Significance
New advances in the treatment of aggressive neuroendocrine lung cancers are needed to improve survival in patients with this class of tumors. The current treatment approach, which has remained unchanged for the past 30 years, involves combination chemotherapy and radiation. To uncover novel drug targets, we identified the transcriptome of achaete-scute homolog 1 (ASCL1), a transcription factor that is both necessary for the proper development of neuroendocrine cells and essential for the growth and survival of neuroendocrine lung cancers. Analysis of downstream targets of ASCL1 has revealed unique molecular vulnerabilities that can be exploited for future therapeutic use.
Keywords: ASCL1 transcriptome, target discovery, personalized therapy
Abstract
Aggressive neuroendocrine lung cancers, including small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), represent an understudied tumor subset that accounts for approximately 40,000 new lung cancer cases per year in the United States. No targeted therapy exists for these tumors. We determined that achaete-scute homolog 1 (ASCL1), a transcription factor required for proper development of pulmonary neuroendocrine cells, is essential for the survival of a majority of lung cancers (both SCLC and NSCLC) with neuroendocrine features. By combining whole-genome microarray expression analysis performed on lung cancer cell lines with ChIP-Seq data designed to identify conserved transcriptional targets of ASCL1, we discovered an ASCL1 target 72-gene expression signature that (i) identifies neuroendocrine differentiation in NSCLC cell lines, (ii) is predictive of poor prognosis in resected NSCLC specimens from three datasets, and (iii) represents novel “druggable” targets. Among these druggable targets is B-cell CLL/lymphoma 2, which when pharmacologically inhibited stops ASCL1-dependent tumor growth in vitro and in vivo and represents a proof-of-principle ASCL1 downstream target gene. Analysis of downstream targets of ASCL1 represents an important advance in the development of targeted therapy for the neuroendocrine class of lung cancers, providing a significant step forward in the understanding and therapeutic targeting of the molecular vulnerabilities of neuroendocrine lung cancer.
Gene expression signatures from large cohorts of lung tumors suggest that cancers with neuroendocrine features appear in ∼10% of pathologically diagnosed non-small cell lung cancers (NSCLCs) (1, 2), whereas small cell lung cancers (SCLCs) compose 15–20% of all lung cancer cases (3). In the United States, this represents nearly 40,000 patients per year presenting with a high-grade neuroendocrine lung tumor. Molecular and functional characterization of these aggressive tumors, along with the development of relevant preclinical models, is needed to rationally develop and test new targeted therapies.
A highly expressed gene in the class of neuroendocrine lung cancers is the lineage-specific transcription factor achaete-scute homolog 1 (ASCL1) (4, 5). ASCL1 is required to establish the lineage of pulmonary neuroendocrine cells (6) and is necessary for the continued survival of SCLCs (7, 8). ASCL1’s appearance in an NSCLC subset, neuroendocrine NSCLC (NE-NSCLC), is a recent and unexplained finding (9), and, importantly, its role as a potential lineage oncogene in lung tumors has been heretofore unexplored. The lineage addiction hypothesis in cancer suggests that certain tumors arise from dysregulation of genes involved in normal development. Hijacking these genes, which are involved in numerous facets of growth, cell division, and differentiation, provides a budding precancerous cell with the framework within which to progress to full tumorigenicity. The transcription factors sex determining region Y-box 2 (SOX2, required for basal cell generation) and thyroid transcription factor 1 (TTF1/NKX2.1, required for distal lung formation) have been implicated as lineage-dependent oncogenes in lung squamous cell carcinoma (10) and adenocarcinoma (11), respectively. ASCL1 may play a comparable role in ASCL1-expressing lung cancers, where, as we show here for NE-NSCLC and others have shown for SCLC, loss of ASCL1 leads to cell death, suggesting an “addiction” to ASCL1 for tumor cell survival. Pursuit of ASCL1 lineage-based drug targets may provide previously unidentified insight into the treatment of high-grade neuroendocrine lung cancers.
To extend previous findings of the dependence of SCLC on ASCL1 expression, we first identified a subset of NSCLC tumors (∼10% of NSCLSs) and cell lines that have neuroendocrine features (NE-NSCLC) and a gene expression signature similar to that of SCLC. With these NE-NSCLC lines, we verified that ASCL1 is expressed in, and required for, their continued growth and survival. We then performed ASCL1-focused ChIP-Seq analysis on ASCL1(+) NE-NSCLC and SCLC lines compared with ASCL1(−) (but neuroendocrine) SCLCs to identify downstream targets of ASCL1, to (i) elucidate the ASCL1 transcriptome in lung cancers expressing ASCL1, (ii) determine the prognostic ability of ASCL1 transcriptional targets, (iii) uncover targets of ASCL1 that are “druggable” to serve as therapeutic interventions, and (iv) provide proof-of-principle studies showing that druggable downstream ASCL1 targets can inhibit ASCL1(+) lung cancer growth. Our findings demonstrate that targeting the ASCL1 pathway is a promising route of therapy for ASCL1-addicted neuroendocrine lung cancers.
Results
Identification of a Neuroendocrine Subset of NSCLCs Expressing ASCL1.
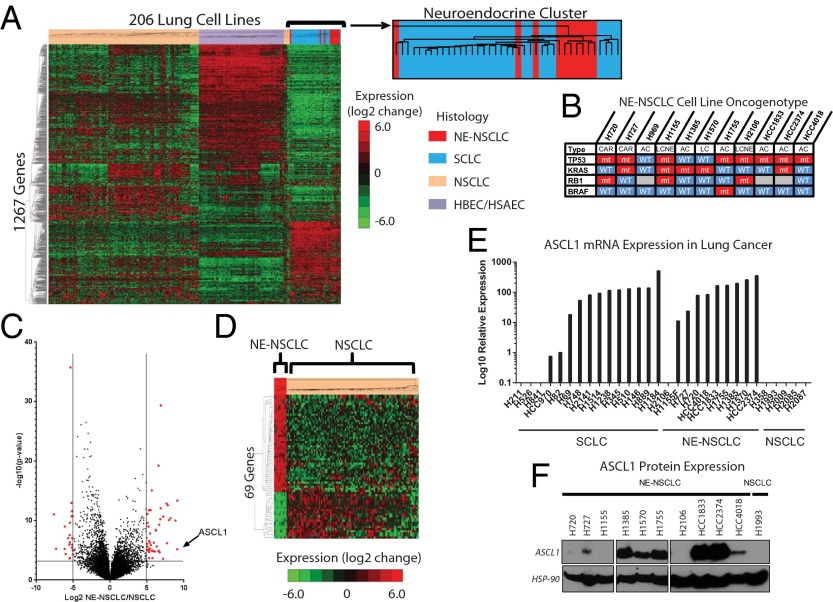
We analyzed genome-wide mRNA expression data collected from 206 human lung cell lines, including NSCLC (n = 118), SCLC (n = 29), and our own normal, immortalized human bronchial and small airway epithelial cells (HBECs/HSAECs; n = 59) (Fig. 1A) (12). An important finding was that 10 of the histologically diagnosed NSCLC cell lines (all NE-NSCLCs) grouped with SCLC, suggesting a common neuroendocrine gene expression phenotype. Only two of the lines were derived from patients with large cell neuroendocrine carcinoma (LCNEC), a neuroendocrine subtype recognized by the World Health Organization (13). It is unlikely that these NE-NSCLC cell lines were developed from patients with misdiagnosed SCLC, owing to the prevalence of KRAS mutations (5/10 lines), which are rarely if ever seen in SCLC, and the absence of RB mutations (7/10 lines), which are seen in the majority of SCLC cases (3) (Fig. 1B).
Fig. 1.
ASCL1 is highly expressed in a subset of NSCLC cell lines. (A) Unsupervised clustering analysis of genome-wide expression analysis performed on 206 lung cell lines including NSCLCs, SCLCs, and normal immortalized HBECs/HSAECs. (B) Known TP53, KRAS, RB1, and EGFR mutations for the putative NE-NSCLC cohort. (C) Log2 expression differences between NE-NSCLC and NSCLC lines plotted against –log10(P value). ASCL1 is noted. A total of 69 genes fall within the cutoff of ± log2 >5.0 and P < 0.001. (D) Supervised clustering of NSCLC cell lines using the 69 most significantly differentially expressed genes. (E) Log10 relative mRNA expression of ASCL1 in lung cancer cell lines measured by qRT-PCR. (F) Protein expression of ASCL1 in lung cancer cell lines. These data were collected from three separate gels.
We performed an extensive mutation correlation analysis between high ASCL1-expressing cell lines as well as high ASCL1-expressing NSCLCs from The Cancer Genome Atlas (TCGA) and compared them with NSCLC lines and resected tumors without ASCL1 expression. Using Fisher’s exact test, we identified nine mutated genes (CLIP1, FSHR, OR4E2, PPRC2B, REG3A, SRPX, TACR3, TROAP, and ZYMYM2) that had a higher probability of mutation in NE-NSCLCs compared with NSCLCs, albeit with low false discovery rate significance (SI Appendix, Table S1). In fact, sushi-repeat containing protein, X-linked (SRPX/DRS) is a tumor-suppressor gene previously reported to have down-regulated expression in highly malignant pulmonary neuroendocrine tumors (14). However, although our data showed increased SRPX mutation frequency, it did not show decreased expression in NE-NSCLCs. The in vitro morphology of the 10 NE-NSCLC lines is varied, including attached, floating, and combined populations, unlike the typical floating spheroids found in SCLCs (15). Whereas SCLCs are very sensitive to platinum and etoposide therapy, no difference was detected between NE-NSCLC and NSCLC cell lines in their response to these and other standard chemotherapy regimens (SI Appendix, Table S2).
A direct comparison of the genes expressed in the 10 NE-NSCLC lines versus those expressed in typical NSCLC lines demonstrated an up-regulation of a panel of well-studied neuroendocrine markers, of which ASCL1 showed the greatest differential expression (SI Appendix, Fig. S1A). A total of 69 genes were differentially expressed between NE-NSCLCs and NSCLCs (log2 > ±5.00; P < 0.001) (Fig. 1C). Those 69 genes, inclusive of ASCL1, were able to cleanly separate NE-NSCLC cell lines from NSCLC lines (Fig. 1D). Quantitative RT-PCR analysis verified ASCL1 expression levels in SCLC, NE-NSCLC, and NSCLC cell lines and was significantly correlated with microarray expression data (R2 = 0.72, P < 0.0001) (Fig. 1E). In addition, the NE-NSCLC cell lines also expressed high levels of delta-like 1 (DLL1), an established ASCL1 target (16), which was correlated with the expression pattern of ASCL1 (SI Appendix, Fig. S1B). ASCL1 protein was detected in 8 of 10 NE-NSCLC cell lines and was not expressed in a typical NSCLC cell line (Fig. 1F). NCI-H1155 expresses ASCL1 mRNA, but not protein, and is regulated by a separate neurogenic transcription factor, NeuroD1 (17). NCI-H2106 does not express ASCL1 mRNA, suggesting that a separate pathway drives the neuroendocrine phenotype in this cell line.
ASCL1 Is Required for Survival of NE-NSCLC Cell Lines.
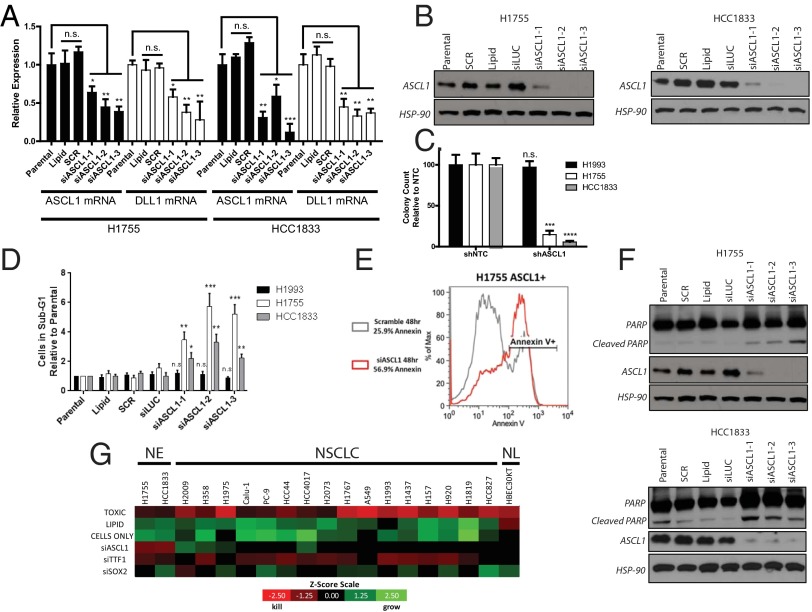
Knockdown of ASCL1 in ASCL1(+) NE-NSCLC lines NCI-H1755 and HCC1833 using three independent siRNAs resulted in significant down-regulation of ASCL1 mRNA and protein (Fig. 2 A and B), with a concomitant reduction in DLL1 mRNA. Lentiviral shRNAs targeting ASCL1 or shRNAs with a nontargeting sequence (NTC) were transfected into ASCL1(+) and ASCL1(−) lung cancer lines and selected with puromycin for 1 wk. Liquid colony-formation assays showed significant inhibition of colony-forming ability by the ASCL1 shRNA in ASCL1(+) lung cancer lines (NCI-H1755 and HCC1833) compared with an ASCL1(−) lung cancer line (NCI-H1993) (Fig. 2C and SI Appendix, Fig. S2 A and B). In a 5-d MTS proliferation assay, siASCL1-3-mediated loss of ASCL1 in NCI-H1755 and HCC1833 reduced growth by 60–70%, an effect comparable to that seen in our “toxic” oligonucleotide siRNA control (SI Appendix, Fig. S2C).
Fig. 2.
ASCL1 is required for the survival of NE-NSCLC cell lines. (A) siRNA-mediated knockdown of ASCL1 reduces relative mRNA expression of ASCL1 and DLL1 in NCI-H1755 and HCC1833 cells, as measured by qRT-PCR. n.s., not significant. n = 3. *P < 0.05; **P < 0.01; ***P < 0.005. (B) siRNA-mediated knockdown reduces ASCL1 protein expression in NCI-H1755 and HCC1833 cells compared with controls. (C) Long-term stable shRNA-mediated ASCL1 knockdown inhibits the colony-forming ability of NE-NSCLC cell lines compared with typical NSCLC lines. Quantification of colony-forming ability after 14 d in culture is shown. n = 3 wells for shNTC and shASCL1. ***P < 0.005; ****P < 0.0001. (D) ASCL1 knockdown induces apoptosis in NCI-H1755 and HCC1833 cell lines, as measured by cell cycle analysis of the sub-G1 population. NCI-H1993 is unaffected by siASCL1. n = 3 for each group; t test performed between SCR and siASCL1-1, -2, and -3. *P < 0.05; **P < 0.01; ***P < 0.005. (E) Loss of ASCL1 in NCI-H1755 increases Annexin-V positivity, as measured by flow cytometry. (F) ASCL1 knockdown induces cleavage of PARP in NCI-H1755 and HCC1833 cells compared with controls. (G) siRNA screen comparing cell death phenotype in NE-NSCLC, NSCLC, and normal HBEC lung cell lines after knockdown of ASCL1, SOX2, and TTF1. The z-scores were calculated based on control experiments and color-coded to visualize cell death and growth, respectively. NE, NE-NSCLC lines; NL, normal immortalized HBEC line.
At 72 h after siRNA transfection, cell cycle analysis showed a twofold to sixfold increase in the number of cells in sub-G1 phase, indicating cell death in siASCL1-treated cells compared with control cells in ASCL1(+) NCI-H1755 and HCC1833, whereas ASCL1(−) NCI-H1993 was unaffected by siASCL1 (Fig. 2D). Apoptosis was verified as the cell death mechanism by induction of cleaved poly-ADP ribose polymerase (PARP) and flow cytometry analysis of annexin-expressing cells posttransfection (Fig. 2 E and F). An siRNA screen was performed to identify the phenotype after siRNA-mediated knockdown of ASCL1 in comparison with the lung lineage oncogenes TTF1 and SOX2 in NE-NSCLC cell lines, NSCLC cell lines, and normal HBECs. Knockdown of TTF1 appeared to be nearly pan-cytotoxic to lung cancer cell lines, inducing cell death in both NE-NSCLC and NSCLC lines while sparing a normal immortalized HBEC cell line (Fig. 2G). Knockdown of SOX2 induced cell death in only one NSCLC cell line, NCI-H2009, whereas ASCL1 knockdown confirms the previously reported results and is cytotoxic to NE-NSCLCs only. Taken together, these results demonstrate that ASCL1 is highly expressed in most NE-NSCLCs, and that its continued expression is required for the growth and survival of NE-NSCLC lines.
ASCL1 Protein Is Expressed in Lung Adenocarcinoma Samples.
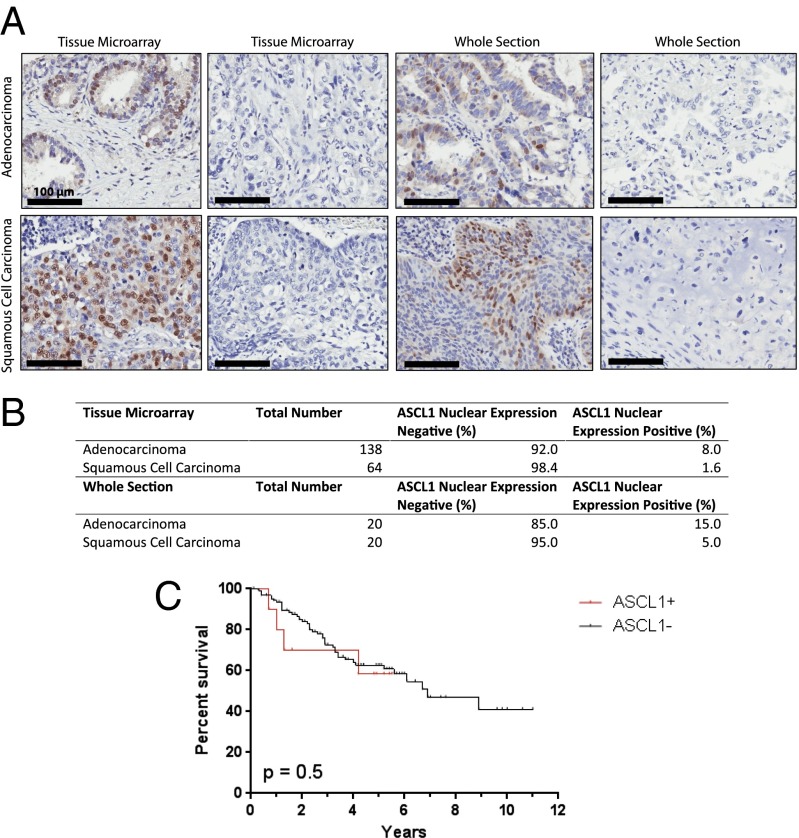
Previous genome-wide mRNA expression analyses of lung adenocarcinoma specimens identified the presence of neuroendocrine tumors in ∼10% of lung cancers histologically diagnosed as adenocarcinomas (1). To determine the frequency of ASCL1 expression in a large cohort of well-characterized, surgically resected tumors that had been diagnosed as NSCLC, we stained 138 lung adenocarcinoma (AC) and 64 lung squamous cell carcinoma (SCC) specimens for ASCL1 on a tissue microarray by immunohistochemistry (IHC) (Fig. 3A). These studies demonstrated ASCL1 protein was expressed in 8% of AC samples, agreeing with prior reports of neuroendocrine incidence in AC. In contrast, only one SCC sample was positive for ASCL1. In addition to tissue microarray studies, whole section staining was performed on 20 randomly selected AC and SCC samples, with three AC samples testing positive for ASCL1 (Fig. 3B). Survival analysis comparing IHC ASCL1(+) ACs and ASCL1(−) ACs found no significant difference in mortality, suggesting that detection of ASCL1 expression alone is not sufficient to determine prognosis in cases of NSCLC (Fig. 3C). Coexpression of the lung lineage oncogenes SOX2 and TTF1 was detected in seven of eight ASCL1(+) tumors, suggesting that these cancers may be dependent on multiple lineage factors (SI Appendix, Table S3).
Fig. 3.
IHC analysis for ASCL1 performed on adenocarcinoma and squamous cell carcinoma samples. (A) Representative slides showing ASCL1 nuclear staining in adenocarcinoma and squamous cell carcinoma samples from tissue microarray and whole- section IHC. Negative samples are displayed as a reference. (B) Table displaying the number of tumor samples staining positive for ASCL1. (C) Overall survival was not significantly different between ASCL1(+) tumors and ASCL1(−) tumors.
ChIP-Seq Analysis Uncovers Downstream Targets of ASCL1 That Predict for Poor Prognosis in NSCLC.
Based on our observation that ASCL1 is highly expressed in high-grade neuroendocrine lung cancers and is required for the survival of NE-NSCLC cell lines, we investigated whether a gene signature composed of ASCL1 target genes could be developed and applied to patient tumor specimens to determine prognosis. To identify downstream targets of ASCL1, we performed ChIP analysis on ASCL1(+) and ASCL1(−) lung cancer cell lines, followed by massively parallel sequencing (ChIP-Seq). The five ASCL1(+) lung cancer cell lines included two NE-NSCLCs (NCI-H1755 and HCC4018) and three SCLCs (NCI-H128, NCI-H1184, and NCI-H2107), with two neuroendocrine ASCL1(−) SCLC lines (NCI-H524 and NCI-H526) serving as controls (SI Appendix, Fig. S3A). SCLC cell lines were included to allow identification of a consensus ASCL1 transcriptome irrespective of histology.
The ASCL1(+) cell lines demonstrated high relative ChIP-Seq peak heights compared with the ASCL1(−) control cell lines (Fig. 4A). Several thousand ASCL1-bound sites were identified in each lung cancer line (Fig. 4B and SI Appendix, Table S4). A de novo motif analysis revealed ASCL1’s DNA-binding site (E-box; CASSTG) as the primary motif in each cell line. A total of 912 consensus peaks were found among the five ASCL1(+) cell lines (Fig. 4B); the binding sites were also enriched with the E-box motif, and these corresponded to 1,330 potential gene targets [analysis performed using GREAT (18); SI Appendix, Fig. S3B]. Among the common ASCL1 targets are the previously identified Notch ligands DLL1 and DLL3 (16) (SI Appendix, Table S5 and Fig. S3C), whereas other gene targets include neuroendocrine markers such as CDH2, GRP, INSM1, and SYT1, suggesting that ASCL1 supports the neuroendocrine phenotype in these cancers (SI Appendix, Fig. S3D). Supervised clustering of the lung cell line panel using the expression of 1,330 ASCL1 target genes resulted in separation of the neuroendocrine and non-neuroendocrine lung cancer cell lines (SI Appendix, Fig. S4).
Fig. 4.
ASCL1 ChIP-Seq analysis delineates downstream targets. (A) Base coverage plot of the 912 consensus peaks in all five ASCL1(+) cell lines. The base coverage in a 1,000-bp window is centered on the summits of the 912 consensus peaks collected at a 10-bp resolution and averaged for each cell line. The vertical axis is the normalized average base coverage of the consensus peaks, and the horizontal axis is the position relative to the summit. (B) Venn diagram showing peak-level overlap for all five ASCL1(+) samples along with the E-box binding site for each sample, as well as the consensus E-box binding site. Peak-level heterogeneity between samples is evident. (C) The 72-gene ASCL1 expression signature clustering analysis groups 9/10 of the NE-NSCLC cell lines separate from the typical NSCLC lines. (D) The ASCL1-associated 72-gene signature correlates with poor survival in retrospective analyses of lung adenocarcinoma patient datasets. Patients more likely to express the gene signature have poorer 5-y outcomes. (Upper) P < 0.0001, Gehan–Breslow–Wilcoxon test. (Lower) P < 0.0302, Gehan–Breslow–Wilcoxon test.
ASCL1-Driven 72-Gene Prognostic Signature.
Seventy-two genes identified in our ChIP-Seq studies also exhibited significantly higher mRNA expression (log2 >2; P < 0.01) in the five neuroendocrine ASCL1(+) lung cancer lines compared with the two neuroendocrine ASCL1(−) lung cancer lines. These 72 genes represent targets whose overexpression likely is directly regulated by ASCL1. Akin to ASCL1 expression being able to stratify typical NSCLC and NE-NSCLC cell lines, a supervised clustering analysis using the 72-gene ASCL1-associated signature performed on the 118 NSCLC lines grouped 9/10 NE-NSCLC cell lines (Fig. 4C and SI Appendix, Table S6). The misclassified cell line, NCI-H2106, does not express ASCL1, indicating that the neuroendocrine phenotype in NCI-H2106 likely is driven through a different pathway.
To determine whether the 72-gene ASCL1 target signature has prognostic capability, we applied the signature to three clinically annotated resected NSCLC mRNA expression datasets: NCI Director’s Consortium (19), Tomida GSE13213 (20), and SPORE GSE41271 (21). Using supervised principal component analysis, we found that patients whose tumors had higher probability of expressing the 72-gene ASCL1-associated gene signature had a significantly worse prognosis, independent of which dataset served as the training set or testing set (Fig. 4D and SI Appendix, Fig. S5). This finding suggests that ASCL1 transcriptional targets can be used to predict prognosis in retrospective patient dataset analyses, and can potentially serve as biomarkers to detect neuroendocrine differentiation in NSCLCs.
B-Cell CLL/Lymphoma 2: A Druggable Downstream Target of ASCL1 Identified by ChIP-Seq.
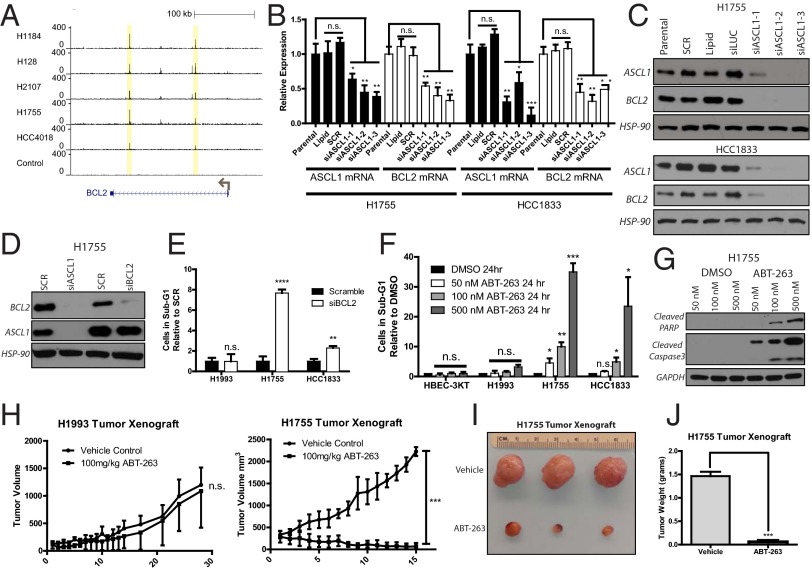
ChIP-Seq data affords the ability to systematically identify downstream targets of ASCL1 that are potentially sensitive to pharmacologic inhibition via induction of apoptotic effects similar to those seen with ASCL1 knockdown. Using a database of druggable genes (DrugBank) (22) to uncover potential therapeutic targets, we narrowed the list of 72 overexpressed ASCL1 target genes to 24 druggable targets (SI Appendix, Table S7). One target present in both the prognostic and druggable target gene lists for both ASCL1(+) SCLC and NE-NSCLC cell lines was the antiapoptotic factor B-cell CLL/lymphoma 2 (BCL2), which contained conserved ASCL1 E-box binding sites in all five ASCL1(+) cell lines (Fig. 5A). BCL2 expression analysis showed high mRNA levels in SCLC and NE-NSCLC lines and low or absent expression in typical NSCLC lines (SI Appendix, Fig. S6). Knockdown of ASCL1 in NE-NSCLC cell lines resulted in reductions of BCL2 mRNA and protein, suggesting a direct transcriptional link (Fig. 5 B and C). In contrast, knockdown of BCL2 did not affect ASCL1 expression, demonstrating that BCL2 acts downstream of ASCL1 (Fig. 5D). Transfection of siBCL2 into NCI-H1755 and HCC1833 induced a twofold to eightfold increase in cell death as measured by cell cycle analysis, but no cell death was detected with siBCL2 transfected into ASCL1(−) NCI-H1993 (Fig. 5E and SI Appendix, Fig. S6).
Fig. 5.
BCL2 is a conserved ASCL1 target with therapeutic implications. (A) Consensus E-box binding sites for ASCL1 are present in all ASCL1(+) samples within the BCL2 second intronic region. (B) Knockdown of ASCL1 by siRNA reduces BCL2 mRNA in NCI-H1755 and HCC1833 cell lines. n = 3. *P < 0.05; **P < 0.01; ***P < 0.005. (C) siRNA-mediated reduction of ASCL1 reduces BCL2 protein in NCI-H1755 and HCC1833. (D) Knockdown of BCL2 does not affect ASCL1 protein expression. (E) BCL2 knockdown by siRNA induces cell death in NE-NSCLCs, as measured by cell cycle analysis of the sub-G1 phase. n = 3 for each group. **P < 0.01; ****P < 0.0001). (F) Treatment of cell lines with BCL2 inhibitor ABT-263 for 24 h demonstrates specificity for ASCL1(+) NE-NSCLC cell lines. n = 3 for each group. *P < 0.05; **P < 0.01; ***P < 0.005. (G) ABT-263 treatment induces dose-dependent cleavage of PARP and Caspase 3 after 12 h of treatment in NCI-H1755. (H) Xenografts established from NCI-H1993 and NCI-H1755 were treated with 100 mg/kg ABT-263 or vehicle control for 14 consecutive days after tumors reached an average size of ∼250 mm3. n = 3 per treatment group per cell line. ***P < 0.001. (I) Photograph of resected s.c. tumors from ABT-263–treated xenografts. (J) Weight of H1993 and H1755 xenograft tumors treated with ABT-263 after resection. ***P < 0.001.
Several BCL2 small molecule inhibitors have activity in SCLCs (23, 24), including ABT-263, which targets BCL2 as well as the related family members BCL-xL and BCL-w. A 24-h treatment of ASCL1(+) NCI-H1755 and HCC1833 cells with ABT-263 resulted in induction of cell death (within 4 h in NCI-H1755) that was 10- to 30-fold greater than that seen in control HBEC-3KT and ASCL1(−) NCI-H1993 cells (Fig. 5F). Cleavage of PARP and Caspase 3 was detected in a dose-dependent manner after 12 h (Fig. 5G). Treatment of mouse xenografts in vivo recapitulated the in vitro results. In these experiments, 1 × 106 NCI-H1993 and NCI-H1755 cells were implanted in the s.c. flank region of female NOD/SCID mice. Once tumors were established, the mice were treated with ABT-263 or vehicle, delivered via i.p. injection, for 14 consecutive days. ASCL1(−) NCI-H1993 cells were insensitive to treatment, whereas ASCL1(+) NCI-H1755 showed dramatic reductions in tumor volume (Fig. 5H), tumor size (Fig. 5I), and tumor weight (Fig. 5J).
Interestingly, two previously described ASCL1 target genes, aldehyde dehydrogenase 1A1 (ALDH1A1) and prominin 1 (CD133/PROM1), do not appear to be conserved transcriptional targets among all ASCL1(+) cell lines as determined by ASCL1 ChIP-Seq analysis (7). ALDH1A1 and CD133/PROM1 are cancer stem cell-associated genes that presumably mark populations of cells with stem-like or tumor initiation capacity. The ALDH1A1 gene showed no enrichment of ASCL1-bound sites, and knockdown of ASCL1 resulted in no reduction of ALDH1A1 mRNA or protein in NCI-H1755 (SI Appendix, Fig. S7). In contrast, CD133/PROM1 demonstrated enrichment of ASCL1-binding sites only in ASCL1(+) SCLC cell lines, suggesting a potentially important functional difference between NE-NSCLC and SCLCs driven by ASCL1 (SI Appendix, Fig. S8).
A third recently described ASCL1 target gene, ret proto-oncogene (RET) (25), demonstrated enrichment of ASCL1-bound sites only for ASCL1(+) NE-NSCLC lines. In contrast, ChIP-Seq analysis of ASCL1(+) SCLC lines failed to show this enrichment (SI Appendix, Fig. S9A). RET was determined to be significantly expressed by qRT-PCR in 9 of 10 NE-NSCLC lines, in 9 of 15 SCLC lines, and in none of 11 NSCLC lines (SI Appendix, Fig. S9B). Knockdown of ASCL1 in NCI-H1755 and HCC1833 resulted in reductions of RET mRNA and protein, suggesting that ASCL1 is a direct transcriptional regulator of the RET gene (SI Appendix, Fig. S9C). siRNA-mediated knockdown of RET in NE-NSCLC cell lines was able to quantifiably induce cell death (SI Appendix, Fig. S9D); however, treatment with a RET inhibitor (cabozantinib) was unable to demonstrate improved sensitivity in the ASCL1(+) NE-NSCLC lines (SI Appendix, Fig. S9E), and the cabozantinib IC50 values were μM, indicating this drug likely will not be of use in ASCL1-dependent tumors.
Discussion
The lack of specific treatment options for patients with high-grade neuroendocrine lung cancers (SCLC and NE-NSCLC) necessitates the discovery of new, rational targeted therapies as well as preclinical models that will encompass this important subtype of lung cancer. Here we describe human lung cancer cell line models for NE-NSCLC that depend on the lineage-specific transcription factor ASCL1 for survival. Previous lineage-dependent models in the lung focused on SOX2 and TTF1 (10, 11). TTF1 is known to be expressed in SCLC (26) and may represent a lineage factor for neuroendocrine carcinomas. Based on our results, TTF1 is important for maintaining the survival of NE-NSCLC cell lines and thus merits further study.
ASCL1 is expressed in 8% of lung adenocarcinomas and in nearly all SCLCs, representing approximately 40,000 cases of lung cancer per year in the United States. ChIP-Seq combined with gene expression analysis of ASCL1(+) NE-NSCLC and SCLC cell lines led us to identify an up-regulated 72-gene ASCL1-associated signature that is predictive of poor prognosis in resected NE-NSCLCs. This was true despite the fact that ASCL1 expression by itself was not prognostic, indicating the importance of ASCL1 downstream targets. In addition, mining of a druggable genome database identified 24 of the 72 genes as novel ASCL1-regulated drug targets.
One of the specific ASCL1 transcriptional targets was the antiapoptotic regulator BCL2. BCL2 is a natural mediator of lineage survival by virtue of its ability to inhibit release of cytochrome C into the cytoplasm, avoiding a critical apoptotic stimulus within the cell. A previous study showed increased BCL2 expression in mouse non-neuroendocrine epithelial cells following the constitutive expression of ASCL1, further linking the two genes (27). The possibility exists that ASCL1 maintains lineage survival of pulmonary neuroendocrine cell progenitors by regulating the expression of BCL2, similar to the mechanism discovered for Mitf in melanocytes (28). Thus, BCL2 is an attractive oncogene for NE-NSCLC cancers to use, and as such represents an acquired vulnerability for therapy of this lung cancer subtype. Treating NE-NSCLC cells in vitro and xenografts in vivo with a BCL2 inhibitor demonstrates dramatic specificity in ASCL1(+)/BCL2(+) lung cancers compared with ASCL1(−)/BCL2(−) lung cancers. BCL2-targeted therapy has previously shown efficacy in SCLC xenograft models (23). This suggests that BCL2 is an acquired vulnerability for high-grade neuroendocrine lung cancers, and that it likely mediates lineage-specific survival of these tumors. Although a BCL2 inhibitor formulated for clinical trials in patients with treatment-refractory SCLC failed to show a major therapeutic benefit (29), combining anti-BCL2 therapy with other therapies, as well as newer anti-BCL2 formulations that are more specific for BCL2 itself, hold promise as therapies for ASCL1-driven tumors (30). Other druggable targets of ASCL1 were identified by our ChIP-Seq analysis, representing additional potential therapeutic interventions (SI Appendix, Table S7). A mouse model exists for LCNE carcinomas, which may enhance the potential for testing therapeutic agents in vivo (31).
Our ASCL1 ChIP-Seq analysis failed to validate the cancer stem cell markers ALDH1A1 and CD133/PROM1 as conserved ASCL1 transcriptional target genes, suggesting that the transcriptional program driven by ASCL1 is quite heterogenous. The wide range of ASCL1-bound peaks detected across the five ASCL1(+) lung cancer lines used for ChIP-Seq analysis demonstrates the complexity of the ASCL1 transcriptome. Although ALDH1A1 showed no enrichment of ASCL1-bound sites in any of the cell lines tested, CD133/PROM1 showed enrichment in only SCLC lines, whereas another cancer stem cell marker, RET, showed significant enrichment in only NE-NSCLC lines. This discovery demonstrates an important functional distinction between SCLC and NE-NSCLC. Although RET inhibition of ASCL1(+) NE-NSCLC lines with cabozantinib failed to show growth inhibition at nanomolar drug levels, it is possible that more specific RET-targeting agents may exhibit improved responses in ASCL1-dependent NE-NSCLCs.
In conclusion, our findings indicate an important role of ASCL1 in NE-NSCLC, whereas our preclinical models and detailed ASCL1 ChIP-Seq analysis provide a path for the systematic development of ASCL1-targeted cancer therapy for NE-NSCLC and SCLC.
Materials and Methods
Cell Lines.
All cell lines used were obtained from the Hamon Cancer Center Collection (University of Texas Southwestern Medical Center), maintained in RPMI-1640 medium (Life Technologies) supplemented with 5% or 10% FCS without antibiotics at 37 °C in a humidified atmosphere containing 5% CO2 and 95% air. Cell lines were DNA-fingerprinted using the PowerPlex 1.2 Kit (Promega), and mycoplasma was tested using an e-Myco Kit (Boca Scientific). Cells were treated with ABT-263 (Selleck) or DMSO control for up to 72 h.
Protein Expression, MTS Proliferation Assay, Transient siRNA Transfection, Stable shRNA Expression, and In Vivo Tumor Xenograft Experiments.
All of these procedures and assays were performed as described in SI Appendix, Materials and Methods. MTS reagent (Promega), primary antibodies against ASCL1 (BD Biosciences), BCL2, PARP, Cleaved PARP, Cleaved Caspase 3, RET, ALDH1A1, and Hsp90 (Cell Signaling) were used in this study. siRNAs (Qiagen) included siASCL1-1–3, siBCL2, siRET, siSOX2, siTTF1, siLUC, and siSCR. Stable pGIPZ lentiviral shRNA constructs targeting ASCL1 and pGIPZ-shNTC (Thermo Scientific) were selected in lung cancer lines with puromycin. The in vivo efficacy of ABT-263 was tested in s.c. flank xenografts of NCI-H1993 and NCI-H1755 in female 5- to 6-wk-old NOD/SCID mice. All animal care was provided in accordance with institutional guidelines and approved Institutional Animal Use and Care Committee protocols. IHC methodology and scoring were performed as reported previously (32).
Microarray Analysis.
Total RNA from cell lines was isolated using RNEasy kit (Qiagen). Gene expression profiling on each sample was performed using Illumina HumanWG-6 V3 BeadArrays (dataset for the 206 lung cell lines deposited as GSE32036) and analyzed as described previously (33).
ChIP, Sequence Library Preparation, Alignment, Peak Calling, and Motif Analysis.
ChIP and analyses were performed as described previously (34) and in SI Appendix, Materials and Methods. The cell lines used for ChIP were H1755, HCC4018, H128, H1184, H2107, and control cell lines H524 and H526. Bowtie was used for DNA sequence alignment after library preparation using reference genome HG19 (35). ChIP-Seq peaks were called using MACS version 1.4.0rc (36). DNA motif analysis was performed using HOMER (37).
Survival Analysis.
The prognostic performance of the 72-gene set was tested on three independent mRNA expression datasets: 442 primary lung adenocarcinomas from the National Cancer Institute Director’s Challenge Consortium study (19); the Tomida dataset, consisting of 119 lung adenocarcinomas (GSE13213) (20); and 209 primary lung adenocarcinomas and squamous cell carcinomas from our SPORE dataset (GSE41271) (21), as described in SI Appendix, Materials and Methods using Kaplan–Meier analysis, the log-rank test, and published standard methods (38).
SWEAVE Documentation.
This documentation is provided for ChIP procedures, correlation with microarray expression data, and survival analyses in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank all of the members of the J.D.M. laboratory for critical discussions and experimental support. This work was supported by National Cancer Institute Specialized Program of Research Excellence in Lung Cancer Grant P50CA70907, National Cancer Institute Grant CTD2N CA176284, and Cancer Prevention and Research Institute of Texas Grants RP110708 (to J.D.M., I.I.W., Y.X., and L.G.) and RP110383 (to J.E.J. and M.H.C.). A.A. is supported by the University of Texas Southwestern Medical Center Medical Scientist Training Program and by a Ruth L. Kirschstein National Research Service Award for Individual Predoctoral MD/PhD Fellows (F30: 1F30CA168264).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: ChIP-Seq and mRNA expression data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE61197).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410419111/-/DCSupplemental.
References
- 1.Bhattacharjee A, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98(24):13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones MH, et al. Two prognostically significant subtypes of high-grade lung neuroendocrine tumours independent of small-cell and large-cell neuroendocrine carcinomas identified by gene expression profiles. Lancet. 2004;363(9411):775–781. doi: 10.1016/S0140-6736(04)15693-6. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez E, Lilenbaum RC. Small cell lung cancer: Past, present, and future. Curr Oncol Rep. 2010;12(5):327–334. doi: 10.1007/s11912-010-0120-5. [DOI] [PubMed] [Google Scholar]
- 4.Guillemot F, et al. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75(3):463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- 5.Johnson JE, Birren SJ, Anderson DJ. Two rat homologues of Drosophila achaete-scute specifically expressed in neuronal precursors. Nature. 1990;346(6287):858–861. doi: 10.1038/346858a0. [DOI] [PubMed] [Google Scholar]
- 6.Borges M, et al. An achaete-scute homologue essential for neuroendocrine differentiation in the lung. Nature. 1997;386(6627):852–855. doi: 10.1038/386852a0. [DOI] [PubMed] [Google Scholar]
- 7.Jiang T, et al. Achaete-scute complex homologue 1 regulates tumor-initiating capacity in human small cell lung cancer. Cancer Res. 2009;69(3):845–854. doi: 10.1158/0008-5472.CAN-08-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osada H, Tatematsu Y, Yatabe Y, Horio Y, Takahashi T. ASH1 gene is a specific therapeutic target for lung cancers with neuroendocrine features. Cancer Res. 2005;65(23):10680–10685. doi: 10.1158/0008-5472.CAN-05-1404. [DOI] [PubMed] [Google Scholar]
- 9.Miki M, Ball DW, Linnoila RI. Insights into the achaete-scute homolog-1 gene (hASH1) in normal and neoplastic human lung. Lung Cancer. 2012;75(1):58–65. doi: 10.1016/j.lungcan.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 10.Bass AJ, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41(11):1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwei KA, et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008;27(25):3635–3640. doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramirez RD, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64(24):9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 13.Brambilla E, Travis WD, Colby TV, Corrin B, Shimosato Y. The new World Health Organization classification of lung tumours. Eur Respir J. 2001;18(6):1059–1068. doi: 10.1183/09031936.01.00275301. [DOI] [PubMed] [Google Scholar]
- 14.Shimakage M, et al. Downregulation of drs tumor suppressor gene in highly malignant human pulmonary neuroendocrine tumors. Oncol Rep. 2009;21(6):1367–1372. doi: 10.3892/or_00000362. [DOI] [PubMed] [Google Scholar]
- 15.Carney DN, et al. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res. 1985;45(6):2913–2923. [PubMed] [Google Scholar]
- 16.Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126(3):525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- 17.Osborne JK, et al. NeuroD1 regulates survival and migration of neuroendocrine lung carcinomas via signaling molecules TrkB and NCAM. Proc Natl Acad Sci USA. 2013;110(16):6524–6529. doi: 10.1073/pnas.1303932110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shedden K, et al. Director’s Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14(8):822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomida S, et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J Clin Oncol. 2009;27(17):2793–2799. doi: 10.1200/JCO.2008.19.7053. [DOI] [PubMed] [Google Scholar]
- 21.Sato M, et al. Human lung epithelial cells progressed to malignancy through specific oncogenic manipulations. Mol Cancer Res. 2013;11(6):638–650. doi: 10.1158/1541-7786.MCR-12-0634-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wishart DS, et al. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34(Database issue):D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shoemaker AR, et al. Activity of the Bcl-2 family inhibitor ABT-263 in a panel of small cell lung cancer xenograft models. Clin Cancer Res. 2008;14(11):3268–3277. doi: 10.1158/1078-0432.CCR-07-4622. [DOI] [PubMed] [Google Scholar]
- 24.Hann CL, et al. Therapeutic efficacy of ABT-737, a selective inhibitor of BCL-2, in small cell lung cancer. Cancer Res. 2008;68(7):2321–2328. doi: 10.1158/0008-5472.CAN-07-5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosari F, et al. ASCL1 and RET expression defines a clinically relevant subgroup of lung adenocarcinoma characterized by neuroendocrine differentiation. Oncogene. 2014;33(29):3776–3783. doi: 10.1038/onc.2013.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kargi A, Gurel D, Tuna B. The diagnostic value of TTF-1, CK 5/6, and p63 immunostaining in classification of lung carcinomas. Appl Immunohistochem Mol Morphol. 2007;15(4):415–420. doi: 10.1097/PAI.0b013e31802fab75. [DOI] [PubMed] [Google Scholar]
- 27.Wang XY, et al. Achaete-scute homolog-1 linked to remodeling and preneoplasia of pulmonary epithelium. Lab Invest. 2007;87(6):527–539. doi: 10.1038/labinvest.3700552. [DOI] [PubMed] [Google Scholar]
- 28.McGill GG, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109(6):707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 29.Rudin CM, et al. Phase II study of single-agent navitoclax (ABT-263) and biomarker correlates in patients with relapsed small cell lung cancer. Clin Cancer Res. 2012;18(11):3163–3169. doi: 10.1158/1078-0432.CCR-11-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Souers AJ, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19(2):202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 31.Cui M, et al. PTEN is a potent suppressor of small cell lung cancer. Mol Cancer Res. 2014;12(5):654–659. doi: 10.1158/1541-7786.MCR-13-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang X, et al. Abnormalities of the TITF-1 lineage-specific oncogene in NSCLC: Implications in lung cancer pathogenesis and prognosis. Clin Cancer Res. 2011;17(8):2434–2443. doi: 10.1158/1078-0432.CCR-10-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Byers LA, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res. 2013;19(1):279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borromeo MD, et al. A transcription factor network specifying inhibitory versus excitatory neurons in the dorsal spinal cord. Development. 2014;141(14):2803–2812. doi: 10.1242/dev.105866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2(4):E108. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.