Carotta et al. show that the interaction between IRF8 and PU.1 controls the propensity of B cells to undergo class-switch recombination and plasma cell differentiation by concurrently promoting the expression of BCL6 and PAX5 and repressing AID and BLIMP-1.
Abstract
Activated B cells undergo immunoglobulin class-switch recombination (CSR) and differentiate into antibody-secreting plasma cells. The distinct transcriptomes of B cells and plasma cells are maintained by the antagonistic influences of two groups of transcription factors: those that maintain the B cell program, including BCL6 and PAX5, and plasma cell–promoting factors, such as IRF4 and BLIMP-1. We show that the complex of IRF8 and PU.1 controls the propensity of B cells to undergo CSR and plasma cell differentiation by concurrently promoting the expression of BCL6 and PAX5 and repressing AID and BLIMP-1. As the PU.1–IRF8 complex functions in a reciprocal manner to IRF4, we propose that concentration-dependent competition between these factors controls B cell terminal differentiation.
The terminal differentiation of B cells into antibody-secreting cells (ASCs) is an essential component of a productive immune response. Upon antigen encounter, B cells undergo multiple rounds of division, initiate Ig class-switch recombination (CSR), and acquire the ability to differentiate into short-lived cycling plasmablasts, long-lived postmitotic plasma cells, or memory B cells (Fairfax et al., 2008). Plasmablasts arise from extrafollicular B cell responses, whereas plasma cells are predominantly germinal center (GC) derived. The GC reaction also produces memory B cells that can rapidly differentiate into ASC after reexposure to antigen.
The process of B cell terminal differentiation can be studied in vitro, as B cells are capable of both CSR and differentiation to ASCs in response to T cell–derived stimuli (CD40L and cytokines) or toll-like receptor–mediated signals (LPS). Quantitative analysis of B cell cultures has revealed a striking relationship between cell division history and CSR and ASC differentiation (Deenick et al., 1999; Hasbold et al., 2004; Nutt et al., 2011). These findings have led to a division-based model of B cell behavior that describes how stochastic decisions taken at a single cell level result in the controlled generation of a variety of differentiated cell types in the population as a whole (Hasbold et al., 2004).
A small number of transcription factors have been identified that guide the developmental program leading to ASC differentiation, with the evidence to date suggesting that this gene regulatory network is dominated by transcriptional repression (Shaffer et al., 2000, 2002, 2004; Shapiro-Shelef et al., 2003). One group of factors, including PAX5, BACH2, and BCL6, are expressed in activated B cells and act predominantly by repressing differentiation (Nutt et al., 2011). PAX5 represses genes associated with the stem cell and non–B lineage programs, as well as several genes involved in ASC differentiation including Prdm1 (the gene encoding BLIMP-1 [B lymphocyte-induced maturation protein-1]) and Igj (J chain; Delogu et al., 2006). BCL6 and BACH2 suppress ASC development in part by repressing Prdm1 (Shaffer et al., 2000; Tunyaplin et al., 2004; Muto et al., 2010).
The molecular changes that overcome this repression and allow ASC formation remain unclear, but it is known that differentiation requires IRF4 and BLIMP-1 (Mittrücker et al., 1997; Shapiro-Shelef et al., 2003), whereas high level Ig secretion is XBP1-dependent (Todd et al., 2009; Taubenheim et al., 2012). IRF4 is present at relatively low amounts in activated B cells, where it regulates CSR and GC formation (Sciammas et al., 2006, 2011; Ochiai et al., 2013; Willis et al., 2014). Upon further differentiation, IRF4 expression markedly increases—an event essential for ASC development (Sciammas et al., 2006).
BLIMP-1 is expressed in ASC where it is required for the generation of a functional ASC compartment and normal serum Ig titers (Shapiro-Shelef et al., 2003; Kallies et al., 2004; Kallies et al., 2007). BLIMP-1 is, however, dispensable for the initiation of the ASC differentiation program, as several early events in the terminal differentiation process, including the down-regulation of PAX5 and BCL6, initiation of Prdm1 transcription, and low level Ig secretion occur in BLIMP-1–deficient B cells (Kallies et al., 2007). This raises the question: what factor, if not BLIMP-1, initiates the terminal differentiation process?
Here, we show that the transcription factors IRF8 and PU.1 together function to negatively regulate ASC differentiation. IRF8 is closely related to IRF4 and is required for many aspects of myelopoiesis and DC development (Belz and Nutt, 2012). Despite the fact that much biochemical evidence has been provided to support a role for IRF8 and PU.1 in B cell development and function, conditional inactivation of either factor in B cells leads to essentially normal humoral responses (Polli et al., 2005; Feng et al., 2011). As PU.1 and IRF proteins (both IRF4 and 8) are well known to cooperatively bind to composite DNA recognition motifs (Pongubala et al., 1992; Eisenbeis et al., 1993; Kanno et al., 2005), we have addressed the importance of this interaction by creating mice in which IRF8 and PU.1 are deleted in B cells. We found that the loss of both factors led to a dramatic enhancement in the rates of CSR and ASC differentiation. IRF8/PU.1 controlled the B cell to ASC transition by simultaneously activating components of the B cell program, including Bcl6, as well as repressing ASC-promoting factors such as Prdm1. The negative regulation of ASC differentiation by IRF8/PU.1 contrasts strikingly with the established role of IRF4 in promoting CSR and ASC differentiation (Mittrücker et al., 1997; Klein et al., 2006; Sciammas et al., 2006). We provide evidence that IRF8 and IRF4 bind to the same sites in critical target genes and propose a model whereby the rate of ASC differentiation is controlled by the relative concentration of these two factors acting in a reciprocal manner.
RESULTS
B cell development in the absence of IRF8 and PU.1
Despite the biochemical evidence for a decisive role of IRF8 and PU.1 in late B cell differentiation and function, genetic deletion of either factor has only minimal consequence for B cell development and no reported impact on B cell function (Polli et al., 2005; Feng et al., 2011). As PU.1, IRF4, and IRF8 are well known to cooperatively bind to a variety of composite elements in the promoters and enhancers of many myeloid and lymphoid genes, we have genetically addressed the importance of this interaction by deleting both IRF8 and PU.1 in B cells. To achieve this, we crossed mice in which PU.1 is conditionally inactivated in B cells (Polli et al., 2005; Spi1fl/flCd19cre/+, termed PU.1cKO) to mice lacking IRF8 (Holtschke et al., 1996). All mice were also crossed to the BLIMP-1/GFP reporter strain (Prdm1gfp/+) to facilitate the identification of ASCs (Kallies et al., 2004).
B lymphopoiesis proceeded relatively normally in mice lacking either IRF8 and PU.1 or both. Flow cytometric analysis of the splenic B cell compartment revealed a twofold reduction in the proportion of follicular B cells in Irf8−/−PU.1cKO mice (Table S1). Enumeration of the ASC compartment using the BLIMP-1/GFP reporter strain revealed a normal distribution of BLIMP-1/GFPlow plasmablasts and BLIMP-1/GFPhigh plasma cells in the spleen of PU.1cKO mice. In contrast, Irf8−/− mice had generally increased ASC numbers, potentially through both B cell–intrinsic and –extrinsic mechanisms (Holtschke et al., 1996). Strikingly, the proportion and number of BLIMP-1/GFPlow plasmablasts was further increased in the absence of both IRF8 and PU.1 (Table S1). The increased proportion of plasmablasts in resting mice suggests a higher production of ASC in the absence of IRF8/PU.1.
IRF8 and PU.1 negatively regulate ASC differentiation
To rigorously examine the function of IRF8 and PU.1 in late B cell differentiation, we made use of quantitative assays for in vitro B cell differentiation (Hasbold et al., 2004). Naive B cells were isolated from the lymph nodes by negative selection, labeled with cell division tracking dyes (CFSE or cell trace violet [CTV]), and cultured for up to 5 d in the presence of various combinations of CD40-ligand (CD40L), T cell–derived cytokines (IL-4 and IL-5), and/or LPS. The exclusive use of lymph node follicular B cells both avoids the complications arising from the alteration of splenic architecture in IRF8-deficient mice and excludes the possibility of marginal zone and B1 B cell contamination in the cultures. The resulting cultures were then assessed for proliferation, Ig CSR, and secretion, as well as the expression of ASC-associated proteins such as CD138 and BLIMP-1/GFP.
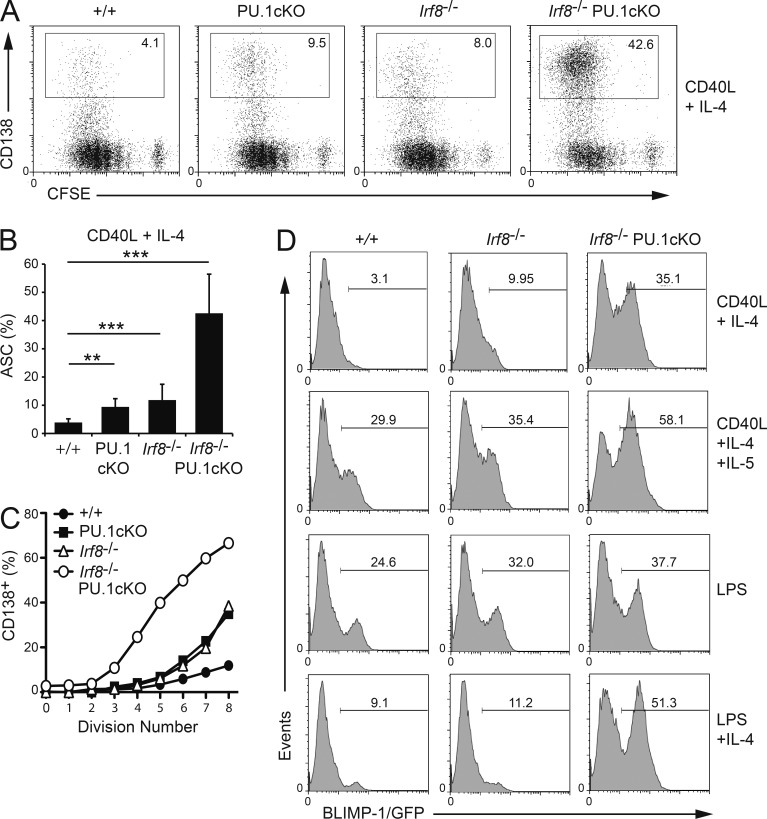
B cells from WT, single mutants, or IRF8/PU.1 double-deficient mice cultured in CD40L+IL-4 revealed similar proliferation characteristics as measured by CFSE dilution and total cell numbers (Fig. 1 A and not depicted). Strikingly, although CD40L+IL-4 was a relatively inefficient inducer of ASC from WT cultures, B cells lacking IRF8 and PU.1 formed CD138+ or BLIMP-1/GFP+ ASCs at a high frequency (Fig. 1, A–D). B cells singularly deficient in either factor generated an intermediate phenotype with a twofold increased proportion of ASCs (Fig. 1, A–D). Analysis of the propensity of B cells to differentiate into ASCs in relation to the number of cell divisions revealed that IRF8/PU.1-deficient B cells initiated the differentiation process at an earlier division and at a higher frequency with each subsequent division (Fig. 1 C). B cells singularly deficient in IRF8 and PU.1 again showed an intermediate phenotype. The increase in ASC generation was not strongly linked to the type or potency of differentiation-inducing stimuli provided in the cultures, as IRF8/PU.1-deficient B cells showed hyper-ASC differentiation under all conditions tested (Fig. 1 D).
Figure 1.
Increased ASC differentiation in the absence of IRF8 and PU.1. (A) Resting lymph node B cells from mice of the indicated genotypes were labeled with CFSE and cultured in the presence of CD40L+IL-4 for 4 d before flow cytometric analyses for CD138 and CFSE dilution. Numbers in boxes are the proportion of CD138+ ASCs. Data are representative of 7 experiments. (B) Quantitation of the numbers of ASCs in cells cultured as in A. Data are the mean ± SD from 7 identical experiments, except for PU.1cKO where n = 3. **, P < 0.01; ***, P < 0.005 for the indicated comparisons using an unpaired Student’s t test. (C) Graph showing the proportion of CD138+ cells present in each cell division peak at 4 d of the culture. Data are representative of 3 experiments. (D) Resting B cells from the lymph nodes of mice of the indicated genotypes and carrying the BLIMP-1/GFP reporter allele were cultured in the presence of CD40L+IL-4 ±IL-5 or LPS ±IL-4 for 4 d and examined for the expression of BLIMP-1/GFP. Numbers indicate the proportion of BLIMP-1/GFP+ ASC in each culture. Data are representative of at least 3 experiments for each genotype.
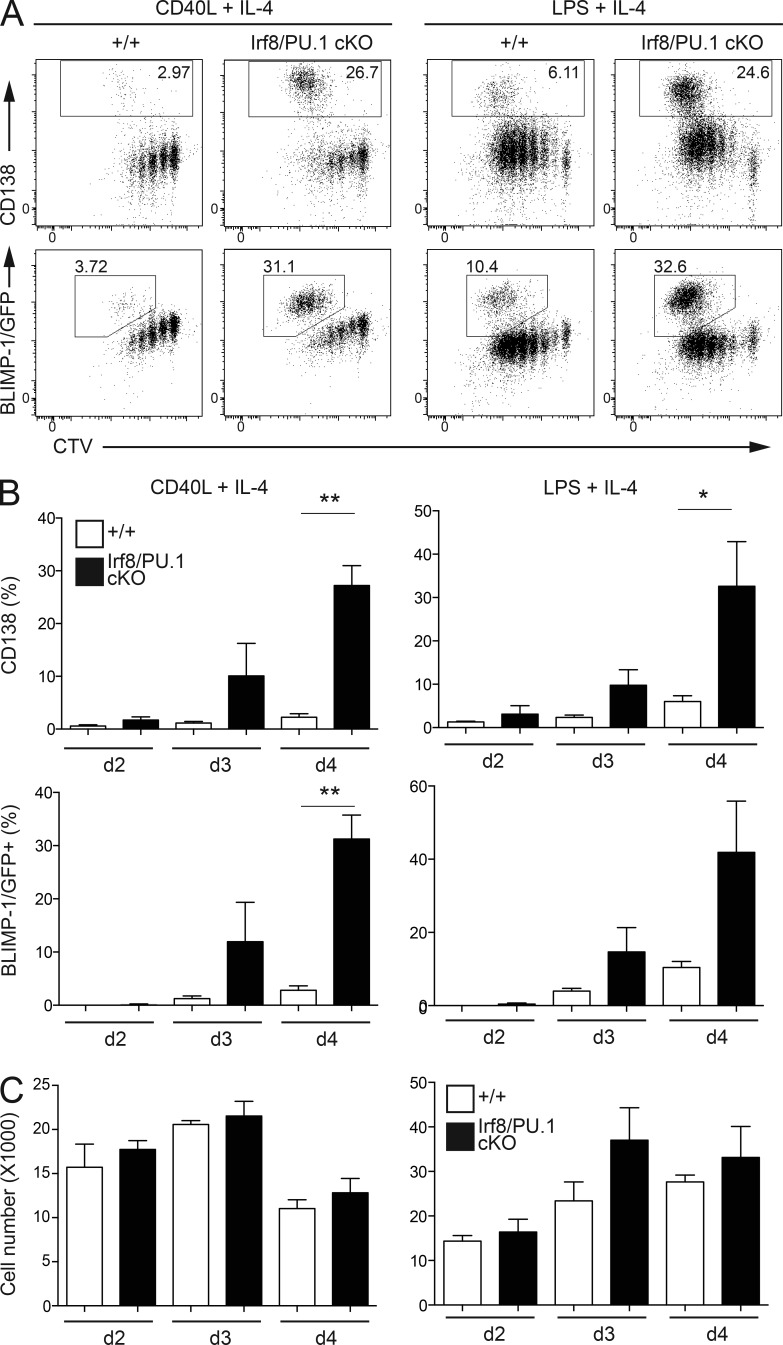
To further demonstrate the B cell–intrinsic nature of the hyper-ASC differentiation phenotype, we generated mice that lack both IRF8 and PU.1 only in B cells (Irf8fl/flSpi1fl/flCd19cre/+, termed IRF8/PU.1cKO). These mice displayed normal resting frequency of splenic follicular and marginal zone B cells, plasmablasts, and plasma cells, confirming that the altered, potentially proinflammatory environment in mice harboring a null allele of Irf8 was required to elicit the increased plasmablast numbers seen in double knockout mice (Table S1). Most importantly, culture of mature resting B cells from the lymph nodes of these mice in either CD40L+IL-4 or LPS+IL-4 resulted in a dramatic increase in the frequency of CD138+ and BLIMP-1/GFP+ ASCs at days 3 and 4 after stimulation, without any impact on the cell division rate or the number of viable cells (Fig. 2, A–C). Collectively these results demonstrate that IRF8 and PU.1 act together to repress ASC differentiation in a B cell–intrinsic manner.
Figure 2.
The increased ASC differentiation in the absence of IRF8 and PU.1 is B cell intrinsic. (A) Resting lymph node B cells from mice of the indicated genotypes and carrying the BLIMP-1/GFP reporter allele were labeled with CTV and cultured in the presence of CD40L+IL-4 or LPS+IL-4 for 4 d before flow cytometric analyses for CD138, BLIMP-1/GFP, and CTV dilution. Numbers in boxes are the proportion of CD138+ or BLIMP-1/GFP+ ASCs. Data are representative of 3 experiments. (B) Quantitation of the numbers of ASC cells cultured as in A for 2, 3, and 4 d. Data are the mean ± SD from 3 identical experiments. *, P < 0.05; **, P < 0.01 for the indicated comparisons using an unpaired Student’s t test. (C) Quantitation of the numbers of viable cells cultured as in A for 2, 3, and 4 d. Data are the mean ± SD from 3 identical experiments.
IRF8 and PU.1 repress immunoglobulin CSR
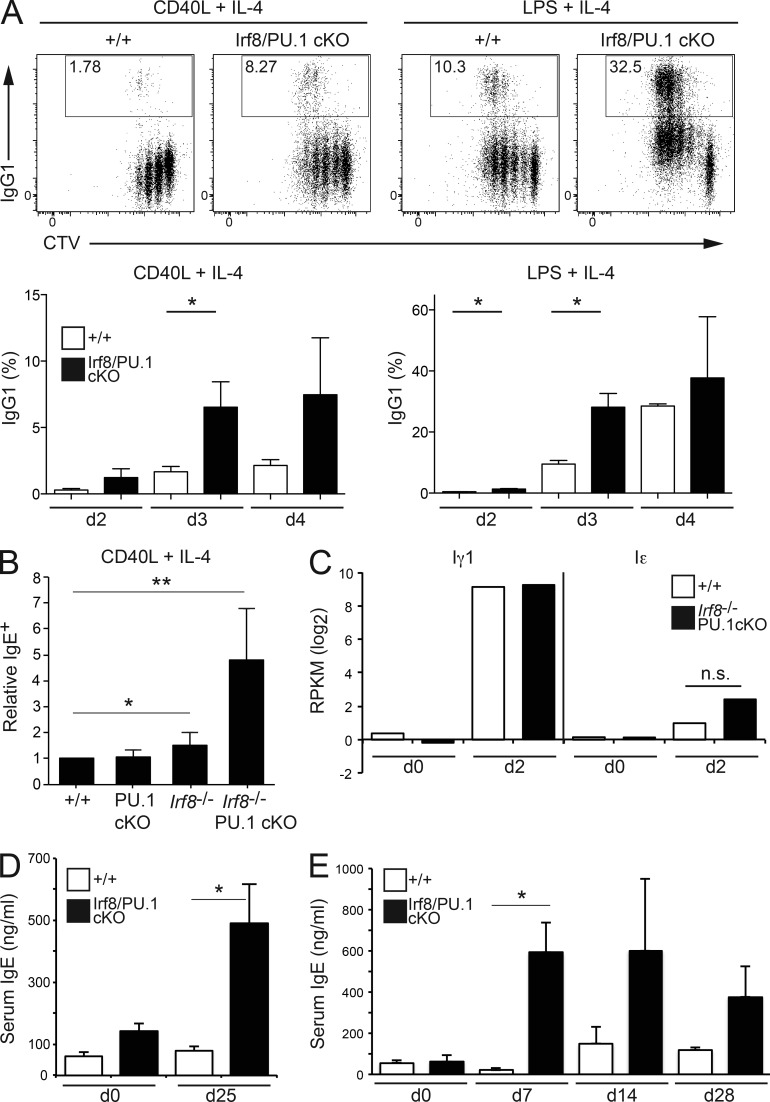
Activated B cells also undergo Ig CSR, a process which is completely shut off in ASC due to BLIMP-1–dependent repression of Aicda (Shaffer et al., 2002). The cellular decisions to undergo CSR or to differentiate into ASCs are considered to be independently regulated events (Hasbold et al., 2004; Duffy et al., 2012). B cells cultured in CD40L+IL-4 typically show pronounced CSR to IgG1 and a lower rate of switching to IgE. IRF8/PU.1-deficient B cells showed hyper-CSR to IgG1 (Fig. 3 A) and IgE (Fig. 3 B). Interestingly, the increased CSR did not result from altered accessibility of the Igh loci, as the Iγ1 and Iε sterile transcripts were normally induced by CD40L and IL-4 in the absence of IRF8 and PU.1 (Fig. 3 C). The hyper-CSR was not restricted to CD40L+IL-4 stimulation, as cells cultured in LPS+IL-4 or LPS+IFN-γ also produced markedly more IgG1 or IgG2c, respectively (Fig. 3, A and B; and not depicted). To assess whether this hyper-CSR also occurs in vivo, we immunized IRF8/PU.1cKO mice with the T cell–dependent antigens Ovalbumin (Fig. 3 D) and 4(hydroxy-3-nitrophenyl) acetyl (NP)–KLH (Fig. 3 E) and measured serum antibody titers. Although the canonical IgG1 response of the double-deficient B cells was equivalent to WT controls (not depicted), the level of IgE was markedly increased at each time point analyzed for both antigens (Fig. 3, D and E). Together, these data show that despite the very high rate of ASC differentiation that was expected to truncate the developmental window available for CSR (Muto et al., 2010), IRF8/PU.1-deficient cells displayed a pronounced increase in their propensity to undergo CSR.
Figure 3.
Increased immunoglobulin isotype switching in the absence of IRF8 and PU.1. (A) Resting lymph node B cells from mice of the indicated genotypes were cultured in the presence of CD40L+IL-4 or LPS+IL-4 for 2, 3, or 4 d, fixed and permeabilized, and examined for the expression of IgG1. Numbers in boxes are the proportion of IgG1+ cells at day 3. Data in graphs are the mean ± SD of 3 experiments. *, P < 0.05 for the indicated comparisons using an unpaired Student’s t test. (B) Resting lymph node B cells from mice of the indicated genotypes were cultured in the presence of CD40L+IL-4 for 4 d, fixed and permeabilized, and examined for the expression of IgE. Data are the mean ± SD of 2 (PU.1cKO) and 5 (all other genotypes) experiments and are normalized to 1 for the WT sample. *, P < 0.05; **, P < 0.01 for the indicated comparisons using an unpaired Student’s t test. (C) Resting B cells from WT (+/+) or Irf8−/−PU.1cKO mice were either lysed immediately (d0) or cultured in the presence of CD40L+IL-4 for 48 h (d2). RNA was extracted from all samples and subjected to whole transcriptome RNA-Seq analysis. Data are the mean reads per kilobase per million (RPKM) for the Iε and Iγ1 sterile transcripts, from two independent experiments. n.s., not significant P > 0.05. (D and E) Serum IgE (ng/ml) from mice of the indicated genotypes, either non-immunized (d0) or immunized with Ovalbumin in alum (d25; D) or NP-KLH in alum (E; d7, 14, and 28). Data are the mean ± SEM of 5–6 samples (D) or 3–5 samples (E) per genotype and time point. *, P < 0.05 for the indicated comparisons using an unpaired Student’s t test.
Expression of IRF4, IRF8, and PU.1 during B cell differentiation
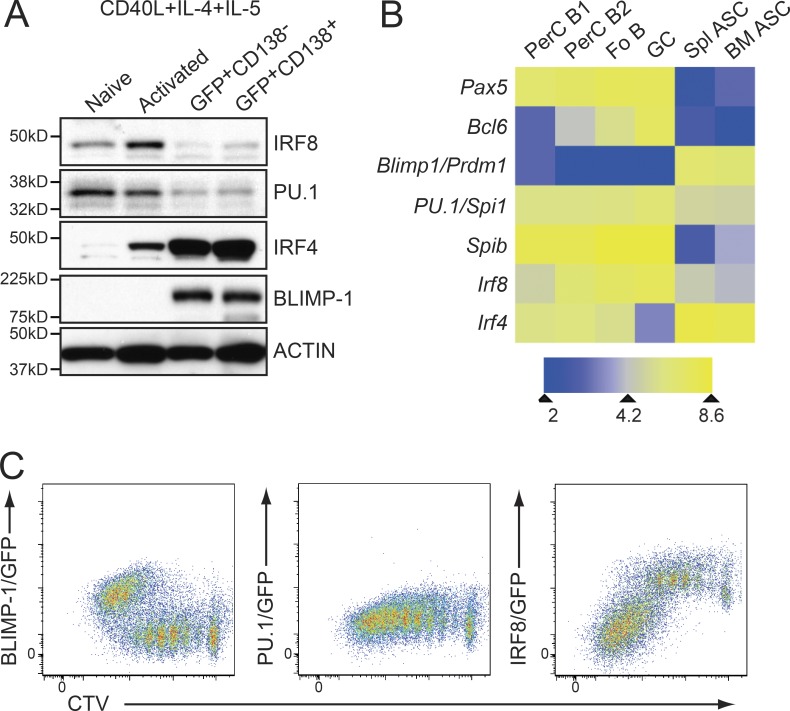
The potent role of IRF8 and PU.1 in suppressing CSR and ASC differentiation contrasts with the requirement for IRF4 for both functions (Sciammas et al., 2006). Therefore, we determined the expression pattern of these regulators in late B cell differentiation in vitro and in vivo. PU.1 concentration is relatively low in all B cell subsets and is further down-regulated, but not completely silenced, in ASCs (Fig. 4, A–C). The related ETS family member SpiB is also expressed in mature B cells and GC B cells before being strongly down-regulated in ASCs (Fig. 4 B). IRF8 is constitutively expressed in all B cells but is further induced in activated and GC B cells (Fig. 4, A–C; and not depicted). IRF8 is then rapidly silenced in ASCs. This expression pattern is further supported by the analysis of knock-in mice harboring eGFP fused to the C terminus of the IRF8 protein (Wang et al., 2014), where the loss of IRF8 expression in the late divisions of cells cultured in CD40L+IL-4+IL-5 corresponded temporally to the induction of BLIMP-1/GFP expression (Fig. 4 C). As has previously been reported, IRF4 had the reciprocal expression pattern to IRF8, being weakly expressed on a protein level in naive cells, induced to an intermediate concentration in activated cells, and very highly expressed in ASCs (Sciammas et al., 2006; Ochiai et al., 2013; Willis et al., 2014;Fig. 4, A and B). Collectively the expression profiles of IRF4, IRF8, and PU.1 are fully compatible with the cellular data presented above that suggests these proteins have opposing roles during the early activation events that lead to the terminal differentiation of B cells.
Figure 4.
Expression of IRF8, IRF4, and PU.1 during late B cell differentiation. (A) Resting lymph node B cells from Prdm1gfp/+ mice were either lysed immediately or cultured in the presence of CD40L+IL-4+IL-5 for 4 d. At day 4, cells were sorted into activated (CD138−BLIMP-1/GFP−) B cells, CD138−BLIMP-1/GFP+, or CD138+BLIMP-1/GFP+ ASC compartments and subjected to Western blotting for the indicated proteins. Actin serves as a control for protein loading. Positions of molecular mass markers are shown on the left. (B) Heat map of the expression of key transcription factors in the indicated populations. Data were derived by whole transcriptome RNA-Seq analysis. PerC B1 (peritoneal B1 cells, B220lowCD23−Mac1+), PerC B2 (conventional B2 cells in peritoneal cavity, B220+CD23+Mac1−), Fo B (follicular B cells, small size, B220+CD23+), GC (GC B cells, B220+Fas+PNA+), Spl ASC (spleen CD138+BLIMP-1/GFP+ ASC), and BM ASC (BM CD138+BLIMP-1/GFP+ ASC). Data are the mean of two experiments. (C) Expression of BLIMP-1/GFP (Prdm1gfp/+), PU.1/GFP (Spi1gfp/gfp), and IRF8/GFP (Irf8gfp/gfp) alleles during ASC differentiation. Resting splenic B cells were labeled with CTV and cultured as in A for 4 d. Data in A and C are representative of 3 experiments.
Maintenance of the B cell program by IRF8 and PU.1
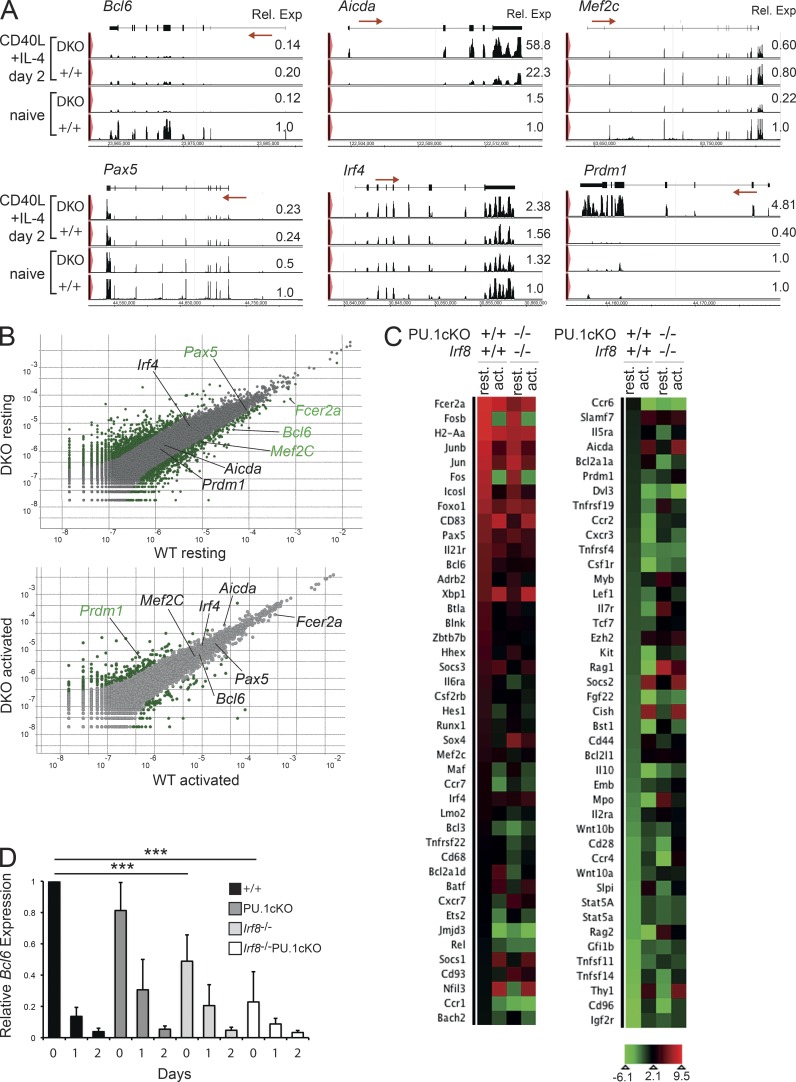
For a global perspective of IRF8 and PU.1 function in late B cell differentiation, we performed RNA sequencing analyses using RNA derived from either WT or IRF8/PU.1-deficient B cells, either naive or activated in the presence of CD40L+IL-4 for 2 d. These data demonstrated that 1,053 annotated gene transcripts were differentially expressed between naive WT and IRF8/PU.1-deficient B cells (>2-fold expression change, 1% false discovery rate, 411 genes decreased and 642 increased in IRF8/PU.1-deficient B cells). This analysis did reveal that a selection of genes associated with pre–B cells, such as Il7r and Rag1/2, were weakly up-regulated in the absence of IRF8/PU.1, probably reflecting the mildly impaired maturation or more rapid turnover of the double-deficient B cells (Fig. 5 C and Table S2). More importantly, there was no differential expression between naive control and mutant cells in the expression of a suite of activation and differentiation markers including Aicda, Prdm1, Nfil3, and Xbp1, demonstrating that the vast majority of IRF8/PU.1-deficient cells were indeed naive in phenotype (Fig. 5 and Table S2). IRF8/PU.1-dependent genes in naive B cells included known PU.1 targets such as Tlr9 (Schroder et al., 2007) and Fcer2a (encoding CD23; DeKoter et al., 2010), as well as several novel targets including Zbtb7b (ThPok), Socs3, Il6ra, Jmjd3, and three members of the anti-apoptotic A1 gene cluster Bcl2a1a,b,d (Table S2). IRF8/PU.1-deficient naive B cells expressed normal amounts of a suite of transcription factors implicated in late B cell differentiation, including Irf4, Bach2, Spib, and Ets2 but, strikingly, displayed reduced expression of Bcl6, Pax5, and Mef2c, factors that are required to maintain the B cell program (Fig. 5, A–C; and Table S2). The dependence of Bcl6, Pax5, and Mef2c on IRF8/PU.1 was independently confirmed by real-time PCR (Fig. 5 D and not depicted).
Figure 5.
Maintenance of the B cell program by IRF8 and PU.1. (A) Resting B cells from WT (+/+) or Irf8−/−PU.1cKO mice were either lysed immediately or cultured in the presence of CD40L+IL-4 for 48 h. RNA was extracted from all samples and subjected to whole transcriptome RNA-Seq analysis. The read coverage for representative genes is shown mapped to the exon–intron structure. Numbers indicate the mean normalized read counts for each sample (set to 1.0 for naive +/+ cells) from two independent experiments for each condition. Arrows indicate the direction of transcription. (B) Gene expression comparison between resting (top) and activated (bottom) +/+ and DKO B cells. Differentially expressed genes are shown in green, with some key genes highlighted. A cutoff of twofold differential regulation and 1% false discovery rate has been applied. (C) Heat map showing normalized expression (Log2) of selected genes found to be differentially regulated between +/+ resting versus activated or between +/+ and DKO B cells. A cutoff of twofold differential regulation and 1% false discovery rate has been applied. Data in A–C are the mean normalized expression derived from two independent experiments for each condition. (D) Quantitative RT-PCR analysis of Bcl6 transcripts in resting B cells from the indicated mice cultured in the presence of CD40L+IL-4 for 0, 1, or 2 d. Results are presented relative to the expression of Hmbs and are the mean ± SEM of triplicate measurements. Data are representative of 3 independent experiments. ***, P < 0.001 for the indicated comparisons using an unpaired Student’s t test.
B cell activation resulted in considerable change in the WT transcriptome, with heightened expression of 1,590 genes and reduced expression of 1,177 (>2-fold expression change, 1% false discovery rate). Genes encoding components of the B cell program, such as Pax5, Bcl6, and Spib, were significantly down-regulated (Fig. 5, A–C; Table S2; and not depicted). Other genes showed a striking increase in expression in the absence of PU.1/IRF8. Genes in this category included Aicda, providing a likely explanation for the increased CSR observed in the IRF8/PU.1-deficient B cells (Fig. 6, A and B), as well as Emb and Cd28, Pax5-repressed genes which are up-regulated in ASCs (Delogu et al., 2006). A prominent gene in this category was Prdm1, whose premature expression provides one mechanism whereby the plasma cell fate is enhanced in the absence of IRF8/PU.1 (Fig. 5, A and B).
Figure 6.
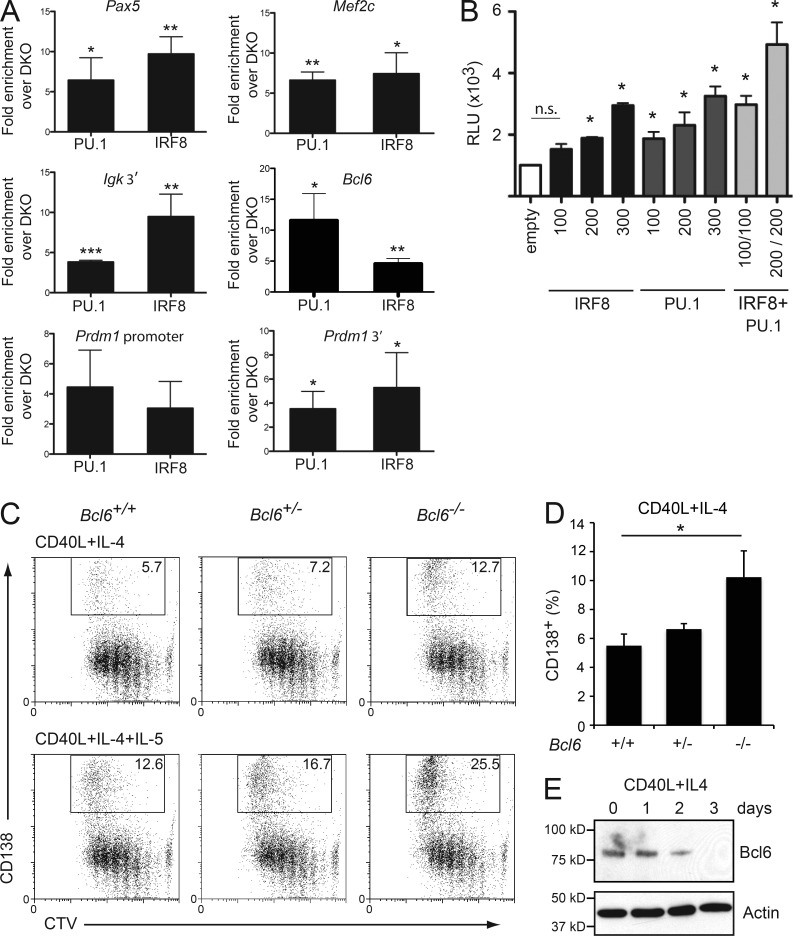
Direct regulation of the B cell differentiation program by IRF8 and PU.1. (A) ChIP assay of PU.1 and IRF8 binding to the regulatory regions of Pax5, Mef2c, Igκ, Bcl6, and Prdm1. Resting B cells from either WT (+/+) or Irf8−/−PU.1cKO mice were cultured in the presence of either CD40L+IL-4 for 2 d before being fixed and subjected to ChIP using either an anti-PU.1 or anti-IRF8 antibody. Negative control ChIP lacked the specific antibody, whereas Irf8−/−PU.1cKO B cells served as an additional control. ChIP products were amplified by real-time PCR using primers that span the putative IRF8 and PU.1 binding sites using primers described in Table S4. Numbers indicate the mean fold enrichment from +/+ material over the binding in Irf8−/−PU.1cKO B cells after subtraction of no antibody control ± SD from 3–4 experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.005 for the indicated sample compared with binding in Irf8−/−PU.1cKO B cells, using an unpaired Student’s t test. (B) Cooperative activation of the Bcl6 promoter by IRF8 and PU.1. A plasmid containing the Bcl6 promoter linked to firefly luciferase were transiently transfected into 293T cells along with the indicated concentrations (ng) of expression plasmids encoding IRF8 and PU.1. Results are expressed as the mean relative luminescent units (RLU) ± SEM from 4 independent experiments. *, P < 0.05 for the indicated sample compared with empty vector alone, using an unpaired Student’s t test. n.s., not significant P > 0.05. (C) Enhanced formation of ASC in the absence of BCL6. Radiation chimeras were generated with WT, Bcl6+/−, or Bcl6−/− fetal liver cells and analyzed after >8 wk. Resting lymph node B cells from the indicated chimeras were labeled with CTV and cultured in the presence of CD40L+IL-4 ±IL-5 for 4 d and analyzed for CD138 expression. Number in boxes denotes the proportion of CD138+ ASC in each culture. Data are representative of 4 experiments. (D) Quantitation of the mean of CD138+ ASC generated in CD40L+IL-4 from cultures described in C. Data are the mean ± SD from 4 independent experiments. *, P < 0.05 for the indicated comparisons using an unpaired Student’s t test. (E) Resting lymph node B cells from WT C57BL/6 mice were either lysed immediately (d0) or cultured in the presence of either CD40L+IL-4 for 1, 2, or 3 d. Samples were subjected to Western blotting for Bcl6. Actin serves as a control for protein loading. Positions of molecular mass markers are shown on the left. Data are representative of 2 experiments.
IRF8 and PU.1 together bind to key genes that regulate B cell identity and plasma cell differentiation
The clear differential expression of several key regulators of ASC differentiation in double-deficient B cells raised the possibility that the IRF8–PU.1 complex directly regulates these genes. To examine this possibility, we performed chromatin immunoprecipitation (ChIP) with antibodies specific for IRF8 or PU.1 on WT B cells activated in the presence of CD40L+IL-4 for 48 h. In keeping with previously published data that derived from transformed cell lines and early B cell progenitors, PU.1 bound to regions of the enhancers of Igκ and Pax5 and the promoter of Mef2c, which harbor consensus ETS-IRF sites (Pongubala et al., 1992; Decker et al., 2009; Stehling-Sun et al., 2009; Fig. 6 A). Importantly, IRF8 also bound to the same region of all these potential target genes. These results support earlier ChIP-chip analyses demonstrating extensive coincidence of IRF8 and PU.1 binding in GC-derived B cell lymphomas in humans and mice (Shin et al., 2011).
Both IRF4 and IRF8 have been reported to bind to the promoter of BCL6 in human and mouse B cells, respectively (Lee et al., 2006; Saito et al., 2007), whereas IRF4 was recently reported to activate mouse Bcl6 through binding to an upstream enhancer (Ochiai et al., 2013). We analyzed the published PU.1 ChIPseq data from myeloid cells (Ghisletti et al., 2010; Heinz et al., 2010) and found PU.1 peaks in macrophages 15,758 bp upstream of the mouse Bcl6 gene. Examination of the sequences defined by the major peak revealed an ETS-IRF composite site (GAAAAGCGGAA). Importantly, ChIP analysis confirmed that both IRF8 and PU.1 bound to Bcl6 locus in activated B cells (Fig. 6 A). Although IRF4 has previously been reported to repress the Bcl6 promoter (Saito et al., 2007), luciferase assays using the same Bcl6 promoter reporter constructs showed a concentration-dependent activation of the same Bcl6 regulatory elements by IRF8 and PU.1, thus demonstrating that the IRF8–PU.1 complex can act in a reciprocal manner to IRF4 (Fig. 6 B).
BCL6 is known to be involved in modulating late B cell differentiation by repression of Prdm1 (Shaffer et al., 2000) and is expressed in the early time points of the CD40L+IL-4 cultures (Fig. 6 E). To assess the functional relevance of the reduced BCL6 expression to the production of ASC in our system, we generated fetal liver chimeras that carried hematopoietic cells that were WT, heterozygous, or null for Bcl6. Stimulation of naive B cells with CD40L+IL-4 ±IL-5 resulted in an approximately twofold increase in the generation of CD138+ ASCs in the absence of Bcl6, whereas Bcl6+/− cells showed a nonsignificant trend toward increased ASC numbers (Fig. 6, C and D). These data demonstrated that reduced expression of BCL6, while contributing to the IRF8/PU.1-deficient phenotype, was not sufficient to explain the increased propensity of mutant B cells to undergo ASC differentiation. Because Pax5 expression was also reduced by 50% in IRF8/PU.1-deficient cells (Fig. 5 A), we generated Bcl6−/− chimeras that also lacked one copy of Pax5. No further increase in the generation of ASCs beyond that associated with a deficiency in BCL6 alone was observed, indicating that loss of BCL6 and reduction of PAX5 are not the only changes enhancing differentiation of IRF8/PU.1-deficient B cells (unpublished data).
IRF4 has also been reported to bind to multiple target sites in the Prdm1 locus, including an IL-21–responsive element downstream of the coding region (Sciammas et al., 2006; Kwon et al., 2009). ChIP analysis revealed that IRF8 and PU.1 bound to both a conserved composite site in the Prdm1 promoter and in the 3′ regulatory element (Fig. 6 A). Collectively, the gene expression profiling and ChIP analyses suggest that both IRF8 and PU.1 can bind to the regulatory regions of genes that encode both activators and inhibitors of ASC generation, suggesting a model where the IRF8–PU.1 complex coordinates the differentiation process by regulating the gateway between the B cell and ASC programs.
Recently, ChIPseq analysis has been used to determine the binding sites for PU.1 in B cell cultures activated for 1 or 3 d in LPS+IL-4 (Ochiai et al., 2013). A stringent reanalysis of this data revealed that of the 1,053 differentially expressed genes between WT and IRF8/PU.1-deficient B cells at day 0, 198 genes (18.8%) had PU.1 binding sites in the region between −2 and + 1 kb from the transcriptional start site (Ochiai et al., 2013), suggesting that a significant proportion of the identified differentially expressed genes are directly regulated by PU.1.
IRF8 and IRF4 bind the similar DNA sequences during ASC differentiation
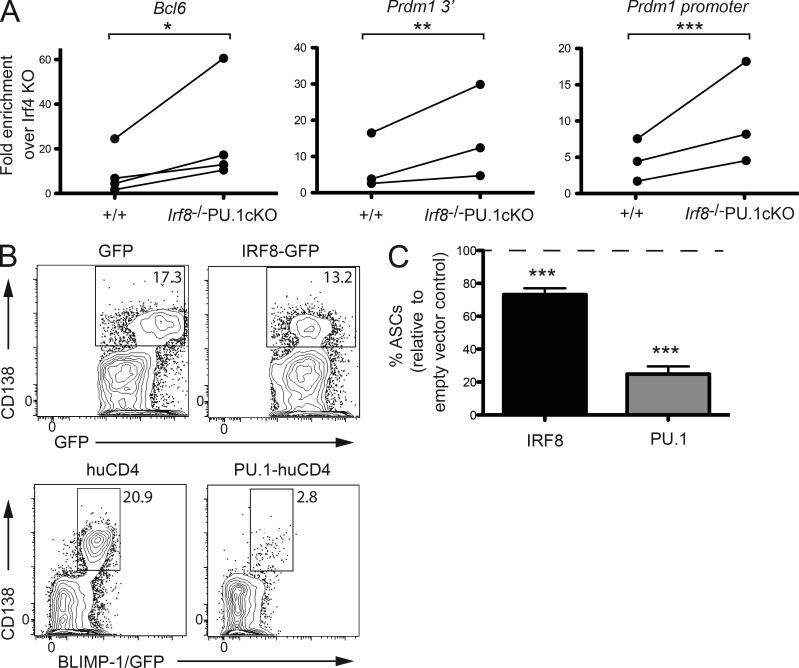
The current understanding is that IRF4 and IRF8 can both bind to identical recognition sequences and interact equivalently with PU.1 (Kanno et al., 2005). Based on the mirror-image phenotypes observed when IRF8/PU.1 or IRF4 are inactivated in B cells, we considered the possibility that IRF4 and IRF8 compete for binding to critical target sequences in activated B cells, which co-express all three factors, and regulate these genes in a reciprocal manner. A prediction from such a model is that in the absence of the relatively high concentration of IRF8 and PU.1 normally found in activated B cells, IRF4, which at this point is expressed at an intermediate level, would be unrestrained in the IRF8/PU.1 mutants and could bind prematurely to targets resulting in a different transcriptional outcome to that normally observed. To directly test this model, we used ChIP to examine the binding of IRF4 in IRF8/PU.1-deficient B cell cultured in CD40L+IL-4 at a time point before the appearance of BLIMP-1/GFP+ ASCs. In keeping with the prediction of our competition model, we found that IRF4 binding was increased in the mutant at the sites in Bcl6 and Prdm1 that we previously showed bound PU.1 and IRF8 in WT cell (Fig. 7 A).
Figure 7.
Competition between IRF family members controls ASC differentiation. (A) ChIP assay of IRF4 binding to the regulatory regions of Bcl6 and the Prdm1 promoter and 3′ enhancer. Resting B cells from WT (+/+) or Irf8−/−PU.1cKO mice were cultured in the presence of CD40L+IL-4 for 2d before being fixed and subjected to ChIP using an anti-IRF4 antibody. ChIP products were amplified by real time PCR using primers that span the IRF4 binding sites. Data are the fold enrichment over Irf4−/− B cells for the +/+ and Irf8−/−PU.1cKO genotypes. Individual data points are indicated by dots and paired experiments are joined by a line. *, P < 0.05; **, P < 0.01; ***, P < 0.005 compare the fold enrichment from the +/+ and Irf8−/−PU.1cKO genotypes using a paired Student’s t test. (B) Enforced overexpression of IRF8 or PU.1 impairs ASC differentiation. Resting B cells from either WT or Prdm1gfp/+ mice were transduced with retroviruses expressing IRF8 (and GFP), PU.1 (and truncated human CD4), or empty vector controls. After 5 d in CD40L+IL-4+IL-5, transduced cells (gated on either GFP or human CD4) were assessed for CD138 (IRF8) or CD138 and BLIMP-1/GFP (PU.1). Plots show representative of 4 (PU.1) and 7 (IRF8) experiments. Numbers are the frequency of ASCs in the boxed area. (C) Quantitation of the relative number of CD138+ ASCs generated in CD40L+IL-4 from cultures described in B. Data are the mean proportion of ASCs ± SEM compared with vector only controls (set to 100%) from 4 (PU.1) and 7 (IRF8) independent experiments. ***, P < 0.005 compares the indicated sample to vector only controls using an unpaired Student’s t test.
A second prediction from the competition model is that increasing the concentration (or preventing the down-regulation) of IRF8 and/or PU.1 would impede ASC differentiation. To test this prediction, WT or BLIMP-1/GFP reporter B cells were activated with CD40L+IL-4+IL-5 for 1 d and transduced with a retrovirus that drives the expression of bicistronic mRNA producing either PU.1 and a truncated human CD4 reporter or IRF8 and GFP. 4 d later, the cells were examined for the frequencies of transduction and the proportion of cells that had undergone ASC differentiation. Increasing PU.1 expression resulted in a dramatic reduction in the frequency of BLIMP-1/GFP+ ASCs, whereas overexpressing IRF8, which is already highly expressed in activated B cells (Fig. 4 C), resulted in a more modest but statistically significant decrease in ASC numbers (Fig. 7, B and C). In keeping with their identical DNA binding specificity, overexpression of SpiB also blocked ASC differentiation (unpublished data). Together with the published literature (Sciammas et al., 2006, 2011) and our own unpublished observations that IRF4 is a dose-dependent activator of ASC generation, these data provide further support for the concept that IRF8 and PU.1 act as antagonists of IRF4 in regulating the transcriptional program of late B cell differentiation.
DISCUSSION
The differentiation of mature B cells into antibody-secreting plasma cells is both essential for a productive humoral immune response and a leading model system to study the terminal differentiation process. The utility of this system for understanding terminal differentiation is highlighted by the stark functional and transcriptional differences that distinguish B cells and ASCs. B cells express cell surface Ig, have high proliferative capacity, and differentiate into both ASCs and memory cells. In contrast, ASCs down-regulate the expression of many B cell genes, including cell surface receptors and lineage-defining transcription factors, retain limited or no proliferative capacity, and secrete huge amounts of antibody. In the current study, we show that the rate of differentiation of activated B cells into ASC is regulated by interactions between the Ets family transcription factor PU.1 and the IRF family member IRF8.
IRF8 and PU.1 are required for multiple aspects of hematopoiesis, including early granulocyte, macrophage, early lymphocyte, and DC differentiation (Lu et al., 2003; Wang et al., 2008; Wang and Morse, 2009; Carotta et al., 2010; Belz and Nutt, 2012). There is also evidence of cooperative DNA binding of these two factors in lymphoid and myeloid cells (Shin et al., 2011; Ochiai et al., 2013). Despite this biochemical data, the approach presented here provides the first genetic evidence that the IRF8–PU.1 complex plays a specific role in hematopoiesis beyond that played by each factor individually. Conditional deletion of IRF8 and PU.1 in B cells resulted in a modest increase in splenic B cells in the case of IRF8 (Feng et al., 2011) and essentially normal humoral responses in the case of PU.1 (Polli et al., 2005). In this study, we observed no change in peripheral B cell numbers or maturation after the removal of both IRF8 and PU.1 specifically from B cells, demonstrating that differentiation up to the mature B cell stage proceeds independently of these factors. IRF4-deficient mice also display relatively normal B cell development, but importantly, IRF4 is essential for GC B cells and ASCs (Klein et al., 2006; Sciammas et al., 2006; Ochiai et al., 2013; Willis et al., 2014).
Despite relatively normal development observed in the absence of any individual factor, this group of genes has important functions in BM pre–B cell development that are masked by a complex set of redundancies. This view is based on the observation that mice doubly deficient for IRF4/IRF8 (Lu et al., 2003), PU.1/SpiB (Sokalski et al., 2011), and PU.1/IRF4 (unpublished data) all show a profound developmental block at the pre–B cell stage. In contrast to the redundant roles of these factors in BM development, the current study showed that IRF8/PU.1-deficient B cells have markedly heightened CSR and ASC differentiation, demonstrating that these factors normally act to restrict B cell terminal differentiation. Heightened ASC differentiation may also help to explain the strong association between single nucleotide polymorphisms in IRF8, and to a lesser degree PU.1, with autoimmune diseases such as lupus erythematosus (Cunninghame Graham et al., 2011; Hikami et al., 2011), systemic sclerosis (Gorlova et al., 2011), and multiple sclerosis (De Jager et al., 2009).
The first descriptions of the genetic network controlling B cell terminal differentiation were made by the Calame and Staudt laboratories (Shaffer et al., 2000, 2002, 2004; Shapiro-Shelef and Calame, 2005). These models were built on the finding that transcriptional repression reinforces mutually exclusive expression programs in B cells and ASCs. In this model, B cell factors, such as BCL6, PAX5, and BACH2, directly repress the expression of the ASC regulators BLIMP-1, XBP1, and IRF4, respectively. BLIMP-1 in turn represses the expression of BCL6 and PAX5. The finding that IRF4 is critical for ASC differentiation by both repressing BCL6 and activating BLIMP-1 expression has provided strong support for this concept (Sciammas et al., 2006; Saito et al., 2007). The model does, however, have some limitations. First, it is not apparent how plasma cell differentiation is initiated, as we previously showed that neither BLIMP-1 (Kallies et al., 2007) nor XBP1 (Taubenheim et al., 2012) is essential for this process, and we show here that loss of BCL6 resulted in only a twofold increase in ASC numbers. In contrast, we have previously shown that inhibition of PAX5 function is a key step in initiating plasma cell differentiation, although the molecular mechanism underlying this inhibition is currently unknown (Kallies et al., 2007). A second limitation of the model is that it does not readily explain the probabilistic and cell division–linked nature of the differentiation process (Hasbold et al., 2004). IRF8 and PU.1 represent excellent candidate regulators of the division-linked differentiation process, as they function to prevent premature plasma cell differentiation to allow sufficient clonal expansion of activated B cells and CSR. Further support for this concept comes from our finding that both PU.1 and, more strikingly, IRF8 and SpiB are down-regulated precisely at the point of ASC commitment. In contrast to ASC differentiation, we have no evidence that the increased CSR and Aicda expression we observed in the absence of IRF8 and PU.1 is direct, and as IRF4 is a known activator of Aicda (Sciammas et al., 2006), we propose that this aspect of the phenotype may reflect unrestrained IRF4 activity without IRF8/PU.1.
The expression of IRF4 was unaltered in IRF8/PU.1-deficient B cells, suggesting that it is not a critical target of this complex. IRF4 and IRF8 are closely related and both factors recognize the same DNA sequence, a finding which has led many researchers to conclude that both factors are likely to function in a redundant manner to regulate expression of the same target genes (Lu et al., 2003). However, the contrasting patterns of IRF8 and IRF4 expression and function suggest a more antagonistic model in which these factors bind to the same regulatory regions of critical target genes, resulting in opposite transcriptional outcomes. Specifically, we propose that the IRF8–PU.1 complex is more highly expressed in B cells and predominates at ETS-IRF composite sites, whereas IRF4 expression remains relatively low. Upon B cell activation, IRF4 expression increases, allowing a period of coexpression of all three factors that ultimately results in decreased IRF8 and PU.1, allowing the IRF4-driven ASC differentiation to proceed. Due to the lack of high quality ChIP antibodies, our model at present does not incorporate SpiB, the ETS family member most related to PU.1, whose expression pattern is very similar to IRF8, being expressed in naive, activated, and GC B cells before being silenced in ASCs (Fig. 5 B). Spib expression was unaltered in the absence of IRF8 and PU.1, suggesting that the enhanced ASC differentiation seen in the double-deficient mice occurred in the presence of SpiB. Although redundancy of SpiB with PU.1 and/or differential interaction with IRF proteins remain distinct possibilities, particularly in the GC where SpiB is abundantly expressed, new reagents and mouse models will be required to assess the exact role of SpiB in B cell terminal differentiation.
We have tested two key predictions of our model of B cell terminal differentiation. First, we showed that in the absence of IRF8/PU.1, IRF4 has greater access to shared target genes and thus can promote the ASC fate. Second, we showed that enforced expression of PU.1, and to a lesser extent IRF8, blocks ASC differentiation. This latter finding is in agreement with studies showing that PU.1 expression is under microRNA control in B cells and that increased PU.1 impairs CSR (Vigorito et al., 2007) and terminal differentiation (Lu et al., 2014). It remains to be determined whether the reciprocal functions of IRF4 and IRF8 described here also occur elsewhere in the immune system where IRF proteins play a role, often with alternative partners such as members of the BATF family (Ciofani et al., 2012; Glasmacher et al., 2012; Li et al., 2012; Tussiwand et al., 2012).
MATERIALS AND METHODS
Mice.
Spi1fl-gfp (the floxed allele that also contains GFP knock-in into the 3′ untranslated region) and Spi1fl/flCd19Cre (PU.1cKO) were generated by our group previously (Nutt et al., 2005; Polli et al., 2005). Irf8−/− (Holtschke et al., 1996), Irf8fl/fl (IRF8cKO; Feng et al., 2011), Prdm1gfp/+ (Kallies et al., 2004), Bcl6−/− (Dent et al., 1997), Pax5−/− (Urbánek et al., 1994), and Irf4−/− (Mittrücker et al., 1997) mouse strains have been previously reported. Irf8gfp/gfp mice have eGFP fused to the C terminus of the endogenous IRF8 (Wang et al., 2014). To overcome the lethality of BCL6-deficient mice, fetal liver transplants were performed as follows: E14 embryos from inter-crossed Bcl6+/− (C57BL/6 Ly5.2) mice were genotyped and fetal liver chimeras generated in Rag1−/− (C57BL/6 Ly5.1) recipients and analyzed after >8 wk as described. All mice were in a C57BL/6 genetic background. Animal experiments were conducted according to the protocols approved by the Walter and Eliza Hall Institute animal ethics committee.
Flow cytometry and ELISA.
mAb against mouse B220 (RA3-6B2), CD19 (1D3), IgM (331.12), IgD (1126C), CD21 (7G6), CD23 (B3B4), MacI (M1/70), and human CD4 (OKT4) were purified from hybridoma supernatant and conjugated in the authors’ laboratory. Anti-CD138 (281–2) and Fas (JO2) were obtained from BD and peanut agglutinin (PNA) from Vector Laboratories. Cells were analyzed on LSRII or FACSCanto flow cytometers and cell sorting was performed using FACSDiVa or Aria flow cytometers (BD). For intracellular transcription factor and Ig determination, cells were fixed in paraformaldehyde as previously described (Hasbold et al., 2004) and stained with anti–mouse IgG1 (A85.1) and IgE (23G3; BD). Serum and supernatant Ig amounts were measured using ELISA as described previously (Hasbold et al., 2004).
Cell culture.
Naive lymph node B cells were purified by negative selection using the B cell Isolation kit (Miltenyi Biotec) and cultured as previously described (Hasbold et al., 2004). Cultures were seeded at 105/ml with optimal concentrations of recombinant CD40L (1:200, from Sf21 cells [Hasbold et al., 2004] or 100 ng/ml from R&D Systems), IL-4 (10 ng/ml; R&D Systems), and IL-5 (2-5 ng/ml; R&D Systems). 4 × 105 cells/ml were used for 20 µg/ml LPS ± IL-4 stimulation. For cell division, tracking cells were labeled with CFSE or CTV (Molecular Probes).
Immunization.
Mice were immunized with NP-KLH as previously described (Willis et al., 2014). Ovalbumin immunizations used 20 µg Ovalbumin plus 2.25 mg Aluminum hydroxide (Alum) on days 1 and 14. 1 wk after the second injection, mice then received an aerosol challenge containing Ovalbumin for ∼15 min/d on days 21–24 and blood was taken at day 25.
Western blotting.
The monoclonal antibodies against Bcl6 (7D1), IRF4 (3E4; Kallies et al., 2011), and BLIMP-1 (6D3 or 5E7; Kallies et al., 2004) were previously described. Polyclonal antibodies against IRF8 (sc-6058), PU.1 (T-21), and β-actin (sc-2030) were from Santa Cruz Biotechnology, Inc. Protein extracts corresponding to equal cell numbers were loaded onto the gel subjected to Western blotting using standard techniques.
RT-PCR analysis.
Naive or cultured B cells were sorted and total RNA was prepared with the RNeasy kit (QIAGEN). cDNA was synthesized from total RNA with random hexamers and SuperScript III reverse transcription (Invitrogen). Real-time PCR was performed using GoTaq qPCR SYBR Green Master Mix (Promega). Analyses were done in triplicate and mean normalized expression was calculated with the Q-Gene application with Hmbs as the reference gene. Primer sequences are supplied in Table S3.
Whole transcriptome analysis.
RNA was isolated from either naive or activated (48 h in CD40L+IL-4) B cells, or from flow cytometrically sorted B cell and ASC populations from the Prdm1gfp/+ mice using the RNeasy kit (QIAGEN). Sorted populations were peritoneal B1 cells (B220lowCD23−Mac1+), conventional B2 cells in peritoneal cavity (B220+CD23+Mac1−), follicular B cells (small size, B220+CD23+), GC B cells (B220+Fas+PNA+), spleen ASCs (CD138+BLIMP-1/GFP+), and BM ASCs (CD138+BLIMP-1/GFP+).
Two biological replicates were generated and sequenced for each sample. For all samples, 5 µg RNA was subjected to transcriptome resequencing using 90–100 bp paired-end sequencing on a HiSeq2000 (Illumina) at the Beijing Genomics Institute (Shenzhen, China) or the Australian Genome Research Facility (Melbourne, Australia). Between 12 and 190 million reads were analyzed per sample. Each sample’s read set was aligned to the NCBI37/mm9 build of the Mus musculus genome using the Subread (v1.3.0) and TopHat (v1.0.14) aligners with default parameters (Trapnell et al., 2009; Liao et al., 2013). A raw expression measure for each gene in each sample was calculated by summing the read counts across all annotated RefSeq exons using featureCounts program (Liao et al., 2014). Read sequenced from Ig genes accounted for ∼70% of all mapped reads in ASC samples. These reads were removed from the analysis to allow normalization with all other samples. The gene level raw expression measures were then analyzed using limma or edgeR. Differential expression was assessed using the classic exact test in edgeR (McCarthy et al., 2012), or using empirical Bayes moderated t test for voom normalized data in limma. Multiple testing was controlled using the Benjamini and Hochberg approach.
ChIP.
For ChIP analysis naive B cells were cultured with CD40L and IL-4 for 2 d. ChIP was performed on 5 × 107 B cells with 10 µg antibodies against PU.1 (T-21), IRF8, or IRF4 (all from Santa Cruz Biotechnology, Inc.), as previously described (Boyer et al., 2006). To measure enrichment, quantitative PCR was performed with primers listed in Table S4. Samples processed in the absence of the primary antibody and B cells deficient for the target transcription factor in the presence of the primary antibody served as negative controls. Standard curves were generated from serial dilutions of whole cell extract DNA. Reactions were performed in triplicate with SYBR green PCR master mix (Invitrogen) on an ABI 7900HT machine.
Luciferase assays.
HeLa cells were maintained in DME with 10% FCS. For transient transfection, cells were plated in 6-well plates and transfected using FuGENE 6 according to the manufacturer’s instructions (Roche). The Bcl6 reporter vector encoded the firefly luciferase gene under control of the Bcl6 promoter as previously reported (Lee et al., 2006). 1,200 ng of reporter was used, together with 100–300 ng of expression vector encoding full-length murine cDNAs for IRF8 or PU.1. The total amount of DNA per transfection was normalized using an empty vector (pCDNA3.1). Luciferase activity was determined 24 h after transfection using a Dual-Luciferase assay kit (Promega) and measured with a LUMIstar galaxy luminometer (BMG Labtech).
Retroviral infection.
The MSCV-IRF8-ires-GFP, MSCV-PU.1-ires-hCD4 and control retroviral vectors were generated by transient transfection of 293T cells and used to transduce primary cells as previously reported (Carotta et al., 2010). Purified lymph node B cells were activated with CD40L+IL-4+IL-5 for 30 h (as described above) and used for retroviral transduction. For retroviral transduction, cells were resuspended in 50 µl of media and spun at 2,400 g for 1.5 h in the presence of 1 ml of viral supernatant and 1.25 µg/ml polybrene. After spinning, cells were placed in the same medium for 4 d. Cells were analyzed by flow cytometry at day 5.
Online supplemental material.
Table S1 shows cellularity of splenic B cell populations. Table S2 lists differentially expressed genes during B cell activation. Table S3 shows primers used for quantitative real time PCR. Table S4 shows primers used for quantitative ChIP PCR. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20140425/DC1.
Supplementary Material
Acknowledgments
We thank Axel Kallies and Rhys Allan for helpful discussions, Rebecca Thong, Jamie Leahy, and Nadia Iannarella for technical assistance, and Riccardo Dalla-Favera for plasmid constructs.
This work was supported by a National Health and Medical Research Council (NHMRC) of Australia Program grant (575500 and APP1054925 to D.M. Tarlinton, L.M. Corcoran, P.D. Hodgkin, and S.L. Nutt), Victorian State Government Operational Infrastructure Support, Australian Government NHMRC IRIIS, and the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases (to H.C. Morse III). S. Carotta was supported by an NHMRC Career Development Award, S.H.M. Pang by the Leukaemia Foundation, M. Inouye and S.N. Willis by NHMRC Post-doctoral Fellowships, S.L. Nutt by an Australian Research Council Future Fellowship, and D.M. Tarlinton, L.M. Corcoran, and P.D. Hodgkin by NHMRC Research Fellowships.
The authors declare no competing financial interests.
Author contributions: S. Carotta, S.N. Willis, J. Hasbold, S.H.M. Pang, D. Emslie, A. Light, H. Wang, H.C. Morse III, D.M. Tarlinton, L.M. Corcoran, P.D. Hodgkin, and S.L. Nutt designed and performed experiments; M. Inouye, M. Chopin, and W. Shi provided bioinformatics analysis; and S. Carotta and S.L. Nutt wrote the manuscript.
Footnotes
Abbreviations used:
- ASC
- antibody-secreting cell
- ChIP
- chromatin immunoprecipitation
- CSR
- class-switch recombination
- CTV
- cell trace violet
- GC
- germinal center
References
- Belz, G.T., and Nutt S.L.. 2012. Transcriptional programming of the dendritic cell network. Nat. Rev. Immunol. 12:101–113. 10.1038/nri3149 [DOI] [PubMed] [Google Scholar]
- Boyer, L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 441:349–353. 10.1038/nature04733 [DOI] [PubMed] [Google Scholar]
- Carotta, S., Dakic A., D’Amico A., Pang S.H., Greig K.T., Nutt S.L., and Wu L.. 2010. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity. 32:628–641. 10.1016/j.immuni.2010.05.005 [DOI] [PubMed] [Google Scholar]
- Ciofani, M., Madar A., Galan C., Sellars M., Mace K., Pauli F., Agarwal A., Huang W., Parkurst C.N., Muratet M., et al. 2012. A validated regulatory network for Th17 cell specification. Cell. 151:289–303. 10.1016/j.cell.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunninghame Graham, D.S., Morris D.L., Bhangale T.R., Criswell L.A., Syvänen A.C., Rönnblom L., Behrens T.W., Graham R.R., and Vyse T.J.. 2011. Association of NCF2, IKZF1, IRF8, IFIH1, and TYK2 with systemic lupus erythematosus. PLoS Genet. 7:e1002341. 10.1371/journal.pgen.1002341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager, P.L., Jia X., Wang J., de Bakker P.I., Ottoboni L., Aggarwal N.T., Piccio L., Raychaudhuri S., Tran D., Aubin C., et al. International MS Genetics Consortium. 2009. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat. Genet. 41:776–782. 10.1038/ng.401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker, T., Pasca di Magliano M., McManus S., Sun Q., Bonifer C., Tagoh H., and Busslinger M.. 2009. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 30:508–520. 10.1016/j.immuni.2009.01.012 [DOI] [PubMed] [Google Scholar]
- Deenick, E.K., Hasbold J., and Hodgkin P.D.. 1999. Switching to IgG3, IgG2b, and IgA is division linked and independent, revealing a stochastic framework for describing differentiation. J. Immunol. 163:4707–4714 [PubMed] [Google Scholar]
- DeKoter, R.P., Geadah M., Khoosal S., Xu L.S., Thillainadesan G., Torchia J., Chin S.S., and Garrett-Sinha L.A.. 2010. Regulation of follicular B cell differentiation by the related E26 transformation-specific transcription factors PU.1, Spi-B, and Spi-C. J. Immunol. 185:7374–7384. 10.4049/jimmunol.1001413 [DOI] [PubMed] [Google Scholar]
- Delogu, A., Schebesta A., Sun Q., Aschenbrenner K., Perlot T., and Busslinger M.. 2006. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 24:269–281. 10.1016/j.immuni.2006.01.012 [DOI] [PubMed] [Google Scholar]
- Dent, A.L., Shaffer A.L., Yu X., Allman D., and Staudt L.M.. 1997. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 276:589–592. 10.1126/science.276.5312.589 [DOI] [PubMed] [Google Scholar]
- Duffy, K.R., Wellard C.J., Markham J.F., Zhou J.H., Holmberg R., Hawkins E.D., Hasbold J., Dowling M.R., and Hodgkin P.D.. 2012. Activation-induced B cell fates are selected by intracellular stochastic competition. Science. 335:338–341. 10.1126/science.1213230 [DOI] [PubMed] [Google Scholar]
- Eisenbeis, C.F., Singh H., and Storb U.. 1993. PU.1 is a component of a multiprotein complex which binds an essential site in the murine immunoglobulin lambda 2-4 enhancer. Mol. Cell. Biol. 13:6452–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairfax, K.A., Kallies A., Nutt S.L., and Tarlinton D.M.. 2008. Plasma cell development: from B-cell subsets to long-term survival niches. Semin. Immunol. 20:49–58. 10.1016/j.smim.2007.12.002 [DOI] [PubMed] [Google Scholar]
- Feng, J., Wang H., Shin D.M., Masiuk M., Qi C.F., and Morse H.C. III. 2011. IFN regulatory factor 8 restricts the size of the marginal zone and follicular B cell pools. J. Immunol. 186:1458–1466. 10.4049/jimmunol.1001950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti, S., Barozzi I., Mietton F., Polletti S., De Santa F., Venturini E., Gregory L., Lonie L., Chew A., Wei C.L., et al. 2010. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 32:317–328. 10.1016/j.immuni.2010.02.008 [DOI] [PubMed] [Google Scholar]
- Glasmacher, E., Agrawal S., Chang A.B., Murphy T.L., Zeng W., Vander Lugt B., Khan A.A., Ciofani M., Spooner C.J., Rutz S., et al. 2012. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. 338:975–980. 10.1126/science.1228309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlova, O., Martin J.E., Rueda B., Koeleman B.P., Ying J., Teruel M., Diaz-Gallo L.M., Broen J.C., Vonk M.C., Simeon C.P., et al. Spanish Scleroderma Group. 2011. Identification of novel genetic markers associated with clinical phenotypes of systemic sclerosis through a genome-wide association strategy. PLoS Genet. 7:e1002178. 10.1371/journal.pgen.1002178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasbold, J., Corcoran L.M., Tarlinton D.M., Tangye S.G., and Hodgkin P.D.. 2004. Evidence from the generation of immunoglobulin G-secreting cells that stochastic mechanisms regulate lymphocyte differentiation. Nat. Immunol. 5:55–63. 10.1038/ni1016 [DOI] [PubMed] [Google Scholar]
- Heinz, S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikami, K., Kawasaki A., Ito I., Koga M., Ito S., Hayashi T., Matsumoto I., Tsutsumi A., Kusaoi M., Takasaki Y., et al. 2011. Association of a functional polymorphism in the 3′-untranslated region of SPI1 with systemic lupus erythematosus. Arthritis Rheum. 63:755–763. 10.1002/art.30188 [DOI] [PubMed] [Google Scholar]
- Holtschke, T., Löhler J., Kanno Y., Fehr T., Giese N., Rosenbauer F., Lou J., Knobeloch K.P., Gabriele L., Waring J.F., et al. 1996. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 87:307–317. 10.1016/S0092-8674(00)81348-3 [DOI] [PubMed] [Google Scholar]
- Kallies, A., Hasbold J., Tarlinton D.M., Dietrich W., Corcoran L.M., Hodgkin P.D., and Nutt S.L.. 2004. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J. Exp. Med. 200:967–977. 10.1084/jem.20040973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies, A., Hasbold J., Fairfax K., Pridans C., Emslie D., McKenzie B.S., Lew A.M., Corcoran L.M., Hodgkin P.D., Tarlinton D.M., and Nutt S.L.. 2007. Initiation of plasma-cell differentiation is independent of the transcription factor Blimp-1. Immunity. 26:555–566. 10.1016/j.immuni.2007.04.007 [DOI] [PubMed] [Google Scholar]
- Kallies, A., Carotta S., Huntington N.D., Bernard N.J., Tarlinton D.M., Smyth M.J., and Nutt S.L.. 2011. A role for Blimp1 in the transcriptional network controlling natural killer cell maturation. Blood. 117:1869–1879. 10.1182/blood-2010-08-303123 [DOI] [PubMed] [Google Scholar]
- Kanno, Y., Levi B.Z., Tamura T., and Ozato K.. 2005. Immune cell-specific amplification of interferon signaling by the IRF-4/8-PU.1 complex. J. Interferon Cytokine Res. 25:770–779. 10.1089/jir.2005.25.770 [DOI] [PubMed] [Google Scholar]
- Klein, U., Casola S., Cattoretti G., Shen Q., Lia M., Mo T., Ludwig T., Rajewsky K., and Dalla-Favera R.. 2006. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 7:773–782. 10.1038/ni1357 [DOI] [PubMed] [Google Scholar]
- Kwon, H., Thierry-Mieg D., Thierry-Mieg J., Kim H.-P., Oh J., Tunyaplin C., Carotta S., Donovan C.E., Goldman M.L., Tailor P., et al. 2009. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity. 31:941–952. 10.1016/j.immuni.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.H., Melchers M., Wang H., Torrey T.A., Slota R., Qi C.F., Kim J.Y., Lugar P., Kong H.J., Farrington L., et al. 2006. Regulation of the germinal center gene program by interferon (IFN) regulatory factor 8/IFN consensus sequence-binding protein. J. Exp. Med. 203:63–72. 10.1084/jem.20051450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P., Spolski R., Liao W., Wang L., Murphy T.L., Murphy K.M., and Leonard W.J.. 2012. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 490:543–546. 10.1038/nature11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y., Smyth G.K., and Shi W.. 2013. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41:e108. 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y., Smyth G.K., and Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lu, R., Medina K.L., Lancki D.W., and Singh H.. 2003. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev. 17:1703–1708. 10.1101/gad.1104803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, D., Nakagawa R., Lazzaro S., Staudacher P., Abreu-Goodger C., Henley T., Boiani S., Leyland R., Galloway A., Andrews S., et al. 2014. The miR-155/PU.1 axis acts on Pax-5 to enable efficient terminal B cell differentiation. J. Exp. Med. 211:2183–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, D.J., Chen Y., and Smyth G.K.. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40:4288–4297. 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittrücker, H.W., Matsuyama T., Grossman A., Kündig T.M., Potter J., Shahinian A., Wakeham A., Patterson B., Ohashi P.S., and Mak T.W.. 1997. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 275:540–543. 10.1126/science.275.5299.540 [DOI] [PubMed] [Google Scholar]
- Muto, A., Ochiai K., Kimura Y., Itoh-Nakadai A., Calame K.L., Ikebe D., Tashiro S., and Igarashi K.. 2010. Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. EMBO J. 29:4048–4061. 10.1038/emboj.2010.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt, S.L., Metcalf D., D’Amico A., Polli M., and Wu L.. 2005. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J. Exp. Med. 201:221–231. 10.1084/jem.20041535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt, S.L., Taubenheim N., Hasbold J., Corcoran L.M., and Hodgkin P.D.. 2011. The genetic network controlling plasma cell differentiation. Semin. Immunol. 23:341–349. 10.1016/j.smim.2011.08.010 [DOI] [PubMed] [Google Scholar]
- Ochiai, K., Maienschein-Cline M., Simonetti G., Chen J., Rosenthal R., Brink R., Chong A.S., Klein U., Dinner A.R., Singh H., and Sciammas R.. 2013. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity. 38:918–929. 10.1016/j.immuni.2013.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polli, M., Dakic A., Light A., Wu L., Tarlinton D.M., and Nutt S.L.. 2005. The development of functional B lymphocytes in conditional PU.1 knock-out mice. Blood. 106:2083–2090. 10.1182/blood-2005-01-0283 [DOI] [PubMed] [Google Scholar]
- Pongubala, J.M., Nagulapalli S., Klemsz M.J., McKercher S.R., Maki R.A., and Atchison M.L.. 1992. PU.1 recruits a second nuclear factor to a site important for immunoglobulin kappa 3′ enhancer activity. Mol. Cell. Biol. 12:368–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, M., Gao J., Basso K., Kitagawa Y., Smith P.M., Bhagat G., Pernis A., Pasqualucci L., and Dalla-Favera R.. 2007. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 12:280–292. 10.1016/j.ccr.2007.08.011 [DOI] [PubMed] [Google Scholar]
- Schroder, K., Lichtinger M., Irvine K.M., Brion K., Trieu A., Ross I.L., Ravasi T., Stacey K.J., Rehli M., Hume D.A., and Sweet M.J.. 2007. PU.1 and ICSBP control constitutive and IFN-γ-regulated Tlr9 gene expression in mouse macrophages. J. Leukoc. Biol. 81:1577–1590. 10.1189/jlb.0107036 [DOI] [PubMed] [Google Scholar]
- Sciammas, R., Shaffer A.L., Schatz J.H., Zhao H., Staudt L.M., and Singh H.. 2006. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 25:225–236. 10.1016/j.immuni.2006.07.009 [DOI] [PubMed] [Google Scholar]
- Sciammas, R., Li Y., Warmflash A., Song Y., Dinner A.R., and Singh H.. 2011. An incoherent regulatory network architecture that orchestrates B cell diversification in response to antigen signaling. Mol. Syst. Biol. 7:495. 10.1038/msb.2011.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer, A.L., Yu X., He Y., Boldrick J., Chan E.P., and Staudt L.M.. 2000. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 13:199–212. 10.1016/S1074-7613(00)00020-0 [DOI] [PubMed] [Google Scholar]
- Shaffer, A.L., Lin K.I., Kuo T.C., Yu X., Hurt E.M., Rosenwald A., Giltnane J.M., Yang L., Zhao H., Calame K., and Staudt L.M.. 2002. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 17:51–62. 10.1016/S1074-7613(02)00335-7 [DOI] [PubMed] [Google Scholar]
- Shaffer, A.L., Shapiro-Shelef M., Iwakoshi N.N., Lee A.H., Qian S.B., Zhao H., Yu X., Yang L., Tan B.K., Rosenwald A., et al. 2004. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 21:81–93. 10.1016/j.immuni.2004.06.010 [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef, M., and Calame K.. 2005. Regulation of plasma-cell development. Nat. Rev. Immunol. 5:230–242. 10.1038/nri1572 [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef, M., Lin K.I., McHeyzer-Williams L.J., Liao J., McHeyzer-Williams M.G., and Calame K.. 2003. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 19:607–620. 10.1016/S1074-7613(03)00267-X [DOI] [PubMed] [Google Scholar]
- Shin, D.M., Lee C.H., and Morse H.C. III. 2011. IRF8 governs expression of genes involved in innate and adaptive immunity in human and mouse germinal center B cells. PLoS ONE. 6:e27384. 10.1371/journal.pone.0027384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokalski, K.M., Li S.K., Welch I., Cadieux-Pitre H.A., Gruca M.R., and DeKoter R.P.. 2011. Deletion of genes encoding PU.1 and Spi-B in B cells impairs differentiation and induces pre-B cell acute lymphoblastic leukemia. Blood. 118:2801–2808. 10.1182/blood-2011-02-335539 [DOI] [PubMed] [Google Scholar]
- Stehling-Sun, S., Dade J., Nutt S.L., DeKoter R.P., and Camargo F.D.. 2009. Regulation of lymphoid versus myeloid fate ‘choice’ by the transcription factor Mef2c. Nat. Immunol. 10:289–296. 10.1038/ni.1694 [DOI] [PubMed] [Google Scholar]
- Taubenheim, N., Tarlinton D.M., Crawford S., Corcoran L.M., Hodgkin P.D., and Nutt S.L.. 2012. High rate of antibody secretion is not integral to plasma cell differentiation as revealed by XBP-1 deficiency. J. Immunol. 189:3328–3338. 10.4049/jimmunol.1201042 [DOI] [PubMed] [Google Scholar]
- Todd, D.J., McHeyzer-Williams L.J., Kowal C., Lee A.H., Volpe B.T., Diamond B., McHeyzer-Williams M.G., and Glimcher L.H.. 2009. XBP1 governs late events in plasma cell differentiation and is not required for antigen-specific memory B cell development. J. Exp. Med. 206:2151–2159. 10.1084/jem.20090738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C., Pachter L., and Salzberg S.L.. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 25:1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunyaplin, C., Shaffer A.L., Angelin-Duclos C.D., Yu X., Staudt L.M., and Calame K.L.. 2004. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J. Immunol. 173:1158–1165. 10.4049/jimmunol.173.2.1158 [DOI] [PubMed] [Google Scholar]
- Tussiwand, R., Lee W.L., Murphy T.L., Mashayekhi M., Kc W., Albring J.C., Satpathy A.T., Rotondo J.A., Edelson B.T., Kretzer N.M., et al. 2012. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature. 490:502–507. 10.1038/nature11531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbánek, P., Wang Z.Q., Fetka I., Wagner E.F., and Busslinger M.. 1994. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 79:901–912. 10.1016/0092-8674(94)90079-5 [DOI] [PubMed] [Google Scholar]
- Vigorito, E., Perks K.L., Abreu-Goodger C., Bunting S., Xiang Z., Kohlhaas S., Das P.P., Miska E.A., Rodriguez A., Bradley A., et al. 2007. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 27:847–859. 10.1016/j.immuni.2007.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., and Morse H.C. III. 2009. IRF8 regulates myeloid and B lymphoid lineage diversification. Immunol. Res. 43:109–117. 10.1007/s12026-008-8055-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Lee C.H., Qi C., Tailor P., Feng J., Abbasi S., Atsumi T., and Morse H.C. III. 2008. IRF8 regulates B-cell lineage specification, commitment, and differentiation. Blood. 112:4028–4038. 10.1182/blood-2008-01-129049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Yan M., Sun J., Jain S., Yoshimi R., Abolfath S.M., Ozato K., Coleman W.G.J. Jr, Ng A.P., Metcalf D., et al. 2014. A reporter mouse reveals lineage-specific and heterogeneous expression of IRF8 during lymphoid and myeloid cell differentiation. J. Immunol. 193:1766–1777. 10.4049/jimmunol.1301939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis, S.N., Good-Jacobson K.L., Curtis J., Light A., Tellier J., Shi W., Smyth G.K., Tarlinton D.M., Belz G.T., Corcoran L.M., et al. 2014. Transcription factor IRF4 regulates germinal center cell formation through a B cell-intrinsic mechanism. J. Immunol. 192:3200–3206. 10.4049/jimmunol.1303216 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.