Significance
Quorum sensing (QS) is a coordinated gene-regulation system that controls bacterial social behaviors, such as virulence, motility, biofilm formation, and toxin production, in response to cell density. Acyl-homoserine lactone-mediated QS coordinates cooperativity between individual cells of many Proteobacteria species. QS may also control nutrient acquisition and help maintain the homeostatic primary metabolism of individuals in a cooperative population. Here, we show that QS restricts glucose uptake and slows primary metabolism of individual cells in crowded conditions. QS-deficient cells experienced serious physiological challenges under similar conditions, indicating that QS functions as a means to ensure efficient energy and resource utilization of individuals in crowded environments.
Abstract
Acyl-homoserine lactone (AHL)-mediated quorum sensing (QS) controls the production of numerous intra- and extracellular products across many species of Proteobacteria. Although these cooperative activities are often costly at an individual level, they provide significant benefits to the group. Other potential roles for QS include the restriction of nutrient acquisition and maintenance of metabolic homeostasis of individual cells in a crowded but cooperative population. Under crowded conditions, QS may function to modulate and coordinate nutrient utilization and the homeostatic primary metabolism of individual cells. Here, we show that QS down-regulates glucose uptake, substrate level and oxidative phosphorylation, and de novo nucleotide biosynthesis via the activity of the QS-dependent transcriptional regulator QsmR (quorum sensing master regulator R) in the rice pathogen Burkholderia glumae. Systematic analysis of glucose uptake and core primary metabolite levels showed that QS deficiency perturbed nutrient acquisition, and energy and nucleotide metabolism, of individuals within the group. The QS mutants grew more rapidly than the wild type at the early exponential stage and outcompeted wild-type cells in coculture. Metabolic slowing of individuals in a QS-dependent manner indicates that QS acts as a metabolic brake on individuals when cells begin to mass, implying a mechanism by which AHL-mediated QS might have evolved to ensure homeostasis of the primary metabolism of individuals under crowded conditions.
Acyl-homoserine lactone (AHL)-mediated quorum sensing (QS) controls diverse behaviors, including virulence, biofilm formation, and motility, in many Proteobacteria (1–3). Such QS-dependent activities are the result of density-dependent expression of both intra- and extracellular gene products important for survival in crowded conditions (4–7). Other roles for bacterial QS have been proposed, including allowing bacteria to control nutrient uptake and maintain individual metabolic homeostasis within a crowded but cooperative population. The metabolic status of bacteria is usually defined as the average metabolic activity of individual cells in a population; however, the concepts of population biology have often been ignored in this context.
Little is known regarding whether or not individual cells change their primary metabolism under crowded, but cooperative, conditions or how they maintain metabolic homeostasis at the population level. In addition to the feedback inhibition circuits characteristic of many biochemical processes (8–10), we hypothesized that QS might control both glucose uptake and metabolic homeostasis of individual cells in crowded populations based upon earlier analyses of QS-dependent gene expression in Burkholderia glumae and the role of QS in regulating the respiration of Burkholderia thailandensis (11, 12). The model organism used in this study, B. glumae, is an important agricultural pathogen due to its ability to cause rice panicle blight. Compared with other closely related pathogenic bacteria, B. glumae is relatively easy to handle in the laboratory and contains only a single LuxI-R-type QS system, signal synthase (TofI), synthesizing octanoyl-homoserine lactone (C8-HSL), and the cognate receptor TofR (13), making this organism an ideal model for the study of QS in bacteria. C8-HSL binds to TofR, activating expression of an isocitrate lyase regulator R (IclR)-type transcriptional regulator QsmR, which in turn activates a series of genes associated with the production of intra- and extracellular products important for survival in crowded conditions (14–16).
Here, we address two major questions regarding the individual cellular response to QS in B. glumae. First, we investigate whether individual cells restrict nutrient acquisition under crowded conditions, as a QS-mediated cooperative activity. Next, we ask whether QS mechanisms control the primary metabolism of individual cells as a means of maintaining metabolic homeostasis within the group. We found that QS activity directly modulates glucose uptake and slows primary metabolism at the level of substrate metabolism, oxidative phosphorylation, and de novo nucleotide biosynthesis within individual cells as a means of ensuring homeostatic metabolism under crowded but cooperative conditions.
Results
QS Modulates Glucose Uptake.
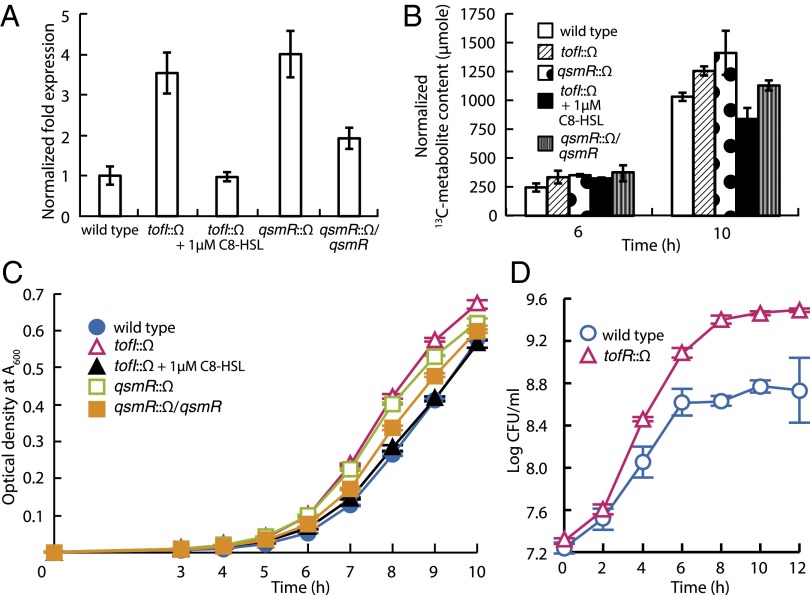
Based on a previous analysis of QS-dependent gene expression in B. glumae (11), we hypothesized that QS might affect glucose uptake in individual cells. To determine the effect of QS on glucose uptake, we chose ptsI (bglu_1g31820), one of two phosphoenolpyruvate-protein phosphotransferase genes and a representative gene in the multicomponent phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS), based on previously published RNAseq results (11) and measured the expression levels in wild type (BGR1), the tofI mutant BGS2, and the qsmR mutant BGS9. Expression of ptsI was significantly higher in the QS mutants than the wild type (Fig. 1A). The levels of transported d-glucose-1-[13C] were significantly higher in the QS mutants than the wild type, as assessed using [13C]-NMR spectroscopy (Fig. 1B and SI Appendix, Fig. S1). Levels of glucose uptake in the tofI mutant BGS2 were restored to wild-type levels by addition of 1 µM exogenous C8-HSL (Fig. 1B). Together, these results show that the rate of glucose uptake is regulated in part by the QS response. However, the mutation of ptsI did not cause a significant change in growth rate compared with wild type (SI Appendix, Fig. S2).
Fig. 1.
Down-regulation of glucose uptake mediated by QS and outgrowth of QS mutants. (A) Expression of the phosphoenolpyruvate-protein phosphotransferase-encoding gene (ptsI) in B. glumae. The expression levels of ptsI were higher in the QS-null mutants than the wild type, as revealed by quantitative reverse transcription-PCR. (B) Total amounts of [13C] after feeding of d-glucose-1-[13C] in the wild type and QS mutants of B. glumae. The total cellular amounts of [13C] were higher in QS mutants than the wild type as assessed by integration of peaks generated by [13C]-NMR spectroscopy. Error bars represent the error ranges of triplicate experiments. (C) Difference of growth rate between the wild-type BGR1 and the QS mutants. The QS mutants grew more rapidly at the early exponential stage than did the wild type, and statistical analysis of the data are presented in SI Appendix, Fig. S4. (D) Competition assays between the tofR mutant BGS1 and the wild-type BGR1. The tofR mutant BGS1 outcompeted the wild-type strain in coculture.
Growth of QS Mutants.
To investigate whether the growth rate of QS mutants might be different from that of the wild type, we monitored the growth of wild-type cells (BGR1), the tofI mutant BGS2, the tofR mutant BGS1, and the qsmR mutant BGS9 in Luria–Bertani (LB) medium. The QS mutants grew more rapidly at the early exponential stage than did the wild type (Fig. 1C and SI Appendix, Figs. S3 and S4). The growth rate of BGS2 was recovered to the wild-type level by the exogenous addition of 1 µM C8-HSL (Fig. 1C). A complemented strain (S9C17) of the qsmR mutant BGS9 exhibited a growth phenotype similar to that of BGR1 (Fig. 1C). The addition of 1 µM C8-HSL to BGS1 did not change the growth pattern (SI Appendix, Figs. S3 and S4). To explore the competitive fitness of the QS mutants, we cocultured BGS1 and BGR1 and monitored the colony-forming units (cfus) of each strain. We found that the tofR mutant BGS1 outcompeted the wild-type strain in coculture (Fig. 1D).
Primary Metabolic Activity Is Higher in the QS Mutants than the Wild Type.
To demonstrate that QS acts as a regulator of metabolic homeostasis, we quantitatively analyzed the levels of 75 cationic and anionic core primary metabolites of substrate-level and oxidative phosphorylation, the pentose phosphate pathway, and de novo purine and pyrimidine biosynthesis. We performed capillary electrophoresis time-of-flight mass spectrometry (CE-TOFMS) 6 h and 10 h after bacterial subculture in LB or buffered LB (BLB) broth, reflecting the early and midstages of the QS response, respectively. The average levels of all metabolites in wild-type BGR1, tofI mutant BGS2, and qsmR mutant BGS9 were 109,616 pmol/109 cells, 124,620 pmol/109 cells, and 134,059 pmol/109 cells, respectively, at 6 h after subculture in LB media. These levels increased to 126,419 pmol/109 cells, 209,024 pmol/109 cells, and 186,800 pmol/109 cells, respectively, at 10 h (SI Appendix, Table S1). The differences between the 10- and 6-h totals reflect a ratio of 1.15 in the wild-type strain, compared with 1.39 and 1.68 in the QS mutants. These data indicate that the level of primary metabolic activity was higher in the QS mutants than the wild type.
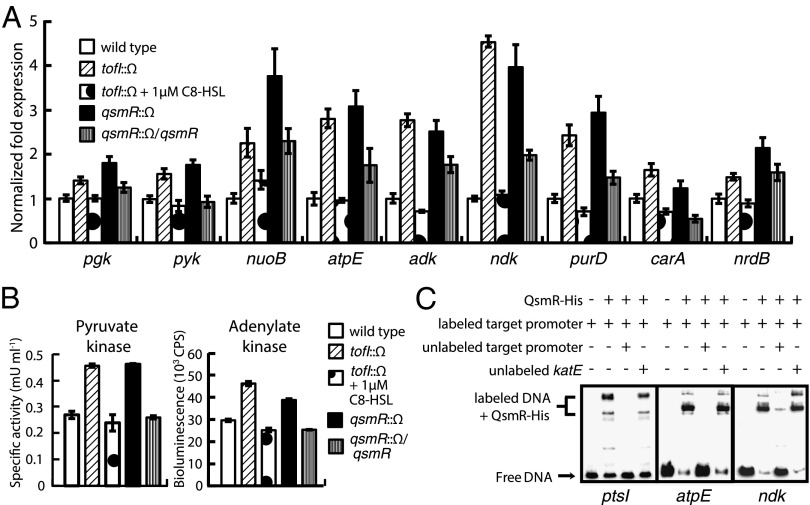
We found that significant differences in the levels of key metabolites of substrate-level and oxidative phosphorylation were evident between the wild-type strain and the QS mutants (SI Appendix, Table S1). For example, glucose-6-phosphate, 3-phosphoglycerate, and phosphoenolpyruvate levels were all significantly higher in the tofI mutant BGS2 than in the wild-type strain 10 h after subculture (Fig. 2 and SI Appendix, Fig. S5 and Table S1). These results are consistent with repression of glucose uptake by a QS-dependent mechanism. When we assessed the expression levels of the phosphoglycerate kinase (pgk) and pyruvate kinase (pyk) genes, using our earlier RNAseq data, we found that both were higher in the QS mutants than the wild type (Fig. 3A). In addition, the level of pyruvate kinase activity was higher in the QS mutants than the wild type (Fig. 3B). In terms of oxidative phosphorylation, the expression levels of the NADH dehydrogenase (nuoB) and F0F1ATP synthase C (atpE) genes were significantly higher in the QS mutants than the wild type (Fig. 3A). QsmR was shown to bind directly to the putative promoter regions of ptsI, pgk, pyk, nuoB, and atpE (Fig. 3C and SI Appendix, Fig. S6).
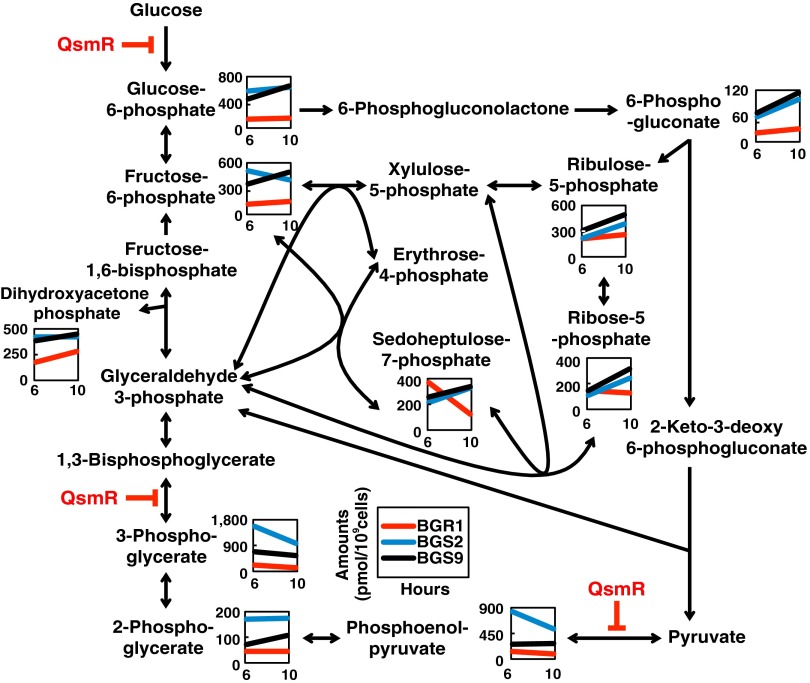
Fig. 2.
QS-dependent changes in the levels of metabolites involved in central carbon metabolism. The levels of metabolites of the Entner–Doudoroff and pentose phosphate pathways were plotted on pathway maps. The x and y axes refer to two different incubation times and show metabolite concentrations (in pmol per 109 cells), respectively. Red, blue, and black lines indicate wild-type BGR1, the tofI mutant BGS2, and the qsmR mutant BGS9, respectively. Blunt arrows in red indicate QsmR-mediated repression of target genes. Each value is an average of data from three independent experiments, and statistical analysis of the data are presented in SI Appendix, Fig. S5.
Fig. 3.
Repression of substrate-level and oxidative phosphorylation, and de novo nucleotide biosynthesis, by QsmR. (A) The expression levels of nine genes involved in substrate-level and oxidative phosphorylation, and de novo nucleotide biosynthesis, in the wild type and QS mutants of B. glumae. The expression levels were determined by quantitative reverse transcription-PCR. (B) The activities of pyruvate kinase and adenylate kinase were higher in the QS mutants than wild-type B. glumae. (C) Direct binding of QsmR to the promoter regions of the ptsI, atpE, and ndk genes. The electrophoretic mobility-shift assay confirmed direct control of ptsI, atpE, and ndk by QsmR. Error bars represent the error ranges of triplicate experiments.
Regulation of Pentose Phosphate Pathway by QS.
As both uptake and utilization of glucose were elevated in the QS mutants, we next compared the levels of metabolites associated with the pentose phosphate pathway because B. glumae uses the Entner–Doudoroff pathway rather than glycolysis. Concentrations of both ribose-5-phosphate and ribulose-5-phosphate increased gradually over time in the QS mutants but remained relatively constant in the wild type (Fig. 2). However, levels of sedoheptulose 7-phosphate were dramatically decreased in the wild type but showed slight increases over time in the QS mutants (Fig. 2). Furthermore, because the pentose phosphate pathway is the main source of bacterial NADPH, we also measured NADP+ concentrations. NADP+ concentrations were significantly higher in the QS mutants at 10 h after subculture compared with the wild type (SI Appendix, Fig. S5 and Table S1), indicative of higher levels of pathway activation in the QS mutants.
Regulation of de Novo Nucleotide Biosynthesis by QS.
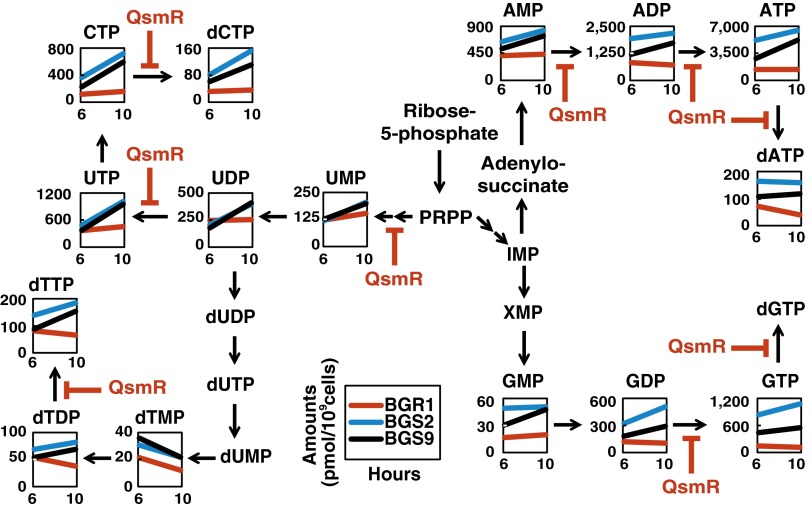
To determine whether the suppressive effects of QS on glucose metabolism were also reflected in the biosynthesis of ribonucleoside 5′-triphosphates (NTPs) and 5′-diphosphates (NDPs), we compared levels of NTPs and NDPs in the wild type and QS mutants. The concentrations of all NTPs were significantly higher in the QS mutants compared with the wild type 10 h after subculture (Fig. 4 and SI Appendix, Table S1). The ATP/ADP ratio remained relatively constant in the wild type but increased from 2.0 to 3.0 over time in the QS mutants (SI Appendix, Table S1). The ATP level was therefore homeostatic in the wild type, but not in the QS mutants. Another line of evidence was afforded by analysis of the differential expression of the adenylate kinase (adk) and nucleoside-diphosphate kinase (ndk) genes involved in de novo purine biosynthesis (Fig. 3A). Adenylate kinase activity was elevated in the QS mutants compared with the wild type (Fig. 3B). To explain why the other NTPs were also unbalanced, we estimated the expression levels of the phosphoribosylglycinamide synthetase D (purD) and carbamoyl phosphate synthase A (carA) genes involved in de novo purine and pyrimidine biosynthesis, respectively, as suggested by our earlier RNAseq data (11). The expression levels of both genes were higher in QS mutants than in the wild type (Fig. 3A), explaining the observed imbalances in other NTPs in the QS mutants. We also observed imbalances in the levels of deoxyribonucleoside 5′-triphosphates (dNTPs) and higher-level expression of the ribonucleotide reductase B (nrdB) gene, whose product catalyzes formation of deoxyribonucleotides from ribonucleotides, in the QS mutants compared with the wild type (Fig. 3A and SI Appendix, Table S1). Direct binding of QsmR to the putative promoter regions of the adk, ndk, purD, carA, and nrdB genes supported the notion that these genes were regulated by QsmR (Fig. 3C and SI Appendix, Fig. S6).
Fig. 4.
Down-regulation of nucleotide metabolism by QsmR. The levels of intermediates of nucleotide metabolism were plotted on pathway maps. The x and y axes refer to two different incubation times and show metabolite concentrations (in pmol per 109 cells), respectively. Red, blue, and black lines indicate wild-type BGR1, the tofI mutant BGS2, and the qsmR mutant BGS9, respectively. Blunt arrows in red indicate QsmR-mediated repression of target genes. Each value is an average of data from at least three independent experiments, and statistical analyses of the data are presented in SI Appendix, Fig. S5.
Discussion
There is accumulating evidence that QS controls cooperative activities of bacteria and confers benefits upon individuals in the group. However, among QS-mediated cooperative activities, one long-standing idea that has not yet been addressed is whether QS mechanisms can regulate the nutrient uptake of individual cells as a form of cooperative activity. We believe that we have uncovered a previously unknown cooperative activity of AHL-mediated QS in B. glumae. QS-mediated repression of multiple intracellular products to maintain energy and nucleotide homeostasis represents a case where QS functions as a metabolic brake. QS-mediated control of glucose utilization and primary metabolism adds another tier of regulation in primary metabolism, functioning alongside a variety of other systems, including feedback inhibition in bacteria.
QS did not affect the metabolic diversity in phenotype-microarray experiments of B. thailandensis; however, it did have a negative influence on the respiration rate using several carbon and nitrogen sources (12). One possible explanation for this difference is that the metabolic costs of producing QS-induced factors lead to the slower growth of the wild-type strain relative to the QS mutant. Likewise, we believe that the metabolic costs to produce QS-induced factors may have led to the metabolite imbalance in B. glumae. The effects of such costs on the general metabolism and growth of QS mutants might be an issue if nutrients are limiting in terms of growth. However, it is not clear whether metabolic costs seriously affect general metabolism upon growth in a nutrient-rich medium such as LB. Alternatively, we explored the possibility that QS might repress primary metabolism directly in this study. We showed that multiple genes encoding enzymes associated with glucose uptake and primary metabolism are directly repressed by QS-dependent QsmR.
The phosphoenolpyruvate-dependent sugar PTS is a common method for transporting sugars into bacteria (17, 18). Our data showed that QsmR represses the expression of ptsI, indicating that QsmR directly represses the phospho-transfer of glucose in B. glumae. To our knowledge, this is the first report that QS or QS-dependent transcriptional regulators directly control nutrient acquisition of individual cells in crowded conditions. This restriction of glucose consumption of individual cells in a cooperative population is somewhat similar to calorie restriction in mammalian cells (19) and is consistent with a report demonstrating a negative influence of QS on the rate of respiration in B. thailandensis in the presence of various carbon and nitrogen sources (12). However, the growth rate of the ptsI mutant was similar to that of the wild type. No difference of growth rate of the ptsI mutant compared to the wild type may be due to genetic redundancy; alternatively, the phosphoenolpyruvate-dependent sugar PTS might not be crucial for growth in LB, which is a glucose-limited medium.
Repression of the pgk and pyk genes by QsmR would affect the cellular concentration of ATP because phosphoglycerate kinase and pyruvate kinase are two key enzymes involved in the substrate-level phosphorylation that generates ATP. The ATP imbalance observed in the QS mutants may also be attributable to elevated expression of nuoB and atpE, which are involved in oxidative phosphorylation and proton-motive force-driven ATP biosynthesis, respectively, in those mutants. Furthermore, high levels of ribose-5-phosphate in QS mutants could also cause an imbalance in nucleotide concentration because intracellular ribose-5-phosphate is the primary regulator of de novo purine and pyrimidine biosynthesis (20). Further evidence shows that the nucleotide imbalance resulted from dysregulation of de novo nucleotide biosynthesis by QsmR.
Imbalance in the ATP/ADP ratio may be due to elevated adenylate kinase and nucleoside-diphosphate kinase activities in the QS mutants compared with wild type because these enzymes are essential for maintaining equilibrium among the various nucleoside triphosphates (21, 22). These data therefore show that the QS mutants suffer from energy and nucleotide imbalance because ATP is the general energy donor of metabolism. Furthermore, we observed a dNTP imbalance in the QS mutants, suggesting that a QS deficiency may negatively affect the genetic stability of B. glumae because dNTP imbalances have been shown to trigger high-frequency mutation in Escherichia coli (23, 24).
Elevated expression of nuoB in QS mutants would result in higher levels of NAD+ in the QS mutants compared with the wild type (SI Appendix, Table S1). Similarly, higher levels of NADP+ in the QS mutants may be attributable to elevation of the activity of the pentose phosphate pathway, a major source of bacterial NADPH. Together with the higher levels of oxidized divalent glutathione evident in QS mutants (SI Appendix, Table S1), the data suggest that QS mutants are likely exposed to more oxidative stress than the wild type because NADPH and reduced monovalent glutathione are the principal reducing agents protecting cells against toxic reactive oxygen species. These results are consistent with our previous report that the QS mutants have lower catalase activity (25).
In AHL QS-mediated gene regulation, a LuxR-type regulator can act as either a transcriptional activator or a repressor (26–28) although evidence of simultaneous activation and repression is relatively rare. According to genome-wide AHL-dependent analyses of Pseudomonas aeruginosa and other bacteria, there are indications that QS negatively influences gene expression (29, 30). More recently, a QS-dependent transcriptome analysis of B. thailandensis found that the expression of genes encoding HPr, cytidylate kinase, and ribonucleotide diphosphate reductase, which are involved in glucose uptake and nucleotide metabolism, respectively, was down-regulated by QS (31). It should be noted that the strains of B. thailandensis were grown in LB supplemented with 50 mM 3-(N-morpholino) propanesulfonic acid, which is different from the growth conditions we used for the growth of B. glumae. Nonetheless, these results indicate that QS-mediated metabolic slowing is not an unusual phenotype in B. glumae.
We report that QS-dependent QsmR represses multiple genes encoding important intracellular products in B. glumae. Repression of multiple genes by QsmR is not unusual because IclR-type transcriptional regulators have been shown to function as both activators and repressors in other bacteria (26). Repression of multiple enzymes involved in glucose uptake and primary metabolism by QsmR is therefore a normal aspect of QS, but also the result of indirect control of QS in B. glumae.
One might argue that imbalance of primary metabolism in QS mutants occurred as a result of direct control of glucose uptake by QsmR. However, it should be noted that d-glucose-1-[13C] was added to LB media for all glucose uptake assays, but not for metabolomic analyses. Considering that LB is a glucose-limited medium (32), it is unlikely that different glucose uptake rates between the wild type and QS mutants seriously affected the balance of primary metabolism. Nevertheless, we cannot rule out a possibility of different uptake rates for amino acids present in LB medium, which might affect primary carbon metabolism.
By repressing the synthesis of multiple intracellular products, QS responses benefit both individual cells as well as the group by placing more restrictive controls on nutrient utilization and cellular energy consumption under high-cell-density conditions. It is plausible that the repression of multiple intracellular products by QS as a mechanism of cooperativity might provide survival benefits under conditions of nutrient limitation brought on by crowding. Our systematic analysis of the levels of core primary metabolites and the molecular and biochemical evidence supporting these claims clearly show that QS acts as a metabolic brake on individuals within a group and reveal that individuals in a crowded but noncooperative population experience serious physiological challenges. In conclusion, QS slows metabolic functions to ensure homeostasis of the primary metabolism of individual cells in crowded situations and is a form of cooperative activity.
Materials and Methods
Strains and growth conditions are described in SI Appendix, SI Materials and Methods. Primers are listed in SI Appendix, Table S3. Detailed protocols for statistical analysis, RNA isolation, quantitative reverse transcription-PCR, and electrophoretic mobility-shift assay are described in SI Appendix, SI Materials and Methods.
Growth Monitoring and Competition Assay.
BGR1 (wild type), three QS mutants, BGS1 (BGR1 tofR::Ω), BGS2 (BGR1 tofI::Ω), and BGS9 (BGR1 qsmR::Ω), and KJ1120 (BGR1 ptsI::mini-Tn5rescue) were grown overnight and diluted to ∼1 × 106 CFU/mL for subculture in LB. The optical density (OD) at A600 was recorded every hour from 3 h until 11 h after subculture. For the competition experiments, the tofR mutant BGS1 and wild type were grown individually overnight and diluted to OD = 0.05 at A600 to give ∼2 × 107 cfu/mL. Cells of each strain were mixed at a 1:1 ratio. After coculture, the cells were spotted onto LB agar plates and LB agar plates supplemented with 100 μg/mL spectinomycin from serial dilutions. Colonies grown on LB agar plates containing spectinomycin were counted as the tofR mutant BGS1; these numbers were subtracted from the cell numbers on LB agar plates to calculate the wild-type cell number.
Quantitative Analysis of d-glucose-1-[13C] Uptake Using NMR Spectroscopy.
Seed cultures of B. glumae BGR1 and two QS-defective mutants, BGS2 (BGR1 tofI::Ω) and BGS9 (BGR1 qsmR::Ω), were subcultured for 6 h and 10 h. Cells (8.0 × 108 and 3.0 × 109 at 6 h and 10 h, respectively) were harvested from 10-mL subcultures, washed thoroughly in sterilized distilled water to remove residual d-glucose-1-[13C], and resuspended in 0.5 mL of 99% (vol/vol) D2O. As a reference, 0.5 µL of 0.5 M trimethylsilyl propanoic acid (TSP) (100 µmol) was added to all samples. Pulsed [13C]-NMR spectra were obtained on a Bruker Avance 600 MHz spectrometer with a probe temperature of 27 °C, operating at 150.90 MHz, using the following parameters: a 30° pulse angle, a spectral width of 42.373 kHz, 65-K data points, an acquisition time of 0.7733 s, and a 2.0-s relaxation delay. Typically, 10,240 scans were accumulated, and line broadening of 3 Hz was used during data processing. Expression levels were assigned to the integrated values of individual peaks, which were identified by reference to the chemical shift of the reference, TSP, at 0 ppm.
Metabolome Analysis.
Intracellular metabolites of B. glumae BGR1 and two QS-defective mutants, BGS2 (BGR1 tofI::Ω) and BGS9 (BGR1 qsmR::Ω), were extracted as follows. Cells were grown in either LB or BLB medium at 37 °C. Culture aliquots (2 mL) were collected 6 h and 10 h after subculture, diluted to 1 × 109 cells per mL, and passed through a 0.45-μm filter. Cells were washed with 20-mL Milli-Q water to remove any residual LB medium, and the filter was then placed into sealable Petri dishes containing a 2-mL methanolic solution of internal standards (5 μM), and sonicated for 30 s to resuspend the cells. Next, 1.6-mL of chloroform and 640-μL of Milli-Q water were added, and the suspension was mixed thoroughly. The aqueous layer (375 μL) was then collected and filtered via centrifugation through a Millipore 5-kDa-cutoff filter to remove soluble proteins. The filtrate was dried under vacuum and dissolved in 50 μL of Milli-Q water. Samples were then analyzed by CE-TOFMS; intracellular metabolites were quantified as described previously (33, 34). All experiments were performed using at least three independent replicates.
Pyruvate Kinase Assay.
Pyruvate kinase activity was measured using a fluorometric assay kit (Biovision) according to the manufacturer’s instructions. In this assay, phosphoenolpyruvate and ADP react to form pyruvate and ATP, in a reaction catalyzed by pyruvate kinase. The resulting pyruvate is then oxidized by pyruvate oxidase to produce a colored fluorescent product. We measured fluorescence using Ex/Em wavelengths of 535/587 nm every 5 min for 30 min at 25 °C. Pyruvate kinase activity was calculated using a standard curve, and the ΔF and ΔT values were obtained from the region of linear signal increase.
Adenylate Kinase Assay.
Adenylate kinase activity was measured using a ToxiLight bioassay kit (Lonza) according to the manufacturer’s instructions. ADP is converted to ATP by adenylate kinase. Luciferase catalyzes the formation of light from ATP and luciferin. When the two reactions are combined, the emitted light intensity is linearly related to adenylate kinase concentration and is measured using a multilabel plate reader (PerkinElmer).
Supplementary Material
Acknowledgments
This work was supported by the Creative Research Initiatives Programs (2010-0018280) of the National Research Foundation of Korea.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412431111/-/DCSupplemental.
References
- 1.Fuqua C, Greenberg EP. Listening in on bacteria: Acyl-homoserine lactone signalling. Nat Rev Mol Cell Biol. 2002;3(9):685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 2.Parsek MR, Greenberg EP. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol. 2005;13(1):27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuster M, Sexton DJ, Diggle SP, Greenberg EP. Acyl-homoserine lactone quorum sensing: From evolution to application. Annu Rev Microbiol. 2013;67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- 5.Darch SE, West SA, Winzer K, Diggle SP. Density-dependent fitness benefits in quorum-sensing bacterial populations. Proc Natl Acad Sci USA. 2012;109(21):8259–8263. doi: 10.1073/pnas.1118131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dandekar AA, Chugani S, Greenberg EP. Bacterial quorum sensing and metabolic incentives to cooperate. Science. 2012;338(6104):264–266. doi: 10.1126/science.1227289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pai A, Tanouchi Y, You L. Optimality and robustness in quorum sensing (QS)-mediated regulation of a costly public good enzyme. Proc Natl Acad Sci USA. 2012;109(48):19810–19815. doi: 10.1073/pnas.1211072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savageau MA. Optimal design of feedback control by inhibition: Dynamic considerations. J Mol Evol. 1975;5(3):199–222. doi: 10.1007/BF01741242. [DOI] [PubMed] [Google Scholar]
- 9.Kell DB, Westerhoff HV. Metabolic control theory: Its role in microbiology and biotechnology. FEMS Microbiol Rev. 1986;39(4):305–320. [Google Scholar]
- 10.Reaves ML, Young BD, Hosios AM, Xu YF, Rabinowitz JD. Pyrimidine homeostasis is accomplished by directed overflow metabolism. Nature. 2013;500(7461):237–241. doi: 10.1038/nature12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goo E, et al. Bacterial quorum sensing, cooperativity, and anticipation of stationary-phase stress. Proc Natl Acad Sci USA. 2012;109(48):19775–19780. doi: 10.1073/pnas.1218092109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandler JR, et al. Mutational analysis of Burkholderia thailandensis quorum sensing and self-aggregation. J Bacteriol. 2009;191(19):5901–5909. doi: 10.1128/JB.00591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J, et al. Quorum sensing and the LysR-type transcriptional activator ToxR regulate toxoflavin biosynthesis and transport in Burkholderia glumae. Mol Microbiol. 2004;54(4):921–934. doi: 10.1111/j.1365-2958.2004.04338.x. [DOI] [PubMed] [Google Scholar]
- 14.Kim J, et al. Regulation of polar flagellum genes is mediated by quorum sensing and FlhDC in Burkholderia glumae. Mol Microbiol. 2007;64(1):165–179. doi: 10.1111/j.1365-2958.2007.05646.x. [DOI] [PubMed] [Google Scholar]
- 15.Goo E, Kang Y, Kim H, Hwang I. Proteomic analysis of quorum sensing-dependent proteins in Burkholderia glumae. J Proteome Res. 2010;9(6):3184–3199. doi: 10.1021/pr100045n. [DOI] [PubMed] [Google Scholar]
- 16.Kim H, Goo E, Kang Y, Kim J, Hwang I. Regulation of universal stress protein genes by quorum sensing and RpoS in Burkholderia glumae. J Bacteriol. 2012;194(5):982–992. doi: 10.1128/JB.06396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Postma PW, Lengeler JW, Jacobson GR. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57(3):543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70(4):939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bishop NA, Guarente L. Genetic links between diet and lifespan: Shared mechanisms from yeast to humans. Nat Rev Genet. 2007;8(11):835–844. doi: 10.1038/nrg2188. [DOI] [PubMed] [Google Scholar]
- 20.Turnbough CL, Jr, Switzer RL. Regulation of pyrimidine biosynthetic gene expression in bacteria: Repression without repressors. Microbiol Mol Biol Rev. 2008;72(2):266–300. doi: 10.1128/MMBR.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noda L. In: The Enzymes. Boyer PD, editor. Academic; New York: 1973. pp. 279–305. [Google Scholar]
- 22.Ray NB, Mathews CK. Nucleoside diphosphokinase: A functional link between intermediary metabolism and nucleic acid synthesis. Curr Top Cell Regul. 1992;33:343–357. doi: 10.1016/b978-0-12-152833-1.50025-3. [DOI] [PubMed] [Google Scholar]
- 23.Hofer A, Crona M, Logan DT, Sjöberg BM. DNA building blocks: Keeping control of manufacture. Crit Rev Biochem Mol Biol. 2012;47(1):50–63. doi: 10.3109/10409238.2011.630372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nordman J, Wright A. The relationship between dNTP pool levels and mutagenesis in an Escherichia coli NDP kinase mutant. Proc Natl Acad Sci USA. 2008;105(29):10197–10202. doi: 10.1073/pnas.0802816105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chun H, et al. The quorum sensing-dependent gene katG of Burkholderia glumae is important for protection from visible light. J Bacteriol. 2009;191(13):4152–4157. doi: 10.1128/JB.00227-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molina-Henares AJ, Krell T, Eugenia Guazzaroni M, Segura A, Ramos JL. Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors. FEMS Microbiol Rev. 2006;30(2):157–186. doi: 10.1111/j.1574-6976.2005.00008.x. [DOI] [PubMed] [Google Scholar]
- 27.Castang S, Reverchon S, Gouet P, Nasser W. Direct evidence for the modulation of the activity of the Erwinia chrysanthemi quorum-sensing regulator ExpR by acylhomoserine lactone pheromone. J Biol Chem. 2006;281(40):29972–29987. doi: 10.1074/jbc.M601666200. [DOI] [PubMed] [Google Scholar]
- 28.von Bodman SB, Majerczak DR, Coplin DL. A negative regulator mediates quorum-sensing control of exopolysaccharide production in Pantoea stewartii subsp. stewartii. Proc Natl Acad Sci USA. 1998;95(13):7687–7692. doi: 10.1073/pnas.95.13.7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: A transcriptome analysis. J Bacteriol. 2003;185(7):2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: Effects of growth phase and environment. J Bacteriol. 2003;185(7):2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majerczyk C, et al. Global analysis of the Burkholderia thailandensis quorum sensing-controlled regulon. J Bacteriol. 2014;196(7):1412–1424. doi: 10.1128/JB.01405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sezonov G, Joseleau-Petit D, D’Ari R. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol. 2007;189(23):8746–8749. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soga T, et al. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J Proteome Res. 2003;2(5):488–494. doi: 10.1021/pr034020m. [DOI] [PubMed] [Google Scholar]
- 34.Ishii N, et al. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science. 2007;316(5824):593–597. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.