Abstract
Summary: We present Vacceed, a highly configurable and scalable framework designed to automate the process of high-throughput in silico vaccine candidate discovery for eukaryotic pathogens. Given thousands of protein sequences from the target pathogen as input, the main output is a ranked list of protein candidates determined by a set of machine learning algorithms. Vacceed has the potential to save time and money by reducing the number of false candidates allocated for laboratory validation. Vacceed, if required, can also predict protein sequences from the pathogen’s genome.
Availability and implementation: Vacceed is tested on Linux and can be freely downloaded from https://github.com/sgoodswe/vacceed/releases (includes a worked example with sample data). Vacceed User Guide can be obtained from https://github.com/sgoodswe/vacceed.
Contact: John.Ellis@uts.edu.au
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Several subunit vaccines against prokaryotic pathogens have been identified (Ariel et al., 2002; Montigiani et al., 2002; Ross et al., 2001) using reverse vaccinology (Rappuoli, 2000). Vaxign (He et al., 2010) and NERVE (Vivona et al., 2006) are examples of vaccine discovery tools for prokaryotes, but there is currently no equivalent tool for eukaryotes. Freely available bioinformatics tools and an unprecedented volume of –omics data now present an opportunity for in silico vaccine discovery for eukaryotic pathogens. A general approach is to use several tools to predict and gather evidence for protein characteristics. From this potential evidence, the researcher makes an informed decision as to a protein’s vaccine candidacy suitability. Determining which tools are appropriate, as well as how to use them, presents the first of many challenges. A further challenge, especially to a researcher with limited programming ability, is to extract and gather the pertinent evidence distributed within large-scale outputs. The subsequent and more imposing challenge is that the evidence is mainly in different formats, contradicting and inaccurate. Poor evidence reliability arises because some of the input data to the tools (e.g. protein sequences and training data) are inaccurate or missing. Moreover, tools used to predict protein characteristics are, in general, inaccurate.
Vaccine candidates identified in silico can only be validated in a laboratory. Validation should provide feedback to inform and improve vaccine candidacy decision making. The repetitive nature for this ideal in silico approach is in need of automation. Furthermore, an automated process must accommodate an ever-increasing choice of new or improved prediction programs that inevitably replace existing ones.
We have developed Vacceed to address the challenges raised here i.e. to provide a flexible, automated process to predict worthy vaccine candidates from large volumes of superfluous, disseminated and noisy data. Vacceed is the collective name for a framework of linked bioinformatics programs, Perl scripts, R functions and Linux shell scripts. A previous published study provided guidance in development (Goodswen et al., 2013b).
2 DISTINCTIVE FEATURES
A detailed description of all aspects of Vacceed is provided in a comprehensive user guide provided as Supplementary Information. The focus of this article is to introduce Vacceed via a selection of distinctive features.
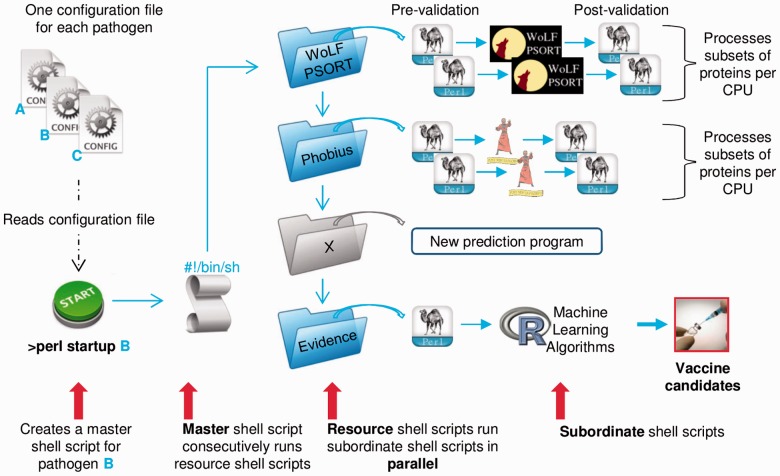
The Vacceed framework is built around the concept of linked resources (see Fig. 1). Each resource, in this context, is built from a central Linux shell script encapsulating all programs needed to perform specific but related tasks. Typical tasks include predicting a particular protein characteristic as well as pre- and post-validation. A resource can be executed as an independent modular unit. This flexible design allows for scalability and easy maintenance. Any prediction program can be integrated within an existing or new resource if it meets the following criteria: runs in a Linux environment, has high-throughput capability, is applicable to eukaryotes, can be trained or has trained data specific to target pathogen and provides consistent text output. From a user’s perspective, all the work involved in the complexity of linking tasks and resources into a seamless continuous pipeline has already been resolved in Vacceed. The only time a user must be concerned with the contents of a resource is when adding a new one. There is a template resource script and generic Perl scripts to ease this process.
Fig. 1.
Vacceed framework. A set hierarchal structure exists for the execution of all Vacceed scripts e.g. startup → master script → resource script → subordinate script (only three resources are shown to maintain clarity)
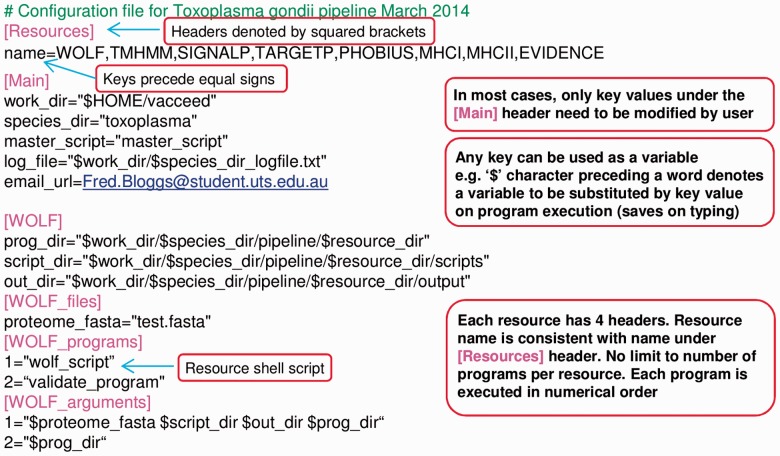
Core to Vacceed are user-definable configuration files (see Fig. 2). These files are in effect the user’s interface to configuring each resource, if desired, and consequently controlling the outcome of the entire pipeline. For example, by altering names in a list, the user can determine the resources to be run and their order. The expectation is to have one configuration file for each target pathogen. The command-line syntax to invoke Vacceed is ‘perl startup xx’, where xx determines the appropriate configuration file. Specifying a code allows for multiple instances of Vacceed to process different species or resource combinations. No other user input is needed. An e-mail with attached log file is sent on successful completion or immediately following an error.
Fig. 2.
Extract of a Vacceed configuration file defined by a header-key format (only one resource, WoLF PSORT, is shown for brevity)
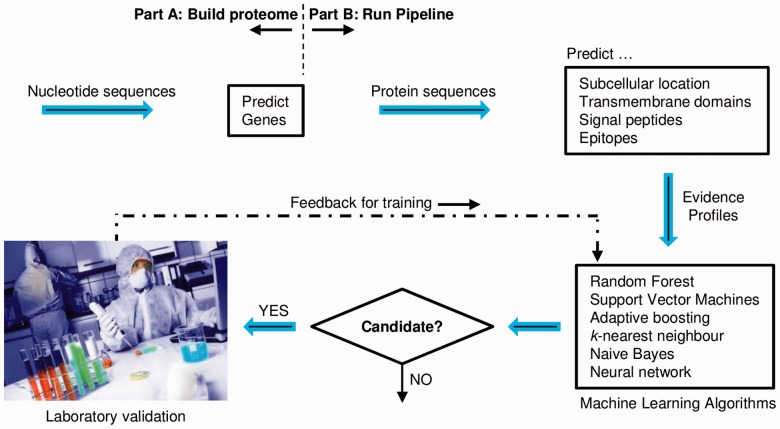
The framework is organized into two major parts referred henceforth as part A—build proteome, and part B—run pipeline (see Fig. 3). Table 1 lists the programs currently integrated in each part. A starting prerequisite for part B is a file containing amino acid sequences for proteins from the target eukaryotic pathogen i.e. the proteome. Known protein sequences for many pathogens can be downloaded from public databases. Part A is used, only if required, to predict novel protein sequences and/or collect evidence to support the existence of known proteins. Part A resources typically predict genes, which is one among multiple tasks within linked resources involved in building the proteome. Examples of other tasks are validating gene start and end sequences (e.g. ATG, TAA, TAG or TGA), predicting exon locations relative to gene start, converting predictions to amino acid sequences and homology searching.
Fig. 3.
Schematic of data flow in Vacceed
Table 1.
Programs currently integrated in Vacceed
| Name | Function | URL (last viewed May 2014) |
|---|---|---|
| Part A—Build proteome | ||
| Augustus | Ab initio gene predictor | http://bioinf.uni-greifswald.de/augustus |
| GlimmerHMM | Ab initio gene predictor | http://ccb.jhu.edu/software/glimmerhmm |
| BLAT | Aligns expressed sequence tags (ESTs) to DNA | http://genome.ucsc.edu/FAQ/FAQblat.html |
| GMAP | Aligns expressed sequence tags (ESTs) to DNA | http://research-pub.gene.com/gmap |
| N-Scan | Ab initio gene predictor supported by genome comparison | http://mblab.wustl.edu/software.html |
| BLASTN | Finds regions of similarity between nucleotide sequences | http://www.ncbi.nlm.nih.gov |
| BLASTP | Finds regions of similarity between protein sequences | http://www.ncbi.nlm.nih.gov |
| Part B—Run pipeline (vaccine candidate discovery) | ||
| WoLf PSORT | Protein subcellular localization prediction | http://wolfpsort.seq.cbrc.jp |
| SignalP | Predicts presence and location of signal peptide cleavage sites | http://www.cbs.dtu.dk/services/SignalP |
| TargetP | Protein subcellular localization prediction | http://www.cbs.dtu.dk/services/TargetP |
| Phobius | Combined transmembrane topology and signal peptide predictor | http://phobius.binf.ku.dk/instructions.html |
| TMHMM | Prediction of transmembrane helices in proteins | http://www.cbs.dtu.dk/services/TMHMM |
| MHC I-binding | Peptide binding to MHC class I molecules | http://tools.immuneepitope.org/mhci/download |
| MHC II-binding | Peptide binding to MHC class II molecules | http://tools.immuneepitope.org/mhcii/download |
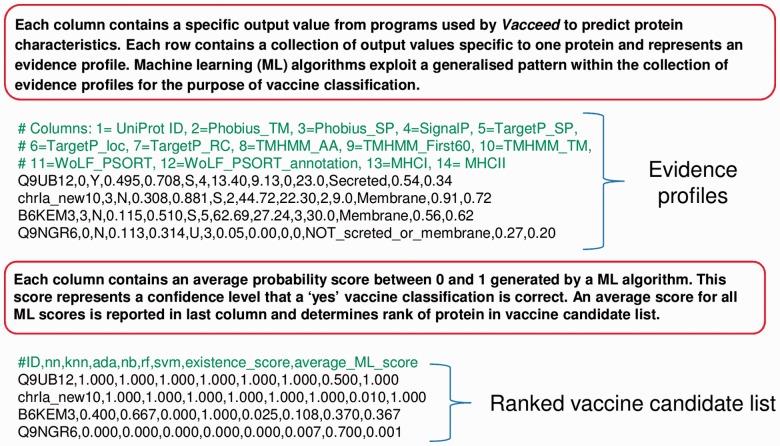
Part B resources predict protein characteristics. One resource called ‘Evidence’, however, parses output files and collates relevant protein characteristics (referred henceforth as an evidence profile). A typical profile is a mixture of data types corresponding to an accuracy measure or score for the predicted characteristic (see Fig. 4). A crucial feature of the resource is a set of supervised machine learning algorithms for binary classification executed via Rscript. The ensemble of classifiers constitutes the heart of Vacceed’s decision making. The main output is a ranked vaccine candidate list of all proteins in the target pathogen based on an average probability of individual classifier predictions (see Fig. 4). Machine learning algorithms are the key to overcoming the challenge that an unknown percentage of evidence is questionable in each profile.
Fig. 4.
Examples of evidence profiles and a ranked vaccine candidate list (only four proteins out of potentially thousands constituting the target pathogen are shown for brevity)
Resources encapsulate, for the most part, a large number of independent computation-intensive tasks. Vacceed takes advantage of multi-core processors. Part A processes one chromosome per CPU in parallel. Chromosomes are queued if there are more chromosomes than CPUs. The user, however, can specify the number of chromosomes to process in parallel. Part B internally splits the proteins by the number of CPUs and processes each subset in parallel. Alternatively, the user can specify the split value.
Proof of concept: There is no program yet to evaluate in silico vaccine candidates in a host–vaccine interaction. The best interim option is to validate the in silico process by predicting candidates using experimentally validated proteins with known immunogenicity characteristics i.e. compare predicted with expected to determine sensitivity and specificity of the process. Using a mixed dataset of 140 published proteins observed to induce or not induce immune responses, we demonstrated in an earlier study (Goodswen et al., 2013a) Vacceed’s decision making potential by effectively distinguishing expected true from expected false vaccine candidates, with an average sensitivity and specificity of 0.97 and 0.98, respectively.
Funding: PhD scholarship from Zoetis (Pfizer) Animal Health.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Ariel N, et al. Search for potential vaccine candidate open reading frames in the Bacillus anthracis virulence plasmid pXO1: in silico and in vitro screening. Infect. Immun. 2002;70:6817–6827. doi: 10.1128/IAI.70.12.6817-6827.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodswen SJ, et al. A novel strategy for classifying the output from an in silico vaccine discovery pipeline for eukaryotic pathogens using machine learning algorithms. BMC Bioinformatics. 2013a;14:315. doi: 10.1186/1471-2105-14-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodswen SJ, et al. A guide to in silico vaccine discovery for eukaryotic pathogens. Brief. Bioinform. 2013b;14:753–774. doi: 10.1093/bib/bbs066. [DOI] [PubMed] [Google Scholar]
- He Y, et al. Vaxign: the first web-based vaccine design program for reverse vaccinology and applications for vaccine development. J. Biomed. Biotechnol. 2010:297505. doi: 10.1155/2010/297505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montigiani S, et al. Genomic approach for analysis of surface proteins in Chlamydia pneumoniae. Infect. Immun. 2002;70:368–379. doi: 10.1128/IAI.70.1.368-379.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R. Reverse vaccinology. Curr. Opin. Microbiol. 2000;3:445–450. doi: 10.1016/s1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- Ross BC, et al. Identification of vaccine candidate antigens from a genomic analysis of Porphyromonas gingivalis. Vaccine. 2001;19:4135–4142. doi: 10.1016/s0264-410x(01)00173-6. [DOI] [PubMed] [Google Scholar]
- Vivona S, et al. NERVE: new enhanced reverse vaccinology environment. BMC Biotechnol. 2006;6:35. doi: 10.1186/1472-6750-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.