Abstract
Background and Aims
DEFICIENS (DEF)- and GLOBOSA (GLO)-like proteins constitute two sister clades of floral homeotic transcription factors that were already present in the most recent common ancestor (MRCA) of extant angiosperms. Together they specify the identity of petals and stamens in flowering plants. In core eudicots, DEF- and GLO-like proteins are functional in the cell only as heterodimers with each other. There is evidence that this obligate heterodimerization contributed to the canalization of the flower structure of core eudicots during evolution. It remains unknown as to whether this strict heterodimerization is an ancient feature that can be traced back to the MRCA of extant flowering plants or if it evolved later during the evolution of the crown group angiosperms.
Methods
The interactions of DEF- and GLO-like proteins of the early-diverging angiosperms Amborella trichopoda and Nuphar advena and of the magnoliid Liriodendron tulipifera were analysed by employing yeast two-hybrid analysis and electrophoretic mobility shift assay (EMSA). Character-state reconstruction, including data from other species as well, was used to infer the ancestral interaction patterns of DEF- and GLO-like proteins.
Key Results
The yeast two-hybrid and EMSA data suggest that DEF- and GLO-like proteins from early-diverging angiosperms both homo- and heterodimerize. Character-state reconstruction suggests that the ability to form heterodimeric complexes already existed in the MRCA of extant angiosperms and that this property remained highly conserved throughout angiosperm evolution. Homodimerization of DEF- and GLO-like proteins also existed in the MRCA of all extant angiosperms. DEF-like protein homodimerization was probably lost very early in angiosperm evolution and was not present in the MRCA of eudicots and monocots. GLO-like protein homodimerization might have been lost later during evolution, but very probably was not present in the MRCA of eudicots.
Conclusions
The flexibility of DEF- and GLO-like protein interactions in early-diverging angiosperms may be one reason for the highly diverse flower morphologies observed in these species. The results strengthen the hypothesis that a reduction in the number of interaction partners of DEF- and GLO-like proteins, with DEF–GLO heterodimers remaining the only DNA-binding dimers in core eudicots, contributed to developmental robustness, canalization of flower development and the diversification of angiosperms.
Keywords: Flower development, DEFICIENS, GLOBOSA, APETALA3, PISTILLATA, protein–protein interaction, yeast two-hybrid, EMSA, character-state evolution, MADS-domain protein, floral homeotic gene, early-diverging angiosperms, basal angiosperms
INTRODUCTION
Depending on their partner proteins, transcription factors may affect the regulation of certain genes or developmental pathways in very different ways. MIKC-type MADS-domain proteins are a good case in point. In higher eudicots, virtually all MIKC-type MADS-domain proteins constitute dimers with several different partners (Immink et al., 2003; de Folter et al., 2005; Leseberg et al., 2008). These dimers bind to cis-regulatory DNA elements termed CArG-boxes [consensus 5′-CC(A/T)6GG-3′]. Combinatorial dimer formation is assumed to be of vital importance for the ability of MADS-domain proteins to regulate a plethora of developmental processes (de Folter et al., 2005; Kaufmann et al., 2005, 2010). For example, the floral homeotic protein SEPALLAT3 (SEP3) from Arabidopsis thaliana may interact with APETALA1 (AP1), another floral homeotic protein, to control floral meristem identity (Gregis et al., 2009). Later during development, SEP3 interacts with the floral homeotic protein AGAMOUS (AG) to determine carpel identity and, even later, SEP3 forms complexes with SHATTERPROOF1 and SHATTERPROOF2 to control ovule development (de Folter et al., 2005; Immink et al., 2009).
Although combinatorial dimer formation emerges as a common property among MIKC-type MADS-domain proteins, the subfamily of DEFICIENS (DEF)- and GLOBOSA (GLO)-like proteins [also known as APETALA3 (AP3)- and PISTILLATA (PI)-like proteins, respectively] constitutes a remarkable exception from this rule. DEF- and GLO-like transcription factors are highly conserved homeotic selector proteins that determine petal and stamen identity in probably all angiosperms (Kim et al., 2005; Zahn et al., 2005b; Litt and Kramer, 2010). In almost all core eudicots, DEF- and GLO-like proteins form DNA-binding dimers exclusively with each other, and do not form homodimers or DNA-binding heterodimers with other MADS-domain proteins (Riechmann et al., 1996a; Leseberg et al., 2008; Liu et al., 2010). This strict (or obligate) DEF–GLO heterodimerization is accompanied by a positive autoregulatory feedback loop in which DEF–GLO heterodimers foster the expression of their own transcripts (Schwarz-Sommer et al., 1992; Goto and Meyerowitz, 1994; McGonigle et al., 1996; Lenser et al., 2009). After the initial activation of DEF- and GLO-like genes (by factors that are not further considered here), there is an interdependence of DEF- and GLO-like protein expression in core eudicots; only if both of the partner proteins are expressed is the heterodimer formed and can in turn activate DEF- and GLO-like genes (Schwarz-Sommer et al., 1992; Goto and Meyerowitz, 1994).
DEF- and GLO-like proteins of core eudicots are usually expressed in the second and third whorl of the flower, in the primordia of which petals and stamens develop (for reviews, see Zahn et al., 2005b; Theißen and Melzer, 2007; Litt and Kramer, 2010). The interdependence in expression of DEF- and GLO-like proteins stabilizes this expression pattern; the misexpression of either a DEF- or a GLO-like protein alone would remain without consequences for floral organ identity as the appropriate partner would be missing (Winter et al., 2002b). It has therefore been proposed that the strict heterodimerization in conjunction with positive feedback regulation enhances developmental robustness of floral organ identity specification and contributed to the standardization of the floral structure (Winter et al., 2002b; Lenser et al., 2009). This raises the question as to when during evolution heterodimerization between DEF- and GLO-like proteins was established. DEF- and GLO-like genes originated by duplication in a common ancestor of all extant angiosperms (Kim et al., 2004). Extant gymnosperms, the closest living relatives of angiosperms, possess gene subfamilies (GGM2- and DAL12- and CJMADS1-like genes) that are ancestral to both DEF- and GLO-like genes (Winter et al., 2002a), whereas the sister to all other extant angiosperms, Amborella trichopoda, has both DEF- and GLO-like genes, indicating that the duplication that generated DEF- and GLO-like genes occurred in the lineage that led to extant angiosperms after the lineage that led to extant gymnosperms had already branched off (Aoki et al., 2004; Kim et al., 2004). In contrast to many angiosperm DEF- and GLO-like proteins, gymnosperm GGM2-like proteins form homodimers (Sundström and Engström, 2002; Winter et al., 2002b). It appears likely therefore that the heterodimerization between DEF- and GLO-like proteins originated from a homodimerizing ancestor (Winter et al., 2002b).
To understand better how DEF- and GLO-like proteins evolved towards the strictly heterodimerizing proteins in core eudicots, analysis of the orthologous proteins from early-diverging angiosperms is required. Here, we analyse the interaction of DEF- and GLO-like proteins from the early-diverging angiosperms A. trichopoda and Nuphar advena as well as from the magnoliid Liriodendron tulipifera. These branched off successively very early during angiosperm evolution, thus forming a grade in the phylogenetic tree (Fig. 1) (APG III, 2009). Our data suggest that DEF-like and GLO-like proteins from early diverging angiosperms heterodimerize with each other but also have the ability to homodimerize as well as to interact weakly with a number of other MADS-domain proteins. Character-state reconstruction revealed that DEF- and GLO-like proteins possessed the ability to form heterodimeric complexes with each other in the most recent common ancestor (MRCA) of extant angiosperms and that this property remained highly conserved throughout angiosperm evolution. In contrast, homodimerization and interactions with proteins from other subfamilies appear much less conserved.
Fig. 1.

Simplified seed plant phylogeny. The phylogeny is mainly based on analyses of the angiosperm phylogeny group (APG III, 2009). The phylogenetic position of A.trichopoda, N. advena and L. tulipifera is indicated. Major groups of seed plants are highlighted in different colours.
MATERIALS AND METHODS
Plant material
Flower buds from Liriodendron tulipifera were collected in the Park an der Ilm, Weimar, Germany. Flower buds of Nuphar advena were collected in the Old Botanical Garden of Göttingen, Germany. Male flower buds of Amborella trichopoda were collected in the Botanical Garden Bonn, Germany. The collected material was placed immediately into liquid nitrogen and stored at –80 °C until further use.
cDNA sequences used in this study
Partial coding sequences of LtAP3, LtPI, Nu.ad.AP3·1, Nu.ad.AP3·2 and Nu.ad.AGL2 have been published previously (Kramer et al., 1998; Kim et al., 2005; Zahn et al., 2005a). Full-length coding sequences of these cDNAs were obtained using 5′-RACE (rapid amplification of cDNA ends). The partial coding sequence of AMtrAGL9 (Zahn et al., 2005a) was completed by alignment with an expressed sequence tag (EST) sequence (FD432914·1). The predicted transcript was subsequently PCR amplified.
LtPI2 was isolated using 3′- and 5′-RACE. Nu.ad.PI1 and Nu.ad.AGL6·1 were derived from ESTs. Nu.ad.PI2 and Nu.ad.AGL6·2 were isolated by PCR with primers derived from Nu.ad.PI1 and Nu.ad.AGL6·1, respectively. Nu.ad.AGL6·1 is, except for three nucleotide differences, identical to a previously published AGL6-like sequence from N. advena (accession no. GU048649) (Kim et al., 2013).
The AmAP3 and AmPI cDNA sequences used here are very similar or identical to Am.tr.AP3·1 and Am.tr.PI1, which were recently reported (Amborella Genome Project, 2013)
For a complete list of cDNA sequences used, see Supplementary Data Table S1. Protein and gene names as assigned in previous publications were used throughout the manuscript.
cDNA sequences used for yeast two-hybrid and electrophoretic mobility shift assay (EMSA) experiments were provided by the Floral Genome Project (http://fgp.bio.psu.edu/) or by Seishiro Aoki (University of Tokyo), or were synthesized from RNA. For RNA isolation from L. tulipifera, total RNA isolation reagent (Biomol) was used. RNA from N. advena was extracted with the RNeasy Plant Mini Kit. For RNA extraction from A. trichopoda, we used a combination of a CTAB (cetyltrimethylammonium bromide) DNA extraction method and the RNeasy Plant Mini Kit (Kim et al., 2004). cDNA was synthesized using an oligo(dT) primer with MuLV reverse transcriptase.
For yeast two-hybrid experiments, full-length cDNA sequences were cloned into pGADT7 and pGBKT7. Full-length and C-terminal deleted cDNA sequences were cloned into pSPUTK for EMSA experiments. Nu.ad.PI2 was amplified with primers originally designed to amplify Nu.ad.PI1. As the primers used for amplification covered part of the coding sequence, the C-terminal end as well as the beginning of the MADS-domain were identical in the Nu.ad.PI1 and Nu.ad.PI2 clones used for EMSA and yeast two-hybrid analyses. This identity also applies to Nu.ad.AGL6·1 and Nu.ad.AGL6·2.
Yeast two-hybrid studies
Yeast two-hybrid assays were carried out essentially as described (Wang et al., 2010). For assaying an interaction, similar amounts of yeast cells were dissolved in water and 10-fold serially diluted up to 1:10 000 in water. Afterwards, the diluted yeast cells were spotted on selective medium lacking histidine, leucine and tryptophan containing 3 mM 3-amino-1,2,4-triazole. The plates were incubated for up to 14 d at 22 °C. All interactions were tested with at least two independent matings.
Electrophoretic mobility shift assay
The EMSA experiments were conducted essentially as described (Melzer et al., 2009). Proteins were produced by in vitro translation using the SP6 TNT Quick Coupled Transcription/Translation mix (Promega). After in vitro translation, proteins were shock frozen in liquid nitrogen and stored at –80 °C until use.
cDNA sequences of the C-terminal deleted constructs used are listed in Supplementary Data Table S2.
The sequence of the DNA probe used was 5′-CGTTC CATAC TTTCC TTATT TGGAA TATAA TTAAA TTTCG-3′ (the CArG-box is underlined). The concentration of the labelled DNA probe was generally approx. 3 nm. Usually 4 μL of in vitro translated protein were used per binding reaction.
Phylogenetic analysis and ancestral character-state reconstructions
Phylogenetic trees for DEF- and GLO-like genes shown in Figs 5 and 6, and in Supplementary Data Figs S5, S6, S9–S12 were drawn manually according to the APG III (2009) tree topology with the following modifications. Gymnosperm sequences of Gnetum gnemon and Picea abies were included as outgroup representatives. Gymnosperm branching was implemented as described (Winter et al., 2002b). Branching of the DEF-like genes within Asparagales was based on Mondragon-Palomino et al. (2009). The TM6/euAP3 split was arranged according to phylogenies in Hernandez-Hernandez et al. (2007) and Lee and Irish (2011). The GLO1/GLO2 split within the core lamiids was arranged according to Lee and Irish (2011).
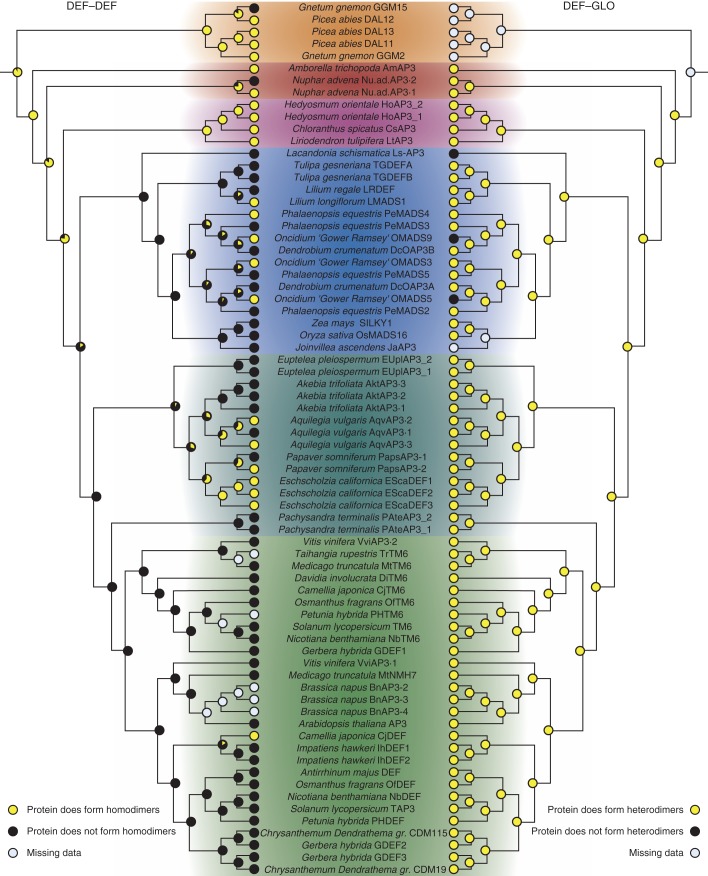
Fig. 5.
Ancestral character-state reconstruction for the ability of DEF-like proteins to form homo- and heterodimeric complexes. Trees are in general based on the APG III phylogeny as described in the Materials and Methods. The tree on the left depicts character-state reconstruction for the homodimerization capability of DEF-like proteins. The tree on the right has the same topology as the tree on the left but shows character-state reconstruction for heterodimerization of DEF-like proteins with GLO-like proteins. Yellow circles at the terminal positions indicate the presence of an interaction, and black circles indicate the absence of an interaction. Grey circles indicate that interaction data are not available for that particular protein. The likelihood of an interaction at internal nodes is indicated by pie charts. Proteins from different plant groups are highlighted in colours as in Fig. 1. Colour coding is as follows: gymnosperms, orange; early-diverging angiosperms, red; magnoliids, purple; monocots, blue; early-diverging eudicots, dark green; core eudicots, light green
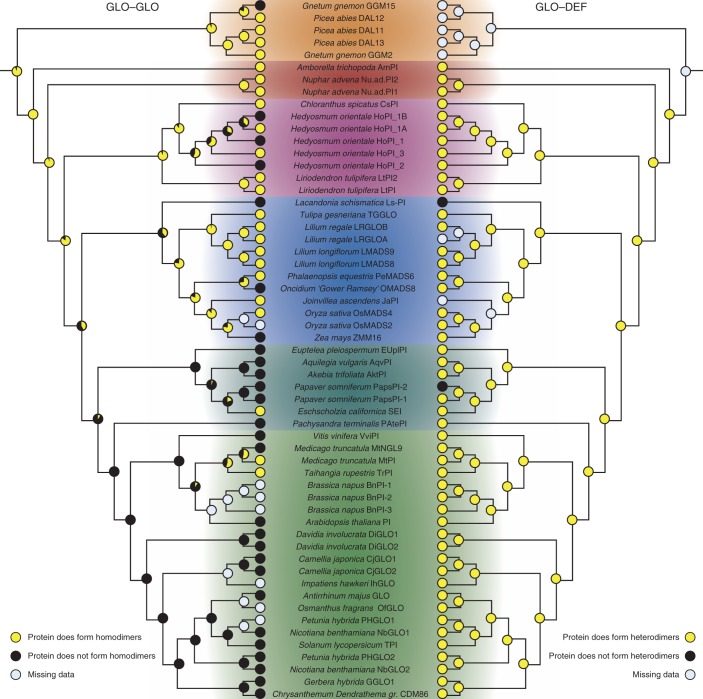
Fig. 6.
Ancestral character-state reconstruction for the ability of GLO-like proteins to form homo- and heterodimeric complexes. The trees on the left and right depict character state reconstructions for homodimerization of GLO-like proteins and heterodimerization of GLO-like with DEF-like proteins, respectively. For details of the labelling, see Fig. 5.
In addition to the species-based phylogeny described above, ancestral character-state reconstruction was also done with phylogenetic trees inferred from the DEF- and GLO-like sequences under study (Supplementary Data Figs S1–S4, S7, S8). cDNA sequences used for this were either obtained in this study or downloaded from the NCBI nucleotide collection (http://www.ncbi.nlm.nih.gov/nuccore). To create a codon alignment, all cDNA sequences were first translated to amino acid sequences using ExPASy Translate (http://web.expasy.org/translate/). The amino acid sequences were aligned with MAFFT 7 applying the E-INS-i strategy (Katoh and Standley, 2013). Using the respective cDNA sequences, the resulting amino acid alignment was converted into a codon alignment with RevTrans 1·4 (http://www.cbs.dtu.dk/services/RevTrans/). The quality of the codon alignment was examined in Seaview 4 (Gouy et al., 2010). Phylogenetic trees were calculated using the Bayesian inference method in MrBayes 3 (Ronquist and Huelsenbeck, 2003). Because of high sequence diversity that led to uncertain alignments of the C-terminal domain, only MADS-, I- and K-domains were considered for the calculation of the phylogenetic trees. The analyses were run for 4 000 000 generations applying the 4by4 nucleotide model. The first 25 % of the calculated trees were discarded. Gymnosperm relatives of DEF- and GLO-like genes were defined as the outgroup.
The interaction data used for the ancestral character-state reconstruction were based on yeast two-hybrid studies and EMSAs either obtained in this study or published previously. A complete list of publications from which interaction data were extracted can be found in Supplementary Data Table S3. All proteins included in the ancestral-state reconstruction were analysed for their homodimerization ability and their ability to form heterodimers with the respective DEF- or GLO-like partner proteins, as well as with proteins from a clade consisting of AGAMOUS-LIKE6-like (AGL6-like), LOFSEP- and SEP3-like proteins (AGL6/LOFSEP/SEP3 clade) (Malcomber and Kellogg, 2005; Zahn et al., 2005a; Kim et al., 2013). A DEF- or GLO-like protein was defined as ‘interacting’ with the other subfamily if interaction with at least one member of the other clade was detected, and as ‘non-interacting’ if none of the tested interactions showed a positive result. In general, these rules were also applied for interactions of DEF- and GLO-like proteins with AGL6/LOFSEP/SEP3-like proteins. However, in these cases, to be designated as ‘not interacting’, we required a protein to be negatively tested with members of at least two of the three sub-clades of AGL6/LOFSEP/SEP3-like proteins. This constraint was incorporated to minimize the risk of including false negatives. If different publications or methods yielded contradictory interaction data, the protein was defined as ‘interacting’ if at least one study showed the respective interaction.
The ancestral character-state reconstructions were performed in Mesquite 2·75 (Maddison and Maddison, 2011) following a maximum likelihood approach with a Markov one-parameter model (i.e. rates for gains and losses of interactions are identical). We used the one-parameter model as this is usually preferred over a two-parameter model (in which different rates for gains vs. losses are allowed) for medium-sized data sets like those used here (Mooers and Schluter, 1999). Asymmetry likelihood ratio tests as implemented in Mesquite 2·75 also favoured a one-parameter model over a two-parameter model for the majority of the data sets (P > 0·05). For the trees drawn manually and constrained to the species phylogeny, a branch length of one was uniformly assigned. For the trees based on the gene phylogeny, branch lengths as obtained from the phylogenetic reconstructions were taken.
Alignments and phylogenetic trees have been deposited at TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S15503).
RESULTS
Interactions among MADS-domain proteins from early-diverging angiosperms
To investigate protein–protein interactions of MADS-domain proteins from A. trichopoda, N. advena and L. tulipifera, a GAL-4-based yeast two-hybrid system was employed. DEF- and GLO-like proteins from each species were tested bidirectionally (i.e. proteins were tested as fusions with the GAL4 DNA-binding domain as well as with the GAL4 transcription activation domain) in an ‘all against all’ fashion. Orthologues of other floral homeotic proteins were also tested for comparison.
In addition to the yeast two-hybrid assays, EMSAs were conducted to characterize further the interactions between the MADS-domain proteins and to study their DNA-binding abilities. MADS-domain proteins are well known to bind as dimers to CArG-boxes (Schwarz-Sommer et al., 1992; Huang et al., 1996; Riechmann et al., 1996b). Using a CArG-box derived from the regulatory intron of AG from A. thaliana, we assayed the formation of protein–DNA complexes. In EMSAs, the formation of heteromeric protein complexes bound to DNA was inferred when the co-incubation of two different proteins with the DNA probe yielded a complex with an electrophoretic mobility different from that of the respective homomeric complexes (Huang et al., 1996; Melzer et al., 2009; Wang et al., 2010). If the homomeric complexes possessed similar electrophoretic mobilities, C-terminal deleted versions of one of the proteins were used to distinguish a homomeric from a heteromeric complex, as described previously (Wang et al., 2010).
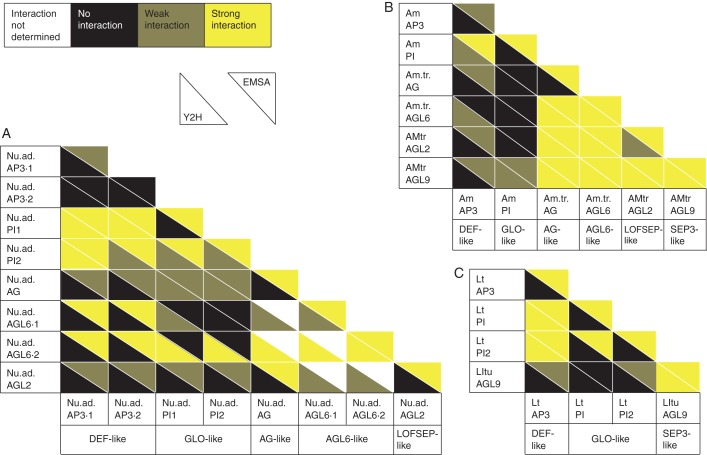
From all three species, DEF-, GLO- and LOFSEP- or SEP3-like proteins were assayed using yeast two-hybrid analyses and EMSAs. From A. trichopoda and N. advena, AGL6- and AG-like proteins were also analysed. The results for A. trichopoda are shown in Figs 2 and 3, and those for N. advena and L. tulipifera in Supplementary Data Figs S13–S15. Results are summarized for all species analysed in Fig. 4. Homomeric interactions were detected in only a few instances in yeast two-hybrid assays. This is in contrast to the EMSA results, according to which probably all of the proteins except Nu.ad.AP3·2 formed homomeric complexes bound to DNA, although homomeric AmAP3 and Nu.ad.AP3·1 protein–DNA complexes were only very weakly and not consistently detected (Figs 2–4). With respect to heteromeric complexes, extensive interactions between LOFSEP-, SEP3-, AGL6- and AG-like proteins were detected. At least in EMSAs, but in many cases also in yeast two-hybrid assays, LOFSEP-, SEP3-, AGL6- and AG-like proteins interacted in all tested combinations with each other (Fig. 4). Also DEF- and GLO-like proteins reliably formed DNA-binding heterodimers with each other (Figs 3 and 4). This was in stark contrast to interactions of DEF- and GLO-like proteins with proteins from the LOFSEP-, SEP3-, AGL6- or AG-like clade. In these cases, interactions were often undetectable, were detected only in either yeast two-hybrid assays or EMSAs and/or were so weak that reliable detection was difficult (Fig. 4). For example, the interactions that were very weakly and/or not consistently observed in EMSAs all involved at least one DEF-like or one GLO-like protein (Fig. 4).
Fig. 2.
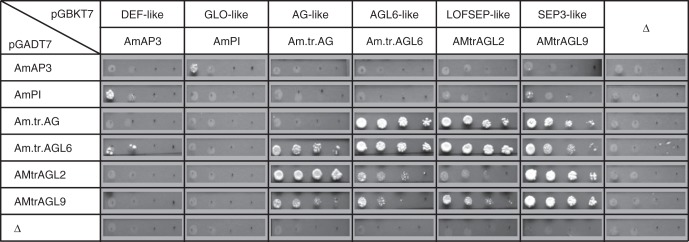
Representative yeast two-hybrid results for MADS-domain proteins from A. trichopoda. Photographs show colony growth on selective –Leu/–Trp/–His media with yeasts grown at 22 °C. For each interaction tested, yeast cells were spotted in a 10-fold serial dilution (from left to right). Proteins that were expressed as fusions with the GAL4 DNA-binding domain (vector pGBKT7) are shown horizontally; proteins expressed as GAL4 activation domain fusions (vector pGADT7) are shown vertically. Δ indicates negative controls in which empty vectors that did not contain a MADS-box gene cDNA insert were used.
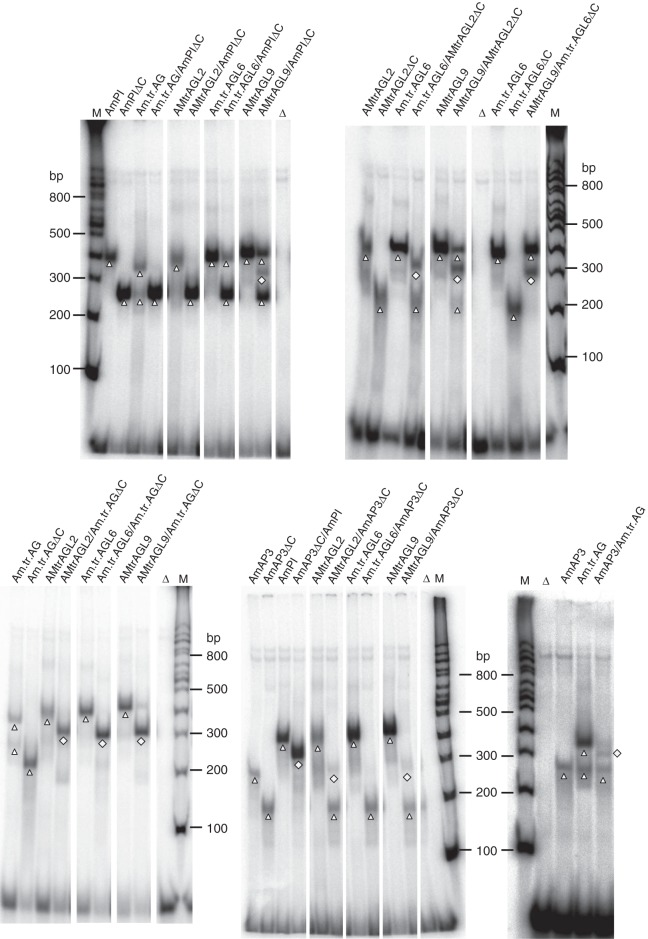
Fig. 3.
EMSA results for MADS-domain proteins from A. trichopoda. In vitro translated proteins were incubated together with a radioactively labelled DNA probe which carried one CArG-box. Proteins applied are noted above the gel. ‘ΔC’ is used to indicate C-terminal deleted proteins. Triangles highlight homomeric DNA-bound complexes; squares highlight heteromeric DNA-bound complexes. Free DNA is seen at the bottom of the gels. Homomeric complexes were not always visible when heteromer formation was tested, possibly because the vast majority of protein was assembled into DNA-bound heteromeric complexes (AmAP3ΔC/AmPI, for example) or in heteromeric complexes not or only very weakly binding to DNA (AMtrAGL9/AmAP3ΔC, for example). Note that certain potential heteromeric complexes were very weak and sometimes difficult to distinguish from homomeric complexes (AMtrAGL2/AmAP3ΔC, for example). For unknown reasons, the homomeric AmAP3–DNA complex possessed an unsusually high electrophoretic mobility and some proteins (Am.tr.AG, for example) formed two distinct protein–DNA complexes even in the absence of a partner. ‘Δ’ indicates negative controls in which in vitro translation lysate programmed with a vector that did not contain a cDNA insert was added. ‘M’ denotes lanes in which a radioactively labelled DNA marker (NEB 100 bp DNA ladder) was applied (bp = base pairs).
Fig. 4.
Summary of the interaction patterns of MADS-domain proteins from (A) N. advena, (B) A. trichopoda and (C) L. tulipifera. Yeast two-hybrid interactions were scored as strong if yeast growth was observed in all dilutions in at least one direction, and weak if yeast growth was observed but not in all dilutions tested. EMSA results were designated as strong if a protein–DNA complex was reliably observed, and as weak if a protein–DNA complex was not reproducibly observed, and/or if the protein–DNA complex was detected as a faint band only (that was sometimes difficult to distinguish from homomeric complexes).
In some cases, co-translation of two proteins failed to detect one of the corresponding homomeric complexes but also failed to yield a strong heteromeric complex (compare AMtrAGL9 and AMtrAGL9/AmAP3ΔC, in Fig. 3, for example). We do not have a satisfactory explanation for this observation. However, it could be that heteromeric complexes that either did not bind or only weakly bound to DNA were reconstituted in these cases. Furthermore, the protein–DNA complexes reconstituted in EMSAs span a considerable range of electrophoretic mobilities (compare homomeric AmAP3 with homomeric AmAG complexes in Fig. 3, for example), although the molecular masses and charges of the proteins are very similar. It is unclear whether the different mobilities resulted from different conformations of the proteins or of the protein–DNA complexes or from differences in the stoichiometry of binding.
Reconstruction of the ancestral DEF- and GLO-like protein interaction behaviour
Interaction patterns among DEF- and GLO-like proteins have previously been determined for several monocot and eudicot species (Davies et al., 1996; Riechmann et al., 1996a; Winter et al., 2002b; Immink et al., 2003; Kanno et al., 2003; Yang et al., 2003; Vandenbussche et al., 2004; Whipple et al., 2004; Kramer et al., 2007; Yao et al., 2008; Liu et al., 2010, to mention but a few). Using published data along with the newly identified interactions from early-diverging angiosperms obtained in this study, we aimed at understanding the trajectories of interactions of DEF- and GLO-like proteins during angiosperm evolution employing character-state reconstruction. The EMSA and yeast two-hybrid data were combined for these analyses. Character-state reconstruction was conducted using (1) a phylogenetic tree drawn manually according to the species phylogeny as reported by APG III (Fig. 1) (APG III, 2009) and (2) phylogenetic trees based on the phylogenetic relationships among the genes. Tree topologies were similar for the two approaches, and the character-state reconstructions yielded essentially the same results (compare Figs 5, 6 and Supplementary Data Figs S5, S6 with Figs S1–S4, S7, S8). Interestingly, almost all of the DEF- and GLO-like proteins assayed so far are capable of interacting with a partner from the other subfamily, i.e. of forming DEF–GLO-like protein complexes (Figs 5 and 6; Supplementary Data Figs S1, S3). This result strongly indicates that heterodimerization between DEF- and GLO-like proteins was established at the base of extant angiosperms and remained highly conserved throughout angiosperm evolution (Figs 5 and 6).
Several DEF-like and GLO-like proteins not only constitute DEF–GLO heterodimers but also possess the ability to homodimerize (Winter et al., 2002a; Liu et al., 2010). Character-state reconstruction suggests that homodimerization of both DEF- and GLO-like proteins was probably present in the MRCA of extant angiosperms (Figs 5 and 6). The analyses further suggest that homodimerization of DEF-like proteins was lost relatively early during angiosperm evolution and regained in several eudicot and monocot lineages (Fig. 5). Character-state reconstruction using the gene phylogeny of DEF-like sequences indicates that DEF-like protein homodimerization was lost independently in eudicots and monocots. However, this result can very probably be attributed to the unsusual placement of magnoliid DEF-like genes as being closely related to eudicot DEF-like genes (Supplementary Data Fig. S2).
For GLO-like proteins, the situation was slightly different; GLO-like protein homodimerization prevails in the extant monocots analysed, although it is not clear whether homodimerization was preserved at the base of the monocots or lost and re-established later during monocot evolution (Fig. 6; Supplementary Data Fig. S4). In contrast, only a very limited number of eudicot species possess homodimerizing GLO-like proteins (Fig. 6; Supplementary Data Fig. S4), strongly indicating that the MRCA of eudicots did not possess a homodimerizing GLO-like protein.
We also conducted character-state reconstructions for interactions of DEF- and GLO-like proteins with proteins from the clade of AGL6/LOFSEP/SEP3-like proteins (Supplementary Data Figs S5–S8). These analyses were included because DEF- and GLO-like proteins act in tetrameric complexes with AGL6/LOFSEP/SEP3-like proteins to determine petal and stamen identity (Theißen, 2001; Theißen and Saedler, 2001; Wang et al., 2010). Interactions of DEF-like as well as of GLO-like proteins with AGL6/LOFSEP/SEP3-like proteins appear to be relatively scattered across the angiosperm phylogeny. For example, about half of the eudicot DEF-like proteins analysed interact with at least one AGL6/LOFSEP/SEP3-like protein (Supplementary Data Figs S5, S7). However, the pattern emerges that DEF–AGL6/LOFSEP/SEP3 as well as GLO–AGL6/LOFSEP/SEP3 interactions were already present early in angiosperm evolution. Also the MRCA of DEF- and GLO-like proteins at the base of the seed plants probably already possessed the respective interaction (Supplementary Data Figs S5–S8).
For the above-described character-state reconstruction, proteins were designated as interacting when a positive result had been reported either from yeast two-hybrid assays or from EMSAs. For 15 DEF-like and 12 GLO-like proteins from angiosperms, homodimerization data from both techniques (EMSA and yeast two-hybrid assays) were available (Supplementary Data Figs S9, S11). Of these 27 cases, 16 yielded identical results in yeast two-hybrid assays and EMSAs with respect to homodimerization (i.e. proteins did or did not form homodimers in both assays). For nine interactions, homodimerization was detected in EMSAs but not in yeast two-hybrid assays. The converse case, i.e. detection of homodimerization in yeast two-hybrid assays but not in EMSAs, was observed only for two proteins (Supplementary Data Figs S9, S11). In contrast, heterodimerization among DEF- and GLO-like proteins was, with one exception, consistently observed in EMSAs as well as yeast two-hybrid assays (Supplementary Data Figs S10, S12). This confirms previous observations that homodimerization, in particular of MADS-domain proteins, is more readily detected in EMSAs than in yeast two-hybrid assays (Wang et al., 2010). To account for the different results that these techniques yielded, ancestral character-state reconstruction for homo- and heterodimerization of DEF- and GLO-like proteins was conducted separately for data sets based on yeast two-hybrid and EMSA results (Supplementary Data Figs S9–S12). Heterodimerization between DEF- and GLO-like proteins was inferred to be ancestral and highly conserved for all data sets (Supplementary Figs S10, S12). Also homodimerization was still inferred to be ancestral for DEF- and GLO-like proteins from flowering plants when only EMSA data were considered. In contrast, when only yeast two-hybrid data were taken into account, homodimerization was inferred to be absent in the MRCA of extant flowering plants.
DISCUSSION
Conservation of the MADS-domain protein interaction pattern during angiosperm evolution
In core eudicots and monocots, certain dimers of MADS-domain proteins are involved in floral organ specification. Dimers of DEF- and GLO-like proteins function in petal and stamen specification, and dimers of an AGL6/LOFSEP/SEP3-like protein and an AG-like protein are involved in stamen and carpel development (Schwarz-Sommer et al., 1992; Huang et al., 1996; Riechmann et al., 1996a; Theißen, 2001; de Folter et al., 2005; Rijpkema et al., 2009; Thompson et al., 2009; Liu et al., 2010). Comparison of the interaction patterns obtained from A. trichopoda, N. advena and L. tulipifera with those from model eudicots and monocots (i.e. A. thaliana, Petunia hybrida and Oryza sativa) revealed that these heterodimeric interactions important for organ specification in derived angiosperms are also detected in early diverging angiosperms (Fig. 7), pointing towards a high degree of conservation of these interactions during flowering plant evolution. Recently published yeast two-hybrid data from A. trichopoda (Amborella Genome Project, 2013) largely agree with the interactions reported here. However, we detected some additional interactions (AMtrAGL9/AMtrAGL2, for example), probably because we used milder assay conditions (e.g. growing yeast cells at 22 °C instead of using higher temperatures).
Fig. 7.
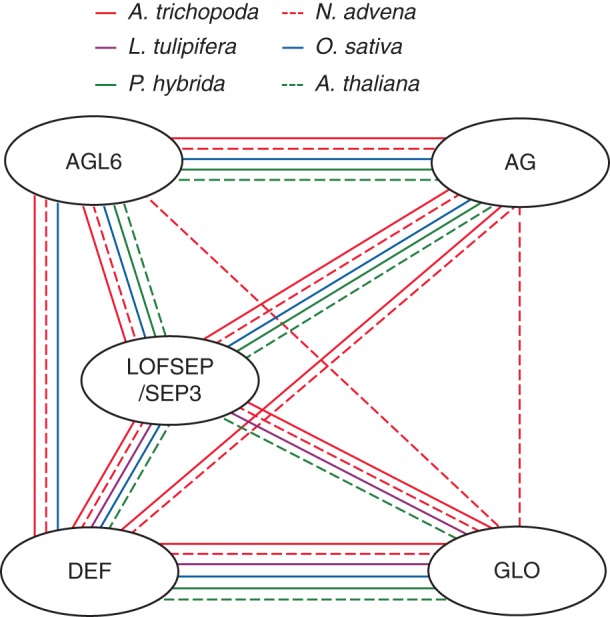
Conservation of interactions among MADS-domain proteins. The different subfamilies are depicted by ovals; lines between the ovals indicate interactions in the different species as colour coded in the key. Only interactions between proteins of different subfamilies are depicted. Not all protein combinations have been tested in all species. The absence of an interaction here does not therefore necessarily mean that the proteins do not interact. Publications used to depict interactions among proteins from A. thaliana, P.hybrida and O. sativa are listed in Supplementary Data Table S3.
Our data suggest that the interactions governing flower development in core eudicots were already established at the base of extant angiosperms and remained highly conserved since then. Specifically, our results indicate that the heterodimerization between DEF-like and GLO-like proteins was already present in the MRCA of extant angiosperms and was virtually never rewired. Alternative scenarios in which heterodimerization between DEF- and GLO-like proteins was established independently in eudicots and monocots would invoke multiple losses and gains of DEF–GLO heterodimerization and therefore appear less likely.
We observed only three DEF-like (OMADS5 and OMADS9 from Oncidium ‘Gower Ramsey’ and Ls-AP3 from Lacandonia schismatica) (Alvarez-Buylla et al., 2010; Chang et al., 2010) and two GLO-like proteins (PapsPI-2 from Papaver somniferum and Ls-PI from L. schismatica) (Drea et al., 2007; Alvarez-Buylla et al., 2010) for which heterodimerization with a partner from the other subfamily was not detected (Figs 5 and 6). However, in O. ‘Gower Ramsey’ and P. somniferum, at least one additional DEF- or GLO-like protein exists that can constitute a DEF–GLO heterodimer and thus may compensate for the loss of heterodimerization in the respective paralogue. In contrast, Ls-AP3 and Ls-PI are the only DEF- and GLO orthologues that have been isolated from L. schismatica. Intriguingly, L. schismatica deviates from the basic floral ‘bauplan’ in that carpels surround stamens, which are positioned in the centre of the flower (Marquez-Guzman et al., 1989; Alvarez-Buylla et al., 2010), a feature that is otherwise only known from the genus Trithuria (Rudall et al., 2009) and thus is extremely rare in angiosperms. It is interesting to note that the loss of DEF–GLO heterodimerization coincides with the modification of the floral bauplan in L. schismatica, although it remains elusive whether there is a causal relationship between these two observations.
Use of the yeast two-hybrid system to detect homodimerization of proteins
The available data clearly suggest that homodimerization of DEF- and GLO-like proteins is under-represented in yeast two-hybrid assays as compared with EMSAs. Indeed, character-state reconstructions using only yeast two-hybrid data infer that homodimerization of DEF- or GLO-like proteins was not present in the MRCA of flowering plants, whereas the opposite is predicted when using only EMSA data or a combination of both data sets (Figs 5, 6; Supplementary Data Figs S9, S11). One explanation for the discrepancy between yeast two-hybrid and EMSA results might be that homodimerzation of DEF- and GLO-like proteins is stabilized by DNA binding, as has also been proposed for other MIKC-type MADS-domain proteins (Wang et al., 2010). In addition, it has been described that homodimerization is in general difficult to detect with the yeast two-hybrid system (Smirnova et al., 1999; Newman et al., 2000). One plausible explanation for that is that the GAL4 DNA-binding domain used in the yeast two-hybrid experiments is already capable of homodimerization. This homodimerization in turn elevates the local concentration of the proteins fused to the GAL4 DNA-binding domain (Hu, 2000; Newman et al., 2000), thereby favouring the formation of homodimers between hybrid proteins containing a GAL4 DNA-binding domain, at the expense of interactions with hybrid proteins containing the GAL4 activation domain. We therefore assume that EMSA data or a combination of yeast two-hybrid and EMSA data are better suited to trace the ancestral character state of homodimerizing proteins than yeast two-hybrid data alone.
The developmental relevance of DEF- and GLO-like protein interactions beyond DEF–GLO heterodimerization
The functional relevance of DEF–GLO heterodimers for determining petal and stamen identity is well established (Schwarz-Sommer et al., 1992; Lenser et al., 2009). However, one may ask what the function of the occasionally observed DEF- and GLO-like protein homodimers or dimers of DEF- or GLO-like proteins with other floral homeotic proteins is. The frequent occurrence of GLO-like protein homodimers in particular has fostered speculation that these protein complexes are of developmental relevance (Winter et al., 2002b). This possibility is supported by expression studies showing that in monocots and early diverging angiosperms in particular, expression of GLO-like genes is sometimes not associated with DEF-like gene expression and is also observed in organs other than petals and stamens (see, for example, Münster et al., 2001; Kim et al., 2005).
However, to the best of our knowledge, a developmental function of GLO-like protein homodimers or DEF-like protein homodimers has not yet been described. A detailed analysis of these possibilities would require downregulation or knockout of DEF- and GLO-like genes in species in which DEF- or GLO-like proteins homodimerize. Respective data are only available from very few species, including O. sativa (Nagasawa et al., 2003; Ronai et al., 2003; Yao et al., 2008), Aquilegia vulgaris (Kramer et al., 2007) and P. somniferum (Drea et al., 2007). In these cases, downregulation of the complete set of DEF-like genes resulted in homeotic transformations that very much resembled or were identical to those obtained when the concomitant GLO-like genes were downregulated. These observations are in line with the assumption that DEF- and GLO-like proteins function as heterodimers during development and that potential DEF- or GLO-like protein homodimers cannot substitute for the developmental role of DEF–GLO heterodimers. However, it should be taken into consideration that the formation of GLO-like or DEF-like protein homodimers may lower the concentration of protein monomers available for heterodimer formation, thus perhaps indirectly affecting the activity of DEF–GLO heterodimers. Also, in several species, DEF- or GLO-like protein homodimers possess DNA-binding activity (Winter et al., 2002b; Kanno et al., 2003; Ronai et al., 2003; Whipple and Schmidt, 2006). These dimers may compete with DEF–GLO heterodimers for target gene occupancy. Mechanisms may thus have evolved to prevent the interference between homodimers and heterodimers of DEF- and GLO-like proteins. Further studies involving quantitative analyses of interaction strengths will reveal whether and how such mechanisms are employed.
The functional relevance of homodimers in determining floral organ identity notwithstanding, it remains to be noted that DEF- and GLO-like protein homodimerization is especially prevalent in early-diverging angiosperms. In these species, flower morphology is considerably less canalized than in core eudicots and monocots; very often the transition between different floral organ types is gradual and not as distinct as in more derived angiosperms (Buzgo et al., 2004; Soltis et al., 2006, 2007, 2009). It has been proposed that this gradual transition is related to gradients of floral homeotic gene expression (Buzgo et al., 2004; Soltis et al., 2006, 2007, 2009). According to the ‘fading border’ model, weak expression of floral homeotic genes may lead to the establishment of organs of intermediate identity. For example, weak expression of DEF- and/or GLO-like genes in the outer floral organs of A. trichopda may give rise to sepaloid tepals. An increase of DEF- and GLO-like gene expression towards the centre of the flower may result in a more petaloid appearance of tepals. It could well be that GLO- or DEF-like protein homodimers contributed to the broadening of the expression domain and to the gradual transition between floral organs. The evolutionary establishment of obligate heterodimerization, possibly in conjunction with autoregulatory control, may have sharpened the expression boundaries of DEF- and GLO-like proteins and hence contributed to the origin of distinct organ types within the flower. Unfortunately, mutant analyses in the early-diverging angiosperms investigated here to test hypotheses on the functional relevance of homodimers of DEF- or GLO-like proteins are not yet possible.
It is also noteworthy that circumstantial evidence for a role for DEF-like protein homodimers during development was provided by the analysis of the orchid DEF-like gene OMADS3 (Hsu and Yang, 2002). OMADS3 forms homodimers and does not interact with AP3 or PI from Arabidopsis. However, when ectopically expressed in Arabidopsis, precocious flowering is observed. This was taken as evidence that OMADS3 homodimers have a function in floral induction (Hsu and Yang, 2002). Future experiments may further substantiate whether such a novel function of DEF-like proteins in orchids does indeed exist.
Beyond homodimerization and interactions with each other, some DEF- and GLO-like proteins occasionally interact with AGL6/LOFSEP/SEP3-like proteins. These interactions may be considered in conjunction with the ability of floral homeotic MADS-domain proteins to form tetrameric complexes termed floral quartets (Theißen, 2001; Theißen and Saedler, 2001; Smaczniak et al., 2012). These tetrameric complexes are constituted by two dimers of MADS-domain proteins that are bound in the vicinity of each other on the DNA and interact with each other. Specific floral quartets are proposed to confer the identity of each of the different floral organ types. DNA-bound heterodimers of DEF- and GLO-like proteins are implicated to be part of the quartets determining petal and stamen identity. In A. thaliana, for example, these are AP3-PI/SEP3–AP1 and AP3-PI/SEP3–AG complexes, respectively (Honma and Goto, 2001; Theißen, 2001; Theißen and Saedler, 2001). Most of the heterodimeric interactions between DEF- or GLO-like and AGL6/LOFSEP/SEP3-like proteins may therefore reflect the interactions operating between DNA-bound dimers within the quartet. This hypothesis is supported by the notion that in the (admittedly few) eudicot species tested so far, DNA-binding activity of heterodimers consisting of a DEF-like or a GLO-like protein and an AGL6/LOFSEP/SEP3-like protein has not been detected (Davies et al., 1996; Geuten et al., 2006). Co-operative interactions between DNA-bound dimers are usually very weak in nature. Depending on the actual experimental set-up, they may escape detection with the yeast two-hybrid assay. That may explain why interactions between DEF- or GLO-like and AGL6/LOFSEP/SEP3-like proteins appear to be scattered across the angiosperm phylogeny. However, it remains to be noted that at the base of the angiosperms DNA-binding dimers between DEF- or GLO-like and AGL6/LOFSEP/SEP3-like proteins may have existed. Remnants of this property are detected in early-diverging angiosperms. Also, a gymnosperm orthologue of DEF- and GLO-like proteins, GGM2, forms DNA-binding dimers with AGL6-like proteins (Wang et al., 2010). We hypothesize that the ability of DEF- and GLO-like proteins to form DNA-binding dimers with proteins from other subfamilies was quickly lost during angiosperm evolution, as the respective complexes are difficult to detect in early-diverging angiosperms.
The evolutionary and developmental relevance of obligate heterodimerization
Taken together, our data indicate that a DEF–GLO heterodimer is the only functional DNA-binding dimer in the vast majority of angiosperms. As summarized elsewhere, this obligate heterodimerization may have contributed to the canalization of flower development as well as to developmental robustness (Winter et al., 2002b; Lenser et al., 2009). However, the question remains as to why specifically DEF- and GLO-like proteins and not other floral homeotic MADS-domain proteins have undergone such a drastic reduction of interaction partners.
The evolution of interactions among floral homeotic proteins can probably not be understood without taking tetramer formation into account. In eudicots, tetramer formation and hence floral organ development largely depends on AGL6/LOFSEP/SEP3-like proteins (or the closely related SQUA-like proteins) (Immink et al., 2009, 2010), and this dependence may effectively prevent the development of floral organs outside the floral context (Melzer et al., 2010). This dependence may in turn have contributed to the concerted development of petals, stamens and carpels in close proximity to each other and thus to the evolutionary origin and developmental stabilization of the flower as a reproductive entity (Melzer et al., 2010). However, if one protein of the AGL6/LOFSEP/SEP3 clade is always part of the tetramers determining organ identity, an obligate heterodimer beyond the DEF–GLO heterodimer cannot be constituted for stoichiometric reasons. For example, at the dimeric level, it is plausible to assume that a hypothetical obligate heterodimer of two different AG-like proteins could buffer developmental perturbations and contribute to the canalization of development within the flower in a similar way to as proposed for a DEF–GLO heterodimer. It may prevent, for example, misexpression of AG-like proteins outside the two inner floral whorls. In the context of tetramer formation, stamen development would in this hypothetical scenario be controlled by a tetramer constituted of a DEF–GLO heterodimer and a heterodimer of two AG-like proteins. However, AGL6/LOFSEP/SEP3-like proteins would not be part of such a complex. Therefore, selection may have acted against the formation of such complexes as these may have negatively affected the robustness of the flower as a single reproductive entity.
Concluding remarks
The evolution of DEF- and GLO-like interaction patterns is a fascinating example of how the reduction of molecular interactions may have contributed to developmental robustness that in turn may have led to species diversity. It is clear, however, that much remains to be learned about the pattern of protein interactions of these two subfamilies of floral homeotic MADS-domain proteins. Of particular interest is the functional relevance of dimers other than DEF–GLO heterodimers in early-diverging angiosperms and gymnosperms. Eventually, analysis of loss-of-function mutants of def- and glo-like genes in these species may be necessary to substantiate the evolutionary dynamics of DEF- and GLO-like protein interactions.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Seishiro Aoki (University of Tokyo) and Charlie Scutt (ENS Lyon) for providing cDNAs of A. trichopoda MADS-box genes, and Michael Schwerdtfeger (Botanical Garden Göttingen) and Wilhelm Barthlott (Botanical Garden Bonn) for providing plant material. Very special thanks to Chrissi Gafert for excellent technical support. We are grateful to Kerstin Kaufmann for valuable discussions during the planning phase of the project. R.M. is grateful to the University of Leipzig for general support. This research was supported by DFG grants to G.T. (TH417/5–1) and to G.T. and R.M. (TH417/5–2). R.M. was supported by a post-doctoral fellowship of the Carl-Zeiss-Foundation.
LITERATURE CITED
- Alvarez-Buylla ER, Ambrose BA, Flores-Sandoval E, et al. B-function expression in the flower center underlies the homeotic phenotype of Lacandonia schismatica (Triuridaceae) The Plant Cell. 2010;22:3543–3559. doi: 10.1105/tpc.109.069153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amborella Genome Project. The Amborella genome and the evolution of flowering plants. Science. 2013;342:11. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- Aoki S, Uehara K, Imafuku M, Hasebe M, Ito M. Phylogeny and divergence of basal angiosperms inferred from APETALA3- and PISTILLATA-like MADS-box genes. Journal of Plant Research. 2004;117:229–244. doi: 10.1007/s10265-004-0153-7. [DOI] [PubMed] [Google Scholar]
- APG III. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society. 2009;161:105–121. [Google Scholar]
- Buzgo M, Soltis PS, Soltis DE. Floral developmental morphology of Amborella trichopoda (Amborellaceae) International Journal of Plant Sciences. 2004;165:925–947. [Google Scholar]
- Chang YY, Kao NH, Li JY, et al. Characterization of the possible roles for B class MADS box genes in regulation of perianth formation in orchid. Plant Physiology. 2010;152:837–853. doi: 10.1104/pp.109.147116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B, EgeaCortines M, Silva ED, Saedler H, Sommer H. Multiple interactions amongst floral homeotic MADS box proteins. EMBO Journal. 1996;15:4330–4343. [PMC free article] [PubMed] [Google Scholar]
- Drea S, Hileman LC, De Martino G, Irish VF. Functional analyses of genetic pathways controlling petal specification in poppy. Development. 2007;134:4157–4166. doi: 10.1242/dev.013136. [DOI] [PubMed] [Google Scholar]
- de Folter S, Immink RGH, Kieffer M, et al. Comprehensive interaction map of the Arabidopsis MADS box transcription factors. The Plant Cell. 2005;17:1424–1433. doi: 10.1105/tpc.105.031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuten K, Becker A, Kaufmann K, et al. Petaloidy and petal identity MADS-box genes in the balsaminoid genera Impatiens and Marcgravia. The Plant Journal. 2006;47:501–518. doi: 10.1111/j.1365-313X.2006.02800.x. [DOI] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes and Development. 1994;8:1548–1560. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Dorca-Fornell C, Kater MM. The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. The Plant Journal. 2009;60:626–637. doi: 10.1111/j.1365-313X.2009.03985.x. [DOI] [PubMed] [Google Scholar]
- Hernandez-Hernandez T, Martinez-Castilla LP, Alvarez-Buylla ER. Functional diversification of B MADS-box homeotic regulators of flower development: adaptive evolution in protein–protein interaction domains after major gene duplication events. Molecular Biology and Evolution. 2007;24:465–481. doi: 10.1093/molbev/msl182. [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001;409:525–529. doi: 10.1038/35054083. [DOI] [PubMed] [Google Scholar]
- Hsu HF, Yang CH. An orchid (Oncidium Gower Ramsey) AP3-like MADS gene regulates floral formation and initiation. Plant and Cell Physiology. 2002;43:1198–1209. doi: 10.1093/pcp/pcf143. [DOI] [PubMed] [Google Scholar]
- Hu JC. A guided tour in protein interaction space: coiled coils from the yeast proteome. Proceedings of the National Academy of Sciences; USA. 2000. pp. 12935–12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Tudor M, Su T, Zhang Y, Hu Y, Ma H. DNA binding properties of two Arabidopsis MADS domain proteins: binding consensus and dimer formation. The Plant Cell. 1996;8:81–94. doi: 10.1105/tpc.8.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink RGH, Ferrario S, Busscher-Lange J, et al. Analysis of the petunia MADS-box transcription factor family. Molecular Genetics and Genomics. 2003;268:598–606. doi: 10.1007/s00438-002-0781-3. [DOI] [PubMed] [Google Scholar]
- Immink RG, Tonaco IA, de Folter S, et al. SEPALLATA3: the ‘glue’ for MADS box transcription factor complex formation. Genome Biology. 2009;10:R24. doi: 10.1186/gb-2009-10-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink RG, Kaufmann K, Angenent GC. The ‘ABC’ of MADS domain protein behaviour and interactions. Seminars in Cell and Developmental Biology. 2010;21:87–93. doi: 10.1016/j.semcdb.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Kanno A, Saeki H, Kameya T, Saedler H, Theißen G. Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana) Plant Molecular Biology. 2003;52:831–841. doi: 10.1023/a:1025070827979. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Melzer R, Theissen G. MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene. 2005;347:183–198. doi: 10.1016/j.gene.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Pajoro A, Angenent GC. Regulation of transcription in plants: mechanisms controlling developmental switches. Nature Reviews Genetics. 2010;11:830–842. doi: 10.1038/nrg2885. [DOI] [PubMed] [Google Scholar]
- Kim ST, Yoo MJ, Albert VA, Farris JS, Soltis PS, Soltis DE. Phylogeny and diversification of B-function MADS-box genes in angiosperms: evolutionary and functional implications of a 260-million-year-old duplication. American Journal of Botany. 2004;91:2102–2118. doi: 10.3732/ajb.91.12.2102. [DOI] [PubMed] [Google Scholar]
- Kim S, Koh J, Yoo MJ, et al. Expression of floral MADS-box genes in basal angiosperms: implications for the evolution of floral regulators. The Plant Journal. 2005;43:724–744. doi: 10.1111/j.1365-313X.2005.02487.x. [DOI] [PubMed] [Google Scholar]
- Kim S, Soltis PS, Soltis DE. AGL6-like MADS-box genes are sister to AGL2-like MADS-box genes. Journal of Plant Biology. 2013;56:315–325. [Google Scholar]
- Kramer EM, Dorit RL, Irish VF. Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics. 1998;149:765–783. doi: 10.1093/genetics/149.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Holappa L, Gould B, Jaramillo MA, Setnikov D, Santiago PM. Elaboration of B gene function to include the identity of novel floral organs in the lower eudicot Aquilegia. The Plant Cell. 2007;19:750–766. doi: 10.1105/tpc.107.050385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HL, Irish VF. Gene duplication and loss in a MADS box gene transcription factor circuit. Molecular Biology and Evolution. 2011;28:3367–3380. doi: 10.1093/molbev/msr169. [DOI] [PubMed] [Google Scholar]
- Lenser T, Theissen G, Dittrich P. Developmental robustness by obligate interaction of class B floral homeotic genes and proteins. PLoS Computational Biology. 2009;5:e1000264. doi: 10.1371/journal.pcbi.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leseberg CH, Eissler CL, Wang X, Johns MA, Duvall MR, Mao L. Interaction study of MADS-domain proteins in tomato. Journal of Experimental Botany. 2008;59:2253–2265. doi: 10.1093/jxb/ern094. [DOI] [PubMed] [Google Scholar]
- Litt A, Kramer EM. The ABC model and the diversification of floral organ identity. Seminars in Cell and Developmental Biology. 2010;21:129–137. doi: 10.1016/j.semcdb.2009.11.019. [DOI] [PubMed] [Google Scholar]
- Liu CJ, Zhang JA, Zhang N, et al. Interactions among proteins of floral MADS-box genes in basal eudicots: implications for evolution of the regulatory network for flower development. Molecular Biology and Evolution. 2010;27:1598–1611. doi: 10.1093/molbev/msq044. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. 2011. Version 2.75 http://mesquiteproject.org .
- Malcomber ST, Kellogg EA. SEPALLATA gene diversification: brave new whorls. Trends in Plant Science. 2005;10:427–4335. doi: 10.1016/j.tplants.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Marquez-Guzman J, Engleman M, Martinez-Mena A, Martinez E, Ramos C. Reproductive anatomy of Lacandonia schismatica (Lacandoniaceae) Annals of the Missouri Botanical Garden. 1989;76:124–127. [Google Scholar]
- McGonigle B, Bouhidel K, Irish VF. Nuclear localization of the Arabidopsis APETALA3 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes and Development. 1996;10:1812–1821. doi: 10.1101/gad.10.14.1812. [DOI] [PubMed] [Google Scholar]
- Melzer R, Verelst W, Theissen G. The class E floral homeotic protein SEPALLATA3 is sufficient to loop DNA in ‘floral quartet’-like complexes in vitro. Nucleic Acids Research. 2009;37:144–157. doi: 10.1093/nar/gkn900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer R, Wang YQ, Theissen G. The naked and the dead: the ABCs of gymnosperm reproduction and the origin of the angiosperm flower. Seminars in Cell and Developmental Biology. 2010;21:118–128. doi: 10.1016/j.semcdb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- Mondragon-Palomino M, Hiese L, Harter A, Koch MA, Theissen G. Positive selection and ancient duplications in the evolution of class B floral homeotic genes of orchids and grasses. BMC Evolutionary Biology. 2009;9:81. doi: 10.1186/1471-2148-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooers AO, Schluter D. Reconstructing ancestor states with maximum likelihood: support for one- and two-rate models. Systematic Biology. 1999;48:623–633. [Google Scholar]
- Münster T, Wingen LU, Faigl W, Werth S, Saedler H, Theißen G. Characterization of three GLOBOSA-like MADS-box genes from maize: evidence for ancient paralogy in one class of floral homeotic B-function genes of grasses. Gene. 2001;262:1–13. doi: 10.1016/s0378-1119(00)00556-4. [DOI] [PubMed] [Google Scholar]
- Nagasawa N, Miyoshi M, Sano Y, et al. SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development. 2003;130:705–718. doi: 10.1242/dev.00294. [DOI] [PubMed] [Google Scholar]
- Newman JR, Wolf E, Kim PS. A computationally directed screen identifying interacting coiled coils from Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences, USA; 2000. pp. 13203–13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Krizek BA, Meyerowitz EM. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proceedings of the National Academy of Sciences, USA; 1996a. pp. 4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Wang MQ, Meyerowitz EM. DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Research. 1996b;24:3134–3141. doi: 10.1093/nar/24.16.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijpkema AS, Zethof J, Gerats T, Vandenbussche M. The petunia AGL6 gene has a SEPALLATA-like function in floral patterning. The Plant Journal. 2009;60:1–9. doi: 10.1111/j.1365-313X.2009.03917.x. [DOI] [PubMed] [Google Scholar]
- Ronai Z, Wang Y, Khandurina J, et al. Transcription factor binding study by capillary zone electrophoretic mobility shift assay. Electrophoresis. 2003;24:96–100. doi: 10.1002/elps.200390037. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rudall PJ, Remizowa MV, Prenner G, Prychid CJ, Tuckett RE, Sokoloff DD. Nonflowers near the base of extant angiosperms? Spatiotemporal arrangement of organs in reproductive units of Hydatellaceae and its bearing on the origin of the flower. American Journal of Botany. 2009;96:67–82. doi: 10.3732/ajb.0800027. [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Hue I, Huijser P, et al. Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens – evidence for DNA-binding and autoregulation of its persistent expression throughout flower development. EMBO Journal. 1992;11:251–263. doi: 10.1002/j.1460-2075.1992.tb05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C, Immink RG, Angenent GC, Kaufmann K. Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development. 2012;139:3081–3098. doi: 10.1242/dev.074674. [DOI] [PubMed] [Google Scholar]
- Smirnova E, Shurland DL, Newman-Smith ED, Pishvaee B, van der Bliek AM. A model for dynamin self-assembly based on binding between three different protein domains. Journal of Biological Chemistry. 1999;274:14942–14947. doi: 10.1074/jbc.274.21.14942. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE, Kim S, Chanderbali A, Buzgo M. Expression of floral regulators in basal angiosperms and the origin and evolution of ABC-function. Advances in Botanical Research. 2006;44:483–506. [Google Scholar]
- Soltis DE, Chanderbali AS, Kim S, Buzgo M, Soltis PS. The ABC model and its applicability to basal angiosperms. Annals of Botany. 2007;100:155–163. doi: 10.1093/aob/mcm117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis PS, Brockington SF, Yoo MJ, et al. Floral variation and floral genetics in basal angiosperms. American Journal of Botany. 2009;96:110–128. doi: 10.3732/ajb.0800182. [DOI] [PubMed] [Google Scholar]
- Sundström J, Engström P. Conifer reproductive development involves B-type MADS-box genes with distinct and different activities in male organ primordia. The Plant Journal. 2002;31:161–169. doi: 10.1046/j.1365-313x.2002.01343.x. [DOI] [PubMed] [Google Scholar]
- Theißen G. Development of floral organ identity: stories from the MADS house. Current Opinion in Plant Biology. 2001;4:75–85. doi: 10.1016/s1369-5266(00)00139-4. [DOI] [PubMed] [Google Scholar]
- Theißen G, Melzer R. Molecular mechanisms underlying origin and diversification of the angiosperm flower. Annals of Botany. 2007;100:603–619. doi: 10.1093/aob/mcm143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theißen G, Saedler H. Plant biology. Floral quartets. Nature. 2001;409:469–471. doi: 10.1038/35054172. [DOI] [PubMed] [Google Scholar]
- Thompson BE, Bartling L, Whipple C, et al. bearded-ear encodes a MADS box transcription factor critical for maize floral development. The Plant Cell. 2009;21:2578–2590. doi: 10.1105/tpc.109.067751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M, Zethof J, Royaert S, Weterings K, Gerats T. The duplicated B-class heterodimer model: whorl-specific effects and complex genetic interactions in Petunia hybrida flower development. The Plant Cell. 2004;16:741–754. doi: 10.1105/tpc.019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YQ, Melzer R, Theissen G. Molecular interactions of orthologues of floral homeotic proteins from the gymnosperm Gnetum gnemon provide a clue to the evolutionary origin of ‘floral quartets. The Plant Journal. 2010;64:177–190. doi: 10.1111/j.1365-313X.2010.04325.x. [DOI] [PubMed] [Google Scholar]
- Whipple CJ, Schmidt RJ. Genetics of grass flower development. Advances in Botanical Research. 2006;44:385–424. [Google Scholar]
- Whipple CJ, Ciceri P, Padilla CM, Ambrose BA, Bandong SL, Schmidt RJ. Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development. 2004;131:6083–6091. doi: 10.1242/dev.01523. [DOI] [PubMed] [Google Scholar]
- Winter KU, Saedler H, Theißen G. On the origin of class B floral homeotic genes: functional substitution and dominant inhibition in Arabidopsis by expression of an orthologue from the gymnosperm Gnetum. The Plant Journal. 2002a;31:457–475. doi: 10.1046/j.1365-313x.2002.01375.x. [DOI] [PubMed] [Google Scholar]
- Winter KU, Weiser C, Kaufmann K, et al. Evolution of class B floral homeotic proteins: obligate heterodimerization originated from homodimerization. Molecular Biology and Evolution. 2002b;19:587–596. doi: 10.1093/oxfordjournals.molbev.a004118. [DOI] [PubMed] [Google Scholar]
- Yang YZ, Fanning L, Jack T. The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. The Plant Journal. 2003;33:47–59. doi: 10.1046/j.0960-7412.2003.01473.x. [DOI] [PubMed] [Google Scholar]
- Yao SG, Ohmori S, Kimizu M, Yoshida H. Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development. Plant and Cell Physiology. 2008;49:853–857. doi: 10.1093/pcp/pcn050. [DOI] [PubMed] [Google Scholar]
- Zahn LM, King HZ, Leebens-Mack JH, et al. The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics. 2005a;169:2209–2223. doi: 10.1534/genetics.104.037770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn LM, Leebens-Mack J, Depamphilis CW, Ma H, Theißen G. To B or not to B a flower: the role of DEFICIENS and GLOBOSA orthologs in the evolution of the angiosperms. Journal of Heredity. 2005b;96:225–240. doi: 10.1093/jhered/esi033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.