Process (17.2%), Transport (11.9%), Translation (9.6%), and Response to Stress (7.3%). A detailed description of the constituent proteins of each functional category is beyond the scope of this article, and the discussion will therefore be restricted to some examples relevant to three critical aspects of stigma biology: stigma development, pollen–stigma interactions, and stigma responses to biotic and abiotic stress.
Fig. 3.
Functional distribution of triticale mature stigma proteins identified using different methods of analysis and their combined total. 2D, 2D LC-MS/MS; 1D, 1D LC-MS/MS; OGE, OGE LC-MS/MS.
Proteins involved in stigma development
Very little is known regarding the identity and function of the proteins underlying stigma development in the Triticeae; however, it is possible to infer a potential role for the triticale stigma proteins identified in this study (Supplementary Table S1) based on homology to proteins shown to play a role in development, primarily in model species. Transcription factors (TFs) are evident candidates as potential regulators of stigma development, and some of the TFs found within the triticale mature stigma included orthologues to TFs known to have a role in gyneocium development including DROOPING LEAF (DL)/CRABS CLAW (CRC), which is involved in determining carpel identity and mid-rib formation (Li et al., 2011), GT2, KNOTTED-LIKE, ARABIDOPSIS THALIANA3 (KNAT3), which, although shown to be involved in embryo sac development in Arabidopsis (Pagnussat et al., 2007), was also found in the triticale stigma as a transcript (Tran et al., 2013), and SHORT INTERNODES (SHI)/STYLISH (STY), which regulates auxin-mediated stigma and style development by interacting with YUCCA4 (YUC4) (Larsson et al., 2013). Auxin has been proposed to play a role in stigma and style development and function (Ståldal and Sundberg, 2009), and microarray analysis has shown that auxin-signalling components are over-represented in the rice stigma (Li et al., 2007) and that several auxin response factors show enriched expression in the triticale stigma (Tran et al., 2013). A role for auxin in triticale stigma development is further supported by the presence of orthologues to the Arabidopsis proteins HEMIVENATA (HVE), a CULLIN-ASSOCIATED-NEDD8-DISSOCIATED1 (CAND1) protein that negatively regulates ubiquitin-mediated auxin signalling and plays a role in leaf venation and cell division (Pérez-Pérez et al., 2011), and AUXIN RESISTANT1 (AXR1), a RUB/NEDD8-activating enzyme E1 regulatory subunit that plays a role in auxin signalling and in development (Nakasone et al., 2012), as well as the ABC transporter ABCB15 shown to transport auxin (Kaneda et al., 2011).
A variety of additional proteins were identified that may also function in triticale stigma development and these comprised several CELL DIVISION CONTROL (CDC) 48-like proteins, which are transitional endoplasmic reticulum ATPases involved in protein turnover and shown in Arabidopsis to affect cell division, expansion, and differentiation, as well as pollen-tube growth (Park et al., 2008). Other triticale stigma proteins with homologues shown to have developmental roles included FLORAL ORGAN REGULATOR-LIKE1 (FORL1) and FORL2, which are related to OsFOR1 involved in regulating floral organ number in rice (Jang et al., 2003), TASSELSEED2 (TS2), which when absent leads to the feminization of maize tassels (DeLong et al., 1993), POINTED FIRST LEAF (PFL), which represents ribosomal proteins that can affect growth and development (Ito et al., 2000), a RAN GTPase-activating protein (RanGAP1) with a conserved function in cell division and important to female gametophyte development in Arabidopsis (Rodrigo-Peiris et al., 2011), ABNORMAL INFLORESCENCE MERISTEM1 (AIM1), a fatty acid β-oxidation multifunctional protein (Richmond and Bleecker, 1999), and PEPPER (PEP), a protein containing KH RNA-binding domains that regulates gynoecium development including that of the stigma (Ripoll et al., 2006).
A number of proteins highlight the importance of epigenetic mechanisms in triticale stigma development such as orthologues to the Arabidopsis METHYL-CPG-BINDING DOMAIN PROTEIN11 (MBD11), which when downregulated causes developmental defects including abnormal flower positions, delayed flowering, and reduced fertility (Berg et al., 2003), and MULTI-SUBUNIT SUPPRESSOR OF IRA1 (MSI1) involved in gene silencing during female gametogenesis and early seed development (Hennig and Derkacheva, 2009), as well as GLABRA2 EXPRESSION MODULATOR (GEM), which, via histone modification, inhibits the expression of the transcription factor GLABRA2 (GL2) implicated in epidermal cell division and cell fate (Qing and Aoyama, 2012) and which may play a role in Triticeae stigma papillae formation. Lastly, the conservation of mechanisms underlying monocot floral patterning is indicated by the presence of an orthologue to the Arabidopsis HISTONE DEACETYLASE19 (HDA19) shown to be part of a repressor complex with APETAL2 (AP2) and TOPLESS (TPL) regulating floral organ identity genes (Krogan et al., 2012).
A role for protein phosphorylation in stigma development is supported by the presence of Arabidopsis orthologues CYCLIN-DEPENDENT KINASE F;1 (CDKF;1), a major regulator of cell proliferation and expansion (Takatsuka et al., 2009), different SNF1-related protein kinase (SNRK) regulatory subunits, which can interact with NAC-domain TFs to positively regulate development (Kleinow et al., 2009), CASEIN KINASE2 (CK2) implicated in multiple developmental pathways (Mulekar and Huq, 2014), and 2A PHOSPHATASE ASSOCIATED PROTEIN OF 46KD (TAP46) with roles in development via the TARGET OF RAPAMYCIN (TOR) and ABI5 regulatory pathways (Hu et al., 2014). At present, the great majority of the many protein phosphatases identified in this study could not be assigned a putative role.
The Triticeae stigma is a non-green tissue enclosed within the floret and hence it will be interesting to investigate the role of a number Arabidopsis orthologues associated with photomorphogenesis including different subunits of the CONSTITUTIVE MORPHOGENESIS9 (COP9) SIGNALOSOME (CSN), which although involved in photomorphogenesis also regulates many different cellular processes by regulating the activity of CULLIN-RING E3 ubiquitin ligases (Stratmann and Gusmaroli, 2011), TF Z-BOX BINDING FACTOR3 (ZBF3)/CALMODULIN7 (CAM7) that integrates light signals with seedling development (Gangappa et al., 2013), and THYLAKOID FORMATION1 (THF1) whose rice orthologue NYC4 was recently shown to play a role in chlorophyll–protein complex degradation during leaf senescence (Yamatani et al., 2013), as well as SCHLEPPERLESS (SLP)/CHAPERONIN60 (CPN60) and chaperones part of the chloroplast multi-subunit Clp protease such as DE-REGULATED CAO ACCUMULATION1 (DCA1)/ClpC1 and ALBINO OR PALE GREEN6 (APG6)/ClpB3, which are all involved in chloroplast development (Apuya et al., 2001; Sjögren et al., 2004; Myouga et al., 2006).
These few examples illustrate (mostly for the first time) the occurrence of proteins involved in the regulation of Triticeae stigma development and other such examples can be found in Supplementary Table S1.
Proteins involved in pollen–stigma interactions
The landing of a compatible pollen grain onto the stigma surface marks the beginning of pollen–pistil interactions enabling pollen recognition, adhesion, germination, tube growth, and finally fertilization. Little is known of the molecules involved in the recognition process within the grasses (Klaas et al., 2011). The first layers of contact are the protein pellicle and then the cuticle covering the dry stigma, both of which the pollen tube must breach. The triticale mature stigma expressed many proteases (Supplementary Table S1), and some may facilitate penetration of the protein pellicle. A cutinase responsible for the breakdown of the stigma cuticle has not been identified, but a GDSL lipase was recently shown to fit the description (Takahashi et al., 2010) and one of the GDSL lipases expressed in the triticale stigma could have a similar role. A significant number of other triticale stigma proteins within the Lipid Metabolic Process category are likely to be involved in stigma cuticle formation, and these include orthologues to the Arabidopsis FIDDLEHEAD (FDH), which has been shown to play a role in pollen–stigma interactions (Lolle and Cheung, 1993), DEFECTIVE IN CUTICULAR RIDGES (DCR), a diacylglycerol acyltransferase involved in cuticle formation, epidermal cell differentiation, and the regulation of water flow (Panikashvili et al., 2009), a GDSL lipase shown to function in cutin biosynthesis (Shi et al., 2011), an ABC transporter similar to the barley EIBI1, and the Arabidopsis ATP BINDING CASSETTEG32 (ABCG32) likely to export cutin precursors from the epidermal cells (Bessire et al., 2011), ECERIFERUM8 (CER8)/LONG-CHAIN ACYL-COA SYNTHETASE1 (LACS1) proteins involved in wax and cutin synthesis (Lü et al., 2009), and ANTHOCYANINLESS2 (ANL2), an HD-START TF that plays a role in epidermal cell development as well as anthocyanin and cutin accumulation (Wu et al., 2011). HD-START TFs represented a major category of TFs enriched within the stigma transcriptome (Tran et al., 2013). Finally, several non-specific lipid transfer proteins were found within the stigma proteome. Lipid transfer proteins have been ascribed a number of different roles in development, signalling, and biotic and abiotic stresses, as well as cutin biosynthesis (Edstam et al., 2011). As with many of the proteins described below, it will be interesting to investigate whether many of these proteins play a role primarily in maturing stigma development/growth or pollen germination/tube growth, or are involved in both activities.
The pollen tube penetrates the stigma cell wall and grows within it, thus necessitating proteins for both cell growth and cell wall degradation and synthesis. The triticale stigma proteome exhibited numerous cell wall-modifying proteins including, for example, many glycosyl hydrolases and several expansins, as well as pectin-modifying enzymes like pectinesterases and pectate lyase (Supplementary Table S1). Other proteins of interest included lipoxygenases, which are known to affect papillae expansion (Caldelari et al., 2011), TASSELSEED1 (TS1), a lipoxygenase necessary for maize pistil primordial abortion possibly via the jasmonic acid pathway (Acosta et al., 2009), dynamin-related proteins, which have a role in cytokinesis and cell expansion including cell wall formation (Collings et al., 2008) and were shown to be involved in Arabidopsis stigma papillae expansion (Kang et al., 2003), two CELLULOSE SYNTHASE-INTERACTING (CSI) proteins shown in Arabidopsis to bridge cellulose synthase to cortical microtubules, a process underlying directional growth (Li et al., 2012), and multiple versions of UDP-ARABINOPYRANOSE MUTASE (UAM)/REVERSIBLY GLYCOSYLATED POLYPEPTIDE (RGP), which generates UDP-arabinofuranosyl associated with arabinoxylan, an important component of grass primary and secondary cell walls (Konishi et al., 2011). In addition, four different fasciclin-like arabinogalactan proteins were identified; arabinogalactan proteins are known to be abundant in transmitting tissue and are a major constituent of lily stigma exudate (Rejón et al., 2013), while the fasciclin domain involved in protein–protein interactions was shown to be involved in cell adhesion and could also mediate extracellular interactions (Ellis et al., 2010). Lastly, orthologues to the Arabidopsis FERONIA (FER) and HERCULES (HERK) were identified within the triticale stigma proteome, and these receptor-like protein kinases are important to cell wall metabolism and cell growth (Lindner et al., 2012).
The growth of the pollen tube through the stigma eventually becomes auxotrophic and relies on support from the surrounding tissue, and this in turn signifies a requirement for the transport of various nutrients and signals. Accordingly, transport is the third largest functional category of the triticale stigma proteins (Fig. 3), consistent with the enrichment of transport-related transcripts in the mature stigma (Xu et al., 2012; Tran et al., 2013). A role for vesicle-mediated secretion of stigma resources has been suggested (Samuel et al., 2011), and a high proportion of the transport proteins of the triticale stigma proteome belong to relevant proteins such as annexins, coatomer subunits, and SEC proteins (Supplementary Table S1). GTPase-mediated regulation of membrane trafficking is also reflected by the high number of associated proteins detected in the triticale stigma proteome including ADP-RIBOSYLATION FACTORS (ARFs), GDP DISSOCIATION INHIBITORS (GDIs), ARF GTPASE-ACTIVATING PROTEINS (ARFGAPs), and PRENYLATED RAB ACCEPTORS (PRAs), as well as many GTPases, most of which are either dynamin-related or Rab GTPases, with the most highly represented GTPase being SECRETION-ASSOCIATED RAS1 (SAR1). The role of Rab GTPases in membrane trafficking and signalling has recently been reviewed (Rehman and Di Sansebastiano, 2014), and characterizing the role of this process in cereal pollen–pistil interactions will be a significant future challenge. The next largest group of proteins in the Transport category was ATPases mainly V-type and P-type proton ATPases important in the creation of gradients underlying membrane potential and ion transport during pollen-tube growth (Hepler et al., 2013), as well as AAA+ ATPases, which are essentially molecular chaperones with a diversity of functions revolving around processes such as protein unfolding or disassembly of protein complexes (Ogura and Wilkinson, 2001).
Stigmas can accumulate high amounts of reactive oxygen species, as well as H2O2 (McInnis et al., 2006), and these are known to function as signalling molecules in a wide range of processes in plants including stress responses (Foyer and Noctor, 2013). Furthermore, the role of redox regulation in pollen–stigma interactions is becoming increasingly evident (Dresselhaus and Franklin-Tong, 2013; Traverso et al., 2013). The Metabolic Process category contained a high proportion of triticale stigma proteins associated with redox processes (Supplementary Table S1), with examples including many enzymes involved in glutathione metabolism such as glutathione S-transferases, peroxidases, reductases, and synthetases, as well as numerous redoxins and enzymes such as superoxide dismutases, ascorbate peroxidases, catalases, peroxidases, malate dehydrogenases, malic enzymes, 6-phosphogluconate dehydrogenases, and isocitrate dehydrogenases. The role of these proteins in Triticeae pollen–stigma interactions remains to be elucidated.
The continuous interaction between the pollen grain/tube and the stigma implies a high degree of communication, and numerous additional triticale stigma proteins likely to play a significant role in signalling were identified including several 14-3-3 proteins, which regulate an ever-growing number of processes such as cell growth and responses to hormones, light, and stress via phosphorylation-dependent associations (Denison et al., 2011), as well as proteins linked to calcium signalling such as calmodulins, calreticulins, CaLB domain-containing proteins, calcium-dependent protein kinases (CDPKs) and several phospholipase D (PLDs), which have been shown to be involved for example in cytoskeletal dynamics, membrane remodelling, and biotic and abiotic stress responses (Wang et al., 2014). The triticale stigma also expressed RECEPTOR FOR ACTIVATED C KINASE 1A (RACK1A) proteins, which are scaffolding proteins that interact with signalling proteins in a variety of signalling pathways associated with development, hormonal, and stress responses (Dongping et al., 2013), and several inositol-3-phosphate synthases responsible for the first step of inositol biosynthesis, a key substrate in the phosphoinositide signalling pathway affecting processes such as growth and development, as well as stress responses (Boss and Im, 2012).
As exemplified above, the interaction between the pollen grain/tube and the stigma operates at multiple levels, and elucidation of the cellular processes involved will be of great interest.
Proteins involved in biotic and abiotic stress
The stigma and style are meant to be breached by, and provide nutrients to, the growing pollen tube while preventing this from happening with pathogens and this represents a remarkable challenge to the plant. Furthermore, the next generation must also be preserved against abiotic stresses. Accordingly, these tissues often express relatively high numbers of stress-related genes and indeed it has been found that considerable functional overlap exists between reproductive and stress-responsive genes (Li et al., 2007). In this study, this is supported by the fact that many of the proteins involved in stigma development and pollen–stigma interactions discussed above such as AIM1, HDA19, FORL, SNRK, CK2, THF1, APG6, COP9, and PLDs, as well as many of the redox proteins, are known to also play a role in the stress response. The triticale mature stigma expressed a number of proteins with homology to protein kinases known to be involved in the defence and stress responses including ZmCPK11 (Ding et al., 2013), PROTEIN KINASE INTERACTOR1 (PTI1) (Forzani et al., 2011), OsCDPK7 (Saijo et al., 2000), STRESS-ACTIVATED PROTEIN KINASE7 (SAPK7) (Kobayashi et al., 2004), and AtMPK1 (Hwa and Yang, 2008). The TF AtNF-YC2, which is activated by oxidative stress and is involved in flowering (Hackenberg et al., 2012), was also identified in the triticale stigma proteome.
The major class of stress-associated proteins found in the triticale stigma consisted of heat-shock-related proteins represented mainly by HEAT SHOCK COGNATE70 (HSC70), HEAT SHOCK PROTEIN70 (HSP70), and HSP90 proteins (Supplementary Table S1). In fact, nearly half of the Protein Metabolic Process category was made up of proteins involved in chaperone activities also including many PROTEIN DISULFIDE ISOMERASES, PEPTIDYL PROLYL ISOMERASES, and CHAPERONIN60 (CPN60)/T-COMPLEX PROTEIN1 (TCP1) family proteins, all of which may play a role in the stress response. The next largest group of the Protein Metabolic Process category was made up of proteins associated with the proteasome, which not only mediates many developmental processes but also plays a major role in plant responses to stress (Lyzenga and Stone, 2012). One possible example is the triticale orthologue to the Arabidopsis TF ABA BINDING FACTOR3 (ABF3), which, like many other proteins in this study, implicates abscisic acid in stress tolerance and was recently shown to be regulated via proteasome-mediated proteolysis (Chen et al., 2013). Stress-related proteins, which were found repeatedly, also included the UNIVERSAL STRESS PROTEIN (USP) family proteins and UVB-RESISTANCE8 (UVR8) proteins (Supplementary Table S1).
Lastly, defence proteins, which appeared often within the triticale mature stigma proteome, included HYPERSENSITIVE-INDUCED REACTION (HIR) proteins, PATHOGENESIS-RELATED (PR) proteins, thaumatin-like proteins, chitinases, ricin B-like proteins, allene oxide oxidases, as well as HEAT repeat-containing ILITYHIA (ILA)-like proteins required for disease resistance in Arabidopsis (Monaghan and Li, 2010) and ARGONAUTE4 (AGO4) proteins involved in RNA-directed DNA methylation shown to be involved in bacterial and viral disease resistance (Seo et al., 2013).
Therefore, this study not only confirmed the presence of a high proportion of proteins involved in both biotic and abiotic stress in the stigma, but also showed the occurrence of a considerable functional overlap in many stress and developmental proteins.
Comparison of the stigma and leaf proteome
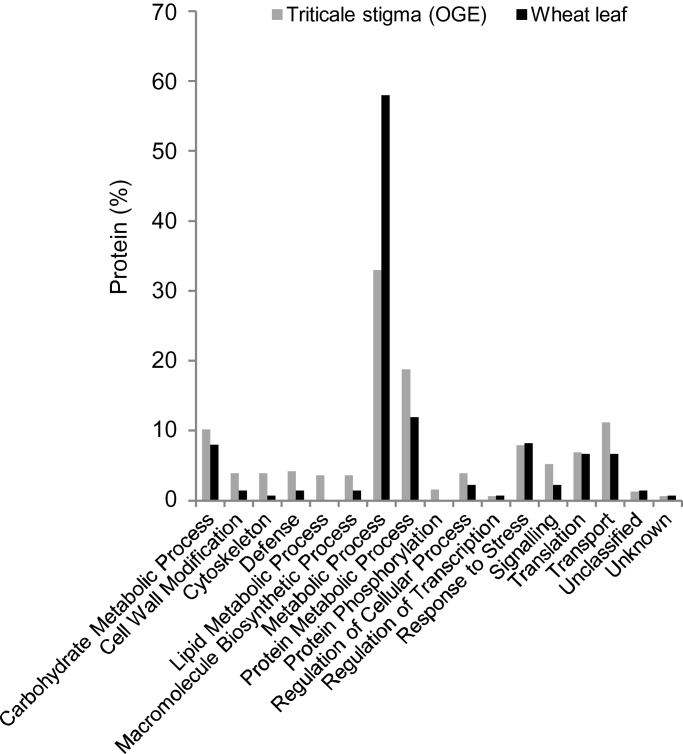
In the hope of highlighting functional categories that may be under- or over-represented within the stigma, the functional distribution of mature stigma proteins (OGE) was compared with that of the wheat leaf proteome obtained in a different project (Fig. 4). The Metabolic Process category represented a much larger proportion of the leaf proteome, and this reflects the greater number of leaf proteins associated with photosynthesis (Supplementary Table S1). In contrast, the stigma proteome was noticeably enriched in a number of categories including Cell Wall Modification, Cytoskeleton, Defence, Lipid Metabolic Process, Protein Metabolic Process, Signalling and Transport. These results are only in partial agreement with the functional classification of the transcripts enriched in the mature triticale stigma (Tran et al., 2013), and this may be due to the fact that the gene expression enrichment analysis was much more comprehensive and compared all vegetative and reproductive triticale tissues. Considerably more proteomic data from Triticeae tissues will be required for a thorough comparison.
Fig. 4.
Comparison of the functional distribution of triticale mature stigma and wheat leaf proteins. OGE, OGE LC-MS/MS.
Stigma proteins with a signal peptide
Many of the interactions between the stigma and the pollen grain/tube are extracellular and, although not all extracellular proteins possess signal peptides (Drakakaki and Dandekar, 2013), the presence of a signal peptide is a reliable indicator of protein secretion (Lum and Min, 2011) and may point to stigma proteins involved in extracellular interactions. We identified 158 proteins predicted to contain a signal peptide representing 7.9% of the total number of stigma proteins with a start codon (Supplementary Table S1). As the proportion of all the triticale proteins with a signal peptide is unknown, it is impossible to determine whether this percentage represents a possible enrichment in the stigma. However, as a comparison, about 14% of all the maize proteins are predicted to have a signal peptide (Xu et al., 2012), so it seems possible that the low proportion of secreted proteins identified in this study represent specialized proteins that need to be present for the initial interactions with the pollen whereas additional proteins are generated as the pollen tube grows through the stigma. The great majority of the triticale stigma proteins with a signal peptide were found within only five functional categories: Protein Metabolic Process (36%), Carbohydrate Metabolic Process (21%), Cell Wall Modification (18%), Defence (10%), and Transport (8%) (Supplementary Fig. S2 at JXB online). The Protein Metabolic Process category consisted primarily of proteins such as heat-shock proteins/luminal-binding proteins and protein disulfide isomerases, which are probably important in maintaining protein integrity (Supplementary Table S1). It also included several protease inhibitors and proteases and, although proteolytic enzymes can have significant roles in reproduction such as defence, signalling, and programmed cell death (Radlowski, 2005), their potential function in the triticale stigma remains unknown. Many of the proteins in the Carbohydrate Metabolic Process class were also found in the Cell Wall Modification category, and the need for their extracellular localization is in line with a role in allowing and supporting pollen-tube growth. The proportion of extracellular proteins in the Defence category was higher than the value for the total stigma proteome (Fig. 3), again indicating a requirement for these proteins at the site of pollen-tube penetration. The Transport category included more than half of the lipid transfer proteins and this could be due for example to requirements for cuticle synthesis and maintenance.
Correlation between triticale mature stigma mRNA and protein expression
Based on the assumption that the 2184 proteins identified in the current study are biased towards the most abundant proteins (Feng et al., 2009), their functional distribution was compared with that of the 2184 most abundantly expressed stigma genes identified in a microarray analysis (Tran et al., 2013) and the results are shown in Fig. 5A. Of the 2184 genes, 238 showed enriched expression in the pistil when compared with all other triticale tissues, and 179 of those genes showed enriched expression in the stigma (Supplementary Table S1). While similarities were observed in the number of expressed genes and proteins found in certain functional categories, differences were also evident; for example, the proportion of proteins associated with the categories Cytoskeleton and Metabolic Process was considerably higher than that of the associated genes, whereas the reverse trend was observed for Protein Phosphorylation, Regulation of Transcription, Regulation of Cellular Process and Transport (Fig. 5A). These observations are in general agreement with reports describing metabolic and structural genes as being stable and having a high protein/mRNA ratio, unlike proteins involved in regulatory processes, which may require more rapid production and turnover in reaction to a stimulus (Vogel and Marcotte, 2012). Given that the mature stigma has essentially stopped growing and stands in readiness for pollination, it is possible that it accumulates mRNA encoding regulatory and transport genes to be rapidly translated upon pollen adhesion, hydration, germination, and tube growth.
Fig. 5.
(A) Comparison of the functional distribution of the 2184 proteins and most abundantly expressed genes in the triticale mature stigma. (B) Comparison of triticale mature stigma protein and mRNA abundance. Pearson correlation coefficient, R 2 = 0.267.
In order to explore further the relationship between production and degradation of individual mRNAs and proteins, 180 proteins from the 1D LC-MS/MS analysis that were found to specifically match genes present in the microarray analysis (Tran et al., 2013) were used to compare corresponding mRNA and protein levels (Supplementary Table S1). There was very little correlation between the level of mRNA and that of the corresponding proteins (R 2=0.267) within this relatively small dataset (Fig. 5B). Such a low correlation has also been described for the Arabidopsis floral proteome (Feng et al., 2009) and not only emphasizes the importance of both post-transcriptional and post-translational processes but also confirms the importance of investigating both the transcriptome and the proteome.
Conclusion
To our knowledge, this is first investigation of the Triticeae stigma global proteome. Numerous proteins likely to play significant roles in stigma development, function, and protection of the next generation were revealed and should provide the basis for future research aimed at a better understanding of cereal pollen–stigma interactions.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. 1D SDS-PAGE of triticale mature stigma proteins indicating bands excised for 1D LC-MS/MS analysis.
Supplementary Fig. S2. Functional distribution of triticale mature stigma proteins with a predicted signal peptide.
Supplementary Table S1. Triticale mature stigma proteins identified by 1D LC-MS/MS, 2D LC-MS/MS, and OGE LC-MS/MS. List of top 2184 genes expressed in the triticale mature stigma. List of proteins used in the protein/mRNA quantitative comparison. List of wheat leaf proteins.
Glossary
Abbreviations:
- 1D
one-dimensional
- 2D
two-dimensional
- DTT
dithiothreitol
- IEF
isoelectric focusing
- LC-MS/MS
liquid chromatography/tandem mass spectrometry
- MS
mass spectrometry
- OGE
OFFGEL electrophoresis
- RP-HPLC
reversed-phase high-pressure liquid chromatography
- TF
transcription factor.
References
- Acosta IF, Laparra H, Romero SP, Schmelz E, Hamberg M, Mottinger JP, Moreno MA, Dellaporta SL. 2009. tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination in maize. Science 323, 262–265 [DOI] [PubMed] [Google Scholar]
- Apuya NR, Yadegari R, Fischer RL, Harada JJ, Zimmerman JL, Goldberg RB. 2001. The Arabidopsis embryo mutant schlepperless has a defect in the chaperonin-60α gene. Plant Physiology 126, 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg A, Meza TJ, Mahić M, Thorstensen T, Kristiansen K, Aalen RB. 2003. Ten members of the Arabidopsis gene family encoding methyl-CpG-binding domain proteins are transcriptionally active and at least one, AtMBD11, is crucial to normal development. Nucleic Acids Research 31, 5291–5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessire M, Borel S, Fabre G, et al. 2011. A member of the PLEIOTROPIC DRUG RESISTANCE family of ATP binding cassette transporters is required for the formation of a functional cuticle in Arabidopsis . Plant Cell 23, 1958–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss WF, Im YJ. 2012. Phosphoinositide signaling. Annual Review of Plant Biology 63, 409–429 [DOI] [PubMed] [Google Scholar]
- Caldelari D, Wang G, Farmer EE, Dong X. 2011. Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Molecular Biology 75, 25–33 [DOI] [PubMed] [Google Scholar]
- Chalivendra SC, Lopez-Casado G, Kumar A, et al. 2013. Developmental onset of reproductive barriers and associated proteome changes in stigma/styles of Solanum pennellii . Journal of Experimental Botany 64, 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman BD, Salmon CD, Dyson D, Blackley K. 2005. Triticale production and utilization manual, spring and winter triticale for grain, forage and value-added. Alberta Agriculture, Food and Rural Development. http://www1.agric.gov.ab.ca/$department/deptdocs.nsf/all/fcd10535
- Chen Y, Liu H, Stone S, Callis J. 2013. ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3. The Plant Journal 75, 965–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampolini F, Shivanna KR, Cresti M. 2001. Organization of the stigma and transmitting tissue of rice, Oryza sativa (L). Plant Biology 3, 149–155 [Google Scholar]
- Collings DA, Gebbie LK, Howles PA, Hurley UA, Birch RJ, Cork AH, Hocart CH, Arioli T, Williamson RE. 2008. Arabidopsis dynamin-like protein DRP1A: a null mutant with widespread defects in endocytosis, cellulose synthesis, cytokinesis, and cell expansion. Journal of Experimental Botany 59, 361–376 [DOI] [PubMed] [Google Scholar]
- Davidson RM, Hansey CN, Gowda M, et al. 2011. Utility of RNA sequencing for analysis of maize reproductive transcriptomes. Plant Genome 4, 191–203 [Google Scholar]
- DeLong A, Calderon-Urrea A, Dellaporta SL. 1993. Sex determination gene TASSELSEED2 of maize encodes a short-chain alcohol dehydrogenase required for stage-specific floral organ abortion. Cell 74, 757–768 [DOI] [PubMed] [Google Scholar]
- Denison FC, Paul A, Zupanska AK, Ferl RJ. 2011. 14-3-3 proteins in plant physiology. Seminars in Cell Developmental Biology 22, 720–727 [DOI] [PubMed] [Google Scholar]
- Ding Y, Cao J, Ni L, Zhu Y. 2013. ZmCPK11 is involved in abscisic acid-induced antioxidant defence and functions upstream of ZmMPK5 in abscisic acid signalling in maize. Journal of Experimental Botany 64, 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongping Z, Li C, Bing LV, Jiansheng L. 2013. The scaffolding protein RACK1: a platform for diverse functions in the plant kingdom. Journal of Plant Biology & Soil Health , 1, 7 [Google Scholar]
- Drakakaki G, Dandekar A. 2013. Protein secretion: how many secretory routes does a plant cell have? Plant Science 203/204, 74–78 [DOI] [PubMed] [Google Scholar]
- Dresselhaus T, Franklin-Tong N. 2013. Male-female cross-talk during pollen germination, tube growth and guidance, and double fertilization. Molecular Plant 6, 1018–1036 [DOI] [PubMed] [Google Scholar]
- Dwivedi RC, Spicer V, Harder M, Antonovici M, Ens W, Standing KG, Wilkins JA, Krokhin OV. 2008. Practical implementation of 2D HPLC scheme with accurate peptide retention prediction in both dimensions for high-throughput bottom-up proteomics. Analytical Chemistry 80, 7036–7042 [DOI] [PubMed] [Google Scholar]
- Edstam M, Viitamen L, Salminen TA, Edqvist J. 2011. Evolutionary history of the non-specific lipid transfer proteins. Molecular Plant 4, 947–964 [DOI] [PubMed] [Google Scholar]
- Ellis M, Egelund J, Schultz CJ, Bacic A. 2010. Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiology 153, 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Li L, Zhou X, Stanley B, Ma H. 2009. Analysis of the Arabidopsis floral proteome: detection of over 2000 proteins and evidence for posttranslational modifications. Journal of Integrative Plant Biology 51, 207–223 [DOI] [PubMed] [Google Scholar]
- Forzani C, Carreri A, de la Fuente van Bentem S, Lecourieux D, Lecourieux F, Hirt H. 2011. The Arabidopsis protein kinase Pto-interacting 1–4 is a common target of the oxidative signal-inducible 1 and mitogen-activated protein kinases. FEBS Journal 278, 1126–1136 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. 2013. Redox signaling in plants. Antioxidants & Redox Signaling 18, 2087–2090 [DOI] [PubMed] [Google Scholar]
- Fujita M, Horiuchi Y, Ueda Y, Mizuta Y. 2010. Rice expression atlas in reproductive development. Plant and Cell Physiology 51, 2060–2081 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Srivastava AK, Maurya JP, Ram H, Chattopadhyay S. 2013. Z-box binding transcription factors (ZBFs): a new class of transcription factors in Arabidopsis seedling development. Molecular Plant 6, 1758–1768 [DOI] [PubMed] [Google Scholar]
- Hackenberg D, Keetman U, Grimm B. 2012. Homologous NF-YC2 subunit from Arabidopsis and tobacco is activated by photooxidative stress and induces flowering. International Journal of Molecular Sciences 13, 3458–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Derkacheva M. 2009. Diversity of polycomb group complexes in plants: same rules, different players? Trends in Genetics 25, 414–423 [DOI] [PubMed] [Google Scholar]
- Hepler PK, Rounds CM, Winship LJ. 2013. Control of cell wall extensibility during pollen tube growth. Molecular Plant 6, 998–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison Y, Shivanna KR. 1977. The receptive surface of the angiosperm stigma. Annals of Botany 41, 1233–1258 [Google Scholar]
- Hu R, Zhu Y, Shen G, Zhang H. 2014. TAP46 plays a positive role in ABSCISIC ACID INSENSITIVE5-regulated gene expression in Arabidopsis. Plant Physiology 164, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwa C, Yang X. 2008. The AtMKK3 pathway mediates ABA signaling in Arabidopsis . Acta Physiologiae Plantarum 30, 277–286 [Google Scholar]
- Ito T, Kim G, Shinozaki K. 2000. Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. The Plant Journal 22, 257–264 [DOI] [PubMed] [Google Scholar]
- Jang S, Lee B, Kim C, Kim S, Yim J, Han J, Lee S, Kim S, An G. 2003. The OsFOR1 gene encodes a polygalacturonase-inhibiting protein (PGIP) that regulates floral organ number in rice. Plant Molecular Biology 53, 357–369 [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, Kristen U. 1987. Developmental aspects of ultrastructure, histochemistry and receptivity of the stigma of Nicotiana sylvestris . Annals of Botany 60, 427–437 [Google Scholar]
- Kaneda M, Schuetz M, Lin BSP, Chanis C, Hamberger B, Western TL, Ehlting J, Samuels AL. 2011. ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. Journal of Experimental Botany 62, 2063–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B, Busse JS, Bednarek SY. 2003. Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell 15, 899–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical Chemistry 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- Klaas M, Yang B, Bosch M, Thorogood D, Manzanares C, Armstead IP, Franklin FCH, Barth S. 2011. Progress towards elucidating the mechanisms of self-incompatibility in the grasses: further insights from studies in Lolium . Annals of Botany 108, 677–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinow T, Himbert S, Krenz B, Jeske H, Koncz C. 2009. NAC domain transcription factor ATAF1 interacts with SNF1-related kinases and silencing of its subfamily causes severe developmental defects in Arabidopsis . Plant Science 177, 360–370 [Google Scholar]
- Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T. 2004. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 16, 1163–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T, Aohara T, Igasaki T, Hayashi N, Miyazaki Y, Takahashi A, Hirochika H, Iwai H, Satoh S, Ishii T. 2011. Down-regulation of UDP-arabinopyranose mutase reduces the proportion of arabinofuranose present in rice cell walls. Phytochemistry 72, 1962–1968 [DOI] [PubMed] [Google Scholar]
- Krogan NT, Hogan K, Long JA. 2012. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139, 4180–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson E, Franks RG, Sundberg E. 2013. Auxin and the Arabidopsis thaliana gynoecium. Journal of Experimental Botany 64, 2619–2627 [DOI] [PubMed] [Google Scholar]
- Lausser A, Dresselhaus T. 2010. Sporophytic control of pollen tube growth and guidance in grasses. Biochemical Society Transactions 38, 631–634 [DOI] [PubMed] [Google Scholar]
- Li H, Liang W, Yin C, Zhu L, Zhang D. 2011. Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy. Plant Physiology 156, 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Xu W, Yang W, Kong Z, Xue Y. 2007. Genome-wide gene expression profiling reveals conserved and novel molecular functions of the stigma in rice. Plant Physiology 144, 1797–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lei L, Somerville CR, Gu Y. 2012. Cellulose synthase interactive protein 1 (CSI1) links microtubules and cellulose synthase complexes. Proceedings of the National Academy of Sciences, USA 109, 185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H, Müller LM, Boisson-Dernier A, Grossnicklaus U. 2012. CrRLK1L receptor-like kinases: not just another brick in the wall. Current Opinion in Plant Biology 15, 1–11 [DOI] [PubMed] [Google Scholar]
- Lolle SJ, Cheung AY. 1993. Promiscuous germination and growth of wildtype pollen from Arabidopsis and related species on the shoot of the Arabidopsis mutant, fiddlehead . Developmental Biology 155, 250–258 [DOI] [PubMed] [Google Scholar]
- Lü S, Song T, Kosma DK, Parsons EP, Rowland O, Jenks MA. 2009. Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. The Plant Journal 59, 553–564 [DOI] [PubMed] [Google Scholar]
- Lum G, Min XJ. 2011. Plant secretomes: current status and future perspectives. Plant Omics Journal 4, 114–119 [Google Scholar]
- Lyzenga WJ, Stone SL. 2012. Abiotic stress tolerance mediated by protein ubiquitination. Journal of Experimental Botany 63, 599–616 [DOI] [PubMed] [Google Scholar]
- Ma XF, Gustafson JP. 2008. Allopolyploidization-accommodated genomic sequence changes in triticale. Annals of Botany 101, 825–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnis SM, Desikan R, Hancock JT, Hiscock SJ. 2006. Production of reactive oxygen species and reactive nitrogen species by angiosperm stigmas and pollen: potential signalling crosstalk? New Phytologist 172, 221–228 [DOI] [PubMed] [Google Scholar]
- Monaghan J, Li X. 2010. The HEAT repeat protein ILITYHIA is required for plant immunity. Plant and Cell Physiology 51, 742–553 [DOI] [PubMed] [Google Scholar]
- Mulekar JJ, Huq E. 2014. Expanding roles of protein kinase CK2 in regulating plant growth and development. Journal of Experimental Botany 65, 2883–2893 [DOI] [PubMed] [Google Scholar]
- Myouga F, Motohashi R, Kuromori T, Nagata N, Shinozaki K. 2006. An Arabidopsis chloroplast-targeted Hsp101 homologue, APG6, has an essential role in chloroplast development as well as heat-stress response. The Plant Journal 48, 249–260 [DOI] [PubMed] [Google Scholar]
- Nakasone A, Fujiwara M, Fukao Y, Biswas KK, Rahman A, Kawai-Yamada M, Narumi I, Uchimiya H, Oono Y. 2012. SMALL ACIDIC PROTEIN1 acts with RUB modification components, the COP9 signalosome, and AXR1 to regulate growth and development in Arabidopsis. Plant Physiology 160, 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T, Wilkinson AJ. 2001. AAA+ superfamily ATPases: common structure—diverse function. Genes to Cells 6, 575–597 [DOI] [PubMed] [Google Scholar]
- Osaka M, Matsuda T, Sakazono S, et al. 2013. Cell type-specific transcriptome of Brassicaceae stigmatic papilla cells from a combination of laser microdissection and RNA sequencing. Plant and Cell Physiology 54, 1894–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu H, Sundaresan V. 2007. Cell-fate switch of synergid to egg cell in Arabidopsis oestre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 19, 3578–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikashvili D, Shi JX, Schreiber L, Aharoni A. 2009. The Arabidopsis DCR encoding a soluble BADH acyltransferase is required for cutin polyester formation and seed hydration properties. Plant Physiology 151, 1773–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Rancour DM, Bednarek SY. 2008. In planta analysis of the cell cycle-dependent localization of AtCDC48A and its critical roles in cell division, expansion, and differentiation. Plant Physiology 148, 246–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejin DJ, Mojović LV, Pejin JD, Grujić OS, Markov SL, Nikolić SB, Marković MN. 2012. Increase in bioethanol production yield from triticale by simultaneous saccharification and fermentation with application of ultrasound. Journal of Chemical Technology and Biotechnology 87, 170–176 [Google Scholar]
- Pérez-Pérez JM, Rubio-Díaz S, Dhondt S, Hernández-Romero D, Sánchez-Soriano J, Beemster GTS, Ponce MR, Micol JL. 2011. Whole organ, venation and epidermal cell morphological variations are correlated in the leaves of Arabidopsis mutants. Plant, Cell & Environment 34, 2200–2211 [DOI] [PubMed] [Google Scholar]
- Prenni JE, Vidal M, Olver CS. 2012. Preliminary characterization of the murine membrane reticulocyte proteome. Blood Cells, Molecules, and Diseases 49, 74–82 [DOI] [PubMed] [Google Scholar]
- Qing L, Aoyama T. 2012. Pathways for epidermal cell differentiation via the homeobox gene GLABRA2: update on the roles of the classic regulator. Journal of Integrative Plant Biology 54, 729–737 [DOI] [PubMed] [Google Scholar]
- Radlowski M. 2005. Proteolytic enzymes from generative organs of flowering plants (Angiospermae). Journal of Applied Genetics 46, 247–257 [PubMed] [Google Scholar]
- Rampitsch C, Day J, Subramaniam R, Walkowiak S. 2013. Comparative secretomes analysis of Fusarium graminearum and two of its non-pathogenic mutants upon deoxynivalenol induction in vitro. Proteomics 13, 1913–1921 [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using Stage Tips. Nature Protocols 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- Rehman RUI, Di Sansebastiano G. 2014. Plant Rab GTPases in membrane trafficking and signalling. In: Rehman Hakeem K, Rehman RUI, Tahir I. eds. Plant signaling: understanding the molecular crosstalk. New Delhi: Springer, 51–73 [Google Scholar]
- Rejón JD, Delalande F, Schaeffer-Reiss C, Carapito C, Zienkiewicz K, de Dios Alché J, Rodríguez-Garcia MI, Van Dorsselaer A, Castro AJ. 2013. Proteomics profile reveals novel proteins and functions of the plant stigma exudate. Journal of Experimental Botany 64, 5695–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TA, Bleecker AB. 1999. A defect in β-oxidation causes abnormal inflorescence development in Arabidopsis. Plant Cell 11, 1911–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll JJ, Ferrándiz C, Martínez-Laborda A, Vera A. 2006. PEPPER, a novel K-homology domain gene, regulates vegetative and gynoecium development in Arabidopsis . Developmental Biology 289, 346–359 [DOI] [PubMed] [Google Scholar]
- Rodrigo-Peiris T, Xu XM, Zhao Q, Wang H, Meier I. 2011. RanGAP is required for post-meiotic mitosis in female gametophyte development in Arabidopsis thaliana . Journal of Experimental Botany 62, 2705–2714 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K. 2000. Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. The Plant Journal 23, 319–327 [DOI] [PubMed] [Google Scholar]
- Samuel MA, Tang W, Jamshed M, Northey J, Patel D, Smith D, Siu KWM, Muench DG, Wang Z, Goring DR. 2011. Proteomic analysis of Brassica stigmatic proteins following the self-incompatibility reaction reveals a role of microtubule dynamics during pollen responses. Molecular and Cellular Proteomics 10, M111.011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang YL, Xu M, Ma FF, Chen H, Xu XH, Gao X, Zhang XS. 2012. Comparative proteomics analysis reveals similar and distinct features of proteins in dry and wet stigma. Proteomics 12, 1983–1998 [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan S, Jamshed M, Deb S, Chatfield-Reed K, Kwon EG, Chua G, Samuel MA. 2013. Deciphering the stigmatic transcriptional landscape of compatible and self-incompatible pollinations in Brassica napus reveals a rapid stigma senescence response following compatible pollination. Molecular Plant 6, 1988–1991 [DOI] [PubMed] [Google Scholar]
- Seo J, Wu J, Lii Y, Jin H. 2013. Contribution of small RNA pathway components in plant immunity. Molecular Plant–Microbe Interactions 26, 617–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi JX, Malitsky S, De Oliveira S, Branigan C, Franke RB, Schreiber L, Aharoni A. 2011. SHINE transcription factors act redundantly to pattern the archetypal surface of Arabidopsis flower organs. PLoS Genet 7, e1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren LLE, MacDonald TM, Sutinen S, Clarke AK. 2004. Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiology 136, 4114–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståldal V, Sundberg E. 2009. The role of auxin in style development and apical-based patterning of the Arabidopsis thaliana gynoecium. Plant Signaling and Behavior 4, 83–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann JW, Gusmaroli G. 2011. Many jobs for one good cop—the COP9 signalosome guards development and defense. Plant Science 185–6, 50–64 [DOI] [PubMed] [Google Scholar]
- Swanson R, Clark T, Preuss D. 2005. Expression profiling of Arabidopsis stigma tissue identifies stigma-specific genes. Sexual Plant Reproduction 18, 163–171 [Google Scholar]
- Takahashi K, Shimada T, Kondo M, Tamai A, Mori M, Nishimura M, Hara-Nishimura I. 2010. Ectopic expression of an esterase, which is a candidate for the unidentified plant cutinase, causes cuticular defects in Arabidopsis thaliana . Plant Cell Physiology 51, 123–131 [DOI] [PubMed] [Google Scholar]
- Takatsuka H, Ohno R, Umeda M. 2009. The Arabidopsis cyclin-dependent kinase-activating kinase CDKF;1 is a major regulator of cell proliferation and cell expansion but is dispensable for CDKA activation. The Plant Journal 59, 475–487 [DOI] [PubMed] [Google Scholar]
- Tran F, Penniket C, Patel RV, Provart NJ, Laroche A, Rowland O, Robert LS. 2013. Developmental transcriptional profiling reveals key insights into Triticeae reproductive development. The Plant Journal 74, 971–988 [DOI] [PubMed] [Google Scholar]
- Traverso JA, Pulido A, Rodríguez-Garía MI, Alché JD. 2013. Thiol-based redox regulation in sexual plant reproduction: new insights and perspectives. Frontiers in Plant Science 4, 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM. 2012. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nature Reviews Genetics 13, 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Guo L, Wang G, Li M. 2014. PLD: phospholipase Ds in plant signaling. In: Wang X. ed. Phospholipases in plant signalling. Heidelberg: Springer, 3–26 [Google Scholar]
- Wasiak S, Legendre-Guillemin V, Puertollano R, Blondeau F, Girard M, de Heuvel E, Boismenu D, Bell AW, Bonifacino JS, McPherson PS. 2002. Enthoprotin: a novel clathrin-associated protein identified through subcellular proteomics. Journal of Cell Biology 158, 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Mann M. 2009. Combination of FASP and stage tip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. Journal of Proteome Research 8, 5674–5678 [DOI] [PubMed] [Google Scholar]
- Wolters-Arts M, Lush WM, Mariani C. 1998. Lipids are required for directional pollen-tube growth. Nature 392, 818–821 [DOI] [PubMed] [Google Scholar]
- Wu R, Li S, He S, Waßmann F, Yu C, Qin G, Schreiber L, Qu L, Gu H. 2011. CFL1, a WW domain protein, regulates cuticle development by modulating the function of HDG1, a class IV homeodomain transcription factor, in rice and Arabidopsis. Plant Cell 23, 3392–3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XH, Chen H, Sang YL, Wang F, Ma JP, Gao X, Zhang XS. 2012. Identification of genes specifically or preferentially expressed in maize silk reveals similarity and diversity in transcript abundance of different dry stigmas. BMC Genomics 10.1186/1471–2164–13–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XH, Wang F, Chen H, Sun W, Zhang XS. 2013. Transcript profile analyses of maize silks reveal effective activation of genes involved in microtubule-based movement, ubiquitin-dependent protein degradation, and transport in the pollination process. PLoS One 8, e53545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamatani H, Sato Y, Masuda Y, et al. 2013. NYC4, the rice ortholog of Arabidopsis THF 1, is involved in the degradation of chlorophyll-protein complexes during leaf senescence. The Plant Journal 74, 652–662 [DOI] [PubMed] [Google Scholar]
- Zinkl GM, Zwiebel BI, Grier DG, Preuss D. 1999. Pollen–stigma adhesion in Arabidopsis: a species-specific interaction mediated by lipophilic molecules in the pollen exine. Development 126, 5431–5440 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.