Abstract
Malondialdehyde (MDA), an endogenous genotoxic product formed upon lipid peroxidation and prostaglandin biosynthesis, can react with DNA to form stable adducts. These adducts may contribute to the development of such inflammation-mediated diseases as cancer and cardiovascular and neurodegenerative diseases. The predominant MDA-derived DNA adduct formed under physiological conditions is 3-(2-deoxy-β-d-erythro-pentafuranosyl)pyrimido[1,2-α]purin-10(3H)-one deoxyguanosine (M1dG). In this study, we developed a novel liquid chromatography (LC)–nanoelectrospray ionization (NSI)–high-resolution tandem mass spectrometry (HRMS/MS) method for the analysis of M1dG in human leukocyte DNA. After enzymatic hydrolysis of DNA, M1dG and the added internal standard [13C3]M1dG were reduced to their 5,6-dihydro derivatives by addition of sodium borohydride to the hydrolysate and purified by solid-phase extraction and column chromatography. The 5,6-dihydro derivatives in the purified samples were analyzed by LC–NSI–HRMS/MS using higher-energy collisional dissociation (HCD) fragmentation, isolation widths of 1 Da for both the analyte and internal standard, and a resolution of 50 000. The detection limit of the developed method is 5 amol on-column, and the limit of quantitation is 0.125 fmol/mg DNA starting with 200 μg of DNA. Method accuracy and precision were characterized. The developed method was further applied to the analysis of leukocyte DNA from 50 human subjects. M1dG was detected in all samples and ranged from 0.132 to 275 fmol/mg DNA, or 0.004 to 9.15 adducts per 108 bases. This unique and highly sensitive HRMS/MS-based method can be used in future studies investigating the pathophysiological role of M1dG in human diseases.
Introduction
Chronic inflammation, a key mechanism in the pathogenesis of cancer and cardiovascular disease,1−4 induces lipid peroxidation, which in turn generates a spectrum of reactive electrophiles capable of causing extensive damage to DNA and proteins, resulting in toxic and mutagenic events.5,6 Malondialdehyde is the principal and most studied product of lipid peroxidation (Figure 1).7 Under physiological conditions, malondialdehyde can react with DNA to form adducts mainly to deoxyguanosine and deoxyadenosine, with the predominant one being 3-(2-deoxy-β-d-erythro-pentafuranosyl)pyrimido[1,2-α]purin-10(3H)-one deoxyguanosine (M1dG, Figure 1).7 M1dG is a premutagenic lesion and has been shown to induce G → T and G → A mutations in DNA;8 these mutations are believed to be important steps in carcinogenesis, contributing to the etiology of human cancer.
Figure 1.
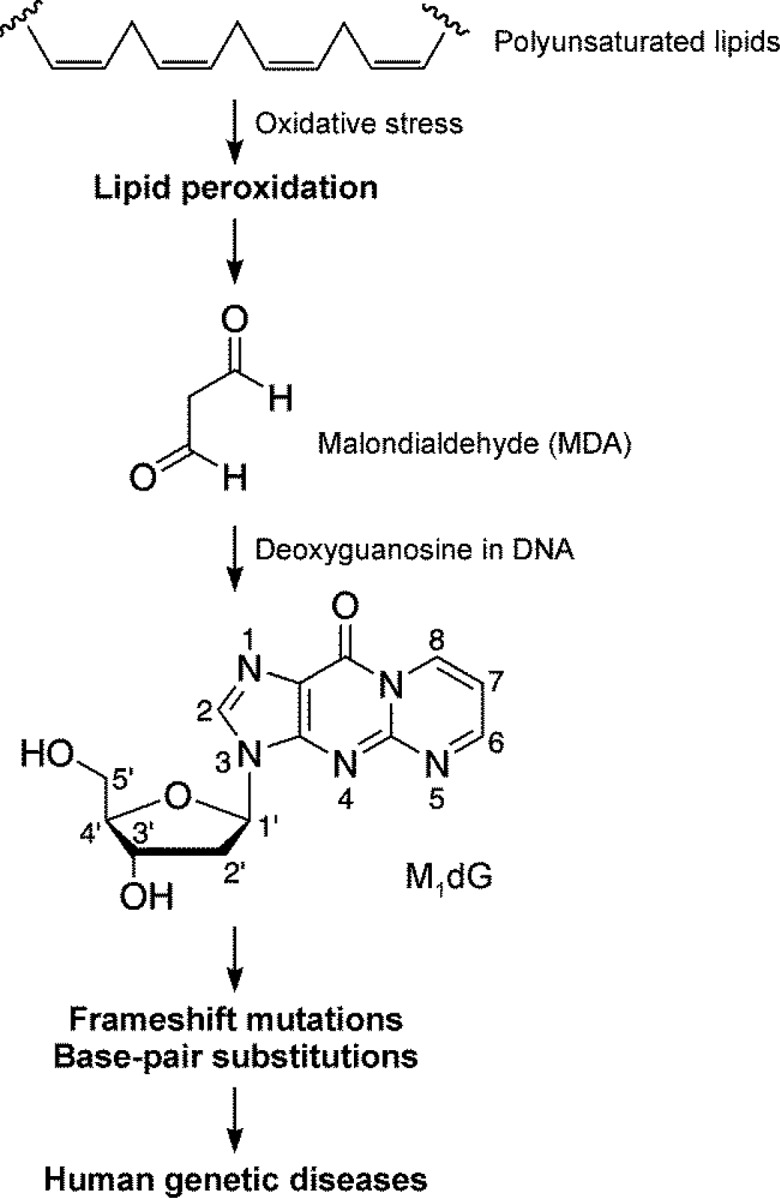
Formation of 3-(2-deoxy-β-d-erythro-pentafuranosyl)pyrimido[1,2-α]purin-10(3H)-one deoxyguanosine (M1dG) via lipid peroxidation.
While M1dG may serve as a valuable biomarker in studies of the role of inflammation in human disease, its sensitivity to changes in inflammation-inducing exposures is not clear. For instance, cigarette smoke contains high levels of pro-oxidants, such as reactive oxygen and nitrogen species,9 and it is plausible to expect that levels of M1dG would be elevated in DNA from smokers as compared to nonsmokers. Indeed, many studies demonstrated that levels of urinary F2-isoprostane 8-epi-PGF2α, a reliable urinary biomarker of oxidative stress, are elevated in smokers,12−15 and chronic inflammation is a major contributing factor in the pathogenesis of cigarette smoke-associated diseases, including lung cancer.10,11 However, existing reports on M1dG levels in smokers and nonsmokers are inconsistent: some studies demonstrate that levels of M1dG are modestly elevated in smokers,16−18 whereas other studies show no differences.19,20 These inconsistencies may be, at least in part, due to differences in the methodologies used by different research groups.
Analytical methods that have been used to detect and quantify M1dG in human biological samples include gas chromatography–mass spectrometry,19 immunoslot blot,21 and 32P-postlabeling technique22 as well as liquid chromatography–tandem mass spectrometry (LC–MS/MS).23,24 By using these methods, M1dG has been detected in liver, pancreas, breast, leukocytes, and lymphocytes from human subjects, with the levels of this adduct ranging from 1 to 120 adducts/108 nucleotides, depending on the sample type, subject characteristics or exposures, and applied methodologies for M1dG analysis.5,7 One of the potential issues with some methodologies could be overestimation of M1dG levels, due to either lack of specificity or artifactual formation.5,7 Thus, to better understand the role of M1dG in human diseases, there is a need for a robust, specific, and sensitive method that can be applied in large population studies investigating relationships between relevant exposures and health outcomes.
It should be noted that the latest LC–MS/MS-based methods for M1dG measurement provide high sensitivity and selectivity; however, the published assays involve the use of custom-made immunohistochemistry columns to purify samples prior to analysis.23,24 Therefore, robust transfer of this methodology to other laboratories is not practical. Our goal in this study was to develop a new robust and sensitive mass spectrometry-based method for the analysis of M1dG in human leukocyte DNA, with the initial primary focus on modifying the sample preparation procedure to exclude the immunohistochemistry-based purification step.
Experimental Procedures
Chemicals and Enzymes
M1dG and [13C3]M1dG were purchased from Toronto Research Chemicals (North York, Ontario, Canada). Reagents and enzymes for DNA isolation were obtained from Qiagen Sciences (Germantown, MD). Calf thymus DNA was purchased from Worthington Biochemical Corporation (Lakewood, NJ). All other chemicals and solvents were purchased from Sigma-Aldrich Chemical Co. (Milwaukee, WI).
Synthesis of 5,6-Dihydro-M1dG and 5,6-Dihydro-[13C3]M1dG
The synthesis of 5,6-dihydro-M1dG was performed by reacting M1dG with NaBH4, employing a previously described protocol.23 Briefly, M1dG (5 mg) was dissolved in 2 mL of a MeOH/H2O (2:8, v/v) solution, 1.3 mg of NaBH4 was added, and the mixture was stirred for 30 min at room temperature. The final product was isolated and purified on a Phenomenex Bondclone C18 column (3.9 × 300 mm, 10 μm). 1H NMR (d6-DMSO, 500 MHz) of 5,6-dihydro-M1dG was consistent with that previously reported:16 δ 7.93 (s, 1H, H2), 7.29 (dt, J = 8.4, 1.9 Hz, 1H, H8), 6.08 (dd, J = 7.8, 6.1 Hz, 1H, H1′), 5.48 (m, 1H, H7), 4.34 (m, 2H, H6), 4.05 (br s, 1H, H3′), 3.81 (m, 1H, H4′), 3.50 (m, 2H, H5′), 2.48–2.17 (m, 2H, H2′). ESI–MS: MH+, m/z 306. A similar procedure was employed to synthesize 5,6-dihydro-[13C3]M1dG from [13C3]M1dG, and 1H NMR similar to that of 5,6-dihydro-M1dG was obtained for the product; MH+m/z 309. The yields for 5,6-dihydro-M1dG and 5,6-dihydro-[13C3]M1dG were 65% and 55%, respectively.
Subjects and Blood Collection
Blood samples from 25 smokers and 25 nonsmokers were obtained from the “Methodology and Development of Tobacco Related Biomarkers” biorepository at the Masonic Cancer Center and Tobacco Research Programs. Collection of these samples was approved by the University of Minnesota Human Research Protection Programs Institutional Review Board (IRB Study no. 0908M70881). All subjects were at least 18 years old, not pregnant or breastfeeding, and were in good physical and mental health. Additional criteria for smokers included smoking at least 10 cigarettes per day (CPD), having been a smoker for at least 5 years with no change greater than 50% in CPD or brand in the last year, and not using any other tobacco products in the last 6 months. Nonsmokers were required to have smoked less than 100 cigarettes in their lifetime and were not using any tobacco products regularly. Smoking status was confirmed by expired carbon monoxide (CO) levels.
DNA Isolation from Human Leukocytes
DNA isolation from human blood samples was performed using the commercial protocol for DNA purification from buffy coat (Qiagen, Valencia, CA) with several modifications. Briefly, 3 mL of red blood cell lysis solution was added to 1 mL of buffy coat prepared from 10 mL of blood. The leukocyte pellet was collected by centrifugation (3000 × g, 10 min) and mixed with 5 mL of cell lysis solution. Proteinase K (2 μL of 20 mg/mL solution) was added, and the mixture was incubated at room temperature overnight with gentle shaking. The following day, 50 μL of RNase A solution (4 mg/mL) was added, and the sample was incubated at room temperature for 2 h. Protein precipitation solution (1.5 mL) was added to the cell lysate, and the sample was vortex-mixed for 30 s and centrifuged (3000 × g, 10 min) to remove proteins. DNA was precipitated from the supernatant by the addition of 5 mL of isopropanol. The DNA pellet was washed with 2 mL of 70% ethanol in H2O and then 2 mL of 100% ethanol. DNA was dried under a stream of nitrogen and stored at −20 °C until use. Potential RNA contamination was assessed by HPLC analysis of enzymatically hydrolyzed DNA samples for uridine. No uridine was detected. In a previously published study, addition of antioxidants during DNA isolation did not affect the measured M1dG levels, suggesting that artifactual formation of M1dG is minimal during this step.19
DNA Hydrolysis and Adduct Enrichment
The DNA samples were dissolved in 1 mL of 25 μM Tris-HCl (pH 7.4) buffer containing 5 μM CaCl2 and 5 μM MgCl2. The resulting solution was mixed with 25 fmol [13C3]M1dG (internal standard), followed by the addition of micrococcal nuclease (1 unit), phosphodiesterase I (0.003 units), and alkaline phosphatase (0.25 units). The mixed solution was then incubated overnight at 37 °C. The next day, 25 μL of hydrolysate was taken for the analysis of deoxyguanosine by HPLC,25 which was used to calculate the amount of DNA as described previously.26 The remaining volume of hydrolysate was incubated with 50 μL of NaBH4 (2 mg/mL) at room temperature for 30 min to reduce M1dG and [13C3]M1dG to their corresponding 5,6-dihydro derivatives. After the incubation, samples were loaded on Bond Elut PBA cartridges (100 mg, Agilent Technologies, Lake Forest, CA) activated with 1 mL of MeOH and 1 mL of H2O. The cartridges were washed with 1 mL of H2O and 1 mL of 3% MeOH sequentially and finally eluted with 2 mL of 25% MeOH. The 25% MeOH fraction containing analytes was collected and concentrated to dryness in a centrifugal evaporator. The residue was redissolved in 20 μL of deionized H2O and subjected to column purification on an Agilent 1100 HPLC system equipped with a Zorbax SB C18 column (5 μm, 150 × 0.5 mm, Agilent Technologies, Wilmington, DE). The mobile phase consisted of 15 mM NH4OAc and CH3CN, with a gradient from 4 to 27% CH3CN within 15 min, increased to 33% CH3CN over 2 min, then returned to 4% CH3CN in 1 min and held for 15 min at this composition, at a flow rate of 12 μL/min. The detection wavelength was set at 254 nm, and the column temperature was maintained at 25 °C. Benzamide (2 μg/mL), which has similar retention time (∼15.5 min) to that of 5,6-dihydro M1dG under the described HPLC conditions, was used as a UV marker. The fraction eluting at 15–17 min was collected (Supporting Information, Figure S1), evaporated to dryness, and redissolved in H2O prior to analysis by LC–NSI–HRMS/MS.
To assess possible artifactual M1dG formation during the sample preparation, 5 fmol of 15N5-deoxyguanosine was added to calf thymus DNA, and the sample was enzymatically hydrolyzed and purified as described above. No 5,6-dihydro-[15N5]M1dG was detected (Supporting Information, Figure S2).
LC–Electrospray Ionization (ESI)–(MS/MS)
The LC–ESI–MS/MS analysis was carried out on a TSQ Vantage triple quadrupole mass spectrometer (Thermo Scientific, Waltham, MA) interfaced with an Agilent 1100 capillary HPLC system (Agilent, Palo Alto, CA). Analysis was performed on an Agilent Zorbax SB C18 column at a flow rate of 12 μL/min with the temperature maintained at 30 °C. Sample injection volume was 8 μL. The mobile phase consisted of 15 mM NH4OAc and CH3CN with a linear gradient from 4 to 35% CH3CN over a period of 18 min and then returned to 4% CH3CN followed by 15 min re-equilibration. The ESI source was operated in positive ion mode, monitoring m/z 306.2 [M + H]+ → 190.2 [C8H8N5O]+ for 5,6-dihydro-M1dG and corresponding ions at m/z 309.2 → 193.2 for 5,6-dihydro-[13C3]M1dG. The collision gas was Ar at 1 mTorr with collision energy of 12 eV. The quadrupoles were operated at a resolution of 0.2 (Q1) and 0.7 (Q3) Da.
LC–Nanoelectrospray Ionization (NSI)–High-Resolution (HR) MS/MS
The LC–NSI–HRMS/MS was performed on an LTQ Orbitrap Velos instrument (Thermo Scientific, Waltham, MA) interfaced with a Nano2D–LC HPLC (Eksigent, Dublin, CA) system using nanoelectrospray ionization. The analysis was performed using a capillary column (75 μm i.d., 10 cm length, 15 μm orifice) created by hand packing a commercially available fused-silica emitter (New Objective, Woburn, MA) with Luna C18 bonded separation media (Phenomenex, Torrance, CA). The mobile phase consisted of 5 mM NH4OAc and CH3CN. A 5 μL injection loop was used, and the sample (3 μL) was loaded onto the capillary column with a 1000 nL/min flow under the initial conditions for 5.5 min. Separation on the capillary column was performed using a linear gradient at a flow rate of 300 nL/min with increasing CH3CN from 2 to 33% over 12 min, followed by ramping to 98% CH3CN within 1 min and holding at this composition for additional 2 min. The gradient was then returned to 2% CH3CN (initial condition) in 1 min, and the system was re-equilibrated at this mobile phase composition for 6 min at 1000 nL/min before next injection. The nanoelectrospray source voltage was set at 1.6 kV. The capillary temperature was 350 °C, and the S-Lens RF Level was set at 40%. The analysis was performed using accurate mass extracted ion chromatograms of m/z 190.0723 [C8H8N5O]+ and 117.0546 [C5H9O3]+ (parent ion m/z 306.1) for 5,6-dihydro-M1dG and corresponding fragments (m/z 193.0824 and 117.0546) for 5,6-dihydro-[13C3]M1dG with a mass tolerance of 2 ppm. The scan events were performed using higher-energy collisional dissociation (HCD) fragmentation with a normalized collision energy of 20 units, isolation widths of 1 Da for both the analyte and internal standard, and product ion spectra acquisition at a resolution of 50 000. The quantitation of M1dG was based on the peak area ratio of 5,6-dihydro-M1dG (m/z 306.1 → 190.0723) to 5,6-dihydro-[13C3]M1dG (m/z 309.1 → 193.0824), the constructed calibration curves, and the amount of internal standard added.
A calibration curve was constructed before each analysis using a series of standard solutions of 5,6-dihydro-M1dG and 5,6-dihydro-[13C3]M1dG. The calibration standard solutions contained a constant amount of 5,6-dihydro-[13C3]M1dG (2.5 fmol on column) and varying amounts of 5,6-dihydro-M1dG (0.005, 0.025, 0.1, 0.5, 2.5, and 5 fmol on column).
Method Characterization and Sample Analysis
Accuracy was determined by adding different amounts of M1dG (0.025, 0.1, 0.5, 2.5 fmol) and 2.5 fmol of internal standard to 200 μg of calf thymus DNA in 1 mL of 25 μM Tris-HCl (pH 7.4) buffer containing 5 μM CaCl2 and 5 μM MgCl2, followed by hydrolysis and purification as described above. Samples at each level of added M1dG were analyzed in triplicate.27 To characterize method precision, 0.5 fmol of M1dG and 2.5 fmol of internal standard were added to 200 μg of calf thymus DNA, followed by the described hydrolysis and purification protocol. The precision was determined as intraday and interday coefficients of variation (% CV), which were calculated based on the analyses of three aliquots of the samples on three separate days. Trace levels of M1dG were present in calf thymus DNA; these levels were quantified and subtracted from the levels of M1dG measured in the samples during the method characterization.
The limit of detection (LOD) was determined using standard solutions of 5,6-dihydro-M1dG. The limit of quantitation (LOQ) was established in calf thymus DNA samples by adding M1dG (0.005, 0.01, 0.025, and 0.05 fmol) and internal standard (2.5 fmol) to calf thymus DNA samples, followed by hydrolysis and purification, and analyzing each sample in triplicate. The LOQ was defined by identification of the lowest M1dG level that produced a coefficient of variation (CV) lower than 5%.28
Recovery was determined by comparing the results of samples to which [13C3]M1dG (2.5 fmol) was added to 200 μg of calf thymus DNA at the beginning and at the end of sample preparation procedure.29 All data are presented as mean ± standard deviation (SD). Two-tailed unpaired Student’s t-test was used for two group comparison. A p value less than 0.05 was considered significant.
Results
Development of the Analytical Procedure
The purpose of this study was to develop a robust and sensitive analytical procedure for the analysis of M1dG by LC–MS/MS. The developed protocol is outlined in Scheme 1. The addition of NaBH4 after enzymatic hydrolysis was used to increase the detection sensitivity by reducing M1dG to 5,6-dihydro-M1dG.23 In agreement with the previous report,23 the detection sensitivity of 5,6-dihydro-M1dG increased 12-fold as compared to that of M1dG. Because it has been previously reported that M1dG can form a conjugate with Tris buffer,30 we tested the stability of M1dG upon overnight incubation in Tris-HCl and in other two buffers commonly used in DNA hydrolysis: MOPS and sodium succinate buffer. The subsequent reduction with NaBH4 and quantification of the formed 5,6-dihydro-M1dG showed no difference among the three tested buffers, suggesting no effect of Tris-HCl on the sensitivity of our assay. This could be due to the reported instability of Tris–M1dG conjugate in aqueous solutions at room temperature30 or due to deconjugation of this product during the NaBH4 reduction step.
Scheme 1. Analytical Scheme for the Analysis of M1dG in Human Leukocyte DNA.
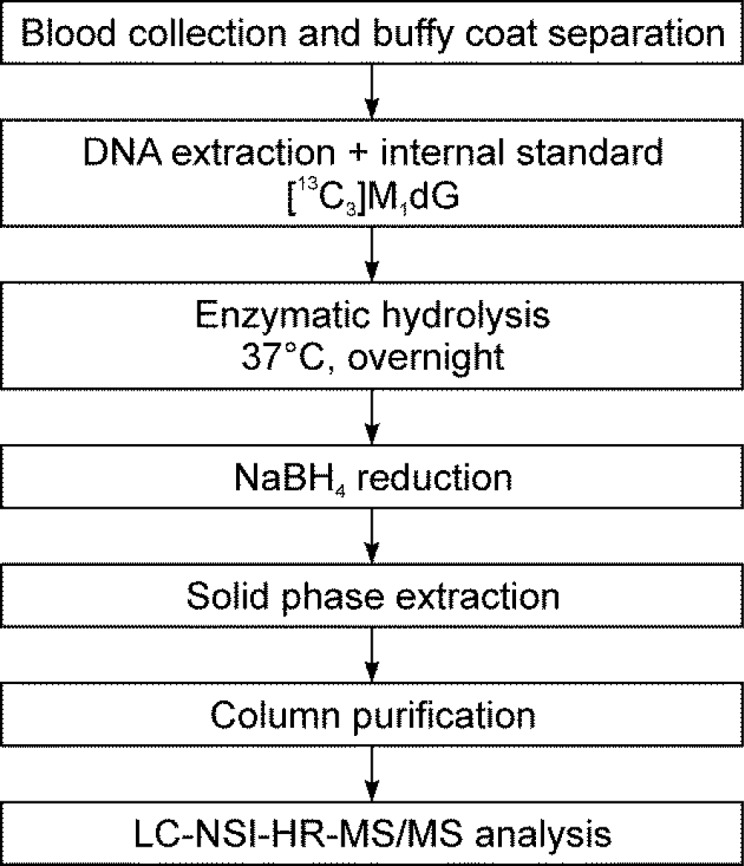
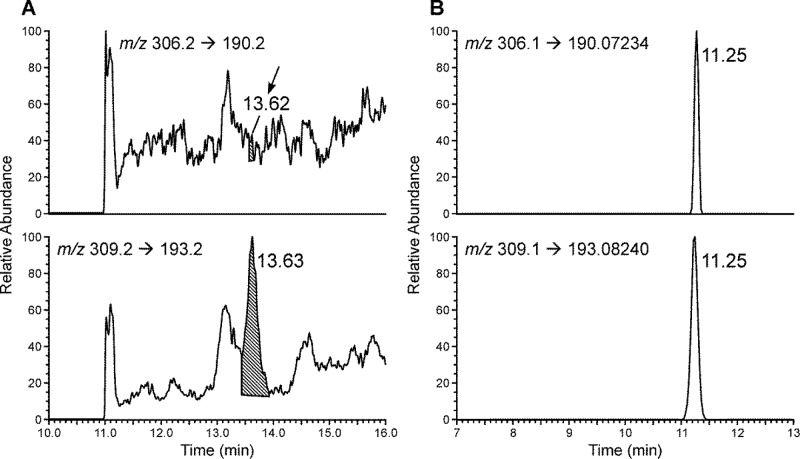
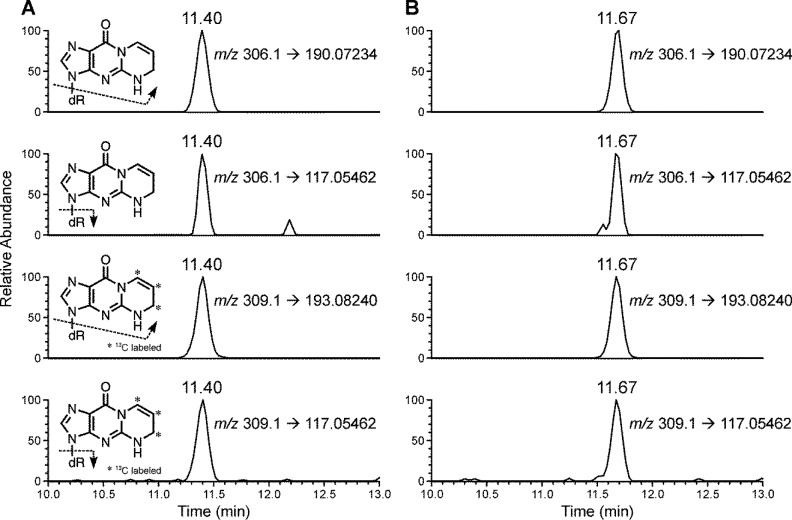
We initially used low-resolution triple quadrupole mass spectrometry (LC–ESI–MS/MS), monitoring m/z 306.2 → 190.2 for 5,6-dihydro-M1dG and m/z 309.2 → 193.2 for 5,6-dihydro-[13C3]M1dG. By using this method, an LOQ of 3.3 fmol (on-column) was achieved in calf thymus DNA (data not shown). However, when the method was applied to human leukocyte DNA, high noise levels and co-eluting peaks were observed for both 5,6-dihydro-M1dG and its internal standard, with no obvious peak being detected at the retention time of 5,6-dihydro-M1dG (Figure 2A). Analysis of the same human leukocyte DNA sample using the high resolving and accurate mass capabilities of the Orbitrap detector produced clear peaks for both 5,6-dihydro-M1dG and its internal standard without any baseline noise or coeluting peaks (Figure 2B). The product scans of the analytes using HCD fragmentation generated two major fragments for 5,6-dihydro-M1dG at m/z 190.0723 and m/z 117.0546 and two major fragments for 5,6-dihydro-[13C3]M1dG at m/z 193.0824 and m/z 117.0546 (Supporting Information, Figure S3). Because of the higher signal intensities, the transitions m/z 306.1 → 190.0723 and m/z 309.1 → 193.0824 were selected for quantitation of 5,6-dihydro-M1dG and 5,6-dihydro-[13C3]M1dG, respectively; the peak area ratios between the two major fragments of 5,6-dihydro-M1dG were used to confirm its identity, and the corresponding ratio was used to confirm the identity of the internal standard (Supporting Information, Figure S3).
Figure 2.
Chromatograms obtained upon analysis of M1dG in the same human leukocyte DNA sample by using (A) LC–ESI–MS/MS and (B) LC–NSI–HRMS/MS.
Method Characteristics
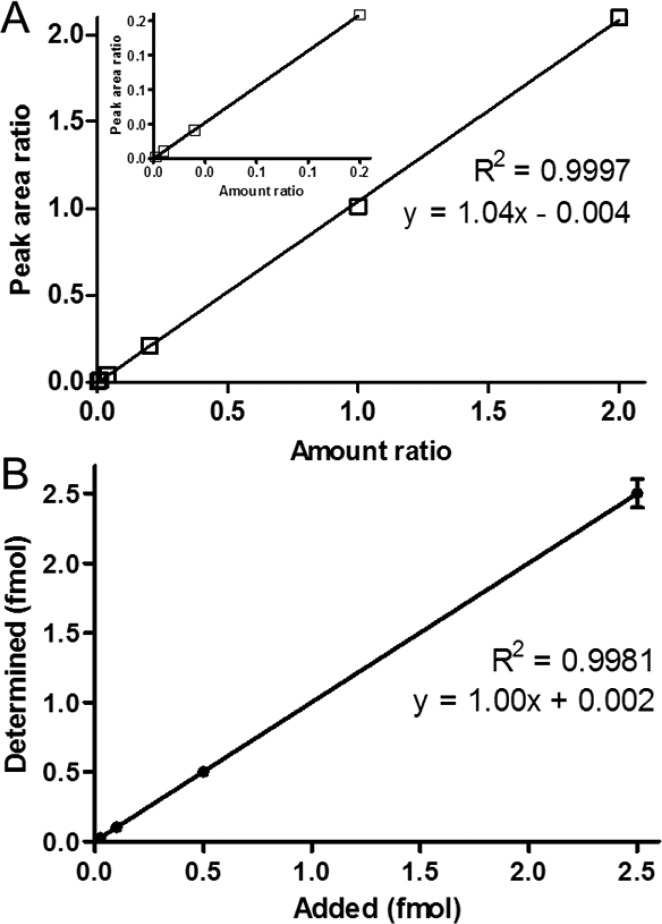
The calf thymus DNA that was used for the method development contained 13 fmol of M1dG/mg DNA. This value was subtracted during the analysis of data obtained for the samples used for method characterization. By using the developed LC–NSI–HRMS/MS method, an LOD of 5 amol (on-column) was obtained. The instrument response and the 5,6-dihydro-M1dG/5,6-dihydro-[13C3]M1dG ratio were linear in the 0.005–5 fmol (on-column) range of M1dG (R2 = 0.9997, Figure 3A). The LOQ was 25 amol on-column based on a CV of 3.21%. The accuracy of measured levels of 5,6-dihydro-M1dG (expressed as % of added M1dG) at 0.025, 0.1, 0.5, and 2.5 fmol was 99.6, 103, 100, and 100%, respectively, exhibiting excellent linearity (R2 = 0.9981, Figure 3B). The interday CV was 6.0%. The recovery of the assay was 41.3 ± 3.47% (n = 5).
Figure 3.
Method characteristics. (A) Linearity of 5,6-dihydro-M1dG/5,6-dihydro-[13C3]M1dG peak area ratio at constant level of 5,6-dihydro-[13C3]M1dG (2.5 fmol on-column) and varying levels of 5,6-dihydro-M1dG (from 5 to 5000 amol on-column). (B) Relationship between added M1dG and measured 5,6-dihydro-M1dG in calf thymus DNA (R2 = 0.998) in the range from 0.025 to 2.5 fmol of M1dG per 200 μg of DNA; M1dG originally present in the calf thymus DNA was determined and subtracted from each value.
Quantitation of M1dG Adducts in Human Leucocyte DNA
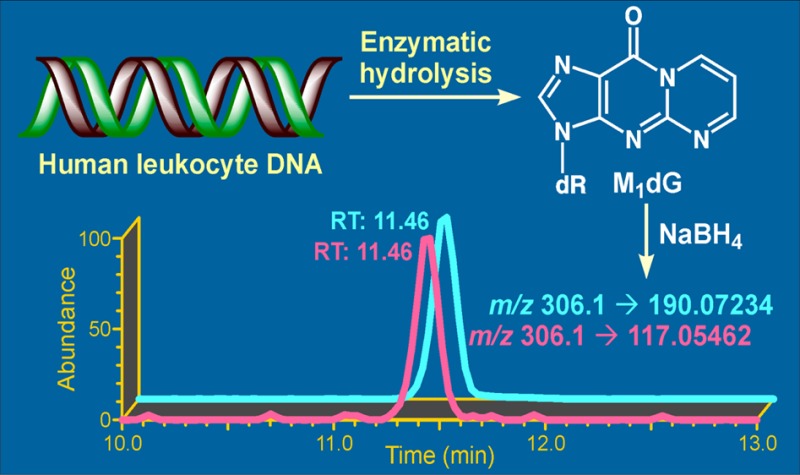
The method was applied to the analysis of leukocyte DNA samples from 50 human subjects. To investigate the potential effect of cigarette smoking on levels of M1dG in human leukocyte DNA, samples from 25 smokers and 25 nonsmokers were selected for this analysis. Typical chromatograms upon analysis of M1dG in human leukocytes from a smoker and a nonsmoker are presented in Figure 4. The results of M1dG levels from the 50 subjects are summarized in Table 1. The yield of DNA in this study averaged 136 ± 55 μg, which was sufficient to detect and quantify the M1dG adduct in all samples. The levels of M1dG in the analyzed samples ranged from 0.004 to 9.15 adducts/108 nucleotides, averaging (±SD) 2.02 ± 2.17 adducts/108 nucleotides. Levels of M1dG in leukocyte DNA from smokers averaged 64.9 ± 71.9 fmol/mg DNA and in nonsmokers, 56.5 ± 58.8 fmol/mg DNA, or 2.16 ± 2.40 and 1.89 ± 1.96 adducts/108 nucleotides, respectively. Although the average M1dG level in smokers was slightly higher than that in nonsmokers, this difference was not significant.
Figure 4.
Typical LC–NSI–HRMS/MS chromatograms obtained upon analysis of 5,6-dihydro-M1dG in human leukocyte DNA from a (A) smoker and (B) nonsmoker.
Table 1. Levels of M1dG in Smokers’ and Nonsmokers’ Leukocyte DNA.
| smokers |
nonsmokers |
||||
|---|---|---|---|---|---|
| subject no. | DNA yield | M1dG | subject no. | DNA yield | M1dG |
| (μg) | (fmol/mg DNA) | (μg) | (fmol/mg DNA) | ||
| 1 | 128 | 2.14 | 1 | 61.1 | 72.1 |
| 2 | 196 | 6.06 | 2 | 49.4 | 47.9 |
| 3 | 90.7 | 18.5 | 3 | 79.4 | 37.0 |
| 4 | 200 | 14.0 | 4 | 46.8 | 75.8 |
| 5 | 164 | 14.8 | 5 | 54.6 | 95.3 |
| 6 | 99.1 | 106 | 6 | 94.9 | 132 |
| 7 | 69.4 | 275 | 7 | 67.3 | 79.9 |
| 8 | 90.2 | 79.4 | 8 | 125 | 135 |
| 9 | 149 | 125 | 9 | 87.8 | 134 |
| 10 | 71.3 | 139 | 10 | 124 | 30.3 |
| 11 | 89.4 | 220 | 11 | 168 | 93.0 |
| 12 | 132 | 57.3 | 12 | 194 | 0.714 |
| 13 | 139 | 48.7 | 13 | 209 | 2.22 |
| 14 | 165 | 102 | 14 | 162 | 55.9 |
| 15 | 122 | 120 | 15 | 236 | 27.9 |
| 16 | 160 | 5.49 | 16 | 244 | 2.82 |
| 17 | 165 | 13.0 | 17 | 69.8 | 9.83 |
| 18 | 93.1 | 0.132 | 18 | 113 | 17.7 |
| 19 | 183 | 16.0 | 19 | 199 | 7.89 |
| 20 | 223 | 1.14 | 20 | 166 | 251 |
| 21 | 213 | 3.19 | 21 | 155 | 28.0 |
| 22 | 39.2 | 73.3 | 22 | 140 | 12.5 |
| 23 | 170 | 43.1 | 23 | 182 | 17.3 |
| 24 | 204 | 117 | 24 | 158 | 21.4 |
| 25 | 166 | 22.9 | 25 | 78.7 | 26.6 |
| mean ± SD | 141 ± 50.2 | 64.9 ± 71.9 | mean ± SD | 130 ± 60.3 | 56.5 ± 58.8 |
Discussion
Measurement of lipid peroxidation-induced DNA damage in populations with varying environmental and occupational exposures and lifestyle and dietary habits could greatly advance our understanding of the role of these factors in the induction of chronic inflammation and the associated diseases. In this study, we developed a novel LC–NSI–HRMS/MS method for the analysis of M1dG, the major DNA adduct derived from the lipid peroxidation product malondialdehyde, in human leukocyte DNA. This robust and sensitive method was successfully applied to the analysis of leukocyte DNA from 25 smokers and 25 nonsmokers. Studies are underway to investigate how the levels of this highly mutagenic adduct in humans are related to various exposures and disease risk.
In the process of method development, we initially explored the use of low-resolution triple quadrupole mass spectrometry and were able to quantify M1dG in calf thymus DNA at levels as low as 3.3 fmol, which was comparable to the previously developed LC–MS method.24 However, application of the method to human leukocyte DNA produced high background noise and co-eluting peaks, leading to inaccurate quantitation of the adduct levels in these samples. As an alternative, we employed an accurate mass high-resolution Orbitrap mass spectrometer, which proved to be highly sensitive and selective in a previous study on another DNA adduct, 7-ethylguanine.29
After the transition to HRMS/MS, both capillary-ESI at a flow rate of 10 μL/min and nano-ESI at a flow rate of 300 nL/min were investigated, with nano-ESI showing at least 50-fold increase in sensitivity compared to that of capillary-ESI. Moreover, nano-ESI has been shown to be more tolerant of salt contamination than conventional ESI.31 Therefore, nano-ESI was finally adopted. However, it has been reported that substantial ion suppression can occur at flow rates as low as 50 nL/min.32 In our developed method, a flow rate of 300 nL/min is used and therefore the analysis could be susceptible to signal suppression if sufficient purification of the sample is not performed. Indeed, analysis of M1dG in samples that underwent only one purification step, extraction on Bond Elut PBA cartridges, showed a 4-fold signal decrease due to ion suppression effect (data not shown) compared to samples that were also subjected to a column purification step (Scheme 1). We also investigated mass tolerance, which reflects measurement and calibration errors of the Orbitrap instrument. Changing the mass tolerance setting for ion extraction from 10 to 5 to 2 ppm did not alter the peak area of the analyte (Supporting Information, Figure S4). Consequently, the mass tolerance was set at 2 ppm to achieve better selectivity and accuracy. Overall, the transition to high-resolution MS/MS improved the sensitivity of the method by approximately 130-fold compared with the originally used low-resolution MS/MS. Moreover, the baseline noise and occurrence of co-eluting peaks were completely eliminated, greatly improving selectivity of the method (Figures 2 and 3). The average M1dG level determined in our study was 2.02 ± 2.17 adducts/108 nucleotides, which is comparable to the previously reported levels.19 The lowest DNA yield among the analyzed human leukocyte DNA samples was 39 μg, and M1dG was reliably quantified in that sample. The sensitivity and selectivity of the developed method indicates that measurement of M1dG in even lower amounts of DNA is possible in future studies.
Although the average M1dG levels were slightly different between smokers and nonsmokers in this study (Table 1), the observed difference was not statistically significant. This is consistent with the results of several studies that compared M1dG levels in leukocyte DNA from smokers and nonsmokers. For instance, Peluso et al. reported that M1dG levels per 108 nucleotides were 4.8 ± 0.4 in leukocyte DNA of current smokers, 4.2 ± 0.7 in ex-smokers, and 3.7 ± 0.4 in nonsmokers; however, the differences were not statistically significant.17 Another study investigating the effect of formaldehyde exposure on leukocyte M1dG levels in a group of Italian pathologists found that the levels of M1dG per 108 nucleotides tended to increase in smokers compared to nonsmokers, 4.5 ± 1.3 vs 3.8 ± 0.9, respectively, but without reaching statistical significance.18 Cigarette smoke is a rich source of exposure to free radicals capable of inducing oxidative damage to DNA and promoting oxidative stress in smokers.9−11 However, the levels of M1dG in humans may also be affected by such factors as age, gender, diet, environmental or occupational exposures, alcohol consumption, and inflammatory diseases. For instance, M1dG has been reported to be lower in leukocyte DNA of women as compared to men: 5.1 ± 0.4 adducts/108 nucleotides vs 6.7 ± 1.1 adducts/108 nucleotides, respectively.19 The potential contribution of the diet to the measured M1dG is exemplified by the findings of a study in which levels of this adduct in leukocyte DNA of female subjects who consumed a diet rich in polyunsaturated fatty acids were nearly 20-fold higher than in the control group.33 In addition to the potential contribution of the mentioned demographic and lifestyle factors, a small sample size in the studies that compared M1dG in smokers and nonsmokers, including the present study, may have prevented detection of statistically significant differences between these two groups. Furthermore, M1dG analysis in different cell types that are characterized by different lifespans may potentially reveal differences between smokers and nonsmokers and should be considered in future studies.
Availability of a validated and accurate method for the detection and quantitation of M1dG in humans may be extremely useful not only for investigations of the role of environmental exposures or lifestyle factors in health outcomes but also for the prevention, prognosis, and diagnosis of diseases associated with inflammation and oxidative stress. For instance, Wang et al. reported that M1dG levels in the normal breast tissue of women with breast cancer were increased 2- to 3-fold compared to the normal tissue of women without breast cancer.34 In another study, M1dG levels in lymphocyte DNA of thalassemia patients were 4-fold higher than in healthy control subjects, which indicates elevated oxidative stress and LPO-induced DNA damage in internal organs.35 Further applications of the developed methodology to measure M1dG levels in specific cohorts could facilitate our understanding of the importance of malondialdehyde-induced DNA damage in these and other diseases.
In summary, we developed a novel LC–NSI–HRMS/MS method for the quantitation of M1dG in human leukocyte DNA, and successfully applied this method to the analysis of M1dG in leukocyte DNA from 50 human subjects. Our approach features a unique application of high-resolution mass spectrometry to achieve the requisite sensitivity and specificity. The method can be used in future studies aimed at understanding the pathophysiological role of M1dG in humans.
Acknowledgments
We thank Adam Zarth for his help with the analysis of NMR data and Tobacco Research Programs, University of Minnesota, for providing human samples collected through the “Methodology and Development of Tobacco Related Biomarkers” biorepository. We also thank Bob Carlson for editorial assistance.
Glossary
Abbreviations
- CPD
cigarettes per day
- HCD
higher energy collisional dissociation
- LOD
limit of detection
- LOQ
limit of quantitation
- LPO
lipid peroxidation
- M1dG
3-(2-deoxy-β-d-erythro-pentafuranosyl)pyrimido[1,2-α]purin-10(3H)-one deoxyguanosine
- MDA
malondialdehyde
Supporting Information Available
Chromatogram of a standard mixture of benzamide and 5,6-dihydro-M1dG during column chromatography purification, chromatograms obtained upon analysis of possible artifactual M1dG production during the sample preparation, product ion spectra of 5,6-dihydro-M1dG and 5,6-dihydro-[13C3]M1dG, and extracted ion chromatograms for 5,6-dihydro-M1dG at mass tolerances of 10, 5, and 2 ppm using the Orbitrap mass analyzer. This material is available free of charge via the Internet at http://pubs.acs.org.
This study was supported by startup funds to I.S. from the Masonic Cancer Center, University of Minnesota, via NCI grant P30 CA077598 and support from Minnesota Masonic Charities. Mass spectrometry analyses were carried out in the Analytical Biochemistry Shared Resource of the Masonic Cancer Center, supported in part by grant CA-77598 from the NCI.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Coussens L. M.; Werb Z. (2002) Inflammation and cancer. Nature 420, 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S. P.; Harris C. C. (2007) Inflammation and cancer: an ancient link with novel potentials. Int. J. Cancer 121, 2373–2380. [DOI] [PubMed] [Google Scholar]
- Black P. H.; Garbutt L. D. (2002) Stress, inflammation and cardiovascular disease. J. Psychosom. Res. 52, 1–23. [DOI] [PubMed] [Google Scholar]
- Pearson T. A.; Mensah G. A.; Alexander R. W.; Anderson J. L.; Cannon R. O. III; Criqui M.; Fadl Y. Y.; Fortmann S. P.; Hong Y.; Myers G. L.; Rifai N.; Smith S. C. Jr.; Taubert K.; Tracy R. P.; Vinicor F. (2003) Markers of inflammation and cardiovascular disease: application to clinical and public health practice: A statement for healthcare professionals from the Centers for Disease Control and Prevention and the American Heart Association. Circulation 107, 499–511. [DOI] [PubMed] [Google Scholar]
- Nair U.; Bartsch H.; Nair J. (2007) Lipid peroxidation-induced DNA damage in cancer-prone inflammatory diseases: a review of published adduct types and levels in humans. Free Radical Biol. Med. 43, 1109–1120. [DOI] [PubMed] [Google Scholar]
- Bartsch H.; Nair J. (2005) Accumulation of lipid peroxidation-derived lesions: potential lead markers for chemoprevention of inflammation driven malignancies. Mutat. Res. 591, 34–44. [DOI] [PubMed] [Google Scholar]
- Marnett L. J. (1999) Lipid peroxidation—DNA damage by malondialdehyde. Mutat. Res. 424, 83–95. [DOI] [PubMed] [Google Scholar]
- Fink S. P.; Reddy G. R.; Marnett L. J. (1997) Mutagenicity in Escherichia coli of the major DNA adduct derived from the endogenous mutagen malondialdehyde. Proc. Natl. Acad. Sci. U.S.A. 94, 8652–8657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor W. A.; Hales B. J.; Premovic P. I.; Church D. F. (1983) The radicals in cigarette tar: their nature and suggested physiological implications. Science 220, 425–427. [DOI] [PubMed] [Google Scholar]
- Church D. F.; Pryor W. A. (1985) Free radical chemistry of cigarette smoke and its toxicological implications. Environ. Health Perspect. 64, 111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla D. K.; Hirata F.; Rishi A.; Gairola C. G. (2009) Cigarette smoke, inflammation, and lung injury: a mechanistic perspective. J. Toxicol. Environ. Health, Part B 12, 45–64. [DOI] [PubMed] [Google Scholar]
- Yan W.; Byrd G. D.; Ogden M. W. (2007) Quantitation of isoprostane isomers in human urine from smokers and nonsmokers by LC–MS/MS. J. Lipid. Res. 48, 1607–1617. [DOI] [PubMed] [Google Scholar]
- Hecht S. S.; Yuan J.-M.; Hatsukami D. K. (2010) Applying tobacco carcinogen and toxicant biomarkers in product regulation and cancer prevention. Chem. Res. Toxicol. 23, 1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roethig H. J.; Munjal S.; Feng S.; Liang Q.; Sarkar M.; Walk R. A.; Mendes P. E. (2009) Population estimates for biomarkers of exposure to cigarette smoke in adult U.S. cigarette smokers. Nicotine Tob. Res. 11, 1216–1225. [DOI] [PubMed] [Google Scholar]
- Frost-Pineda K.; Liang Q.; Liu J.; Rimmer L.; Jin Y.; Feng S.; Kapur S.; Mendes P.; Roethig H.; Sarkar M. (2011) Biomarkers of potential harm among adult smokers and nonsmokers in the total exposure study. Nicotine Tob. Res. 13, 182–193. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Chen S. Y.; Hsu T.; Santella R. M. (2002) Immunohistochemical detection of malondialdehyde–DNA adducts in human oral mucosa cells. Carcinogenesis 23, 207–211. [DOI] [PubMed] [Google Scholar]
- Peluso M.; Srivatanakul P.; Munnia A.; Jedpiyawongse A.; Ceppi M.; Sangrajrang S.; Piro S.; Boffetta P. (2010) Malondialdehyde–deoxyguanosine adducts among workers of a Thai industrial estate and nearby residents. Environ. Health Perspect. 118, 55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono R.; Romanazzi V.; Munnia A.; Piro S.; Allione A.; Ricceri F.; Guarrera S.; Pignata C.; Matullo G.; Wang P.; Giese R. W.; Peluso M. (2010) Malondialdehyde–deoxyguanosine adduct formation in workers of pathology wards. The role of air formaldehyde exposure. Chem. Res. Toxicol. 23, 1342–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzer C. A.; Chaudhary A. K.; Nokubo M.; Ferguson D. M.; Reddy G. R.; Blair I. A.; Marnett L. J. (1997) Analysis of the malondialdehyde-2′-deoxyguanosine adduct pyrimidopurinone in human leukocyte DNA by gas chromatography/electron capture/negative chemical ionization/mass spectrometry. Chem. Res. Toxicol. 10, 181–188. [DOI] [PubMed] [Google Scholar]
- Singh R.; Kaur B.; Kalina I.; Popov T. A.; Georgieva T.; Garte S.; Binkova B.; Sram R. J.; Taioli E.; Farmer P. B. (2007) Effects of environmental air pollution on endogenous oxidative DNA damage in humans. Mutat. Res. 620, 71–82. [DOI] [PubMed] [Google Scholar]
- Leuratti C.; Singh R.; Lagneau C.; Farmer P. B.; Plastaras J. P.; Marnett L. J.; Shuker D. E. (1998) Determination of malondialdehyde-induced DNA damage in human tissues using an immunoslot blot assay. Carcinogenesis 19, 1919–1924. [DOI] [PubMed] [Google Scholar]
- Yi P.; Sun X.; Doerge D. R.; Fu P. P. (1998) An improved 32P-postlabeling/high performance liquid chromatography method for the analysis of the malondialdehyde-derived 1,2-propanodeoxyguanosine DNA adducts in animal and human tissues. Chem. Res. Toxicol. 11, 1032–1041. [DOI] [PubMed] [Google Scholar]
- Otteneder M.; Daniels J. S.; Voehler M.; Marnett L. J. (2003) Development of a method for determination of the malondealdehyde–deoxyguanosine adduct in urine using liquid chromatography-tandem mass spectrometry. Anal. Biochem. 315, 147–151. [DOI] [PubMed] [Google Scholar]
- Hoberg A. M.; Otteneder M.; Marnett L. J.; Poulsen H. E. (2004) Measurement of the malondialdehyde-2′-deoxyguanosine adduct in human urine by immuno-extraction and liquid chromatography/atmospheric pressure chemical ionization tandem mass spectrometry. J. Mass Spectrom. 39, 38–42. [DOI] [PubMed] [Google Scholar]
- Stepanov I.; Hecht S. S. (2009) Mitochondrial DNA adducts in the lung and liver of F344 rats chronically treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and (S)-4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol. 22, 406–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao Y.; Yu N.; Kassie F.; Villalta P. W.; Hecht S. S. (2007) Formation and accumulation of pyridyloxobutyl DNA adducts in F344 rats chronically treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol. 20, 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2001) U.S. FDA Guidance for Industry: Bioanalytical Method Validation, U.S. Department of Health and Human Services. http://www.fda.gov/downloads/Drugs/Guidances/ucm070107.pdf.
- Dolan J. W. (2009) Calibration curves, part II: What are the limits?. LCGC North Am. 27, 306–312. [Google Scholar]
- Balbo S.; Villalta P. W.; Hecht S. S. (2011) Quantitation of 7-ethylguanine in leukocyte DNA from smokers and nonsmokers by liquid chromatography-nanoelectrospray-high resolution tandem mass spectrometry. Chem. Res. Toxicol. 24, 1729–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer L. J.; Riley M.; Schnetz-Boutaud N.; Sanduwaran G.; Chaudhary A. K.; Reddy G. R.; Marnett L. J. (1997) Temperature-dependent formation of a conjugate between tris(hydroxymethyl)aminomethane buffer and the malondialdehyde–DNA adduct pyrimidopurinone. Chem. Res. Toxicol. 10, 556–561. [DOI] [PubMed] [Google Scholar]
- Juraschek R.; Dulcks T.; Karas M. (1999) Nanoelectrospray—more than just a minimized-flow electrospray ionization source. J. Am. Soc. Mass Spectrom. 10, 300–308. [DOI] [PubMed] [Google Scholar]
- Schmidt A.; Karas M.; Dulcks T. (2003) Effect of different solution flow rates on analyte ion signals in nano-ESI MS, or: when does ESI turn into nano-ESI?. J. Am. Soc. Mass Spectrom. 14, 492–500. [DOI] [PubMed] [Google Scholar]
- Fang J. L.; Vaca C. E.; Valsta L. M.; Mutanen M. (1996) Determination of DNA adducts of malondialdehyde in humans: effects of dietary fatty acid composition. Carcinogenesis 17, 1035–1040. [DOI] [PubMed] [Google Scholar]
- Wang M.; Dhingra K.; Hittleman W. N.; Liehr J. G.; De Andrade M.; Li D. (1996) Lipid peroxidation-induced putative malondialdehyde-DNA adducts in human breast tissues. Cancer Epidemiol., Biomarkers Prev. 5, 705–710. [PubMed] [Google Scholar]
- Meerang M.; Nair J.; Sirankapracha P.; Thephinlap C.; Srichairatanakool S.; Arab K.; Kalpravidh R.; Vadolas J.; Fucharoen S.; Bartsch H. (2009) Accumulation of lipid peroxidation-derived DNA lesions in iron-overloaded thalassemic mouse livers: comparison with levels in the lymphocytes of thalassemia patients. Int. J. Cancer 125, 759–766. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.