Abstract
A hallmark of protein conformational disease, exemplified by neurodegenerative disorders, is the expression of misfolded and aggregated proteins. The relationship between protein aggregation and cellular toxicity is complex, and various models of experimental pathophysiology have often yielded conflicting or controversial results. In this study, we examined the biophysical properties of amyotrophic lateral sclerosis (ALS)-linked mutations of Cu/Zn superoxide dismutase 1 (SOD1) expressed in human tissue culture cells. Fluorescence correlation spectroscopy (FCS) and Forster resonance energy transfer (FRET) analyses revealed that changes in proteasome activity affected both the expression of FCS- and FRET-detected oligomers and cellular toxicity. Under normal conditions, highly aggregation-prone mutant SOD1 exhibited very little toxicity. However, when the activity of the proteasome was transiently inhibited, only upon recovery did we observe the appearance of ordered soluble oligomers, which were closely correlated with cellular toxicity. These results shed light on the importance of balance in proteostasis and suggest that transient shifts of activity in the cellular machinery can alter the course of protein conformational transitions and dysregulate modulation of proteasome activity. In neurodegenerative disorders including ALS, such changes may be a risk factor for pathogenesis.
Introduction
Amyotrophic lateral sclerosis (ALS) is a severe neurodegenerative disorder characterized by the loss of motor neurons (Cleveland & Rothstein 2001; Bruijn et al. 2004). Approximately 10% of ALS cases are familial, and approximately 20% of these cases are caused by mutations in the gene encoding SOD1 (Rosen et al. 1993). The majority of ALS-linked mutations of SOD1 lead to folding instability and misfolding, with aggregates of mutant SOD1 detected in the spinal cord of affected individuals (Bruijn et al. 1998). Accumulation of mutant SOD1 aggregates has been proposed to lead to neuronal cell death by gain-of-function toxicity and inhibition of a multitude of cellular functions including axonal transport, mitochondrial function, and protein homeostasis (Williamson et al. 2000; Bruijn et al. 2004; Matsumoto et al. 2005; Gidalevitz et al. 2009). Similarly, sporadic ALS is associated with mutations and aggregation of TDP-43 and FUS (Da Cruz & Cleveland 2011). Although the molecular mechanisms underlying the cytotoxicity of mutant SOD1, TDP-43, and FUS are poorly understood, it is possible that misfolding and aggregation interfere with many cellular processes, either directly by co-sequestration (Olzscha et al. 2011) or by chaperone competition and proteostatic collapse (Gidalevitz et al. 2006, 2010).
The balance between folding and misfolding is regulated by proteostatic pathways, which include the chaperone network and the clearance machineries of the proteasome and autophagy (Holmberg et al. 2004; Vabulas & Hartl 2005; Vilchez et al. 2012). Genetic screens and proteomics have identified key components involved in proteostasis, as well as the networks that are affected by protein conformational disease or can alter the course of such diseases (Holmberg et al. 2004; Vilchez et al. 2012). Expression of misfolded and aggregation-prone proteins has multiple consequences, including direct effects, such as inhibition of proteasome activity, and indirect effects, such as interference with the degradation of other proteins. Mutant SOD1 is polyubiquitinated and degraded by the ubiquitin-proteasome system (Johnston et al. 2000; Niwa et al. 2007). Consequently, inhibition of proteasome activity increases the accumulation of mutant but not wild-type SOD1 into aggregates (Johnston et al. 2000; Niwa et al. 2007). Mutant SOD1 aggregates to form amorphous structures comprised of mobile components that can exchange with soluble mutant SOD1 in the cytosol (Matsumoto et al. 2005). However, little is known about the properties of the aggregated and soluble states of mutant SOD1 or their relevance to toxicity. Here, we show that the toxicity of mutant SOD1 aggregates expressed in cultured human cells is determined by the state and activity of the proteasome. Using spectroscopic approaches with single-molecule sensitivity, we demonstrated the persistence of soluble and ordered oligomeric species of SOD1 mutants during the inhibition of proteasome activity. Furthermore, we found that marked cellular toxicity was only observed during recovery from proteasome inhibition.
Results
Dissociation of mutant SOD1 from aggresomes during recovery of proteasome activity
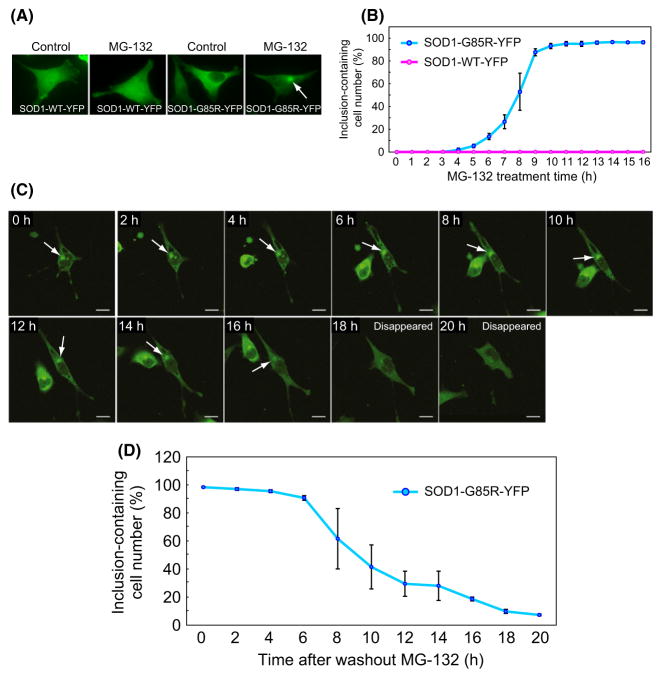
We examined the aggregation and toxicity of mutant SOD1 expressed in a HeLa cell line engineered for conditional expression of tagged wild-type SOD1 (SOD1-WT-YFP) or an ALS-linked SOD1 mutant with a G85R mutation (SOD1-G85R-YFP), using a tetracycline-regulated promoter under the control of doxycycline. After 16-h treatment with the proteasome inhibitor MG-132 (2 μM), SOD1-G85R-YFP formed perinuclear inclusions in ~96% of cells, whereas SOD1-WT-YFP was unaffected (Fig. 1A,B). The accumulation of aggregated mutant SOD1 in MG-132-treated cells was confirmed by centrifugal fractionation (Fig. S1A), filter-trap assay (Fig. S1B), and immunostaining analysis (Fig. S1D) using antibodies against aggresome markers (Johnston et al. 2000; Kopito 2000).
Figure 1.
Disaggregation of mutant SOD1 from inclusions to the cytosol. (A) HeLa cell lines stably expressing SOD1-WT-YFP or SOD1-G85R-YFP were treated with MG-132 or DMSO (used as a negative control). The white arrow indicates a perinuclear inclusion. (B) Numbers of cells containing SOD1-YFP inclusions during 16-h treatment with MG-132 (mean ± SD, n > 600). (C) Disappearance of inclusions during recovery of proteasome activity. After 16-h treatment with MG-132, cells were transferred to a recovery culture without MG-132 and incubated for the indicated periods. Time-lapse images were taken using a confocal microscope. White arrows indicate inclusions. (D) Numbers of cells containing inclusions during the recovery culture (mean ± S.D., n > 600).
We next examined the fate of mutant SOD1 inclusions during recovery of proteasome activity. MG-132-treated cells expressing mutant SOD1 were transferred to recovery medium lacking MG-132, and proteasome activity was measured using a proteasome-targeted GFP substrate (GFPu) (Bence et al. 2001). After incubation in MG-132-free medium, proteasome activity was recovered within 4 h (Fig. S2). Time-lapse observations of individual cells revealed that SOD1-G85R-YFP inclusions were no longer detectable after 20 h of recovery (Fig. 1C); inclusions had a half-life of ~9 h (Fig. 1D). No significant morphological changes in the cells were observed during these processes. Likewise, the detergent-insoluble fraction of SOD1-G85R-YFP aggregates decreased by 50% during recovery (Fig. S1, A, C). Similar results were observed in cells treated with the specific proteasome inhibitor epoxomicin, although the half-life of inclusions was slightly extended relative to cells treated with MG-132 (Fig. S1E). The decrease in mutant SOD1 inclusions was also observed in MG-132-treated cells allowed to recover in the presence of an autophagy/lysosome inhibitor, bafilomycin A1 (0.1 μM) (Fig. S1F). LC3 accumulation, a standard indicator of autophagosome formation, was clearly observed after treatment with bafilomycin A1 at the same concentration (Fig. S1F inset), suggesting that autophagy is not essential for disaggregation. These results indicate that living cells possess a mechanism for disaggregation of proteins that is associated with the restoration of proteasome activity.
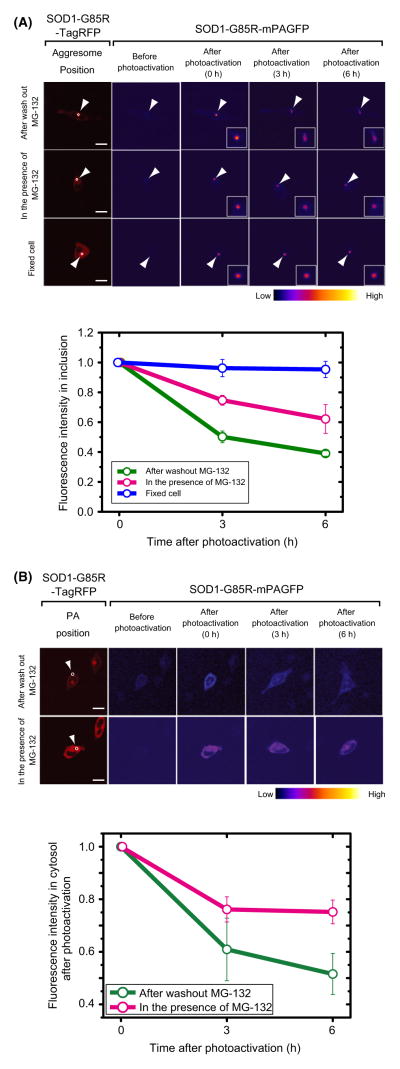
To examine whether mutant SOD1 that dissociates from inclusions redistributes into the soluble state in the cytoplasm, we carried out dynamic imaging experiments using a monomeric variant of photoactivatable green fluorescent protein (mPAGFP). This variant can be converted from a dark state to a bright green fluorescent state by irradiation with violet light (Patterson & Lippincott-Schwartz 2002; Lippincott-Schwartz & Patterson 2003). In cells expressing SOD1-G85R-mPAGFP, ROIs in the inclusion or cytosol were converted to the bright state by 0.45 s irradiation with violet light. In control cells, fluorescence intensity of photoactivated inclusions decreased rapidly, reaching 50% after 3 h of recovery in the absence of MG-132, and the aggresome structures containing photoactivated SOD1 gradually expanded (Fig. 2A). By contrast, in the presence of MG-132, fluorescence intensity decreased by only 25% during the same period, and the shapes of aggresome structures were unchanged (Fig. 2A). Photoactivation of cytosolic SOD1-G85R-mPAGFP also revealed a faster decrease in fluorescence intensity under recovery conditions than in the presence of MG-132 (Fig. 2B), suggesting that mutant SOD1 is degraded in the cytosol. These results suggest that mutant SOD1 dissociates from inclusions and enters the cytosol, where it is degraded.
Figure 2.
Photoactivation analysis of SOD1-G85R-mPAGFP during disappearance of inclusions, in inclusions (A) and in the cytosol (B). Red images in the far left column show the distribution of cotransfected SOD1-G85R-TagRFP. Pseudo-colored images indicate the intensity of photoactivated green fluorescence from SOD1-G85R-mPAGFP. White circles show photoactivated regions. White arrowheads indicate inclusions. Bar = 10 mm. Graphs show quantified fluorescence intensities after photoactivation. The relative cytosolic fluorescence intensity during chase periods (3 and 6 h) was normalized against the intensity immediately after photoactivation (0 h; n = 3–4).
Formation and disappearance of soluble oligomers of mutant SOD1, depending on proteasome activity
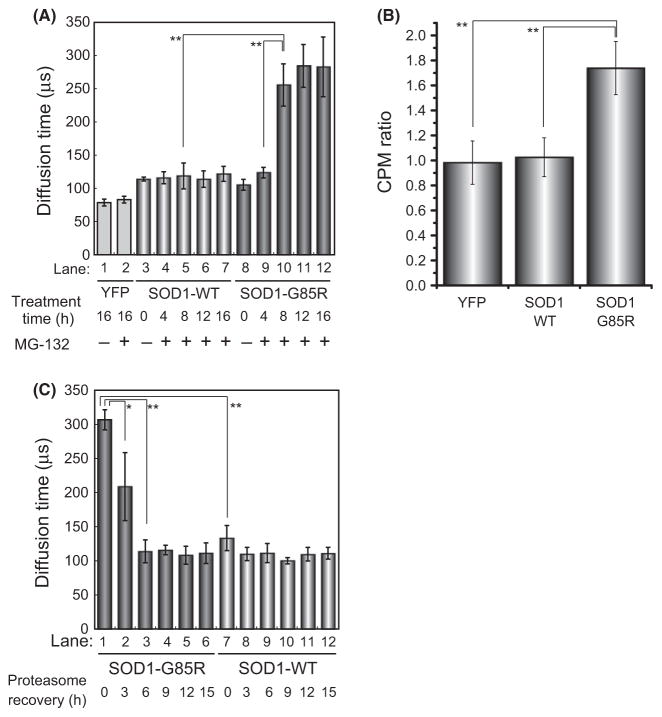
We next determined the oligomeric state of the soluble mutant SOD1 that appears upon increase in proteasome activity. To this end, we used FCS, a method developed to analyze rapid movements of fluorescent molecules at single-molecule sensitivity (Rigler et al. 1993; Lippincott-Schwartz et al. 2001; Kitamura et al. 2006). Cells expressing SOD1-YFP were lysed, and the soluble fraction was recovered and analyzed by FCS. The size of SOD1-WT-YFP, calculated from the diffusion time, indicated the presence of a dimeric species (~79 kDa, Fig. 3A, lanes 3–7). By contrast, in the absence of MG-132, mutant SOD1-G85R-YFP exhibited a diffusion time intermediate between those of monomers and dimers (~60 kDa, Fig. 3A, lane 8), suggesting that both monomers and dimers were present under these conditions, as previously reported (Johnston et al. 2000; Wang et al. 2009). In the presence of MG-132 treatment, FCS revealed the presence of high molecular weight species (~1000 kDa) of mutant SOD1 (Fig. 3A, lane 10–12). These high molecular weight species were significantly smaller than previously reported soluble aggregates of expanded polyglutamine (>100 MDa) (Kitamura et al. 2006). We also confirmed the formation of high molecular weight species of SOD1-G85R-YFP during inhibition of proteasome activity by sucrose density-gradient ultracentrifugation analysis (Fig. S5). Furthermore, we also observed the accumulation of polyubiquitinated SOD1-G85R-YFP during the inhibition of proteasome activity (Fig. S6). In FCS analysis, counts per molecule (CPM), a mean brightness of the particles, is a useful indicator of the formation of high molecular weight species such as oligomer formation. CPM ratio of SOD1-G85R-YFP was significantly increased during inhibition of proteasome activity compared with SOD1-WT-YFP and YFP monomer (Fig. 3B). Thus, we concluded that the high molecular weight species of SOD1-G85R-YFP detected by CPM ratio are composed of soluble oligomers. In contrast, after transfer to recovery medium, the SOD1-G85R-YFP oligomers disappeared over a period of 6 h (Fig. 3C, lanes 1–3). The size of SOD1-G85R-YFP oligomers decreased after treatment with 100 mM dithiothreitol, a strong reducing agent (Fig. S4A, lanes 12–16; and Fig. S4B, lanes 7–12), in agreement with the observation that mutant SOD1 can form disulfide-bond-dependent aggregates (Niwa et al. 2007; Furukawa et al. 2008; Karch & Borchelt 2008).
Figure 3.
Fluorescence correlation spectroscopy (FCS) analysis of soluble mutant SOD1 oligomers during aggregation and disaggregation. (A and C) Diffusion time of wild-type and mutant SOD1. Lysates were prepared from cells expressing YFP, SOD1-wt-YFP, or SOD1-G85R-YFP and then treated with MG-132 for the indicated periods (A), followed by transfer to recovery media for the indicated periods (C). (B) CPM ratio of wild-type and mutant SOD1. Significant differences were determined using Student’s t-test: ★P < 0.05, ★★P < 0.01.
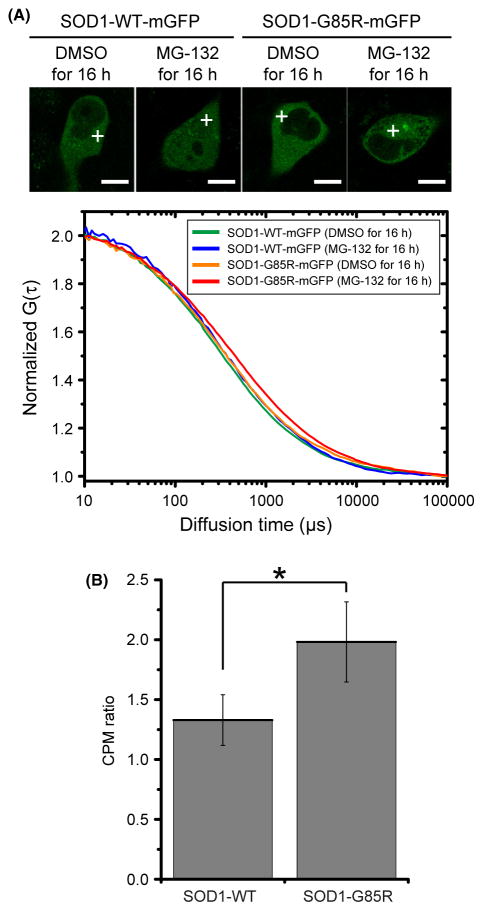
We quantitatively determined the diffusion coefficients of SOD1-mGFPs in live cells using two-component curve fitting analysis of the autocorrelation function measured by FCS (Fig. 4A and Table 1). The fast diffusion coefficient of SOD1-G85R-mGFP in the presence of MG-132 (16.9 ± 3.8 μm2/s) was significantly smaller than that of G85R in the absence of MG-132 (Table 1) and that of the WT protein in the presence or absence of MG-132 (Table 1). SOD1-G85R-mGFP had the smallest slow diffusion coefficient (0.52 ± 0.40 μm2/s) (Table 1). The ratios of fast-diffusing to slow-diffusing molecules exhibited no significant differences (Table 1). These data clearly showed that the diffusion rate of SOD1-G85R-mGFP in the presence of MG-132 was slower than that of the G85R mutant in the absence of MG-132 and that of the wild-type protein under either condition. In general, diffusion rate in the cytoplasm decreases as molecular weight increases (see also the Stokes–Einstein relation described in Experimental procedures). In addition, CPM ratio of SOD1-G85R-YFP was increased compared with SOD1-WT-YFP (Fig. 4B). Therefore, the slow diffusion of SOD1-G85R-mGFP in the presence of MG-132 is a consequence of oligomerization. As these findings are in agreement with previously reported diffusion coefficients obtained by FRAP analysis (Matsumoto et al. 2005), it is clear that FCS can detect the oligomeric state of mutant SOD1.
Figure 4.
FCS analysis using live cells expressing SOD1-mGFPs. (A) FCS measurements were taken in the cytosolic positions indicated by white crosses. Cells were analyzed under the indicated conditions (top), and the quantified data are presented as averages of the normalized autocorrelation function (n = 3–4) (bottom). (B) CPM ratio in live cells expressing SOD1-mGFPs. Significant differences were determined using Student’s t-test: ★P < 0.01.
Table 1.
Diffusion coefficients and contents of SOD1-YFPs fractions estimated from the results of FCS analysis of live cells. Results are shown after curve fitting of the autocorrelation function, which were analyzed using a translational diffusion mathematical model comprising two components. Dfast and Dslow denote diffusion coefficients of fast and slow fractions of the two components, respectively. The diffusion coefficient is the diffusion area of molecules per time unit. All D values and contents of fractions were determined from three to five independent experiments (mean ± SD)
| Types of SOD1 | Reagent | DFast (μm2/s) | Fast fraction (%) | Dslow (μm2/s) | Slow fraction (%) | χ2 | Cell numbers | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT-mGFP | DMSO for 16 h | 22.5 ± 3.1 |
 * *
|
87.8 ± 6.6 | 1.9 ± 2.3 |
 ** **
|
12.6 ± 6.3 | <10−7 | 16 | ||||
| WT-mGFP | MG-132 for 16 h | 23.7 ± 3.9 |
|
82.5 ± 11.6 | 2.8 ± 3.2 |
|
17.5 ± 11.6 | <10−7 | 18 | ||||
| G85R-mGFP | DMSO for 16 h | 21.2 ± 7.0 |
|
84.4 ± 9.0 | 2.4 ± 2.4 |
|
15.6 ± 9.0 | <10−7 | 21 | ||||
| G85R-mGFP | MG-132 for 16 h | 16.9 ± 3.8 | 85.6 ± 5.1 | 0.52 ± 0.4 | 14.4 ± 5.1 | <10−7 | 14 | ||||||
P < 0.01,
P < 0.05.
χ2 denotes the results of a chi-squared test for curve fitting.
Dynamic conformational transition of aggregated mutant SOD1
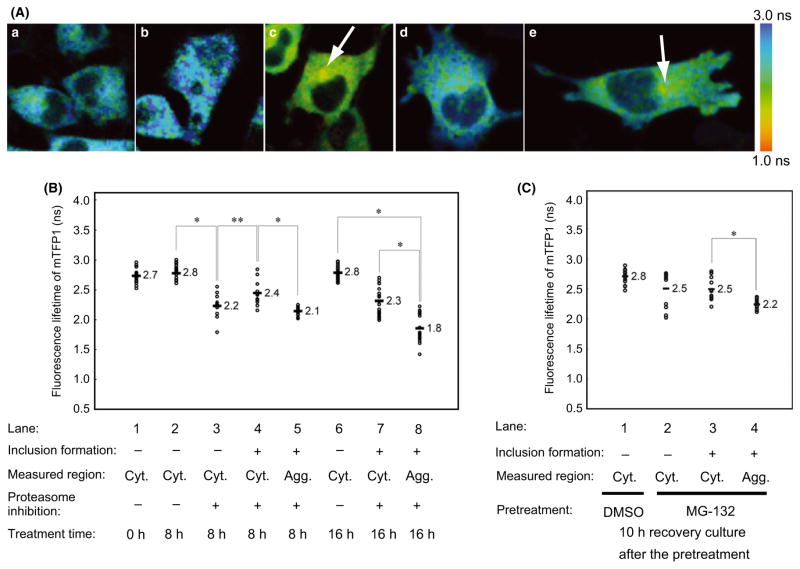
We next examined the structural basis of the mutant SOD1 oligomers using intermolecular Forster resonance energy transfer (FRET), which detects molecular interactions (including distance and orientation) between fluorophores (Lippincott-Schwartz et al. 2001). For quantitative FRET analysis, we used fluorescence lifetime imaging microscopy (FLIM) (Yasuda 2006), in which the fluorescence lifetime of a donor fluorophore (τd) becomes shorter in response to energy transfer. We chose mTFP1 as a donor, because it is a bright monomeric cyan fluorescent protein with a single-component fluorescence lifetime (Ai et al. 2006), and conventional mVenus, a bright monomeric yellow protein, as the acceptor. As previously reported, when both donor and acceptor fluorescent proteins were tagged with SOD1-G85R, no significant FRET was observed, even in the presence of MG-132 (Fig. S7A) (Matsumoto et al. 2006). We hypothesized that the absence of FRET could be due to an inappropriate orientation between the two fluorophores. Therefore, we constructed several circularly permutated mVenus proteins, which exhibit altered rotational orientations between the fluorophores (Nagai et al. 2004), and tested these as acceptors. Among them, one of the mVenus variants, cp173m Venus, exhibited significant FRET (τd = 1.8 ± 0.23 ns) in inclusions (Fig. 5A,C; Fig. 5B, lane 8). By contrast, no significant FRET (τd = 2.8 ± 0.13 ns, Fig. 5A,b; Fig. 5B, lane 6) was observed in the presence of DMSO (used as a control). Likewise, no significant FRET was observed from cells expressing either SOD1-WT-mTFP1 (Fig. S7D) or mTFP1 alone (Fig. S7C). Moreover, the τd of SOD1-G85R-mTFP1 in the cytosol of MG-132-treated cells was significantly shorter (2.4 ± 0.19 ns at 8 h, Fig. 5B, lane 4; and 2.3 ± 0.23 ns at 16 h, Fig. 5B, lane 7) than in the absence of proteasome inhibition (Fig. 5B, lanes 2 and 6). These findings suggest that MG-132 treatment induced oligomer formation in the cytosol. After 8 h of MG-132 treatment, we noticed that τd in the cytosol of inclusion-harboring cells (2.4 ± 0.19 ns, Fig. 5B, lane 4) was significantly longer than in the cytosol of inclusion-free cells (2.2 ± 0.22 ns, Fig. 5B, lane 3). These results suggest that the misfolded mutant SOD1 initially forms ordered cytosolic oligomers that are gradually sequestered into the inclusions.
Figure 5.
FRET-FLIM analysis of mutant SOD1 in inclusions and cytosol during aggregation and disaggregation. (A) Pseudo-color fluorescence lifetime images of cells expressing both SOD1-G85R-mTFP1 and SOD1-G85R-cp173mVenus. White arrows indicate inclusions. Cells were treated for 16 h with no reagents (a), DMSO (b), or MG-132 (c). After MG-132 treatment for 16 h, cells were transferred to the recovery culture and incubated for 10 h (d and e). Images of cells without (d) or with (e) inclusion structures are shown. (B, C) Comparison of the fluorescence lifetime of the FRET donor SOD1-G85R-mTFP1 after MG-132 treatment for 8 h (B, lanes 2–5) or 16 h (B, lanes 6–8), or in recovery culture for 10 h (C, lanes 1–4). Fluorescence lifetime values of individual cells are shown as open circles (n = 10–20), and average values are shown with bars. Significant differences were determined using Student’s t-test: ★P < 0.01, ★★P < 0.05.
Next, we carried out FRET-FLIM analysis during recovery from proteasome inhibition. After MG-132 treatment, cells were transferred to recovery medium and incubated for 10 h. The τd of SOD1-G85R-mTFP1 was shorter in the cytosol in the presence of MG-132 (2.5 ± 0.43 ns for inclusion-free cells, Fig. 5A, d, Fig. 5C, lane 2; 2.5 ± 0.27 ns for inclusion-harboring cells, Fig. 5A, e, Fig. 5C, lanes 3) compared with the DMSO control (2.8 ± 0.15 ns, Fig. 5C, lane 1), suggesting that ordered oligomers persist in the cytosol after disaggregation from inclusions. The τd for SOD1-G85R-mTFP1 in inclusions was significantly longer during recovery (2.2 ± 0.12 ns, Fig. 5C, lane 4) than before recovery (1.8 ± 0.23 ns, Fig. 5B, lane 8). Based on these observations, we propose that mutant SOD1 in inclusions undergoes a molecular reorganization during the recovery of proteasome activity, resulting in the dissociation of mutant SOD1 from the inclusions and the formation of ordered oligomers.
Increased cytotoxicity of mutant SOD1 during the recovery of proteasome activity
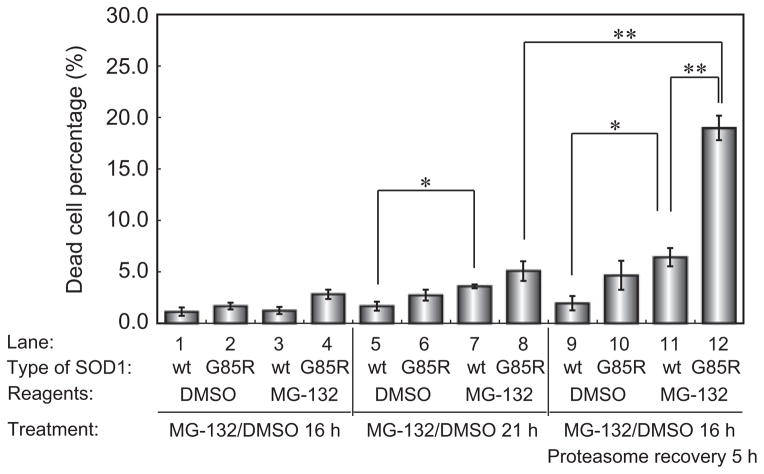
To examine the cytotoxicity of mutant SOD1 during the aggregation and disaggregation phases, we assessed viability in cells expressing SOD1-WT-YFP or SOD1-G85R-YFP (Fig. 6). After 16 h of MG-132 treatment, only 2.8 ± 0.45% and 1.2 ± 0.35% of cells expressing SOD1-G85R-YFP and SOD1-WT-YFP, respectively, were nonviable (lane 4 and lane 3). However, SOD1-G85R-YFP-expressing cells treated with MG-132 and allowed to recover in the absence of MG-132 exhibited a 15-fold increase in cell death (19.0 ± 1.2%, lane 12). This corresponds to a ~4-fold increase in cell death relative to SOD1-G85R-YFP-expressing cells maintained in the proteasome-inhibited state (5.0 ± 0.94%, lane 8) or to cells expressing SOD1-WT-YFP during recovery (6.4 ± 0.89%, lane 11). These results indicated that the cytotoxicity of mutant SOD1 increased sharply during the recovery of proteasome activity, in contrast to the period of treatment with proteasome inhibitor (Fig. 7).
Figure 6.
Cytotoxicity of mutant SOD1 during the disaggregation process. Number of PI-stained cells under the indicated conditions (n = 3). Error bars represent SD. Significant differences were determined using Student’s t-test: ★P < 0.01, ★★P < 0.001.
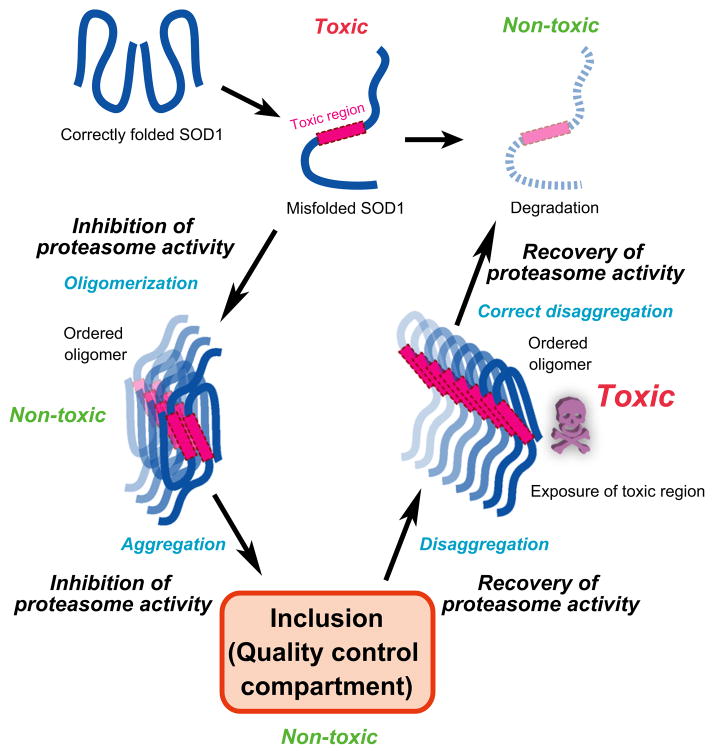
Figure 7.
Model for the conversion among the possible conformational states of mutant SOD1 and the appearance of toxic species.
Discussion
Here, we have shown that the formation of mutant SOD1 aggregates and the toxicity of the mutant protein can be regulated by the modulation of proteasome activity. The appearance of aggregate species and cellular toxicity appear to be inversely related: Under normal conditions of expression, mutant SOD1 is highly aggregation-prone but exhibits very little toxicity. However, when the activity of the proteasome is transiently inhibited, only upon recovery do we observe the appearance of ordered soluble oligomers and associated cellular toxicity.
Over the past years, multiple types of cellular inclusions have been identified in various prokaryotic and eukaryotic organisms, and the biological relevance of inclusion formation has been debated. A recent study reported that misfolded cytosolic proteins are partitioned into either a ‘juxta nuclear quality control compartment (JUNQ)’ or an ‘insoluble protein deposit (IPOD)’ (Kaganovich et al. 2008). Bagola and Sommer pointed out that the classical aggresome structure shares features in common with JUNQ: Both are formed under stress, such as inhibition of the proteasome, although they are distinguished by the presence or absence of a vimentin cage (Bagola & Sommer 2008). The data presented here indicate that following proteasome inhibition, mutant SOD1 is sequestered into inclusions, which correspond to a subcellular compartment for quality control, such as JUNQ. We have also clearly shown that during recovery of proteasomal activity, mutant SOD1 can be dissociated from inclusions as soluble oligomers (Figs. 1–4). These findings suggest the existence of a cytosolic quality control system for misfolded SOD1, which includes disaggregation from a quality control compartment and the degradation of released misfolded proteins. Moreover, FRET-FLIM analysis revealed that the intermolecular orientation of mutant SOD1 in inclusions can be rearranged during disaggregation (Fig. 5B, lane 8; Fig. 5C, lane 4), suggesting that sequestered proteins in the quality control compartment are not likely to be static; instead, molecular rearrangement may slowly occur as a result of modulation of proteasome activity. Molecular chaperones including Hsp70 and Hsp110 cooperate in the disaggregation process of aggregate-prone proteins (Winkler et al. 2012); likewise, several molecular chaperones may be orchestrated and involved in the disaggregation process of mutant SOD1. Therefore, it will be important to elucidate not only the mechanism by which misfolded proteins are sorted into specific compartment, but also the details of the dissociation process.
With respect to the relationship between cytotoxicity and aggregate formation, it has been controversial whether and how aggregation of misfolded protein leads to cellular toxicity. For example, the formation of inclusions containing mutant huntingtin has been hypothesized to improve survival and reduce the level of toxicity in neuronal cells (Arrasate et al. 2004), whereas aggregates of mutant SOD1 are correlated directly with neuronal cell death (Matsumoto et al. 2005). Here, we showed that the cellular toxicity of mutant SOD1 is exerted when proteasomal activity recovers upon removal of a proteasome inhibitor (Fig. 6). There is a likely explanation for these unexpected observations. When mutant SOD1 is misfolded, the toxic region of mutant SOD1 is sequestered through oligomerization; however, during the recovery of proteasome activity, the dissociation of inclusions could expose a toxic interface of oligomers (Fig. 7). Although the average size of mutant SOD1 oligomers and the intermolecular distance of mutant SOD1 within oligomers are equal during inhibition and recovery of proteasome activity (Fig. 3 and 5), the intermolecular orientations of mutant SOD1 in inclusions could be dynamically exchanged between these two states (Fig. 2A; Fig. 5B, lanes 5 and 8; Fig. 5C, lane 4). Furthermore, mutation-dependent structural polymorphism of SOD1 aggregates has been suggested (Furukawa et al. 2010). We therefore hypothesize that the structure of mutant SOD1 aggregates fluctuates and the dynamic exchange in inclusions may cause the production of diverse structure of oligomers. In familial ALS, cell death of motor neurons appears to result from a gain-of-function toxic phenotype of SOD1 (Bruijn et al. 1998; Furukawa et al. 2006; Bosco et al. 2010). Also, many reports demonstrate the relationship between conformational change of SOD1 and cytotoxicity (Lilley & Ploegh 2005; Oda et al. 2006; Nishitoh et al. 2008; VandeVelde et al. 2008; Ilieva et al. 2009; Fujisawa et al. 2012). For example, conformation-dependent binding of mutant SOD1 to Derlin-1 (Lilley & Ploegh 2005; Oda et al. 2006) inhibits endoplasmic reticulum (ER)-associated degradation (ERAD) and thereby generates ER stress (Nishitoh et al. 2008; Ilieva et al. 2009; Fujisawa et al. 2012). Mutant SOD1 is thought to damage mitochondria by being deposited on the cytoplasmic surface of the outer membrane (VandeVelde et al. 2008). Thus, the toxic region(s) of mutant SOD1 oligomers are likely to interact with and inactivate a number of endogenous functional proteins.
Moreover, mutant SOD1 interacts with Hsc70 in the mouse spinal cord during early development, and Hsp110 associates with soluble oligomers of mutant SOD1 during aging, when mutant SOD1 exerts neurotoxicity (Wang et al. 2009). Several groups, including ours, report that the cytosolic chaperonin CCT/TRiC, which inhibits the aggregation of hydrophobic β-sheet-containing proteins (Kubota et al. 2006; Hartl & Hayer-Hartl 2009), reduces polyglutamine toxicity by altering the state of soluble aggregates (Behrends et al. 2006; Kitamura et al. 2006; Tam et al. 2006). CCT/TRiC may inhibit the toxicity of mutant SOD1 during modulation of proteasome activity in the same way. Alternatively, the molecular characteristics of the diverse oligomeric species liberated from mutant SOD1 inclusions may differ with respect to their structures and modifications. Post-translational modifications of mutant SOD1 have been implicated in the transition from monomeric to aggregated states (Fujiwara et al. 2007; Furukawa et al. 2008); consequently, such modifications could also affect the soluble oligomeric structures, which closely correlate with cytotoxicity (Holmberg et al. 2004). For example, in sporadic ALS, aggregation of an overoxidized form of SOD1 damages mitochondria in lymphoblasts (Guareschi et al. 2012). Indeed, oxidization of mutant SOD1 also causes toxicity in motor neurons (Kabashi et al. 2007). These reports imply that post-translational modifications of SOD1 can influence both conformational changes and toxicity.
Thus, we have established that the modulation of proteasome activity induces a conformational change of oligomeric misfolded SOD1, which is closely related to cellular toxicity. Our findings suggest that fluctuation of proteasome activity during aging and/or under transient stress may cause the emergence of toxic species and be a risk factor for neuronal cell death in neurodegenerative disorders. Further detailed analyses of the mechanism by which the modulation of proteasome activity changes conformations of mutant SOD1 in cells will enhance our understanding of neuronal cell death and pathophysiology. Such understanding will assist in the design of therapeutic strategies for ALS caused by SOD1 carrying aggregate-prone mutations and perhaps for other neurodegenerative diseases.
Experimental procedures
Construction of plasmids
Plasmids for producing stable tet-off cell lines that express YFP-tagged wild-type (pTRE-SOD1-WT-YFP) and mutant SOD1 (pTRE-SOD1-G85R-YFP) under the control of doxycycline were described previously (Matsumoto et al. 2005, 2006). To construct a vector for doxycycline-regulated expression of EYFP alone (pTRE-YFP), an NheI-NotI fragment containing EYFP was excised from pEYFP-N1 (Clontech, Mountain View, CA) and subcloned into pTRE2hyg (Clontech). Vectors expressing SOD1 tagged with other fluorescent proteins were constructed as follows. Human wild-type SOD1 (SOD1-WT) or G85R-mutant SOD1 (SOD1-G85R) cDNAs were amplified by PCR using the primers 5′-GCATGAATTCCACCATGGCGACGAAGGCCGTGTGCGTGCTG-3′ and 5′-GCTACCGCGGTTGGGCGATCCCAATTACACCACAAGCC-3′ (Life Technologies, Carlsbad, CA). Amplified DNA fragments were subcloned into pT7Blue (Novagen, San Diego, CA) and subcloned into the EcoRI-SacII site of pEGFP-N1 (Clontech). NheI-NotI fragments encoding SOD1-WT-EGFP or SOD1-G85R-EGFP were subcloned into pTRE2hyg for transient doxycycline-regulated expression in tet-off cell lines. When it was necessary to replace the EGFP sequence with other fluorescent proteins, the AgeI-NotI fragment was replaced with the appropriate sequences. Fluorescent protein sequences used for replacement included mGFP (monomeric A206K variant of EGFP) (Zacharias et al. 2002), mPAGFP (monomeric A206K variant of PAGFP, which can be converted from a dark state to bright green fluorescent state by irradiation with violet light; kindly provided from Dr. Jennifer Lippincott-Schwartz, National Institutes of Health, Bethesda, MD) (Patterson & Lippincott-Schwartz 2002), mTFP1 (a bright monomeric cyan fluorescent protein with a single-component fluorescence lifetime; Allele Biotechnology, San Diego, CA) (Ai et al. 2006), mVenus (a bright monomeric yellow protein; kindly provided from Dr. Atsushi Miyawaki in RIKEN, Japan) (Nagai et al. 2002), cp173mVenus (a circularly permutated mVenus, which exhibits altered rotational orientations between the fluorophores; kindly provided by Dr. Takeharu Nagai, Osaka University, Japan) (Nagai et al. 2004), and TagRFP (a monomeric red fluorescent protein; Evrogen, Moscow, Russia) (Merzlyak et al. 2007), yielding the expression vectors pTRE-SOD1-WT-mGFP, pTRE-SOD1-G85R-mGFP, pTRE-SOD1-G85R-mPAGFP, pTRE-SOD1-WT-mTFP1, pTRE-SOD1-G85R-mTFP1, pTRE-SOD1-WT-cp173mVenus, pTRE-SOD1-G85R-cp173mVenus, pTRE-SOD1-WT-mVenus, pTRE-SOD1-G85R-mVenus, pTRE-SOD1-WT-TagRFP, and pTRE-SOD1-G85R-TagRFP. To avoid translational initiation from an internal ATG codon within the SOD1–fluorescent protein fusions, all methionines (ATG) in the N-terminal regions of the fluorescent proteins were converted to alanines (GCC) using a one-primer quick-change method (Miyawaki et al. 2003) or PCR amplification. For proteasome activity measurements, a proteasomal degron CL1 tag (ACKNWFSSLSHFVIHL) (Bence et al. 2001) was inserted at the carboxyl terminus of EGFP (pEGFP-C1; CLONTECH) to yield the constitutively degraded variant GFPu.
Cell culture and establishment of stable cell lines
HeLa tet-off cell lines (Clontech), in which expression from pTRE vectors is regulated by doxycycline, were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum (FBS; J R Scientific, Woodland, CA), 200 μg/ml G418 (Nacalai Tesque, Kyoto, Japan), 100 units/ml penicillin G (Sigma-Aldrich), and 100 μg/ml streptomycin (Sigma-Aldrich) in a humidified 5% CO2/95% air atmosphere at 37°C. HeLa tet-off cell lines for doxycycline-regulated expression of YFP, SOD1-WT-YFP, or SOD1-G85R-YFP were selected in medium supplemented with 500 μg/ml HygroGold (Invivo-Gen, San Diego, CA) and 1.0 μg/ml doxycycline (Sigma-Aldrich). For transfections, cells were seeded on 3.5-cm dishes (BD, Franklin Lakes, NJ) or 3.5-cm glass-based dishes (Asahi-Technoglass, Tokyo, Japan) 1 day before transfection. All constructs were transfected using Effectene (Qiagen, Dusseldorf, Germany). For photoactivation analysis, mixtures of pTRE-SOD1-G85R-mPAGFP and pTRE-SOD1-G85R-TagRFP constructs (1:1) were transfected into HeLa tet-off cells. For FRET-FLIM assays, pTRE-SOD1-G85R-cp173mVenus or the control pTRE-SOD1-G85R-mVenus was cotransfected with pTRE-SOD1-G85R-mTFP1 at a ratio of 3:1.
Proteasome inhibitor treatment and recovery of proteasome activity
To induce SOD1 expression, cells were washed with HBSS (Sigma-Aldrich) and cultured for up to 16 h in medium lacking doxycycline and supplemented with 2 μM MG-132 (Peptide Institute, Osaka, Japan) to inhibit proteasome activity. As a negative control for proteasome inhibition, 0.02% DMSO (Nacalai Tesque) was added instead of MG-132. Cells were then washed three times with HBSS and cultured in medium lacking MG-132, to recover proteasome activity, but supplemented with 1.0 μg/ml doxycycline, to inhibit transcription of SOD1 genes. Treatment with 50 nM epoxomicin (Sigma-Aldrich) was carried out in the same manner as treatment with MG-132. Inhibition of autophagy and lysosomal protein degradation was carried out in medium supplemented with 0.1 μM bafilomycin A1 (Sigma-Aldrich) after 16-h treatment in medium supplemented with 2 μM MG-132.
Immunofluorescence analysis
Cells were cultivated on acid-washed and type I collagen-coated coverslips (0.14–0.18 mm) (Matsumami Glass Ind., Ltd., Osaka, Japan) and then fixed with 4% paraformaldehyde at 37°C (for staining for ubiquitin, Hsp70, Hsc70, vimentin, and the 20S proteasome) or in ice-cold methanol at −20°C (for α-tubulin staining). Cells were washed with TBS and permeabilized in the presence of 0.5% (v/v) Triton X-100 (Sigma-Aldrich) and 0.5% (w/v) saponin (Nacalai Tesque). After blocking nonspecific binding activity in blocking buffer containing 5% normal goat serum (DAKO, Glostrup, Denmark) and 0.02% (v/v) Triton X-100 in PBS, cells were incubated for 1 h at room temperature in blocking buffer supplemented with primary antibodies against the following proteins: ubiquitin (Z0458, DAKO), Hsc70 (SPA815, Stressgen, Ann Arbor, MI), Hsp70 (SPA810, Stressgen), α-tubulin (DM1A, Cedarlane Laboratories, Ontario, Canada), 20S proteasome (PW8155, Biomol, Butler Pike Plymouth Meeting, PA), or vimentin (V6630, Sigma-Aldrich). Cells were then incubated with anti-mouse, anti-rabbit, or anti-rat IgG conjugated with Alexa Fluor 647 (Life Technologies) in blocking buffer for 1 h at room temperature. Subsequently, cells were incubated with anti-GFP antibody conjugated with Alexa Fluor 488 (A21311, Life Technologies) as a fluorescence enhancer for the EYFP tag. Cells stained on coverslips were mounted with ProLong Gold (Life Technologies), and images were captured on an LSM 510 META microscope (Carl Zeiss, Jena, Germany) equipped with a Plan-Apochromat 63 × /1.4 NA DIC oil-immersion objective. Alexa Fluor 488 was excited at 488-nm with an Ar+ gas laser, and Alexa Fluor 647 was excited at 633 nm with a He-Ne gas laser. The captured images were processed with Photoshop 6.0J (Adobe Systems, Tokyo, Japan).
Cell lysis and Western blotting
Cells were lysed in lysis buffer (50 mM HEPES/KOH [pH 7.4], 150 mM NaCl, 1% (v/v) Triton X-100, 5 mM EDTA, and 1% (v/v) protease-inhibitor cocktail [Sigma-Aldrich]). After centrifugation (15 000 g, 15 min, 4°C), the supernatant and pellet were recovered. The supernatant protein concentration was determined using the Bradford Ultra reagent (Novexin Ltd, Cambridge, UK), and concentrations were adjusted by dilution. Pellets were washed with PBS and solubilized by sonication in lysis buffer containing 1% (w/v) SDS. Supernatant and pellet samples were boiled in SDS-PAGE sample buffer containing 25 mM dithiothreitol and then separated by SDS-PAGE using a 12.5% ePAGEL gel (Atto, Tokyo, Japan). Proteins were transferred onto Hybond-P PVDF membranes (GE Healthcare), and membranes were blocked in PBS containing 5% (w/v) skim milk and 0.05% (v/v) Tween 20. After incubation with anti-GFP antibody (GF200, Nacalai Tesque, for detection of SOD1 tagged with YFP, mGFP, or mPAGFP) or anti-GAPDH antibody (6C5, HyTest Ltd., Turku, Finland), membranes were incubated with anti-mouse or anti-rabbit IgG conjugated with alkaline phosphatase. Specific binding of antibodies was visualized using nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl phosphate toluidine salt solution (Sigma-Aldrich) as a chromogenic substrate.
Filter-trap assay
Cells expressing SOD1-YFPs were suspended in PBS containing 1% protease-inhibitor cocktail and lysed by freeze-thawing. After determination of protein concentrations, cell lysates (300 μg protein) were diluted in PBS containing 1% SDS and sonicated. Samples were loaded onto a cellulose-acetate membrane (0.2 μm pore size; Advantec Toyo, Ltd., Tokyo, Japan) on a Bio-Dot SF vacuum blotter (Bio-Rad Laboratories, Hercules, CA). Trapped SOD1-YFP aggregates were detected by Western blotting using anti-GFP antibody.
Photoactivation analysis in living cells
Cells expressing both SOD1-G85R-mPAGFP and SOD1-G85R-TagRFP were cultured in a glass-based dish with printed grid patterns (Asahi-Technoglass). Cells were observed on a LSM 510 META microscope using a C-Apochromat 40 × /1.2NA UV-VIS-IR DIC water-immersion objective. After a region of interest (ROI) was determined using the red channel, mPAGFPs in the ROI were photoactivated by 30 iterations (0.45 s) of irradiation with a 405-nm diode laser at 15% power. Fluorescence images of activated mPAGFP in living cells were captured 0, 3, or 6 h after photoactivation, and fluorescence intensities were determined using ImageJ ver. 1.41o (NIH).
Fluorescence correlation spectroscopy (FCS) analysis
Fluorescence correlation spectroscopy measurements (Rigler et al. 1993; Kitamura et al. 2006) were taken on a ConfoCor 2 system and C-Apochromat 40 × /1.2NA UV-VIS-IR Korr. water-immersion objective (Carl Zeiss). EGFP and EYFP were excited at 488 nm and 514 nm, respectively. Confocal pinhole diameter was adjusted to 70 μm at 488 nm or 74 μm at 514 nm. Emission signals were detected with a 505-nm long-pass filter for EGFP or a 530–600-nm bandpass filter for EYFP. Cells were suspended in 0.2 ml PBS supplemented with 1% protease-inhibitor cocktail (Sigma) and lysed by passage through a 27-gauge needle. The supernatant was recovered after centrifugation (1000 g, 2 min) and diluted appropriately. Supernatant fluorescence signals were recorded using Lab-Tek 8-well chamber slides (NUNC, Rochester, NY) at 25°C. For live-cell analysis, cells were cultured on glass-based 3.5-cm dishes (Asahi-Technoglass) in phenol red-free medium (Life Technologies) supplemented with 25 mM HEPES/NaOH (pH 7.4) and 10% FBS and measured at 37 °C in a 5% CO2/95% air-humidified atmosphere.
The fluorescence autocorrelation functions, G(τ), from which the average residence time (τ) and the absolute number of fluorescent proteins in the detection volume were calculated, were obtained as follows:
| (1) |
where I(t + τ) is the fluorescence intensity obtained by the single-photon counting method in a detection volume at a delay time τ (angular brackets denote ensemble averages). Curve fitting for the multicomponent model is given by:
| (2) |
where Fi and τi are the fraction and diffusion time of component i, respectively; N is the average number of fluorescent molecules in the detection volume defined by the beam waist w0 and the axial radius z0; s is the structure parameter representing the ratio of w0 and z0; T is the triplet fraction; and τt is the relaxation time of the triplet state. G(τ)s in aqueous solutions were measured twenty times for 15 s, whereas G(τ)s in live cells were measured ten times for 15 s. After pinhole adjustment, diffusion time and structure parameter were determined using a 10−7 M rhodamine 6G (Rh6G) solution as a standard before measurements. The values of structural parameters were 5.0–10. The diffusion coefficients of fluorescent molecules (Dsample) were calculated from the published diffusion coefficient of Rh6G, DRh6G (280 μm2/s), and the measured diffusion times of Rh6G under the condition (τRh6G) and probe proteins (τsample) as follows:
| (3) |
For the diffusion coefficients, we applied the Stokes–Einstein equation:
| (4) |
where the Boltzmann constant k, the viscosity of the solution η, and temperature T all remain constant throughout our studies. To yield a relationship between the diffusion coefficient and the Stokes’ radius of the molecule, , we assumed that the molecule in question is a sphere with a Stokes’ radius rH and a volume proportional to its molecular weight. Thus, we can relate the diffusion coefficient to molecular weight with D ∝ M−1/3. Count per molecule (CPM) was determined as mean brightness of measured sample divided by the number of molecules determined by FCS analysis. CPM ratio was calculated as a CPM value of the sample in addition of MG-132 divided by that in addition of DMSO as a control.
Polyubiquitination analysis
Supernatants of cell lysates were prepared in the same procedure as FCS analysis. The concentration of cell lysate was adjusted to 600 μg followed by determination of protein concentration using Bradford Ultra reagent (Novexin Ltd.). Supernatants of cell lysate supplemented with 1% TritonX-100 were incubated with rat monoclonal anti-GFP antibody-conjugated agarose beads (D153-8, MBL, Nagoya, Japan) for 1 h at 4 °C. After washing the beads three times with PBS supplemented with 1% TritonX-100, precipitated proteins were solubilized in Laemmli sample buffer. Protein samples were loaded in a 5–20% gradient gel (ePAGEL) and then transferred on Hybond-P PVDF membranes (GE healthcare) in a mini-transblot cell (BioRad). To detect ubiquitin, SOD1-YFP, and GAPDH, rabbit polyclonal anti-ubiquitin antibody (Z0458, Dako), mouse monoclonal anti-GFP antibody (GF200, Nacalai), and mouse monoclonal anti-GAPDH antibody (6C5, HyTest) were used, respectively. After incubation with horseradish peroxidase-conjugated anti-immunoglobulin antibodies as a secondary antibody, membranes were treated with ECL plus reagent (GE Healthcare). Chemiluminescent signals were detected in LAS4000 (Fujifilm, Tokyo, Japan).
FRET-FLIM analysis
Cells were fixed in 4% paraformaldehyde buffered with 100 mM HEPES-KOH (pH 7.5) for 30 min at 37 °C. After washing four times with TBS, cells were mounted in TBS, and FLIM measurements were taken using a combination of laser scanning microscopy and the time-correlated single-photon counting (TCSPC) principle. A SP5 confocal laser scanning unit (Leica, Wetzar, Germany) was connected to an inverted microscope DMI6000 (Leica). Fluorescence signals were corrected using a C-Apochromat 63 × /1.2 NA UV-VIS-IR Korr. water-immersion objective (Carl Zeiss) through an RMSA1 mount adaptor (Thorlabs, Newton, NJ). mTFP1 was excited with a 405-nm pulse laser (Pico-Quant, Berlin, Germany), and fluorescence signals were separated through a 440–530-nm band-pass slit and detected in a photomultiplier tube (PMT) for single-photon counting. TCSPC was carried out on an SPC-830 PCI slot board (Becker and Hickl, Berlin, Germany) in a PC/AT-compatible PC (DELL, Round Rock, TX) under the control of the SPCM software (Becker and Hickl) in Microsoft Windows XP SP3. Raw data obtained from the SPCM were exported to the SPCImage analysis software (Becker and Hickl).
Fluorescence lifetime measurements typically involve donor fluorescence only. The time course of donor fluorescence I(t), after a short pulse of excitation, is obtained as follows:
| (5) |
where τd is the fluorescence lifetime of the donor. When FRET occurs, the lifetime of the donor excited state is shortened.
Sucrose density-gradient fractionation
Supernatants of cell lysates containing SOD1-G85R-YFP were prepared as for FCS measurements. Lysates were applied to 10–60% sucrose gradients containing 25 mM HEPES/KOH (pH 7.5). Gradients were centrifuged at 157 000 g for 16 h in a SW41 swing rotor (Beckman Coulter, Indianapolis, IN) at 4 °C. Laemmli sample buffer was added to each fraction, and the amount of SOD1-G85R-YFP was detected by SDS-PAGE followed by Western blotting using anti-GFP antibody (GF200, Nacalai).
Dead-cell analysis
Cells were cultured on glass-based dishes in phenol red-free DMEM supplemented with 25 mM HEPES and 10% FBS. Dead cells were stained with 1.0 μg/ml propidium iodide (PI) solution (Life Technologies) for 5 min. Images of YFP and PI channels were captured with an LSM 510 META microscope through a Plan-Neofluar 10 × /0.3NA objective at 37 °C. Pinhole size was opened to maximum. Numbers of YFP- and PI-positive cells were counted using ImageJ, and the percentages of dead cells were determined from the number of PI-positive cells divided by the number of YFP-positive cells.
Supplementary Material
Acknowledgments
We thank the Nagata and Kinjo lab members for helpful discussions; Y. Fukuda, Y. Moriyama, and M. Uchida for technical assistance; Y. Ishida, C.G. Pack, M. Iwai, K. Yanagitani, and I. Wada for kind suggestions; and J. Lippincott-Schwartz, H. Fujii, T. Nagai, and A. Miyawaki for providing the constructs for expression of fluorescent proteins. A.K. was supported by a fellowship (204474) from the Japan Society for Promotion of Science (JSPS), supported by Grants-in-Aid for Scientific Research for Young Scientists (23770215) from JSPS. N.I. was supported by Grants-in-Aid for Scientific Research for a Plant Graduate Student from NAIST, supported by the Ministry of Education, Culture, Sports, Science, and Technology (MEXT). H.K., G.M., M.K., and K.N. were supported by Grants-in-Aid for Creative Scientific Research (19G50314) and for Scientific Research (19058008). R.I.M. was supported by grants from the National Institutes of Health (NIGMS, NIA, and NINDS) and the HDSA Coalition for the Cure.
Footnotes
Additional Supporting Information may be found in the online version of this article at the publisher’s web site:
Figure S1 Inclusion formation by YFP-tagged mutant SOD1 in HeLa cells under proteasome-inhibiting conditions.
Figure S2 Recovery of proteasome activity.
Figure S3 Expression and solubility of SOD1 tagged with mPAGFP (A) or mGFP (B).
Figure S4 Fluorescence correlation spectroscopy (FCS) analysis of soluble mutant SOD1 oligomers during treatment with MG-132 for the indicated periods (A), followed by transfer to recovery media for the indicated periods (B).
Figure S5 Sucrose density-gradient fractionation of cell lysates containing SOD1-G85R-YFP in the presence or absence of 2 μM MG-132.
Figure S6 Poly-ubiquitination analysis of SOD1-WT-YFP and SOD1-G85R-YFP in the presence or absence of 2 μM MG-132.
Figure S7 Fluorescence lifetime of FRET donors in FRET-FLIM analysis of controls.
References
- Ai HW, Henderson JN, Remington SJ, Campbell RE. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem J. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Bagola K, Sommer T. Protein Quality Control: on IPODs and Other JUNQ. Curr Biol. 2008;18:R1020. doi: 10.1016/j.cub.2008.09.036. [DOI] [PubMed] [Google Scholar]
- Behrends C, Langer CA, Boteva R, Bottcher UM, Stemp MJ, Schaffar G, Rao BV, Giese A, Kretzschmar H, Siegers K, Hartl FU. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell. 2006;23:887–897. doi: 10.1016/j.molcel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Bosco DA, Morfini G, Karabacak NM, et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat Neurosci. 2010;13:1396–1403. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruijn LI, Houseweart MK, Kato S, Anderson KL, Anderson SD, Ohama E, Reaume AG, Scott RW, Cleveland DW. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281:1851–1854. doi: 10.1126/science.281.5384.1851. [DOI] [PubMed] [Google Scholar]
- Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat Rev Neurosci. 2001;2:806–819. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- Da Cruz S, Cleveland DW. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol. 2011;21:904–919. doi: 10.1016/j.conb.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa T, Homma K, Yamaguchi N, Kadowaki H, Tsuburaya N, Naguro I, Matsuzawa A, Takeda K, Takahashi Y, Goto J, Tsuji S, Nishitoh H, Ichijo H. A novel monoclonal antibody reveals a conformational alteration shared by amyotrophic lateral sclerosis-linked SOD1 mutants. Ann Neurol. 2012;72:739–749. doi: 10.1002/ana.23668. [DOI] [PubMed] [Google Scholar]
- Fujiwara N, Nakano M, Kato S, Yoshihara D, Ookawara T, Eguchi H, Taniguchi N, Suzuki K. Oxidative modification to cysteine sulfonic acid of Cys111 in human copper-zinc superoxide dismutase. J Biol Chem. 2007;282:35933–35944. doi: 10.1074/jbc.M702941200. [DOI] [PubMed] [Google Scholar]
- Furukawa Y, Fu R, Deng HX, Siddique T, O’Halloran TV. Disulfide cross-linked protein represents a significant fraction of ALS-associated Cu, Zn-superoxide dismutase aggregates in spinal cords of model mice. Proc Natl Acad Sci USA. 2006;103:7148–7153. doi: 10.1073/pnas.0602048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa Y, Kaneko K, Yamanaka K, Nukina N. Mutation-dependent polymorphism of Cu, Zn-Superoxide dismutase aggregates in the familial form of amyotrophic lateral sclerosis. J Biol Chem. 2010;285:22221–22231. doi: 10.1074/jbc.M110.113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa Y, Kaneko K, Yamanaka K, O’Halloran TV, Nukina N. Complete loss of post-translational modifications triggers fibrillar aggregation of SOD1 in the familial form of amyotrophic lateral sclerosis. J Biol Chem. 2008;283:24167–24176. doi: 10.1074/jbc.M802083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- Gidalevitz T, Kikis EA, Morimoto RI. A cellular perspective on conformational disease: the role of genetic background and proteostasis networks. Curr Opin Struct Biol. 2010;20:23–32. doi: 10.1016/j.sbi.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T, Krupinski T, Garcia S, Morimoto RI. Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS Genet. 2009;5:e1000399. doi: 10.1371/journal.pgen.1000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guareschi S, Cova E, Cereda C, Ceroni M, Donetti E, Bosco DA, Trotti D, Pasinelli P. An over-oxidized form of superoxide dismutase found in sporadic amyotrophic lateral sclerosis with bulbar onset shares a toxic mechanism with mutant SOD1. Proc Natl Acad Sci USA. 2012;109:5074–5079. doi: 10.1073/pnas.1115402109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 2009;16:574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- Holmberg CI, Staniszewski KE, Mensah KN, Matouschek A, Morimoto RI. Inefficient degradation of truncated polyglutamine proteins by the proteasome. EMBO J. 2004;23:4307–4318. doi: 10.1038/sj.emboj.7600426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilieva H, Polymenidoe M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. 2009;187:761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JA, Dalton MJ, Gurney ME, Kopito RR. Formation of high molecular weight complexes of mutant Cu, Zn-superoxide dismutase in a mouse model for familial amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2000;97:12571–12576. doi: 10.1073/pnas.220417997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Rouleau GA. Oxidized/misfolded superoxide dismutase-1: the cause of all amyotrophic lateral sclerosis? Ann Neurol. 2007;62:553–559. doi: 10.1002/ana.21319. [DOI] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch CM, Borchelt DR. A limited role for disulfide cross-linking in the aggregation of mutant SOD1 linked to familial amyotrophic lateral sclerosis. J Biol Chem. 2008;283:13528–13537. doi: 10.1074/jbc.M800564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura A, Kubota H, Pack CG, Matsumoto G, Hirayama S, Takahashi Y, Kimura H, Kinjo M, Morimoto RI, Nagata K. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat Cell Biol. 2006;8:1163–1170. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- Kubota S, Kubota H, Nagata K. Cytosolic chaperonin protects folding intermediates of Gb from aggregation by recognizing hydrophobic beta-strands. Proc Natl Acad Sci USA. 2006;103:8360–8365. doi: 10.1073/pnas.0600195103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL. Multi protein complexes that link dislocation, ubiquitination, and extraction of mis-folded proteins from the endoplasmic reticulum membrane. Proc Natl Acad Sci USA. 2005;102:14296–14301. doi: 10.1073/pnas.0505014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Patterson GH. Development and use of fluorescent protein markers in living cells. Science. 2003;300:87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- Matsumoto G, Kim S, Morimoto RI. Huntingtin and mutant SOD1 form aggregate structures with distinct molecular properties in human cells. J Biol Chem. 2006;281:4477–4485. doi: 10.1074/jbc.M509201200. [DOI] [PubMed] [Google Scholar]
- Matsumoto G, Stojanovic A, Holmberg CI, Kim S, Morimoto RI. Structural properties and neuronal toxicity of amyotrophic lateral sclerosis-associated Cu/Zn superoxide dismutase 1 aggregates. J Cell Biol. 2005;171:75–85. doi: 10.1083/jcb.200504050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, Fradkov AF, Gaintzeva A, Lukyanov KA, Lukyanov S, Gadella TW, Chudakov DM. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- Miyawaki A, Mizuno H, Nagai T, Sawano A. Development of genetically encoded fluorescent indicators for calcium. Methods Enzymol. 2003;360:202–225. doi: 10.1016/s0076-6879(03)60111-4. [DOI] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci USA. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitoh H, Kadowaki H, Nagai A, Maruyama T, Yokota T, Fukutomi H, Noguchi T, Matsuzawa A, Takeda K, Ichijo H. ALS-linked mutant SOD1 induces ER stress- and ASK1-dependent motor neuron death by targeting Derlin-1. Genes Dev. 2008;22:1451–1464. doi: 10.1101/gad.1640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa J, Yamada S, Ishigaki S, Sone J, Takahashi M, Katsuno M, Tanaka F, Doyu M, Sobue G. Disulfide bond mediates aggregation, toxicity, and ubiquitylation of familial amyotrophic lateral sclerosis-linked mutant SOD1. J Biol Chem. 2007;282:28087–28095. doi: 10.1074/jbc.M704465200. [DOI] [PubMed] [Google Scholar]
- Oda Y, Okada T, Yoshida H, Kaufman RJ, Nagata K, Mori K. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J Cell Biol. 2006;172:383–393. doi: 10.1083/jcb.200507057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144:67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- Rigler R, Mets U, Widengren J, Kask P. Fluorescence correlation spectroscopy with high count rate and low-background analysis of translational diffusion. Eur Biophys J Biophys Lett. 1993;22:169–175. [Google Scholar]
- Rosen DR, Siddique T, Patterson D, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Tam S, Geller R, Spiess C, Frydman J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat Cell Biol. 2006;8:1155–1162. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabulas RM, Hartl FU. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science. 2005;310:1960–1963. doi: 10.1126/science.1121925. [DOI] [PubMed] [Google Scholar]
- VandeVelde C, Miller TM, Cashman NR, Cleveland DW. Selective association of misfolded ALS-linked mutant SOD1 with the cytoplasmic face of mitochondria. Proc Natl Acad Sci USA. 2008;105:4022–4027. doi: 10.1073/pnas.0712209105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, Rodrigues APC, Manning G, Dillin A. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489:263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- Wang J, Farr GW, Zeiss CJ, Rodriguez-Gil DJ, Wilson JH, Furtak K, Rutkowski D, Kaufman RJ, Ruse CI, Yates JR, Perrin S, Feany MB, Horwich AL. Progressive aggregation despite chaperone associations of a mutant SOD1-YFP in transgenic mice that develop ALS. Proc Natl Acad Sci USA. 2009;106:1392–1397. doi: 10.1073/pnas.0813045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson TL, Corson LB, Huang L, Burlingame A, Liu J, Bruijn LI, Cleveland DW. Toxicity of ALS-linked SOD1 mutants. Science. 2000;288:399. doi: 10.1126/science.288.5465.399a. [DOI] [PubMed] [Google Scholar]
- Winkler J, Tyedmers J, Bukau B, Mogk A. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J Cell Biol. 2012;198:387–404. doi: 10.1083/jcb.201201074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda R. Imaging spatiotemporal dynamics of neuronal signaling using fluorescence resonance energy transfer and fluorescence lifetime imaging microscopy. Curr Opin Neurobiol. 2006;16:551–561. doi: 10.1016/j.conb.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.