Abstract
The model pathogen Parastagonospora nodorum is a necrotroph and the causal agent of the wheat disease Septoria nodorum blotch (SNB). The sequenced P. nodorum genome has revealed that the fungus harbours a large number of secondary metabolite genes. Secondary metabolites are known to play important roles in the virulence of plant pathogens, but limited knowledge is available about the SM repertoire of this wheat pathogen. Here, we review the secondary metabolites that have been isolated from P. nodorum and related species of the same genus and provide an in-depth genome-wide overview of the secondary metabolite gene clusters encoded in the P. nodorum genome. The secondary metabolite gene survey reveals that P. nodorum is capable of producing a diverse range of small molecules and exciting prospects exist for discovery of novel virulence factors and bioactive molecules.
Keywords: Parastagonospora nodorum, secondary metabolites, plant pathogen, polyketide synthase, nonribosomal peptide synthetase, terpene synthase
Introduction
Filamentous fungi are prolific producers of bioactive small molecules, known as secondary metabolites (SMs). Fungi are an important source for drug discovery, examples of which include important drugs like the antibiotic penicillin and the cholesterol-lowering statins; yet many SMs are mycotoxins that are harmful to humans, such as aflatoxins, trichothecenes and fumonisins. In plant pathogenic fungi, the SMs often play an important role in plant infection and virulence. Fungi are the major causal agents of diseases in crop plants. SMs that lead to the damage or killing of plants are also known as phytotoxins, and their modes of action are diverse (Möbius et al. 2009). These phytotoxins include plant-damaging photosensitizers (e.g. cercosporin) (Daub et al. 2005), inhibitors of plant cellular functions (e.g. ATPase inhibitor tentoxin) (Meiss et al. 2008), membrane disruptants (e.g. beticollin 0) (Goudet et al. 2000) or triggers of plant apoptosis (Möbius et al. 2009). Some of these phytotoxins are host specific while others have activities against a broader range of plant hosts. Some well-known SM host-specific toxins (HSTs) include HC-toxin (Walton 2006), victorin (Lorang et al. 2012) and T-toxins (Turgeon & Baker 2007). These HSTs are known to target plant hosts by interacting with a specific susceptibility gene in an inverse manner of the classical gene-for-gene system, similar to the proteinaceous effectors in necrotrophic pathogens (Oliver & Solomon 2010; Stergiopoulos et al. 2013).
Other SMs that are not directly involved in damaging the plants could play important role in protection against environmental stress (e.g. melanin) (Eisenman & Casadevall 2012), mineral uptake (e.g. siderophores) (Johnson 2008), interfering host hormone signalling (e.g. giberrellins) (Johnson 2008) and suppressing plant defence (e.g. supprescins and brefeldin) (Tietjen & Matern 1984; Wada et al. 1995). As individual pathogenic fungi and their hosts are not isolated in the environment, the pathogens will be constantly interacting with other organisms, including other competing microbes, endophytes and fungivores. Therefore, it is not surprising that many SMs from plant pathogenic fungi also possess antibacterial, antifungal and cytotoxic activities.
Although there are increasing numbers of SMs being isolated and characterized from plant pathogenic fungi, our understanding of SM production and their functional roles in these pathogens is still largely incomplete. This is especially true for many wheat pathogens, which are currently the focus of the research in our laboratory. Wheat (Triticum aestivum) is among the most important staple food crops. Like most crop plants, fungi are the biggest threat among the causal agents of diseases in wheat, causing significant yield loss annually. Among the most important wheat fungal diseases are the yellow (tan) spot caused by Pyrenophora tritici-repentis, Septoria nodorum blotch caused by Parastagonospora nodorum, Septoria tritici blotch caused by Zymoseptoria tritici, stripe rust caused by Puccinia striiformis f. sp. tritici, crown rot caused by Fusarium pseudograminearum and the head blight caused by Fusarium graminearum (Murray & Brennan 2009).
Recent advances in genome sequencing technology have led to an unprecedented surge of genomic information that was previously unavailable. These whole genome sequencing efforts have revealed that fungal genomes encode for immense biosynthetic potential that far surpasses the chemical diversity that we have previously perceived. This holds true as well for the major wheat pathogens mentioned above. The availability of genome sequences for all these wheat pathogens allows a preview to their genetic potential for SM biosynthesis and it shows that all of these wheat pathogens, except the rust (basidiomycete) fungus Puccinia striiformis f.sp. tritici, are rich in SM biosynthesis genes. This review will focus on the model pathogen Parastagonospora nodorum, a necrotroph that cause Septoria nodorum blotch (SNB) in wheat. Here, we review the SMs that are known to be produced by this fungus and provide an in-depth genomic examination of the SM biosynthesis genes in this wheat pathogen, which we hope to serve as the basis for future investigation into the secondary metabolome of P. nodorum.
Parastagonospora nodorum pathobiology and secondary metabolites
Parastagonospora nodorum falls within the order Pleosporales in the class Dothideomycetes (Oliver et al. 2012). More commonly known as Stagonospora nodorum Berk. (teleomorph Phaeosphaeria nodorum), this fungus has been recently re-named as Parastagonospora nodorum (Berk.) Quaedvlieg, Verkley & Crous in a molecular taxonomy study due to the clustering of the type of the fungal genus Stagonospora (Stagonospora paludosa) with Massarinaceae and not Phaeosphaeriaceae (Quaedvlieg et al. 2013). Furthermore, it was shown in the same study that P. nodorum does not cluster with or near the type of the genus Phaeosphaeria (Phaeosphaeria oryzae). SNB (Figure 1) leads to > $100 million yield loss per annum in Australia (Murray & Brennan 2009) and is one of the major plant pathogens in North America and worldwide. It is genetically tractable and has well-annotated genome sequence (Hane et al. 2007; Oliver et al. 2012). The availability of simple in vitro (detached-leaf) and whole plant (spray) virulence assays offer an excellent model for studying the functional roles of SMs in plant pathogens. We will discuss briefly the pathogen biology, lifecycle and modes of nutrition of P. nodorum, and how SMs may play a role in their different stages of growth.
Figure 1.
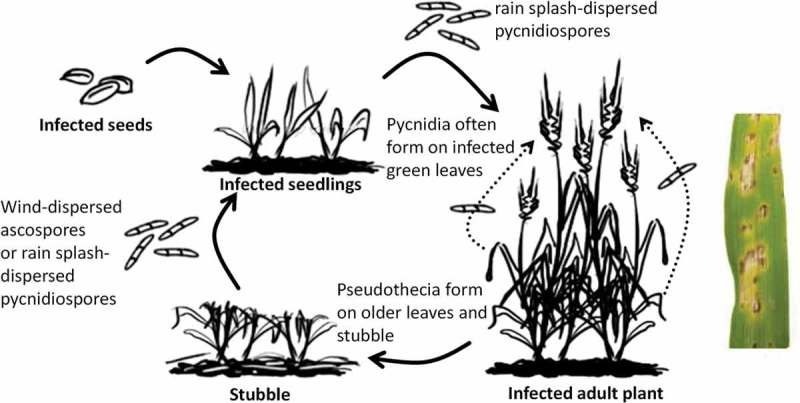
Parastagonospora nodorum causing Septoria nodorum blotch (SNB) on wheat. Left: the P. nodorum lifecycle. Right: the disease symptom of SNB on wheat leaf.
Parastagonospora nodorum is a necrotrophic pathogen, thus its lifecycle can be divided into a parasitic and saprophytic stage (Figure 1). During the parasitic phase, it can infect all above-ground plant parts with the asexual pycnidiospores and sexual ascospores dispersed to upper plant parts, including wheat heads, primarily via rain splashes (Griffiths & Ao 1976; Brennan et al. 1985). However, the sexual ascospores produced by the perithecia are capable of long-distance wind dispersal (Sanderson & Hampton 1978; Bathgate & Loughman 2001). The ascospore-producing perithecia are present during both the parasitic and saprophytic phase. During the saprophytic phase, the pathogen overwinters on wheat straw and stubble until the parasitic phase starts again (Shaner 1981). Both infected seeds and ascospores are considered the major inocula of this fungal wheat disease (Solomon et al. 2006).
Parastagonospora nodorum produces proteinaceous necrotrophic effectors as major pathogenicity factors, where each interacts with a specific wheat susceptibility gene, not unlike the gene-for-gene relationship observed in plant disease resistance to biotrophs (Oliver et al. 2012). Three such effectors, SnTox1, SnToxA and SnTox3, which induce necrosis in specific genotype of wheat, have been identified from P. nodorum (Oliver et al. 2012). Recent studies suggest that there may be more necrotrophic effectors in P. nodorum that remain to be identified (Francki et al. 2011; Crook et al. 2012; Friesen et al. 2012; Tan et al. 2013).
Despite the importance of P. nodorum among wheat diseases, the SM repertoire of this fungus is poorly understood. The SM molecules that have been identified so far include septorines (Devys et al. 1978, 1982, 1992), melleins and mycophenolic acids (Devys et al. 1980, 1994) (Figure 2). Even though Devys et al. have continually worked on isolating SMs from P. nodorum for over a decade, apparently only analogues of these three above groups of compounds from P. nodorum can be identified via traditional compound isolation approaches. It is possible that P. nodorum only produces these compounds above as major metabolites under the laboratory culture conditions tested whilst other metabolites are either not produced or only present in minor quantities. This is not surprising given that many fungal SM pathways are known to be silent and only expressed in response to specific environmental stimuli (Brakhage & Schroeckh 2011).
Figure 2.
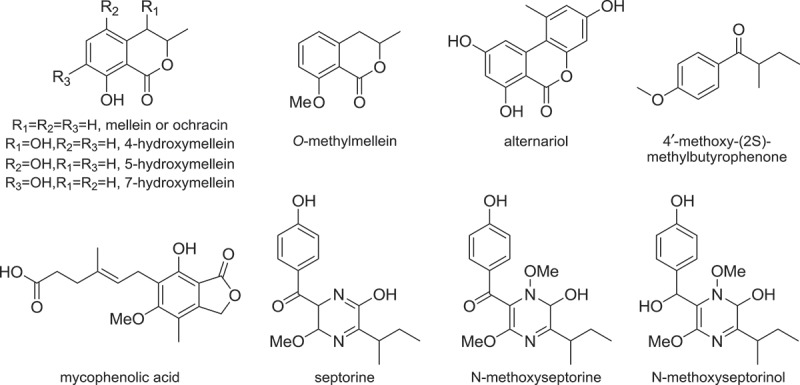
Secondary metabolites isolated from Parastagonospora nodorum.
More recently, alternariol has been identified in a metabolomics study of a P. nodorum mutant lacking a short-chain dehydrogenase (Sch1) (Tan et al. 2009). The study found that the concentration of alternariol in the culture of an sch1 mutant strain is 200-fold greater than the wild-type P. nodorum, although the relationship between sch1 and alternariol biosynthesis is still unknown. Another new compound, (+)-4ʹ-methoxy-(2S)-methylbutyrophenone (Figure 2), was isolated from P. nodorum recently by supplementing epigenetic modifiers to the culture medium (Yang et al. 2013). These two most recent P. nodorum SM studies have highlighted that both genetic and epigenetic approaches can be used to induce the expression of otherwise silent SM pathways in laboratory culture conditions (Brakhage & Schroeckh 2011).
Among the known P. nodorum SMs, mellein and septorines have been shown to exhibit phytotoxic activities (Venkatasubbaiah & Chilton 1990; Parisi et al. 1993). To date, no SM-type effector has been identified in P. nodorum. The SMs in P. nodorum are most likely to have auxiliary roles in virulence against wheat, but the presence of such host-specific effectors cannot be excluded (Syme et al. 2013). It can also be speculated that SMs are likely to play various other roles, such as in fungal development (e.g. sporulation) and inter-species competition (e.g. during saprophytic stage), at the different stages of P. nodorum lifecycle.
Since limited information is available for SMs from P. nodorum, we have surveyed the literature for compounds produced by fungi of the same genus (Phaeosphaeria or Stagonospora) to obtain some insights into the diversity of SM compounds that might be produced by P. nodorum (see Supplemental Table 1). The literature survey revealed that species in the related genera are capable of producing diverse structures with the SM compounds dominated by polyketides followed by a small number of terpenoid compounds and alkaloids. Some of these compounds will be related to the P. nodorum SM biosynthetic genes to be discussed in the following section.
Table 1.
SM backbone enzymes found in Parastagonospora nodorum genome.
| SM backbone enzymes | |
|---|---|
| Polyketide synthase | 23 |
| Non-reducing PKS | 7 |
| Partially reducing PKS | 1 |
| Highly reducing PKS | 14 |
| Hybrid PKS-NRPS | 1 |
| Type III PKS | 1 |
| Nonribosomal peptide synthetase | 14 |
| Multimodular NRPS | 6 |
| Dimodular NRPS | 1 |
| Monomodular NRPS-like | 7 |
| Sesquiterpene synthases (class I) | 3 |
| Diterpene synthase (class II) | 1 |
| DMATS-type Prenyltransferase | 2 |
| UbiA-like Prenyltransferase | 3 |
Genomic outlook for the SM biosynthetic potential of Parastagonospora nodorum
The identification of the genes involved in fungal SM pathways has been partly facilitated by the likelihood that genes belonging to a single pathway will cluster together on chromosomes (Hoffmeister & Keller 2007). The recent progress in our understanding of the molecular genetics of fungal secondary metabolism has been further propelled by the advances in genomic technologies with an increasing number of SMs now being linked to their biosynthetic genes or gene clusters. Consequently, we now have the ability to identify the genes within fungal genomes that encode for SM biosynthetic enzymes and provide clues to the structural class of the compounds that may be produced based on the type of backbone biosynthetic enzymes. For example, the structurally diverse polyketides are produced by the large multifunctional iterative type I polyketide synthases (PKSs), which utilize acetate/malonate units as building blocks. The carbon backbones of aromatic compounds, such as aflatoxins and griseofulvin, are synthesized by non-reducing PKSs (NR-PKSs), while the aliphatic compounds (acyclic and cyclic), such as fumonisins and lovastatin, are produced by highly reducing PKSs (HR-PKSs) (Cox 2007; Chooi & Tang 2012). Besides iterative Type I PKSs, some fungi also harbour Type III PKSs, which are more commonly found in plants (e.g. chalcone synthase) (Seshime et al. 2005). In contrast, the non-ribosomal peptide compounds, such as the cyclicpeptides HC-toxin and echinocandins, are produced by large multimodular enzymes called non-ribosomal peptide synthases (NRPSs), which utilize proteinogenic and non-proteinogenic amino acids as building blocks (Finking & Marahiel 2004). Terpenoids/isoprenoids, on the other hand, are synthesized from multimers of five-carbon isoprene units assembled by isopropanoid synthases (ISs) and prenyltransferases (PTs) folded by various terpene synthases (TSs) into diverse structures (Christianson 2008), e.g. trichothecenes, botrydial. Fungal SMs can also be produced from mixed pathways; for example, tenuazonic acid and cytochalasin are generated from polyketide-nonribosomal peptide pathways, whereas sirodesmin is produced from nonribosomal peptide-isoprenoid pathway and fumagillin originates from polyketide-isoprenoid pathways. Typically, the backbone SM genes are clustered with multiple genes encode for tailoring enzymes such as oxygenases, methyltransferases, acyltransferases, etc., that further diversify the chemical structures.
The genome sequencing of P. nodorum SN15 was completed in 2007 and was the first fungal genome in the large Dothideomycete class to be sequenced (Hane et al. 2007). More recently, the resequencing and comparative genomics of P. nodorum have further improved the assembly of this genome and revealed additional SM genes after sequence correction (Syme et al. 2013). The 37 Mb genome of P. nodorum encodes 23 PKSs, 14 NRPSs, 4 TSs and 5 PTases (Table 1). More than one SM backbone biosynthetic genes are found in some of the P. nodorum SM gene clusters, forming hybrid gene clusters, thus the total number of SM gene clusters is lower than the number of SM backbone biosynthetic genes. SMURF SM gene cluster prediction software detected a total of 29 SM gene clusters in P. nodorum (Khaldi et al. 2010). However, we can identify a total of 38 SM gene clusters in P. nodorum by including the TSs, type III PKSs, and UbiA-like PTases not considered in the SMURF software and some PKS and NRPS genes left out in the analysis (Supplemental Table 2). Nevertheless, only five groups of SM molecules have so far been identified in P. nodorum (Figure 2), and they therefore only represent about 13% of the SM biosynthesis potential of P. nodorum. Furthermore, none of these identified SMs have been linked to the genes in P. nodorum.
Table 2.
Domain architecture of PKSs encoded in Parastagonospora nodorum genome.
| PKS locus | Domain architecture | Closest characterized BLAST hit/polyketide product |
|---|---|---|
| NR-PKSs | ||
| SNOG_06682 | SAT-KS-AT-PT-ACP-CM-TE/CLC | Penicillium brevicompactum MpaC (38%)/mycophenolic acid |
| SNOG_07020 | SAT-KS-AT-PT-ACP-ACP-CM-TE | Monascus purpurus CtnPKS (29%)/citrinin |
| SNOG_08274 | SAT-KS-AT-PT-ACP-ACP-TE/CLC | Nectria haematococca PKSN (52%)/unknown red pigment |
| SNOG_08614 | SAT-KS-AT-PT-ACP-TE/CLC | Cercospora nicotianae CTB1 (56%)/cercosporin |
| SNOG_09932 | SAT-KS-AT-PT-ACP-TE/CLC | Aspergillus nidulans wA (42%)/naphthopyrone YWA1 |
| SNOG_11981 | SAT-KS-AT-PT-ACP-ACP-TE/CLC | Alternaria alternata ALM1 (80%)/melanin |
| SNOG_15829 | SAT-KS-AT-PT-ACP | Penicillium aethiopicum GsfA (63%)/griseofulvin |
| HR-PKSs | ||
| SNOG_02561 | KS-AT-DH-CM-ER-KR-ACP | Phoma sp. SQTKS (35%)/squalestatin tetraketide |
| SNOG_04868 | KS-AT-DH-CM-ER-KR-ACP | Leptosphaeria maculans PKS2 (90%)/phomenoic acid |
| SNOG_05791 | KS-AT-DH-CM-ER-KR-ACP | Alternaria solani PKSN (82%)/alternapyrone |
| SNOG_06676 | KS-AT-DH-ER-KR-ACP | Alternaria brassicicola DEP5 (66%)/depudecin |
| SNOG_07866 | KS-AT-DH-CM-KR-ACP-R | C. heterostrophus PKS16 (74%)/uncharacterized |
| SNOG_09490 | KS-AT-DH-ER-KR-ACP | Botryiotinia fuckeliana BcBOA9 (38%)/botcinic acid |
| SNOG_09623 | KS-AT-DH-CM-KR-ACP | Fusarium verticillioides Fum1p (40%)/fumonisin |
| SNOG_11066 | KS-AT-DH-ER-KR-ACP | L. maculans LEMA_P006610 (54%)/uncharacterized |
| SNOG_11076 | KS-AT-DH-CM-ER-KR-ACP | Fusarium verticillioides Fum1p (38%)/fumonisin |
| SNOG_11272 | KS-AT-DH-ER-KR-ACP | Botryitis cinerea BcBOA9 (50%)/botcinic acid |
| SNOG_12897 | KS-AT-DH-ER-KR-ACP | Chaetomium chiversii CcRADS1 (45%)/radicicol starter unit |
| SNOG_13032 | KS-AT-DH-CM-ER-KR-ACP | Fusarium verticillioides Fum1p (38%)/fumonisin |
| SNOG_14927 | KS-AT-DH-KR-ACP | Fusarium verticillioides Fum1p (41%)/fumonisin |
| SNOG_15965 | KS-AT-DH-ER-KR-ACP | Fusarium verticillioides Fum1p (35%)/fumonisin |
| PKS-NRPS | ||
| SNOG_00308 | KS-AT-DH-CM-ER°-KR-ACP-C-A-T-R | Aspergillus clavatus CcsA (46%)/cytochalasin E and K |
| PR-PKSs | ||
| SNOG_00477 | Aspergillus terreus 6MSAS (54%)/6-methylsalicylic acid | |
| Type III PKSs | ||
| SNOG_09622 | KS-AT-TH-KR-ACP | Neurospora crassa ORAS (36%)/2ʹ-oxoalkylresorcylate |
Notes: PKS domain abbreviation – SAT, starter unit; ACP transacylase; KS, β-ketoacyl synthase; MAT, malonyl-CoA:ACP transacylase; PT, product template; ACP, acyl-carrier protein; TE/CLC, thioesterase/Claisen cyclase; CM, C-methyltransferase; DH, dehydratase; ER, enoyl reductase; KR, ketoreductase; TH, thiohydrolase. See Chooi and Tang (2012) review for detail description of the PKS functional domains. Closest BLAST hit is included when there is no characterized PKS in the top 100 hits.
Polyketide synthases (PKSs)
All the common types of fungal type I PKSs (NR-, HR- and PR-PKSs) as well as the less common type III PKSs can be found in P. nodorum (Table 1). The number of PKS genes (total 24) in P. nodorum is comparable to those found in the genome of Aspergillus spp., which are often regarded as highly prolific SM producers (Sanchez et al. 2012). Some of these P. nodorum PKSs showed significant homology to characterized PKS genes, inferring that they may produce compounds of similar structures. Other PKSs though have the domain architecture and/or accessory enzymes encoded in their gene clusters that match the biosynthetic requirements required to make certain compounds isolated previously from P. nodorum and related species. The domain architectures of the PKSs encoded by these genes and their corresponding homologues (closest characterized homologue and/or closest BLASTP homologue) are shown in Table 2. We will discuss selected PKS (NR-PKS, HR-PKS, PR-PKS, type III PKS) genes/gene clusters with the aim to provide an overview of the chemical structural diversity encoded in the genome of P. nodorum. In each section, the genes are listed in the order from those having the highest similarity (at the protein level) to another known gene, followed by those with more tentative matches.
Among the NR-PKSs, SNOG_11981 shares 80% protein identity with the Alternaria alternata ALM1, which is responsible for biosynthesis of DHN (1,8-dihydroxynaphthalene) melanin precursor 1,3,6,8-tetrahydroxynaphthalene (Figure 3) (Kimura & Tsuge 1993), suggesting that it may play a similar role in P. nodorum. The biosynthesis of DHN melanin may proceed either via the pentaketide route followed by deacetylation (by NR-PKSs such as Colletotrichum lagenarium PKS1) or via the heptaketide route followed by chain-shortening (by NR-PKSs such as Aspergillus nidulans WA and Aspergillus fumigatus Alb1) to form the key precursor 1,3,6,8-tetrahydroxynaphthalene (Tsai et al. 2001; Fujii et al. 2004; Wheeler et al. 2008; Vagstad et al. 2012). The similarity of SNOG_11981 and A. alternata ALM1 to C. lagenarium PKS1 (46% and 47% identity, respectively) and A. nidulans WA (44%% and 46% identity, respectively) are comparable, thus it remains to be determined if P. nodorum and A. alternata biosynthesize melanin via the pentaketide or heptaketide pathway. Interestingly, P. nodorum has also been shown to synthesize melanin via the L-DOPA (l-3,4-dihydroxyphenylalanine) pathway (Solomon et al. 2004). Other NR-PKSs that may involve in pigment biosynthesis are SNOG_08274 and SNOG_09932, which are homologues of the Nectria haematococcoa PKSN involved in red perithecia pigment biosynthesis (Graziani et al. 2004) and A. nidulans WA heptaketide napthopyrone synthase (Fujii et al. 2001), respectively (Table 2). It is yet to be determined if SNOG_08274 or SNOG_09932 is responsible for the pink pigmentation of P. nodorum conidia during their extrusion from the pycnidia.
Figure 3.
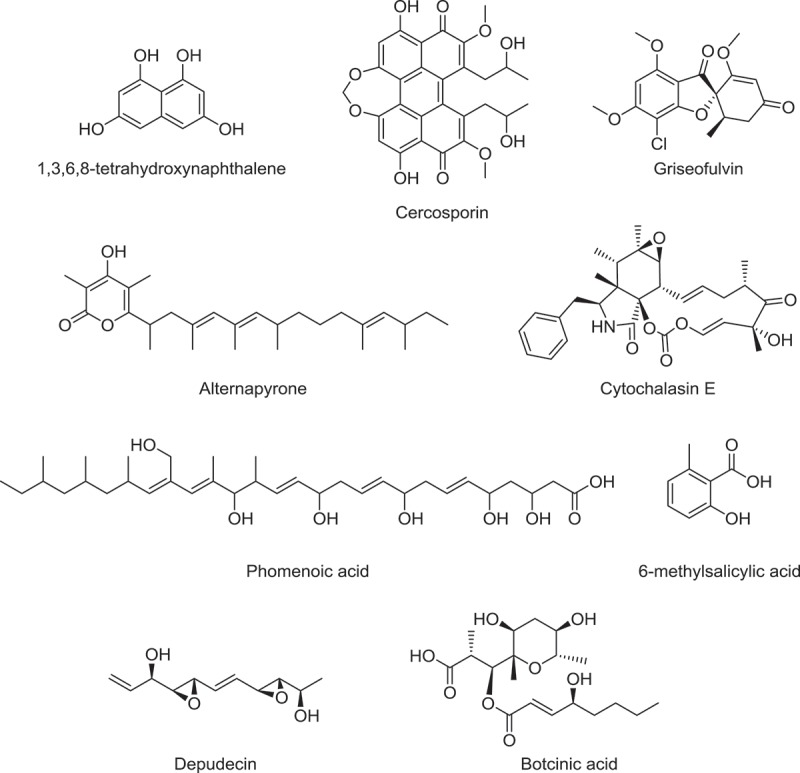
Polyketide compounds synthesized by putative homologues of Parastagonospora nodorum PKSs.
Another P. nodorum NR-PKS with significant similarity to characterized PKSs is SNOG_15829, which is 63% identical to griseofulvin PKS GsfA from Penicillium aethiopicum – the closest characterized homologue (Chooi et al. 2010). Like GsfA, SNOG_15829 lacks the C-terminal thioesterase (TE) releasing domain often required for product release either by Claisen cyclization or hydrolysis (Crawford & Townsend 2010; Chooi & Tang 2012). Most of these ‘TE-less’ NR-PKSs require a standalone metallo-β-lactamase-like TE for releasing the mature polyketide products (Awakawa et al. 2009; Li et al. 2011). Nonetheless, GsfA alone is capable of catalysing both aldol cyclization and Claisen cyclization of a heptaketide intermediate and release norlichexanthone as the immediate product (Cacho et al. 2013). The heptaketide chain length of griseofulvin (Figure 3) corresponds to the polyketide backbone of the mycotoxin alternariol (Thomas 1961), suggesting that SNOG_15829 is a promising candidate for alternariol production in P. nodorum.
Parastagonospora nodorum also contains a NR-PKS (SNOG_08614) that shares 56% identity to cercosporin synthase CTB1 from Cercospora nicotianae (Choquer et al. 2005). Homologues of O-methyltransferase CTB2 in the cercosporin pathway can also be found in the vicinity of SNOG_08614 locus (Chen et al. 2007) (Supplemental Table 2, cluster 19). Cercosporin (Figure 3) is a host non-selective photoactivated phytotoxin that has been demonstrated to contribute to the virulence of the plant pathogenic Cercospora species (Daub 1982; Daub et al. 2005). Similar perylenequinone compounds (compound 27-40 in Supplemental Table 1) have been isolated from an endolichenic Phaeosphaeria sp. (Li et al. 2012), but not in P. nodorum. Whether P. nodorum produces such perylenequinone compounds and their involvement in pathogenesis of wheat are questions that warrant further investigation.
SNOG_06682 and SNOG_07020 belong to a subclass of NR-PKSs that contain a C-methyltransferase domain. Both NR-PKSs also contain a TE domain at the C-terminal, instead of the thioreductase domain commonly found in azaphilone-type PKS gene clusters (Zabala et al. 2012). NR-PKSs with similar domain architecture to SNOG_06682 and SNOG_07020 have been implicated in biosynthesis of dimethylorsellinic acid (DMBA)-type meroterpenoids (Itoh et al. 2012; Lo et al. 2012) as well as mycophenolic acid (Figure 3) (Hansen et al. 2011, 2012; Regueira et al. 2011). Hansen et al. (2012) noted that P. nodorum SNOG_06682 gene cluster shared multiple homologues with the mycophenolic acid mpa cluster (mpaC, mpaD and mpaE) in Penicillium brevicompactum, suggesting that both clusters shared a common ancestry (Supplemental Table 2, cluster 13). Interestingly, mycophenolic acid has been reported to be isolated from P. nodorum. However, SNOG_06682 shares only 38% identity with the PKS MpaC in the mpa cluster and the corresponding gene cluster does not contain a UbiA-like membrane-bound long-chain prenyltransferase MpaA that was assumed to transfer a farnesyl chain to the phthalide intermediate. Instead, the SNOG_06682 gene cluster encodes a prenyltransferase belonging to the DMATS (dimethylallyl tryptophan synthase)-type short-chain prenyltransferase family, which are soluble α/β-fold proteins (Li 2009). A HR-PKS gene (SNOG_6676) is also found in vicinity of SNOG_06682 NR-PKS gene. It is unknown if the sequenced P. nodorum SN15 strain produces mycophenolic acid using a slightly different enzyme repertoire or if it produces a structural analogue of mycophenolic acid.
Among the P. nodorum HR-PKSs, SNOG_05791 shared 82% identity with the alternapyrone synthase PKSN from Alternaria solani (Fujii et al. 2005). However, alternapyrone (Figure 3), the direct product of PKSN, is unlikely to be the final product of the gene cluster as three cytochrome P450 genes and one oxidase gene are found in the vicinity of pksN (Fujii et al. 2005). Likewise, a flavin-dependent oxidase gene and a cytochrome P450 gene can be found in the vicinity of SNOG_05791; the two clusters may produce similar products. Since A. solani is also a plant pathogen (causes tomato and potato early blight disease), it would be interesting to examine whether the products of these two clusters are implicated in plant diseases. The PKS gene cluster of an antimicrobial compound, phomenoic acid, has recently been identified in the canola pathogen Leptosphaeria maculans (Elliott et al. 2013). Elliott et al. (2013) identified that P. nodorum encodes a close PKS homologue of L. maculans pks2, which is SNOG_04868 (90% identity to PKS2). Along with the presence of phomenoic acid tailoring genes homologues in the SNOG_04868 cluster, this suggests a high likelihood that P. nodorum also produces phomenoic acid (Supplemental Table 2, cluster 10). Interestingly, no phomenoic acid can be detected in P. nodorum under the same culture condition for L. maculans, showing that the regulation of this PKS gene differs from that in P. nodorum (Elliott et al. 2013).
SNOG_06676 shares 66% protein identity with the Alternaria brassicicola DEP5 (AbPKS5) HR-PKS, which synthesizes the histone deacetylase inhibitor depudecin (Wight et al. 2009). Depudecin appears to play a small role in the virulence of A. brassicicola on cabbage. Another HR-PKS SNOG_11272 shares 50% identity with Botrytis cinerea BcBOA9 involved in the production of the phytotoxin botcinic acid (Dalmais et al. 2011). It is unknown if P. nodorum SNOG_06676 and SNOG_11272 produce related phytotoxic molecules. Other similar HR-PKSs in P. nodorum may also be responsible for production of macrolide compounds such as the phytotoxic stagonolides from Stagonospora cirsii (Evidente, Cimmino, Berestetskiy, Andolfi, et al. 2008; Evidente, Cimmino, Berestetskiy, Mitina, 2008) (Supplemental Table 1).
The hybrid PKS-NRPSs are HR-PKSs that have a single NRPS module appended at the C-terminal. P. nodorum has one such hybrid PKS-NRPS, SNOG_00308. The closest homologue of SNOG_00308 is the cytochalasin (Figure 3) PKS CcsA (46% protein identity) from Aspergillus clavatus (Qiao, Chooi, et al. 2011). Qiao, Chooi, et al. (2011) identified homologues of ccsC, ccsD, ccsE, ccsF and ccsG in the P. nodorum genome clustering with SNOG_00308, suggesting that P. nodorum may produce related cytochalasin compounds (Supplemental Table 2, cluster 2). Cytochalasins are actin polymerization inhibitors and are known to be produced by plant-associated fungi, including dothidiomycete pathogens from the genera Phoma, Aschochyta and Drechslera (Scherlach et al. 2010) and from many other kinds of ascomycete fungi. Some cytochalasins have been reported to be phytotoxic (Evidente et al. 2002; Berestetskiy et al. 2008), while others have demonstrated that cytochalasins inhibit the actin-related plant defence mechanism, allowing non-host pathogens to penetrate plant cells (Kobayashi, Kobayashi, et al. 1997; Kobayashi, Yamada, 1997). However, involvement of cytochalasins in pathogenicity of P. nodorum has not been demonstrated. Compounds isolated from other Stagonospora species that may be produced by a PKS-NRPS include the tetramic acid antifungal, pramanicin (Schwartz et al. 1994) (Supplemental Table 1).
Mellein has been reported to be a major metabolite of P. nodorum along with the O-methyl and hydroxylated analogues (Devys et al. 1980). Mellein isolated from Diplodia pinea (=Sphaeropsis sapinea) was shown to be phytotoxic against pine and tomato (Cabras et al. 2006). Interestingly, mellein is also a metabolite of actinomycete bacteria, including Saccharopolyspora erythraea. The molecule has been shown to be produced by a PR-PKS in S. erythraea (Sun et al. 2012). So far, all characterized fungal PR-PKSs were shown to produce 6-methylsalicylic acid (Figure 3). Since no 6-methylsalicylic acid has been isolated from P. nodorum and SNOG_00477 is the only PR-PKS in P. nodorum, this makes SNOG_00477 a promising candidate PKS gene for mellein production (Yang et al. 2013). SNOG_14927 is another possible candidate PKS suggested by Yang et al. (2013); it has the identical domain architecture as SNOG_00477 and 6-methylsalicylic acid PR-PKSs. However, SNOG_14927 shares higher sequence similarity to other HR-PKSs, such as F. verticillioides Fum1p (Table 2), suggesting SNOG_14927 may have diverged from an ancestral HR-PKS more recently via losing its ER domain. It is to be determined which of the two PKS above is responsible for production of mellein and its derivatives in P. nodorum.
Besides the iterative type I PKSs typical to fungi, P. nodorum harbours a type III PKS gene (SNOG_09622), which shares 36% protein identity to Neurospora crassa 2ʹ-oxoalkylresorcylic acid synthase (ORAS) (Funa et al. 2007). Yang et al. suggested that SNOG_09622 along with the neighbouring HR-PKS gene SNOG_09623 may be responsible for the production (+)-4ʹ-methoxy-(2S)-methylbutyrophenone (Yang et al. 2013). Presumably, SNOG_09623 synthesizes the methylated reduced diketide as a starter unit, which is then incorporated by SNOG_09622 into (+)-4ʹ-methoxy-(2S)-methylbutyrophenone.
Nonribosomal peptide synthetases (NRPSs)
A minimum NRPS module typically consists of an adenylation (A) domain, thiolation domain (T) and a condensation (C) domain. Based on the typical collinearity rule of NRPSs (Finking & Marahiel 2004), the number of modules in an NRPS usually hints at the number of peptide residues in its final product. The domain architecture of P. nodorum NRPSs and their corresponding homologues (closest characterized homologue and/or closest BLASTP homologue) are shown in Table 3. P. nodorum has a total of six NRPSs with more than two modules. Except for SNOG_14834, all of these multimodular NRPSs end with a C-terminal condensation domain. The C-terminal condensation domain (CT domain) of fungal NRPSs has been shown to be involved in peptide cyclization (Gao, Haynes, et al. 2012); thus, these multimodular NRPSs in P. nodorum may be responsible for the production of cyclic peptides. We will discuss the possible function of the P. nodorum NRPSs below, beginning with the multimodular NRPSs, followed by the dimodular and monomodular NRPSs.
Table 3.
Domain architecture of selected NRPSs encoded in Parastagonospora nodorum genome.
| NRPS locus | Domain architecture | Closest BLAST hit(s) |
|---|---|---|
| Multimodular NRPSs* | ||
| SNOG_01105 | A-T-C-A-T-C-A-T-C-A-T-C-A-T-C | Pyrenophora teres PTT_10812 (79%) |
| SNOG_02134 | A-T-C-A-T-C-A-T-C-T-C-T-C | Pyrenophora teres PTT_13461 (55%) |
| SNOG_09081 | A-T-C-T-C-A-T-C-A-T-C-A-T-C-C | Metarhizium anisopliae MAA_01639 (41%) |
| SNOG_09488 | C-A-T-C-A-T-C-A-T-C | Sphaerulina musiva SEPMU_134958 (35%) |
| SNOG_14098 | A-T-C-A-T-C-A-T-C-A-T-C-A-C | Metarhizium acridum MAC_04974 (43%) |
| SNOG_14834 | T-C-A-T-C-A-T-C-A-T-C-A-T-C-A-T | Pyrenophora teres PTT_10907 (66%) |
| Dimodular NRPSs | ||
| SNOG_14923 | A-T-C-A-T-C | Bipolaris maydis NPS5 (75%) |
| Monomodular NRPSs | ||
| SNOG_03620 | A-T-TE | L. maculans LEMA_P067200.1 (73%) |
| SNOG_03771 | A-T-R | Pyrenophora teres PTT_06870 (70%) |
| SNOG_04863 | C-A | Pyrenophora teres PTT_16992 (62%) |
| SNOG_07021 | A-T-C | Pyrenophora teres PTT_13470 (69%) |
| SNOG_07126 | A-T-R-R | L. maculans Maa1 (85%) |
| SNOG_14368 | A-T-C-T-T-C | L. maculans LEMA_P070680 (71%) |
Notes: NRPS domain abbreviation – A, adenylation; T, thiolation; C, condensation; TE, thioesterase, R, reductase. See Finking and Marahiel (2004) review for detail description of the NRPS functional domains. *For multimodular NRPSs, the percentage protein identity values shown reflect the portion of the protein that has the highest match to the homologue (i.e. the value taken from a BLAST search) and may not represent 100% head-to-tail coverage (see ‘Methods’).
Siderophores are iron-sequestering cyclic peptides that play significant roles in virulence and fungal-host interactions (Johnson 2008). Interestingly, almost all known hydroxamate siderophore-synthesizing NRPSs do not follow the collinearity rule typical of NRPSs, but the iterative use of A domains (activating an identical amino acid more than once in an NRPS catalytic cycle) is the norm, with additional T-C domains as partial modules that extend the NRP products beyond the number of complete A-T-C modules in the NRPSs (Schwecke et al. 2006; Johnson 2008). SNOG_02134 is one such NRPS in P. nodorum, which consists of three A-T-C modules and two additional T-C partial modules. Recently, the SidN homologue from Epichloe festucae has been shown to be responsible for the production of a new siderophore, epichloënin A (Figure 4) (Johnson et al. 2013). To examine if P. nodorum encodes NRPSs with A domain that activate Nδ-acyl-Nδ-hydroxy-L-ornithine, the hydroxamate-containing residues commonly found in siderophores, we performed a BLAST search of the P. nodorum genome with the SidN-A3 domain as the query sequence (Lee et al. 2010). The closest blast hits to SidN-A3 domain is the first and third domain of SNOG_02134 NRPS, which shared 32% and 33% identity respectively with SidN-A3 domain. A putative L-ornithine N5-oxygenase known to be involved in hydroxamate siderophore biosynthesis (Hissen et al. 2005) is also encoded in the SNOG_02134 gene cluster (Supplemental Table 2, cluster 5). Thus, it is worth investigating whether SNOG_02134 encodes for hydroxamate siderophore biosynthesis in P. nodorum.
Figure 4.
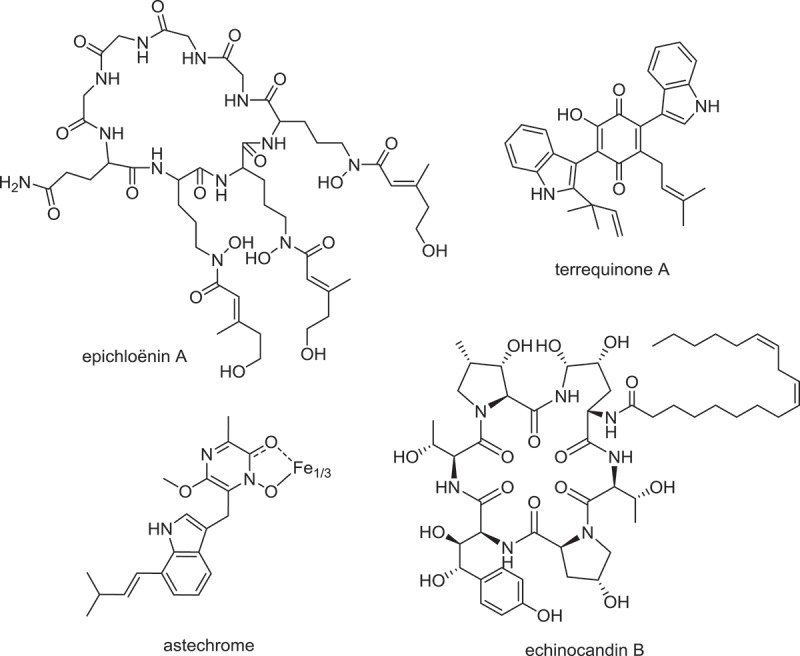
Nonribosomal peptide compounds synthesized by putative homologues of Parastagonospora nodorum NRPSs.
SNOG_14834 encodes a four module NRPSs with a T domain at the N-terminus. Fungal NRPSs with an N-terminal T domain have been shown to be involved in the biosynthesis of bioactive lipopeptides, such as emericellamide (Chiang et al. 2008), echinocandin (Figure 4) (Cacho et al. 2012) and pneumocandin (Chen et al. 2013). Interestingly, Pyrenophora teres f. teres genome contains an orthologue of SNOG_14834 (PTT_10907, 66% protein identity), however it appears that the orientation of the last two domains of the NRPSs are in reverse order compared to SNOG_14834, with PTT_10907 terminates with a C domain. It is yet to be determined if SNOG_14834 and PTT_10907 encode for cyclic lipopeptides. One of the P. nodorum multimodular NRPS genes (SNOG_09488) is located in close vicinity of a HR-PKS gene (SNOG_09490), suggesting the two genes may be part of a SM gene cluster (Supplemental Table 2, cluster 21). SNOG_09488 NRPS begins with an N-terminal C domain. Although such NRPSs have not been characterized in fungi, bacterial NRPSs with N-terminal C domain (or starter C domain) have been shown to be involved in biosynthesis of lipopeptides, where the starter C domain is responsible for incorporation of fatty acid or polyketide derived acyl-chain as a NRPS starter unit (Chooi & Tang 2010; Kraas et al. 2010, 2012). Therefore, it may be possible that the SNOG_09488 NRPS utilizes the polyketide chain synthesized by SNOG_09490 HR-PKS as starter unit to produce similar lipopeptide compound(s).
Septorine and its N-methoxy derivatives are phytotoxic pyrazine-containing molecules that have been isolated from P. nodorum (Devys et al. 1978, 1982, 1992). Thiopyrazine-synthesizing monomodular NRPS from Aspergillus terreus has been characterized previously (Qiao, Zhou, et al. 2011). However, the two oxygen substitutions in addition to the acyl substitutions at the pyrazine ring of septorines suggest that they are more similar to another class of NRPS molecule called diketopiperazine, which include sirodesmin (Gardiner et al. 2004), gliotoxin (Balibar & Walsh 2006; Cramer et al. 2006), and brevianamide (Maiya et al. 2006). More recently, another group of tryptophan-derived diketopiperazines, including hexadehydro-astechrome, with similar methoxy substitution as septorine has been isolated from Aspergillus fumigatus (Yin et al. 2013). Hexadehydro-astechrome (Figure 4) has the tendency to form iron complex and could be acting as a siderophore in A. fumigatus. As septorine, like most diketopiperazines natural products, is derived from the condensation of two amino acids (a tyrosine and an l-isoleucine), it is most likely synthesized by a dimodular NRPS. SNOG_14923 appears to be the only dimodular NRPS in P. nodorum.
Many single modular NRPS-like proteins do not contain the typical A-T-C module architecture, but often consist of A-T didomain followed by a variety of C-terminal domains (Bushley & Turgeon 2010). These monomodular NRPS-like proteins may or may not be involved in SM biosynthesis; for example, the aminoadipate reductase LYS2 in lysine biosynthesis pathway (terminates with thioester reductase domain). One of the monomodular NRPS that may be involved in SM biosynthesis is SNOG_03620 (33% identity to A. nidulans TdiA), which terminates with a thioesterase (TE) domain. These A-T-TE monomodular NRPSs are known to synthesize 1,4-benzoquinone compounds from condensation of two identical α-keto acids derived from amino acids; they include SM such as terrequinone (Figure 4) by TdiA (Balibar et al. 2007; Schneider et al. 2007) and atromentin by AtrA (Schneider et al. 2008; Wackler et al. 2012). More recently, such A-T-TE NRPS-like enzyme AN3396 (33% identity to SNOG_03620) has also been shown to produce a furanone compound, named microperfuranone (Yeh et al. 2012). No such α-keto acid-derived benzoquinone of furanone compounds have been discovered in P. nodorum so far. Interestingly, the canola pathogen L. maculans harbours a close homologue of SNOG_03620 (73% protein identity) with identical domain architecture. Besides SNOG_03620, most P. nodorum monomodular NRPSs appear to be conserved among the plant pathogens of the dothideomycete class as well (Table 3). Conservation of monomodular NRPSs with unknown functions among the dothideomycete was previously noted and it was proposed that some of these may play important roles in cellular metabolism (Bushley & Turgeon 2010).
Terpene synthases (TSs) and prenyltransferases (PTases)
Some terpenoid compounds are known to play significant roles in plant pathogenic fungi. The isoprenoid backbones of these compounds are synthesized by terpene synthases (TSs). The classification of various terpene synthases and their catalytic mechanisms have recently been reviewed (Gao, Honzatko, et al. 2012). Notable plant pathogen examples where the TSs in the pathway have been characterized include trichothecene family of mycotoxins (Cane et al. 1995; Rynkiewicz et al. 2002), and the non-host-specific phytotoxin botrydial (Pinedo et al. 2008). Although no terpenoid SMs have been reported from P. nodorum, the genome revealed that it encodes three putative sesquiterpene synthases (class I), which include SNOG_03562, SNOG_04807, and SNOG_10024. The diterpene plant hormone gibberellins are known to be produced by a Phaeosphaeria sp. strain L487 (Supplemental Table 1), where the ent-kaurene synthase that catalyses the key step in gibberellin biosynthesis has been characterized (Kawaide et al. 1997). A BLAST search using the ent-kaurene synthase from Phaeophaeria sp. L487 and Gibberella fujikuroi (Tudzynski et al. 1998) did not detect any significant homologue in P. nodorum suggesting that this fungus is incapable of producing gibberellins. Nevertheless, a gene (SNOG_120607) encoding a class II diterpene cyclase can be found in the P. nodorum genome adjacent to a cytochrome P450 monooxygenase (Supplemental Table 2, cluster 28). Thus, the gene cluster may be responsible for the production of a diterpene molecule.
Fungal aromatic prenyltransferases (PTases) belongs to the dimethylallyltryptophan synthase (DMATS) family are the key enzymes to the biosynthesis of many indole alkaloids (Li 2009). The most well-known examples among the plant-associated fungi are the ergot alkaloids (Wallwey & Li 2011). More recently, these DMATS-type PTases have also been implicated in biosynthesis of meroterpenoid compounds (polyketide-terpenoid hybrid compounds), such as viridicatumtoxin (Chooi et al. 2012) and prenyl xanthones (Sanchez et al. 2011). Two PTases were found in the P. nodorum genome, SNOG_06682, which clustered together with a NR-PKS gene (SNOG_06680), and SNOG_008527. Besides mycophenolic acid (Devys et al. 1980), which was mentioned above, no meroterpenoid or indole alkaloid compounds have been identified from P. nodorum. Two prenylated compounds that have been isolated from related species are spartinoxide and 4-hydroxy-3-prenyl-benzoic acid (Elsebai et al. 2010) (Supplemental Table 1).
Other than the DMATS-type PTases, P. nodorum also harbours five genes encoding UbiA-like membrane-bound PTases, which is responsible for transfer of long prenyl chains, such as the E. coli UbiA and S. cerevisiae COQ2 (p-hydroxybenzoate:polyprenyl transferase) of the ubiquinone pathway. UbiA-like PTases are also known to be involved in biosynthesis of SMs, such as the pyripyropene A (Itoh et al. 2010) and 3,5-dimethylorsellinic acid (DMOA)-derived meroterpenoids (Itoh et al. 2012; Lo et al. 2012). More recently, a novel membrane-bound class I terpene cyclase, which shares structural and sequence homology to UbiA-like PTases, has been shown to cyclise farnesyl-diphosphate to β-trans-bergamotene in the fumagillin pathway (Lin et al. 2013). Two of the five P. nodorum UbiA-like PTases (SNOG_00816 and 05304) showed overall high protein identity (>80%) with other ascomycetes, and are likely to be involved in primary metabolism. The other three UbiA-like PTases (SNOG_00008, 07120 and 09915) share lower similarity to those found in other ascomycete genomes and may be involved in SM biosynthesis. The SNOG_09915 gene is clustered with a HR-PKS gene (SNOG_09932) and maybe responsible for biosynthesis of a meroterpenoid of mixed polyketide-isoprenoid origin (Supplemental Table 2, cluster 23). On the other hand, the SNOG_07120 locus is in vicinity of a NRPS gene (SNOG_07126) and thus may be a terpene alkaloid gene cluster (Supplemental Table 2, cluster 15).
Future perspectives
In summary, the whole-genome survey of the SM gene inventory of P. nodorum shows that the wheat pathogen harbours significant unexplored SM biosynthetic potential. However, none of the SM products from these gene clusters are known and only a handful of metabolites (representing only about one-tenth of the total number of gene clusters in P. nodorum) have been identified from this important wheat pathogen. Several SM backbone genes/gene clusters appear to be conserved across multiple plant pathogens; these SM genes may play important roles that are common to their pathogenic life style. Apart from uncovering novel SM virulence factors, the P. nodorum genome presents a unique opportunity for natural product discovery with some SM gene clusters potentially encoding analogues of known bioactive molecules amongst others that are novel.
Methods
In silico analysis of P. nodorum SM genes
P. nodorum PKS genes were retrieved from the P. nodorum genome on the NCBI GenBank and DOE-JGI database by using an arbitrary fungal KS domain (e.g. P. aethiopicum VrtA) as a BLASTP query. The PKS domains are predicted using the NCBI Conserved Domain Search and antiSMASH 2.0 (Blin et al. 2013). The SAT and PT domains of NR-PKSs not detected by the automated analyses above were determined by protein sequence alignment with Aspergillus parasiticus norsoloronic acid synthase (PksA) and manually search for the conserved motifs described in the previous work (Crawford et al. 2006, 2008). The closest characterized BLAST hits were retrieved by BLASTP program on NCBI website using P. nodorum PKSs as query sequences.
P. nodorum NRPS genes were retrieved from the P. nodorum genome by using an arbitrary fungal A domain (e.g. P. aethiopicum TqaA-A1domain) as a BLASTP query. The NRPS domains were predicted using the NCBI Conserved Domain Search and antiSMASH2.0 (Blin et al. 2013). Due to the large size of these proteins and their tendency to rearrange/truncate during evolution, a good head-to-tail pairwise alignment was not possible unless there was a close homologue with identical domain architecture. Thus, for quick and simple comparison, the percentage protein identity values was taken directly from the BLASTP analysis and they reflect the portion of the protein that has the highest match to the homologue and may not represent 100% head-to-tail coverage, especially those that shown a lower percentage identity scores. Nevertheless, the values are still useful for comparison as the portion of protein that has a good match is often the conserved domain (C, A and T domains) and may reflect the substrate specificities.
P. nodorum PTase and TS genes were retrieved from the genome by using an arbitrary fungal PTase (e.g. P. aethiopicum VrtC) and TS (e.g. trichodiene synthase and aristolochene synthase) respectively as BLASTP query.
A list of all the SM backbone genes retrieved using the methods above was generated and compared with the list of P. nodorum gene clusters predicted by SMURF (Khaldi et al. 2010). The loci where the SM backbone genes were not detected by SMURF were manually inspected for neighbouring biosynthetic genes on the DOE-JGI genome browser. These additional P. nodorum SM gene clusters were added to the list in Supplemental Table 2.
Acknowledgements
YHC is supported by an Australian Research Council Discovery Early Career Researcher Award (DECRA) fellowship. MJMG is a recipient of an Australian Government Endeavour Award and a Mexican CONACyT scholarship. PSS is an Australian Research Council Future Fellow.
Supplemental data
Supplemental data for this article can be accessed here.
References
- Awakawa T, Yokota K, Funa N, Doi F, Mori N, Watanabe H, Horinouchi S Physically discrete β-lactamase-type thioesterase catalyzes product release in atrochrysone synthesis by iterative type I polyketide synthase. Chem Biol. 2009;16:613–623. doi: 10.1016/j.chembiol.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Balibar CJ, Howard-Jones AR, Walsh CT Terrequinone A biosynthesis through L-tryptophan oxidation, dimerization and bisprenylation. Nat Chem Biol. 2007;3:584–592. doi: 10.1038/nchembio.2007.20. [DOI] [PubMed] [Google Scholar]
- Balibar CJ, Walsh CT Glip, a multimodular nonribosomal peptide synthetase in Aspergillus fumigatus, makes the diketopiperazine scaffold of gliotoxin. Biochemistry. 2006;45:15029–15038. doi: 10.1021/bi061845b. [DOI] [PubMed] [Google Scholar]
- Bathgate J, Loughman R Ascospores are a source of inoculum of Phaeosphaeria nodorum, P. avenaria f. sp. avenaria and Mycosphaerella graminicola in Western Australia. Australas Plant Pathol. 2001;30:317–322. doi: 10.1071/AP01043. [DOI] [Google Scholar]
- Berestetskiy A, Dmitriev A, Mitina G, Lisker I, Andolfi A, Evidente A Nonenolides and cytochalasins with phytotoxic activity against Cirsium arvense and Sonchus arvensis: a structure-activity relationships study. Phytochemistry. 2008;69:953–960. doi: 10.1016/j.phytochem.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T Antismash 2.0 – a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41:W204–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakhage AA, Schroeckh V Fungal secondary metabolites – strategies to activate silent gene clusters. Fungal Genet Biol. 2011;48:15–22. doi: 10.1016/j.fgb.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Brennan R, Fitt BD, Taylor G, Colhoun J Dispersal of Septoria nodorum pycnidiospores by simulated raindrops in still air. J Phytopathol. 1985;112:281–290. doi: 10.1111/j.1439-0434.1985.tb00805.x. [DOI] [Google Scholar]
- Bushley KE, Turgeon BG Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthetases and their evolutionary relationships. BMC Evol Biol. 2010;10:26. doi: 10.1186/1471-2148-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabras A, Mannoni MA, Serra S, Andolfi A, Fiore M, Evidente A Occurrence, isolation and biological activity of phytotoxic metabolites produced in vitro by Sphaeropsis sapinea, pathogenic fungus of Pinus radiata . Eur J Plant Pathol. 2006;115:187–193. doi: 10.1007/s10658-006-9006-7. [DOI] [Google Scholar]
- Cacho RA, Chooi YH, Zhou H, Tang Y Complexity generation in fungal polyketide biosynthesis: a spirocycle-forming P450 in the concise pathway to the antifungal drug griseofulvin. ACS Chem Biol. 2013;8:2322–2330. doi: 10.1021/cb400541z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacho RA, Jiang W, Chooi YH, Walsh CT, Tang Y Identification and characterization of the echinocandin B biosynthetic gene cluster from Emericella rugulosa NRRL 11440. J Am Chem Soc. 2012;134:16781–16790. doi: 10.1021/ja307220z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cane DE, Yang G, Xue Q, Shim JH Trichodiene synthase. Substrate specificity and inhibition. Biochemistry. 1995;34:2471–2479. doi: 10.1021/bi00008a010. [DOI] [PubMed] [Google Scholar]
- Chen HQ, Lee MH, Daub ME, Chung KR Molecular analysis of the cercosporin biosynthetic gene cluster in Cercospora nicotianae . Mol Microbiol. 2007;64:755–770. doi: 10.1111/j.1365-2958.2007.05689.x. [DOI] [PubMed] [Google Scholar]
- Chen L, Yue Q, Zhang X, Xiang M, Wang C, Li S, Che Y, Ortiz-Lopez FJ, Bills GF, Liu X Genomics-driven discovery of the pneumocandin biosynthetic gene cluster in the fungus Glarea lozoyensis . BMC Genomics. 2013;14:339. doi: 10.1186/1471-2164-14-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YM, Szewczyk E, Nayak T, Davidson AD, Sanchez JF, Lo HC, Ho WY, Simityan H, Kuo E, Praseuth A Molecular genetic mining of the Aspergillus secondary metabolome: discovery of the emericellamide biosynthetic pathway. Chem Biol. 2008;15:527–532. doi: 10.1016/j.chembiol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chooi YH, Cacho R, Tang Y Identification of the viridicatumtoxin and griseofulvin gene clusters from Penicillium aethiopicum . Chem Biol. 2010;17:483–494. doi: 10.1016/j.chembiol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chooi YH, Tang Y Adding the lipo to lipopeptides: do more with less. Chem Biol. 2010;17:791–793. doi: 10.1016/j.chembiol.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Chooi YH, Tang Y Navigating the fungal polyketide chemical space: from genes to molecules. J Org Chem. 2012;77:9933–9953. doi: 10.1021/jo301592k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chooi YH, Wang P, Fang J, Li Y, Wu K, Wang P, Tang Y Discovery and characterization of a group of fungal polycyclic polyketide prenyltransferases. J Am Chem Soc. 2012;134:9428–9437. doi: 10.1021/ja3028636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquer M, Dekkers KL, Chen HQ, Cao L, Ueng PP, Daub ME, Chung KR The CTB1 gene encoding a fungal polyketide synthase is required for cercosporin biosynthesis and fungal virulence of Cercospora nicotianae . Mol Plant Microbe Interact. 2005;18:468–476. doi: 10.1094/MPMI-18-0468. [DOI] [PubMed] [Google Scholar]
- Christianson DW Unearthing the roots of the terpenome. Curr Opin Chem Biol. 2008;12:141–150. doi: 10.1016/j.cbpa.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RJ Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org Biomol Chem. 2007;5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- Cramer RA Jr., Gamcsik MP, Brooking RM, Najvar LK, Kirkpatrick WR, Patterson TF, Balibar CJ, Graybill JR, Perfect JR, Abraham SN Disruption of a nonribosomal peptide synthetase in Aspergillus fumigatus eliminates gliotoxin production. Eukaryot Cell. 2006;5:972–980. doi: 10.1128/EC.00049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JM, Dancy BC, Hill EA, Udwary DW, Townsend CA Identification of a starter unit acyl-carrier protein transacylase domain in an iterative type I polyketide synthase. Proc Natl Acad Sci USA. 2006;103:16728–16733. doi: 10.1073/pnas.0604112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JM, Thomas PM, Scheerer JR, Vagstad AL, Kelleher NL, Townsend CA Deconstruction of iterative multidomain polyketide synthase function. Science. 2008;320:243–246. doi: 10.1126/science.1154711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JM, Townsend CA New insights into the formation of fungal aromatic polyketides. Nat Rev Microbiol. 2010;8:879–889. doi: 10.1038/nrmicro2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook AD, Friesen TL, Liu ZH, Ojiambo PS, Cowger C Novel necrotrophic effectors from Stagonospora nodorum and corresponding host sensitivities in winter wheat germplasm in the southeastern United States. Phytopathology. 2012;102:498–505. doi: 10.1094/PHYTO-08-11-0238. [DOI] [PubMed] [Google Scholar]
- Dalmais B, Schumacher J, Moraga J, LEP P, Tudzynski B, Collado IG, Viaud M The Botrytis cinerea phytotoxin botcinic acid requires two polyketide synthases for production and has a redundant role in virulence with botrydial. Mol Plant Pathol. 2011;12:564–579. doi: 10.1111/j.1364-3703.2010.00692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub ME Cercosporin, a photosensitizing toxin from Cercospora species. Phytopathology. 1982;72:370–374. doi: 10.1094/Phyto-72-370. [DOI] [Google Scholar]
- Daub ME, Herrero S, Chung KR Photoactivated perylenequinone toxins in fungal pathogenesis of plants. FEMS Microbiol Lett. 2005;252:197–206. doi: 10.1016/j.femsle.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Devys M, Barbier M, Bousquet JF, Kollmann A Isolation of the (-)-(3r)-5-hydroxymellein from the fungus septoria nodorum . Phytochemistry. 1994;35:825–826. doi: 10.1016/S0031-9422(00)90617-4. [DOI] [Google Scholar]
- Devys M, Barbier M, Kollmann A, Bousquet JF Septorine and n-methoxy septorine, substituted pyrazines from the fungus Septoria nodorum berk. Tetrahedron Lett. 1982;23:5409–5412. [Google Scholar]
- Devys M, Barbier M, Kollmann A, Bousquet JF N-methoxy septorinol, a substituted pyrazine from the fungus Septoria nodorum . Phytochemistry. 1992;31:4393–4394. doi: 10.1016/0031-9422(92)80492-W. [DOI] [Google Scholar]
- Devys M, Bousquet JF, Kollmann A, Barbier M Isolation of a new pyrazine, septorine, from culture medium of phyto-pathogen fungus Septoria nodorum berk. C R Acad Sci. 1978;286:457–458. [Google Scholar]
- Devys M, Bousquet JF, Kollmann A, Barbier M Dihydroisocoumarins and mycophenolic-acid of culture-medium of a phytopathogenic fungus, Septoria nodorum . Phytochemistry. 1980;19:2221–2222. doi: 10.1016/S0031-9422(00)82234-7. [DOI] [Google Scholar]
- Eisenman HC, Casadevall A Synthesis and assembly of fungal melanin. Appl Microbiol Biotechnol. 2012;93:931–940. doi: 10.1007/s00253-011-3777-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott CE, Callahan DL, Schwenk D, Nett M, Hoffmeister D, Howlett BJ A gene cluster responsible for biosynthesis of phomenoic acid in the plant pathogenic fungus, Leptosphaeria maculans . Fung Genet Biol. 2013;53:50–58. doi: 10.1016/j.fgb.2013.01.008. [DOI] [PubMed] [Google Scholar]
- Elsebai MF, Kehraus S, Gutschow M, Konig GM Spartinoxide, a new enantiomer of A82775C with inhibitory activity toward HLE from the marine-derived fungus Phaeosphaeria spartinae . Nat Prod Commun. 2010;5:1071–1076. [PubMed] [Google Scholar]
- Evidente A, Andolfi A, Vurro M, Zonno MC, Motta A Cytochalasins Z1, Z2 and Z3, three 24-oxa[14]cytochalasans produced by Pyrenophora semeniperda . Phytochemistry. 2002;60:45–53. doi: 10.1016/S0031-9422(02)00071-7. [DOI] [PubMed] [Google Scholar]
- Evidente A, Cimmino A, Berestetskiy A, Andolfi A, Motta A Stagonolides G-I and modiolide A, nonenolides produced by Stagonospora cirsii, a potential mycoherbicide for Cirsium arvense . J Nat Prod. 2008;71:1897–1901. doi: 10.1021/np800415w. [DOI] [PubMed] [Google Scholar]
- Evidente A, Cimmino A, Berestetskiy A, Mitina G, Andolfi A, Motta A Stagonolides B-F, nonenolides produced by Stagonospora cirsii, a potential mycoherbicide of Cirsium arvense . J Nat Prod. 2008;71:31–34. doi: 10.1021/np0703038. [DOI] [PubMed] [Google Scholar]
- Finking R, Marahiel MA Biosynthesis of nonribosomal peptides. Annu Rev Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- Francki MG, Shankar M, Walker E, Loughman R, Golzar H, Ohm H New quantitative trait loci in wheat for flag leaf resistance to Stagonospora nodorum blotch. Phytopathology. 2011;101:1278–1284. doi: 10.1094/PHYTO-02-11-0054. [DOI] [PubMed] [Google Scholar]
- Friesen TL, Chu C, Xu SS, Faris JD Sntox5-snn5: a novel Stagonospora nodorum effector-wheat gene interaction and its relationship with the SnToxA-Tsn1 and SnTox3-Snn3-B1 interactions. Mol Plant Pathol. 2012;13:1101–1109. doi: 10.1111/j.1364-3703.2012.00819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii I, Watanabe A, Sankawa U, Ebizuka Y Identification of claisen cyclase domain in fungal polyketide synthase WA, a naphthopyrone synthase of Aspergillus nidulans . Chem Biol. 2001;8:189–197. doi: 10.1016/S1074-5521(00)90068-1. [DOI] [PubMed] [Google Scholar]
- Fujii I, Yasuoka Y, Tsai HF, Chang YC, Kwon-Chung KJ, Ebizuka Y Hydrolytic polyketide shortening by ayg1p, a novel enzyme involved in fungal melanin biosynthesis. J Biol Chem. 2004;279:44613–44620. doi: 10.1074/jbc.M406758200. [DOI] [PubMed] [Google Scholar]
- Fujii I, Yoshida N, Shimomaki S, Oikawa H, Ebizuka Y An iterative type I polyketide synthase PKSN catalyzes synthesis of the decaketide alternapyrone with regio-specific octa-methylation. Chem Biol. 2005;12:1301–1309. doi: 10.1016/j.chembiol.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Funa N, Awakawa T, Horinouchi S Pentaketide resorcylic acid synthesis by type III polyketide synthase from Neurospora crassa . J Biol Chem. 2007;282:14476–14481. doi: 10.1074/jbc.M701239200. [DOI] [PubMed] [Google Scholar]
- Gao X, Haynes SW, Ames BD, Wang P, Vien LP, Walsh CT, Tang Y Cyclization of fungal nonribosomal peptides by a terminal condensation-like domain. Nat Chem Biol. 2012;8:823–830. doi: 10.1038/nchembio.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Honzatko RB, Peters RJ Terpenoid synthase structures: a so far incomplete view of complex catalysis. Nat Prod Rep. 2012;29:1153–1175. doi: 10.1039/c2np20059g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner DM, Cozijnsen AJ, Wilson LM, Pedras MS, Howlett BJ The sirodesmin biosynthetic gene cluster of the plant pathogenic fungus Leptosphaeria maculans . Mol Microbiol. 2004;53:1307–1318. doi: 10.1111/j.1365-2958.2004.04215.x. [DOI] [PubMed] [Google Scholar]
- Goudet C, Milat ML, Sentenac H, Thibaud JB Beticolins, nonpeptidic, polycyclic molecules produced by the phytopathogenic fungus Cercospora beticola, as a new family of ion channel-forming toxins. Mol Plant Microbe Interact. 2000;13:203–209. doi: 10.1094/MPMI.2000.13.2.203. [DOI] [PubMed] [Google Scholar]
- Graziani S, Vasnier C, Daboussi MJ Novel polyketide synthase from Nectria haematococca . Appl Environ Microbiol. 2004;70:2984–2988. doi: 10.1128/AEM.70.5.2984-2988.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths E, Ao HC Dispersal of Septoria nodorum spores and spread of glume blotch of wheat in the field. Trans Br Mycol Soc. 1976;67:413–418. doi: 10.1016/S0007-1536(76)80166-0. [DOI] [Google Scholar]
- Hane JK, Lowe RG, Solomon PS, Tan KC, Schoch CL, Spatafora JW, Crous PW, Kodira C, Birren BW, Galagan JE Dothideomycete plant interactions illuminated by genome sequencing and EST analysis of the wheat pathogen Stagonospora nodorum . Plant Cell. 2007;19:3347–3368. doi: 10.1105/tpc.107.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen BG, Mnich E, Nielsen KF, Nielsen JB, Nielsen MT, Mortensen UH, Larsen TO, Patil KR Involvement of a natural fusion of a cytochrome P450 and a hydrolase in mycophenolic acid biosynthesis. Appl Environ Microbiol. 2012;78:4908–4913. doi: 10.1128/AEM.07955-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen BG, Salomonsen B, Nielsen MT, Nielsen JB, Hansen NB, Nielsen KF, Regueira TB, Nielsen J, Patil KR, Mortensen UH Versatile enzyme expression and characterization system for Aspergillus nidulans, with the Penicillium brevicompactum polyketide synthase gene from the mycophenolic acid gene cluster as a test case. Appl Environ Microbiol. 2011;77:3044–3051. doi: 10.1128/AEM.01768-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hissen AH, Wan AN, Warwas ML, Pinto LJ, Moore MM The Aspergillus fumigatus siderophore biosynthetic gene sida, encoding l-ornithine n5-oxygenase, is required for virulence. Infect Immun. 2005;73:5493–5503. doi: 10.1128/IAI.73.9.5493-5503.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeister D, Keller NP Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep. 2007;24:393–416. doi: 10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- Itoh T, Tokunaga K, Matsuda Y, Fujii I, Abe I, Ebizuka Y, Kushiro T Reconstitution of a fungal meroterpenoid biosynthesis reveals the involvement of a novel family of terpene cyclases. Nat Chem. 2010;2:858–864. doi: 10.1038/nchem.764. [DOI] [PubMed] [Google Scholar]
- Itoh T, Tokunaga K, Radhakrishnan EK, Fujii I, Abe I, Ebizuka Y, Kushiro T Identification of a key prenyltransferase involved in biosynthesis of the most abundant fungal meroterpenoids derived from 3,5-dimethylorsellinic acid. Chembiochem. 2012;13:1132–1135. doi: 10.1002/cbic.201200124. [DOI] [PubMed] [Google Scholar]
- Johnson L Iron and siderophores in fungal-host interactions. Mycol Res. 2008;112 doi: 10.1016/j.mycres.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Johnson LJ, Koulman A, Christensen M, Lane GA, Fraser K, Forester N, Johnson RD, Bryan GT, Rasmussen S An extracellular siderophore is required to maintain the mutualistic interaction of epichloë festucae with lolium perenne. Plos Pathog. 2013;9:e1003332. doi: 10.1371/journal.ppat.1003332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaide H, Imai R, Sassa T, Kamiya Y Ent-kaurene synthase from the fungus Phaeosphaeria sp. L487. cDNA isolation, characterization, and bacterial expression of a bifunctional diterpene cyclase in fungal gibberellin biosynthesis. J Biol Chem. 1997;272:21706–21712. doi: 10.1074/jbc.272.35.21706. [DOI] [PubMed] [Google Scholar]
- Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura N, Tsuge T Gene cluster involved in melanin biosynthesis of the filamentous fungus Alternaria alternata . J Bacteriol. 1993;175:4427–4435. doi: 10.1128/jb.175.14.4427-4435.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kobayashi I, Funaki Y, Fujimoto S, Takemoto T, Kunoh H Dynamic reorganization of microfilaments and microtubules is necessary for the expression of non-host resistance in barley coleoptile cells. Plant J. 1997;11:525–537. doi: 10.1046/j.1365-313X.1997.11030525.x. [DOI] [Google Scholar]
- Kobayashi Y, Yamada M, Kobayashi I, Kunoh H Actin microfilaments are required for the expression of nonhost resistance in higher plants. Plant Cell Physiol. 1997;38:725–733. doi: 10.1093/oxfordjournals.pcp.a029226. [DOI] [Google Scholar]
- Kraas FI, Giessen TW, Marahiel MA Exploring the mechanism of lipid transfer during biosynthesis of the acidic lipopeptide antibiotic CDA. FEBS Lett. 2012;586:283–288. doi: 10.1016/j.febslet.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Kraas FI, Helmetag V, Wittmann M, Strieker M, Marahiel MA Functional dissection of surfactin synthetase initiation module reveals insights into the mechanism of lipoinitiation. Chem Biol. 2010;17:872–880. doi: 10.1016/j.chembiol.2010.06.015. [DOI] [PubMed] [Google Scholar]
- Lee TV, Johnson LJ, Johnson RD, Koulman A, Lane GA, Lott JS, Arcus VL Structure of a eukaryotic nonribosomal peptide synthetase adenylation domain that activates a large hydroxamate amino acid in siderophore biosynthesis. J Biol Chem. 2010;285:2415–2427. doi: 10.1074/jbc.M109.071324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Wang H, Zhu R, Sun L, Wang L, Li M, Li Y, Liu Y, Zhao Z, Lou H Phaeosphaerins A-F, cytotoxic perylenequinones from an endolichenic fungus, Phaeosphaeria sp. J Nat Prod. 2012;75:142–147. doi: 10.1021/np200614h. [DOI] [PubMed] [Google Scholar]
- Li SM Evolution of aromatic prenyltransferases in the biosynthesis of indole derivatives. Phytochemistry. 2009;70:1746–1757. doi: 10.1016/j.phytochem.2009.03.019. [DOI] [PubMed] [Google Scholar]
- Li Y, Chooi YH, Sheng Y, Valentine JS, Tang Y Comparative characterization of fungal anthracenone and naphthacenedione biosynthetic pathways reveals an α-hydroxylation-dependent claisen-like cyclization catalyzed by a dimanganese thioesterase. J Am Chem Soc. 2011;133:15773–15785. doi: 10.1021/ja206906d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HC, Chooi YH, Dhingra S, Xu W, Calvo AM, Tang Y The fumagillin biosynthetic gene cluster in Aspergillus fumigatus encodes a cryptic terpene cyclase involved in the formation of beta-trans-bergamotene. J Am Chem Soc. 2013;135:4616–4619. doi: 10.1021/ja312503y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo HC, Entwistle R, Guo CJ, Ahuja M, Szewczyk E, Hung JH, Chiang YM, Oakley BR, Wang CC Two separate gene clusters encode the biosynthetic pathway for the meroterpenoids austinol and dehydroaustinol in aspergillus nidulans. J Am Chem Soc. 2012;134:4709–4720. doi: 10.1021/ja209809t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang J, Kidarsa T, Bradford CS, Gilbert B, Curtis M, Tzeng SC, Maier CS, Wolpert TJ Tricking the guard: exploiting plant defense for disease susceptibility. Science. 2012;338:659–662. doi: 10.1126/science.1226743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiya S, Grundmann A, Li SM, Turner G The fumitremorgin gene cluster of Aspergillus fumigatus: identification of a gene encoding brevianamide F synthetase. Chembiochem. 2006;7:1062–1069. doi: 10.1002/cbic.200600003. [DOI] [PubMed] [Google Scholar]
- Meiss E, Konno H, Groth G, Hisabori T Molecular processes of inhibition and stimulation of ATP synthase caused by the phytotoxin tentoxin. J Biol Chem. 2008;283:24594–24599. doi: 10.1074/jbc.M802574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möbius N, Hertweck C Fungal phytotoxins as mediators of virulence. Curr Opin Plant Biol. 2009;12:390–398. doi: 10.1016/j.pbi.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Murray GM, Brennan JP Estimating disease losses to the Australian wheat industry. Australas Plant Pathol. 2009;38:558–570. doi: 10.1071/AP09053. [DOI] [Google Scholar]
- Oliver RP, Friesen TL, Faris JD, Solomon PS Stagonospora nodorum: from pathology to genomics and host resistance. Annu Rev Phytopathol. 2012;50:23–43. doi: 10.1146/annurev-phyto-081211-173019. [DOI] [PubMed] [Google Scholar]
- Oliver RP, Solomon PS New developments in pathogenicity and virulence of necrotrophs. Curr Opin Plant Biol. 2010;13:415–419. doi: 10.1016/j.pbi.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Parisi A, Piattelli M, Tringali C, Di San Lio GM Identification of the phytotoxin mellein in culture fluids of Phoma tracheiphila . Phytochemistry. 1993;32:865–867. doi: 10.1016/0031-9422(93)85221-C. [DOI] [Google Scholar]
- Pinedo C, Wang CM, Pradier JM, Dalmais B, Choquer M, Le Pêcheur P, Morgant G, Collado IG, Cane DE, Viaud M Sesquiterpene synthase from the botrydial biosynthetic gene cluster of the phytopathogen Botrytis cinerea . ACS Chem Biol. 2008;3:791–801. doi: 10.1021/cb800225v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao K, Chooi YH, Tang Y Identification and engineering of the cytochalasin gene cluster from Aspergillus clavatus NRRL 1. Metab Eng. 2011;13:723–732. doi: 10.1016/j.ymben.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao KJ, Zhou H, Xu W, Zhang WJ, Garg N, Tang Y A fungal nonribosomal peptide synthetase module that can synthesize thiopyrazines. Org Lett. 2011;13:1758–1761. doi: 10.1021/ol200288w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaedvlieg W, Verkley GJ, Shin HD, Barreto RW, Alfenas AC, Swart WJ, Groenewald JZ, Crous PW Sizing up septoria. Stud Mycol. 2013;75:307–390. doi: 10.3114/sim0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regueira TB, Kildegaard KR, Hansen BG, Mortensen UH, Hertweck C, Nielsen J Molecular basis for mycophenolic acid biosynthesis in Penicillium brevicompactum . Appl Environ Microbiol. 2011;77:3035–3043. doi: 10.1128/AEM.03015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rynkiewicz MJ, Cane DE, Christianson DW X-ray crystal structures of D100E trichodiene synthase and its pyrophosphate complex reveal the basis for terpene product diversity. Biochemistry. 2002;41:1732–1741. doi: 10.1021/bi011960g. [DOI] [PubMed] [Google Scholar]
- Sanchez JF, Entwistle R, Hung JH, Yaegashi J, Jain S, Chiang YM, Wang CC, Oakley BR Genome-based deletion analysis reveals the prenyl xanthone biosynthesis pathway in Aspergillus nidulans . J Am Chem Soc. 2011;133:4010–4017. doi: 10.1021/ja1096682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez JF, Somoza AD, Keller NP, Wang CC Advances in Aspergillus secondary metabolite research in the post-genomic era. Nat Prod Rep. 2012;29:351–371. doi: 10.1039/c2np00084a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson F, Hampton J Role of the perfect states in the epidemiology of the common septoria diseases of wheat. New Zeal J Agr Res. 1978;21:277–281. doi: 10.1080/00288233.1978.10427411. [DOI] [Google Scholar]
- Scherlach K, Boettger D, Remme N, Hertweck C The chemistry and biology of cytochalasans. Nat Prod Rep. 2010;27:869–886. doi: 10.1039/b903913a. [DOI] [PubMed] [Google Scholar]
- Schneider P, Bouhired S, Hoffmeister D Characterization of the atromentin biosynthesis genes and enzymes in the homobasidiomycete Tapinella panuoides . Fungal Genet Biol. 2008;45:1487–1496. doi: 10.1016/j.fgb.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Schneider P, Weber M, Rosenberger K, Hoffmeister D A one-pot chemoenzymatic synthesis for the universal precursor of antidiabetes and antiviral bis-indolylquinones. Chem Biol. 2007;14:635–644. doi: 10.1016/j.chembiol.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Schwartz RE, Helms GL, Bolessa EA, Wilson KE, Giacobbe RA, Tkacz JS, Bills GF, Liesch JM, Zink DL, Curotto JE Pramanicin, a novel antimicrobial agent from a fungal fermentation. Tetrahedron. 1994;50:1675–1686. doi: 10.1016/S0040-4020(01)80843-7. [DOI] [Google Scholar]
- Schwecke T, Göttling K, Durek P, Dueñas I, Käufer NF, Zock-Emmenthal S, Staub E, Neuhof T, Dieckmann R, von Döhren H Nonribosomal peptide synthesis in schizosaccharomyces pombe and the architectures of ferrichrome-type siderophore synthetases in fungi. Chembiochem. 2006;7:612–622. doi: 10.1002/cbic.200500301. [DOI] [PubMed] [Google Scholar]
- Seshime Y, Juvvadi PR, Fujii I, Kitamoto K Discovery of a novel superfamily of type III polyketide synthases in Aspergillus oryzae . Biochem Biophys Res Commun. 2005;331:253–260. doi: 10.1016/j.bbrc.2005.03.160. [DOI] [PubMed] [Google Scholar]
- Shaner G Effect of environment on fungal leaf blights of small grains. Annu Rev Phytopathol. 1981;19:273–296. doi: 10.1146/annurev.py.19.090181.001421. [DOI] [Google Scholar]
- Solomon PS, Lowe RG, Tan KC, Waters OD, Oliver RP Stagonospora nodorum: cause of stagonospora nodorum blotch of wheat. Mol Plant Pathol. 2006;7:147–156. doi: 10.1111/j.1364-3703.2006.00326.x. [DOI] [PubMed] [Google Scholar]
- Solomon PS, Tan KC, Sanchez P, Cooper RM, Oliver RP The disruption of a galpha subunit sheds new light on the pathogenicity of Stagonospora nodorum on wheat. Mol Plant Microbe Interact. 2004;17:456–466. doi: 10.1094/MPMI.2004.17.5.456. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos I, Collemare J, Mehrabi R, De Wit PJ Phytotoxic secondary metabolites and peptides produced by plant pathogenic dothideomycete fungi. FEMS Microbiol Rev. 2013;37:67–93. doi: 10.1111/j.1574-6976.2012.00349.x. [DOI] [PubMed] [Google Scholar]
- Sun H, Ho CL, Ding F, Soehano I, Liu XW, Liang ZX Synthesis of (r)-mellein by a partially reducing iterative polyketide synthase. J Am Chem Soc. 2012;134:11924–11927. doi: 10.1021/ja304905e. [DOI] [PubMed] [Google Scholar]
- Syme RA, Hane JK, Friesen TL, Oliver RP Resequencing and comparative genomics of Stagonospora nodorum: sectional gene absence and effector discovery. G3 (Bethesda) 2013;3:959–969. doi: 10.1534/g3.112.004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K-C, Trengove RD, Maker GL, Oliver RP, Solomon PS Metabolite profiling identifies the mycotoxin alternariol in the pathogen Stagonospora nodorum . Metabolomics. 2009;5:330–335. doi: 10.1007/s11306-009-0158-2. [DOI] [Google Scholar]
- Tan K-C, Waters O, Rybak K, Antoni E, Furuki E, Oliver R Sensitivity to three Parastagonospora nodorum necrotrophic effectors in current Australian wheat cultivars and the presence of further fungal effectors. Crop Pasture Sci. 2013;65:150–158. http://dx.doi.org/10.1071/CP13443 [Google Scholar]
- Thomas R Studies in the biosynthesis of fungal metabolites. 2. The biosynthesis of alternariol and its relation to other fungal phenols. Biochem J. 1961;78:748–758. doi: 10.1042/bj0780748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietjen KG, Matern U Induction and suppression of phytoalexin biosynthesis in cultured cells of safflower, Carthamus tinctorius L., by metabolites of Alternaria carthami chowdhury. Arch Biochem Biophys. 1984;229:136–144. doi: 10.1016/0003-9861(84)90138-3. [DOI] [PubMed] [Google Scholar]
- Tsai HF, Fujii I, Watanabe A, Wheeler MH, Chang YC, Yasuoka Y, Ebizuka Y, Kwon-Chung KJ Pentaketide melanin biosynthesis in Aspergillus fumigatus requires chain-length shortening of a heptaketide precursor. J Biol Chem. 2001;276:29292–29298. doi: 10.1074/jbc.M101998200. [DOI] [PubMed] [Google Scholar]
- Tudzynski B, Kawaide H, Kamiya Y Gibberellin biosynthesis in Gibberella fujikuroi: cloning and characterization of the copalyl diphosphate synthase gene. Curr Genet. 1998;34:234–240. doi: 10.1007/s002940050392. [DOI] [PubMed] [Google Scholar]
- Turgeon BG, Baker SE Genetic and genomic dissection of the Cochliobolus heterostrophus tox1 locus controlling biosynthesis of the polyketide virulence factor t-toxin. Adv Genet. 2007;57:219–261. doi: 10.1016/S0065-2660(06)57006-3. [DOI] [PubMed] [Google Scholar]
- Vagstad AL, Hill EA, Labonte JW, Townsend CA Characterization of a fungal thioesterase having claisen cyclase and deacetylase activities in melanin biosynthesis. Chem Biol. 2012;19:1525–1534. doi: 10.1016/j.chembiol.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatasubbaiah P, Chilton WS Phytotoxins of Botryosphaeria obtusa . J Nat Prod. 1990;53:1628–1630. doi: 10.1021/np50072a044. [DOI] [Google Scholar]
- Wackler B, Lackner G, Chooi YH, Hoffmeister D Characterization of the Suillus grevillei quinone synthetase GreA supports a nonribosomal code for aromatic α-keto acids. Chembiochem. 2012;13:1798–1804. doi: 10.1002/cbic.201200187. [DOI] [PubMed] [Google Scholar]
- Wada M, Kato H, Malik K, Sriprasertsak P, Ichinose Y, Shiraishi T, Yamada T A supprescin from a phytopathogenic fungus deactivates transcription of a plant defense gene encoding phenylalanine ammonia-lyase. J Mol Biol. 1995;249:513–519. doi: 10.1006/jmbi.1995.0313. [DOI] [PubMed] [Google Scholar]
- Wallwey C, Li SM Ergot alkaloids: structure diversity, biosynthetic gene clusters and functional proof of biosynthetic genes. Nat Prod Rep. 2011;28:496–510. doi: 10.1039/c0np00060d. [DOI] [PubMed] [Google Scholar]
- Walton JD Hc-toxin. Phytochemistry. 2006;67:1406–1413. doi: 10.1016/j.phytochem.2006.05.033. [DOI] [PubMed] [Google Scholar]
- Wheeler MH, Abramczyk D, Puckhaber LS, Naruse M, Ebizuka Y, Fujii I, Szaniszlo PJ New biosynthetic step in the melanin pathway of Wangiella (Exophiala) dermatitidis: evidence for 2-acetyl-1,3,6,8-tetrahydroxynaphthalene as a novel precursor. Eukaryot Cell. 2008;7:1699–1711. doi: 10.1128/EC.00179-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wight WD, Kim KH, Lawrence CB, Walton JD Biosynthesis and role in virulence of the histone deacetylase inhibitor depudecin from Alternaria brassicicola . Mol Plant Microbe Interact. 2009;22:1258–1267. doi: 10.1094/MPMI-22-10-1258. [DOI] [PubMed] [Google Scholar]
- Yang XL, Awakawa T, Wakimoto T, Abe I Induced biosyntheses of a novel butyrophenone and two aromatic polyketides in the plant pathogen Stagonospora nodorum . Nat Prod Bioprosp. 2013;3:141–144. doi: 10.1007/s13659-013-0055-2. [DOI] [Google Scholar]
- Yeh HH, Chiang YM, Entwistle R, Ahuja M, Lee KH, Bruno KS, Wu TK, Oakley BR, Wang CC Molecular genetic analysis reveals that a nonribosomal peptide synthetase-like (NRPS-like) gene in Aspergillus nidulans is responsible for microperfuranone biosynthesis. Appl Microbiol Biotechnol. 2012;96:739–748. doi: 10.1007/s00253-012-4098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin WB, Baccile JA, Bok JW, Chen Y, Keller NP, Schroeder FC A nonribosomal peptide synthetase-derived iron(iii) complex from the pathogenic fungus Aspergillus fumigatus . J Am Chem Soc. 2013;135:2064–2067. doi: 10.1021/ja311145n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabala AO, Xu W, Chooi YH, Tang Y Characterization of a silent azaphilone gene cluster from Aspergillus niger ATCC 1015 reveals a hydroxylation-mediated pyran-ring formation. Chem Biol. 2012;19:1049–1059. doi: 10.1016/j.chembiol.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


