Abstract
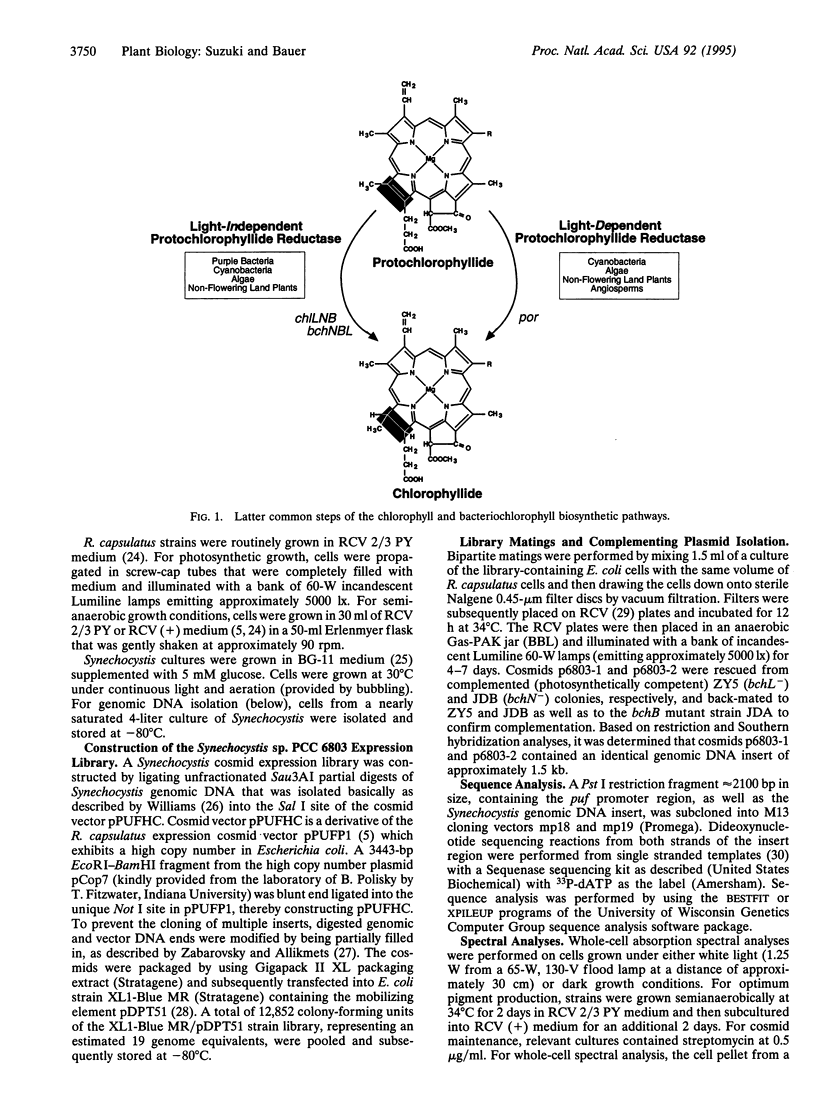
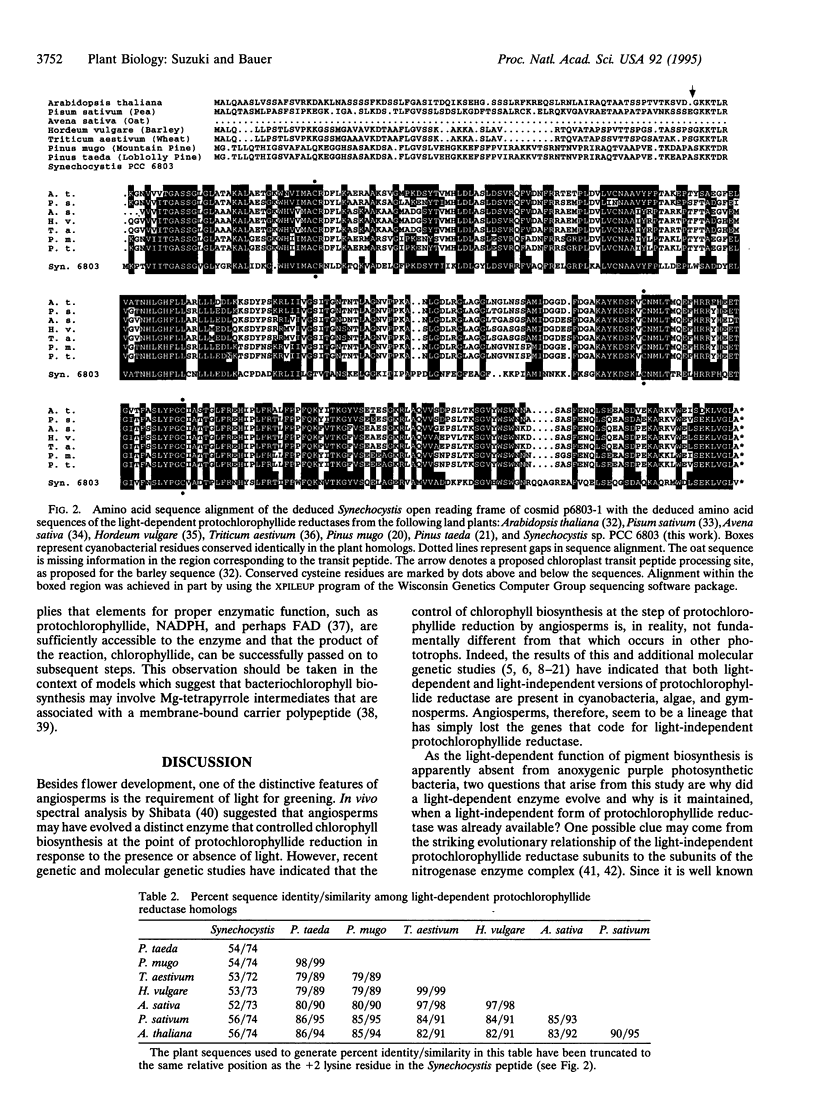
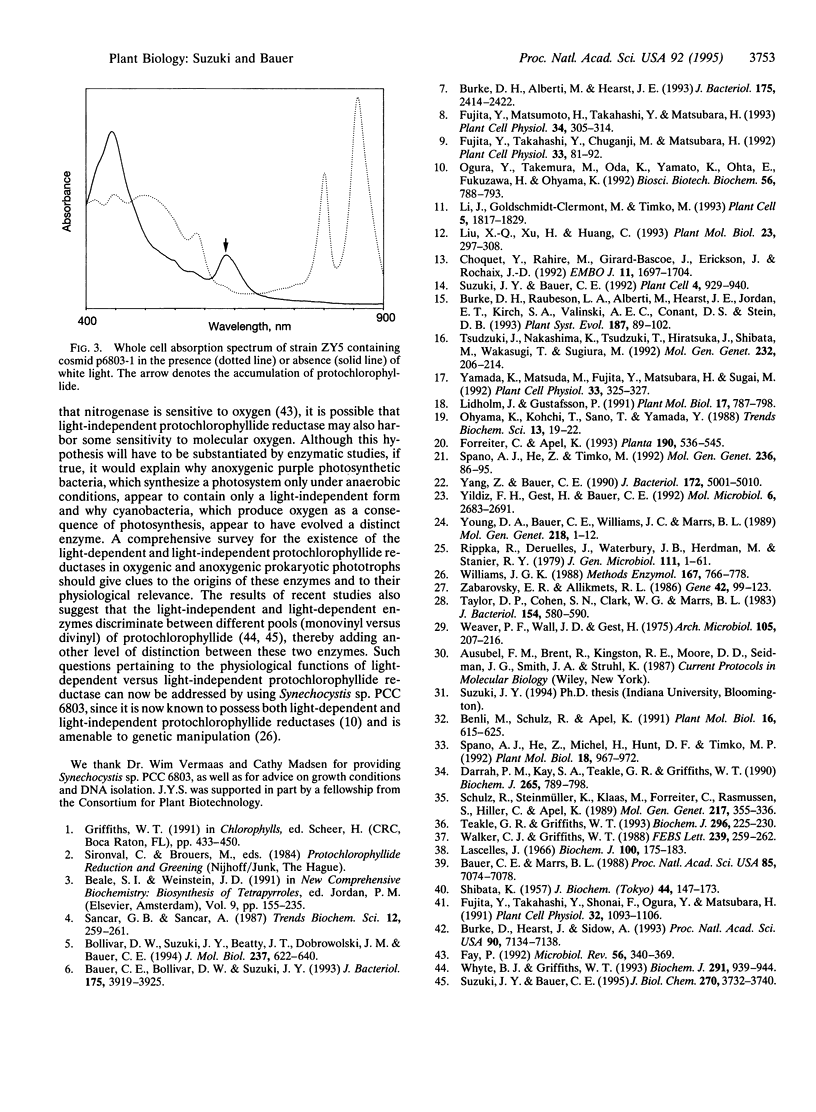
Flowering plants require light for chlorophyll synthesis. Early studies indicated that the dependence on light for greening stemmed in part from the light-dependent reduction of the chlorophyll intermediate protochlorophyllide to the product chlorophyllide. Light-dependent reduction of protochlorophyllide by flowering plants is contrasted by the ability of nonflowering plants, algae, and photosynthetic bacteria to reduce protochlorophyllide and, hence, synthesize (bacterio) chlorophyll in the dark. In this report, we functionally complemented a light-independent protochlorophyllide reductase mutant of the eubacterium Rhodobacter capsulatus with an expression library composed of genomic DNA from the cyanobacterium Synechocystis sp. PCC 6803. The complemented R. capsulatus strain is capable of synthesizing bacteriochlorophyll in the light, thereby indicating that a chlorophyll biosynthesis enzyme can function in the bacteriochlorophyll biosynthetic pathway. However, under dark growth conditions the complemented R. capsulatus strain fails to synthesize bacteriochlorophyll and instead accumulates protochlorophyllide. Sequence analysis demonstrates that the complementing Synechocystis genomic DNA fragment exhibits a high degree of sequence identity (53-56%) with light-dependent protochlorophyllide reductase enzymes found in plants. The observation that a plant-type, light-dependent protochlorophyllide reductase enzyme exists in a cyanobacterium indicates that light-dependent protochlorophyllide reductase evolved before the advent of eukaryotic photosynthesis. As such, this enzyme did not arise to fulfill a function necessitated either by the endosymbiotic evolution of the chloroplast or by multicellularity; rather, it evolved to fulfill a fundamentally cell-autonomous role.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bauer C. E., Bollivar D. W., Suzuki J. Y. Genetic analyses of photopigment biosynthesis in eubacteria: a guiding light for algae and plants. J Bacteriol. 1993 Jul;175(13):3919–3925. doi: 10.1128/jb.175.13.3919-3925.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C. E., Marrs B. L. Rhodobacter capsulatus puf operon encodes a regulatory protein (PufQ) for bacteriochlorophyll biosynthesis. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7074–7078. doi: 10.1073/pnas.85.19.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benli M., Schulz R., Apel K. Effect of light on the NADPH-protochlorophyllide oxidoreductase of Arabidopsis thaliana. Plant Mol Biol. 1991 Apr;16(4):615–625. doi: 10.1007/BF00023426. [DOI] [PubMed] [Google Scholar]
- Bollivar D. W., Suzuki J. Y., Beatty J. T., Dobrowolski J. M., Bauer C. E. Directed mutational analysis of bacteriochlorophyll a biosynthesis in Rhodobacter capsulatus. J Mol Biol. 1994 Apr 15;237(5):622–640. doi: 10.1006/jmbi.1994.1260. [DOI] [PubMed] [Google Scholar]
- Burke D. H., Alberti M., Hearst J. E. bchFNBH bacteriochlorophyll synthesis genes of Rhodobacter capsulatus and identification of the third subunit of light-independent protochlorophyllide reductase in bacteria and plants. J Bacteriol. 1993 Apr;175(8):2414–2422. doi: 10.1128/jb.175.8.2414-2422.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D. H., Hearst J. E., Sidow A. Early evolution of photosynthesis: clues from nitrogenase and chlorophyll iron proteins. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):7134–7138. doi: 10.1073/pnas.90.15.7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y., Rahire M., Girard-Bascou J., Erickson J., Rochaix J. D. A chloroplast gene is required for the light-independent accumulation of chlorophyll in Chlamydomonas reinhardtii. EMBO J. 1992 May;11(5):1697–1704. doi: 10.1002/j.1460-2075.1992.tb05220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrah P. M., Kay S. A., Teakle G. R., Griffiths W. T. Cloning and sequencing of protochlorophyllide reductase. Biochem J. 1990 Feb 1;265(3):789–798. doi: 10.1042/bj2650789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay P. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol Rev. 1992 Jun;56(2):340–373. doi: 10.1128/mr.56.2.340-373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forreiter C., Apel K. Light-independent and light-dependent protochlorophyllide-reducing activities and two distinct NADPH-protochlorophyllide oxidoreductase polypeptides in mountain pine (Pinus mugo). Planta. 1993;190(4):536–545. doi: 10.1007/BF00224793. [DOI] [PubMed] [Google Scholar]
- Fujita Y., Matsumoto H., Takahashi Y., Matsubara H. Identification of a nifDK-like gene (ORF467) involved in the biosynthesis of chlorophyll in the cyanobacterium Plectonema boryanum. Plant Cell Physiol. 1993 Mar;34(2):305–314. [PubMed] [Google Scholar]
- Lascelles J. The accumulation of bacteriochlorophyll precursors by mutant and wild-type strains of Rhodopseudomonas spheroides. Biochem J. 1966 Jul;100(1):175–183. doi: 10.1042/bj1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Goldschmidt-Clermont M., Timko M. P. Chloroplast-encoded chlB is required for light-independent protochlorophyllide reductase activity in Chlamydomonas reinhardtii. Plant Cell. 1993 Dec;5(12):1817–1829. doi: 10.1105/tpc.5.12.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidholm J., Gustafsson P. Homologues of the green algal gidA gene and the liverwort frxC gene are present on the chloroplast genomes of conifers. Plant Mol Biol. 1991 Oct;17(4):787–798. doi: 10.1007/BF00037061. [DOI] [PubMed] [Google Scholar]
- Liu X. Q., Xu H., Huang C. Chloroplast chlB gene is required for light-independent chlorophyll accumulation in Chlamydomonas reinhardtii. Plant Mol Biol. 1993 Oct;23(2):297–308. doi: 10.1007/BF00029006. [DOI] [PubMed] [Google Scholar]
- Ogura Y., Takemura M., Oda K., Yamato K., Ohta E., Fukuzawa H., Ohyama K. Cloning and nucleotide sequence of a frxC-ORF469 gene cluster of Synechocystis PCC6803: conservation with liverwort chloroplast frxC-ORF465 and nif operon. Biosci Biotechnol Biochem. 1992 May;56(5):788–793. doi: 10.1271/bbb.56.788. [DOI] [PubMed] [Google Scholar]
- Ohyama K., Kohchi T., Sano T., Yamada Y. Newly identified groups of genes in chloroplasts. Trends Biochem Sci. 1988 Jan;13(1):19–22. doi: 10.1016/0968-0004(88)90013-8. [DOI] [PubMed] [Google Scholar]
- Schulz R., Steinmüller K., Klaas M., Forreiter C., Rasmussen S., Hiller C., Apel K. Nucleotide sequence of a cDNA coding for the NADPH-protochlorophyllide oxidoreductase (PCR) of barley (Hordeum vulgare L.) and its expression in Escherichia coli. Mol Gen Genet. 1989 Jun;217(2-3):355–361. doi: 10.1007/BF02464904. [DOI] [PubMed] [Google Scholar]
- Spano A. J., He Z., Michel H., Hunt D. F., Timko M. P. Molecular cloning, nuclear gene structure, and developmental expression of NADPH: protochlorophyllide oxidoreductase in pea (Pisum sativum L.). Plant Mol Biol. 1992 Mar;18(5):967–972. doi: 10.1007/BF00019210. [DOI] [PubMed] [Google Scholar]
- Spano A. J., He Z., Timko M. P. NADPH: protochlorophyllide oxidoreductases in white pine (Pinus strobus) and loblolly pine (P. taeda). Evidence for light and developmental regulation of expression and conservation in gene organization and protein structure between angiosperms and gymnosperms. Mol Gen Genet. 1992 Dec;236(1):86–95. [PubMed] [Google Scholar]
- Suzuki J. Y., Bauer C. E. Altered monovinyl and divinyl protochlorophyllide pools in bchJ mutants of Rhodobacter capsulatus. Possible monovinyl substrate discrimination of light-independent protochlorophyllide reductase. J Biol Chem. 1995 Feb 24;270(8):3732–3740. [PubMed] [Google Scholar]
- Suzuki J. Y., Bauer C. E. Light-independent chlorophyll biosynthesis: involvement of the chloroplast gene chlL (frxC). Plant Cell. 1992 Aug;4(8):929–940. doi: 10.1105/tpc.4.8.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. P., Cohen S. N., Clark W. G., Marrs B. L. Alignment of genetic and restriction maps of the photosynthesis region of the Rhodopseudomonas capsulata chromosome by a conjugation-mediated marker rescue technique. J Bacteriol. 1983 May;154(2):580–590. doi: 10.1128/jb.154.2.580-590.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teakle G. R., Griffiths W. T. Cloning, characterization and import studies on protochlorophyllide reductase from wheat (Triticum aestivum). Biochem J. 1993 Nov 15;296(Pt 1):225–230. doi: 10.1042/bj2960225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsudzuki J., Nakashima K., Tsudzuki T., Hiratsuka J., Shibata M., Wakasugi T., Sugiura M. Chloroplast DNA of black pine retains a residual inverted repeat lacking rRNA genes: nucleotide sequences of trnQ, trnK, psbA, trnI and trnH and the absence of rps16. Mol Gen Genet. 1992 Mar;232(2):206–214. doi: 10.1007/BF00279998. [DOI] [PubMed] [Google Scholar]
- Weaver P. F., Wall J. D., Gest H. Characterization of Rhodopseudomonas capsulata. Arch Microbiol. 1975 Nov 7;105(3):207–216. doi: 10.1007/BF00447139. [DOI] [PubMed] [Google Scholar]
- Whyte B. J., Griffiths W. T. 8-vinyl reduction and chlorophyll a biosynthesis in higher plants. Biochem J. 1993 May 1;291(Pt 3):939–944. doi: 10.1042/bj2910939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. M., Bauer C. E. Rhodobacter capsulatus genes involved in early steps of the bacteriochlorophyll biosynthetic pathway. J Bacteriol. 1990 Sep;172(9):5001–5010. doi: 10.1128/jb.172.9.5001-5010.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz F. H., Gest H., Bauer C. E. Conservation of the photosynthesis gene cluster in Rhodospirillum centenum. Mol Microbiol. 1992 Sep;6(18):2683–2691. doi: 10.1111/j.1365-2958.1992.tb01445.x. [DOI] [PubMed] [Google Scholar]
- Young D. A., Bauer C. E., Williams J. C., Marrs B. L. Genetic evidence for superoperonal organization of genes for photosynthetic pigments and pigment-binding proteins in Rhodobacter capsulatus. Mol Gen Genet. 1989 Jul;218(1):1–12. doi: 10.1007/BF00330558. [DOI] [PubMed] [Google Scholar]
- Zabarovsky E. R., Allikmets R. L. An improved technique for the efficient construction of gene libraries by partial filling-in of cohesive ends. Gene. 1986;42(1):119–123. doi: 10.1016/0378-1119(86)90158-7. [DOI] [PubMed] [Google Scholar]


