Abstract
Novel cellular behaviors and characteristics can be obtained by coupling engineered gene networks to the cell's natural regulatory circuitry through appropriately designed input and output interfaces. Here, we demonstrate how an engineered genetic circuit can be used to construct cells that respond to biological signals in a predetermined and programmable fashion. We employ a modular design strategy to create Escherichia coli strains where a genetic toggle switch is interfaced with: (i) the SOS signaling pathway responding to DNA damage, and (ii) a transgenic quorum sensing signaling pathway from Vibrio fischeri. The genetic toggle switch endows these strains with binary response dynamics and an epigenetic inheritance that supports a persistent phenotypic alteration in response to transient signals. These features are exploited to engineer cells that form biofilms in response to DNA-damaging agents and cells that activate protein synthesis when the cell population reaches a critical density. Our work represents a step toward the development of “plug-and-play” genetic circuitry that can be used to create cells with programmable behaviors.
Keywords: heterologous gene expression, synthetic biology, Escherichia coli
The engineering of gene regulatory networks is a cornerstone of synthetic biology (1, 2) and has been instrumental in elucidating basic principles that govern the dynamics of small gene networks (3-14) and the origins and consequences of stochasticity in gene expression (13-18). In addition, gene circuits designed to perform sophisticated computational tasks, such as memory storage and logical operations, may support biotechnological and biomedical applications where they “program” cellular behaviors (19-22). However, most networks of this type are designed to respond to nonendogenous, externally applied stimuli. To make full use of the customizable computational capabilities of engineered gene networks in programmable cells, such networks must be designed to respond to endogenously generated signals and be coupled directly to the regulatory circuitry of the cell.
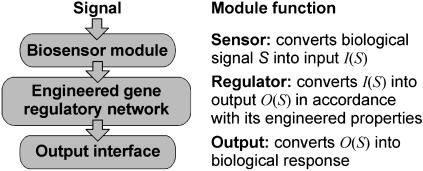
Many cell regulatory systems are organized as modules (23-25) and a similar design strategy may allow the construction of cells with desired behaviors and characteristics. We envision that engineered gene networks can be used as regulatory modules and interfaced with the cell's genetic circuitry as “plug-and-play” devices to execute specific programs in response to particular biological signals. The simplest programmable cell obtained with this design strategy would be comprised of three distinct modules (Fig. 1): (i) a signaling pathway (the biosensor module) that detects relevant signals and interfaces these signals to a regulatory circuit, (ii) an artificial genetic module (the regulatory circuit) capable of responding to the signals transmitted by the biosensor module, and directing output signals according to its engineered properties, and (iii) an output interface that converts the signals transmitted by the regulatory circuit into a biological response. The behavior of the programmed cell is then determined by the dynamical and logical properties of the regulatory module and by the signaling pathways that are used as input and output interfaces.
Fig. 1.
The modular structure of a simple programmable cell.
As concrete demonstrations of this modular design strategy, we have created the four Escherichia coli strains listed in Table 1. In these strains, a genetic toggle switch (4) is interfaced with two different signaling pathways: (i) the SOS signaling pathway (strains A1 and A2), which detects single-stranded DNA after DNA damage (26-28), and (ii) a transgenic quorum sensing signaling pathway from Vibrio fischeri (29-31) that detects acyl-homoserine lactone (AHL) molecules (strains B1 and B2). The V. fischeri signaling pathway, which has been exploited to engineer whole-cell biosensors (32, 33) and cell-cell communication (5, 10, 34) systems in E. coli, is used to program a strain (B2) that synthesizes a target protein when the cell population reaches a critical density. We mainly employ GFP as a quantifiable biological response, but also demonstrate that the design strategy can be used to control a natural phenotype by creating a strain (A2) that enters a biofilm-forming state in response to transient activation of the SOS pathway. Although the engineering of cellular behavior is not novel, most existing programmed cells are designed for specific purposes, such as whole-cell biosensing (35-39), programmed self-destruction (40-43), and protein synthesis controlled by excess glucolytic flux (44) or cell density (33, 45). The modular approach that we propose would facilitate a standardization of genetic circuits that, in analogy to electronic circuit modules, could be used as general components in the construction of programmable cells for a variety of biotechnological and bioengineering applications.
Table 1. The circuit components and characteristics of the four E. coli strains constructed for this study.
| Strain | Circuit components | Characteristics |
|---|---|---|
| A1 | Sensor: the SOS pathway | Detects and retains memory of DNA damage |
| Regulator: toggle switch plasmid pTSMa | ||
| Output: GFP reporter plasmid pCIRa | ||
| A2 | Sensor: the SOS pathway | Forms biofilm in response to DNA damage |
| Regulator: toggle switch plasmid pTSMa | ||
| Output: biofilm plasmid pBFR | ||
| B1 | Sensor: AHL inducible plasmid pAHLa | Detects and retains memory of quorum sensing molecules |
| Regulator: toggle switch plasmid pTSMb1 | ||
| Output: polycistronic GFP expression | ||
| B2 | Sensor: AHL self-inducible plasmid pAHLb | Density dependent protein synthesis |
| Regulator: toggle switch plasmid pTSMb2 | ||
| Output: GFP reporter plasmid pCIRb |
Experimental Procedures
Strains, Plasmids, and Genes. The four strains listed in Table 1 were obtained by transforming parental E. coli strains (JM 2.300 for strains A1, B1, and B1) with the indicated plasmids. The A2 strain was obtained by using a modified K-12 parental strain (see supporting information, which is published on the PNAS web site). All plasmids were derived from the published pTAK plasmids (4) and pZ expression vectors (46) by using standard cloning techniques. A full description of plasmids and genes is given in the supporting information.
Fluorescence Measurements. GFP expression was quantified by using a FACSCalibur flow cytometer. Samples were prepared by pelleting cells from 1 ml of culture followed by resuspension in phosphate-buffered saline.
DNA Damage. Cells were grown aerobically in LB medium containing the appropriate antibiotics at 37°C and 300 rpm. Colonies were picked from selective plates and grown for 17-24 h, followed by an additional 16 h in medium containing 2 mM isopropyl-β-thiogalactopyranoside (IPTG). DNA damage was induced with mitomycin C (MMC) or UV irradiation. In the experiments with MMC treatment, the IPTG-containing culture was used to inoculate fresh LB medium with different MMC concentrations and grown for 15 h. The MMC-treated cells were grown for 3-56 h with dilutions every 12 h to keep the cells in the logarithmic growth phase. In the experiments with UV treatment, cells were plated and incubated for 2 h at 30°C before being exposed to irradiation (Stratalinker 2400) for 1-10 s. Cells were subsequently collected and grown in fresh medium for 4 h before being filtered (0.22-μm Millipore Millex-GV membrane filter) and assayed.
Biofilm Formation. Cells were grown aerobically in M63 minimal medium [1.052 g/liter KH2PO4/5.613 g/liter K2HPO4/2.0 g/liter (NH4)2SO4/0.50 mg FeSO4(H2O)7/1.0 mmol MgSO4, pH 7.2] containing 0.2% glucose and appropriate antibiotics at 37°C and 300 rpm. After exposure to MMC or UV irradiation, a small number of cells were used to inoculate 100-μl fresh M63 loaded into 96-well polystyrene plates. The plates were incubated for 24 h before the level of biofilm was quantified by using a crystal violet staining assay (47). Absorbance at 600 nm was measured by using a TECAN SPECTRAfluor Plus plate reader. Microfermentor experiments were carried out by using 20-ml continuous-flow fermentors (flow rate, 13 ml/h), stirred by aeration with sterile air and containing submerged Corning glass plates as the substratum for the biofilm. The fermentors were inoculated with 10 μl of culture treated with MMC as described above. Digital pictures were taken 48 h later.
AHL-Dependent Expression. All experiments involving the strains B1 and B2 were carried out in LB medium at 30°C unless otherwise stated. Cells were kept in the logarithmic growth phase by dilutions at appropriate intervals. AHL used to induce strain B1 [N-(β-ketocaproyl)-l-homoserine lactone] was obtained from Sigma. Cells with high and low initial GFP expression were obtained by growth in medium containing 2 mM IPTG for 12 h and growth at 42°C for 12 h, respectively. The cells were subsequently washed and used to inoculate fresh medium. The density-dependent expression experiment was carried out by growing the transformed cells on selective plates containing 2 mM IPTG, followed by growth at very low cell densities for 8 h in LB containing 2 mM IPTG. Cells were subsequently pelleted, washed three times, and used to inoculate batch cultures at various initial cell densities. The absorbance (cell density) of the cultures at 600 nm (A600) was determined with a SPECTRAfluor Plus plate reader.
Results
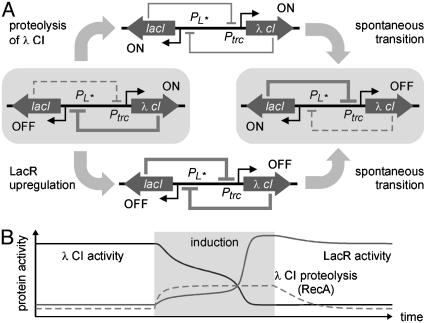
Rational Design of Interface Modules. When interfacing an engineered gene network into the genetic circuitry of the cell, the first step is to achieve an in-depth understanding of the network's dynamic properties. In our case, the regulatory circuit (a genetic toggle switch) is comprised of two genes, lacI and λ cI, that encode the transcriptional regulator proteins, LacR and λ CI. The lacI gene is expressed from a modified PL promoter, PL*, which is repressed by λ CI. The λ cI gene is expressed from a promoter, Ptrc, which is repressed by LacR. This design endows cells with two distinct phenotypic states (4): one where the λ CI activity is high and the expression of lacI is low, and one where the activity of LacR is high and the expression of λ cI is low (Fig. 2A).
Fig. 2.
Transitions in the genetic toggle switch. (A) The network has two stable expression states (indicated by gray boxes) where λ CI represses lacI expression and LacR represses λ cI expression, respectively. Transition from the high λ CI state can be induced by degrading λ CI or by introducing additional LacR molecules. (B) Simulated transition induced when the rate of λ CI proteolysis is temporarily increased under inducing conditions.
There are two ways perturbations can cause a transition from one stable expression state to the other: (i) the activity of the protein that is highly expressed can be decreased, or (ii) the activity of the protein whose expression is repressed can be increased. These transitions are illustrated in Fig. 2 A for the cases where the perturbations cause a transition from the high λ CI/low LacR state to the high LacR/low λ CI state. When λ CI activity is decreased, lacI expression is derepressed and LacR activity increases. This represses λ cI expression, which decreases λ CI activity and further increases LacR activity. The same result can be achieved with a perturbation that increases the activity of LacR. In both cases, a transition from one stable state to the other occurs if the perturbation is sufficiently large to bring the system across a certain threshold (see supporting information).
Transitions from one stable state to the other can be induced by high-amplitude random fluctuations, referred to as noise-induced transitions (48), or by signals that temporarily change the parameters of the system. In bistable gene circuits, noise-induced transitions can cause individual cells to change expression state at random (49). The result is the emergence of a mixed population consisting of cells in different expression states, which appears as a bimodal population distribution when protein levels are measured in single cells (7, 13, 50).
The genetic toggle switch is a robust bistable system, and noise-induced transitions are rare (4). In such systems, transitions from one stable state to the other can be induced by a signal that temporarily brings the system out of the region of bistability. A mathematical analysis (see supporting information) indicates that transitions from the high λ CI state to the high LacR state can be induced by a signal that temporarily increases (i) the λ CI decay rate or (ii) the LacR basal synthesis rate. The simulated response of a single cell to such signals is shown in Fig. 2B. It illustrates how a cell initially in the high λ CI state switches to the high LacR state as a result of a transient increase in λ CI proteolysis. Increasing the basal LacR synthesis rate gives a similar response (see supporting information). In both cases, a transition to the high LacR state occurs when the signal reaches a threshold value where the high λ CI state is destabilized. Because individual cells have slightly different threshold values, due, for instance, to variability in plasmid copy number, and because the probability of a noise-induced transition increases as the bifurcation parameter approaches the threshold value (48), it is expected that intermediate signals will give rise to bimodal population distributions.
Guided by the mathematical analysis, we interfaced the toggle switch with a natural signaling pathway that increases the rate of λ CI decay and an engineered signaling pathway that increases the rate of LacR synthesis, respectively. The signaling pathway that degrades λ CI in strains A1 and A2 (Table 1) is the SOS-response pathway, where the RecA coprotease is activated in the presence of single-stranded DNA (24). Activated RecA cleaves the λ CI repressor protein, causing derepression of the PL promoter (51). The signaling pathway that increases the basal expression of the lacI gene in strains B1 and B2 (Table 1) is based on the quorum sensing pathway V. fischeri (29-31). In this pathway, the regulator protein of the lux operon, LuxR, is induced by AHL, and the induced LuxR protein activates expression from the lux promoter, PluxI. By placing the lacI gene downstream of PluxI, the rate of LacR synthesis is increased when AHL molecules are present in the environment.
Strain A1: Interfacing the SOS Pathway. Interfacing the genetic toggle switch (the regulatory circuit) with the SOS network (the biosensor module) required a series of alterations of the original pTAK plasmid (4). The toggle switch plasmid (pTSMa, see Fig. 3A) was made by replacing the cI857 gene, which encodes a λ CI variant that is cleaved inefficiently by RecA (52), with wild-type λ cI, and by changing the origin of replication to decrease the plasmid copy number. This was required to achieve compatibility between the biosensor module and the regulatory circuit (see below). As the output interface, we used a medium-copy number reporter plasmid (pCIRa), carrying a fusion of PL* and the gfp gene (Fig. 3A).
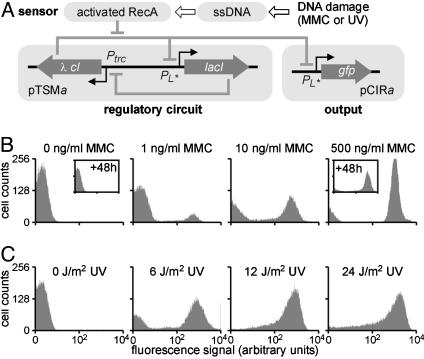
Fig. 3.
Interfacing the SOS signaling pathway in strain A1. (A) Diagram of the engineered genetic circuitry. The genetic toggle switch module (pTSMa) controls the expression of GFP from plasmid pCIRb in response to DNA damage. (B) Induction of GFP expression after exposure to MMC. (C) Induction of GFP expression after 1-10 s of UV irradiation.
To evaluate the ability of the modified toggle switch to respond to activation of the SOS pathway, we quantified GFP expression in single cells 3-6 h after exposure to various concentrations of MMC for 15 h. In the absence of MMC, all cells exhibited little or no GFP expression (Fig. 3B). Nearly all of the cells expressed GFP after treatment with 500 ng/ml MMC. The high and low GFP expression states remained unchanged after 48 h of additional growth without MMC (see Fig. 3B Insets). These findings confirm that the two expression states coexist in the modified toggle switch and that these states are robust against noise-induced transitions. Bimodal distributions were observed at intermediate MMC concentrations (Fig. 3B), probably because of variability in plasmid copy number and the resultant differences in cellular λ CI concentrations giving rise to variability in induction threshold.
The A1 strain can also detect brief exposures (<10 s) to UV irradiation (Fig. 3C). As in the experiment with MMC, UV irradiation at intermediate intensities induces a binary cellular response, resulting in bimodal population distributions. In both MMC- and UV-treated cells, the feedback architecture of the toggle switch module prevents expression of the λ cI gene, even after the damaged DNA has been repaired and cells resume their pretreatment activities (see Fig. 2B). This allows cells to retain memory of DNA damage over successive generations, as demonstrated by the high expression state >48 h (corresponding to 50-60 generations) after the removal of MMC (see Fig. 3B).
Detecting DNA Damage with Strain A1. Strain A1 is a highly sensitive sensor of DNA damaging agents. Treatment with 1 ng/ml and 10 ng/ml MMC (Fig. 3B) gave a 1.9-fold and 19-fold increase in the population-averaged fluorescence signal (geometric mean), respectively. For comparison, the two sensor strains developed by Vollmer et al. (36) showed a 1.8-fold and 5.0-fold increase in the detected signal in response to 10 ng/ml MMC, whereas Kostrzynska et al. (37) reported a minimum detection limit of 4 ng/ml MMC (0.012 μM). In addition, the response of the A1 strain to UV irradiation at 6 J/m2 and 12 J/m2 was a 44-fold and 250-fold increase in average fluorescence (Fig. 3C). This represents a significant improvement in yield compared to previous reports of 4- to 5-fold increases in signal intensity at 10 J/m2 (37, 38).
To evaluate how the architecture of the regulatory circuit affects the ability of the A1 strain to detect DNA damage, we tested the response to MMC treatment of a strain that contains a regulatory circuit identical to the pTSMa toggle switch, except that it lacks the lacI feedback gene (plasmid pCIE). Fluorescence could not be detected after 15-h treatments at concentrations <1,000 ng/ml. A relatively weak fluorescence signal was detected when pCIE/pCIRa cells were assayed 30-60 min after the removal of MMC at concentrations between 1,000 and 4,000 ng/ml. The poor sensitivity and yield are probably due to the cellular activity of RecA being unable to cleave λ CI at a sufficient rate (see supporting information for further discussion). However, GFP expression could not be detected in cells assayed 3 h after the removal of MMC. This indicates that the PL* promoter is active only for a limited time period after DNA damage in the circuit lacking the lacI gene. Comparing these results with those obtained from the A1 strain demonstrates that the feedback architecture of the genetic toggle switch provides at least a 1,000-fold improvement in sensitivity and enables readout of a detection event long after the DNA-damaging agent is removed. The latter could significantly improve the signal-to-noise ratio, because this feature allows for long signal integration. The disadvantages of a toggle switch-based biosensor include a loss of temporal information and a requirement of resetting, i.e., application of IPTG (4), between detection events.
Strain A2: Permanent Phenotypic Alteration. The above experiments indicate that the epigenetic inheritance capabilities of the genetic toggle switch might enable a permanent phenotypic change in response to a transient signal. To demonstrate this feature, we transferred the control of biofilm formation from the cell's natural circuitry to the genetic toggle switch in strain A2. This was done by deleting the traA gene (53) from the genome of the host strain and by constructing a biofilm-forming output plasmid (pBFR) where the expression of the traA gene is controlled by the PL* promoter. The engineered regulatory circuits of the A2 strain are illustrated in Fig. 4A. In this strain, the traA gene is constitutively expressed when the cells are in the high LacR/low λ CI state. As a result, the strain is programmed to produce biofilm only when it has been subjected to DNA damage.
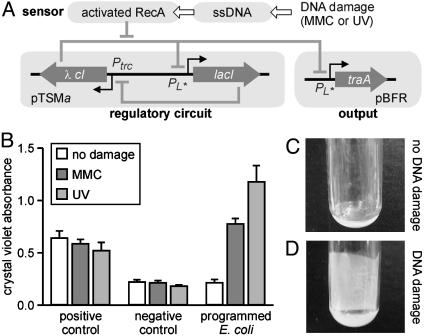
Fig. 4.
Example of programmed phenotype in strain A2. (A) Diagram of the engineered genetic circuitry. The genetic toggle switch module (pTSMa) controls the expression of traA from plasmid pBFR in response to DNA damage. (B) Biofilm formation quantified by crystal violet staining in cultures of strain K12/AK4 (positive control), strain K12/AK3 (negative control), and strain A2 (programmed E. coli). (C and D) Pictures of microfermentors incubated with untreated cells (C) or cells treated with MMC (D).
Biofilm formation experiments were carried out by using a strain that has the traA gene and a strain that lacks the traA gene as the positive and negative controls, respectively. The level of biofilm was measured quantitatively by using a crystal violet microtiter absorbance assay (see Experimental Procedures) for untreated cells, cells treated with 100 ng/ml MMC for 15 h, and cells exposed to 8 J/m2 UV irradiation before inoculation of the microplate. The strain lacking the traA gene (the negative control) and the strain with the traA gene (the positive control) gave low and high absorbance signals, respectively, regardless of DNA damage (Fig. 4B). The A2 strain with the toggle switch-controlled traA gene generated a high signal indicative of biofilm formation only after exposure to MMC or UV irradiation (Fig. 4B). We confirmed this observation by using microfermentor experiments (Fig. 4 C and D) where the biofilm formed after MMC treatment can be detected visually (Fig. 4D). We also confirmed that prolonged traA expression, i.e., a persistent phenotypic alteration, is necessary for biofilm formation. In separate control experiments, biofilm was only observed if traA was expressed for >4 h (see supporting information). Such sustained expression after a brief signal, e.g., a 2-s UV pulse, is enabled by the memory property of the genetic toggle switch.
Strain B1: Interfacing General Input Signals. The experiments described above demonstrate that a natural signaling pathway can be interfaced with an engineered gene network. However, those studies exploit a preexisting molecular compatibility: the λ CI protein is naturally cleaved upon the activation of the SOS pathway. As indicated by the mathematical analysis (supporting information), a transition between stable expression states in the genetic toggle switch can also occur if the expression of the repressed transcription factor protein is increased in response to an incoming signal. Thus, in principle, any cellular signal that activates the expression from a bacterial promoter might be used to couple the genetic toggle switch to natural regulatory circuits.
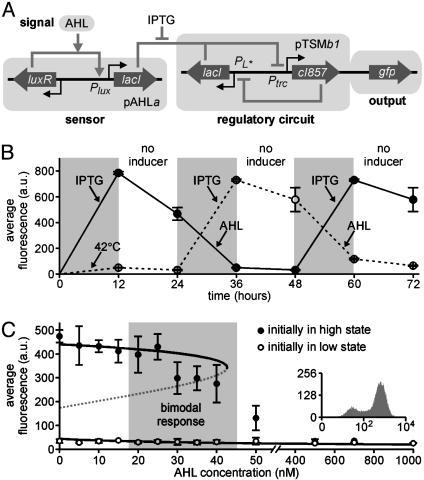
To demonstrate the generality of the input interface, and the plug-and-play features of the design strategy, we created a strain (B1) where a biosensor of AHL molecules interacts with the genetic toggle switch via the lacI gene. The engineered regulatory circuitry in the B1 strain (Fig. 5A) consists of a low-copy number AHL sensor plasmid (pAHLa), carrying a fusion of the lacI gene and the luxR-PluxI fragment from the V. fischeri lux operon, and a medium-copy number toggle switch plasmid (pTSMb1). In this strain, the toggle switch plasmid carries a copy of the gfp gene, such that cells fluoresce in the high λ CI/low LacR state.
Fig. 5.
Interfacing an AHL biosensor module in strain B1. (A) Diagram of the engineered genetic circuitry. (B) Repeated activation and deactivation of GFP expression by using IPTG and AHL, respectively. Cultures were induced for 12 h, as indicated, followed by growth for 12 h without inducers. (C) Population-averaged fluorescence signal in the presence of AHL. The cell population is partially induced (bimodal response, see Inset) at intermediate AHL concentrations. Open and closed circles in B and C indicate fluorescence measured in populations initially grown at 42°C and treated with IPTG, respectively.
The architecture of the regulatory circuitry in strain B1 (Fig. 5A) means that GFP expression should be activated by transient treatment with IPTG and deactivated by transient exposure to AHL. Fig. 5B shows the result of repeated treatments with IPTG or AHL for 12 h followed by a 12-h period (corresponding to 12-15 generations) where the inducing signals were absent. Over a 72-h time period, the cells were successfully switched back and forth between expression states three times, confirming that the engineered circuits remain functional over many cell generations. The partial decrease in fluorescence observed 12 h after removal of IPTG (Fig. 5B) reflects the relaxation from a state where LacR is completely inactive to a stable state where λ CI is the dominant repressor, but LacR still has some basal activity. The stability of the distinct expression states was confirmed in a separate control experiment where stable expression was observed for up to 50 h (corresponding to 50-60 generations) after the removal of the inducing factor (see supporting information).
To evaluate the switching dynamics and the sensitivity of the B1 strain, we conducted a series of experiments where cells initially in the high or low GFP expression states were exposed to AHL at various concentrations for 24 h (Fig. 5C). Regardless of the concentration of AHL, cells that were initially in the high LacR state (low GFP expression, open circles in Fig. 5C) remained in this state. Cells initially in the high λ CI state (high GFP expression, closed circles in Fig. 5C) remained in that state at AHL concentrations <20 nM. All cells switched to the low GFP state when treated with AHL at 50 nM concentration or higher. Bimodal population distributions (Fig. 5C Inset) were observed at AHL concentrations between 20 and 50 nM. It is clear from Fig. 5 B and C that the long-term stability of the two expression states and the switching properties of the A1 strain (see Fig. 3B) are preserved in the B1 strain.
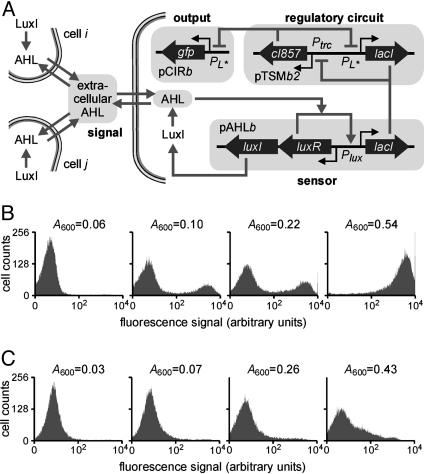
Strain B2: Density-Dependent Gene Activation. AHL is a natural biological signal secreted by Gram-negative bacteria as a means of coordinating cellular activity with the cell population density (29-31). To enable the E. coli population to measure its own density through AHL, we created the plasmid pAHLb where the luxI gene from V. fischeri is expressed polycistronically with the luxR gene and lacI is expressed from the PluxI promoter (Fig. 6A). The protein encoded by luxI is a synthetase that converts common precursor metabolites into AHL signaling molecules (29-31), and the extracellular concentration of AHL correlates with the cell density in cultures of cells that carry the luxI gene. As a result, LuxR should be activated, and lacI expression from the pAHLb plasmid increased, when the cell density increases.
Fig. 6.
Density-dependent gene activation in strain B2. (A) Diagram of the engineered genetic circuitry. The luxI and luxR genes are expressed constitutively. (B) Cell density-dependent expression of GFP. (C) The expression of GFP in cells that lack luxI and are unable to produce AHL.
To construct a strain where the expression of a target gene is induced at a critical cell density, we cotransformed three different plasmids to create the B2 strain (Table 1): the low-copy pAHLb plasmid regulating lacI expression, the medium-copy toggle switch plasmid pTSMb2, and the high-copy reporter plasmid pCIRb (Fig. 6A). A strain lacking the luxI gene (plasmid pAHLa) was used as a negative control. To evaluate the dependence of GFP expression on cell density, we inoculated cultures with different numbers of cells, and assayed them after 14 h of growth. In cultures with low or high densities (A600 = 0.06 and A600 = 0.56), all cells were observed to express GFP at very low or very high levels, respectively (Fig. 6B). At intermediate cell densities (A600 = 0.10 and A600 = 0.22), the population distribution contains two peaks (i.e., a bimodal response). Negative control experiments showed that GFP synthesis remained repressed at all densities in cultures of cells lacking the luxI gene (Fig. 6C). The high GFP expression observed in Fig. 6B can thus be attributed to the engineered circuit sensing that the cell population has reached a critical density. Experiments where cultures were inoculated with equal numbers of cells but incubated for different time periods gave similar results (data not shown).
Because of the modular design of this system, density-dependent synthesis of any protein can be achieved simply by replacing the gfp gene on the high-copy number reporter plasmid with a gene of interest. For example, programmed population control could be achieved by replacing gfp with a killer gene, as it was recently shown (34), by fusing the ccdB gene to the PluxI promoter and synthesizing LuxR and LuxI constitutively inside E. coli cells. Moreover, the sharp switching threshold of our system might be useful in industrial-scale production of proteins that inhibit cell growth because the target protein is synthesized only when the population has reached a high density.
Discussion
This study has demonstrated how programmable cells can be constructed by designing appropriate interfaces that couple engineered gene networks to the regulatory circuitry of the cell. An engineered genetic toggle switch (4) was used as a signal processing circuit to construct strains with binary switching responses and persistent changes in gene expression patterns to biological signals. The memory capability of the toggle switch, which allows cells to indefinitely store a record of a detection event for later interrogation, was exploited to “program” a strain with a specific phenotypic response (i.e., biofilm formation). We also constructed a strain where the expression of a target gene is controlled in a binary on/off fashion when the cell density reaches a critical value. In this strain, it is the sharp switching dynamics of the genetic toggle switch, rather than its epigenetic memory, that is important for the desired cell function. Our work shows that programmable cells can be assembled by using a modular design strategy, paving the way for the development of “plug-and-play” genetic circuit devices.
Our investigations also revealed some of the current challenges in constructing artificial gene circuits with sophisticated dynamical and computational properties. Interfacing these circuits with natural signaling pathways (or with each other) requires that the signals (e.g., activating or repressing transcription factors) are appropriately adjusted to allow effective information transmission between circuit modules while, at the same time, maintaining the proper function of the system as a whole. In many cases, the properties of the system must be optimized rather than those of the individual components (see supplemental information). In this respect, the modular design strategy could benefit significantly from the development of directed evolution technologies (54, 55) that can select for nontrivial dynamical behaviors. Moreover, more complex gene regulatory modules and interfaces need to be constructed to fully realize the capabilities of modular genetic control circuits. This could enable sophisticated processing capabilities, including event counting and signal integration.
Acknowledgments
This work was supported by the Department of Energy, the National Science Foundation, the Defense Advanced Research Program Agency, the Army Research Laboratory, and the Danish Research Agency.
Abbreviations: IPTG, isopropyl-β-thiogalactopyranoside; MMC, mitomycin C; AHL, acylhomoserine lactone.
References
- 1.Benner, S. A. (2003) Nature 421, 118. [DOI] [PubMed] [Google Scholar]
- 2.Ferber, D. (2004) Science 303, 158-161. [DOI] [PubMed] [Google Scholar]
- 3.Elowitz, M. B. & Leibler, S. (2000) Nature 403, 335-338. [DOI] [PubMed] [Google Scholar]
- 4.Gardner, T. S., Cantor, C. R. & Collins, J. J. (2000) Nature 403, 339-342. [DOI] [PubMed] [Google Scholar]
- 5.Tchuraev, R. N., Stupak, I. V., Tropynina, T. S. & Stupak, E. E. (2000) FEBS Lett. 486, 200-202. [DOI] [PubMed] [Google Scholar]
- 6.Weiss, R. & Knight, T. F., Jr. (2000) in Sixth International Meeting on DNA-Based Computers, DNA6, eds. Condon, A. & Rozenberg, G. (Springer, Heidelberg), pp. 1-16.
- 7.Becskei, A., Seraphin, B. & Serrano, L. (2001) EMBO J. 20, 2528-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guet, C. C., Elowitz, M. B., Hsing, W. & Leibler, S. (2002) Science 296, 1466-1470. [DOI] [PubMed] [Google Scholar]
- 9.Rosenfeld, N., Elowitz, M. B. & Alon, U. (2002) J. Mol. Biol. 323, 785-793. [DOI] [PubMed] [Google Scholar]
- 10.Basu, S., Karig, D. & Weiss, R. (2002) in Eighth International Meeting on DNA-Based Computers, DNA8, eds. Hagiya, M. & Ohuchi, A. (Springer, Heidelberg), pp. 61-72.
- 11.Atkinson, M. R., Savageau, M. A., Myers, J. T. & Ninfa, A. J. (2003) Cell 113, 597-607. [DOI] [PubMed] [Google Scholar]
- 12.Mangan, S., Zaslaver, A. & Alon, U. (2003) J. Mol. Biol. 334, 197-204. [DOI] [PubMed] [Google Scholar]
- 13.Isaacs, F. J., Hasty, J., Cantor, C. R. & Collins, J. J. (2003) Proc. Natl. Acad. Sci. USA 100, 7714-7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaern, M., Blake, W. J. & Collins, J. J. (2003) Annu. Rev. Biomed. Eng. 5, 179-206. [DOI] [PubMed] [Google Scholar]
- 15.Becskei, A. & Serrano, L. (2000) Nature 405, 590-593. [DOI] [PubMed] [Google Scholar]
- 16.Ozbudak, E. M., Thatti, M., Kurtser, I., Grossman, A. D. & van Oudenaarden, A. (2002) Nat. Genet. 31, 69-73. [DOI] [PubMed] [Google Scholar]
- 17.Elowitz, M. B., Levine, A. J., Siggia, E. D. & Swain, P. S. (2002) Science 297, 1183-1186. [DOI] [PubMed] [Google Scholar]
- 18.Blake, W. J., Kaern, M., Cantor, C. R. & Collins, J. J. (2003) Nature 422, 633-637. [DOI] [PubMed] [Google Scholar]
- 19.Simpson, M. L., Sayler, G. S., Fleming, J. T. & Applegate, B. (2001) Trends Biotechnol. 19, 317-321. [DOI] [PubMed] [Google Scholar]
- 20.Hasty, J., McMillen, D. & Collins, J. J. (2002) Nature 420, 224-230. [DOI] [PubMed] [Google Scholar]
- 21.Weber, W. & Fussenegger, M. (2002) J. Biotechnol. 98, 161-187. [DOI] [PubMed] [Google Scholar]
- 22.Weiss, R., Basu, S., Hooshangi, S., Kalmbach, A., Karig, D., Mehreja, R. & Netravali, I. (2003) Nat. Comput. 2, 47-84. [Google Scholar]
- 23.Hartwell, L. H., Hopfield, J. J., Leibler, S. & Murray, A. W. (1999) Nature 402, 47-52. [DOI] [PubMed] [Google Scholar]
- 24.Wolf, D. M. & Arkin, A. P. (2003) Curr. Opin. Microbiol. 6, 125-134. [DOI] [PubMed] [Google Scholar]
- 25.Rives, A. W. & Galitski, T. (2003) Proc. Natl. Acad. Sci. USA 100, 1128-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walter, G. C. (1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology, eds. Neidhardt, F. C., Curtiss, R. & Lin, E. C. (Am. Soc. Microbiol. Press, Washington, DC), pp. 1400-1416.
- 27.Ronen, M., Rosenberg, R., Shraiman, B. I. & Alon, U. (2002) Proc. Natl. Acad. Sci. USA 99, 10555-10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gardner, T. S., di Bernardo, D., Lorenz, D. & Collins, J. J. (2003) Science 301, 102-105. [DOI] [PubMed] [Google Scholar]
- 29.Miller, M. B. & Bassler, B. L. (2001) Annu. Rev. Microbiol. 55, 165-199. [DOI] [PubMed] [Google Scholar]
- 30.Fuqua, C. & Greenberg, E. P. (2002) Nat. Rev. Mol. Cell Biol. 3, 685-695. [DOI] [PubMed] [Google Scholar]
- 31.Taga, M. E. & Bassler, B. L. (2003) Proc. Natl. Acad. Sci. USA 100, 14549-14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winson, M. K., Swift, S., Fish, L., Thorup, J. P., Jørgensen, F., Chhabra, S. R., Bycroft, B. W., Williams, P. & Steward, G. S. A. B. (1998) FEMS Microbiol. Lett. 163, 185-192. [DOI] [PubMed] [Google Scholar]
- 33.Andersen, J. B., Heydorn, A., Hentzer, M., Eberl, L., Geisenberger, O., Christensen, B. B., Molin, S. & Givskov, M. (2001) Appl. Environ. Microbiol. 67, 575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.You, L., Cox, R. S., III, Weiss, R. & Arnold, F. H. (2004) Nature 428, 868-871. [DOI] [PubMed] [Google Scholar]
- 35.Toman, Z., Dambly-Chaudiere, C., Tenenbaum, L. & Radman, M. (1985) J. Mol. Biol. 186, 97-105. [DOI] [PubMed] [Google Scholar]
- 36.Vollmer, A. C., Belkin, S., Smulski, D. R., Van Dyk, T. K. & LaRossa, R. A. (1997) Appl. Environ. Microbiol. 63, 2566-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elasri, M. O. & Miller, R. V. (1998) Appl. Microbiol. Biotechnol. 50, 455-458. [DOI] [PubMed] [Google Scholar]
- 38.Kostrzynska, M., Leung, K. T., Lee, H. & Trevors, J. T. (2002) J. Microbiol. Methods 48, 43-51. [DOI] [PubMed] [Google Scholar]
- 39.Nivens, D. E., McKnight, T. E., Moser, S. A., Osbourn, S. J., Simpson, M. L. & Sayler, G. S. (2004) J. Appl. Microbiol. 96, 33-46. [DOI] [PubMed] [Google Scholar]
- 40.Szafranski, P., Mello, C. M., Sano, T., Smith, C. L., Kaplan, D. L. & Cantor, C. R. (1997) Proc. Natl. Acad. Sci. USA 94, 1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ronchel, M. C. & Ramos, J. L. (2001) Appl. Environ. Microbiol. 67, 2649-2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torres, B., Jaenecke, S., Timmis, K. N., Garcia, J. L. & Diaz, E. (2003) Microbiology 149, 3595-3601. [DOI] [PubMed] [Google Scholar]
- 43.Steidler, L., Neirynck, S., Huyghebaert, N., Snoeck, V., Vermeire, A., Goddeeris, B., Cox, E., Remon, J. P. & Remaut, E. (2003) Nat. Biotechnol. 21, 785-789. [DOI] [PubMed] [Google Scholar]
- 44.Farmer, W. R. & Liao, J. C. (2000) Nat. Biotechnol. 18, 533-537. [DOI] [PubMed] [Google Scholar]
- 45.Bulter, T., Lee, S.-G., Wong, W. W., Fung, E., Connor, M. R. & Liao, J. C. (2004) Proc. Natl. Acad. Sci. USA 101, 2299-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lutz, R. & Bujard, H. (1997) Nucleic Acids Res. 25, 1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Toole, G. A. & Kolter, R. (1998) Mol. Microbiol. 28, 449-461. [DOI] [PubMed] [Google Scholar]
- 48.Horsthemke, W. & Lefever, R. (1984) Noise-Induced Transitions: Theory and Applications in Physics, Chemistry, and Biology (Springer, Berlin).
- 49.Hasty, J., Pradines, J., Dolnik, M. & Collins, J. J. (2000) Proc. Natl. Acad. Sci. USA 97, 2075-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ozbudak, E. M., Thatti, M., Lim, H. N., Shraiman, B. I. & van Oudenaarden, A. (2004) Nature 427, 737-740. [DOI] [PubMed] [Google Scholar]
- 51.Ptashne, M. (1992) A Genetic Switch: Phage λ and Higher Organisms (Cell Press & Blackwell, Oxford).
- 52.Petranovic, M., Salaj-Smic, E., Petranovic, D. & Trogovcevic, Z. (1979) J. Bacteriol. 140, 848-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghigo, J. M. (2001) Nature 412, 442-445. [DOI] [PubMed] [Google Scholar]
- 54.Yokobayashi, Y., Weiss, R. & Arnold, F. H. (2002) Proc. Natl. Acad. Sci. USA 99, 16587-16591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Francois, P. & Hakim, V. (2004) Proc. Natl. Acad. Sci. USA 101, 580-585. [DOI] [PMC free article] [PubMed] [Google Scholar]