Abstract
Eukaryotic genomes contain long stretches of repetitive DNA sequences, which are the preferred sites for the assembly of heterochromatin structures. The formation of heterochromatin results in highly condensed chromosomal domains that limit the accessibility of DNA to the transcription and recombination machinery to maintain genome stability. Heterochromatin has the tendency to spread, and the formation of boundaries that block heterochromatin spreading is required to maintain stable gene expression patterns. Recent work has suggested that noncoding RNAs are involved in regulating boundary formation in addition to their well-established roles in chromatin regulation. Here, we present a review of our current understanding of the involvement of noncoding RNA at the boundaries of heterochromatin, highlighting their mechanisms of action in different settings.
Introduction
While known and predicted protein-coding genes make up a small percentage of the human genome,1 transcriptome analysis suggests that over half of the human genome is transcribed to some degree.2, 3 Similar studies conducted in model eukaryotes suggest that widespread transcription of the non-coding genome is a conserved feature.4–6 Although a large proportion of noncoding transcription may represent transcriptional noise rather than serve a specific biological function,7 a growing list of non-coding RNAs (ncRNAs) have been identified as key players in diverse cellular processes. RNA molecules, once thought to function solely as intermediates carrying the genetic information required to build a functional protein from the nucleus to the cytoplasm, are now well recognized for their structural, catalytic, and regulatory roles.
Identified ncRNAs are typically classified according to length, with those longer than 200 base pairs termed long non-coding RNAs (lncRNAs), and shorter ones classified as small noncoding RNAs. Both long and short ncRNAs play critical roles in regulating gene expression and genome function by participating in packaging the linear genome into chromatin, the differential compaction of which influences the accessibility of DNA to transcription, replication, DNA damage repair machineries crucial for genome function and maintenance.8 In this article, we will focus our discussion on the role of ncRNAs in modulating the boundaries between different chromatin domains.
The establishment and spreading of heterochromatin
In general, chromatin domains are classified according to degree of compaction and expression levels of resident genes. Euchromatin is typically gene-rich, less condensed, and is characterized by higher expression of resident genes, while heterochromatin is gene-poor, highly condensed, and exhibits lower levels of gene expression.9 Heterochromatin has the tendency to spread to surrounding regions, thus interfering with gene expression of neighboring euchromatic regions.10 Classic examples of heterochromatin spreading include position effect variegation in Drosophila and telomere position effects in budding yeast, in which cases genes inserted near heterochromatic regions are variably silenced. To maintain stable gene expression patterns, the spreading of heterochromatin needs to be precisely regulated, and many specialized DNA elements form boundaries to block the spreading of heterochromatin.11, 12
Key to defining the identity of different chromatin domains is the nucleosome, the basic unit of chromatin composed of about 147bp of DNA wrapped around a core histone octamer, which are subjected to a variety of posttranslational modifications that regulate chromatin compaction.13 Each chromatin state is associated with a specific set of histone tail modifications. For example, the histone tails of euchromatic regions are mostly hyper-acetylated and methylated at histone H3 lysine 4 (H3K4me), whereas those of heterochromatic regions are typically hypoacetylated and trimethylated at histone H3 lysine 9 (H3K9me).14–16
The formation of heterochromatin has long been considered a paradigm for the study of chromatin organization due to the coordinated recruitment of diverse histone modifying enzymes and chromatin binding proteins. This process is usually divided into the establishment stage, when histone-modifying activities are initially recruited to specific locations of the genome, and the spreading stage, when the heterochromatin-associated histone modifications spread into neighboring regions in a sequence-independent manner and, in many cases, without involvement of the initial recruitment signal.9 While the mechanisms of heterochromatin establishment have been extensively studied, the mechanisms by which heterochromatin spreads are less well-understood. A simplified model is that heterochromatin spreads by repeated cycles of chromatin proteins recruiting histone modifying enzymes, leading to the binding of more chromatin proteins, and thus the recruitment of more histone-modifying enzymes, ultimately leading to the “oozing” of histone modifications from nucleation centers to surrounding regions (Fig. 1), although other spreading mechanisms might exist in different situations.10 In some organisms, the DNA within heterochromatin regions is also heavily methylated. However, given that the role of DNA methylation in heterochromatin spreading is not well defined, we will not address the role DNA methylation in regulating heterochromatin assembly in this article.
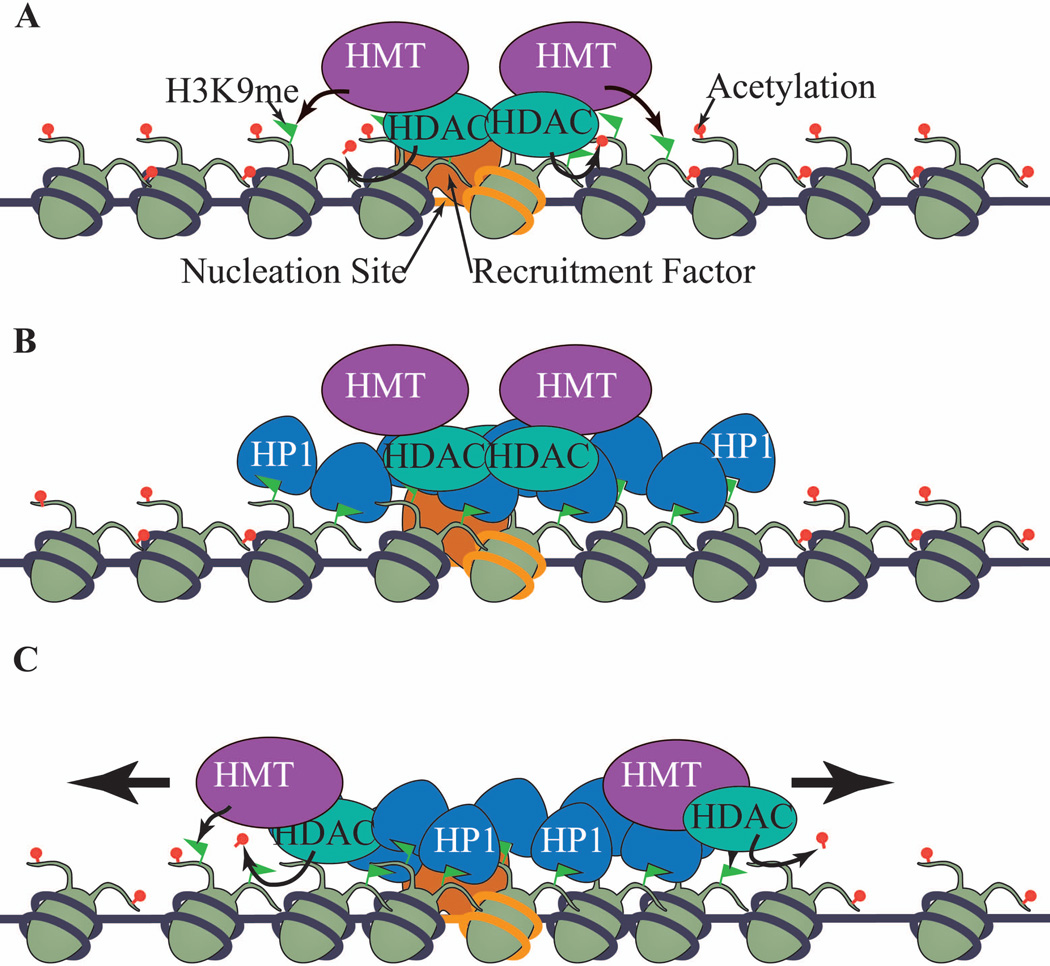
Figure 1. The establishment and spreading of heterochromatin.
(A) In fission yeast and other organisms, specific DNA sequences (in orange) recruit heterochromatin-associated histone modifying enzymes, such as histone methyltransferases (HMTs) that preferentially methylate histone H3K9, and histone deacetylases (HDACs). These enzymes act on histones at the nucleation site to establish heterochromatic modifications. (B) H3K9me is recognized by chromodomain-containing HP1 (Swi6 in fission yeast). (C) HP1/Swi6 recruits HMTs and HDACs to adjacent nucleosomes to reiterate the process of H3K9me and HP1/Swi6 binding, continuous cycles of which lead to the long-range spreading of heterochromatin.
One of the best-studied examples of heterochromatin formation is in budding yeast, Saccharomyces cerevisiae, where a relatively simple form of heterochromatin is present at telomeres and the silent mating-type region.17, 18 Heterochromatin assembly is controlled by the Silent Information Regulator (SIR) complex, consisting of Sir2, Sir3 and Sir4. Sir2 is a highly conserved NAD+-dependent histone deacetylase with preference for acetylated histone H4 lysine 16 (H4K16ac);19–21 Sir3 and Sir4 preferentially interact with histone tails and nucleosomes without H4K16ac;22–24 and Sir4 also directly links Sir2 and Sir3.25 Heterochromatin establishment starts when the Sir2-Sir4 complex is recruited by sequence-specific DNA binding proteins to the telomere repeats or silencers at the mating-type region and deacetylates histone H4K16. These deacetylated histones have higher affinity for Sir3 and Sir2-Sir4, and thus enable deacetylation of adjacent nucleosomes. The reiteration of this deacetylation and the Sir complex binding cycle results in the spreading of heterochromatin across large chromosomal domains.17, 18, 26
Heterochromatin regions in fission yeast, Schizosaccharomyces pombe, and higher eukaryotes are more complex.9, 27 They are located near structurally important chromosomal sites such as the centromeres, which often contain long stretches of DNA repeats. Histones within these regions are generally hypoacetylated and methylated at H3 lysine 9 (H3K9me), a modification that recruits chromodomain proteins of the HP1 family.28–30 In fission yeast, in which the step-wise heterochromatin assembly mechanism is best studied, the establishment of heterochromatin starts with targeting of the H3K9 methyltransferase Clr4 to DNA repeats through the RNA interference (RNAi) pathway or through sequence-specific DNA binding proteins to initiate H3K9me, which leads to recruitment of the HP1 family protein Swi6.31–36 Several histone deacetylases, such as Clr3, Clr6, and Sir2, also function in concert with Clr4 to establish heterochromatin.30, 37–42 Because Swi6 is required for the spreading of H3K9me across large chromosome domains, including regions that are unable to recruit Clr4 by themselves, it is proposed that Swi6 recruits Clr4, leading to the methylation of adjacent nucleosomes and the spread of heterochromatin.31, 36 In addition, the chromodomain of Clr4 interacts with H3K9me3 to stimulate Clr4 enzymatic activity, which also contributes to the step-wise heterochromatin spreading.43, 44 In addition to histone methyltransferases and HP1 proteins, histone deacetylases Clr3 and Sir2 are also required for heterochromatin spreading.37, 39, 41, 45 Given that the chromodomain is a conserved feature of the SUV39 family of H3K9 methyltransferases and that HP1 associates with the H3K9 methyltransferases in higher eukaryotes,46–48 it is likely that such step-wise spreading is conserved in higher eukaryotes.
In metazoans, the silencing of developmentally regulated genes, often in a cell-type and developmental stage-specific manner, requires the formation of large chromosome domains containing nucleosomes trimethylated at H3 lysine 27 (H3K27me3).49 The establishment of these facultative heterochromatin domains begins with the binding of transcription factors to specific DNA sequences, such as Polycomb Response Elements (PREs), and the recruitment of the Polycomb Repressive Complex 2 (PRC2), which harbors the Enhancer of Zeste family of histone methyltransferases (E(Z) in flies and EZH2 in mammals).49 PRC2 trimethylates H3K27,50–53 which recruits the Polycomb Repressive Complex 1 (PRC1) through the interaction between the chromodomain of polycomb (PC) and H3K27me3.54, 55 It was generally believed that H3K27me3 spreads in a manner similar to that of H3K9me, with repeated cycles of H3K27 methylation coupled with binding of PRC2, given that PRC2 subunit EED directly interacts with H3K27me3.56, 57 However, while H3K27me3 is present in large chromosome domains,58, 59 PRC2 occupancy is limited to the sites of recruitment, suggestive of a more complicated spreading mechanism than a step-wise model.58 It is proposed that H3K27me3 spreads when the PRC2 complex bound at PREs diffuses from these nucleation sites or by the formation of chromosome loops.10, 60
Heterochromatin boundaries are typically defined as DNA sequences or regions that act to constrain heterochromatin spreading to the appropriate domain and prevent it from encroaching into euchromatic domains (Fig. 2A). In certain cases, insulators, which block the communication between distant chromosomal regions, can also function as boundary elements.12, 61 While boundaries and insulators show considerable diversity in DNA sequence, they appear to share similar mechanisms of function. First, boundary elements can recruit histone-modifying factors that promote the opening of chromatin and counteract the spread of silencing-associated histone modifications and chromatin compaction (Fig. 2B).45, 62–65 Additionally, boundaries associate with nuclear structures, such as the nuclear envelope, and cluster together to loop out the intervening chromatin domains and control the three-dimensional arrangement of the genome (Fig. 2C).66, 67 These mechanisms are probably not mutually exclusive, and might even cooperate to establish a functional boundary. The transcription of ncRNAs observed at heterochromatin boundaries appears to either enforce or disrupt the mechanisms described above, or function via novel mechanisms involving the ability of transcriptional interference and ncRNA-protein interactions to inhibit the spread of heterochromatin.
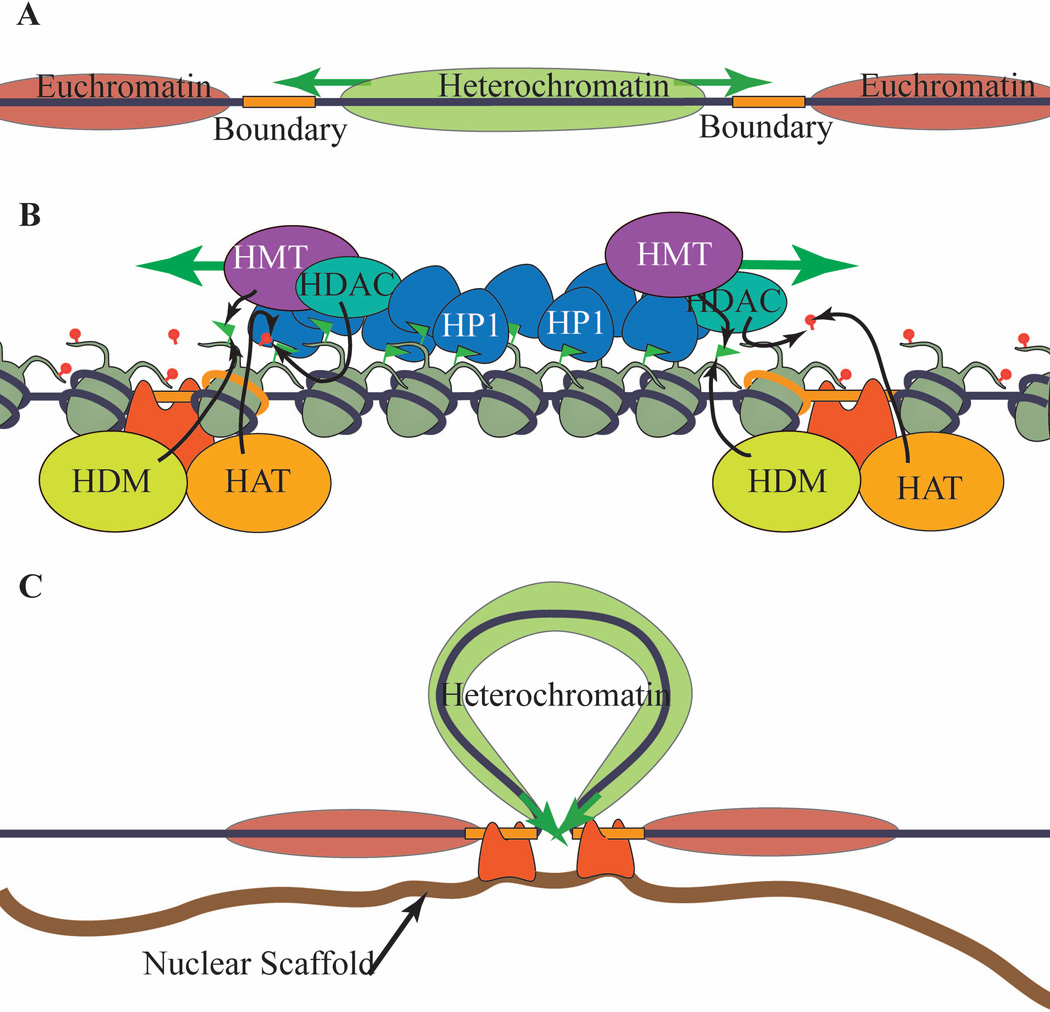
Figure 2. Heterochromatin boundaries.
(A) The transitions between heterochromatin and euchromatin are marked by DNA sequences termed boundary elements (orange) that prevent heterochromatin (green) from spreading into neighboring euchromatin regions (red). (B) One mode of boundary function is the recruitment of euchromatin-associated histone modifying enzymes, such as histone acetyltransferases (HATs) and histone demethylases (HDM) to the boundary region to antagonize the addition of heterochromatin-associated histone modifications to block heterochromatin spreading. (C) Another mechanism by which boundaries function is to spatially separate heterochromatin and euchromatin domains through specific protein complexes that interact with nuclear structures.
Non-coding RNAs and heterochromatin assembly
Short ncRNAs, mostly products of the RNA interference pathway, are involved in heterochromatin assembly at repetitive DNA elements in diverse organisms by targeting histone modifying activities to DNA repeats through effector complexes.68–70 The mechanism of small RNA-induced heterochromatin assembly is best studied in fission yeast (Fig. 3A). In short, ncRNAs transcribed from repeat sequences, such as those surrounding the centromeres, are converted into double-stranded RNAs and processed by the RNAi pathway into small interfering RNAs (siRNAs).32, 71 These siRNAs are loaded into the RITS (RNA-induced Initiation of Transcriptional gene Silencing) complex and direct RITS to nascent transcripts through base pairing between the nascent transcripts and siRNAs.35, 72, 73 RITS associates with the H3K9 methyltransferase Clr4 to initiate H3K9 methylation and the recruitment of HP1 proteins at repeats.74 Other small RNA-induced chromatin modifications, such as RNA-directed DNA methylation in plants and piRNA-mediated silencing of transposons in Drosophila and mammals are also believed to use small RNAs as guides to target chromatin-modifying activities to specific genomic locations.68–70
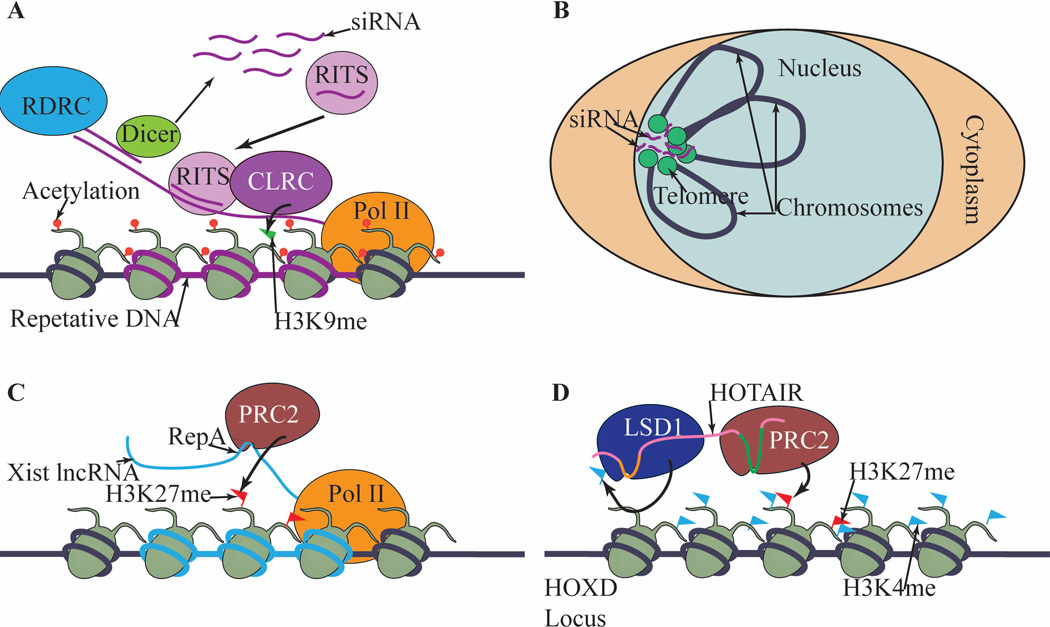
Figure 3. Noncoding RNAs and heterochromatin chromatin assembly.
(A) RNAi-mediated heterochromatin assembly in fission yeast. Repetitive DNA elements are transcribed and processed by the RNAi pathway into siRNAs, which are used by effector protein complex RITS to target histone methyltransferase CLRC to nascent transcripts. (B) siRNAs might mediate the clustering of distant loci, such as telomeres, at the nuclear envelope. (C) Xist interacts directly with PRC2 through a repeat (RepA) and recruits PRC2 to initiate inactivation of the X chromosome. (D) HOTAIR binds both LSD1 and PRC2 via different sequences on the RNA, acting as a scaffold for histone modifying enzymes to coordinate their activities.
Small RNAs have also been implicated in the three-dimensional organization of chromosomal loci (Fig. 3B). For example, the RNAi machinery is required for the clustering of telomeres in fission yeast and the clustering of insulator and Polycomb response elements in Drosophila.75–77 Although these data suggest a possible role for small RNAs in orchestrating long-range chromatin interactions, possibly by serving as a scaffold, the direct involvement of small RNAs in the clustering of these loci has not been clearly demonstrated.
The mechanisms by which lncRNAs regulate chromatin modifications are less well understood. Much of our understanding of lncRNAs function in gene silencing has come through their interaction with PRC2. Similar to small RNAs, lncRNAs are required for the targeting of PRC2 to chromatin. For example, Xist, a 17 kb lncRNA that is transcribed from the X inactivation center of one of the female X chromosomes and coats the chromosome in cis, initiates the inactivation of the entire chromosome to maintain the equivalent dosage of X chromosome expression as in male cells.78 The repeat A regions of Xist directly interact with PRC2 components in vitro,79–82 suggesting that Xist directly recruits PRC2 to initiate H3K27me3 at the inactive X chromosome (Fig. 3C). However, the contribution of such interactions to PRC2 recruitment in vivo is still not fully resolved.83 Other lncRNAs, such as COLDAIR in plants and ANRIL in humans also mediate the recruitment of Polycomb proteins in cis.84–86 Interestingly, the HOTAIR lncRNA operates in trans to aid the recruitment of PRC2 to diverse locations, suggesting a mechanism different from targeting.87 Recent studies demonstrate that HOTAIR directly interacts with both PRC2 and H3K4 demethylase LSD1 through different regions of the RNA and thus facilitates the interaction between these two enzymes, indicating a structural role for this lncRNA88 (Fig. 3D). Genome wide studies also identified large number of ncRNAs associate with PRC2 and mediates its recruitment to target loci,81, 89, 90 suggesting that ncRNA-mediated recruitment of PRC2 is a prevalent mechanism.
Non-coding RNAs and boundary function
Non-coding RNAs regulate boundary function in several ways, some of which are fundamentally similar to their involvement in heterochromatin assembly, such as controlling the targeting of protein complexes and serving structural roles. Recent findings have also expanded the role of ncRNAs to include regulating transcription at boundaries as well as directly counteracting heterochromatin spreading (Fig. 4).
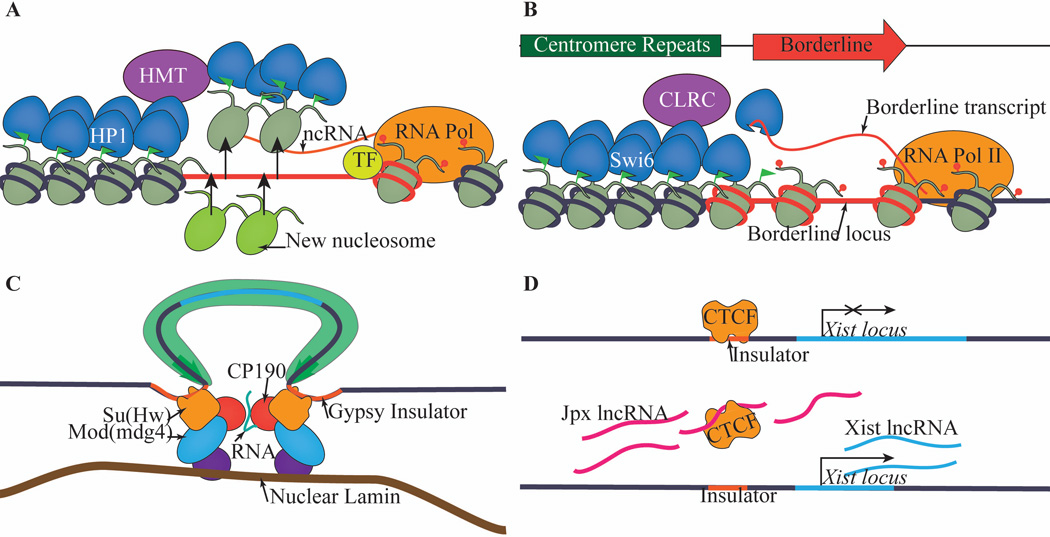
Figure 4. Noncoding RNAs and heterochromatin boundaries.
(A) Many known or predicted heterochromatin boundary elements, like the SINE and tRNA genes, are sites of noncoding RNA transcription. Transcription-mediated histone modifications and histone turn over might contribute to heterochromatin boundary formation at these elements. (B) In fission yeast, BORDERLINE evicts Swi6 in cis from chromatin by directly binding to Swi6 and interfering with its ability to interact with H3K9me, thus halting Swi6-dependent spreading. (C) In Drosophila, ncRNAs serve a scaffolding function to mediate the clustering of insulator bodies. (D) In mammals, ncRNAs directly regulate the binding of CTCF to insulator DNA sequences.
tRNA genes as boundaries
It is well established that genes for noncoding transfer RNAs (tRNAs) function as boundary elements in organisms ranging from yeast to mammals.91–94 Transcription of tRNA genes is mediated by A and B box sequences that recruit the transcription factors TFIIIC, TFIIIB and RNA polymerase (Pol) III.95 In fission yeast, the boundary function of the silent mating-type regions requires B-box sequences present in the boundary element, which recruit TFIIIC, but not Pol III.67 Therefore, the mechanism by which tRNAs function as boundary elements, at least at the fission yeast mating-type region, can be independent of tRNA transcription. TFIIIC helps cluster DNA sequences at the nuclear periphery, indicating that the 3-D organization of chromatin enforced by TFIIIC might be responsible for tRNA gene boundary function.67 In mammals, TFIIIC mediates the relocation of neuronal genes to transcription factories, suggesting that the role of TFIIIC in genome organization might be conserved.96
Short interspersed nuclear elements (SINE) in mammals
Investigation of the chromatin structure of the mouse Growth Hormone (GH) locus during pituitary development revealed that a Short Interspersed Nuclear Element (SINE) B2 retrotransposon is located at the transition zone between an H3K9me3 domain and a euchromatic domain. This repeat functions as a boundary to block the spreading of adjacent heterochromatin, and is consequently required for developmentally regulated GH activation.97 This element is derived from a tRNA gene and is bi-directionally transcribed by both the RNA Pol II and III machinery.98 Further analysis revealed that ongoing transcription from both the RNA Pol II and Pol III promoters is required for boundary function.97 The precise mechanism by which transcription of this SINE B2 element leads to barrier function remains unknown. Although it is possible that the ncRNAs are directly involved in boundary function, it is more likely that the transcription process changes the histone modifications or nuclear positioning in a way that efficiently blocks the spreading of heterochromatin (Fig. 4A).
Further supporting the concept that ncRNA transcription is involved in boundary formation, computational analysis of the genome-wide distribution of active and repressive chromatin modifications in human CD4+ cells led to the observation that a subset of predicted boundary elements coincided with regions of high expression of ncRNA genes, transcription factor binding, RNA Pol III localization, and open chromatin.99 While the abundance of ncRNA transcription may merely correlate with the opening of chromatin or the binding of an insulating complex and not serve a specific function at boundaries, the above studies indicate that transcriptional activity may play an active role in maintaining the borders between chromatin domains.
BORDERLINE RNA in fission yeast
Recently, it was shown that ncRNAs in fission yeast directly mediate boundary function via a novel mechanism by which the RNA molecules bind heterochromatin factors to directly block the spreading of heterochromatin100 (Fig. 4B). The fission yeast pericentric heterochromatin is surrounded by tRNA genes and, in some cases, inverted repeats termed IRCs101. Several lncRNAs, termed BORDERLINE RNAs, are transcribed from the IRC boundary and are then processed by the RNAi pathway into small RNAs.100 Interestingly, replacing the IRC transcribed sequence with a ura4+ reporter gene also resulted in the formation of a boundary in a transcription-dependent manner. Swi6 binds RNA in a sequence independent manner, and RNA interferes with the binding of chromo domain to H3K9me.102 Thus, a plausible hypothesis is that binding of BORDERLINE RNAs to Swi6 results in the release of Swi6 from chromatin and effectively blocks heterochromatin spreading. The fact that mutations in the Swi6 RNA binding region result in heterochromatin spreading further supports such a hypothesis.100 However, given that RNA binding to HP1 in mammals seems to promote rather than disrupt its association with chromatin,103, 104 it is unclear whether the mechanism of HP1 eviction by ncRNA in boundary formation would apply in mammalian systems.
It should be noted that transcription of BORDERLINE RNAs is not the only mechanism by which IRCs function as boundaries. IRCs also recruit a complex composed of a JmjC domain protein Epe1 and a double bromodomain-containing factor Bdf2, which binds acetylated histone tails and directly antagonizes Sir2-mediated deacetylation of H4K16 to block heterochromatin spreading.45, 105–107 Interestingly, loss of Bdf2 or Epe1 results in decreased IRC transcript levels,45, 105 raising the possibility that the Epe1-Bdf2 complex promotes the transcription of IRC to establish heterochromatin boundaries. However, the fact that artificial tethering of Bdf2 to chromatin is sufficient to establish a heterochromatin boundary suggests that Bdf2 has transcription-independent contributions to boundary function.45 Thus transcription-dependent and independent mechanisms might function redundantly to establish heterochromatin boundaries at IRCs.
Noncoding RNAs regulate the action of the gypsy insulator complex in Drosophila
While transcription might be one mechanism by which DNA sequences form boundaries, ncRNAs can also interact with insulating proteins to regulate their functions. The gypsy insulator is a well-characterized retrotransposon-derived sequence in Drosophila that can buffer transgenes from position effects and block long-range enhancer-promoter interactions.108 This insulator is bound by a protein complex composed of suppressor of Hairy-wing [su(Hw)], Modifier of mdg4 [Mod(mdg4)], and Centrosomal protein 190 (CP190) (Fig. 4C). These proteins cluster together and form a relatively small number of nuclear foci anchored at the nuclear periphery, suggesting that their insulator function might be tied to their ability to mediate chromatin domain looping.109, 110 High throughput sequencing of RNAs that immunoprecipitated with su(Hw) and CP190 revealed significant enrichment of su(Hw) and CP190 mRNAs.111 Interestingly, T7 polymerase-driven expression of the su(Hw) and CP190 mRNAs, which are not processed or translated, enhances boundary function and the formation of insulator bodies in vivo, suggesting that these mRNAs contribute to the insulator function of gypsy sequences in a manner independent of their coding function111. It is likely that these RNAs act as scaffolds that stabilize the interactions of insulator components to regulate genome organization.
Noncoding RNA and CTCF-mediated chromatin insulation
CTCF (CCCTC binding Factor) is an important and highly conserved transcription factor that binds to insulators and blocks the communication between enhancers and promoters to regulate gene expression.112, 113 Genome-wide analyses of CTCF and chromatin modifications demonstrate that a large portion of CTCF binding sites also flank repressive H3K27me3-containing chromosome domains, often in a cell-type specific manner, indicating that CTCF binding sites might also function as boundaries to regulate the spreading of facultative heterochromatin domains.59, 114 CTCF seems to execute its insulating function by regulating the 3D organization of the genome to control interactions between distant loci.115–122 Several recent studies have also implicated noncoding RNA in the regulation of CTCF function.
Some ncRNAs enhance the ability of CTCF to regulate insulator function by acting as scaffolds that stabilize the interaction between CTCF and other factors that mediate looping. For example, CTCF associates with P68, a DEAD box RNA binding protein that associates with a ncRNA, Seroid Receptor RNA Activator (SRA).123 Knockdown of SRA resulted in decreased binding of P68 to CTCF, reduced enhancer-blocking function, and also disrupted long-range interaction of CTCF binding sites as determined by chromatin conformation capture (3C) analysis.124 Thus SRA might play a scaffolding role to mediate interaction between insulator components, similar to the requirement of RNAs for stabilizing gypsy insulator interactions in Drosophila (Fig. 4C).
Conversely, other ncRNAs interact with CTCF to interfere with the binding of CTCF to DNA in order to regulate gene expression. For example, Lipopolysaccharide (LPS)-induced expression of the lysozyme gene during the innate immune response requires the eviction of CTCF from an insulator site to allow the C/EBPβ transcription factor to bind its enhancer and activate lysozyme expression.125 An antisense ncRNA, LINoCR, is transcribed from a promoter element 1.9 kb upstream of the lysozyme promoter, whose expression correlated with nucleosome remodeling and lysozyme expression. Transcription of this ncRNA leads to the eviction of CTCF and chromatin remodeling at nearby enhancers, leading to upregulated lysozyme expression.125 Because LINoCR functions in cis, it is unclear whether this ncRNA directly binds CTCF to evict it or merely activating transcription through this locus is sufficient to abolish insulator function.
In contrast, the function of Jpx ncRNA in binding to CTCF in trans demonstrates a direct effect of this ncRNA on CTCF function126 (Fig. 4D). In mammals, transcription of Xist ncRNA, which is required for X chromosome inactivation, is repressed by CTCF before XCI occurs.78 At the onset of XCI, the Jpx ncRNA is upregulated, binds CTCF directly, and subsequently evicts it from Xist promoter,126 which correlates with changes in the nuclear compartmentalization of the X chromosomes, suggesting a direct role for Jpx RNA in regulating CTCF localization.
Thus ncRNA regulates the binding of CTCF to DNA through either directly binding to CTCF or transcription interference. Understanding the details of the CTCF-RNA interaction is critical for dissecting the mechanism by which these ncRNAs regulate the targeting of CTCF to insulators. Although only the insulation and transcription functions of CTCF have been examined in these cases, it is possible that such regulation also functions at CTCF-associated heterochromatin boundaries.
Conclusions
The borders of heterochromatin in many cases are marked by boundary elements, or sometimes insulators, which constrain heterochromatin spreading to maintain stable gene expression patterns. Given the widespread transcription of eukaryotic genomes and the well-studied involvement of ncRNAs in chromatin regulation, it is not surprising that recent work has established additional roles for ncRNA in regulating boundary functions in diverse organisms. There are four possible mechanisms by which ncRNAs function at boundaries (Fig. 4). First, as shown at the developmentally regulated SINE B2 repeat,97 transcription of ncRNAs enforces the formation of a heterochromatin boundary through mechanisms possibly involves transcription factor binding and recruitment of RNA polymerases, which eventually lead to changes in histone modifications that antagonize heterochromatin spreading. Second, as shown at the IRC boundaries flanking the fission yeast pericentric heterochromatin,100 ncRNAs directly interact with heterochromatin proteins to evict them from boundary regions to stop heterochromatin spreading. Third, as shown in Drosophila and mammalian CTCF insulators, ncRNAs act as scaffolds to modulate protein-protein interactions and affect the association of boundaries with the nuclear matrix.111, 124 Fourth, as shown in many different systems, ncRNA can directly or indirectly interact with proteins required for boundary function to regulate their localization.126
While the mechanism of ncRNAs in heterochromatin spreading and boundary function is best studied in single-celled organisms such as fission yeast, which allows precise genetic manipulations, the role of ncRNAs in boundary functions in higher eukaryotes is less clearly defined. It is especially challenging to determine the mechanisms by which ncRNAs function in cis, as it is difficult to distinguish the effect of transcription from the direct involvement of the ncRNAs. While many of these ncRNAs are transiently expressed and modulating their sequence and length at the endogenous loci is difficult in vertebrate models, the advent of genome editing technologies127, 128 may make it easier to dissect some of the characteristics of these boundary ncRNAs in order to separate possible influences of the transcriptional machinery and transcriptional interference from direct roles played by the ncRNA molecules or other effector proteins. Additionally, more comprehensive analysis of RNA-protein and RNA-chromatin interactions will shed new light on the biochemical mechanisms by which these ncRNAs can exert such diverse influences on chromatin boundaries.
Acknowledgements
We thank Terese Lawry, Anudari Letian, and other members of the Jia lab for comments on the manuscript. Work in the Jia lab is supported by NIH grant R01-GM085145.
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 3.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 6.Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 8.Sabin LR, Delas MJ, Hannon GJ. Dogma derailed: the many influences of RNA on the genome. Molecular cell. 2013;49:783–794. doi: 10.1016/j.molcel.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grewal SIS, Jia ST. Heterochromatin revisited. Nature Reviews Genetics. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 10.Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nature reviews. Genetics. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- 11.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 12.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nature Reviews Genetics. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 13.Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol. 2013;20:259–266. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- 14.Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- 15.Noma K, Allis CD, Grewal SIS. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 16.Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Molecular cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- 17.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annual review of biochemistry. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 18.Kueng S, Oppikofer M, Gasser SM. SIR proteins and the assembly of silent chromatin in budding yeast. Annu Rev Genet. 2013;47:275–306. doi: 10.1146/annurev-genet-021313-173730. [DOI] [PubMed] [Google Scholar]
- 19.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 20.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanny JC, Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:415–420. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 23.Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Molecular cell. 2007;28:1015–1028. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 25.Moazed D, Kistler A, Axelrod A, Rine J, Johnson AD. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:2186–2191. doi: 10.1073/pnas.94.6.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moazed D. Mechanisms for the inheritance of chromatin states. Cell. 2011;146:510–518. doi: 10.1016/j.cell.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Almouzni G, Probst AV. Heterochromatin maintenance and establishment: lessons from the mouse pericentromere. Nucleus. 2011;2:332–338. doi: 10.4161/nucl.2.5.17707. [DOI] [PubMed] [Google Scholar]
- 28.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 29.Lachner M, O’Carroll N, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 30.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SIS. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 31.Hall IM, Shankaranarayana GD, Noma KI, Ayoub N, Cohen A, Grewal SIS. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 32.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 33.Jia ST, Noma K, Grewal SIS. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304:1971–1976. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- 34.Kim HS, Choi ES, Shin JA, Jang YK, Park SD. Regulation of Swi6/HP1-dependent heterochromatin assembly by cooperation of components of the mitogen-activated protein kinase pathway and a histone deacetylase Clr6. The Journal of biological chemistry. 2004;279:42850–42859. doi: 10.1074/jbc.M407259200. [DOI] [PubMed] [Google Scholar]
- 35.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanoh J, Sadaie M, Urano T, Ishikawa F. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Current biology : CB. 2005;15:1808–1819. doi: 10.1016/j.cub.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 37.Shankaranarayana GD, Motamedi MR, Moazed D, Grewal SI. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Current biology : CB. 2003;13:1240–1246. doi: 10.1016/s0960-9822(03)00489-5. [DOI] [PubMed] [Google Scholar]
- 38.Freeman-Cook LL, Gomez EB, Spedale EJ, Marlett J, Forsburg SL, Pillus L, Laurenson P. Conserved locus-specific silencing functions of Schizosaccharomyces pombe sir2+ Genetics. 2005;169:1243–1260. doi: 10.1534/genetics.104.032714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SI. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Molecular cell. 2005;20:173–185. doi: 10.1016/j.molcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Alper BJ, Job G, Yadav RK, Shanker S, Lowe BR, Partridge JF. Sir2 is required for Clr4 to initiate centromeric heterochromatin assembly in fission yeast. The EMBO journal. 2013 doi: 10.1038/emboj.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buscaino A, Lejeune E, Audergon P, Hamilton G, Pidoux A, Allshire RC. Distinct roles for Sir2 and RNAi in centromeric heterochromatin nucleation, spreading and maintenance. Embo Journal. 2013;32:1250–1264. doi: 10.1038/emboj.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marina DB, Shankar S, Natarajan P, Finn KJ, Madhani HD. A conserved ncRNA-binding protein recruits silencing factors to heterochromatin through an RNAi-independent mechanism. Genes & development. 2013;27:1851–1856. doi: 10.1101/gad.226019.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 44.Al-Sady B, Madhani HD, Narlikar GJ. Division of labor between the chromodomains of HP1 and Suv39 methylase enables coordination of heterochromatin spread. Molecular cell. 2013;51:80–91. doi: 10.1016/j.molcel.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang JY, Tadeo X, Hou HT, Tu PG, Thompson J, Yates JR, Jia ST. Epe1 recruits BET family bromodomain protein Bdf2 to establish heterochromatin boundaries. Genes & Development. 2013;27:1886–1902. doi: 10.1101/gad.221010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3–9 encode centromere-associated proteins which complex with the heterochromatin component M31. The EMBO journal. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. Central role of Drosophila SU(VAR)3–9 in histone H3-K9 methylation and heterochromatic gene silencing. The EMBO journal. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart MD, Li J, Wong J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Molecular and cellular biology. 2005;25:2525–2538. doi: 10.1128/MCB.25.7.2525-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns unknowns. Nature reviews. Molecular cell biology. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 50.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 51.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 52.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes & development. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 54.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes & development. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes & development. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nature cell biology. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 57.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nature genetics. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 59.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 60.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nature reviews. Genetics. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 61.Yang JP, Corces VG. Insulators, long-range interaction, and genome function. Current Opinion in Genetics & Development. 2012;22:86–92. doi: 10.1016/j.gde.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oki M, Valenzuela L, Chiba T, Ito T, Kamakaka RT. Barrier proteins remodel and modify chromatin to restrict silenced domains. Molecular and cellular biology. 2004;24:1956–1967. doi: 10.1128/MCB.24.5.1956-1967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.West AG, Huang S, Gaszner M, Litt MD, Felsenfeld G. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Molecular cell. 2004;16:453–463. doi: 10.1016/j.molcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 64.Huang S, Li X, Yusufzai TM, Qiu Y, Felsenfeld G. USF1 recruits histone modification complexes and is critical for maintenance of a chromatin barrier. Mol Cell Biol. 2007;27:7991–8002. doi: 10.1128/MCB.01326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lan F, Zaratiegui M, Villen J, Vaughn MW, Verdel A, Huarte M, Shi Y, Gygi SP, Moazed D, Martienssen RAS. pombe LSD1 homologs regulate heterochromatin propagation and euchromatic gene transcription. Molecular cell. 2007;26:89–101. doi: 10.1016/j.molcel.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 66.Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109:551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- 67.Noma KI, Cam HP, Maraia RJ, Grewal SIS. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 68.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lejeune E, Allshire RC. Common ground: small RNA programming and chromatin modifications. Curr Opin Cell Biol. 2011;23:258–265. doi: 10.1016/j.ceb.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 70.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription , epigenetics and beyond. Nature reviews Genetics. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 72.Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 73.Irvine DV, Zaratiegui M, Tolia NH, Goto DB, Chitwood DH, Vaughn MW, Joshua-Tor L, Martienssen RA. Argonaute slicing is required for heterochromatic silencing and spreading. Science. 2006;313:1134–1137. doi: 10.1126/science.1128813. [DOI] [PubMed] [Google Scholar]
- 74.Bayne EH, White SA, Kagansky A, Bijos DA, Sanchez-Pulido L, Hoe KL, Kim DU, Park HO, Ponting CP, Rappsilber J, et al. Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell. 2010;140:666–677. doi: 10.1016/j.cell.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hall IM, Noma K, Grewal SI. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci U S A. 2003;100:193–198. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. RNAi components are required for nuclear clustering of polycomb group response elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 77.Lei EP, Corces VG. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet. 2006;38:936–941. doi: 10.1038/ng1850. [DOI] [PubMed] [Google Scholar]
- 78.Lee JT. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA, chromatin control. Nature reviews Molecular cell biology. 2011;12:815–826. doi: 10.1038/nrm3231. [DOI] [PubMed] [Google Scholar]
- 79.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes & development. 2010;24:2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. Short RNAs Are Transcribed from Repressed Polycomb Target Genes and Interact with Polycomb Repressive Complex-2. Molecular Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, Sanglier-Cianferani S, Van Dorsselaer A, Clerc P, Avner P, et al. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS biology. 2010;8:e1000276. doi: 10.1371/journal.pbio.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cerase A, Smeets D, Tang YA, Gdula M, Kraus F, Spivakov M, Moindrot B, Leleu M, Tattermusch A, Demmerle J, et al. Spatial separation of Xist RNA and polycomb proteins revealed by superresolution microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2235–2240. doi: 10.1073/pnas.1312951111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 85.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Molecular cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Molecular cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Donze D, Kamakaka RT. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. Embo j. 2001;20:520–531. doi: 10.1093/emboj/20.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Scott KC, Merrett SL, Willard HF. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Current Biology. 2006;16:119–129. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- 93.Ebersole T, Kim JH, Samoshkin A, Kouprina N, Pavlicek A, White RJ, Larionov V. tRNA genes protect a reporter gene from epigenetic silencing in mouse cells. Cell Cycle. 2011;10:2779–2791. doi: 10.4161/cc.10.16.17092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Raab JR, Chiu J, Zhu JC, Katzman S, Kurukuti S, Wade PA, Haussler D, Kamakaka RT. Human tRNA genes function as chromatin insulators. Embo Journal. 2012;31:330–350. doi: 10.1038/emboj.2011.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Orioli A, Pascali C, Pagano A, Teichmann M, Dieci G. RNA polymerase III transcription control elements: themes and variations. Gene. 2012;493:185–194. doi: 10.1016/j.gene.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 96.Crepaldi L, Policarpi C, Coatti A, Sherlock WT, Jongbloets BC, Down TA, Riccio A. Binding of TFIIIC to sine elements controls the relocation of activity-dependent neuronal genes to transcription factories. PLoS Genet. 2013;9:e1003699. doi: 10.1371/journal.pgen.1003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lunyak VV, Prefontaine GG, Nunez E, Cramer T, Ju BG, Ohgi KA, Hutt K, Roy R, Garcia-Diaz A, Zhu XY, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- 98.Ferrigno O, Virolle T, Djabari Z, Ortonne JP, White RJ, Aberdam D. Transposable B2 SINE elements can provide mobile RNA polymerase II promoters. Nat Genet. 2001;28:77–81. doi: 10.1038/ng0501-77. [DOI] [PubMed] [Google Scholar]
- 99.Wang JR, Lunyak VV, Jordan IK. Genome-wide prediction and analysis of human chromatin boundary elements. Nucleic Acids Research. 2012;40:511–529. doi: 10.1093/nar/gkr750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Keller C, Kulasegaran-Shylini R, Shimada Y, Hotz HR, Buhler M. Noncoding RNAs prevent spreading of a repressive histone mark. Nature Structural & Molecular Biology. 2013;20:994. doi: 10.1038/nsmb.2619. −+ [DOI] [PubMed] [Google Scholar]
- 101.Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SIS. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nature Genetics. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 102.Keller C, Adaixo R, Stunnenberg R, Woolcock KJ, Hiller S, Buhler M. HP1(Swi6) mediates the recognition and destruction of heterochromatic RNA transcripts. Mol Cell. 2012;47:215–227. doi: 10.1016/j.molcel.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 103.Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nature genetics. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 104.Muchardt C, Guilleme M, Seeler JS, Trouche D, Dejean A, Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 2002;3:975–981. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zofall M, Grewal SIS. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Molecular Cell. 2006;22:681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 106.Trewick SC, Minc E, Antonelli R, Urano T, Allshire RC. The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. The EMBO journal. 2007;26:4670–4682. doi: 10.1038/sj.emboj.7601892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Braun S, Garcia JF, Rowley M, Rougemaille M, Shankar S, Madhani HD. The Cul4-Ddb1(Cdt)(2) ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell. 2011;144:41–54. doi: 10.1016/j.cell.2010.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gdula DA, Gerasimova TI, Corces VG. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc Natl Acad Sci U S A. 1996;93:9378–9383. doi: 10.1073/pnas.93.18.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Molecular Cell. 2000;6:1025–1035. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- 110.Byrd K, Corces VG. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J Cell Biol. 2003;162:565–574. doi: 10.1083/jcb.200305013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Matzat LH, Dale RK, Lei EP. Messenger RNA is a functional component of a chromatin insulator complex. EMBO Rep. 2013;14:916–922. doi: 10.1038/embor.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 113.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui KR, Zhao KJ. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Research. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Molecular cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 116.Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes & development. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hou C, Zhao H, Tanimoto K, Dean A. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20398–20403. doi: 10.1073/pnas.0808506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Majumder P, Gomez JA, Chadwick BP, Boss JM. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. The Journal of experimental medicine. 2008;205:785–798. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nature genetics. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 121.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 122.Xu N, Donohoe ME, Silva SS, Lee JT. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nature genetics. 2007;39:1390–1396. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- 123.Watanabe M, Yanagisawa J, Kitagawa H, Takeyama K, Ogawa S, Arao Y, Suzawa M, Kobayashi Y, Yano T, Yoshikawa H, et al. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor alpha coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. Embo j. 2001;20:1341–1352. doi: 10.1093/emboj/20.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 124.Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes & Development. 2010;24:2543–2555. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lefevre P, Witham J, Lacroix CE, Cockerill PN, Bonifer C. The LPS-Induced Transcriptional Upregulation of the Chicken Lysozyme Locus Involves CTCF Eviction and Noncoding RNA Transcription. Molecular Cell. 2008;32:129–139. doi: 10.1016/j.molcel.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. Jpx RNA activates Xist by evicting CTCF. Cell. 2013;153:1537–1551. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in biotechnology. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pennisi E. The CRISPR craze. Science. 2013;341:833–836. doi: 10.1126/science.341.6148.833. [DOI] [PubMed] [Google Scholar]