Abstract
NADPH oxidases (Noxes), transmembrane proteins found in most eukaryotic species, generate reactive oxygen species and are thereby involved in essential biological processes. However, the fact that genes encoding ferric reductases and ferric-chelate reductases share high sequence similarities and domains with Nox genes represents a challenge for bioinformatic approaches used to identify Nox-encoding genes. Further, most studies on fungal Nox genes have focused mainly on functionality, rather than sequence properties, and consequently clear differentiation among the various Nox isoforms has not been achieved. We conducted an extensive sequence analysis to identify putative Nox genes among 34 eukaryotes, including 28 fungal genomes and one Oomycota genome. Analyses were performed with respect to phylogeny, transmembrane helices, di-histidine distance and glycosylation. Our analyses indicate that the sequence properties of fungal Nox genes are different from those of human and plant Nox genes, thus providing novel insight that will enable more accurate identification and characterization of fungal Nox genes.
Keywords: Fungal genome, NADPH oxidase, Nox, Phylogenetics, ROS
NAPDH oxidases (Noxes) are a class of transmembrane (TM) proteins that catalyze the generation of reactive oxygen species by transporting electrons from NADPH to oxygen (O2) [1, 2]. Noxes can be found in most eukaryotic species and are involved in a magnitude of different biological processes. One Nox often mediates more than one reaction. In humans, Noxes have been shown to be involved in several physiological processes, such as inflammation, Ca2+ regulation, growth regulation, and inactivation of microbial virulence factors [1]. Respiratory burst oxidase homologues (Rbohs) are plant Nox homologues associated with signaling functions involved in hypersensitive responses against pathogens, abiotic stress reactions, mediation of symbiotic interactions, and cell developmental regulation [3]. In fungi, Noxes have been shown to play essential roles in cellular signaling, hyphal development, cell differentiation, maintenance of symbiosis, and fungal pathogenicity [4, 5]. NoxA in Aspergillus nidulans is involved in fruiting body differentiation [6], and Nox1 and Nox2 in Neurospora crassa are involved in female sterility and sexual spore germination, respectively [7]. In the rice blast fungus Magnaporthe oryzae, Nox1 and Nox2 play roles in appressorium formation and growth of aerial hyphae, respectively, and are thus responsible for fungal pathogenicity [8]. More interestingly, NoxA of Epichloë festucae is involved in the maintenance of symbiosis with the plant host [9].
Common structural knowledge for Nox isoforms was deduced initially based on human Nox2 (gp91phox) [1]. Since then, several different isoforms of Nox have been identified in humans, plants and other eukaryotes. Until now, however, the identification of Nox genes and isoforms using bioinformatic approaches has been challenging, owing to the fact that genes encoding ferric reductases (Fre) and ferric-chelate reductases (FRO) share high sequence similarities and homologous domains with Nox genes. The currently known basic Nox structure consists of highly conserved domains, such as NADPH- and FAD-binding domains, six predicted α-helices TM domains and four heme-binding histidines, which are located in the third and fifth transmembrane helices (TMHs; two histidine residues in each TMH). The conserved distance between the heme-binding histidines is 13 amino acids (aa) in the TMH 3 and 12 aa in the TMH 5 [2].
Three different Nox isoforms (NoxA, B, and C) have been identified in fungi. NoxA and B possess only the catalytic core domains, whereas NoxC has an additional longer N-terminal domain that contains a putative calcium-binding EF-hand motif. This motif is similar to those found in human Nox5 and plant Rboh. Binding of the regulatory unit NoxR (a homologue of human p67phox) is required for activation of fungal NoxA and B [5]. In contrast, fungal NoxC seems not to require binding of NoxR, because the Ca2+-binding domains may act as regulatory units [2]. At present, the different activation mechanisms are used to distinguish NoxC from NoxA and B, but this method is unable to differentiate between NoxA and B.
The aim of the study was to investigate the sequence characteristics of fungal Nox genes via bioinformatic analyses to provide insight into fungal Nox isoforms. Our results not only showed separation of Nox, Fre and FRO genes by phylogenetic analysis, but also provide several new sequence features that differ among the fungal Nox isoforms. Taken together, our comprehensive sequence analysis provides new insight into fungal Nox isoforms, including identification of characteristics that enable accurate identification of fungal genes encoding Nox.
MATERIALS AND METHODS
Identification of genes encoding Nox and NoxR
The fungal protein sequences belonging to "ancestral NADPH oxidase" were obtained from PeroxiBase [10]. In order to identify putative NoxR-encoding genes, the sequence information of six characterized NoxR genes was obtained from a survey of the scientific literature. The sequences for each gene family were subjected to multiple sequence alignments using T-Coffee [11]. The resulting alignments were subsequently used as input for construction of sequence profiles using HMMER [12], of which method is based on probabilistic models (profile hidden Markov models). Subsequently, sequences which were grouped together with characterized Fre/FRO were discarded. Each set of sequences grouped together with characterized NoxA, B, C, Duox, and Rboh, respectively, was used in construction of second sequence profiles using HMMER [12]. We also created a HMMER sequence profile for NoxR. These sequence profiles were then applied to the search for putative Nox and NoxR sequences within 34 genome sequences, including different fungal lifestyles and taxa and representative plant and animal species (Table 1). The proteome sequences were obtained from the Comparative Fungal Genomics Platform (CFGP 2.0; http://cfgp.snu.ac.kr/) [13].
Table 1.
Numbers of Noxes, NoxR, Fre, and FRO genes identified in this study

N/D, not defined.
Analyses of phylogenies, TMHs, histidine residues and glycosylation
Nox amino acid sequences obtained using HMMER were aligned by ClustalW (ver. 2) and phylogenetic analysis was conducted using the neighbor-joining method [14]. TMHs within the Nox sequences were predicted using TMHMM2 [15], which is based on a hidden Markov model. Analysis of histidine residues was performed manually from sequence alignments using MEGA5 [16]. Domain analysis was conducted using InterPro Scan (ver. 4.8), scanning the query against the twelve protein databases [17]. Prediction of C-, N- and O-glycosylation sites was performed using NetCGly 1.0 [18], NetNGly 1.0 and NetOGlyc 3.1 [19], which adopted a neural network approach.
RESULTS AND DISCUSSION
Phylogenetic analysis of putative Nox genes
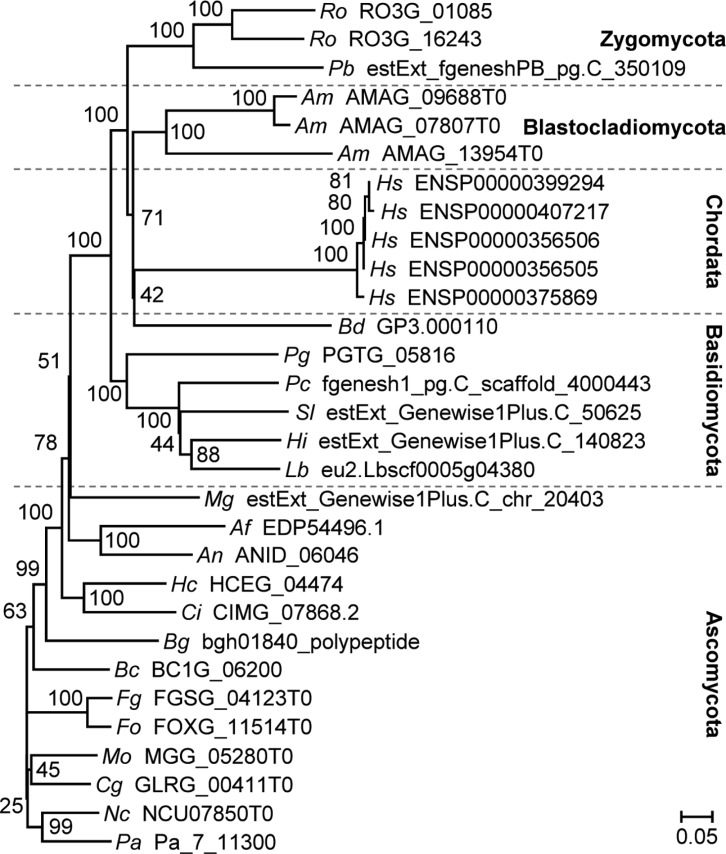
A total of 310 putative Nox amino acid sequences were retrieved from 28 fungal, one Oomycota, two plant and three animal species by sequence profiling using HMMER (Table 1). These sequences were subjected to phylogenetic analysis to reveal relationships among characterized and putative Nox genes. Unexpectedly, however, only 102 sequences were identified as Nox, but more than 200 were identified as Fre or FRO genes (Fig. 1). More Nox genes than Fre or FRO genes are present in plant, but the opposite is true for fungal genomes. Interestingly, no Fre or FRO genes were identified in human, Drosophila melanogaster and Caenorhabditis elegans genomes (Table 1). A total of 102 Nox gene sequences were used for further analyses. This analysis indicated that the HMMER profile based on "ancestral NADPH oxidase" sequences from PeroxiBase potentially predict Fre/FRO sequences as Nox. To provide more stringent information regarding fungal Nox genes, new HMMER profiles were constructed in an attempt to retrieve fungal Nox genes. These sequence profiles were also used in Nox prediction in the fungal peroxidase database (fPoxDB; http://peroxidase.riceblast.snu.ac.kr) [20].
Fig. 1.
Phylogenetic tree for 310 Nox, Fre, and FRO sequences. A neighbor-joining tree was constructed using the 310 sequences. The numbers at each node indicate the bootstrap support with 10,000 replications. Bootstrap values were shown if a node is supported by 50 or more. Clades were named respective to the occurrence of the characterized sequence(s). Two-letter italicized abbreviations represent the species names as follows: Af, Aspergillus fumigatus; Am, Allomyces macrogynus; An, Aspergillus nidulans; At, Arabidopsis thaliana; Bc, Botrytis cinerea; Bd, Batrachochytrium dendrobatidis; Bg, Blumeria graminis; Ca, Candida albicans; Ce, Caenorhabditis elegans; Cg, Colletotrichum graminicola; Ci, Coccidioides immitis; Cn, Cryptococcus neoformans var. grubii; Dm, Drosophila melanogaster; Ec, Encephalitozoon cuniculi; Fg, Fusarium graminearum; Fo, Fusarium oxysporum; Hc, Histoplasma capsulatum; Hi, Heterobasidion irregulare; Hs, Homo sapiens; Lb, Laccaria bicolor; Mg, Mycosphaerella graminicola; Mo, Magnaporthe oryzae; Nc, Neurospora crassa; Os, Oryza sativa; Pa, Podospora anserina; Pb, Phycomyces blakesleeanus; Pc, Phanerochaete chrysosporium; Pg, Puccinia graminis; Pi, Phytophthora infestans; Ro, Rhizopus oryzae; Sc, Saccharomyces cerevisiae; Sl, Serpula lacrymans; Sp, Schizosaccharomyces pombe; Um, Ustilago maydis.
NoxA is present in the greatest number of fungal species, followed by NoxB and C. NoxA and B are present in a wide range of fungal taxa, whereas most of NoxC is present in the phylum Ascomycota. In general, most fungal species possessed one Nox gene for every given isoform, with the exception of Allomyces macrogynus (Table 1). This result is in accordance with experimental observations: only one gene per Nox isoform has been described so far in all fungi evaluated [6,7,8,9,21,22,23,24,25]. In our analysis, no Nox sequence was found in the fungal species of Saccharomyces cerevisiae, Schizosaccharomyces pombe, Candida albicans, Cryptococcus neoformans, Ustilago maydis or Rhizopus oryzae. It has also been reported that yeasts of Ascomycota and dimorphic species of Basidiomycota do not possess Nox genes [8, 23, 26]. More recently, however, Nox functionality has been reported in S. cerevisiae [27], although its sequence similarity was closer to that of Fre than Nox in our analysis. It remains unclear whether Nox genes in these species were lost through the course of evolutionary processes.
No Nox, Fre, or FRO gene was identified in the two fungal species Phycomyces blakesleeanus and Encephalitozoon cuniculi in this analysis. Further analysis using more fungal genomes would provide clues to understanding the contribution of Nox in fungal life cycles. As expected, sequences from the plant species Arabidopsis thaliana and Oryza sativa were found to encode Rboh. Phytophthora infestans, belonging to the phylum Oomycota, also possessed an Rboh sequence similar to plant Rbohs, but lacking the NADPH oxidase respiratory domain (IPR013623).
When these sequences were subjected to phylogenetic analysis, genes encoding NoxA, B, and C isoforms could be distinguished clearly and grouped separately. All metazoan sequences were grouped together in the human Nox1~4, human Nox5, and Duox clades. Human Nox1~4 grouped together with fungal NoxB, whereas human Nox5 and Duox grouped separately. NoxA and B seem to share a common ancestor gene in the phyla Ascomycota and Basidiomycota, as they belong to clades that are close together. NoxC, however, is in a separate clade (Fig. 1). These data suggest that NoxA and B from both fungal phyla were retained during evolutionary development, but NoxC was probably lost recently in some fungal species in both phyla [21]. Furthermore, this would support the hypothesis that NoxA and B are essential for fungal development and physiological processes in most species, whereas NoxC may be essential in fewer species. This is consistent with the fact that NoxA and B mutants from several fungal species are seriously affected developmentally [6,7,8,9,21,22,23,24,25], but NoxC mutants do not show any phenotypic changes [21]. The phylogenetic tree provided not only interesting insight into the overall distribution of Nox isoforms, but also the opportunity to distinguish different Nox isoforms from each other as well as from Fre and FRO.
Transmembrane helices (TMHs)
TMHs are crucial for localizing and anchoring proteins within the membrane, and the number of helices influences the stability and interaction of proteins. Human Nox families generally have six predicted TMHs, except for Duox, which has seven [1, 2]. However, little information is available on TMHs in fungal Nox isoforms. For the genes encoding NoxA, species from the phyla Ascomycota and Basidiomycota have an average of five TMHs, ranging from three (Fusarium oxysporum) to seven (Heterobasidion irregulare). With respect to NoxB, fungi within the phyla Ascomycota and Basidiomycota have seven to eight TMHs, except for F. oxysporum, which has five. NoxC isoforms, which are mostly present in Ascomycota species, have five TMHs. However, NoxB isoforms from A. macrogynus (Blastocladiomycota) and Batrachochytrium dendrobatidis (Chytridiomycota) have fewer and a more variable number of TMHs (Table 2). Most Rbohs in O. sativa and A. thaliana were predicted to have four TMHs, with the exception of a few with five. P. infestans has four Rbohs with three to six TMHs. Interestingly, the numbers of TMHs in human Nox isoforms were found to differ by one to two from those published in the literature [1, 2]. For example, human Duox was predicted to have only six TMHs, which is one less than described previously, whereas C. elegans and D. melanogaster Duox isoforms had seven TMHs (Table 2). This difference may be explained by the usage of different prediction tools to determine the numbers of TMHs. For example, hydropathy analyses predicted four or six TMHs for human Nox2 (gp91phox), from which the structure of other Nox isoforms was deduced [1].
Table 2.
Number of transmembrane helices and glycosylation sites in different Nox isoforms
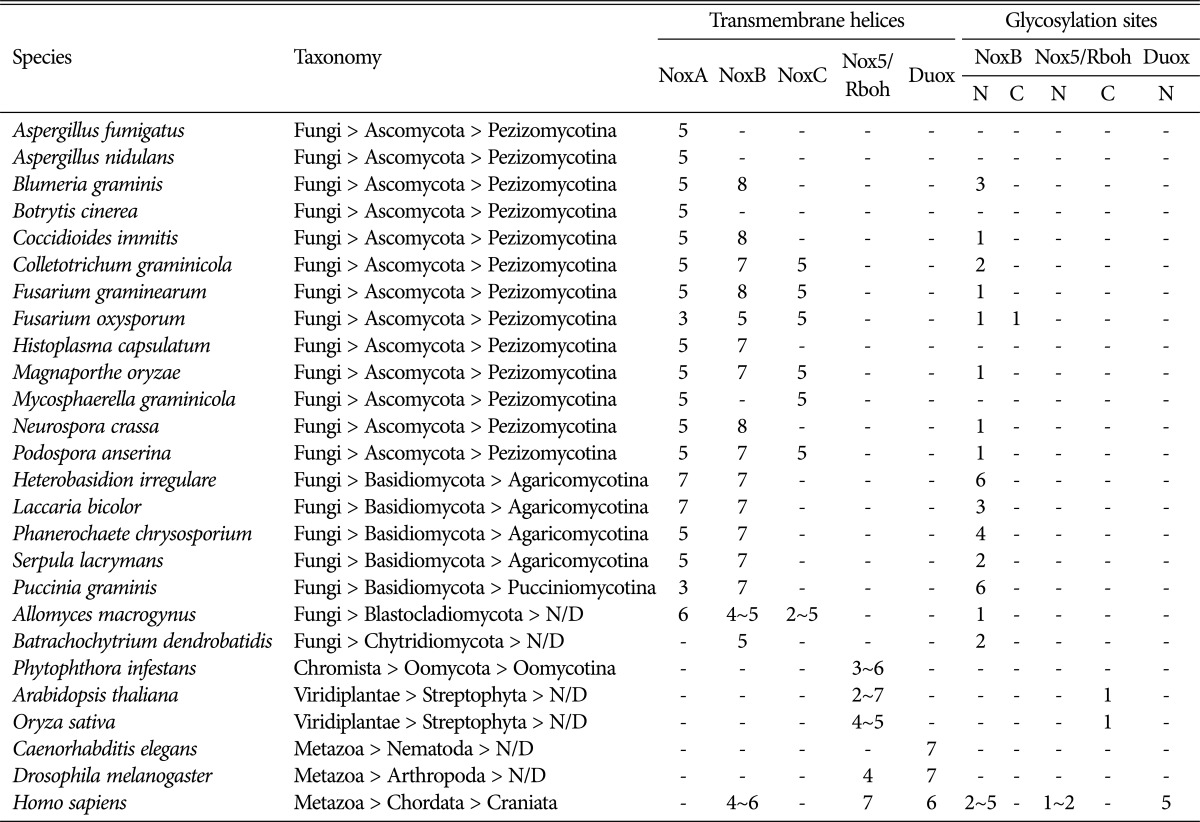
N/D, not defined.
On average, fungal species had one less TMH and one to two more TMHs among their NoxA and B isoforms, respectively, compared with the six TMHs described in the literature [1, 2]. These data indicate that more diverse numbers of TMHs exist among fungal Nox isoforms, compared with human Nox. This may be a consequence of the fungal taxonomic hierarchy or adaptation to different lifestyles.
Distance of histidine residues
The distance between conserved intramembrane di-histidine motifs is critical for heme positioning and localization, and thus crucial for Nox enzymatic activity/functionality. A review from Sumimoto in 2008 [2] describes the distance between histidines in several cytochromes essential for heme binding. For Nox and FRO, the distances between the di-histidine motifs are 13 aa (TMH 3) and 12 aa (TMH 5), whereas both motifs are separated by 13 aa in Fre [2]. Although extensive studies have been conducted on human Nox isoforms, no information is available for fungal Nox.
NoxA in Ascomycota and Basidiomycota species has a histidine distance of only 3 aa in TMH 3 and 11 aa in TMH 5. Most ascomycetes had an additional third dihistidine motif in NoxA, with a spanning distance of 2-8 aa in TMH 1 (Fig. 2). The appearance of a di-histidine motif in TMH 1 of NoxA in the phylum Ascomycota has not been documented. The extents to which the motif is also involved in heme-binding or enzymatic activity are unknown, but it should be considered as an important feature. NoxB isoforms of B. dendrobatidis and of Ascomycota and Basidiomycota species have the same histidine distance scheme as that of NoxA, but with an additional histidine residue in TMH 3, with a distance of 13 aa, for species belonging to the phylum Ascomycota. In addition, a significant difference was observed in the distance of the di-histidine motifs of TMH 3 for NoxA and NoxB from those of Noxes in other organisms. The distance between the only two histidine residues was 3 aa, which is comparably shorter than from the known literature of 13 aa for other Noxes [1, 2]. NoxC had three histidines, separated by distances of 13 aa and 3 aa in TMH 3, while the histidine residues of TMH 5 were 12 aa apart. The Rbohs of P. infestans exhibited the same scheme as NoxC isoforms in Ascomycota species, whereas most plant Rbohs had only one di-histidine motif within TMH 3, with a few exceptions. Interestingly, NoxA and B isoforms (in all fungal species) have a double histidine residue in TMH 5. Although, the contribution of the di-histidine motif to the structure and function of Nox is not known, these results clearly demonstrate the presence of variations in histidine residues within the heme-binding TMHs of fungal Nox isoforms (Fig. 2). This might be a consequence of the complex lifestyles among fungal species and the evolutionarily broad range of taxa.
Fig. 2.
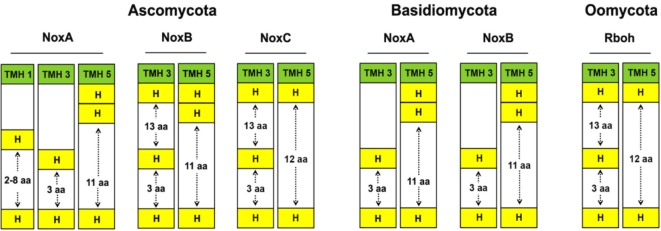
Histidine distribution within transmembrane helices (TMHs) of fungal Nox isoforms. NoxA and B can be found in Ascomycota and Basidiomycota, whereas NoxC is found only in Ascomycota. Distances between histidine residues are depicted by arrows and the number of amino acids.
Glycosylation
Glycosylation of proteins plays an essential role in many biological processes, such as cell-cell recognition and related signal-transduction pathways. N-glycosylation has been reported to occur frequently in human Nox isoforms with differential distribution. It has been demonstrated experimentally that Nox2 and Duox are N-glycosylated [1]. To our knowledge, no reports have been published to date concerning glycosylation site analysis of a broad range of fungal Nox isoforms from different species.
N-glycosylation sites were predicted in fungal NoxB isoforms, but not in NoxA or C (Table 2). C- or O-glycosylation sites were rarely identified in fungal Nox, indicating that glycosylation is a unique feature of fungal NoxB. There was also a significant difference in the number of predicted N-glycosylation sites between the phyla Ascomycota and Basidiomycota (Table 2). The reason higher numbers of glycosylation sites were predicted in Basidiomycota compared with Ascomycota remains to be clarified. Since the prediction tools NetCGly, NetOGly and NetNGlyc were originally designed to predict glycosylation sites in mammalian proteins [18, 19], we cannot completely rule out the possibility that some glycosylation sites were predicted incorrectly in fungal Nox isoforms. However, glycosylation site analysis may be used to differentiate between NoxA and B in Basidiomycota, since they have the same histidine distances (Fig. 2).
NoxR
NoxR proteins are pivotal for the regulation of NoxA and NoxB. This regulatory mechanism was confirmed experimentally using NoxR mutants and NoxA/NoxB double mutants of Botrytis cinerea [24]. However, no information is available concerning the systemic annotation of this gene family in fungi. We created a HMMER sequence profile to retrieve NoxR genes, which were applied to phylogenetic analysis (Fig. 3). Genes encoding NoxR in different phyla were grouped separately. NoxR was found in all species predicted to have NoxA and B, consistent with a regulatory role for NoxR in fungi (Table 1). To date, NoxR has consistently been found in fungi that have NoxA [5]. However, we also found NoxR in B. dendrobatidis, which is predicted to have only NoxB. Furthermore, we found NoxR in two species belong to Zygomycota (R. oryzae and P. blakesleeanus) that do not possess any Nox isoforms. Interestingly, A. macrogynus express three NoxR isoforms, whereas other fungal species possess only one NoxR. It is tempting to speculate that more NoxR isoforms are required to efficiently regulate the one NoxA and three NoxB genes found in this fungal species.
Fig. 3.
Phylogenetic tree for NoxR sequences. A neighbor-joining tree was constructed using the 30 NoxR amino acid sequences. The numbers at each node indicate the bootstrap support with 10,000 replications. Two-letter italicized abbreviations represent the species names as follows: Af, Aspergillus fumigatus; Am, Allomyces macrogynus; An, Aspergillus nidulans; Bc, Botrytis cinerea; Bd, Batrachochytrium dendrobatidis; Bg, Blumeria graminis; Ci, Coccidioides immitis; Cg, Colletotrichum graminicola; Fg, Fusarium graminearum; Fo, Fusarium oxysporum; Hc, Histoplasma capsulatum; Hi, Heterobasidion irregulare; Hs, Homo sapiens; Lb, Laccaria bicolor; Mg, Mycosphaerella graminicola; Mo, Magnaporthe oryzae; Nc, Neurospora crassa; Pa, Podospora anserina; Pc, Phanerochaete chrysosporium; Pg, Puccinia graminis; Ro, Rhizopus oryzae; Sl, Serpula lacrymans.
Taken together, we demonstrated that although Nox, Fre, and FRO have high sequence similarities, they can be differentiated effectively using phylogenetic analysis. Furthermore, our data provide intriguing new insights into the sequence properties of fungal Nox isoforms. The distance between histidine residues within the heme-binding domain is only 3 aa in the TMH 3 of NoxA in the phyla Ascomycota and Basidiomycota. The number of TMHs in fungal Nox isoforms varied among phyla and isoforms. Ascomycota NoxA and NoxC have five TMHs, whereas NoxB has between five and eight. The number of TMHs predicted in NoxA genes from species belonging to the phylum Basidiomycota varied from three to seven, whereas the number in NoxB isoforms was consistently seven. Additionally, our data suggest that N-glycosylation occurs only in NoxB, and that NoxC is restricted mostly to the phylum Ascomycota.
ACKNOWLEDGEMENTS
This work was supported by the Finland Distinguished Professor Program (FiDiPro), from the Academy of Finland (FiDiPro #138116), and a National Research Foundation of Korea grant, funded by the Korean government (2008-0061897 and 2013-003196), and the Next-Generation BioGreen 21 Program of Rural Development Administration in Korea (PJ00821201). We thank Prof. Jari P. T. Valkonen for his valuable suggestions on this work. JC is funded by a graduate fellowship from the Brain Korea 21 Program.
References
- 1.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 2.Sumimoto H. Structure, regulation and evolution of Nox-family NADPH oxidases that produce reactive oxygen species. FEBS J. 2008;275:3249–3277. doi: 10.1111/j.1742-4658.2008.06488.x. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki N, Miller G, Morales J, Shulaev V, Torres MA, Mittler R. Respiratory burst oxidases: the engine of ROS signaling. Curr Opin Plant Biol. 2011;14:691–699. doi: 10.1016/j.pbi.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 4.Heller J, Tudzynski P. Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annu Rev Phytopathol. 2011;49:369–390. doi: 10.1146/annurev-phyto-072910-095355. [DOI] [PubMed] [Google Scholar]
- 5.Scott B, Eaton CJ. Role of reactive oxygen species in fungal cellular differentiations. Curr Opin Microbiol. 2008;11:488–493. doi: 10.1016/j.mib.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Lara-Ortíz T, Riveros-Rosas H, Aguirre J. Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol Microbiol. 2003;50:1241–1255. doi: 10.1046/j.1365-2958.2003.03800.x. [DOI] [PubMed] [Google Scholar]
- 7.Cano-Domínguez N, Alvarez-Delfín K, Hansberg W, Aguirre J. NADPH oxidases NOX-1 and NOX-2 require the regulatory subunit NOR-1 to control cell differentiation and growth in Neurospora crassa. Eukaryot Cell. 2008;7:1352–1361. doi: 10.1128/EC.00137-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egan MJ, Wang ZY, Jones MA, Smirnoff N, Talbot NJ. Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc Natl Acad Sci U S A. 2007;104:11772–11777. doi: 10.1073/pnas.0700574104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka A, Christensen MJ, Takemoto D, Park P, Scott B. Reactive oxygen species play a role in regulating a fungus-perennial ryegrass mutualistic interaction. Plant Cell. 2006;18:1052–1066. doi: 10.1105/tpc.105.039263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koua D, Cerutti L, Falquet L, Sigrist CJ, Theiler G, Hulo N, Dunand C. PeroxiBase: a database with new tools for peroxidase family classification. Nucleic Acids Res. 2009;37:D261–D266. doi: 10.1093/nar/gkn680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, Taly JF, Notredame C. T-Coffee: a web server for multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39:W13–W17. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi J, Cheong K, Jung K, Jeon J, Lee GW, Kang S, Kim S, Lee YW, Lee YH. CFGP 2.0: a versatile web-based platform for supporting comparative and evolutionary genomics of fungi and Oomycetes. Nucleic Acids Res. 2013;41:D714–D719. doi: 10.1093/nar/gks1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 15.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 16.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Julenius K. NetCGlyc 1.0: prediction of mammalian C-mannosylation sites. Glycobiology. 2007;17:868–876. doi: 10.1093/glycob/cwm050. [DOI] [PubMed] [Google Scholar]
- 19.Julenius K, Mølgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–164. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- 20.Choi J, Détry N, Kim KT, Asiegbu FO, Valkonen JP, Lee YH. fPoxDB: fungal peroxidase database for comparative genomics. BMC Microbiol. 2014;14:117. doi: 10.1186/1471-2180-14-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brun S, Malagnac F, Bidard F, Lalucque H, Silar P. Functions and regulation of the Nox family in the filamentous fungus Podospora anserina: a new role in cellulose degradation. Mol Microbiol. 2009;74:480–496. doi: 10.1111/j.1365-2958.2009.06878.x. [DOI] [PubMed] [Google Scholar]
- 22.Giesbert S, Schürg T, Scheele S, Tudzynski P. The NADPH oxidase Cpnox1 is required for full pathogenicity of the ergot fungus Claviceps purpurea. Mol Plant Pathol. 2008;9:317–327. doi: 10.1111/j.1364-3703.2008.00466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malagnac F, Lalucque H, Lepère G, Silar P. Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet Biol. 2004;41:982–997. doi: 10.1016/j.fgb.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 24.Segmüller N, Kokkelink L, Giesbert S, Odinius D, van Kan J, Tudzynski P. NADPH oxidases are involved in differentiation and pathogenicity in Botrytis cinerea. Mol Plant Microbe Interact. 2008;21:808–819. doi: 10.1094/MPMI-21-6-0808. [DOI] [PubMed] [Google Scholar]
- 25.Yang SL, Chung KR. The NADPH oxidase-mediated production of hydrogen peroxide (H2O2) and resistance to oxidative stress in the necrotrophic pathogen Alternaria alternata of citrus. Mol Plant Pathol. 2012;13:900–914. doi: 10.1111/j.1364-3703.2012.00799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aguirre J, Ríos-Momberg M, Hewitt D, Hansberg W. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 2005;13:111–118. doi: 10.1016/j.tim.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 27.Rinnerthaler M, Büttner S, Laun P, Heeren G, Felder TK, Klinger H, Weinberger M, Stolze K, Grousl T, Hasek J, et al. Yno1p/Aim14p, a NADPH-oxidase ortholog, controls extramitochondrial reactive oxygen species generation, apoptosis, and actin cable formation in yeast. Proc Natl Acad Sci U S A. 2012;109:8658–8663. doi: 10.1073/pnas.1201629109. [DOI] [PMC free article] [PubMed] [Google Scholar]