Abstract
We have examined the pattern of striatal messenger RNA expression of over 8000 genes in a rat model of levodopa (L-DOPA)-induced dyskinesia and Parkinson disease (PD). 6-Hydroxydopamine (6-OHDA)-lesioned rats were treated with L-DOPA or physiological saline for 22 days and repeatedly tested for antiakinetic response to L-DOPA and the development of abnormal involuntary movements (AIMs). In a comparison of rats that developed a dyskinetic motor response to rats that did not, we found striking differences in gene expression patterns. In rats that developed dyskinesia, GABA neurons had an increased transcriptional activity, and genes involved in Ca2+ homeostasis, in Ca2+-dependent signaling, and in structural and synaptic plasticity were upregulated. The gene expression patterns implied that the dyskinetic striatum had increased transcriptional, as well as synaptic activity, and decreased capacity for energy production. Some basic maintenance chores such as ribosome protein biosynthesis were downregulated, possibly a response to expended of ATP levels.
Keywords: L-DOPA, Dyskinesia, Parkinson’s disease, 6-Hydroxydop-amine, Gene microarrays, Striatum, Calcium, Ribosomes, ATPases, GABA
Introduction
Parkinson disease (PD) is a progressive neurological disorder characterized by a degeneration of dopamine (DA) neurons in the substantia nigra (Dauer and Przedborski, 2003). DA deficiency in areas innervated by nigral efferent neurons causes the motor symptoms of PD, that is, bradykinesia, rigidity, tremor, and postural instability (Gelb et al., 1999). The etiology of the disease is largely unknown. L-Dihydroxyphenylalanine (levodopa, L-DOPA) is used to treat the symptoms of PD, but it leads to the development of abnormal involuntary movements (AIMs; dyskinesia) complicating long-term treatment (Bezard et al., 2001; Nutt, 2001; Rascol et al., 2003).
We have developed a model of L-DOPA-induced dyskinesia in the rat (Cenci et al., 1998; Lee et al., 2000; Lundblad et al., 2002; Winkler et al., 2002). Rats are lesioned unilaterally with 6-hydroxydopamine (6-OHDA) and subsequently treated with relatively low doses of L- DOPA for a few weeks, during which a majority of the animals develop abnormal involuntary movements (AIMs), while some rats are resistant. This model allows us to study the biochemical and molecular factors involved in dyskinesia in a pharmacologically and genetically controlled environment.
One of the main target areas of nigral DA neurons is the striatum, a structure critically involved in the pathophysiology of parkinsonian motor symptoms (Sian et al., 1999). The striatum is also a prominent site of maladaptive molecular and synaptic plasticity in L-DOPA-induced dyskinesia (Andersson et al., 1999, 2001; Cenci et al., 1998; Johansson et al., 2001; Picconi et al., 2003; Westin et al., 2001). Most of the neurons in the striatum are projection neurons that use GABA as their neurotransmitter. These neurons can be further classified based on their expression of dopamine receptor subtypes and the neuropeptides enkephalin, dynorphin, and substance P (Gerfen, 1992a,b; Le Moine et al., 1991). The striatum has, in addition, interneurons expressing either acetylcholine, parvalbumin, or somatostatin (Kubota and Kawaguchi, 2000; Parent and Hazrati, 1995).
The aim of this study was to define changes in striatal gene expression that are associated with L-DOPA-induced dyskinesia in the rat model. Gene array technology was used to study the expression of over 8000 genes and expressed sequence tags (ESTs) in the striatum. Approximately 3000 of the genes examined were of known or inferred function and were expressed above back-ground levels in at least 20% of our samples. These genes were the focus of our investigation. We present the major differences in striatal gene expression patterns between rats that develop L-DOPA-induced dyskinesia and rats that do not develop dyskinetic side effects in response to L-DOPA, although they show motor improvement. Some of the most interesting genes were further subjected to in situ hybridization histochemistry (ISHH) in the dyskinesia model.
Methods
Subjects
The study was performed in Sprague–Dawley rats (BK Labs, Sweden) weighing 225 g at the beginning of the experiments. Rats were housed three per cage under a 12-h light–dark cycle, with ad libitum access to food and water. Rats from different experimental groups were randomly distributed in the cages. To conform to the procedures used in our previous studies, the experimental subjects were female. Estrus cycle phase was checked once a week in each rat by vaginal mucus analysis, and the different experimental groups had a comparable distribution of the different phases of the cycle. Animal care and experimental procedures conformed to internationally accepted guidelines and had been approved by the Malmö-Lund ethical committee for animal research.
6-OHDA lesions
Rats received unilateral injections of 6-OHDA (Sigma–Aldrich, Sweden AB) into the right nigrostriatal fiber bundle, as described previously (Andersson et al., 1999, 2001; Cenci et al., 1998; Lee et al., 2000). Turning behavior was recorded 2 weeks postlesion in an automated rotometer over a 90-min period after the injection of 2.5 mg/kg dexamphetamine sulfate (Apoteksbolaget, Sweden AB), and rats showing more than five full, ipsilateral turns per minute were selected for the study. This rotational score has been shown to correspond to >95% depletion of striatal DA fiber density (Winkler et al., 2002).
Protocol to induce dyskinesia and behavioral testing
Starting 5–6 weeks after lesioning, rats received single daily injections of methyl L-DOPA/benserazide (6/12 mg/kg i.p.; Sigma–Aldrich) for 22 days. 6-OHDA-lesioned control rats received single daily i.p. injections of saline. The injections were performed around 2:00 PM. Ratings of abnormal involuntary movements (AIMs) were carried out every 2–3 days (eight times total) for 3 h following the daily injection of L-DOPA. The rat dyskinesia scale and rating criteria used in this study are extensively described (Lundblad et al., 2002; Winkler et al., 2002). L-DOPA-induced rat AIMs affect orofacial, trunk, and limb muscles on the side of the body contralateral to the lesion and can be unequivocally distinguished from normal rodent behaviors (e.g., grooming, gnawing, and sniffing). Rat AIMs have the same pharmacological features as L-DOPA-induced dyskinesia in non-human primate models of PD; that is, drugs that reduce dyskinesia in the latter models have the same effect in the rat, and antiparkinsonian agents that have low dyskinesiogenic potential in primates induce little or no AIMs in the rat (Lundblad et al., 2002, 2003). L-DOPA-induced improvement of akinetic motor features was evaluated using a test of spontaneous forelimb use (cylinder test, Lundblad et al., 2002; Schallert et al., 2000). Based on the battery of behavioral tests used in this study, we defined three groups of experimental subjects: (i) saline-injected 6-OHDA-lesioned controls, which showed significant forelimb akinesia in the cylinder test and had no AIMs; these rats provided a model of untreated parkinsonism; (ii) dyskinetic rats, which were treated with L-DOPA and developed severe and disabling AIMs, and (iii) nondyskinetic rats, which were treated with L-DOPA and showed motor improvement in the cylinder test but did not rate on the AIMs scale. This latter group provided a model of treated parkinsonism without motor complications. The groups consisted of six animals each and will be referred to as saline, dyskinetic, and nondyskinetic, respectively.
Rats were sacrificed 18 h after the last injection of L-DOPA or saline, which is a sufficiently long time to avoid potential acute effects of the injection, including stress, while also avoiding effects due to L-DOPA withdrawal. The brains were rapidly extracted, frozen in liquid nitrogen for 20 s, and allowed to freeze thoroughly in powdered dry ice for 2–3 min. The brains were stored at −80°C until dissection. Dorsal striata, ipsilateral to the lesion, were dissected in a cryostat chamber at −18°C.
Sample and array processing
RNA was extracted from approximately 25-mg tissue using the RNAgent kit (Promega, Madison, WI). RNA quality was assessed, and 6 μg total RNA was used for cDNA synthesis with the SuperScript double-stranded cDNA synthesis kit (Invitrogen Corp., Carlsbad, CA). In vitro transcription was performed with the Enzo-IVT kit (Enzo Biochem, Farmingdale, NY). Biotinylated RNA was hybridized to the RG-U34A array (Affymetrix, Santa Clara, CA), and washing and staining were carried out according to company protocol (www.Affymetrix.com). Samples from individual rats were hybridized to individual arrays. The Affymetrix RG-U34A array contains over 8000 genes; each gene is represented by 16 to 20 perfectly matched 25-mer oligonucleotides and the same number of one-mismatch oligonucleotides to provide values for nonspecific binding.
Quality control criteria
Tissue preparation and RNA extractions were performed in a single batch by the same investigator to limit experimental variability. The order of samples was randomized. All striata yielded equal amounts of RNA and biotinylated RNA. An average of 92 ± 14 μg of biotinylated RNA was obtained from the in vitro transcription. All quality control criteria defined by Affymetrix were met by the samples, and no differences between the groups (saline, dyskinesia, or nondyskinesia) were observed. The average percent ‘present’ call across all arrays was 46.6% ± 2.0% and the 3′ /5′GAPDH and β-actin ratios were 1.9 ± 0.3 and 1.6 ± 0.3 respectively. Background (50.0 ± 3.7) and noise (1.6 ± 0.2) were comparable between all groups.
Data analysis
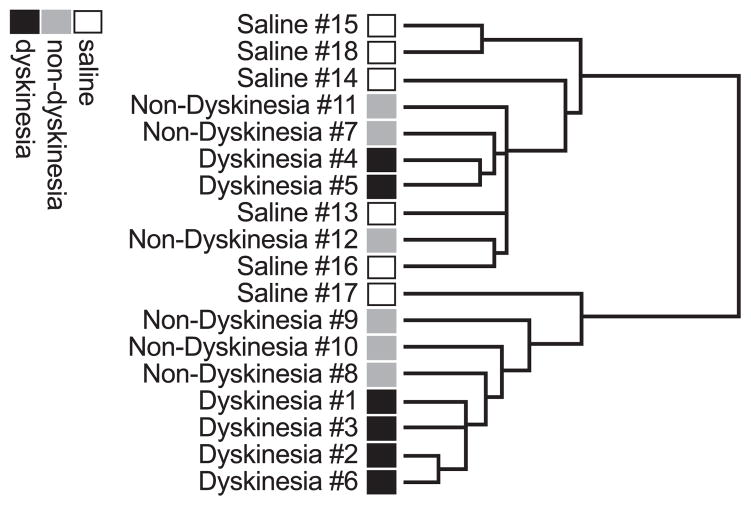
Three different programs were used for data analysis: DNA-Chip Analyzer (dChip version 1.3, http://www.biostat.harvard.edu/complab/dchip/, see also Li and Wong, 2001a,b), Microarray Suite 5.0 (Affymetrix), and Gene Microarray Pathway Profiler (http://www.genmapp.org/, Dahlquist et al., 2002; Doniger et al., 2003). To identify samples with similar profiles, hierarchical clustering was performed with the dChip program, which bases hierarchical clustering on previously published algorithms (Eisen et al., 1998; Golub et al., 1999). All genes with a standard deviation above 3% of the mean of their expression value and above detection limit in at least 20% of all samples were used for clustering (see http://www.biostat.harvard.edu/complab/dchip/filter_gene.htm). Redundant probe sets were excluded from the clustering analysis (Fig. 2).
Fig. 2.
Unsupervised hierarchical clustering of all genes with a standard deviation above 3% of the mean of their expression values and expressed above detection limit in at least 20% of all samples. Significant clustering of dyskinesia samples was observed (P = 0.022).
GenMAPP was used to examine the biological context of the findings. GenMAPP is designed to visualize gene expression data on maps representing either biological pathways or any other grouping of genes defined by the investigator. MAPPFinder calculates the percentage of genes changed in each map and uses this percentage for a z score based on the mean and the standard deviation of the hypergeometric distribution (see http://www.genmapp.org). MAPP-Finder calculates P values based on nonparametric statistics. We list all gene categories with at least two members, a z score above 2, and a permuted P value below 0.05 in Table 2.
Table 2.
MAPPFinder analysis of genes differentially regulated in rats that developed L-DOPA-induced AIMs compared to rats that do not develop AIMs
| Gene categories showing a significant overall change in the comparison between dyskinetic and nondyskinetic rats
| ||||
|---|---|---|---|---|
| Accession no. | Fold change | P value | P call % | |
| I. Ion homeostasis | ||||
| Ca2+ ATPases [4/7; z score 5.96; P = 0.0] | ||||
| ATPase, Ca2+ transporting, SERCA-2 | AA957510 | 1.07 | 0.040 | 100 |
| Plasma membrane calcium ATPase 1 | L04739 | 1.16 | 0.009 | 100 |
| Plasma membrane calcium ATPase 2 | J03754 | 1.15 | 0.048 | 100 |
| Plasma membrane calcium ATPase 3 | M96626 | −1.08 | 0.039 | 100 |
| II. Neurotransmitter synthesis, receptors, and transporters | ||||
| Neuropeptide [3/10; z score 3.38; P = 0.019] | ||||
| Neuropeptide Y | M15880 | −1.12 | 0.030 | 100 |
| Tachykinin 2 | M16410 | 1.61 | 0.000 | 100 |
| Secretogranin II | M93669 | 1.37 | 0.003 | 100 |
| Dopamine receptor [2/5; z score 3.36; P = 0.024] | ||||
| Dopamine D1 receptor (S46131mRNA_r_at) | S46131 | 1.24 | 0.008 | 100 |
| Dopamine D1 receptor (S46131mRNA_s_at) | S46131 | 1.17 | 0.011 | 100 |
| Dopamine D1 receptor | M35077 | 1.16 | 0.023 | 100 |
| Dopamine D1B receptor | M69118 | −1.05 | 0.026 | 100 |
| GABA [4/18; z score 3.06; P = 0.013] | ||||
| GABA-A receptor alpha-2 | L08491 | 1.22 | 0.033 | 100 |
| GABA-A receptor beta-3 | X15468 | 1.25 | 0.030 | 100 |
| Glutamate decarboxylase 1 (GAD67) | X57573 | 1.19 | 0.001 | 100 |
| Vesicular GABA transporter | AF030253 | 1.12 | 0.035 | 100 |
| Transporter-neurotransmitter [2/5; z score 3.38; P = 0.022] | ||||
| Vesicular GABA transporter | AF030253 | 1.12 | 0.035 | 100 |
| Glutamate/aspartate transporter 1 (glial) | S75687 | 1.15 | 0.041 | 100 |
| Glutamate/aspartate transporter 1 (glial) | X63744 | 1.21 | 0.039 | 100 |
| III. Structural and synaptic plasticity | ||||
| Pre-and postsynaptic structures [14/82; z score 4.60; P = 0] | ||||
| Cadherin | D83348 | 1.19 | 0.013 | 100 |
| Integrin alpha 7 | X65036 | 1.26 | 0.002 | 67 |
| SAP-97; Drosophila discs—large tumor suppressor homologue | AI144926 | 1.26 | 0.011 | 92 |
| SAP-97; Drosophila discs—large tumor suppressor homologue | U14950 | 1.2 | 0.020 | 100 |
| Homer 1 | AB003726 | 1.25 | 0.012 | 100 |
| Homer 1 | AF093267 | 1.18 | 0.015 | 100 |
| Homer 1 (AB017140_at) | AB017140 | 1.16 | 0.024 | 100 |
| Homer 1 (AB017140_g_at) | AB017140 | 1.15 | 0.035 | 100 |
| SNAP-25 | AB003992 | 1.15 | 0.034 | 100 |
| Syntaxin 2 | M95735 | 1.2 | 0.003 | 100 |
| Syntaxin 4 | L20821 | 1.26 | 0.005 | 100 |
| Adaptor protein complex AP-2, alpha-2 subunit | X53773 | 1.21 | 0.042 | 100 |
| Adaptor protein complex AP-2, beta-1 subunit | AA964379 | 1.15 | 0.005 | 100 |
| Clathrin assembly protein | AF041373 | 1.1 | 0.031 | 100 |
| Dynamin 2 | L25605 | −1.05 | 0.030 | 100 |
| Densin-180 | U66707 | 1.24 | 0.019 | 100 |
| Neuronal cell adhesion molecule (N-CAM) | X59149 | −1.12 | 0.014 | 100 |
| CRK-associated substrate | D29766 | −1.07 | 0.017 | 100 |
| Actin-related [7/30; z score 4.26; P = 0.001] | ||||
| Actinin alpha-2 associated LIM protein | AF002281 | 1.13 | 0.046 | 100 |
| Coronin, actin-binding protein 1A | AA892506 | −1.24 | 0.048 | 92 |
| SH3 domain-binding protein CR16 | U25281 | 1.14 | 0.006 | 100 |
| Activity-regulated cytoskeletal-associated protein (Arc) | U19866 | 1.23 | 0.043 | 100 |
| Troponin 1, type 3 | M92074 | 1.4 | 0.000 | 100 |
| Tropomodulin 2 | U59240 | 1.13 | 0.024 | 83 |
| Myristoylated, alanine-rich C-kinase substrate (MARCKS) | AA899253 | −1.14 | 0.024 | 100 |
| IV. Kinases and phosphatases | ||||
| Calcium kinases [3/10; z score 3.38; P = 0.01] | ||||
| Protein kinase C-family-related | M15523 | 1.13 | 0.007 | 100 |
| Protein kinase C delta (M18330_at) | M18330 | 1.26 | 0.017 | 75 |
| Protein kinase C delta (M18330_g_at) | M18330 | 1.26 | 0.022 | 100 |
| CaM-kinase II delta | L13406 | 1.1 | 0.029 | 25 |
| IV. Kinases and phosphatases | ||||
| Phosphatase [10/66; z score 3.39; P = 0.003] | ||||
| Protein tyrosine phosphatase, receptor type, O | U28938 | −1.1 | 0.048 | 100 |
| Protein tyrosine phosphatase, receptor type, R | D64050 | −1.11 | 0.043 | 42 |
| Protein tyrosine phosphatase-like N | D38222 | 1.24 | 0.008 | 100 |
| Protein tyrosine phosphatase, nonreceptor type 2 | X58828 | −1.08 | 0.040 | 100 |
| Acid phosphatase 2 | AI234950 | −1.06 | 0.016 | 33 |
| Regulatory subunit of type 1 protein phosphatase | S79213 | 1.17 | 0.036 | 100 |
| Regulatory subunit of protein phosphatase 2A | D14421 | 1.11 | 0.031 | 100 |
| Dual specificity phosphatase 6 (MAP kinase phosphatase 3) | U42627 | 1.18 | 0.038 | 100 |
| Multiple inositol polyphosphate histidine phosphatase 1 | AF012714 | −1.08 | 0.019 | 100 |
| Pyruvate dehydrogenase phosphatase 1 (AF062740_at) | AF062740 | 1.28 | 0.002 | 100 |
| Pyruvate dehydrogenase phosphatase 1 (AF062740_g_at) | AF062740 | 1.31 | 0.002 | 100 |
| V. Energy metabolism, toxicity, and apoptosis | ||||
| Creatine biosynthesis [2/4; z score 3.88; P = 0.01] | ||||
| Guanidinoacetate methyltransferase | J03588 | −1.06 | 0.034 | 83 |
| Ubiquitous mitochondrial creatine kinase (X59737mRNA_at) | X59737 | −1.16 | 0.001 | 100 |
| Ubiquitous mitochondrial creatine kinase (X59737mRNA_g_at) | X59737 | −1.14 | 0.006 | 100 |
| Heat shock protein [2/8; z score 2.4; P = 0.05] | ||||
| Heat shock 20-kDa protein | D29960 | −1.14 | 0.044 | 100 |
| Heat shock 70-kDa protein (S78556_at) | S78556 | 1.23 | 0.018 | 100 |
| Heat shock 70-kDa protein (S78556_g_at) | S78556 | 1.13 | 0.003 | 100 |
| VI. Ribosomal proteins [9/57; z score 3.39; P = 0.005] | ||||
| Ribosomal protein L12 | X53504 | −1.09 | 0.046 | 100 |
| Ribosomal protein L13 | X78327 | −1.1 | 0.026 | 100 |
| Ribosomal protein L17 | X60212 | −1.16 | 0.004 | 100 |
| Ribosomal protein L18a (X14181cds_r_at) | X14181 | −1.12 | 0.006 | 100 |
| Ribosomal protein L18a (X14181cds_s_at) | X14181 | −1.1 | 0.010 | 100 |
| Ribosomal protein L26 | X14671 | −1.09 | 0.047 | 100 |
| Ribosomal protein S15 | E01534 | −1.1 | 0.031 | 100 |
| Phosphoribosyl pyrophosphate synthetase-associated protein 2 | AI231500 | −1.09 | 0.011 | 100 |
| Phosphoribosyl pyrophosphate synthetase, subunit II | X16555 | 1.07 | 0.049 | 100 |
All regulated genes with a P value <0.05 and 20% or more ‘present’ call across all samples were used for the analysis. Two hundred sixty-two probes met the criteria and were included in the analysis. The table shows all gene families that had at least 2 regulated members, reached a z score above 2, and a permuted P value at or below 0.05. The z score was based on an N of 4141 and an R of 233 distinct genes. Positive values indicate higher mRNA levels in the dyskinetic group, negative values indicate higher mRNA levels in the nondyskinetic group.
In situ hybridization histochemistry
We performed ISHH in an independent batch of animals (n = 6 in each group), treated, and selected exactly like the rats for the gene array study. Cryostat sections through the striatum were cut at 16-μm thickness, mounted on Superfrost Plus Adhesion Slides (Electron Microscopy Science, Fort Washington, PA), and stored at −20°C. The oligonucleotide probes used for in situ hybridization in this study are shown in Table 1. ISHH was performed according to a previously published method (Andersson et al., 1999). Oligonucleotides were radioactively labeled at the 3′ end with α35S-dATP and terminal deoxynucleotidyl transferase (Amersham Pharmacia, UK). Hybridization was carried out in a humid chamber at 42°C for 16–18 h, and the slide-mounted sections were washed five times in 1× SSC at 55°C. Sections were exposed to FUJI Imaging plates (Fujifilm Sweden AB) between 9 and 22 h. Plates were scanned in a BAS-5000 phosphorimager (Fujifilm), and the amount of photostimulated luminescence emitted from the hybridized sections was calibrated against 14C standards (Amersham Pharmacia). Densitometric analysis was performed with the TINA software (Fujifilm). For each animal and probe, we analyzed five sections through the main body of the striatum, corresponding to the levels 0.2 to 0.7 mm rostral to bregma (Paxinos and Watson, 1986). Measurements from either side of the striatum were averaged across sections. Statistical analysis was performed with one-way analysis of variance (ANOVA) followed by Fisher post hoc test and significance levels at P <0.05. No significant group differences were found on contralateral side to the lesion, and only results from the DA-denervated side will be reported (see Fig. 3).
Table 1.
Oligonucleotide sequences used for in situ hybridization histochemistry
| Gene | Oligonucleotide composition 5′-3′ | No. of nucleotides |
Corresponding to nucleotides |
Genbank accession no. |
|---|---|---|---|---|
| CamKIV | CGTCGCGTTCACTGTAATAGCCTTTCTCCACAATCCTGTC | 41 | 523–562 | NM_012727 |
| CB1 receptor | GTGGTGATGGTACGGAAGGTGGTGTCTGCAAGGCCATCTAGGATCGAC | 48 | 158–205 | NM_012784 |
| CO1 | GGACGGCCGTAAGTGAGATGAATGAGCCTATAGAGGAGACTG | 42 | 710–751 | S79304.1 |
| D1 receptor | CCCGTTACCTGCGGTGGTCTGGCAATTCTTGGCATGGAC | 39 | 1457–1495 | NM_012546 |
| GABA-A α4 | TCCTGGGCAGCGGTGGTCTTGCTGGAATGGTTCCCCACCTCAG | 43 | 1302–1344 | NM_080587 |
| Homer 1 | GGTCAGTTCCATCTTCTCCTGCGACTTCTCCTTTGCCAG | 39 | 628–666 | NM_031707 |
| NaK-ATPase α1 | GAGTTCGTCCATGTCCCTTTCCTTCTTCGCCTTCTTGCTCTTCTTGTCC | 49 | 294–342 | NM_012504 |
| NF-H | GCAGAGCGCATAGCATCCGTGTTCACTTTGGCTGCCTCTGAGAGTCGG | 48 | 945–992 | NM_012607 |
| PMCA | CGGCATGGTCAATCTCTTCAACATCCTCCGCCAATTCCTCCTCAG | 45 | 3509–3553 | NM_053311 |
| VGAT | GCCGGTGTAGCAGCACACCACCGCGGCGAAGATGATGAGGAACAACC | 47 | 529–575 | AF030253 |
| PKC delta | GAGACAGCTGTCTTCTTCTCGAATCCCTGGTATATTCCGACAGTCTCTG | 49 | 1276–1324 | NM_133307 |
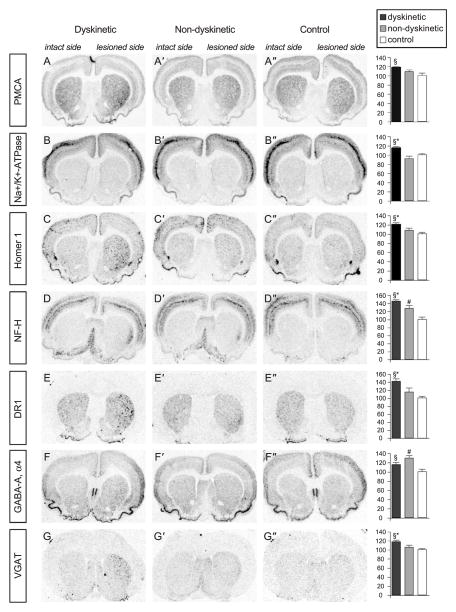
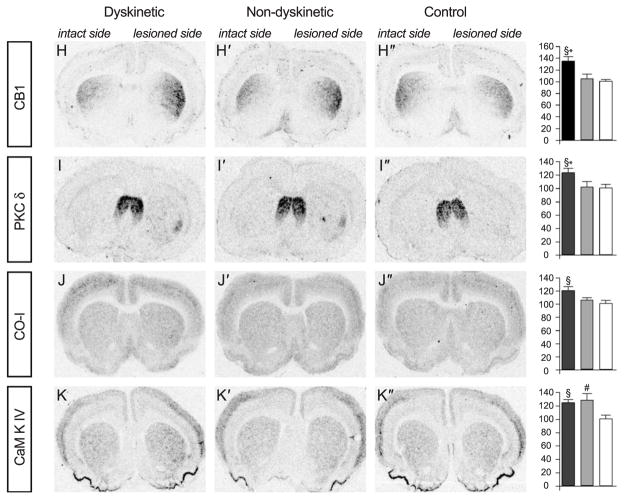
Fig. 3.
Gene changes validated by ISHH. The rows show film autoradiographs of striatal sections from dyskinetic rats, nondyskinetic rats, and control animals. Autoradiographs were obtained with the following probes: A, plasma membrane transporting ATPase 1 (PMCA1); B, the alpha1 subunit of Na+K+-ATPase (NaK-ATPase); C, homer 1; D, neurofilament heavy (NF-H); E, dopamine D1 receptor (DR1); F, α4 subunit of the GABA-A receptor (GABA-A α4); G, the vesicular GABA transporter (VGAT); H, the cannabinoid CB1 receptor (CB1); I, protein kinase C delta (PKCδ); J, cytochrome oxidase I (CO-I); K, calcium-calmodulin kinase IV (CamKIV). The results of the quantitative analysis (percent of control of lesioned side/unlesioned side) are shown in the column at right. P <0.05 for *, dyskinetic versus nondyskinetic; §, dyskinetic versus saline; #, nondyskinetic versus control.
Results
Approximately 4500 mRNAs (3000 known sequences plus ESTs) were above detection limit in at least 20% of the samples and were used for further analysis. At a probability level below 0.05 (P <0.05), 12% of these mRNAs were expressed at altered levels in the dyskinetic striata compared to saline controls, whereas only 5% of all mRNAs of nondyskinetic striata had altered levels (Fig. 1). Samples from dyskinetic rats differed from the non-dyskinetic cases in over 6% of all mRNA species. Of all regulated mRNA species, twice as many were upregulated than down-regulated in dyskinetic as well as nondyskinetic striata, compared to saline-injected controls. The false discovery rate, which calculates the expected proportion of false-positive results in multiple-comparisons analyses (Tusher et al., 2001), was <20% at P ≤ 0.05 and <5% at P ≤ 0.01.
Fig. 1.
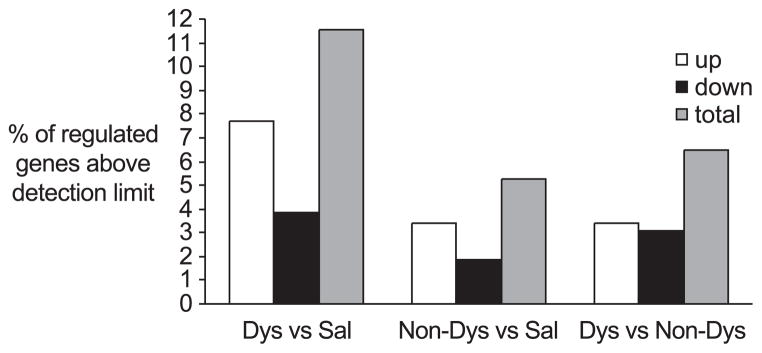
Percent of all mRNAs that differ in the expression levels between the three experimental groups. Bars for upregulation (white), downregulation (black), and total regulation (grey) are presented. Comparisons are presented in experiments versus baseline. Dys indicates rats that developed dyskinesia in response to L-DOPA treatment; Non-dys, rats that did not develop dyskinesia; Sal, rats that were lesioned with 6-OHDA and treated with saline.
Hierarchical clustering shows similar expression patterns in the dyskinetic rats
Unsupervised clustering was used to test if variations in gene expression levels could be explained by the state of dyskinesia. Forty-six genes had sufficient expression levels and variability across groups to be included in the analysis. With these genes, a significant clustering of four dyskinesia samples was observed (Fig. 2; P = 0.022). Samples from nondyskinetic rat striata were spread among lesioned controls and dyskinetic samples, with no significant difference from either group. These data demonstrate that the dyskinetic rats have a unique gene expression profile.
To retrieve information in a biologic context, which is the strength of gene array experiments, we determined if a dyskinetic motor response was accompanied by higher-than-expected regulations in gene categories defined by us a priori. The regulations within gene families coding for individual neuronal-specific structures, processes, or functions were examined with the help of the MAPP (Microarray Pathway Profile)-finder program, which was provided by the GenMAPP group. In this program, we built over 300 MAPPs consisting of genes with expected expression in striatal neurons, grouped according to neurotransmitter systems (e.g., dopamine receptors, and transporters), ion channels (e.g., Ca2+, Na+, or K+ channel proteins), membrane pumps (e.g., ion pumps, ATP pumps), presynaptic proteins (e.g., clathrin-associated proteins), postsynaptic proteins (e.g., receptors and anchoring proteins), structural proteins (e.g., cytoskeletal proteins, ribosomal proteins), proteins involved in energy metabolism (e.g., mitochondrial respiratory chain), immediate early genes, transcription factors, etc. The main results from the comparison between dyskinetic and nondyskinetic cases are shown in Table 2. Categories of genes that showed a significant upregulation in dyskinetic versus nondyskinetic rats included Ca2+ transporting ATPases and Ca2+ kinases, structural and synaptic plasticity genes, GABA receptor subunits, and other GABA-related genes. Significant downregulations were observed in gene categories related to creatine biosynthesis, ribosomal proteins, and tyrosine phosphatases (Table 2).
Calcium transporting ATPases and ion homeostasis
The regulation of Ca2+ transporting ATPases received the highest z score in the MAPPFinder analysis (Table 2). L-DOPA-induced-dyskinesia was associated with an upregulation of genes coding for plasma membrane Ca2+ ATPases (PMCAs) and sarco/ endoplasmic reticulum Ca2+ pumps (SERCAs). PMCA1, PMCA2, and SERCA2 mRNAs were significantly upregulated in the dyskinetic cases compared to both saline-injected animals and L-DOPA-treated nondyskinetic rats. Nondyskinetic rats did not differ from controls. Upregulation of PMCA1 in the dyskinetic striatum was verified with ISHH, which confirmed enhanced levels of this transcript in the lateral caudate-putamen (Fig. 3A). In addition, expression of many other genes involved in ion homeostasis was enhanced in rats treated with L-DOPA, with generally more prominent upregulation in the dyskinetic group (Table 3). With ISHH, we verified the significant upregulation of the alpha1 subunit of Na+K+-ATPase in L-DOPA-treated rats that had developed AIMs. In contrast to the gene array study, the upregulation was restricted to rats that did develop AIMs. Similarly to PMCA1, the change in alpha1 Na+K+-ATPase mRNA was more prominent in the lateral part of the caudate-putamen (Fig 3B). We have previously shown that this region is critical in driving the development of L-DOPA-induced AIMs in the rat, and it exhibits large upregulations of transcription factors and neurotransmitter-encoding mRNAs (Andersson et al., 1999; Cenci et al., 1998).
Table 3.
mRNAs of ion homeostasis and proton transport
| Ion homeostasis and proton transport | Accession no. | Dyskinesia/nondys
|
Dyskinesia/saline
|
Nondys/saline
|
P call %
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold change | P value | Fold change | P value | Fold change | P value | Dyskinesia | Nondys | Saline | |||
| Upregulation | |||||||||||
| Voltage-gated potassium channel Kv1.1 | X12589 | 1.22 | 0.000 | 1.25 | 0.000 | 1.03 | 0.587 | 100 | 100 | 100 | * |
| Voltage-gated potassium channel Kv1.1 | M26161 | 1.12 | 0.003 | 1.15 | 0.009 | 1.03 | 0.527 | 100 | 100 | 100 | * |
| Calcium ATPase, cardiac muscle, slow twitch 2 (SERCA 2) | AA957510 | 1.07 | 0.040 | 1.15 | 0.035 | 1.07 | 0.274 | 100 | 100 | 100 | * |
| Calcium ATPase, cardiac muscle, slow twitch 2 (SERCA 2) | AA799276 | 1.05 | 0.236 | 1.08 | 0.050 | 1.03 | 0.442 | 100 | 100 | 100 | |
| Calcium ATPase, plasma membrane 1 (PMCA 1) | L04739 | 1.16 | 0.009 | 1.21 | 0.009 | 1.04 | 0.474 | 100 | 100 | 100 | * |
| Calcium ATPase, plasma membrane 2 (PMCA 2) | J03754 | 1.15 | 0.048 | 1.24 | 0.012 | 1.07 | 0.221 | 100 | 100 | 100 | * |
| Calcium ATPase, plasma membrane 2 (PMCA 2) | AA943784 | 1.21 | 0.107 | 1.42 | 0.011 | 1.17 | 0.092 | 100 | 100 | 100 | |
| Calcium ATPase, plasma membrane 2 (PMCA 2) | L05557 | 1.14 | 0.496 | 1.48 | 0.039 | 1.30 | 0.184 | 100 | 83 | 66 | |
| Sodium, potassium-ATPase beta-1 | X63375 | 1.08 | 0.123 | 1.37 | 0.002 | 1.27 | 0.015 | 100 | 100 | 66 | |
| Sodium, potassium-ATPase alpha-1 | M74494 | 1.02 | 0.862 | 1.43 | 0.030 | 1.40 | 0.023 | 100 | 100 | 100 | |
| Sodium bicarbonate cotransporter (NBC) | AF004017 | 1.25 | 0.017 | 1.35 | 0.010 | 1.08 | 0.458 | 100 | 100 | 100 | * |
| Sodium-potassium-chloride cotransporter homolog rBSC2 | S82233 | 1.08 | 0.039 | 1.1 | 0.012 | 1.02 | 0.578 | 100 | 100 | 100 | * |
| Sodium-potassium-2chloride cotransporter (Nkcc1) | AF086758 | 1.09 | 0.146 | 1.15 | 0.014 | 1.05 | 0.431 | 100 | 83 | 66 | |
| Calcium channel alpha-1 A | M99222 | 1.01 | 0.647 | 1.15 | 0.019 | 1.13 | 0.028 | 100 | 100 | 66 | |
| Calcium channel alpha-1 A | AI229031 | −1.03 | 0.464 | 1.19 | 0.031 | 1.23 | 0.013 | 100 | 100 | 50 | |
| H+-ATPase, lysosomal (vacuolar proton pump), 16 kDa | D10874 | 1.03 | 0.558 | 1.10 | 0.030 | 1.07 | 0.294 | 100 | 100 | 100 | |
| H+-ATPase, lysosomal (vacuolar proton pump), beta 56/58 kDa | Y12635 | 1.14 | 0.311 | 1.57 | 0.013 | 1.38 | 0.016 | 100 | 100 | 100 | |
| Sodium/hydrogen exchanger 1, (amiloride-sensitive) | M85299 | 1 | 0.956 | 1.06 | 0.040 | 1.06 | 0.108 | 100 | 100 | 83 | |
| Furosemide-sensitive potassium chloride cotransporter (KCC2) | U55816 | −1.02 | 0.456 | 1.14 | 0.009 | 1.16 | 0.005 | 100 | 100 | 100 | |
| Endosulfine alpha (reduces potassium-ATP channel currents) | AJ005984 | 1.21 | 0.040 | 1.19 | 0.098 | −1.02 | 0.865 | 100 | 100 | 100 | * |
| Potassium channel alpha subunit, Kv8.1 | X98564 | 1.14 | 0.166 | 1.29 | 0.033 | 1.13 | 0.141 | 100 | 100 | 100 | |
| Potassium channel ROMK1.1 | AF081365 | 1.07 | 0.071 | −1.05 | 0.3360 | −1.13 | 0.048 | 100 | 83 | 100 | |
| Valosin-containing protein, transitional endoplasmic reticulum ATPase | U11760 | 1.13 | 0.186 | 1.18 | 0.030 | 1.04 | 0.611 | 100 | 100 | 100 | |
| Downregulation | |||||||||||
| Chloride channel (ClC-2) | AF005720 | −1.13 | 0.002 | −1.09 | 0.011 | 1.04 | 0.215 | 100 | 100 | 100 | * |
| Natural resistance-associated macrophage protein 2 (Nramp2) | AF008439 | −1.06 | 0.037 | −1.01 | 0.815 | 1.05 | 0.294 | 100 | 100 | 100 | * |
| Plasma membrane calcium ATPase 3 (PMCA 3) | M96626 | −1.08 | 0.039 | 1.15 | 0.268 | 1.24 | 0.091 | 100 | 100 | 100 | * |
| Amiloride-sensitive cation channel 2, neuronal | AJ006519 | −1.03 | 0.531 | −1.12 | 0.052 | −1.08 | 0.036 | 16 | 33 | 16 | |
Comparisons of gene expression changes between dyskinetic, nondyskinetic, and saline animals. Fold change and P values are presented. Upregulations in dyskinetic animals are listed first, downregulations are underneath. In case of opposite regulations in different comparison groups, the comparison of dyskinetic to nondyskinetic animals decided placement. Percent present call (P call %) is given for each experimental group in the last three columns. The asterisk denotes significant differences between dyskinetic and nondyskinetic animals.
Synaptic function
A large number of genes encoding pre- and postsynaptic structures, such as synaptic scaffold proteins and proteins involved in the docking and fusion of synaptic vesicles, were prominently upregulated in L-DOPA-treated rats that had developed AIMs. Significant changes were found in dyskinetic rats compared to both saline-injected rats and nondyskinetic cases (Table 4). Among the upregulated genes were densin 180, SAP-97, and Homer 1, which constitute part of the molecular scaffold at postsynaptic densities (PSD) of excitatory synapses (for review, see Sheng, 2001). The long isoforms of Homer (Homer 1b/c) physically link type I metabotropic glutamate receptors to the PSD and to calcium release channels in the endoplasmic reticulum (Thomas, 2002). In ISHH, we found significant upregulation of the Homer 1 transcript in L-DOPA-treated rats that had developed AIMs compared to both nondyskinetic rats and saline-injected lesioned controls, thus confirming the gene array data (Fig. 3C).
Table 4.
mRNAs of structural plasticity
| Structural plasticity | Accession no. | Dyskinesia/nondys
|
dyskinesia/saline
|
Nondys/saline
|
P call %
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold change | P value | Fold change | P value | Fold change | P value | Dyskinesia | Nondys | Saline | |||
| Upregulation | |||||||||||
| Homer | AB003726 | 1.25 | 0.012 | 1.09 | 0.343 | −1.15 | 0.147 | 100 | 100 | 100 | * |
| Homer | AF093267 | 1.18 | 0.015 | 1.3 | 0.041 | 1.1 | 0.413 | 100 | 100 | 100 | * |
| Homer (AB017140_at) | AB017140 | 1.16 | 0.024 | 1.19 | 0.140 | 1.03 | 0.817 | 100 | 100 | 100 | * |
| Homer (AB017140_g_at) | AB017140 | 1.15 | 0.035 | 1.21 | 0.054 | 1.05 | 0.564 | 100 | 100 | 100 | * |
| Densin-180 | U66707 | 1.24 | 0.023 | 1.34 | 0.017 | 1.08 | 0.521 | 100 | 100 | 100 | * |
| SAP-97 (discs, large homolog 1) | AI144926 | 1.26 | 0.011 | 1.28 | 0.033 | 1.01 | 0.914 | 100 | 83 | 66 | * |
| SAP-97 (discs, large homolog 1) | U14950 | 1.2 | 0.020 | 1.19 | 0.075 | −1.01 | 0.877 | 100 | 100 | 100 | * |
| SNAP-25 | AB003992 | 1.15 | 0.034 | 1.14 | 0.279 | −1.01 | 0.962 | 100 | 100 | 100 | * |
| Neurofilament, heavy polypeptide | AA818677 | 1.39 | 0.001 | 1.47 | 0.001 | 1.06 | 0.243 | 100 | 100 | 100 | * |
| Neurofilament, heavy polypeptide | X13804 | 1.08 | 0.108 | 1.11 | 0.036 | 1.03 | 0.359 | 100 | 100 | 83 | |
| Neurofilament, light polypeptide | M25638 | −1.05 | 0.472 | 1.29 | 0.020 | 1.35 | 0.006 | 100 | 100 | 100 | |
| Neurofilament, light polypeptide | AF031880 | −1.04 | 0.455 | 1.15 | 0.026 | 1.2 | 0.008 | 100 | 100 | 100 | |
| Gephyrin | X66366 | 1.12 | 0.002 | 1.12 | 0.024 | 1 | 0.995 | 100 | 100 | 100 | * |
| Integrin alpha-7 | X65036 | 1.26 | 0.002 | 1.28 | 0.001 | 1.02 | 0.647 | 100 | 33 | 16 | * |
| Syntaxin 1B | M95735 | 1.2 | 0.003 | 1.13 | 0.073 | −1.05 | 0.455 | 100 | 100 | 100 | * |
| Syntaxin 4 | L20821 | 1.26 | 0.005 | 1.09 | 0.278 | −1.16 | 0.112 | 100 | 100 | 100 | * |
| Syntaxin 6 | U56815 | 1.02 | 0.633 | 1.21 | 0.004 | 1.19 | 0.002 | 83 | 66 | 0 | |
| Neuron-specific protein PEP-19 (Purkinje cell protein 4) | M24852 | 1.17 | 0.004 | 1.08 | 0.107 | −1.09 | 0.088 | 100 | 100 | 100 | * |
| Adaptor protein complex AP-2, beta-1 subunit | AA964379 | 1.15 | 0.005 | 1.02 | 0.822 | −1.13 | 0.235 | 100 | 100 | 100 | * |
| Adaptor protein complex AP-2, alpha-2 subunit | X53773 | 1.21 | 0.042 | 1.35 | 0.008 | 1.12 | 0.167 | 100 | 100 | 83 | * |
| Reelin | AA893471 | 1.26 | 0.006 | 1.5 | 0.000 | 1.19 | 0.030 | 100 | 100 | 100 | * |
| f-Spondin | M88469 | 1.2 | 0.008 | 1.35 | 0.000 | 1.12 | 0.070 | 100 | 100 | 100 | * |
| p47 Protein (AB002086_at) | AB002086 | 1.12 | 0.012 | 1.16 | 0.008 | 1.03 | 0.419 | 100 | 100 | 100 | * |
| P47 Protein (AB002086_g_at) | AB002086 | 1.14 | 0.048 | 1.24 | 0.006 | 1.09 | 0.033 | 100 | 100 | 100 | * |
| Cadherin 22 | D83348 | 1.19 | 0.013 | 1.23 | 0.009 | 1.03 | 0.723 | 100 | 100 | 100 | * |
| Amphiphysin | Y13381 | 1.08 | 0.030 | 1.12 | 0.003 | 1.04 | 0.261 | 100 | 100 | 100 | * |
| Phosphatidylinositol-binding clathrin assembly protein | AF041373 | 1.1 | 0.031 | 1.12 | 0.035 | 1.01 | 0.840 | 100 | 100 | 83 | * |
| Component of rsec6/8 secretory complex p71 | U79417 | 1.16 | 0.037 | 1.3 | 0.006 | 1.12 | 0.135 | 100 | 100 | 100 | * |
| Arc; activity-regulated cytoskeletal- associated protein | U19866 | 1.23 | 0.043 | 1.04 | 0.641 | −1.18 | 0.016 | 100 | 100 | 100 | * |
| Actinin alpha-2 associated LIM protein | AF002281 | 1.13 | 0.046 | −1.01 | 0.921 | −1.14 | 0.055 | 100 | 100 | 83 | * |
| Synapsin 2 (rc_AI145494_g_at) | AI145494 | 1.16 | 0.069 | 1.64 | 0.000 | 1.41 | 0.006 | 100 | 100 | 100 | |
| Synapsin 2 (rc_AI145494_at) | AI145494 | 1.2 | 0.068 | 1.51 | 0.002 | 1.26 | 0.068 | 100 | 100 | 100 | |
| Synapsin 2 | M27925 | 1.06 | 0.423 | 1.33 | 0.001 | 1.25 | 0.023 | 100 | 100 | 100 | |
| Microtubule-associated protein 2 | X17682 | 1.24 | 0.073 | 1.56 | 0.003 | 1.25 | 0.188 | 100 | 100 | 100 | |
| Microtubule-associated protein 2 | X53455 | 1.19 | 0.059 | 1.35 | 0.008 | 1.13 | 0.284 | 100 | 100 | 83 | |
| Latexin | X76985 | 1.14 | 0.172 | 1.38 | 0.005 | 1.21 | 0.114 | 100 | 100 | 100 | |
| GERp95 | H31692 | 1.09 | 0.205 | 1.29 | 0.007 | 1.19 | 0.076 | 100 | 100 | 100 | |
| Narp; neuronal activity-regulated pentraxin | S82649 | 1.06 | 0.231 | 1.17 | 0.009 | 1.1 | 0.123 | 100 | 100 | 100 | |
| Dynamin 2 | AA851887 | 1.03 | 0.724 | 1.31 | 0.011 | 1.27 | 0.011 | 100 | 100 | 100 | |
| Neurexin 1 | AI146018 | 1.07 | 0.226 | 1.26 | 0.012 | 1.17 | 0.080 | 83 | 83 | 50 | |
| Integrin-associated protein (AF017437_at) | AF017437 | 1.02 | 0.787 | 1.26 | 0.015 | 1.23 | 0.006 | 100 | 100 | 100 | |
| Integrin-associated protein (AF017437_g_at) | AF017437 | 1.09 | 0.107 | 1.22 | 0.026 | 1.12 | 0.192 | 100 | 100 | 100 | |
| Downregulation | |||||||||||
| Neural cell adhesion molecule L1 | X59149 | −1.12 | 0.014 | −1.12 | 0.017 | 1.01 | 0.765 | 100 | 100 | 100 | * |
| Dynamin 2 | L25605 | −1.05 | 0.030 | −1.04 | 0.473 | 1.01 | 0.821 | 100 | 100 | 66 | * |
| Vesicle-associated calmodulin- binding protein | L22557 | −1.13 | 0.039 | −1.07 | 0.177 | 1.06 | 0.281 | 100 | 100 | 100 | * |
| Vascular cell adhesion molecule 1 | X63722 | −1.02 | 0.730 | −1.13 | 0.024 | −1.12 | 0.007 | 100 | 100 | 100 | |
Comparisons of gene expression changes between dyskinetic, nondyskinetic, and saline animals. Fold change and P values are presented. Upregulations are listed first, downregulations are underneath. In case of opposite regulations in different comparison groups, the comparison of dyskinetic to nondyskinetic animals decided placement. Percent present call (P call %) is given for each experimental group in the last three columns. The asterisk denotes significant differences between dyskinetic and nondyskinetic animals.
Actin-binding proteins and cytoskeletal constituents
Genes encoding actin-related proteins received a high z score in the MAPPFinder analysis. Most of these genes showed a prominent upregulation in the dyskinetic group compared to both saline-injected controls and nondyskinetic cases. The numerous expression changes of actin-related genes suggest that a process of cellular remodeling is taking place in the dyskinetic striatum. In support of this hypothesis, we found that several genes encoding neuron-specific intermediate filament proteins and microtubule-associated proteins were upregulated in L-DOPA-treated animals that developed dyskinesia (Table 4). We verified the expression of high molecular weight neurofilament (NF-H) mRNA by ISHH (Fig. 3D). NF-H mRNA was upregulated in all L-DOPA-treated rats but significantly more so in the animals that had developed AIMs. Again, the upregulation was particularly prominent in the ventrolateral part of the caudate-putamen.
Neurotransmitter synthesis, receptors, and transporters
Chronic L-DOPA treatment caused altered expression of many genes related to dopamine and GABA transmission (Tables 2, 5). In rats that developed dyskinesia, mRNA levels for the D1 receptor were significantly increased, an effect confirmed by ISHH (Fig. 3E). Downregulations in mRNA levels were observed for the dopamine transporter and for the dopamine D3 and D5 receptors; however, these data were not conclusive because the changes were very small and some transcripts had low expression levels (see, for example, the percent positive calls for the D3 receptor in Table 5). Levels of D2 receptor mRNA were unchanged. The GABA system showed upregulation of various GABA-A receptor subunit mRNAs (alpha-1, -2, -4, beta-3), glutamate decarboxylase 67 (GAD67), and the vesicular GABA transporter. The finding of striatal GAD67 mRNA upregulation in dyskinetic rats is in agreement with previous in situ hybridization studies (Cenci et al., 1998). The increased expression of mRNAs encoding the GABA-A alpha4 subunit and the vesicular GABA transporter in dyskinetic rats was confirmed by ISHH (Figs. 3F and G).
Table 5.
mRNAs of neurotransmitter synthesis, receptors, and transporters
| Neurotransmitter synthesis, receptors and transporters | Accession no. | Dyskinesia/nondys
|
Dyskinesia/saline
|
nondys/saline
|
P call %
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold change | P value | Fold change | P value | Fold change | P value | Dyskinesia | Nondys | Saline | ||||
| Upregulation | ||||||||||||
| Cannabinoid receptor 1 | X55812 | 1.35 | 0.000 | 1.25 | 0.029 | −1.08 | 0.425 | 100 | 100 | 100 | * | |
| Somatostatin receptor 2 | M93273 | 1.15 | 0.016 | 1.14 | 0.022 | −1 | 0.929 | 100 | 100 | 100 | * | |
| Tachykinin 2 (neurokinin B, neuromedin K) | M16410 | 1.61 | 0.000 | 1.43 | 0.004 | −1.13 | 0.316 | 100 | 100 | 100 | * | |
| Secretogranin II | M93669 | 1.37 | 0.003 | 1.34 | 0.005 | −1.02 | 0.855 | 100 | 100 | 100 | * | |
| Cholecystokinin | X01032 | 1.41 | 0.054 | 2.26 | 0.000 | 1.61 | 0.063 | 100 | 100 | 66 | ||
| Glutamate decarboxylase 1 (GAD67) | GABA | X57573 | 1.19 | 0.001 | 1.21 | 0.003 | 1.02 | 0.737 | 100 | 100 | 100 | * |
| GABA-A receptor alpha-1 | L08490 | 1.06 | 0.298 | 1.11 | 0.026 | 1.04 | 0.552 | 100 | 100 | 100 | ||
| GABA-A receptor alpha-2 | L08491 | 1.22 | 0.033 | 1.21 | 0.027 | −1.01 | 0.935 | 100 | 100 | 100 | * | |
| GABA-A receptor alpha-4 | S55933 | 1.31 | 0.073 | 1.83 | 0.002 | 1.4 | 0.057 | 100 | 100 | 100 | ||
| GABA-A receptor alpha-4 | L08493 | 1.08 | 0.129 | 1.1 | 0.025 | 1.02 | 0.605 | 100 | 100 | 100 | ||
| GABA-A receptor beta-3 | X15468 | 1.25 | 0.030 | 1.39 | 0.002 | 1.12 | 0.261 | 100 | 100 | 66 | * | |
| Vesicular GABA transporter | AF030253 | 1.12 | 0.035 | 1.18 | 0.008 | 1.05 | 0.298 | 100 | 100 | 100 | * | |
| Dopamine D1 receptor (S46131mRNA_r_at) | S46131 | 1.24 | 0.008 | 1.04 | 0.656 | −1.2 | 0.053 | 100 | 100 | 100 | * | |
| Dopamine D1 receptor (S46131mRNA_s_at) | S46131 | 1.17 | 0.011 | 1.11 | 0.178 | −1.05 | 0.441 | 100 | 100 | 100 | * | |
| Dopamine D1 receptor | M35077 | 1.16 | 0.023 | 1.08 | 0.445 | −1.07 | 0.536 | 100 | 100 | 100 | * | |
| Glutamate/aspartate transporter 1 (glial) | Glutamate | S75687 | 1.15 | 0.041 | 1.03 | 0.726 | −1.11 | 0.294 | 100 | 100 | 66 | * |
| Glutamate/aspartate transporter 1 (glial) | X63744 | 1.21 | 0.039 | 1.52 | 0.001 | 1.26 | 0.049 | 100 | 100 | 100 | * | |
| Glutamate/aspartate transporter 3 (neuronal) | L35558 | 1.14 | 0.171 | 1.28 | 0.010 | 1.12 | 0.209 | 100 | 100 | 100 | ||
| NMDA receptor NR1 | S39221 | 1.1 | 0.123 | 1.31 | 0.002 | 1.2 | 0.018 | 100 | 100 | 100 | ||
| NMDA receptor NMDA2C | U08259 | −1.04 | 0.426 | 1.07 | 0.147 | 1.11 | 0.021 | 100 | 100 | 83 | ||
| Glutamate receptor, metabotropic 3 | M92076 | 1.11 | 0.074 | 1.16 | 0.022 | 1.05 | 0.404 | 100 | 100 | 100 | ||
| Glutamate receptor, metabotropic 5 | D10891 | 1.05 | 0.513 | 1.24 | 0.042 | 1.18 | 0.099 | 100 | 100 | 83 | ||
| Downregulation | ||||||||||||
| Neuropeptide Y | M15880 | −1.12 | 0.030 | −1.12 | 0.038 | 1 | 0.965 | 100 | 100 | 100 | * | |
| Tachykinin receptor 3 (tachykinin 2 receptor) | J05189 | −1.06 | 0.042 | −1.02 | 0.418 | 1.04 | 0.203 | 83 | 100 | 66 | * | |
| Cholinergic receptor, nicotinic, beta-4 | U42976 | −1.04 | 0.081 | −1.07 | 0.016 | −1.03 | 0.199 | 100 | 100 | 100 | ||
| Dopamine D5 receptor | DA | M69118 | −1.05 | 0.026 | −1.04 | 0.096 | 1.01 | 0.646 | 100 | 100 | 83 | * |
| Dopamine D3 receptor | A17753 | −1.12 | 0.058 | −1.15 | 0.021 | −1.03 | 0.276 | 33 | 66 | 0 | ||
| Dopamine transporter | M80570 | −1.04 | 0.176 | −1.07 | 0.040 | −1.02 | 0.335 | 100 | 100 | 83 | ||
| 5-HT3 receptor | U01227 | 1.01 | 0.781 | −1.06 | 0.044 | −1.07 | 0.034 | 100 | 100 | 100 | ||
| Glutamate receptor, ionotropic, kainate 2 | Z11548 | 1.11 | 0.148 | −1.12 | 0.218 | −1.24 | 0.025 | 100 | 100 | 100 | ||
Comparisons of gene expression changes between dyskinetic, nondyskinetic, and saline animals. Fold change and P values are presented. Upregulations are listed first, downregulations are underneath. In case of opposite regulations in different comparison groups, the comparison of dyskinetic to nondyskinetic animals decided placement. Percent present call (P call %) is given for each experimental group in the last three columns. The asterisk denotes significant differences between dyskinetic and nondyskinetic animals.
Changes in the expression of neuropeptide genes received a high z score (Table. 2). In particular, tachykinin 2 and cholecystokinin were upregulated in dyskinetic rats and unchanged in rats that did not develop AIMs. Both neuropeptides have been previously shown to be upregulated in the 6-OHDA-lesioned striatum after treatment with D1 receptor agonists or L-DOPA (Gerfen, 1992b; Taylor et al., 1992).
In addition to the categories listed in Table 2, other neuro-transmitter systems showed significant gene expression changes in the L-DOPA-treated animals (Table 5). Glutamate receptors are believed to play a central role in the genesis of motor complications during L-DOPA pharmacotherapy (Chase and Oh, 2000; Dunah et al., 2000). In our data set, only a few genes related to the glutamate system showed significant changes with L-DOPA treatment and dyskinesia development. Upregulation of the genes encoding for metabotropic receptor subunits 3 and 5 was seen specifically in the dyskinetic animals. Few changes were observed in genes coding for ionotropic glutamate receptor subunits and sometimes only in the nondyskinetic group of animals (see NMDA 2C and kainate receptor subunit 2 in Table 5). A significant upregulation of the main NMDA receptor subunit, NR1, occurred in both groups of L-DOPA-treated rats compared to saline-injected controls. These results are in line with findings of increased striatal expression of NR1 protein after chronic L-DOPA treatment (Dunah et al., 2000). Indications that striatal glutamate transmission may be altered in dyskinesia came from the analysis of glutamate transporter genes. Both the glial and the neuronal isoforms of the glutamate transporter (i.e., glutamate/aspartate transporter 1 and glutamate/aspartate transporter 3, respectively) showed a prominent upregulation in the dyskinetic rats compared to nondyskinetic cases and/or saline controls.
Among the changes affecting other neurotransmitter systems was a prominent upregulation of cannabinoid receptor CB1 mRNA in the striatum of dyskinetic rats, a finding that was confirmed by ISHH (Fig. 3H). Interestingly, the CB1 receptor has recently emerged as a promising target for antidyskinetic drug therapy (Brotchie, 2003; Ferrer et al., 2003). Genes involved with cannabinoid biosynthesis and degradation such as fatty acid amide hydrolase, oleamide hydrolase and monoacylglycerol (MAG) lipase (Cravatt et al., 1996; Dinh et al., 2002), and phospholipase D (Cadas et al., 1997; Di Marzo et al., 1994) were not altered in the dyskinetic rats (data not shown). This is in agreement with the finding of unchanged endocannabinoid levels in the striatum of 6-OHDA-lesioned rats treated with L-DOPA (Ferrer et al., 2003).
Energy metabolism and mitochondrial enzymes
Genes involved in energy production through glycolysis, oxidative phosphorylation, or creatine biosynthesis were part of the MAPPFinder analysis. Among these categories, “creatine biosynthesis” genes received a high z score (Table 2). Dyskinetic animals showed significant downregulation of genes encoding two key enzymes of the phosphocreatine pathway, guanidinoacetate methyltransferase (GAMT), and ubiquitous mitochondrial creatine kinase (Mi-CK). Dyskinetic rats also had reduced expression of other genes of energy-producing pathways, such as glyceralde-hyde-3-phosphate dehydrogenase and lactate dehydrogenase, both involved in glycolysis (see downregulated genes in Table 6). Of the mitochondrial respiratory chain, some enzymes were upregulated, whereas others were downregulated (Table 6). Considering that twice as many genes were upregulated than downregulated in the dyskinetic striatum, it is noteworthy that genes involved in energy metabolism were more frequently downregulated than upregulated. Dyskinetic rats also showed an upregulated expression of cytochrome oxidase subunit I (CO-I), a mitochondrial gene that is a marker of increased metabolic demands in neurons (for review, see Hirsch et al., 2000). Upregulation of CO-I mRNA in dyskinesia was verified with ISHH (Fig. 3J). These data suggest that L-DOPA-induced dyskinesia is associated with a combination of increased metabolic demands in striatal neurons and reduced capacity for energy production.
Table 6.
mRNAs of energy metabolism, mitochondrial enzymes, and oxidative stress
| Energy metabolism, mitochondrial enzymes and oxidative stress | Accession no. | Dyskinesia/nondys
|
Dyskinesia/saline
|
Nondys/saline
|
P call %
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold change | P value | Fold change | P value | Fold change | P value | Dyskinesia | Nondys | Saline | |||
| Upregulation | |||||||||||
| Pyruvate dehydrogenase phosphatase isoenzyme 1 (AF062740_at) | AF062740 | 1.28 | 0.002 | 1.21 | 0.070 | −1.06 | 0.590 | 100 | 100 | 100 | * |
| Pyruvate dehydrogenase phosphatase isoenzyme 1 (AF062740_g_at) | AF062740 | 1.31 | 0.002 | 1.22 | 0.061 | −1.08 | 0.451 | 100 | 100 | 100 | * |
| Cytochrome C, somatic | AI008815 | 1.16 | 0.022 | 1.22 | 0.058 | 1.04 | 0.670 | 100 | 100 | 100 | * |
| Grp75, mortalin, hsp70-family member (S78556_g_at) | S78556 | 1.13 | 0.003 | 1.11 | 0.037 | −1.02 | 0.702 | 100 | 100 | 100 | * |
| Grp75, mortalin, hsp70-family member (S78556_at) | S78556 | 1.23 | 0.018 | 1.17 | 0.077 | −1.05 | 0.494 | 100 | 100 | 100 | * |
| Stress activated protein kinase alpha II (JNK2), (rc_AI231354_at) | AI231354 | 1.15 | 0.061 | 1.45 | 0.001 | 1.26 | 0.018 | 100 | 100 | 100 | |
| Stress activated protein kinase alpha II (JNK2), (rc_AI231354_g_at) | AI231354 | 1.16 | 0.079 | 1.37 | 0.003 | 1.19 | 0.076 | 100 | 100 | 100 | |
| Stress activated protein kinase alpha II (JNK2) | L27112 | 1.09 | 0.316 | 1.25 | 0.015 | 1.14 | 0.134 | 100 | 100 | 100 | |
| Dihydrolipoamide acetyltransferase | AA892485 | 1.2 | 0.018 | 1.17 | 0.022 | −1.02 | 0.692 | 100 | 100 | 100 | |
| Dihydrolipoamide acetyltransferase | D10655 | 1.11 | 0.114 | 1.17 | 0.042 | 1.05 | 0.445 | 100 | 100 | 100 | |
| Phosphoglycerate mutase type B | S63233 | 1.04 | 0.316 | 1.13 | 0.020 | 1.08 | 0.147 | 100 | 100 | 100 | |
| Adenylate kinase 1 | AA799299 | 1.04 | 0.360 | 1.14 | 0.020 | 1.1 | 0.113 | 100 | 100 | 100 | |
| Malate dehydrogenase mitochondrial (rc_AI010480_g_at) | AI010480 | 1.04 | 0.384 | 1.11 | 0.035 | 1.07 | 0.166 | 100 | 100 | 100 | |
| Malate dehydrogenase mitochondrial (rc_AI010480_at) | AI010480 | 1.03 | 0.450 | 1.1 | 0.043 | 1.07 | 0.169 | 100 | 100 | 100 | |
| Cytochrome oxidase subunit I | S79304 | 1.06 | 0.700 | 1.37 | 0.048 | 1.29 | 0.126 | 100 | 100 | 100 | |
| Glutathione reductase | U73174 | 1.07 | 0.492 | 1.11 | 0.010 | 1.04 | 0.707 | 100 | 100 | 100 | |
| Downregulation | |||||||||||
| Ubiquitous mitochondrial creatine kinase (X59737mRNA_at) | X59737 | −1.16 | 0.001 | −1.16 | 0.002 | −1 | 0.936 | 100 | 100 | 100 | * |
| Ubiquitous mitochondrial creatine kinase (X59737mRNA_g_at) | X59737 | −1.14 | 0.006 | −1.11 | 0.021 | 1.03 | 0.278 | 100 | 100 | 100 | * |
| Acetyl-CoA acyltransferase, 3-oxo acyl-CoA thiolase A | J02749 | −1.05 | 0.029 | −1.01 | 0.456 | 1.03 | 0.118 | 100 | 100 | 100 | * |
| Fatty acid coenzyme A ligase, long chain 5 | AB012933 | −1.14 | 0.042 | −1.12 | 0.072 | 1.01 | 0.674 | 100 | 100 | 100 | * |
| Lactate dehydrogenase B (U07181_g_at) | U07181 | −1.02 | 0.746 | −1.1 | 0.008 | −1.07 | 0.266 | 100 | 100 | 100 | |
| Lactate dehydrogenase B (U07181_at) | U07181 | −1 | 0.939 | −1.1 | 0.030 | −1.1 | 0.040 | 100 | 100 | 100 | |
| Glycerol-3-phosphate dehydrogenase 2 | X78593 | −1.05 | 0.171 | −1.11 | 0.014 | −1.06 | 0.107 | 100 | 100 | 100 | |
| Glyceraldehyde-3-phosphate dehydrogenase | M17701 | −1.06 | 0.377 | −1.17 | 0.018 | −1.11 | 0.047 | 100 | 100 | 100 | |
| ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit (OSCP) | D13127 | 1.09 | 0.217 | −1.03 | 0.637 | −1.11 | 0.041 | 100 | 100 | 100 | |
| Superoxide dismutase 1, soluble | Y00404 | −1.11 | 0.046 | −1.06 | 0.299 | 1.05 | 0.288 | 100 | 100 | 100 | * |
| Glutathione S-transferase, pi 2 | X02904 | −1.11 | 0.049 | −1.17 | 0.031 | −1.05 | 0.360 | 100 | 100 | 100 | * |
| Glutathione S-transferase, pi 2 | AI012589 | −1.09 | 0.057 | −1.15 | 0.036 | −1.05 | 0.355 | 100 | 100 | 100 | |
| Glutathione S-transferase Yc1 | S72505 | −1.03 | 0.638 | −1.17 | 0.017 | −1.14 | 0.018 | 100 | 100 | 100 | |
| Glutathione S-transferase | E01415 | −1.07 | 0.278 | −1.19 | 0.019 | −1.11 | 0.066 | 100 | 100 | 100 | |
| Glutathione S-transferase, mu type 2 (Yb2) | X04229 | −1.15 | 0.280 | −1.43 | 0.024 | −1.24 | 0.046 | 100 | 100 | 100 | |
Comparisons of gene expression changes between dyskinetic, nondyskinetic, and saline animals. Fold change and P values are presented. Upregulations are listed first, downregulations are underneath. In case of opposite regulations in different comparison groups, the comparison of dyskinetic to nondyskinetic animals decided placement. Percent present call (P call %) is given for each experimental group in the last three columns. The asterisk denotes significant differences between dyskinetic and nondyskinetic animals.
Kinases and phosphatases
Upregulation of calcium kinases received a high z score in the MAPPFinder analysis. Messenger RNA levels for protein kinase C delta (PKC-δ) were prominently upregulated in the dyskinetic striatum compared to nondyskinetic cases (Table 7), and this finding was validated with ISHH (Fig. 3I). The similarity between the distribution of PKC-δ mRNA and NF-H mRNA in the striatum of L-DOPA-treated rats (Fig. 3D) is particularly striking since PKCδ is required for neurite outgrowth (Corbit et al., 1999). The beta and delta isoforms of calcium/calmodulin-dependent protein kinase II (CamKII) and calcium/calmodulin-dependent protein kinase IV (CamKIV) were upregulated in the animals that developed dyskinesia more than in the nondyskinetic cases (Table 7 and Fig. 3K). Interestingly, the CaM-kinase II inhibitor alpha was specifically upregulated in the nondyskinetic subgroup of L-DOPA-treated rats, indicating that the inhibition of Ca2+ kinases could help to prevent dyskinesia.
Table 7.
mRNAs of kinases and phosphatases
| Kinases and phosphatases | Accession no. | Dyskinesia/nondys
|
Dyskinesia/saline
|
Nondys/saline
|
P call %
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold change | P value | Fold change | P value | Fold change | P value | Dyskinesia | Nondys | Saline | |||
| Upregulation | |||||||||||
| Protein kinase C-family-related | M15523 | 1.13 | 0.007 | 1.23 | 0.040 | 1.09 | 0.348 | 100 | 100 | 100 | * |
| Protein tyrosine phosphatase-like protein | D38222 | 1.24 | 0.008 | 1.23 | 0.038 | −1 | 0.959 | 100 | 100 | 100 | * |
| Dual specificity Yak1-related kinase | AI104012 | 1.15 | 0.016 | 1.18 | 0.018 | 1.02 | 0.659 | 100 | 83 | 50 | * |
| Protein kinase C delta (M18330_at) | M18330 | 1.26 | 0.017 | 1.18 | 0.060 | −1.07 | 0.238 | 100 | 50 | 66 | * |
| Protein kinase C delta (M18330_g_at) | M18330 | 1.26 | 0.022 | 1.26 | 0.023 | 1 | 0.957 | 100 | 100 | 100 | * |
| Protein kinase C regulatory protein | S55223 | 1.09 | 0.106 | 1.14 | 0.006 | 1.04 | 0.480 | 100 | 100 | 100 | |
| Calcium/calmodulin-dependent protein kinase II delta | L13406 | 1.1 | 0.029 | 1.16 | 0.004 | 1.06 | 0.243 | 50 | 0 | 16 | * |
| Calcium/calmodulin-dependent protein kinase II beta | M16112 | 1.06 | 0.179 | 1.21 | 0.005 | 1.14 | 0.022 | 100 | 100 | 100 | |
| Regulatory subunit of protein phosphatase 2A | D14421 | 1.11 | 0.031 | 1.05 | 0.251 | −1.05 | 0.283 | 100 | 100 | 100 | * |
| Phosphatase inhibitor-2 (regulatory subunit of protein phosphatase 1) | S79213 | 1.17 | 0.036 | 1.33 | 0.021 | 1.14 | 0.232 | 100 | 100 | 100 | * |
| Dual specificity phosphatase 6 (tyrosine phosphatase rVH6) | U42627 | 1.18 | 0.038 | 1.18 | 0.085 | 1 | 0.986 | 100 | 100 | 100 | * |
| Calcineurin B | D14425 | 1.14 | 0.111 | 1.53 | 0.001 | 1.34 | 0.011 | 100 | 100 | 100 | |
| Calcineurin B | D14568 | 1.07 | 0.350 | 1.48 | 0.001 | 1.38 | 0.002 | 100 | 100 | 100 | |
| Calcineurin A-beta, catalytic subunit | M31809 | 1.18 | 0.081 | 1.39 | 0.004 | 1.18 | 0.136 | 100 | 100 | 100 | |
| Mitogen activated protein kinase 1 (ERK2) | M64300 | 1.07 | 0.488 | 1.38 | 0.013 | 1.29 | 0.003 | 100 | 100 | 100 | |
| Protein phosphatase 1A alpha | J04503 | 1.04 | 0.397 | 1.13 | 0.013 | 1.08 | 0.091 | 100 | 100 | 100 | |
| Neurotrophic tyrosine kinase, receptor, type 2 | M55291 | 1.11 | 0.055 | 1.17 | 0.047 | 1.05 | 0.514 | 100 | 100 | 100 | |
| Downregulation | |||||||||||
| Mitogen-activated protein kinase 3 (ERK1) | M61177 | −1.14 | 0.006 | −1.16 | 0.003 | −1.02 | 0.468 | 100 | 100 | 100 | * |
| CaM-kinase II inhibitor alpha | AA858621 | −1.08 | 0.015 | 1.05 | 0.094 | 1.13 | 0.002 | 100 | 100 | 100 | * |
| Acid phosphatase 2 | AI234950 | −1.06 | 0.016 | −1.05 | 0.163 | 1.01 | 0.811 | 33 | 33 | 33 | * |
| Multiple inositol polyphosphate histidine phosphatase 1 | AF012714 | −1.08 | 0.019 | 1.01 | 0.809 | 1.09 | 0.088 | 100 | 100 | 100 | * |
| Protein tyrosine phosphatase, nonreceptor type 2 | X58828 | −1.08 | 0.040 | −1.1 | 0.056 | −1.01 | 0.755 | 100 | 100 | 100 | * |
| Protein tyrosine phosphatase, receptor type, R | D64050 | −1.11 | 0.043 | −1.03 | 0.608 | 1.08 | 0.084 | 33 | 50 | 16 | * |
| Nonreceptor protein kinase (batk) | L34542 | −1.17 | 0.044 | −1.12 | 0.041 | 1.04 | 0.505 | 100 | 100 | 100 | * |
| Protein tyrosine phosphatase, receptor type, O | U28938 | −1.1 | 0.048 | −1.17 | 0.185 | −1.06 | 0.574 | 100 | 100 | 100 | * |
Comparisons of gene expression changes between dyskinetic, nondyskinetic, and saline animals. Fold change and P values are presented. Upregulations are listed first, downregulations are underneath. In case of opposite regulations in different comparison groups, the comparison of dyskinetic to nondyskinetic animals decided placement. Percent present call (P call %) is given for each experimental group in the last three columns. The asterisk denotes significant differences between dyskinetic and nondyskinetic animals.
In the dyskinetic striatum, some regulatory subunits of protein phosphatases were upregulated. Messenger RNAs for various subunits of the calcium-dependent phosphatase, calcineurin, showed enhanced expression in both dyskinetic and nondyskinetic rats. Downregulation was observed with multiple protein tyrosine phosphatases genes in the dyskinetic rat striatum, and this category of genes also received a high z score in the MAPPFinder analysis (Table 2).
Ribosomal proteins
Several genes encoding ribosomal proteins were consistently downregulated in dyskinetic rats compared to nondyskinetic cases, and this functional category of genes received a high z score in the MAPPFinder analysis (Table 2).
G proteins, membrane transducers, and intracellular adaptor proteins
Hundreds of genes coding for membrane transducers and intracellular adaptor proteins were represented on the chips. Because of the large number, this functional category was not included in the MAPPFinder analysis as a whole, but MAPPs were generated for more defined gene families. Some of these families, such as RabGTPases, RasGTPases, and guanine nucleotide-binding proteins (G proteins) were highly affected by L-DOPA treatment, with many significant changes in both dyskinetic and nondyskinetic rats (Table 8).
Table 8.
mRNAs of G proteins, membrane transducers, and intracellular adapter proteins
| G proteins, membrane transducers, and intracellular adapter proteins | Accession no. | Dyskinesia/nondys
|
Dyskinesia/saline
|
Nondys/saline
|
P call %
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold change | P value | Fold change | P value | Fold change | P value | Dyskinesia | Nondys | Saline | ||||
| Upregulation | ||||||||||||
| RAS guanyl releasing protein 1 | Ras-related | AF081196 | 1.38 | 0.008 | 1.51 | 0.001 | 1.09 | 0.443 | 100 | 100 | 100 | * |
| RAS guanyl releasing protein 1 | AF060819 | 1.41 | 0.041 | 1.63 | 0.013 | 1.15 | 0.335 | 100 | 100 | 83 | * | |
| Ras-related protein rab 1A | J02998 | 1.05 | 0.393 | 1.28 | 0.010 | 1.22 | 0.023 | 100 | 100 | 100 | ||
| Ras-related protein rab 5 | AF072935 | 1.03 | 0.632 | 1.3 | 0.005 | 1.27 | 0.011 | 100 | 100 | 100 | ||
| Ras-related protein rab 10 | AA955306 | 1.08 | 0.212 | 1.24 | 0.001 | 1.15 | 0.031 | 100 | 100 | 100 | ||
| Ras-related protein rab 11A (M75153_g_at) | M75153 | 1.11 | 0.208 | 1.34 | 0.015 | 1.21 | 0.085 | 100 | 100 | 100 | ||
| Ras-related protein rab 11A (M75153_at) | M75153 | 1.09 | 0.187 | 1.25 | 0.031 | 1.14 | 0.168 | 100 | 100 | 100 | ||
| Ras-related protein rab 12 | M83676 | 1.01 | 0.805 | 1.16 | 0.030 | 1.14 | 0.069 | 100 | 100 | 100 | ||
| Ras-related protein ral A | L19698 | −1.04 | 0.237 | 1.12 | 0.029 | 1.17 | 0.007 | 100 | 100 | 100 | ||
| Guanosine-3-phosphate-binding protein | E03859 | 1.11 | 0.097 | 1.26 | 0.014 | 1.13 | 0.135 | 100 | 100 | 100 | ||
| Phospholipase C, beta-4 | L15556 | 1.15 | 0.011 | 1.12 | 0.013 | −1.02 | 0.580 | 100 | 100 | 100 | * | |
| Ins(1,4,5)P3-binding protein (130 kDa) | E12159 | 1.16 | 0.030 | 1.13 | 0.162 | −1.03 | 0.722 | 100 | 100 | 100 | * | |
| Phosphatidylinositol transfer protein | M25758 | 1.02 | 0.614 | 1.13 | 0.025 | 1.11 | 0.065 | 100 | 100 | 100 | ||
| Diacylglycerol kinase zeta | D78588 | −1.03 | 0.299 | 1.07 | 0.037 | 1.1 | 0.012 | 100 | 100 | 100 | ||
| Adenylate cyclase 5 | AC | AI044848 | 1.12 | 0.036 | 1.38 | 0.001 | 1.24 | 0.013 | 100 | 100 | 83 | * |
| Adenylate cyclase 5 | M96159 | 1.09 | 0.086 | 1.12 | 0.029 | 1.03 | 0.314 | 100 | 100 | 100 | ||
| Adenylate cyclase 3 | M55075 | 1.07 | 0.189 | 1.24 | 0.011 | 1.15 | 0.064 | 100 | 100 | 100 | ||
| Guanine nucleotide-binding protein beta-1 | G prot | AF022083 | 1.08 | 0.115 | 1.43 | 0.000 | 1.32 | 0.000 | 100 | 100 | 100 | |
| Guanine nucleotide-binding protein, alpha inhibiting 1 | M17527 | −1.02 | 0.809 | 1.36 | 0.004 | 1.39 | 0.006 | 100 | 100 | 100 | ||
| G alpha 12z | S50461 | 1 | 0.943 | 1.42 | 0.003 | 1.42 | 0.004 | 100 | 100 | 100 | ||
| Guanine nucleotide-binding protein, alpha-o | M17526 | −1.03 | 0.333 | 1.1 | 0.069 | 1.14 | 0.020 | 100 | 100 | 100 | ||
| Cyclic GMP stimulated phosphodiesterase | U21101 | 1.06 | 0.116 | 1.11 | 0.007 | 1.04 | 0.293 | 100 | 100 | 100 | ||
| 14-3-3 Protein beta subtype; putative protein kinase C regulatory protein | 14-3-3 | S55223 | 1.09 | 0.106 | 1.14 | 0.006 | 1.04 | 0.480 | 100 | 100 | 100 | |
| 14-3-3 Protein theta subtype | AA942751 | 1.03 | 0.547 | 1.12 | 0.006 | 1.09 | 0.090 | 100 | 100 | 100 | ||
| 14-3-3 Protein zeta subtype | AI180424 | 1.09 | 0.139 | 1.13 | 0.024 | 1.04 | 0.419 | 100 | 100 | 100 | ||
| 14-3-3 Protein zeta subtype; mitochondrial import stimulation factor, S1 subunit | D30740 | 1.11 | 0.081 | 1.14 | 0.028 | 1.02 | 0.674 | 100 | 100 | 100 | ||
| 14-3-3 Protein eta subtype | D17445 | −1.06 | 0.172 | 1.04 | 0.298 | 1.11 | 0.046 | 100 | 100 | 100 | ||
| Downregulation | ||||||||||||
| Guanine nucleotide-binding protein, gamma-8 | L35921 | −1.12 | 0.007 | −1.11 | 0.015 | 1.01 | 0.863 | 100 | 100 | 100 | * | |
| 2′,3′ - Cyclic nucleotide 3′-phosphodiesterase |
L16532 | −1.1 | 0.008 | −1.13 | 0.051 | −1.03 | 0.602 | 100 | 100 | 100 | * | |
| Guanylyl cyclase 1, soluble, alpha-3 | AA849036 | −1.14 | 0.016 | −1.17 | 0.040 | −1.03 | 0.630 | 100 | 100 | 100 | * | |
| Guanylyl cyclase A/atrial natriuretic peptide receptor (GC-A) | J05677 | −1.08 | 0.108 | −1.12 | 0.015 | −1.03 | 0.471 | 83 | 100 | 100 | ||
| Phospholipase C, delta-4 | U16655 | −1.04 | 0.097 | −1.09 | 0.002 | −1.05 | 0.033 | 100 | 100 | 100 | ||
| 14-3-3 Protein epsilon subtype; mitochondrial import stimulation factor, L subunit | D30739 | 1.04 | 0.520 | −1.06 | 0.354 | −1.10 | 0.045 | 100 | 100 | 100 | ||
Comparisons of gene expression changes between dyskinetic, nondyskinetic, and saline animals. Fold change and P values are presented. Upregulations are listed first, downregulations are underneath. In case of opposite regulations in different comparison groups, the comparison of dyskinetic to nondyskinetic animals decided placement. Percent present call (P call %) is given for each experimental group in the last three columns. The asterisk denotes significant differences between dyskinetic and nondyskinetic animals.
14-3-3 Proteins, a group of highly conserved proteins with high abundance in the brain, were mostly upregulated in dyskinesia and received a significant z sore in the MAPPFinder analysis of dyskinetic versus saline-treated rats (z score 3.6) and nondyskinetic versus saline-treated rats (z score 3.52). Although the function of these proteins is not fully elucidated, they seem to act as adaptor molecules involved in protein–protein interactions, and they play a role in signal transduction, apoptosis, and stress response (Berg et al., 2003; Morrison, 1994; Tzivion and Avruch, 2002; van Hemert et al., 2001).
Discussion
Microarray analysis enables an examination of large numbers of mRNAs expressed in different biological systems, and it provides a valuable tool in a discovery process intended to lead to novel hypotheses. Although transcript changes might not be translated into protein levels in every instance, there is generally a good correlation between increases in mRNA and increased protein levels, albeit with some variation in magnitude (Pongrac et al., 2002).
In the present study, microarray analysis was used to define the patterns of striatal gene expression in rats that had developed AIMs, as opposed to rats that, despite motor improvement in response to L-DOPA, remained free of dyskinetic side effects. Both groups of L-DOPA-treated animals were also compared to drug-naive 6-OHDA-lesioned controls. It was demonstrated previously that the rat model of abnormal involuntary movements used in this study shows molecular and biochemical changes similar to those described in nonhuman primate models of L-DOPA-induced dyskinesia and/or PD patients, including an upregulation of ΔFosB-like proteins and opioid precursor mRNAs in the striatum, and changes in opioid receptor binding within the cortico-basal ganglia loop (for review, see Cenci et al., 2002). Our previous studies have indicated that the striatum plays a crucial role in driving the development of abnormal involuntary movements in this model, which can be either exacerbated or reduced by gene knockdown strategies applied to the striatum (Andersson et al., 1999, 2001).
Evidence for increased activity of GABA neurons in dyskinesia
Dyskinesia was accompanied by an upregulation of mRNAs for pre- and postsynaptic proteins, indicating increased synaptic activity, neurotransmitter release, and synaptic remodeling. An increased gene expression of Na+K+-ATPase, as observed in the dyskinesia samples, has been linked to frequent and/or large depolarization events (Mata et al., 1992), suggesting increased neuronal activity within the striatum. Na+K+-ATPase is critical for maintaining the resting membrane potential and for regulating the response to excitatory amino acids (Calabresi et al., 1995). Interestingly, the glial and neuronal glutamate/aspartate transporters were upregulated. Glutamate regulates the expression of glutamate transporters (O’Shea, 2002), and excessive glutamate release in a rat model of temporal lobe epilepsy increased the expression of the neuronal glutamate transporter 3 (Crino et al., 2002). The increased activity of striatal neurons, and the increased expression of glutamate transporters, could therefore be caused by increased glutamate release into the striatum (Calabresi et al., 2000). Alternatively, increased glutamate transporter levels might reflect the need to limit excitation in an otherwise hyperactivated striatum.
The increase in synaptic and neuronal activity seemed to affect predominantly the GABA subpopulation of neurons, since the GABA transporter was upregulated together with the GABA-synthesizing enzyme GAD67. Most of the upregulated mRNA species are expressed in GABA projection neurons, such as the cannabinoid 1 and somatostatin 2 receptors (Allen et al., 2003; Julian et al., 2003), and the GABA-A receptor subtypes alpha-2, alpha-4, and beta-3 (Schwarzer et al., 2001). The upregulated tachykinin 2 mRNA codes for a neuropeptide expressed in a subpopulation of GABA neurons projecting to the substantia innominata (Furuta et al., 2000; Gerfen, 1992b). Furthermore, the GABA subpopulation of D1 receptor-expressing neurons of the “direct pathway” (Gerfen, 1992a,b) seemed to be particularly affected, since D1 (but not D2) receptors were upregulated. This finding is in agreement with previous studies showing significant striatal induction of ΔFosB in prodynorphin-containing cells (i.e., the direct pathway neurons) in the same animal model of L-DOPA-induced dyskinesia (Andersson et al., 1999).
An upregulation of genes involved in Ca2+ homeostasis and Ca2+-dependent signaling indicates increased Ca2+ levels in dyskinesia
Many of the genes that were upregulated in the dyskinetic striatum are either involved in Ca2+ homeostasis or activated by Ca2+. Among the most dramatically upregulated mRNAs were the ones coding for Ca2+ ATPases. Ca2+ ATPases pump Ca2+ out of the cytosol either across plasma membranes (PMCAs) or into the endoplasmic reticulum (SERCAs). They maintain basal levels of intracellular Ca2+, participate in dynamic Ca2+ regulation, and are crucial players of Ca2+ export during normal and pathological conditions (Blaustein et al., 2002; Garcia and Strehler, 1999). Plasma membrane Ca2+ ATPases (PMCAs) are clustered at the active zones and may “reprime” the vesicular release mechanism following activity (Blaustein et al., 2002). The increase in Ca2+ ATPases could therefore be indicative of increased intracellular Ca2+ levels as well as increased neuronal activity.
Messenger RNAs of proteins that are activated by Ca2+ were upregulated as well, such as subunits of Ca2+-activated kinases and the Ca2+-inducible marker of cell stress, mortalin (Massa et al., 1995). Interestingly, whereas Ca2+-activated kinases were upregulated in dyskinetic rats, the CaM-kinase II inhibitor alpha was specifically upregulated in the nondyskinetic subgroup of L-DOPA-treated rats. These data raise the possibility that an upregulation of certain Ca2+-activated kinases might contribute to L-DOPA-induced dyskinesia, whereas inhibition of these Ca2+ kinases might protect against the development of this complication. This hypothesis is supported by the finding that intrastriatal inhibition of CaM kinase II reverses the motor response alterations produced by chronic L-DOPA treatment in 6-OHDA-lesioned rats (Oh et al., 1999).
Overall, the upregulation of this group of mRNAs demonstrates a strong association of dyskinesia with perturbed Ca2+ homeostasis.
An increased consumption of ATP by ion pumps and a downregulation of genes involved in ATP production could lead to energy shortfalls in the dyskinetic striatum
In addition to Ca2+ ATPases, various ion transporters and voltage-gated ion channels were upregulated in the dyskinetic striatum. Because ion pumps and transporters are ATP-dependent (Green and MacLennan, 1989), increased expression of ion pumps might lead to higher ATP consumption (Muller and Gruber, 2003). Despite the potentially increased need for ATP, some aspects of energy production seemed to be impaired in the dyskinetic striatum. The mRNA level for mitochondrial creatine kinase, a key enzyme in brain energy metabolism and ATP generation (Ames, 2000), was downregulated. Phosphocreatine is a source of high-energy phosphates in cells and tissues with high and fluctuating energy demands, such as the brain (Wyss and Kaddurah-Daouk, 2000). Creatine kinase is coupled with glutamate uptake, neurotransmitter release, calcium homeostasis, and the restoration of ion gradients before and after depolarization (Wallimann and Hemmer, 1994). In addition to creatine kinase, enzymes involved in glycolysis were downregulated as well.
Our findings oppose an increased need for ATP with decreased ATP supply. Together, they imply that the dyskinetic striatum may be prone to excitotoxic damage by the combination of impaired energy metabolism and increased levels of Ca2+ and glutamate (Novelli et al., 1988). In agreement with this hypothesis, the microarray data show increased striatal mRNA expression of mortalin and heat shock 70-kDa protein, two notable markers of cell stress (Kregel, 2002; Massa et al., 1995). Moreover, 14-3-3 proteins, a group of proteins with a role in stress response (Berg et al., 2003; van Hemert et al., 2001) were upregulated in dyskinesia (Table 8). The induction of 14-3-3 proteins may have a neuro-protective effect, since some 14-3-3 isoforms can sequester various proapoptotic proteins (e.g., Bad and Bax), thereby preventing apoptosis and promoting survival (Berg et al., 2003; Rosenquist, 2003; van Hemert et al., 2001). There was further indication that antioxidant defense systems were compromised too, since levels of glutathione S-transferase, which plays a role in the protection against reactive oxygen species (Fernandez-Checa, 2003), were significantly reduced in the dyskinetic striatum.
The toxicity of L-DOPA has been widely debated in the literature, but mainly with respect to possible adverse effects of L-DOPA treatment on the survival of nigral DA neurons (Agid, 1998; Melamed et al., 1998). The present data indicate that L-DOPA may be neurotoxic in the striatum and that this phenomenon may be part of the pathogenic cascade leading to the development of motor complications. Indeed, neurodegenerative processes in the striatum have been reported in models of antipsychotic drug-induced, tardive dyskinesia (Andreassen and Jorgensen, 2000; Roberts et al., 1995).
The present data do not allow us to draw any final conclusions about the exact mechanism of the L-DOPA-induced stress response, nor can we establish whether molecular adaptations with neuroprotective value prevail over deleterious ones. However, the data encourage future investigations into the possible role of striatal cell damage in the pathogenesis of L-DOPA-induced motor complications.
Decreased ribosomal proteins in the dyskinetic striatum point toward cellular stress and an inability to keep up with basic chores
Eukaryotic ribosomal protein synthesis has been studied most methodically in yeast. Ribosomal proteins are regulated at the level of transcription, and a tight control between synthesis of mRNAs for ribosomal proteins and nutrient availability has been observed (Warner, 1999). Messenger RNA synthesis for yeast ribosomal proteins is repressed in response to stress conditions such as heat shock, starvation, and defects in the secretory pathway (Planta, 1997; Warner, 1999). A downregulation of mRNAs for ribosomal proteins was described in two different gene array analyses that showed a decrease in genes of the oxidative metabolism/mitochondrial respiratory chain (Konradi et al., 2004; Patti et al., 2003). The downregulation of many ribosomal proteins in dyskinesia could thus be a response to diminished ATP availability or increased cell stress and suggests that the pathogenic process interferes with basic maintenance chores.
In conclusion, the present analysis of striatal gene expression patterns in rats affected by AIMs provides unheralded clues to the dyskinesiogenic action of L-DOPA and offers new directions for future pathophysiological investigations.
Acknowledgments
This work was supported by DA07134 (CK) and by grants from the Elsa and Thorsten Segerfalk Foundation, the Johan and Greta Kock Foundations, the Swedish Association of the Neurologically Disabled, the Swedish Foundation for Parkinson’s Research, and the Swedish National Research Council (MAC). We would like to thank the Gemzeus Foundation, The Marie Curie Host Fellowship Training Program, and the Socrates/Erasmus Program for their support to JEW, MC, and KK, respectively. We thank Hanna Lindgren and Katrin Helgasson for their expert assistance in the in situ hybridization histochemistry part of the study.
References
- Agid Y. Levodopa: is toxicity a myth? Neurology. 1998;50:858–863. doi: 10.1212/wnl.50.4.858. [DOI] [PubMed] [Google Scholar]
- Allen JP, Hathway GJ, Clarke NJ, Jowett MI, Topps S, Kendrick KM, Humphrey PP, Wilkinson LS, Emson PC. Somatos-tatin receptor 2 knockout/lacZ knockin mice show impaired motor coordination and reveal sites of somatostatin action within the striatum. Eur J Neurosci. 2003;17:1881–1895. doi: 10.1046/j.1460-9568.2003.02629.x. [DOI] [PubMed] [Google Scholar]
- Ames A., III CNS energy metabolism as related to function. Brain Res. 2000;34:42–68. doi: 10.1016/s0165-0173(00)00038-2. [DOI] [PubMed] [Google Scholar]
- Andersson M, Hilbertson A, Cenci MA. Striatal fosB expression is causally linked with l-DOPA-induced abnormal involuntary movements and the associated upregulation of striatal prodynorphin mRNA in a rat model of Parkinson’s disease. Neurobiol Dis. 1999;6:461–474. doi: 10.1006/nbdi.1999.0259. [DOI] [PubMed] [Google Scholar]
- Andreassen OA, Jorgensen HA. Neurotoxicity associated with neuroleptic-induced oral dyskinesias in rats. Implications for tardive dyskinesia? Prog Neurobiol. 2000;61:525–541. doi: 10.1016/s0301-0082(99)00064-7. [DOI] [PubMed] [Google Scholar]
- Andersson M, Konradi C, Cenci MA. cAMP response element-binding protein is required for dopamine-dependent gene expression in the intact but not the dopamine-denervated striatum. J Neurosci. 2001;21:9930–9943. doi: 10.1523/JNEUROSCI.21-24-09930.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D, Holzmann C, Riess O. 14-3-3 Proteins in the nervous system. Nat Rev Neurosci. 2003;4:752–762. doi: 10.1038/nrn1197. [DOI] [PubMed] [Google Scholar]
- Bezard E, Brotchie JM, Gross CE. Pathophysiology of levodopa-induced dyskinesia: potential for new therapies. Nat Rev Neurosci. 2001;2:577–588. doi: 10.1038/35086062. [DOI] [PubMed] [Google Scholar]
- Blaustein MP, Juhaszova M, Golovina VA, Church PJ, Stanley EF. Na/Ca exchanger and PMCA localization in neurons and astrocytes: functional implications. Ann N Y Acad Sci. 2002;976:356–366. doi: 10.1111/j.1749-6632.2002.tb04762.x. [DOI] [PubMed] [Google Scholar]
- Brotchie JM. CB1 cannabinoid receptor signalling in Parkinson’s disease. Curr Opin Pharmacol. 2003;3:54–61. doi: 10.1016/s1471-4892(02)00011-5. [DOI] [PubMed] [Google Scholar]
- Cadas H, di Tomaso E, Piomelli D. Occurrence and biosynthesis of endogenous cannabinoid precursor, N-arachidonoyl phosphatidyle-thanolamine, in rat brain. J Neurosci. 1997;17:1226–1242. doi: 10.1523/JNEUROSCI.17-04-01226.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabresi P, Centonze D, Bernardi G. Electrophysiology of dopamine in normal and denervated striatal neurons. Trends Neurosci. 2000;23:S57–S63. doi: 10.1016/s1471-1931(00)00017-3. [DOI] [PubMed] [Google Scholar]
- Calabresi P, De Murtas M, Pisani A, Stefani A, Sancesario G, Mercuri NB, Bernardi G. Vulnerability of medium spiny striatal neurons to glutamate: role of Na+/K+ ATPase. Eur J Neurosci. 1995;7:1674–1683. doi: 10.1111/j.1460-9568.1995.tb00689.x. [DOI] [PubMed] [Google Scholar]
- Cenci MA, Lee CS, Bjorklund A. L-DOPA-induced dyskinesia in the rat is associated with striatal overexpression of prodynorphin- and glutamic acid decarboxylase mRNA. Eur J Neurosci. 1998;10:2694–2706. [PubMed] [Google Scholar]
- Cenci MA, Whishaw IQ, Schallert T. Animal models of neurological deficits: how relevant is the rat? Nat Rev Neurosci. 2002;3:574–579. doi: 10.1038/nrn877. [DOI] [PubMed] [Google Scholar]
- Chase TN, Oh JD. Striatal dopamine- and glutamate-mediated dysregulation in experimental parkinsonism. Trends Neurosci. 2000;23:S86–S91. doi: 10.1016/s1471-1931(00)00018-5. [DOI] [PubMed] [Google Scholar]
- Corbit KC, Foster DA, Rosner MR. Protein kinase Cdelta mediates neurogenic but not mitogenic activation of mitogen-activated protein kinase in neuronal cells. Mol Cell Biol. 1999;19:4209–4218. doi: 10.1128/mcb.19.6.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravatt BF, Giang DK, Mayfield SP, Boger DL, Lerner RA, Gilula NB. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996;384:83–87. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- Crino PB, Jin H, Shumate MD, Robinson MB, Coulter DA, Brooks-Kayal AR. Increased expression of the neuronal glutamate transporter (EAAT3/EAAC1) in hippocampal and neocortical epilepsy. Epilepsia. 2002;43:211–218. doi: 10.1046/j.1528-1157.2002.35001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- Dauer W, Przedborski S. Parkinson’s disease: mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- DiMarzo V, Fontana A, Cadas H, Schinelli S, Cimino G, Schwartz JC, Piomelli D. Formation and inactivation of endogenous cannabinoid anandamide in central neurons. Nature. 1994;372:686–691. doi: 10.1038/372686a0. [DOI] [PubMed] [Google Scholar]
- Dinh TP, Carpenter D, Leslie FM, Freund TF, Katona I, Sensi SL, Kathuria S, Piomelli D. Brain monoglyceride lipase participating in endocannabinoid inactivation. Proc Natl Acad Sci U S A. 2002;99:10819–10824. doi: 10.1073/pnas.152334899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunah AW, Wang Y, Yasuda RP, Kameyama K, Huganir RL, Wolfe BB, Standaert DG. Alterations in subunit expression, composition, and phosphorylation of striatal N-methyl-D-aspartate glutamate receptors in a rat 6-hydroxydopamine model of Parkinson’s disease. Mol Pharmacol. 2000;57:342–352. [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Checa JC. Redox regulation and signaling lipids in mitochondrial apoptosis. Biochem Biophys Res Commun. 2003;304:471–479. doi: 10.1016/s0006-291x(03)00619-3. [DOI] [PubMed] [Google Scholar]
- Ferrer B, Asbrock N, Kathuria S, Piomelli D, Giuffrida A. Effects of levodopa on endocannabinoid levels in rat basal ganglia: implications for the treatment of levodopa-induced dyskinesias. Eur J Neurosci. 2003;18:1607–1614. doi: 10.1046/j.1460-9568.2003.02896.x. [DOI] [PubMed] [Google Scholar]
- Furuta T, Mori T, Lee T, Kaneko T. Third group of neostriatofugal neurons: neurokinin B-producing neurons that send axons predominantly to the substantia innominata. J Comp Neurol. 2000;426:279–296. doi: 10.1002/1096-9861(20001016)426:2<279::aid-cne9>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Garcia ML, Strehler EE. Plasma membrane calcium ATPases as critical regulators of calcium homeostasis during neuronal cell function. Front Biosci. 1999;4:D869–D882. doi: 10.2741/garcia. [DOI] [PubMed] [Google Scholar]
- Gelb DJ, Oliver E, Gilman S. Diagnostic criteria for Parkinson disease. Arch Neurol. 1999;56:33–39. doi: 10.1001/archneur.56.1.33. [DOI] [PubMed] [Google Scholar]
- Gerfen CR. The neostriatal mosaic: multiple levels of compartmental organization. Trends Neurosci. 1992a;15:133–139. doi: 10.1016/0166-2236(92)90355-c. [DOI] [PubMed] [Google Scholar]
- Gerfen CR. The neostriatal mosaic: multiple levels of compartmental organization in the basal ganglia. Annu Rev Neurosci. 1992b;15:285–320. doi: 10.1146/annurev.ne.15.030192.001441. [DOI] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Green NM, MacLennan DH. ATP driven ion pumps: an evolutionary mosaic. Biochem Soc Trans. 1989;17:819–822. doi: 10.1042/bst0170819. [DOI] [PubMed] [Google Scholar]
- Hirsch EC, Perier C, Orieux G, Francois C, Feger J, Yelnik J, Vila M, Levy R, Tolosa ES, Marin C, Trinidad Herrero M, Obeso JA, Agid Y. Metabolic effects of nigrostriatal denervation in basal ganglia. Trends Neurosci. 2000;23:S78–S85. doi: 10.1016/s1471-1931(00)00021-5. [DOI] [PubMed] [Google Scholar]
- Johansson PA, Andersson M, Andersson KE, Cenci MA. Alterations in cortical and basal ganglia levels of opioid receptor binding in a rat model of L-DOPA-induced dyskinesia. Neurobiol Dis. 2001;8:220–239. doi: 10.1006/nbdi.2000.0372. [DOI] [PubMed] [Google Scholar]
- Julian MD, Martin AB, Cuellar B, Rodriguez De Fonseca F, Navarro M, Moratalla R, Garcia-Segura LM. Neuroanatomical relationship between type 1 cannabinoid receptors and dopaminergic systems in the rat basal ganglia. Neuroscience. 2003;119:309–318. doi: 10.1016/s0306-4522(03)00070-8. [DOI] [PubMed] [Google Scholar]
- Konradi C, Eaton ME, MacDonald ML, Walsh J, Benes FM, Heckers S. Molecular evidence for mitochondrial dysfunction in bipolar disorder. Arch Gen Psychiatry. 2004;61:300–308. doi: 10.1001/archpsyc.61.3.300. [DOI] [PubMed] [Google Scholar]
- Kregel KC. Heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. J Appl Physiol. 2002;92:2177–2186. doi: 10.1152/japplphysiol.01267.2001. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Kawaguchi Y. Dependence of GABAergic synaptic areas on the interneuron type and target size. J Neurosci. 2000;20:375–386. doi: 10.1523/JNEUROSCI.20-01-00375.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Moine C, Normand E, Bloch B. Phenotypical characterization of the rat striatal neurons expressing the D1 dopamine receptor gene. Proc Natl Acad Sci U S A. 1991;88:4205–4209. doi: 10.1073/pnas.88.10.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Cenci MA, Schulzer M, Bjorklund A. Embryonic ventral mesencephalic grafts improve levodopa-induced dyskinesia in a rat model of Parkinson’s disease. Brain. 2000;123:1365–1379. doi: 10.1093/brain/123.7.1365. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001a;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001b;2:(research 0032). doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad M, Andersson M, Winkler C, Kirik D, Wierup N, Cenci MA. Pharmacological validation of behavioural measures of akinesia and dyskinesia in a rat model of Parkinson’s disease. Eur J Neurosci. 2002;15:120–132. doi: 10.1046/j.0953-816x.2001.01843.x. [DOI] [PubMed] [Google Scholar]
- Lundblad M, Vaudano E, Cenci MA. Cellular and behavioural effects of the adenosine A2a receptor antagonist KW-6002 in a rat model of l-DOPA-induced dyskinesia. J Neurochem. 2003;84:1398–1410. doi: 10.1046/j.1471-4159.2003.01632.x. [DOI] [PubMed] [Google Scholar]
- Massa SM, Longo FM, Zuo J, Wang S, Chen J, Sharp FR. Cloning of rat grp75, an hsp70-family member, and its expression in normal and ischemic brain. J Neurosci Res. 1995;40:807–819. doi: 10.1002/jnr.490400612. [DOI] [PubMed] [Google Scholar]
- Mata M, Hieber V, Beaty M, Clevenger M, Fink DJ. Activity-dependent regulation of Na+, K(+)-ATPase alpha isoform mRNA expression in vivo. J Neurochem. 1992;59:622–626. doi: 10.1111/j.1471-4159.1992.tb09415.x. [DOI] [PubMed] [Google Scholar]
- Melamed E, Offen D, Shirvan A, Djaldetti R, Barzilai A, Ziv I. Levodopa toxicity and apoptosis. Ann Neurol. 1998;44:S149–S154. doi: 10.1002/ana.410440722. [DOI] [PubMed] [Google Scholar]
- Morrison D. 14-3-3: modulators of signaling proteins? Science. 1994;266:56–57. doi: 10.1126/science.7939645. [DOI] [PubMed] [Google Scholar]
- Muller V, Gruber G. ATP synthases: structure, function and evolution of unique energy converters. Cell Mol Life Sci. 2003;60:474–494. doi: 10.1007/s000180300040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novelli A, Reilly JA, Lysko PG, Henneberry RC. Glutamate becomes neurotoxic via the N-methyl-D-aspartate receptor when intracellular energy levels are reduced. Brain Res. 1988;451:205–212. doi: 10.1016/0006-8993(88)90765-2. [DOI] [PubMed] [Google Scholar]
- Nutt JG. Motor fluctuations and dyskinesia in Parkinson’s disease. Parkinsonism Relat Disord. 2001;8:101–108. doi: 10.1016/s1353-8020(01)00024-4. [DOI] [PubMed] [Google Scholar]
- O’Shea RD. Roles and regulation of glutamate transporters in the central nervous system. Clin Exp Pharmacol Physiol. 2002;29:1018–1023. doi: 10.1046/j.1440-1681.2002.03770.x. [DOI] [PubMed] [Google Scholar]
- Oh JD, Vaughan CL, Chase TN. Effect of dopamine denervation and dopamine agonist administration on serine phosphor-ylation of striatal NMDA receptor subunits. Brain Res. 1999;821:433–442. doi: 10.1016/s0006-8993(99)01121-x. [DOI] [PubMed] [Google Scholar]
- Parent A, Hazrati LN. Functional anatomy of the basal ganglia I. The cortico–basal ganglia–thalamo–cortical loop. Brain Res. 1995;20:91–127. doi: 10.1016/0165-0173(94)00007-c. [DOI] [PubMed] [Google Scholar]
- Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, De Fronzo R, Finlayson J, Kahn CR, Mandarino LJ. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. Academic Press Inc; San Diego: 1986. [Google Scholar]
- Picconi B, Centonze D, Hakansson K, Bernardi G, Greengard P, Fisone G, Cenci MA, Calabresi P. Loss of bidirectional striatal synaptic plasticity in L-DOPA-induced dyskinesia. Nat Neuro-sci. 2003;6:501–506. doi: 10.1038/nn1040. [DOI] [PubMed] [Google Scholar]
- Planta RJ. Regulation of ribosome synthesis in yeast. Yeast. 1997;13:1505–1518. doi: 10.1002/(SICI)1097-0061(199712)13:16<1505::AID-YEA229>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Pongrac J, Middleton FA, Lewis DA, Levitt P, Mirnics K. Gene expression profiling with DNA microarrays: advancing our understanding of psychiatric disorders. Neurochem Res. 2002;27:1049–1063. doi: 10.1023/a:1020904821237. [DOI] [PubMed] [Google Scholar]
- Rascol O, Payoux P, Ory F, Ferreira JJ, Brefel-Courbon C, Montastruc JL. Limitations of current Parkinson’s disease therapy. Ann Neurol. 2003;53:S3–S12. doi: 10.1002/ana.10513. discussion S12–15. [DOI] [PubMed] [Google Scholar]
- Roberts RC, Gaither LA, Gao XM, Kashyap SM, Tamminga CA. Ultrastructural correlates of haloperidol-induced oral dyskinesias in rat striatum. Synapse. 1995;20:234–243. doi: 10.1002/syn.890200307. [DOI] [PubMed] [Google Scholar]
- Rosenquist M. 14-3-3 Proteins in apoptosis. Braz J Med Biol Res. 2003;36:403–408. doi: 10.1590/s0100-879x2003000400001. [DOI] [PubMed] [Google Scholar]
- Schallert T, Fleming SM, Leasure JL, Tillerson JL, Bland ST. CNS plasticity and assessment of forelimb sensorimotor outcome in unilateral rat models of stroke, cortical ablation, parkinsonism and spinal cord injury. Neuropharmacology. 2000;39:777–787. doi: 10.1016/s0028-3908(00)00005-8. [DOI] [PubMed] [Google Scholar]
- Schwarzer C, Berresheim U, Pirker S, Wieselthaler A, Fuchs K, Sieghart W, Sperk G. Distribution of the major gamma-aminobutyric acid(A) receptor subunits in the basal ganglia and associated limbic brain areas of the adult rat. J Comp Neurol. 2001;433:526–549. doi: 10.1002/cne.1158. [DOI] [PubMed] [Google Scholar]
- Sheng M. Molecular organization of the postsynaptic specialization. Proc Natl Acad Sci U S A. 2001;98:7058–7061. doi: 10.1073/pnas.111146298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sian J, Gerlach M, Youdim MB, Riederer P. Parkinson’s disease: a major hypokinetic basal ganglia disorder. J Neural Transm. 1999;106:443–476. doi: 10.1007/s007020050171. [DOI] [PubMed] [Google Scholar]
- Taylor MD, De Ceballos ML, Rose S, Jenner P, Marsden CD. Effects of a unilateral 6-hydroxydopamine lesion and prolonged L-3,4-dihydroxyphenylalanine treatment on peptidergic systems in rat basal ganglia. Eur J Pharmacol. 1992;219:183–192. doi: 10.1016/0014-2999(92)90295-f. [DOI] [PubMed] [Google Scholar]
- Thomas U. Modulation of synaptic signalling complexes by Homer proteins. J Neurochem. 2002;81:407–413. doi: 10.1046/j.1471-4159.2002.00869.x. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzivion G, Avruch J. 14-3-3 Proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J Biol Chem. 2002;277:3061–3064. doi: 10.1074/jbc.R100059200. [DOI] [PubMed] [Google Scholar]
- van Hemert MJ, Steensma HY, van Heusden GP. 14-3-3 Proteins: key regulators of cell division, signalling and apoptosis. Bioessays. 2001;23:936–946. doi: 10.1002/bies.1134. [DOI] [PubMed] [Google Scholar]
- Wallimann T, Hemmer W. Creatine kinase in non-muscle tissues and cells. Mol Cell Biochem. 1994;133–134:193–220. doi: 10.1007/BF01267955. [DOI] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Westin JE, Andersson M, Lundblad M, Cenci MA. Persistent changes in striatal gene expression induced by long-term L-DOPA treatment in a rat model of Parkinson’s disease. Eur J Neurosci. 2001;14:1171–1176. doi: 10.1046/j.0953-816x.2001.01743.x. [DOI] [PubMed] [Google Scholar]
- Winkler C, Kirik D, Bjorklund A, Cenci MA. L-DOPA-induced dyskinesia in the intrastriatal 6-hydroxydopamine model of Parkinson’s disease: relation to motor and cellular parameters of nigrostriatal function. Neurobiol Dis. 2002;10:165–186. doi: 10.1006/nbdi.2002.0499. [DOI] [PubMed] [Google Scholar]
- Wyss M, Kaddurah-Daouk R. Creatine and creatinine metabolism. Physiol Rev. 2000;80:1107–1213. doi: 10.1152/physrev.2000.80.3.1107. [DOI] [PubMed] [Google Scholar]