Background: Carbon metabolism and virulence are often linked in pathogenic bacteria.
Results: Deletion of the catabolite control protein E (CcpE) affects the expression of virulence factors and pathogenicity of Staphylococcus aureus.
Conclusion: Our data suggest that CcpE acts as an attenuator of virulence in S. aureus.
Significance: CcpE may serve to link S. aureus nutritional status to virulence determinant biosynthesis.
Keywords: Bacterial Pathogenesis, Staphylococcus aureus (S. aureus), Transcription Regulation, Tricarboxylic Acid Cycle (TCA Cycle) (Krebs Cycle), Virulence Factor
Abstract
Carbon metabolism and virulence determinant production are often linked in pathogenic bacteria, and several regulatory elements have been reported to mediate this linkage in Staphylococcus aureus. Previously, we described a novel protein, catabolite control protein E (CcpE) that functions as a regulator of the tricarboxylic acid cycle. Here we demonstrate that CcpE also regulates virulence determinant biosynthesis and pathogenesis. Specifically, deletion of ccpE in S. aureus strain Newman revealed that CcpE affects transcription of virulence factors such as capA, the first gene in the capsule biosynthetic operon; hla, encoding α-toxin; and psmα, encoding the phenol-soluble modulin cluster α. Electrophoretic mobility shift assays demonstrated that CcpE binds to the hla promoter. Mice challenged with S. aureus strain Newman or its isogenic ΔccpE derivative revealed increased disease severity in the ΔccpE mutant using two animal models; an acute lung infection model and a skin infection model. Complementation of the mutant with the ccpE wild-type allele restored all phenotypes, demonstrating that CcpE is negative regulator of virulence in S. aureus.
Introduction
Carbon catabolite repression is a common mechanism utilized by bacteria to optimize their transcriptomes in response to the availability of carbon sources (reviewed in Ref. 1). Similarly, many pathogenic bacteria use this same mechanism to link the nutritional status with the transcription of virulence factors (reviewed in Ref. 2). In Staphylococcus aureus, carbon catabolite repression is mediated by several regulators such as the catabolite control protein A (CcpA),5 a glucose-responsive member of the LacI/GalR family of transcriptional regulators (3), CodY, a pleiotropic repressor that responds to GTP and branched-chain amino acids (4), and RpiRc, a putative ribose-responsive regulator that belongs to the RpiR family of transcriptional regulators (5). Recently, we identified CcpE as another potential carbon catabolite responsive element of S. aureus that controls transcription of tricaboxylic acid (TCA) cycle genes (6). In addition to regulating metabolism, CcpA, CodY, and RpiRc also regulate virulence factor expression (3–5).
CcpA regulates the expression of exotoxins, such as α-toxin (encoded by hla) and toxic shock syndrome toxin-1 (encoded by tst), and capsule formation in a glucose-responsive manner (3, 7, 8). In addition, CcpA promotes biofilm formation under in vitro conditions (9) and alters antibiotic susceptibility in methicillin-resistant S. aureus (MRSA) and glycopeptide intermediary resistant S. aureus (3). More recently, CcpA was reported to mediate proline and arginine auxotrophies during in vitro growth (10, 11), and to contribute to infectivity of S. aureus in a murine model of staphylococcal abscess formation (10).
CodY in S. aureus regulates the expression of virulence factors such as the cap operon (encoding proteins required for capsule biosynthesis), coa (encoding coagulase), fnbA (encoding fibronectin-binding protein A), hla, icaADBC (encoding factors required for synthesis of polysaccharide intercellular adhesin), and spa (encoding protein A) (4, 12, 13). Although inactivation of codY did not markedly affect infectivity of S. aureus strain Newman in a murine abscess model, it restored the virulence of a mutant lacking the major (p)ppGpp synthase/hydrolase enzyme RSH to wild-type levels, suggesting that RSH-dependent derepression of CodY-regulated genes is important for virulence of S. aureus (14). More recently, it was found that inactivation of codY decreased the infectivity of the community-associated MRSA (CA-MRSA) USA300 isolate 923 in two murine infection models (15).
The pentose phosphate pathway regulator RpiRc alters the synthesis of several virulence factors such as protein A, capsular polysaccharide, and hemolysins, to decrease the transcription of RNAIII, the regulatory RNA of the agr locus, and a major regulator of virulence factor production in S. aureus, and to promote biofilm formation under in vitro conditions (5). These observations on RpiRc suggest that this regulator might also affect virulence in vivo; however, this has not been tested.
CcpE directly affects transcription of the aconitase-encoding gene citB, increases TCA cycle activity during in vitro growth (6), and decreases pigment production in S. aureus (16). Because both TCA cycle activity (17–22) and pigment production (16, 23, 24) affect virulence determinant synthesis and/or infectivity of S. aureus, it is likely that CcpE modulates the expression of virulence factors and pathogenicity in this medically important pathogen. To test this hypothesis, we assessed the effect of ccpE deletion in S. aureus strain Newman (25) on the transcription of select virulence factors and on its role in infectivity using two unrelated murine infection models. Our data demonstrate that CcpE affects the transcription of virulence determinants, and infectivity of S. aureus in both in vivo infection models.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Strains were grown in Luria-Bertani Lennox (LB-L) medium (BD Biosciences) at 37 °C and aerated at 230 rpm with a flask-to-medium volume ratio of 10:1. The ΔccpE mutants HOM 354 and HOM 355 were obtained by phage transducing the lox66-aphAIII-lox71–tagged ccpE deletion of THa (6) into strains 923 (26) and SA564 (27), respectively.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristic(s)a | Ref. or source |
|---|---|---|
| S. aureus | ||
| 923 | CA-MRSA, clinical isolate of pulsotype USA300, Oxar | 26 |
| HOM 354 | 923 ΔccpE::lox66-aphAIII-lox71, Oxar, Kanr | This study |
| HOM 355 | SA564 ΔccpE::lox66-aphAIII-lox71, Kanr | |
| Newman | Laboratory strain (ATCC 25904); CP-5 producer | 25 |
| SA564 | Low passage human isolate | 27 |
| THa | RN4220 ΔccpE::lox66-aphAIII-lox71, Kanr | 6 |
| TH01 | Newman ΔccpE::lox72 | 6 |
| TH01c | TH01 harboring plasmid pTH2c cis-integrated at the NWMN_0640 locus, leading to a duplication of the NWMN_0640 gene, ccpE+, Tcr | 6 |
| Plasmids | ||
| pSB2035 | Escherichia coli-S. aureus shuttle plasmid, harboring the cat gene conferring chloramphenicol resistance, and a gfp-lux dual reporter system under the control of the agr P3 promoter; Cmr | 32 |
a The following abbreviations were used: CA-MRSA, community associated MRSA; Cmr, chloramphenicol resistant; Kanr, kanamycin resistant; MLST, multi-locus sequence type; Oxar, methicillin/oxacillin resistant; Tcr, tetracycline resistant.
Transcriptional Analyses
For Northern blot experiments, overnight cultures of S. aureus were diluted to an A600 of 0.05 into fresh pre-warmed LB-L and grown at 37 °C with 230 rpm of aeration. Samples were removed from the cultures at the indicated times and centrifuged at 9,000 × g and 4 °C for 2 min, the culture supernatants were discarded, and the cell pellets were snap frozen in liquid nitrogen. Total RNAs were isolated according to Ref. 28, and blotting, hybridization, and labeling were performed as described (29). Primer pairs hla-F/hla-R and RNAIII-F/RNAIII-R (Table 2) were used to generate digoxigenin-labeled hla- and RNAIII-specific probes by PCR labeling, respectively.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′–3′) | |
|---|---|---|
| Northern probe primer | ||
| hla-F | Forward | AGAAAATGGCATGCACAAAAA |
| hla-R | Reverse | TGTAGCGAAGTCTGGTGAAAA |
| RNAIII-F | Forward | GTGATGGAAAATAGTTGATGAG |
| RNAIII-R | Reverse | GTGAATTTGTTCACTGTGTCG |
| Real-time RT PCR primer | ||
| capA | Forward | GAAAATACGCAACTTATCAACATCCA |
| capA | Reverse | TTTTTTCCGAATCTTGTTTATGACC |
| crtM | Forward | ACGCTTTTGACTTGTTACCAGAAGA |
| crtM | Reverse | AAATTGCCCAAACCGCTTT |
| Hla | Forward | GAACCCGGTATATGGCAATCAA |
| Hla | Reverse | GGAAGTTCTCTGCTGCTTTCATAG |
| RNAIII | Forward | AGGAGTGATTTCAATGGCACAAG |
| RNAIII | Reverse | TGTGTCGATAATCCATTTTACTAAGTCA |
| Psma | Forward | ATCAACAACTCATCACTATGTTAAATCAAC |
| Psma | Reverse | GCCATCGTTTTGTCCTCCTGT |
| gyrB | Forward | GACTGATGCCGATGTGGA |
| gyrB | Reverse | AACGGTGGCTGTGCAATA |
| EMSA primer | ||
| agr P2/3 | Forward | CCATCACATCTCTGTGATCTAG |
| agr P2/3 | Reverse | CTCTCCTCACTGTCATTATACG |
| capA p | Forward | ATCATATGATTATAAGCAATAA |
| capA p | Reverse | GTAATACTTCTTTAATTTTTG |
| hla p | Forward | TAATTAATACCCTTTTTCTCTATTTC |
| hla p | Reverse | GTTACTGAGCTGACTATACGTGTTTTCAT |
| psma p | Forward | CAAATTCATGAGCTTAACCTC |
| psma p | Reverse | GAATTGTTCGATTAAGCTTTTG |
For the quantification of transcripts by real-time reverse transcription PCR (qRT-PCR), RNA isolations and qRT-PCRs were carried out essentially as described (30). The cDNA (20 ng/reaction) was used for real-time amplification using the primers listed in Table 2. mRNA levels were normalized against the mRNA level of gyrB, which is constitutively expressed under the conditions analyzed (31). The amounts of transcripts were expressed as the n-fold difference relative to the control gene (2−ΔCT, where ΔCT represents the difference in threshold cycle between the target and control genes).
Electrophoretic Mobility Shift Assays
DNA probes for electrophoretic mobility shift assays (EMSAs) were generated by PCR using S. aureus strain Newman chromosomal DNA as a template, and primer pairs (Table 2) that amplified the DNA regions preceding the capA, hla, hld, purA, and psmα ORFs. The 5′-ends of the double-stranded PCR products were labeled using [γ-32P]ATP and T4 polynucleotide kinase. A typical assay mixture contained (in 20 μl) 10 mm Tris-HCl, pH 7.5, 50 mm KCl, 1 mm dithiothreitol, 5 mm MgCl2, 0.1 μg of nonspecific competitor (poly(dI-dC)), 2.5% (v/v) glycerol, 0.05% (v/v) Igepal, radioactive DNA probe (2000 cpm ml−1) and various amounts (0, 15, 65, 130, and 200 nm) of purified CcpE. After 20 min of incubation at room temperature, 20 μl of this mixture was loaded into a native 5% (w/v) polyacrylamide Tris borate-EDTA Ready Gel (Bio-Rad) and electrophoresed in 1% Tris borate-EDTA (v/v) buffer for 1 h at 100 V cm−1. Radioactive species were detected by autoradiography using direct exposure to films. Radioactivity labeled promoter probes shifting with CcpE were additionally coincubated with increasing amounts of a nonspecific promoter probe and cold competitor, respectively, to demonstrate specificity of the shifting reaction.
Luciferase Assay
For assaying luciferase activities of S. aureus cells harboring plasmid pSB20235 (32), bacteria were cultivated in LB-L supplemented with 10 μg ml−1 chloramphenicol. Luciferase measurements were carried out essentially as described (33). 200-μl samples of the cell suspensions were removed at the time points indicated, and transferred to the wells of a 96-well clear-bottomed black plate (Greiner). The plate was placed in a Wallac Victor2 1420 Multilabel Counter (PerkinElmer Life Sciences), and luminescence readings were taken for 5 s at 37 °C.
Capsular Polysaccharide 5 (CP-5) Production
CP-5 production was determined by indirect immunofluorescence as described (3), using mouse immunoglobulin M monoclonal antibodies to CP-5 (34). Quantification of CP-5-positive cells was done by determining the numbers of 4′,6-diamidino-2-phenylindole (DAPI) and CY-3-positive cells using the software program CellC (35) (Institute of Signal Processing, Tampere University of Technology, Finland). Immune fluorescence intensities were analyzed using the MetaVueTM Research Imaging System (Molecular Devices). Briefly, from each image 80 bacteria detected by DAPI were randomly selected and the regions were transferred to the corresponding CY-3-stained image. Intensities of the single bacteria were measured and the distribution of intensities analyzed by the software program GraphPad Prism (GraphPad Software, Inc.).
Pigment Measurements
Bacteria were harvested after 24 h of growth on tryptic soy agar and carotenoids were extracted as described (36). The optical densities at 465 nm of the methanol extracts were measured and normalized in reference to the values obtained with the wild-type extracts, which were set at 100.
Animal Models
Eight-week-old female C57BL/6N mice were purchased from Charles River Laboratories (Sulzfeld, Germany) and kept under specific pathogen-free conditions according to the regulations of German veterinary law. All animal studies were performed with the approval of the local State Review Boards.
The murine lung infection model was done essentially as described (37). Eight-week-old C57BL/6N mice were slightly anesthetized by intraperitoneal injection of 2.6 mg of ketamin hydrochloride (Pfizer, Berlin, Germany) and 0.18 mg of xylazin hydrochloride (Bayer, Leverkusen, Germany) per mouse and infected intranasally with 5 × 107 colony forming units (cfu) of S. aureus. Twenty-four hours post-infection, the animals were euthanized, the tracheae were cannulated, and a bronchoalveolar lavage was performed (three times with 1 ml of phosphate-buffered saline). The bronchoalveolar lavage fluid (BALF) was centrifuged at 300 × g for 10 min at 4 °C to obtain alveolar cells, which were suspended in 1 ml of PBS. Total cell numbers in BALF were determined using a Neubauer hemocytometer. To identify the bacterial load of the lungs 24 h post-infection, whole lungs were homogenized in 1 ml of PBS, and serial dilutions were plated onto sheep blood agar. CFU were counted after incubation overnight at 37 °C.
The footpad swelling model was carried out as described (38). Age-matched mice were inoculated subcutaneously with 1 × 107 cfu of S. aureus into the left hind footpad, and footpad swelling was measured daily with a micrometric caliper in reference to the uninfected footpad.
Cytokine Determinations
Levels of murine interleukin-1β (IL-1β), keratinocyte-derived chemokine (KC), and granulocyte-colony stimulating factor (G-CSF) in cell-free BALFs and lung homogenates were determined by commercially available sandwich-type ELISAs, according to the manufacturer's instructions (R&D Systems, Wiesbaden-Nordenstadt, Germany).
Statistical Analyses
Statistical significance was assessed using the Mann-Whitney U test. p values <0.05 were considered significant.
RESULTS
Influence of a ccpE Deletion on RNAIII Transcription
Given the importance of the agr locus for virulence determinant production in S. aureus (reviewed in Refs. 39 and 40), we tested whether CcpE affects transcription of this regulatory system. Northern blot analysis revealed that all three strains, Newman, TH01, and TH01c, produced RNAIII transcripts in a growth phase-dependent manner, with a peak transcription rate at the transition from the exponential growth phase to post-exponential growth phase (i.e. 6 h) (Fig. 1). However, deletion of ccpE increased the post-exponential growth phase accumulation of RNAIII transcripts in TH01, suggesting that CcpE negatively affects RNAIII transcription. To test this suggestion, we transformed an RNAIII transcriptional reporter plasmid, pSB2035, (32) into strains Newman, TH01, and TH01c. This plasmid harbors a gfp-luxABCDE dual reporter system under control of the RNAIII transcription-driving agr P3 promoter. Similar to the Northern blot data (Fig. 1B), luciferase activity assays revealed a growth phase-dependent transcription of RNAIII (Fig. 1C). In addition, we observed that deletion of ccpE increased transcription of RNAIII, confirming that the increased amount of RNAIII in the TH01 mutant is due to increased RNAIII transcription and not due to decreased RNAIII degradation. To test whether CcpE might exert this effect via direct binding to the agr P3 promoter, EMSAs were performed with purified CcpE and a radioactively labeled PCR probe covering the agr P2/3 promoter (Fig. 1D). No mobility shifts were observed over a range of protein concentrations, suggesting that CcpE does not directly interact with the agr P3 promoter to modulate RNAIII transcription.
FIGURE 1.
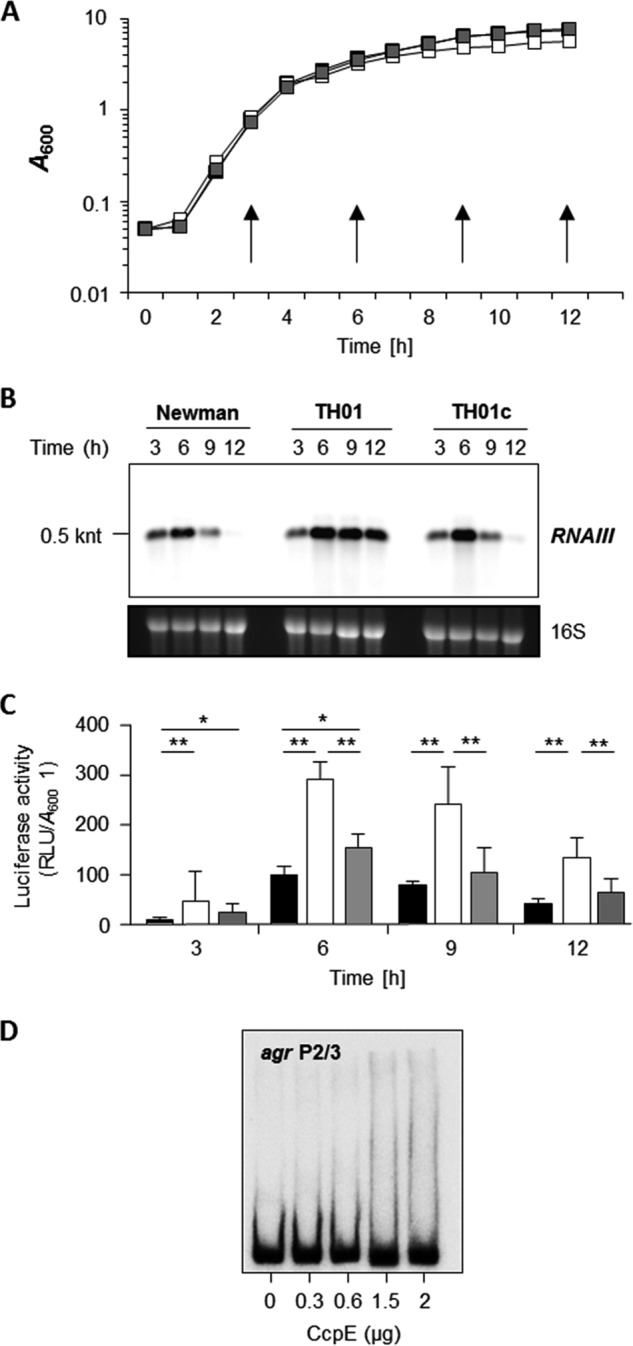
Effect of the ccpE deletion on RNAIII transcription in S. aureus Newman. A, growth characteristics of S. aureus strains Newman (black symbols), TH01 (white symbols), and TH01c (gray symbols) cultured in LB-L at 37 °C and 230 rpm. Time points of sampling for downstream applications (reporter assays, qRT-PCRs) are indicated by arrows. B, Northern blot of RNAIII transcription in strains Newman, TH01 (ΔccpE), and the complemented TH01c during growth in LB-L. Approximate transcript sizes are indicated on the left. Ethidium bromide-stained 16 S rRNA are presented to indicate equivalent RNA loading. C, agr P3 promoter-driven luciferase activities of plasmid pSB2035 harboring derivatives of strains Newman (black bars), TH01 (white bars), and TH01c (gray bars) during growth in LB-L. Luciferase activities were determined at the time points indicated. Data shown are the mean ± S.D. of six independent experiments. Mann-Whitney U test; *, p < 0.05; **, p < 0.01. D, binding activity of CcpE to the agr P2/3 promoter region. The PCR-amplified DNA fragments were radioactively labeled and incubated with the amount of purified CcpE indicated. The results are representative of at least two independent experiments. RLU, relative light units.
CcpE Directly Influences hla Transcription
α-Toxin is a major virulence factor of S. aureus, and its synthesis is regulated at multiple levels, including transcriptional and post-transcriptional mechanisms (3, 41–44). Regulation of hla transcription is also influenced by the carbon catabolite responsive elements CcpA and CodY (3, 4, 15, 45); hence, we hypothesized that hla transcription might be regulated by CcpE as well. Support for this hypothesis can be seen in the Northern blot analysis of hla transcription (Fig. 2A), where hla mRNA levels are much greater in the ΔccpE mutant strain TH01 in all growth phases relative to the wild-type and cis-complemented derivative strain TH01c. To quantify the effect of ccpE deletion on hla transcription, we performed qRT-PCRs on strains Newman, TH01, and TH01c throughout a complete growth cycle (Fig. 2B). Consistent with our Northern blot data, we observed a growth phase-dependent transcription of hla, with a peak in the post-exponential growth phase (9 h). Deletion of ccpE resulted in a massive up-regulation (30 to 60-fold) of hla transcription in strain TH01. Complementation of TH01 with a ccpE wild-type allele restored hla mRNA levels to those seen in the wild-type strain. To exclude that this effect of CcpE was specific for S. aureus strain Newman, we deleted ccpE in two genetically unrelated S. aureus strains, CA-MRSA USA300 isolate 923 (26) and the low passage human isolate SA564 (27), and assessed hla transcription of these strain pairs (Fig. 2C). Deletion of ccpE again strongly increased the transcription of hla in both strains, suggesting that the repressive effect of CcpE on hla transcription is independent of the genetic background. To assess whether CcpE directly regulates transcription of hla, we performed EMSAs with the hla promoter as probe (Fig. 2D). A clear and dose-dependent shift of CcpE with the radioactively labeled hla promoter probe was observed, which was not affected by the addition of a nonspecific promoter probe (data not shown) but was invertable by adding excessive amounts of cold competitor, suggesting that CcpE directly controls transcription of hla.
FIGURE 2.
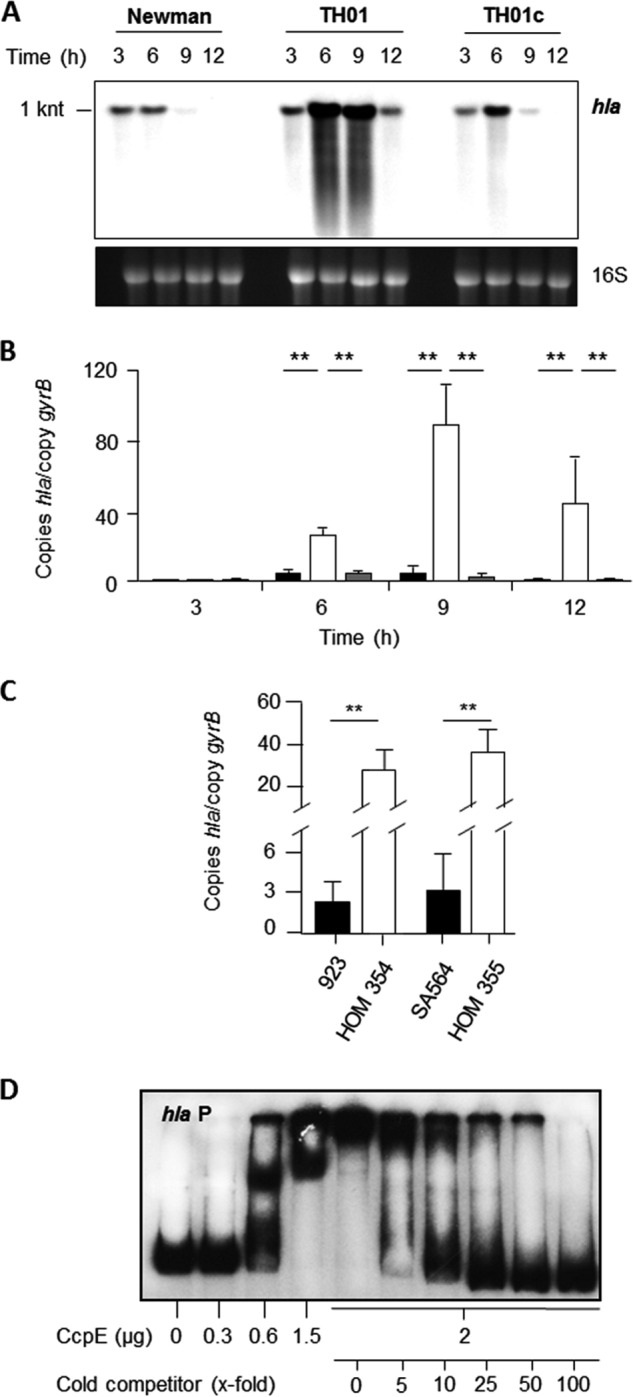
Effect of the ccpE deletion on hla transcription in S. aureus. A, Northern blot of hla transcription in strains Newman, TH01 (ΔccpE), and the complemented TH01c during growth in LB-L. Approximate transcript sizes are indicated on the left. Ethidium bromide-stained 16 S rRNA are presented to indicate equivalent RNA loading. B, quantitative transcript analysis of hla by qRT-PCR of strains Newman (black bars), TH01 (white bars), and TH01c (gray bars) during growth in LB-L. C, quantitative transcript analysis of hla by qRT-PCR of strains 923, HOM 354 (923 ΔccpE), SA564, and HOM 355 (SA564 ΔccpE) after 9 h of growth in LB-L. mRNA levels are expressed relative to gyrase B (in numbers of copies per copy of gyrB). The data presented in B and C are the mean ± S.D. of three independent experiments each determined in duplicate. Mann-Whitney U test: *, p < 0.05; **, p < 0.01. D, binding activity of CcpE to the hla promoter of strain Newman. The PCR-amplified DNA fragments (100 ng/lane) were radioactively labeled and incubated with the amount of purified CcpE in the absence and presence of cold competitor as indicated. The results are representative of at least two independent experiments.
CcpE Promotes Capsule Formation
Capsular polysaccharide is another important virulence factor of S. aureus, whose synthesis is intimately linked to the nutritional status of the bacterium (3, 5, 12). Our results (Fig. 3) demonstrate that in addition to CcpA, CodY, and RpiRc, CcpE also modulates transcription of the cap operon and the elaboration of a capsule. As expected, when S. aureus was cultivated in LB-L, the first gene of the cap operon (capA) was predominantly transcribed during the later stages of growth (Fig. 3A). Deletion of ccpE in TH01 strongly decreased accumulation of capA mRNA throughout the growth cycle. cis-Complementation of TH01 with the wild-type ccpE allele restored capA mRNA levels to those found in the isogenic wild-type strain Newman. Consistent with the transcriptional data, a reduced number of capsular polysaccharide positive cells was observed with the ΔccpE mutant (Fig. 3B). Although about 69 ± 6% of the Newman cells and 75 ± 8% of the TH01c cells incubated with the CP-5 antibodies produced clear fluorescence signals after 24 h of growth in LB-L, in the TH01 cell pool only 35 ± 7% of the cells emitted detectable amounts of fluorescence. Similarly, a ∼10-fold decrease in the mean fluorescence intensity per cell was observed with the ΔccpE mutant (Fig. 3B) when compared with cells of the wild-type and complemented derivative TH01c. To determine whether the CcpE-dependent regulation of capA was due to an interaction with the capA promoter, EMSAs were performed with CcpE and a radioactively labeled probe of the capA promoter. In contrast to the hla promoter, CcpE did not shift the capA promoter probe at any of the CcpE concentrations tested (Fig. 3C), indicating that CcpE indirectly influences cap operon transcription and capsule formation.
FIGURE 3.
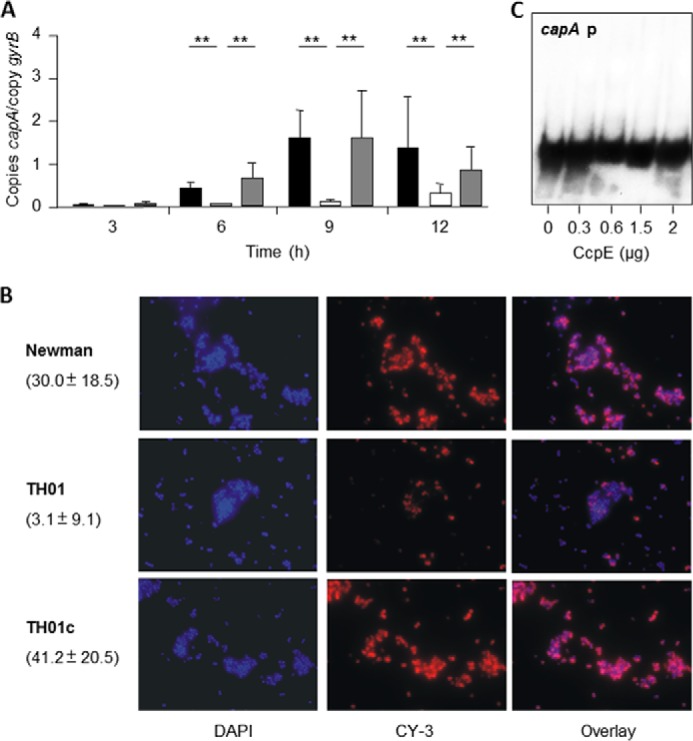
Effect of the ccpE deletion on capsule formation in S. aureus Newman. A, quantitative transcript analysis of capA by qRT-PCR of strains Newman (black bars), TH01 (ΔccpE, white bars), and TH01c (complemented derivative, gray bars) during growth in LB-L. mRNA levels are expressed relative to gyrase B (in numbers of copies per copy of gyrB). The data presented are the mean ± S.D. of three independent experiments each determined in duplicate. Mann-Whitney U test: *, p < 0.05; **, p < 0.01. B, CP-5 expression of strains Newman, TH01, and TH01c during growth in LB-L. Bacteria were grown to an A600 of 0.5, stained with DAPI, marked with CP-5-specific monoclonal antibodies, and stained with Cy-3-conjugated anti-mouse antibodies (CY-3). Numbers in parentheses indicate the mean fluorescence intensities ± S.D. per cell (n = 80). C, binding activity of CcpE to the cap promoter. The PCR-amplified DNA fragments were radioactively labeled and incubated with the amount of purified CcpE indicated. The results in B and C are representative of at least two independent experiments.
CcpE Alters Transcription of the Phenol-soluble Modulin α (psmα) Cluster
Phenol-soluble modulins are a small group of cytolytic and immunomodulating peptides that are important virulence determinants of S. aureus, especially in CA-MRSA USA300 isolates (reviewed in Ref. 46). The S. aureus Newman genome harbors two psm operons, psmα and psmβ, which are transcriptionally affected by regulators such as SarA and AgrA (40, 47). To determine whether CcpE influences psm transcription, we assessed psmα and psmβ transcription using qRT-PCR. Deletion of ccpE had a negligible effect on psmβ transcription (data not shown); however, we observed a significant reduction of psmα transcripts in strain TH01 compared with the wild-type strain (Fig. 4A). Complementation of TH01 with a ccpE wild-type allele restored psmα mRNA levels to that seen in the wild-type strain. EMSAs performed using CcpE and the psmα promoter as a probe failed to shift the radiolabeled probe with any of the protein concentrations tested (Fig. 4B), suggesting an indirect effect of CcpE on psmα transcription.
FIGURE 4.
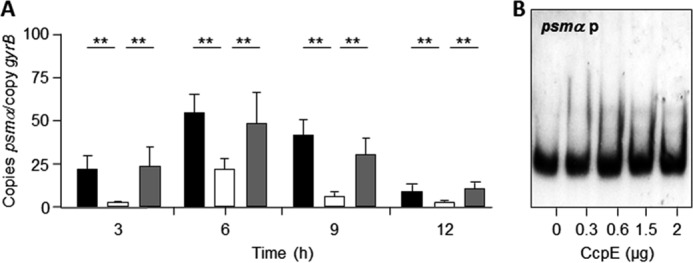
Effect of the ccpE deletion on psmα transcription in S. aureus Newman. A, quantitative transcript analysis of psmα by qRT-PCR of strains Newman (black bars), TH01 (white bars), and TH01c (gray bars) during growth in LB-L. mRNA levels are expressed relative to gyrase B (in numbers of copies per copy of gyrB). The data presented are mean ± S.D. of three independent experiments each determined in duplicate. Mann-Whitney U test: *, p < 0.05; **, p < 0.01. B, binding activity of CcpE to the psmα promoter. The PCR-amplified DNA fragments were radioactively labeled and incubated with the amount of purified CcpE indicated. The results are representative of at least two independent experiments.
CcpE Decreases Pigment Production
Most S. aureus strains produce the carotenoid pigment staphyloxanthin, which is responsible for the yellowish-orange appearance of this bacterium (48). In line with a previous publication (16), we noticed an increase in pigment production after 24 h of growth on tryptic soy agar, and this phenotype was reverted by introducing a functional ccpE into this mutant (Fig. 5). The synthesis of staphyloxanthin is encoded within the crtOPQMN operon (48); hence, to determine whether CcpE affects transcription of crtOPQMN, we assessed crtM mRNA levels in strains Newman, TH01, and TH01c using qRT-PCR. Contrary to the findings reported by Lan and colleagues (16), our results suggest that transcription of crtOPQMN appears to be independent of CcpE (Fig. 5C). Similarly, inactivation of ccpE in strains 923 and SA564 significantly increased the pigment contents of mutant cells compared with wild-type, without affecting crtM transcription, suggesting that this phenomenon is not strain-dependent (Fig. 5).
FIGURE 5.
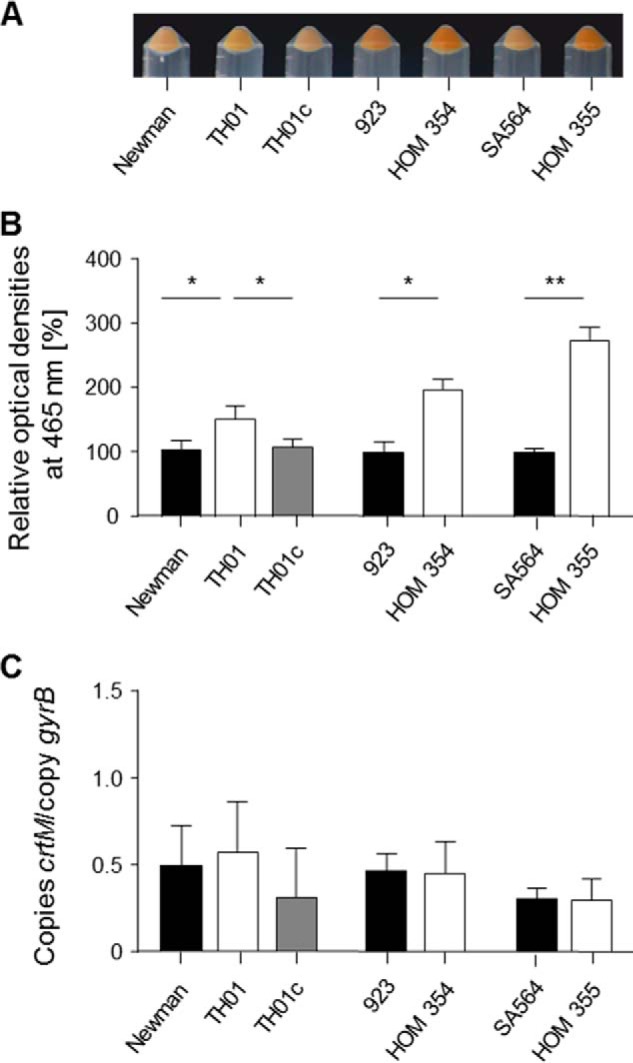
Effect of the ccpE deletion on pigment production in S. aureus. A, pigmentation displays of S. aureus strains grown for 24 h at 37 °C on tryptic soy agar plates. B, measurement of carotenoid pigment contents in S. aureus cells grown for 24 h at 37 °C on tryptic soy agar. The relative optical density units at 465 nm were normalized to those of wild-type, which were set at 100. The data presented are mean ± S.D. of five independent experiments. Mann-Whitney U test: *, p < 0.05; **, p < 0.01. C, quantitative transcript analysis of crtM by qRT-PCR of S. aureus cells grown for 24 h at 37 °C on tryptic soy agar. mRNA levels are expressed relative to gyrase B (in numbers of copies per copy of gyrB). The data presented are mean ± S.D. of three independent experiments each determined in duplicate.
CcpE Attenuates Virulence in Two Murine Infection Models
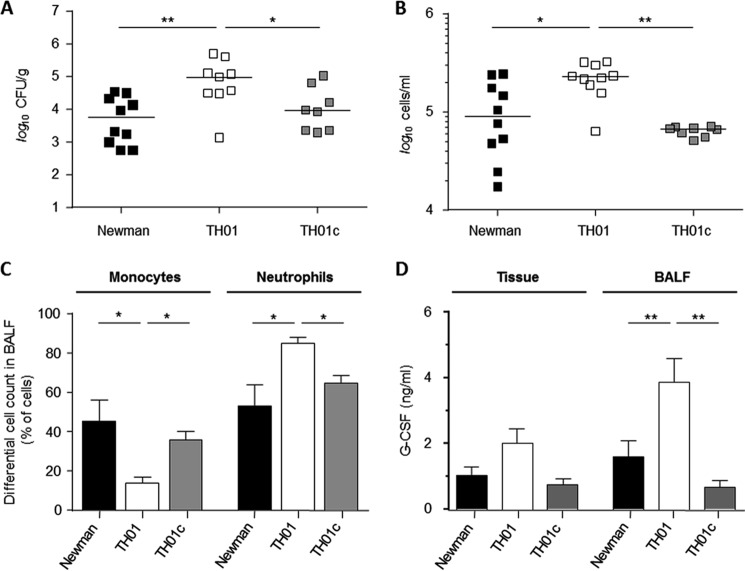
Deletion of ccpE in S. aureus strain Newman augmented transcription of the global virulence regulator RNAIII (Fig. 1) and increased α-toxin (hla) mRNA (Fig. 2). Given the effect of CcpE on virulence factor transcription in vitro, we hypothesized that CcpE might alter infectivity of S. aureus in vivo. To address this hypothesis, we assessed the ability of Newman, TH01, and TH01c strains to cause disease in two different murine infection models. In a murine pneumonia model, C57BL/6N mice were infected intranasally with strains Newman, TH01, or TH01c, and the bacterial load in the lungs and the total amount of eukaryotic cells in BALFs at 24 h post-infection were determined (Fig. 6). Strain TH01 significantly increased the bacterial load in the lungs of mice relative to the wild-type and complemented strains (Fig. 6A). Similarly, we observed a significant increase in total cells in BALFs of the TH01 challenged mice (Fig. 6B), indicating a more severe infection. This increase in total cell numbers correlated with an increased number of neutrophils in BALFs of TH01 challenged mice (Fig. 6C), and this also correlated with increased concentrations of the neutrophil mobilization stimulating factor G-CSF (49) (Fig. 6D). Complementation of the ΔccpE mutant restored all virulence traits back to wild-type levels, confirming that all observed alterations were caused by CcpE. To exclude that this CcpE effect is specific for strain Newman, we additionally infected mice intranasally with strain SA564 and its ΔccpE derivative HOM 355, respectively. In line with our observations made with the strain triplet Newman/TH01/TH01c, we observed significantly increased cfu numbers in the lung tissues of mice that have been infected with the SA564 ccpE mutant (Fig. 7), demonstrating that this virulence diminishing effect of CcpE is not specific for strain Newman.
FIGURE 6.
Effect of the ccpE deletion on infectivity of S. aureus Newman in an acute murine lung infection model. C57BL/6N mice were infected intranasally with 5 × 107 cells of S. aureus strain Newman (black symbols), TH01 (ΔccpE, white symbols), and TH01c (complemented mutant, gray symbols), respectively (n = 8–10 per group). Mice were euthanized 24 h post-infection, BALFs were collected, and lungs were homogenized in PBS to determine the bacterial loads and cytokine concentrations in this tissue. A, bacterial loads in the lungs of infected mice. B, total eukaryotic cell contents in BALFs. Each symbol represents an individual mouse. Horizontal bars indicate the median of all observations. C, ratios of monocytes and neutrophils in BALFs of infected mice. D, G-CSF concentrations in lungs and cell-free BALFs of infected mice. Data are presented as mean ± S.E. (n = 8–10). Mann-Whitney U test: *, p < 0.05; **, p < 0.01.
FIGURE 7.
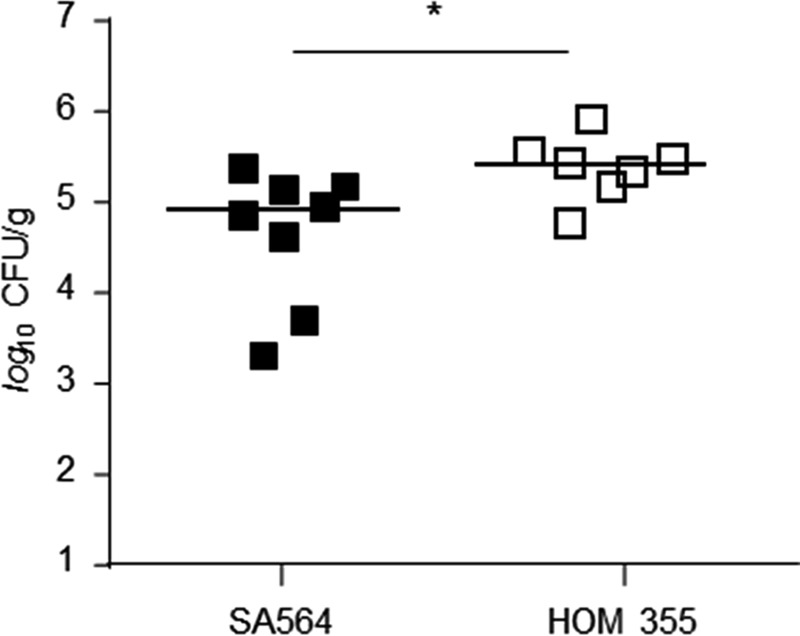
Effect of the ccpE deletion on infectivity of S. aureus SA564 in an acute murine lung infection model. C57BL/6N mice were infected intranasally with 5 × 107 cells of S. aureus strain SA564 (black symbols) and HOM 355 (ΔccpE, white symbols), respectively (n = 7–8 per group). Mice were euthanized 24 h post-infection, and lungs were homogenized in PBS to determine the bacterial loads in this tissue. Each symbol represents an individual mouse. Horizontal bars indicate the median of all observations. Mann-Whitney U test: *, p < 0.05.
To substantiate these findings in another in vivo model, we utilized a murine footpad infection model (38). In this model, bacteria are inoculated into the left hind footpad of mice and footpad swelling ratios are determined on a daily basis for up to 12 days (Fig. 8). Consistent with our observations using a lung infection model, we observed enhanced footpad swelling in mice challenged with the Newman ΔccpE mutant relative to the isogenic wild-type and complemented strains (Fig. 8). Swelling was most significantly increased early in the infection process (days 1 to 4) and in the later stages of the infection (days 8 to 12) when compared with the values obtained with the wild-type and TH01c challenged mice groups.
FIGURE 8.
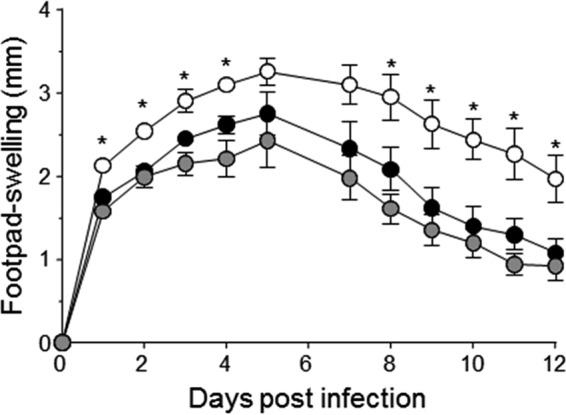
Effect of the ccpE deletion on infectivity of S. aureus Newman in a systemic murine footpad infection model. 1 × 107 cells of S. aureus strain Newman (black symbols) and its derivatives TH01 (ΔccpE, white symbols), and TH01c (complemented mutant, gray symbols) were injected subcutaneously into the footpads of C57BL/6N mice, and swelling of the footpads were measured in reference to the uninfected footpads at the time points indicated. Data shown represent the mean ± S.D. of 8 mice per group. Mann-Whitney U test: *, p < 0.05.
DISCUSSION
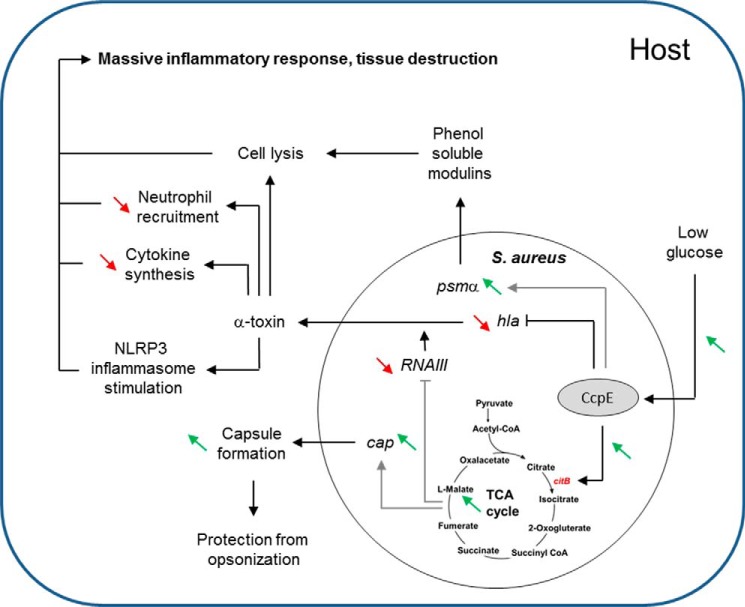
The nosocomial pathogen S. aureus is known to link its virulence factor production with central metabolic pathways (3–5, 8, 9, 13, 22). This linkage is mediated via at least three metabolite responsive regulators; namely, CcpA, (3, 8), CodY (13), and RpiRc (5). Data presented here demonstrate that CcpE represents a fourth regulatory protein that connects virulence factor synthesis with the central metabolism, specifically the TCA cycle (Fig. 9) (6).
FIGURE 9.
Proposed regulatory role of CcpE on virulence factor production and pathogenicity of S. aureus. Under glucose-rich conditions, TCA cycle activity is repressed in S. aureus via CcpA. When glucose concentrations become growth limiting, the transcription of TCA cycle genes is de-repressed in a CcpE-dependent manner, which directly promotes TCA cycle activity via direct transcriptional control of the aconitase encoding gene citB. An active TCA cycle augments capsule formation via an increased transcription of the cap operon. Additionally, it decreases pigment production and transcription of RNAIII, the master virulence regulator of the agr locus. CcpE also directly interferes with hla transcription, leading to reduced α-toxin synthesis, thereby decreasing the synthesis of cytokines, reducing the attraction of neutrophils, and impairing the pathogen-driven stimulation of the NLRP3 inflammasome in lungs. CcpE also enhances transcription of the psmα cluster by a yet unidentified mechanism, thereby increasing the lysis of white blood cells and stimulating an inflammatory response. Experimentally proven positive effects of CcpE are depicted by green diagonal arrows and negative effects by red diagonal arrows. Direct regulation of CcpE is displayed by black connecting lines, and indirect regulation by gray lines. Arrows indicate a stimulatory effect, and perpendicular lines a repressive effect.
Although most effects of CcpE on virulence factor synthesis were indirect, possibly via regulation of TCA cycle activity (22, 27, 50), a direct link between CcpE and hla transcription was established. In a murine pneumonia model, α-toxin is a key virulence determinant involved in the pathogenesis of S. aureus (51, 52); specifically, the level of α-toxin correlated with disease severity in this animal model (53). Mechanistically, α-toxin increases cytokine synthesis, enhances neutrophil recruitment, and stimulates the NLRP3 (NOD-like receptor family, pyrin domain containing 3) inflammasome in lungs, leading to massive inflammatory response and tissue destruction (54, 55). Consistent with these observations, deletion of ccpE increased hla transcription (Fig. 2) and increased the bacterial loads and neutrophil contents in the lungs of mice (Figs. 6 and 7), suggesting that CcpE might influence the virulence of S. aureus during lung infections via transcriptional regulation of hla.
In addition to directly interacting with hla, ccpE deletion increased RNAIII levels, which likely contributed to the altered pathogenesis of the ΔccpE mutant in both animal models. RNAIII is the RNA regulator of the agr locus encoded quorum sensing system (reviewed in Refs. 39 and 40) and it codes for a small lytic peptide called δ-toxin, which is a chemoattractant for neutrophils (50). RNAIII is predominantly transcribed when a threshold level of bacteria is achieved (56, 57). In its regulatory function, RNAIII promotes the expression of many exoproteins including α-toxin, either directly or via control of a repressor protein known as Rot (43, 58). Mutations in agr have been shown to attenuate virulence in several animal models (59–63) including murine models of pneumonia (52, 64) and skin infections (65–67). When the peptide δ-toxin is translated from RNAIII, it is produced in two forms; one without an N-terminal formyl group on the methionine, and one containing a formylated methionine (50). Formylated δ-toxin is a potent neutrophil chemoattractant, suggesting that increased neutrophils in the lungs on TH01-infected mice may be due to an increase in δ-toxin synthesis.
Alterations in the synthesis of virulence factors and RNAIII will likely alter the immune response to the infection. The BALF cytokine profiles of mice infected with strains Newman, TH01, and TH01c were similar with respect to keratinocyte-derived chemokine and IL-1β, however, G-CSF was higher in BALFs and lung homogenates from TH01 challenged mice relative to mice infected with the wild-type strain. G-CSF was originally characterized in hematopoietic cells to stimulate the proliferation and differentiation of neutrophil granulocyte precursors. In addition, G-CSF functions to recruit polymorphonuclear leukocytes to the lung (68), and its expression in lung tissue is stimulated by microbial infections (69–72). Recently, Hua et al. (73) observed in a mouse pneumonia model that preimmunization with an anti-α-toxin antibody significantly decreased the G-CSF contents in BALFs of mice infected with S. aureus. Based on this observation, it is reasonable to speculate that an increase in α-toxin synthesis (Fig. 2) would increase G-CSF production (Fig. 6D), resulting in an increase in neutrophil recruitment (Fig. 6C).
Transcription of RNAIII is primarily promoted by AgrA, the response regulator of the two-component system encoded by the agr locus (74). In addition, AgrA also promotes transcription of the psm operons (40). Because we observed divergent effects of CcpE on RNAIII and psmα transcription (Figs. 1 and 4), we can largely exclude that CcpE modulates RNAIII production via activation of AgrA. Similarly, the agr system promotes capsule synthesis (75–77); however, capA mRNA levels were decreased in the ΔccpE mutant despite an increase in RNAIII transcript levels (Figs. 1 and 3). Interestingly, Somerville and colleagues (20, 27) observed increased RNAIII levels and an impaired capsule biosynthesis in TCA cycle mutants in which the aconitase-encoding gene citB (syn. acnA) was inactivated, demonstrating a link between TCA cycle activity, capsule formation, and RNAIII production. It is possible that CcpE modulates RNAIII transcription and capsule biosynthesis via regulation of TCA cycle activity. However, the effect of TCA cycle inactivation on capsule synthesis is tied to a lack of oxaloacetate for gluconeogenesis (20), and it is still unclear how TCA cycle activity affects transcription of RNAIII. A potential factor might be aconitase itself. This key enzyme of the TCA cycle is reported in Bacillus subtilis to act as a bifunctional protein that possesses enzymatic activity and functions as an RNA-binding regulatory protein (78–80). Similar to B. subtilis, apo-aconitase binds to iron-responsive elements in mRNA,6 raising the possibility of a direct interaction between aconitase and the highly structured RNAIII. Additionally, CcpE might affect RNAIII synthesis and capsule formation via pH alterations. We have recently shown that in vitro cultivation of the ccpE deletion mutant in LB-L led to a significantly reduced alkalinization of the culture medium during later stages of growth (6–12 h) compared with the wild-type culture (6). Alkaline growth conditions were previously reported to repress RNAIII production (81), and to augment capsule formation (82, 83), consistent with our findings of increased RNAIII transcription and decreased capA transcription in TH01 during the later growth stages in LB-L (Figs. 1 and 3).
In conclusion, CcpE modulates the expression of several major virulence factors of S. aureus, which affects its pathogenesis. Given its mostly repressive effect on virulence determinant production, it can be assumed that CcpE serves as an attenuator of virulence in this clinically important pathogen.
Acknowledgment
We thank A. Honecker for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant AI087668 (to G. A. S.) and Grants BI 1350/1-1 and BI 1350/1-2 from the Deutsche Forschungsgemeinschaft (DFG).
G. A. Somerville, unpublished data.
- CcpE
- catabolite control protein E
- TCA
- tricaboxylic acid
- MRSA
- methicillin-resistant S. aureus
- qRT-PCR
- real-time reverse transcription PCR
- CP-5
- Capsular Polysaccharide 5
- BALF
- bronchoalveolar lavage fluid
- G-CSF
- granulocyte-colony stimulating factor.
REFERENCES
- 1. Görke B., Stülke J. (2008) Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6, 613–624 [DOI] [PubMed] [Google Scholar]
- 2. Poncet S., Milohanic E., Mazé A., Nait Abdallah J., Aké F., Larribe M., Deghmane A. E., Taha M. K., Dozot M., De Bolle X., Letesson J. J., Deutscher J. (2009) Correlations between carbon metabolism and virulence in bacteria. Contrib. Microbiol. 16, 88–102 [DOI] [PubMed] [Google Scholar]
- 3. Seidl K., Stucki M., Ruegg M., Goerke C., Wolz C., Harris L., Berger-Bächi B., Bischoff M. (2006) Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 50, 1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Majerczyk C. D., Sadykov M. R., Luong T. T., Lee C., Somerville G. A., Sonenshein A. L. (2008) Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190, 2257–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhu Y., Nandakumar R., Sadykov M. R., Madayiputhiya N., Luong T. T., Gaupp R., Lee C. Y., Somerville G. A. (2011) RpiR homologues may link Staphylococcus aureus RNAIII synthesis and pentose phosphate pathway regulation. J. Bacteriol. 193, 6187–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hartmann T., Zhang B., Baronian G., Schulthess B., Homerova D., Grubmüller S., Kutzner E., Gaupp R., Bertram R., Powers R., Eisenreich W., Kormanec J., Herrmann M., Molle V., Somerville G. A., Bischoff M. (2013) Catabolite control protein E (CcpE) is a LysR-type transcriptional regulator of tricarboxylic acid cycle activity in Staphylococcus aureus. J. Biol. Chem. 288, 36116–36128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Seidl K., Bischoff M., Berger-Bächi B. (2008) CcpA mediates the catabolite repression of tst in Staphylococcus aureus. Infect. Immun. 76, 5093–5099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Seidl K., Müller S., François P., Kriebitzsch C., Schrenzel J., Engelmann S., Bischoff M., Berger-Bächi B. (2009) Effect of a glucose impulse on the CcpA regulon in Staphylococcus aureus. BMC Microbiol. 9, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seidl K., Goerke C., Wolz C., Mack D., Berger-Bächi B., Bischoff M. (2008) Staphylococcus aureus CcpA affects biofilm formation. Infect. Immun. 76, 2044–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li C., Sun F., Cho H., Yelavarthi V., Sohn C., He C., Schneewind O., Bae T. (2010) CcpA mediates proline auxotrophy and is required for Staphylococcus aureus pathogenesis. J. Bacteriol. 192, 3883–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nuxoll A. S., Halouska S. M., Sadykov M. R., Hanke M. L., Bayles K. W., Kielian T., Powers R., Fey P. D. (2012) CcpA regulates arginine biosynthesis in Staphylococcus aureus through repression of proline catabolism. PLoS Pathog. 8, e1003033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Majerczyk C. D., Dunman P. M., Luong T. T., Lee C. Y., Sadykov M. R., Somerville G. A., Bodi K., Sonenshein A. L. (2010) Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 192, 2861–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pohl K., Francois P., Stenz L., Schlink F., Geiger T., Herbert S., Goerke C., Schrenzel J., Wolz C. (2009) CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J. Bacteriol. 191, 2953–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geiger T., Goerke C., Fritz M., Schäfer T., Ohlsen K., Liebeke M., Lalk M., Wolz C. (2010) Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect. Immun. 78, 1873–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Montgomery C. P., Boyle-Vavra S., Roux A., Ebine K., Sonenshein A. L., Daum R. S. (2012) CodY deletion enhances in vivo virulence of community-associated methicillin-resistant Staphylococcus aureus clone USA300. Infect. Immun. 80, 2382–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lan L., Cheng A., Dunman P. M., Missiakas D., He C. (2010) Golden pigment production and virulence gene expression are affected by metabolisms in Staphylococcus aureus. J. Bacteriol. 192, 3068–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chatterjee I., Herrmann M., Proctor R. A., Peters G., Kahl B. C. (2007) Enhanced post-stationary-phase survival of a clinical thymidine-dependent small-colony variant of Staphylococcus aureus results from lack of a functional tricarboxylic acid cycle. J. Bacteriol. 189, 2936–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gaupp R., Schlag S., Liebeke M., Lalk M., Götz F. (2010) Advantage of upregulation of succinate dehydrogenase in Staphylococcus aureus biofilms. J. Bacteriol. 192, 2385–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Massilamany C., Gangaplara A., Gardner D. J., Musser J. M., Steffen D., Somerville G. A., Reddy J. (2011) TCA cycle inactivation in Staphylococcus aureus alters nitric oxide production in RAW 264.7 cells. Mol. Cell. Biochem. 355, 75–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sadykov M. R., Mattes T. A., Luong T. T., Zhu Y., Day S. R., Sifri C. D., Lee C. Y., Somerville G. A. (2010) Tricarboxylic acid cycle-dependent synthesis of Staphylococcus aureus Type 5 and 8 capsular polysaccharides. J. Bacteriol. 192, 1459–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sheldon J. R., Marolda C. L., Heinrichs D. E. (2014) TCA cycle activity in Staphylococcus aureus is essential for iron-regulated synthesis of staphyloferrin A, but not staphyloferrin B: the benefit of a second citrate synthase. Mol. Microbiol. 92, 824–839 [DOI] [PubMed] [Google Scholar]
- 22. Zhu Y., Xiong Y. Q., Sadykov M. R., Fey P. D., Lei M. G., Lee C. Y., Bayer A. S., Somerville G. A. (2009) Tricarboxylic acid cycle-dependent attenuation of Staphylococcus aureus in vivo virulence by selective inhibition of amino acid transport. Infect. Immun. 77, 4256–4264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu C. I., Liu G. Y., Song Y., Yin F., Hensler M. E., Jeng W. Y., Nizet V., Wang A. H., Oldfield E. (2008) A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science 319, 1391–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu G. Y., Essex A., Buchanan J. T., Datta V., Hoffman H. M., Bastian J. F., Fierer J., Nizet V. (2005) Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J. Exp. Med. 202, 209–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Duthie E. S. (1952) Variation in the antigenic composition of staphylococcal coagulase. J. Gen. Microbiol. 7, 320–326 [DOI] [PubMed] [Google Scholar]
- 26. Boyle-Vavra S., Yin S., Daum R. S. (2006) The VraS/VraR two-component regulatory system required for oxacillin resistance in community-acquired methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 262, 163–171 [DOI] [PubMed] [Google Scholar]
- 27. Somerville G. A., Chaussee M. S., Morgan C. I., Fitzgerald J. R., Dorward D. W., Reitzer L. J., Musser J. M. (2002) Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect. Immun. 70, 6373–6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheung A. L., Eberhardt K. J., Fischetti V. A. (1994) A method to isolate RNA from Gram-positive bacteria and mycobacteria. Anal. Biochem. 222, 511–514 [DOI] [PubMed] [Google Scholar]
- 29. McCallum N., Karauzum H., Getzmann R., Bischoff M., Majcherczyk P., Berger-Bächi B., Landmann R. (2006) In vivo survival of teicoplanin-resistant Staphylococcus aureus and fitness cost of teicoplanin resistance. Antimicrob. Agents Chemother. 50, 2352–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chatterjee I., Becker P., Grundmeier M., Bischoff M., Somerville G. A., Peters G., Sinha B., Harraghy N., Proctor R. A., Herrmann M. (2005) Staphylococcus aureus ClpC is required for stress resistance, aconitase activity, growth recovery, and death. J. Bacteriol. 187, 4488–4496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Valihrach L., Demnerova K. (2012) Impact of normalization method on experimental outcome using RT-qPCR in Staphylococcus aureus. J. Microbiol. Methods 90, 214–216 [DOI] [PubMed] [Google Scholar]
- 32. Qazi S. N., Counil E., Morrissey J., Rees C. E., Cockayne A., Winzer K., Chan W. C., Williams P., Hill P. J. (2001) agr expression precedes escape of internalized Staphylococcus aureus from the host endosome. Infect. Immun. 69, 7074–7082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schmitt J., Joost I., Skaar E. P., Herrmann M., Bischoff M. (2012) Haemin represses the haemolytic activity of Staphylococcus aureus in an Sae-dependent manner. Microbiology 158, 2619–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoeger P. H., Lenz W., Boutonnier A., Fournier J. M. (1992) Staphylococcal skin colonization in children with atopic dermatitis: prevalence, persistence, and transmission of toxigenic and nontoxigenic strains. J. Infect. Dis. 165, 1064–1068 [DOI] [PubMed] [Google Scholar]
- 35. Selinummi J., Seppälä J., Yli-Harja O., Puhakka J. A. (2005) Software for quantification of labeled bacteria from digital microscope images by automated image analysis. BioTechniques 39, 859–863 [DOI] [PubMed] [Google Scholar]
- 36. Morikawa K., Maruyama A., Inose Y., Higashide M., Hayashi H., Ohta T. (2001) Overexpression of sigma factor, σ(B), urges Staphylococcus aureus to thicken the cell wall and to resist β-lactams. Biochem. Biophys. Res. Commun. 288, 385–389 [DOI] [PubMed] [Google Scholar]
- 37. Seiler F., Hellberg J., Lepper P. M., Kamyschnikow A., Herr C., Bischoff M., Langer F., Schäfers H. J., Lammert F., Menger M. D., Bals R., Beisswenger C. (2013) FOXO transcription factors regulate innate immune mechanisms in respiratory epithelial cells. J. Immunol. 190, 1603–1613 [DOI] [PubMed] [Google Scholar]
- 38. Nippe N., Varga G., Holzinger D., Löffler B., Medina E., Becker K., Roth J., Ehrchen J. M., Sunderkötter C. (2011) Subcutaneous infection with S. aureus in mice reveals association of resistance with influx of neutrophils and Th2 response. J. Investig. Dermatol. 131, 125–132 [DOI] [PubMed] [Google Scholar]
- 39. Pragman A. A., Schlievert P. M. (2004) Virulence regulation in Staphylococcus aureus: the need for in vivo analysis of virulence factor regulation. FEMS Immunol. Med. Microbiol. 42, 147–154 [DOI] [PubMed] [Google Scholar]
- 40. Queck S. Y., Jameson-Lee M., Villaruz A. E., Bach T. H., Khan B. A., Sturdevant D. E., Ricklefs S. M., Li M., Otto M. (2008) RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32, 150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cheung A. L., Ying P. (1994) Regulation of α- and β-hemolysins by the sar locus of Staphylococcus aureus. J. Bacteriol. 176, 580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ingavale S., van Wamel W., Luong T. T., Lee C. Y., Cheung A. L. (2005) Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect. Immun. 73, 1423–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morfeldt E., Taylor D., von Gabain A., Arvidson S. (1995) Activation of α-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 14, 4569–4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oscarsson J., Kanth A., Tegmark-Wisell K., Arvidson S. (2006) SarA is a repressor of hla (α-hemolysin) transcription in Staphylococcus aureus: its apparent role as an activator of hla in the prototype strain NCTC 8325 depends on reduced expression of sarS. J. Bacteriol. 188, 8526–8533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Leiba J., Hartmann T., Cluzel M. E., Cohen-Gonsaud M., Delolme F., Bischoff M., Molle V. (2012) A novel mode of regulation of the Staphylococcus aureus catabolite control protein A (CcpA) mediated by Stk1 protein phosphorylation. J. Biol. Chem. 287, 43607–43619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peschel A., Otto M. (2013) Phenol-soluble modulins and staphylococcal infection. Nat. Rev. Microbiol. 11, 667–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zielinska A. K., Beenken K. E., Joo H. S., Mrak L. N., Griffin L. M., Luong T. T., Lee C. Y., Otto M., Shaw L. N., Smeltzer M. S. (2011) Defining the strain-dependent impact of the Staphylococcal accessory regulator (sarA) on the α-toxin phenotype of Staphylococcus aureus. J. Bacteriol. 193, 2948–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pelz A., Wieland K. P., Putzbach K., Hentschel P., Albert K., Götz F. (2005) Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J. Biol. Chem. 280, 32493–32498 [DOI] [PubMed] [Google Scholar]
- 49. Suzuki S., Kobayashi M., Chiba K., Horiuchi I., Wang J., Kondoh T., Hashino S., Tanaka J., Hosokawa M., Asaka M. (2002) Autocrine production of epithelial cell-derived neutrophil attractant-78 induced by granulocyte colony-stimulating factor in neutrophils. Blood 99, 1863–1865 [PubMed] [Google Scholar]
- 50. Somerville G. A., Cockayne A., Dürr M., Peschel A., Otto M., Musser J. M. (2003) Synthesis and deformylation of Staphylococcus aureus δ-toxin are linked to tricarboxylic acid cycle activity. J. Bacteriol. 185, 6686–6694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bubeck Wardenburg J., Bae T., Otto M., Deleo F. R., Schneewind O. (2007) Poring over pores: α-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat. Med. 13, 1405–1406 [DOI] [PubMed] [Google Scholar]
- 52. Bubeck Wardenburg J., Patel R. J., Schneewind O. (2007) Surface proteins and exotoxins are required for the pathogenesis of Staphylococcus aureus pneumonia. Infect. Immun. 75, 1040–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bubeck Wardenburg J., Schneewind O. (2008) Vaccine protection against Staphylococcus aureus pneumonia. J. Exp. Med. 205, 287–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bartlett A. H., Foster T. J., Hayashida A., Park P. W. (2008) α-Toxin facilitates the generation of CXC chemokine gradients and stimulates neutrophil homing in Staphylococcus aureus pneumonia. J. Infect. Dis. 198, 1529–1535 [DOI] [PubMed] [Google Scholar]
- 55. Kebaier C., Chamberland R. R., Allen I. C., Gao X., Broglie P. M., Hall J. D., Jania C., Doerschuk C. M., Tilley S. L., Duncan J. A. (2012) Staphylococcus aureus alpha-hemolysin mediates virulence in a murine model of severe pneumonia through activation of the NLRP3 inflammasome. J. Infect. Dis. 205, 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Geisinger E., Chen J., Novick R. P. (2012) Allele-dependent differences in quorum-sensing dynamics result in variant expression of virulence genes in Staphylococcus aureus. J. Bacteriol. 194, 2854–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Novick R. P. (2003) Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48, 1429–1449 [DOI] [PubMed] [Google Scholar]
- 58. Boisset S., Geissmann T., Huntzinger E., Fechter P., Bendridi N., Possedko M., Chevalier C., Helfer A. C., Benito Y., Jacquier A., Gaspin C., Vandenesch F., Romby P. (2007) Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 21, 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kobayashi S. D., Malachowa N., Whitney A. R., Braughton K. R., Gardner D. J., Long D., Bubeck Wardenburg J., Schneewind O., Otto M., Deleo F. R. (2011) Comparative analysis of USA300 virulence determinants in a rabbit model of skin and soft tissue infection. J. Infect. Dis. 204, 937–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Abdelnour A., Arvidson S., Bremell T., Rydén C., Tarkowski A. (1993) The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect. Immun. 61, 3879–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gillaspy A. F., Hickmon S. G., Skinner R. A., Thomas J. R., Nelson C. L., Smeltzer M. S. (1995) Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63, 3373–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cheung G. Y., Wang R., Khan B. A., Sturdevant D. E., Otto M. (2011) Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect. Immun. 79, 1927–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cheung A. L., Eberhardt K. J., Chung E., Yeaman M. R., Sullam P. M., Ramos M., Bayer A. S. (1994) Diminished virulence of a sar−/agr− mutant of Staphylococcus aureus in the rabbit model of endocarditis. J. Clin. Investig. 94, 1815–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Montgomery C. P., Boyle-Vavra S., Daum R. S. (2010) Importance of the global regulators Agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PloS One 5, e15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chua K. Y., Monk I. R., Lin Y. H., Seemann T., Tuck K. L., Porter J. L., Stepnell J., Coombs G. W., Davies J. K., Stinear T. P., Howden B. P. (2014) Hyperexpression of α-hemolysin explains enhanced virulence of sequence type 93 community-associated methicillin-resistant Staphylococcus aureus. BMC Microbiol. 14, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mayville P., Ji G., Beavis R., Yang H., Goger M., Novick R. P., Muir T. W. (1999) Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. U.S.A. 96, 1218–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wright J. S., 3rd, Jin R., Novick R. P. (2005) Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc. Natl. Acad. Sci. U.S.A. 102, 1691–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang P., Bagby G. J., Kolls J. K., Welsh D. A., Summer W. R., Andresen J., Nelson S. (2001) The effects of granulocyte colony-stimulating factor and neutrophil recruitment on the pulmonary chemokine response to intratracheal endotoxin. J. Immunol. 166, 458–465 [DOI] [PubMed] [Google Scholar]
- 69. Koyama S., Sato E., Masubuchi T., Takamizawa A., Kubo K., Nagai S., Izumi T. (1998) Alveolar type II-like cells release G-CSF as neutrophil chemotactic activity. Am. J. Physiol. Lung Cell. Mol. Physiol. 275, L687–L693 [DOI] [PubMed] [Google Scholar]
- 70. Koyama S., Sato E., Nomura H., Kubo K., Miura M., Yamashita T., Nagai S., Izumi T. (2000) The potential of various lipopolysaccharides to release IL-8 and G-CSF. Am. J. Physiol. Lung Cell. Mol. Physiol. 278, L658–L666 [DOI] [PubMed] [Google Scholar]
- 71. Saba S., Soong G., Greenberg S., Prince A. (2002) Bacterial stimulation of epithelial G-CSF and GM-CSF expression promotes PMN survival in CF airways. Am. J. Respir. Cell Mol. Biol. 27, 561–567 [DOI] [PubMed] [Google Scholar]
- 72. Balamayooran G., Batra S., Theivanthiran B., Cai S., Pacher P., Jeyaseelan S. (2012) Intrapulmonary G-CSF rescues neutrophil recruitment to the lung and neutrophil release to blood in Gram-negative bacterial infection in MCP-1−/− mice. J. Immunol. 189, 5849–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hua L., Hilliard J. J., Shi Y., Tkaczyk C., Cheng L. I., Yu X., Datta V., Ren S., Feng H., Zinsou R., Keller A., O'Day T., Du Q., Cheng L., Damschroder M., Robbie G., Suzich J., Stover C. K., Sellman B. R. (2014) Assessment of an anti-α-toxin monoclonal antibody for prevention and treatment of Staphylococcus aureus-induced pneumonia. Antimicrob. Agents Chemother. 58, 1108–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Reyes D., Andrey D. O., Monod A., Kelley W. L., Zhang G., Cheung A. L. (2011) Coordinated regulation by AgrA, SarA, and SarR to control agr expression in Staphylococcus aureus. J. Bacteriol. 193, 6020–6031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gupta R. K., Alba J., Xiong Y. Q., Bayer A. S., Lee C. Y. (2013) MgrA activates expression of capsule genes, but not the α-toxin gene in experimental Staphylococcus aureus endocarditis. J. Infect. Dis. 208, 1841–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. van Wamel W., Xiong Y. Q., Bayer A. S., Yeaman M. R., Nast C. C., Cheung A. L. (2002) Regulation of Staphylococcus aureus type 5 capsular polysaccharides by agr and sarA in vitro and in an experimental endocarditis model. Microb. Pathogen. 33, 73–79 [DOI] [PubMed] [Google Scholar]
- 77. Luong T., Sau S., Gomez M., Lee J. C., Lee C. Y. (2002) Regulation of Staphylococcus aureus capsular polysaccharide expression by agr and sarA. Infect. Immun. 70, 444–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Alén C., Sonenshein A. L. (1999) Bacillus subtilis aconitase is an RNA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 96, 10412–10417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Serio A. W., Pechter K. B., Sonenshein A. L. (2006) Bacillus subtilis aconitase is required for efficient late-sporulation gene expression. J. Bacteriol. 188, 6396–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Pechter K. B., Meyer F. M., Serio A. W., Stülke J., Sonenshein A. L. (2013) Two roles for aconitase in the regulation of tricarboxylic acid branch gene expression in Bacillus subtilis. J. Bacteriol. 195, 1525–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Regassa L. B., Betley M. J. (1992) Alkaline pH decreases expression of the accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 174, 5095–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Anderson K. L., Roux C. M., Olson M. W., Luong T. T., Lee C. Y., Olson R., Dunman P. M. (2010) Characterizing the effects of inorganic acid and alkaline shock on the Staphylococcus aureus transcriptome and messenger RNA turnover. FEMS Immunol. Med. Microbiol. 60, 208–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pané-Farré J., Jonas B., Förstner K., Engelmann S., Hecker M. (2006) The σB regulon in Staphylococcus aureus and its regulation. Int. J. Med. Microbiol. 296, 237–258 [DOI] [PubMed] [Google Scholar]