Abstract
Trade-offs in life-history traits is a central tenet in evolutionary biology, yet their ubiquity and relevance to realized fitness in natural populations remains questioned. Trade-offs in pathogens are of particular interest because they may constrain the evolution and epidemiology of diseases. Here, we studied life-history traits determining transmission in the obligate fungal pathogen, Podosphaera plantaginis, infecting Plantago lanceolata. We find that although traits are positively associated on sympatric host genotypes, on allopatric host genotypes relationships between infectivity and subsequent transmission traits change shape, becoming even negative. The epidemiological prediction of this change in life-history relationships in allopatry is lower disease prevalence in newly established pathogen populations. An analysis of the natural pathogen metapopulation confirms that disease prevalence is lower in newly established pathogen populations and they are more prone to go extinct during winter than older pathogen populations. Hence, life-history trade-offs mediated by pathogen local adaptation may influence epidemiological dynamics at both population and metapopulation levels.
Keywords: Coevolution, disease transmission, epidemiology, host–pathogen interactions, infectivity, trade-offs
Trade-offs between life-history traits, whereby allocation of limited resources to one trait has a negative impact on another trait, is a generally accepted phenomenon in evolutionary biology (Stearns 1989). This trade-off model provides an intuitive framework for explaining how adaptation of populations to new environments may be constrained, and how different life-history strategies evolve, depending on where limited resources are allocated (Maynard Smith 1966; Rausher 1984; Roff 1992). Trade-offs are of fundamental importance in ecology because they prevent the evolution of a Darwinian demon, that is, a hypothetical organism that can maximize all aspects of fitness simultaneously (Law 1979). In general, the most common life-history trade-offs include those between offspring size and offspring number, body size and development time, and survival and reproduction (Roff 2002).
In host–pathogen associations, the concept of life-history trade-offs has been invoked to explain how diversity in host resistance and pathogen infectivity is maintained within populations (Laine and Tellier 2008). A classic model of the genetic interaction between hosts and pathogens is the gene-for-gene (GFG) model, whereby corresponding host resistance and parasite infectivity loci interact to determine infection outcome (Flor 1971). From the late 1950s onward (Flor 1955, 1956), the GFG-hypothesis stimulated a series of deterministic theoretical models exploring how the frequencies of resistance and infectivity genes change over time in host and pathogen populations, respectively (as reviewed in Laine and Tellier 2008). A consistent feature of these models is a high cost associated with infectivity (or resistance), which is required to maintain diversity (Laine and Tellier 2008). Many host–pathogen interactions ranging from plants to insects are shown to conform to the GFG model, resulting in high specificity in host resistance and pathogen infectivity profiles (Thompson and Burdon 1992; Buckling and Rainey 2002; Forde et al. 2004; Bangham et al. 2007). To date, empirical evidence regarding correlations in key interaction traits of hosts and pathogens remain mixed (Bergelson and Purrington 1996; Thrall and Burdon 2003; Tian et al. 2003; Héraudet et al. 2008).
A fundamental but unresolved question is how adaptation influences trade-offs. Although there may be evidence for negative correlations within a particular environment, correlations can shift when populations encounter different environmental conditions (van Noordwijk and de Jong 1986; Sgro and Hoffmann 2004; Vale et al. 2011). Hosts—and the hosts’ defenses—represent the most critical environment for pathogens, as successful host exploitation is a prerequisite of pathogen growth, reproduction, and survival. Consequently, pathogen life-history traits are likely to be under strong selection imposed by the host’s defense system, and pathogens are generally predicted to be locally adapted to their host populations (see Gandon and Michalakis 2002 for conditions favoring the emergence of parasite local adaptation). Indeed, there is abundant evidence for pathogens adapting to their sympatric host populations, although there is also considerable variation in this tendency (as reviewed in Greischar and Koskella 2007; Tack et al. 2012). Notably, if pathogen life-history traits are influenced by both host and pathogen, correlations in these traits may be governed by GHOST × GPATHOGEN interactions (Salvaudon et al. 2005; Pariaud et al. 2012). Hence, if life-history correlations measured on sympatric host genotypes change strength or even shape (from positive to negative or vice versa) on allopatric host genotypes, this could have profound implications for pathogen evolution by constraining the rate at which pathogens adapt to novel host environments. Importantly, we should also expect to see differences in epidemiological dynamics of pathogens in sympatric host populations versus newly colonized host populations in situations where adaptation mediates the shape of trade-offs. A trade-off between infectivity and subsequent stages of growth and transmission has the potential to significantly impede transmission and subsequently disease prevalence. In the interaction between the wild flax and its rust pathogen, a negative trade-off between infectivity and spore production (Thrall and Burdon 2003) may explain low disease prevalence in populations dominated by highly infective pathogen strains (Thrall et al. 2012). To date, studies linking pathogen life-history traits to realized population dynamics are surprisingly scarce given the fundamental importance of the trade-off theory to evolutionary biology.
Here, we study whether the shape of life-history relationships change as pathogens disperse beyond the boundaries of their sympatric host population onto novel host genotypes, and what the epidemiological consequences of this may be. For this purpose, we carried out an inoculation experiment measuring transmission stages of the obligate fungal pathogen Podosphaera plantaginis infecting Plantago lanceolata on sympatric and allopatric host plants originating from the Åland archipelago, southwest of Finland (Fig. S1). In Åland, the pathogen persists as a highly dynamic metapopulation with frequent extinctions and colonizations of local host populations (Laine and Hanski 2006). For the experiment we used strains from seven natural populations to account for potential variation among populations in life-history trait relationships, and potential variation in the degree of local adaptation (Laine 2005). The experimental results revealed that life-history traits are positively associated on sympatric hosts, but on allopatric host genotypes the relationship between infectivity and subsequent transmission traits change shape, becoming even negative. We then analyzed whether dynamics of the natural pathogen metapopulation corresponded with our experimental findings. We predicted that the observed trade-off between infectivity and subsequent asexual growth and propagation detected in allopatry would result in lower disease prevalence of recently colonized host populations. Furthermore, we predicted that at the metapopulation level, these recently established pathogen populations would be more prone to go extinct. Our analyses supported both predictions, suggesting that life-history trade-offs may influence pathogen epidemiology both at the within population and metapopulation levels.
Materials and Methods
Host–Pathogen System
The host plant Pl. lanceolata L. (Plantaginaceae) is a perennial plant that is considered an obligate outcrosser. The seeds of Pl. lanceolata have no special dispersal mechanisms; as they ripen, they are simply dropped to the ground close to the mother plant. Clonally produced side rosettes from the axillary meristems are a common means of reproduction for Pl. lanceolata (Mook et al. 1992).
Podosphaera plantaginis (Castagne; U. Braun and S. Takamatsu) is an obligate powdery mildew fungus in the order Erysiphales within the Ascomycota (Yarwood 1978). The fungus completes its entire life cycle on the surface of the host plant where it is visible as localized (nonsystemic) white powdery lesions. During the growing season, the pathogen is transmitted among hosts by clonally produced dispersal spores, conidia, that are passively carried by wind. The pathogen may pass through approximately 6–8 clonal cycles during the growing season (Ovaskainen and Laine 2006). At the end of the growing season in August, the resting spores (chasmothecia) begin to appear, visible to the eye as black specks roughly 1 mm in diameter. During winter as most host individuals die back to root stock, the local pathogen populations decline.
The interaction between Pl. lanceolata and Po. plantaginis functions in a two-step manner typical of most plant–pathogen associations (Jones and Dangl 2006). First, as the pathogen attempts to infect a new host, the interaction is strain specific as the same host genotype expresses resistance against some strains (i.e., recognition) of the pathogen while being susceptible to others (i.e., nonrecognition; Laine 2007). When a Po. plantaginis strain has successfully established, there is still considerable variation in its development that is affected by both pathogen and host genotype (Laine 2007). The key life-history traits that constitute the asexual transmission stage following successful establishment are: time to germination (measured as the number of days it takes for a spore landing on a leaf to form the first hyphae); time to spore production (measured as the number of days it takes for the established mycelium to begin sporulation), and overall spore production level.
Study of the Natural Pathogen Metapopulation
The Plantago–Podosphaera study system in the Åland Islands comprises approximately 4000 Pl. lanceolata populations. The host populations are typically small (median 0.3 ha), occurring on dry meadows separated by unsuitable habitat consisting of arable land, human settlement, roads, and forest (Nieminen et al. 2004). In early September every year since 2001, all known Pl. lanceolata populations have been checked for the presence of the powdery mildew (for details on the survey, please see Laine and Hanski 2006). These data have demonstrated that Po. plantaginis persists as a highly dynamic metapopulation through extinctions and colonizations of local host populations (Laine and Hanski 2006; Tollenaere et al. 2012). Here, we use these presence/absence data to infer which populations were colonized in 2010, and which populations had gone extinct by September 2011. In year 2010, a subset of the pathogen metapopulation (79 of 176 pathogen populations; Fig. S1) was revisited in mid-September and disease prevalence was recorded (1 ≤ 10% of host plants infected, 2 = 10–25% of host plants infected, 3 = 25–50% of host plants infected, and 4 = 50–100% of host plants infected).
Pathogen Life-History on Sympatric and Allopatric Host Genotypes
We conducted an inoculation experiment to measure possible trade-offs between successive life-history traits that constitute the transmission phase during epidemics—infectivity, time to germination, time to sporulation, and amount of spores produced. Strains of Po. plantaginis were inoculated onto host genotypes originating from their sympatry as well as onto allopatric host genotypes that were the same for all pathogen populations.
In September 2010, 10 infected leaves, each from a different host plant, were collected from seven natural populations of Po. plantaginis (coordinates of the sampled populations are presented in Table S1). The leaves were placed on moist filter paper inside petri dishes. Pure strains were obtained by three consecutive single colony transfers (Nicot et al. 2002). The fungal strains were maintained on detached Pl. lanceolata leaves in 9 cm petri dishes in a growth chamber at 20°C with 16L/8D photoperiod. During purification and maintenance some strains died, and hence, the experiment consisted of the following fungal material (population ID/number of strains): 4/4, 325/5, 511/8, 1062/3, 1413/2, 2220/7, and 9031/4.
Seeds of sympatric plants were collected from the same seven populations of Pl. lanceolata as the fungal strains (altogether from 42 plants; Table S1). The allopatric plants were collected as seeds from five Pl. lanceolata populations (IDs 877, 1915, 1929, 2224, and 3484; coordinates in Table S1) that were not within the typical dispersal range (Laine 2005) of the sympatric pathogen populations chosen for the study. A single plant was used for population 3484, whereas two plants represented the other allopatric host populations. The seeds were sown to 9 × 9 cm pots on 1:1 mixture of garden soil and sand, and the plants were kept in a greenhouse at +20°C. Each plant used in the experiment originated from a distinct mother plant in the natural population.
In the experiment, each of the 33 pathogen strains was inoculated onto six of their sympatric host lines and onto the nine allopatric host lines. We used the same nine allopatric host genotypes for all pathogen strains. A detached leaf from each host plant was exposed to a single pathogen strain. Infection outcome and its timing on detached leaves are similar to leaves still intact with the plant (Laine 2011). The leaf was placed on moist filter paper in a 9 cm Petri dish, and conidial spores from an infected leaf were gently brushed with a fine paint brush over the entire surface of the healthy leaf. Colonies of similar age and size (about 1.0-cm diameter) were used for the inoculations in order to obtain as similar spore densities as possible. This procedure has been shown to produce repeatable infection outcomes (Laine 2004). Inoculated dishes were placed inside a growth chamber at +20°C and with a 16L/8D photoperiod. The resulting infections were observed daily starting at 4th day postinoculation until 12th day. Every day the development of infection was observed under a dissecting microscope, and the level of infection was scaled from 0 to 4 as 0 = only mycelium, 1 = mycelium and conidia visible under microscope, 2 = mycelia visible by naked eye and sparse conidia visible under dissecting microscope, 3 = colonies smaller than 0.5 cm2 with abundant sporulation, and 4 = colonies larger than 0.5 cm2 with abundant sporulation. Infectivity was scored as 0 = no infection and 1 = infection by the 12th day postinoculation. Following inoculation, germination day was marked as the first day mycelia were observed on the leaf surface, and sporulation day was marked as the first day conidial spores were observed. Infection score as measured on day 12 is used is used in the analyses of spore production level.
We included a leaf of a known susceptible host genotype in the inoculations to confirm inoculation success. We repeated the subset of inoculations that failed (2%) immediately after the first trial was completed.
Statistical Analyses
To determine whether pathogen life-history relationships differed on sympatric and allopatric hosts, we analyzed as generalized linear mixed models (GLMM) all pairwise interactions between the life-history traits of infectivity, germination time, sporulation time, and spore production level. In all models, sympatry-allopatry was defined as a fixed explanatory variable, indicating whether the pathogen was tested on sympatric versus allopatric hosts, and we included the interaction term between sympatry-allopatry and each life-history trait. Pathogen population and strain nested within population were defined as random effects. Before analyses, data were averaged for each strain to obtain trait profiles for each strain that included infectivity (consisting of zeros and ones) and measures of pathogen growth and sporulation using only data of the infective responses. The averaged spore production and time to sporulation data were log-transformed and data on infectivity were arcsine-transformed to meet model assumptions of normally distributed errors. All GLMMs were implemented using the MIXED procedure in SAS 9.2. (SAS Institute Inc., Cary, NC). To estimate the significance of slopes separately for sympatry and allopatry, we excluded the main life-history term from our model enabling the covariate part of the model to be nonsingular, thus providing estimates of the slopes (Littell et al. 2006).
We then analyzed whether disease prevalence was lower in newly colonized host populations compared to previously established populations, as would be predicted by the weak or negative relationship found in allopatry between infectivity and other life-history traits (see Results). In the analysis, we included only the subset of the pathogen metapopulation that was surveyed for disease prevalence in year 2010 (N = 79; on a 1–4 scale described in “Study of the natural pathogen metapopulation”) in Åland in 2010. We compared the proportion of populations in each of the disease prevalence classes between pathogen populations colonized in 2010 (N = 30) and pathogen populations that were 2 years or older (N = 49) using a G-test. To see whether there was any metapopulation level effect of the change in trait relationships detected in sympatry versus allopatry, we analyzed whether recently colonized populations had higher extinction probability. We first analyzed for the same 79 pathogen populations whether their probability to go extinct before September 2011 was affected by disease prevalence. We also analyzed whether the extinction probability of all pathogen populations in 2010 (N = 146 of 176, as we only included those host populations that have been surveyed since year 2001) was effected by population age. Both analyses of extinction were logistic regressions with extinction probability (0/1) as the response variable and disease prevalence or age as the explanatory variable. The analyses were implemented in JMP 8.0.2.
Results
Life-History Trade-Offs in Sympatry Versus Allopatry
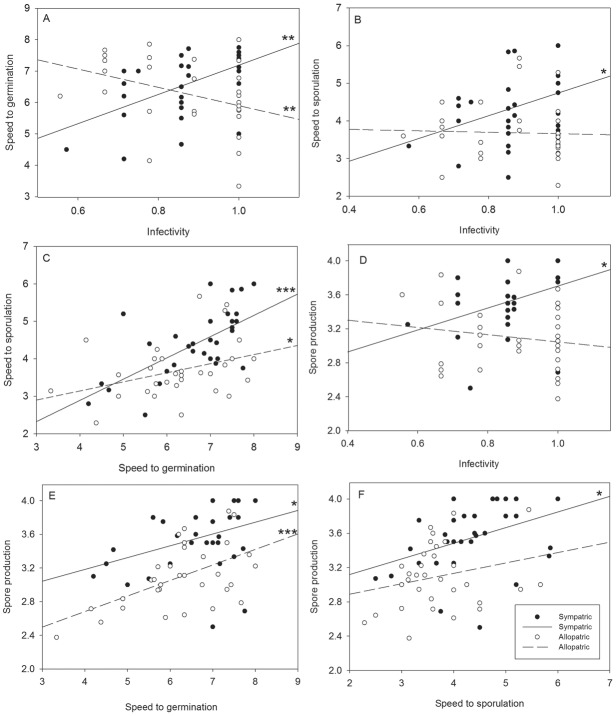
We observed no direct negative relationship between any of the measured life-history traits, indicating that there are no pervasive trade-offs in the life-history traits that constitute the transmission stage in Po. plantaginis (Table 1; Fig. 1). In contrast, there was a trend for positive relationships between many of the measured life-history traits (Fig. 1). Statistically significant positive relationships were found between time to sporulation and time to germination, spore production and time to germination, and spore production and time to sporulation (Table 1; Fig. 1). Sympatry-allopatry explained differences only in time to germination that was significantly shorter on sympatric than on allopatric host plants in the model accounting for infectivity (P = 0.0005; Table 1 and Fig 1A). Time to sporulation was also shorter on sympatric than on allopatric host genotypes in the model accounting for infectivity, but this was only marginally significant (P = 0.0879; Table 1 and Fig. 1B).
Table 1.
Results of the GLMMs analyzing correlations between different life-history stages of Podosphaera plantaginis as measured on sympatric and allopatric host genotypes. Statistically significant results are shown in bold
| Source | F1, 56 | P |
|---|---|---|
| Time to germination | ||
| Infectivity | 0.07 | 0.7872 |
| Sympatry-Allopatry | 13.49 | 0.0005 |
| Infectivity×Sympatry-Allopatry | 18.78 | <0.0001 |
| Time to sporulation | ||
| Infectivity | 2.13 | 0.1501 |
| Sympatry-Allopatry | 2.98 | 0.0897 |
| Infectivity×Sympatry-Allopatry | 9.38 | 0.0034 |
| Time to sporulation | ||
| Time to germination | 27.25 | <0.0001 |
| Sympatry-Allopatry | 5.07 | 0.283 |
| Time to germination×Sympatry-Allopatry | 2.97 | 0.0902 |
| Spore production | ||
| Infectivity | 1.50 | 0.2256 |
| Sympatry-Allopatry | 1.11 | 0.2974 |
| Infectivity×Sympatry-Allopatry | 6.58 | 0.0130 |
| Spore production | ||
| Time to germination | 18.58 | <0.0001 |
| Sympatry-Allopatry | 0.18 | 0.6763 |
| Time to germination×Sympatry-Allopatry | 0.22 | 0.644 |
| Spore production | ||
| Time to sporulation | 8.51 | 0.0051 |
| Sympatry-Allopatry | 0.23 | 0.6358 |
| Time to sporulation×Sympatry-Allopatry | 0.01 | 0.9316 |
Figure 1.
Life-history trait correlations of the 33 Podosphaera plantaginis strains measured on sympatric host genotypes (black symbols, solid line) and allopatric host genotypes (white symbols, dashed line). The significance of the slopes are indicated with asterisks (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001). For those life-history traits that depict timing of events (i.e., speed to germination and speed to sporulation), higher values indicate faster performance.
We discovered that the strength, and even direction, of life-history trait relationships varied depending on whether they were measured on sympatric or allopatric host genotypes (Fig. 1). The highly positive association between time to germination and infectivity measured in sympatry became negative on allopatric host genotypes (Table 1; Fig. 1A). Similarly, the positive relationship between infectivity and time to sporulation disappeared when the strains were infecting allopatric host genotypes (Table 1; Fig. 1B). Most importantly, the relationship between spore production and infectivity varied significantly depending on host origin (infectivity × sympatry–allopatry interaction; P = 0.013). On sympatric hosts, those pathogen strains that were capable of infecting most of the sympatric host genotypes, had also the highest spore production in sympatry. However, on allopatric host genotypes this positive trend disappears (Table 1; Fig. 1D). The estimates of random variables “Population” and “Pathogen Strain” as well as their significance in Wald’s Z-test for all models are reported in Table S2.
Linking Life-History Results with Epidemiological Data
Our prediction that newly established pathogen populations would be smaller than older populations (as predicted by the weak or negative relationship between infectivity and subsequent pathogen growth and sporulation in allopatry; Fig. 1) was confirmed. Pathogen populations established in 2010 were smaller at the end of the growing season than older pathogen populations (G = 9.445, P = 0.0089; Fig. 2A). Although these smaller populations had a higher probability of going extinct by September 2011, the effect of disease prevalence on extinction probability was not statistically significant (χ2 = 2.77, P = 0.2509). Using data from 146 pathogen populations in 2010, we found that population age had a significant impact on extinction probability (χ2 = 17.49, P < 0.0001; Fig. 2B).
Figure 2.
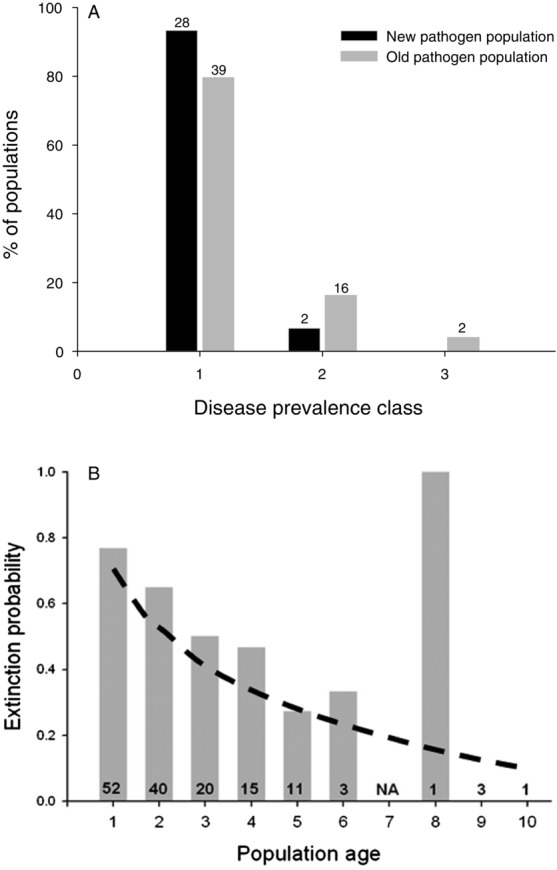
(A) Disease prevalence of Podosphaera plantaginis populations established in 2010 is significantly lower than the prevalence of older pathogen populations. (B) Extinction probability of Po. plantaginis populations between 2010 and 2011 decreases significantly with population age. Numbers indicate the number of populations for each age class.
Discussion
Here we find that infectivity of Po. plantaginis is positively associated with subsequent life-history traits on sympatric host genotypes. However, when the performance of the same pathogen strains is measured on allopatric host genotypes, these relationships change shape, becoming even negative. To our knowledge, this is the first demonstration of pathogen life-history trade-offs being mediated by local adaption. This finding may have significant evolutionary and epidemiological implications as discussed below.
Positive relationships between parasite life-history traits have also been reported elsewhere (Salvaudon et al. 2005; Pariaud et al. 2012). This finding is unintuitive from the perspective of the gene-for-gene models whereby costs of infectivity are required to maintain polymorphism. However, pathogen (meta)populations are generally found to consist of multiple coexisting strains (Tack et al. 2012). In the Plantago–Podosphaera interaction in Åland, local pathogen populations typically consist of several multilocus genotypes (Tollenaere et al. 2012), that differ in their infectivity and transmission efficiency (Laine 2005, 2008). In the absence of trade-offs, genotype-by-environment interactions, metapopulation dynamics, and dormant life-history stages have been proposed as key processes maintaining diversity in local populations (Damgaard 1999; Sasaki 2000; Laine and Tellier 2008; Tellier and Brown 2009; Wolinska and King 2009).
The “big houses–big cars” hypothesis (Reznick et al. 2000) proposes that “super strains,” such as we observe here in sympatry having both high infectivity and transmission capacity, may arise when resources are abundant. Indeed, the qualitative and quantitative composition of available food has a significant impact on life history traits and trade-offs (Lee et al. 2008; Vale et al. 2011; Attisano et al. 2012). Only when food becomes limiting, or conditions otherwise stressful, may trade-offs between life-history stages arise (Reznick et al. 2000). In host–pathogen interactions, local adaptation to host genotypes, that is ability to cope with the hosts’ defenses, may be a direct measure of environmental stress and nutrient availability to pathogens. Here, the positive relationship between infectivity and subsequent transmission stages (time to germination, time to sporulation, and spore production) that we measure on sympatric host genotypes changes shape, becoming even negative on allopatric host genotypes. This finding is most plausibly explained by the limited ability of the pathogens to exploit resources of novel host genotypes and to cope with novel quantitative resistance strategies. Powdery mildew fungi complete their life cycle on the surface of the host plant, using only their feeding roots, haustoria, to penetrate host cells. Quantitative resistance responses following infection including hydrolytic activities on cell walls, and contact toxicity (van Loon et al. 2006), are among the processes that may impede nutrition acquisition and result in reduced transmission efficiency. The loss of trade-off constraints between pathogen life-history traits when tested on their sympatric hosts suggests that adaptation may reduce these limitations, given sufficient time following colonization.
Although putative trade-offs in the life-history of pathogens have received considerable attention, to date we know essentially nothing about how they impact on disease epidemiology (but see Alizon 2009). For a species such as Po. plantaginis, which persists regionally through frequent extinctions and colonizations of local host populations (Laine and Hanski 2006, Soubeyrand et al. 2009), trade-offs between infectivity and transmission traits may have profound epidemiological implications. A pathogen strain emerging into a new host population is initially analogous to an allopatric pathogen encountering novel host genotypes. In these situations, transmission may be slowed down during subsequent growth and reproduction of the strain, despite frequent colonization events enabled by high infectivity rates (Laine and Hanski 2006). Indeed, we found that more recently colonized host populations do have lower disease prevalence. Moreover, we found that the newly established pathogen populations are more prone to go extinct during the first winter than older pathogen populations. These results are observational, and could also reflect other processes. First, in newly colonized host populations, the pathogen may arrive later in the season than in those populations where the pathogen has successfully overwintered. Hence, the newly established pathogen population may simply have less time to increase than the old pathogen populations. However, it should be noted that even established pathogen populations crash during winter and the next epidemic is initiated from few, discrete foci (Ovaskainen and Laine 2006). Tracking epidemiological dynamics throughout the growing season in old and newly established pathogen populations would help disentangle between these processes. Second, our finding that newly colonized host populations are more likely to go extinct than older pathogen populations could also reflect environmental factors that promote persistence in certain areas through time. However, the majority of colonizations take place in close range of established pathogen populations (Laine and Hanski 2006). Our analysis of colonization events between 2001 and 2011 indicate that within these areas the environmental factors favoring the pathogen are similar (A.-L. Laine, pers. comm.). Although we are not able to fully distinguish between the different causes of our population and metapopulation level results, they do offer some of the very first evidence of the potential of pathogen life-history trade-offs to impact on realized epidemiological dynamics.
In conclusions, understanding the conditions that change life-history trait relationships in pathogens is vital to understanding their importance for epidemiology and evolution. Our results confirm that although trade-offs are not detected on sympatric host genotypes, trait relationships change shape, becoming even negative between infectivity and subsequent transmission stages on novel hosts. Our analyses of disease prevalence and metapopulation dynamics suggest that these context-dependent life-history trait relationships may have strong impacts on disease epidemiology. Establishing direct links between the fundamental axes of life-history variation and realized epidemiological dynamics offers an exciting future venue of research, and is needed to truly validate the relevance of the trade-off theory for pathogens.
Associate Editor: J. Wernegreen
Acknowledgments
M. Saastamoinen, Associate Editor J. Wernegreen, and three anonymous reviewers provided helpful comments on the manuscript. The authors thank E. Meyke for producing the maps and A. Tack for statistical advice. This work was supported by funding from the Academy of Finland (Grant Nos. 250444, 136393, and 133499) and European Research Council (PATHEVOL; 281517) to ALL.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher’s website:
Figure S1. Occurrence of Podosphaera plantaginis in the Åland metapopulation in 2010.
Table S1. Coordinates of the pathogen populations sampled for the inoculation study, and of the allopatric host populations.
Table S2. Random estimates of the GLMMs used to analyze life-history correlations in sympatry and allopatry.
Literature Cited
- Alizon S. The price equation framework to study disease within-host evolution. J. Evol. Biol. 2009;22:1123–1132. doi: 10.1111/j.1420-9101.2009.01726.x. [DOI] [PubMed] [Google Scholar]
- Attisano A, Moore AJ. Moore PJ. Reproduction-longevity trade-offs reflect diet, not adaptation. J. Evol. Biol. 2012;25:873–880. doi: 10.1111/j.1420-9101.2012.02476.x. [DOI] [PubMed] [Google Scholar]
- Bangham J, Obbard DJ, Kim KW, Haddrill PR. Jiggins FM. The age and evolution of an antiviral resistant mutation in Drosophila melanogaster. Proc. R. Soc. B. 2007;274:2027–2034. doi: 10.1098/rspb.2007.0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J. Purrington CB. Surveying patterns in the costs of resistance in plants. Am. Nat. 1996;148:536–558. [Google Scholar]
- Buckling A. Rainey PB. Antagonistic coevolution between a bacterium and a bacteriophage. Proc. R. Soc. B. 2002;269:931–936. doi: 10.1098/rspb.2001.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard C. Coevolution of a plant host-pathogen gene-for-gene system in a metapopulation model without cost of resistance or cost of virulence. J. Theor. Biol. 1999;201:1–12. doi: 10.1006/jtbi.1999.1007. [DOI] [PubMed] [Google Scholar]
- Flor HH. Host-parasite interaction in flax rust—its genetics and other implications. Phytopathology. 1955;45:680–685. [Google Scholar]
- Flor HH. The complementary genic systems in flax and flax rust. Adv. Genet. 1956;8:29–54. [Google Scholar]
- Flor HH. The current status of the gene for gene concept. Annu. Rev. Phytopathol. 1971;9:275–296. [Google Scholar]
- Forde SE, Thompson JN. Bohannan BJM. Adaptation varies through space and time in a coevolving host-parasitoid interaction. Nature. 2004;431:841–844. doi: 10.1038/nature02906. [DOI] [PubMed] [Google Scholar]
- Gandon S. Michalakis Y. Local adaptation, evolutionary potential and host-parasite coevolution: interactions between migration, population size and generation time. J. Evol. Biol. 2002;15:451–462. [Google Scholar]
- Greischar MA. Koskella B. A synthesis of experimental work on parasite local adaptation. Ecol. Lett. 2007;10:418–434. doi: 10.1111/j.1461-0248.2007.01028.x. [DOI] [PubMed] [Google Scholar]
- Héraudet V, Salvaudon L. Shykoff JA. Trade-off between latent period and transmission success of a plant pathogen revealed by phenotypic correlations. Evol. Ecol. Res. 2008;10:913–924. [Google Scholar]
- Jones JDG. Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Laine A-L. Resistance variation within and among host populations in a plant-pathogen metapopulation—implications for regional pathogen dynamics. J. Ecol. 2004;92:990–1000. [Google Scholar]
- Laine A-L. Spatial scale of local adaptation in a plant-pathogen metapopulation. J. Evol. Biol. 2005;18:930–938. doi: 10.1111/j.1420-9101.2005.00933.x. [DOI] [PubMed] [Google Scholar]
- Laine A-L. Pathogen fitness components and genotypes differ in their sensitivity to nutrient and temperature variation in a wild plant-pathogen association. J. Evol. Biol. 2007;20:2371–2378. doi: 10.1111/j.1420-9101.2007.01406.x. [DOI] [PubMed] [Google Scholar]
- Laine A-L. Temperature-mediated patterns of local adaptation in a natural plant-pathogen metapopulation. Ecol. Lett. 2008;11:327–337. doi: 10.1111/j.1461-0248.2007.01146.x. [DOI] [PubMed] [Google Scholar]
- Laine A-L. Context dependent effects of induced resistance in mediating subsequent infections in a plant-pathogen interaction. Evol. Appl. 2011;4:696–707. doi: 10.1111/j.1752-4571.2011.00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine A-L. Hanski I. Large-scale spatial dynamics of a specialist plant pathogen in a fragmented landscape. J. Ecol. 2006;94:217–226. [Google Scholar]
- Laine A-L. Tellier A. Heterogeneous selection promotes maintenance of polymorphism in host-parasite interactions. Oikos. 2008;117:1281–1288. [Google Scholar]
- Law R. Optimal life histories under age-specific predation. Am. Nat. 1979;114:399–417. [Google Scholar]
- Lee KP, Simpson SJ, Clissold FJ, Brooks R, Ballard JWO, Taylor PW, Soran N. Raubenheimer D. Lifespan and reproduction in Drosophila: new insights from nutritional geometry. Proc. Natl. Acad. Sci. USA. 2008;105:2498–2503. doi: 10.1073/pnas.0710787105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littell RC, Milliken GA, Stroup WW, Wolfinger RD. Schabenberger O. SAS® for mixed models. 2nd ed. Cary, NC: SAS Institute Inc; 2006. [Google Scholar]
- Maynard Smith J. Sympatric speciation. Am. Nat. 1966;100:637–650. [Google Scholar]
- Mook JH, Haeck J, van der Toorn J. van Tienderen PH. The demographic structure of populations. In: Kuiper PJC, Bos M, editors; Plantago: a multidisciplinary study. Heidelberg: Springer-Verlag; 1992. pp. 69–87. [Google Scholar]
- Nicot PC, Bardin M, Dik AJ. Basic methods for epidemiological studies of powdery mildews: culture and preservation of isolates, production and delivery of inoculum, and disease assessment. In: Bélanger RR, Bushnell RB, Dik AJ, Carver TJW, editors. The powdery mildews: a comprehensive treatise. St. Paul, MN: Am. Phytopathol. Soc; 2002. pp. 83–99. [Google Scholar]
- Nieminen M, Siljander M. Hanski I. Structure and dynamics of Melitaea cinxia metapopulations. In: Ehrlich PR, Hanski I, editors; On the wings of checkerspots: a model system for population biology. Oxford, U.K: Oxford Univ. Press; 2004. pp. 63–91. [Google Scholar]
- Ovaskainen O. Laine A-L. Inferring evolutionary signals from ecological data in a plant-pathogen metapopulation. Ecology. 2006;87:880–891. doi: 10.1890/0012-9658(2006)87[880:iesfed]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Pariaud B, van den Berg F, van den Bosch F, Powers SJ, Kaltz O. Lannou C. Shared influence of pathogen and host genetics on a tradeoff between latent period and spore production capacity in the wheat pathogen, Puccinia triticina. Evol. Appl. 2012;6:303–312. doi: 10.1111/eva.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausher MD. Tradeoffs in performance on different hosts: evidence from within- and between-site variation in the beetle Deloyala guttata. Evolution. 1984;38:582–595. doi: 10.1111/j.1558-5646.1984.tb00324.x. [DOI] [PubMed] [Google Scholar]
- Reznick D, Nunney L. Tessier A. Big houses, big cars, superfleas and the costs of reproduction. Trends Ecol. Evol. 2000;15:421–425. doi: 10.1016/s0169-5347(00)01941-8. [DOI] [PubMed] [Google Scholar]
- Roff DA. The evolution of life histories. London: Chapman and Hall; 1992. [Google Scholar]
- Roff DA. Life history evolution. Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- Salvaudon L, Héraudet V. Shykoff JA. Parasite-host fitness trade-offs change with parasite identity: genotype-specific interactions in a plant-pathogen system. Evolution. 2005;59:2518–2524. [PubMed] [Google Scholar]
- Sasaki A. Host-parasite coevolution in a multilocus gene-for-gene system. Proc. R. Soc. B. 2000;267:2183–2188. doi: 10.1098/rspb.2000.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgro CM. Hoffmann AA. Genetic correlations, tradeoffs and environmental variation. Heredity. 2004;93:241–248. doi: 10.1038/sj.hdy.6800532. [DOI] [PubMed] [Google Scholar]
- Soubeyrand S, Laine A-L, Hanski I. Penttinen A. Spatio-temporal structure of interactions in a host-pathogen metapopulation. Am. Nat. 2009;174:308–320. doi: 10.1086/603624. [DOI] [PubMed] [Google Scholar]
- Stearns SC. Trade-offs in life-history evolution. Funct. Ecol. 1989;3:259–268. [Google Scholar]
- Tack AJM, Thrall PH, Barrett LG, Burdon JJ. Laine A-L. Variation in infectivity and aggressiveness in space and time in wild host–pathogen systems—causes and consequences. J. Evol. Biol. 2012;25:1918–1936. doi: 10.1111/j.1420-9101.2012.02588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellier A. Brown JKM. The influence of perenniality and seed banks on polymorphism in gene-for-gene interactions. Am. Nat. 2009;174:769–779. doi: 10.1086/646603. [DOI] [PubMed] [Google Scholar]
- Thompson JN. Burdon JJ. Gene-for-gene coevolution between plants and parasites. Nature. 1992;360:121–125. [Google Scholar]
- Thrall P. Burdon JJ. Evolution of virulence in a plant host-pathogen metapopulation. Science. 2003;299:1735–1737. doi: 10.1126/science.1080070. [DOI] [PubMed] [Google Scholar]
- Thrall PH, Laine AL, Ravensdale M, Nemri A, Dodds PN, Barrett LG. Burdon JJ. Rapid genetic change underpins antagonistic coevolution in a natural host-pathogen metapopulation. Ecol. Lett. 2012;15:425–435. doi: 10.1111/j.1461-0248.2012.01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Traw MB, Chen JQ, Kreitman M. Bergelson J. Fitness costs of R-gene mediated resistance in Arabidopsis thaliana. Nature. 2003;423:74–77. doi: 10.1038/nature01588. [DOI] [PubMed] [Google Scholar]
- Tollenaere C, Susi H, Nokso-Koivisto J, Koskinen P, Tack A, Auvinen P, Paulin L, Frilander MJ, Lehtonen R. Laine A-L. SNP design from 454 sequencing of Podosphaera plantaginis transcriptome reveals a genetically diverse pathogen metapopulation with high levels of mixed-genotype infection. PLoS ONE. 2012;7:e52492. doi: 10.1371/journal.pone.0052492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale PF, Wilson AJ, Best A, Boots M. Little TJ. Epidemiological, evolutionary, and coevolutionary implications of context-dependent parasitism. Am. Nat. 2011;177:510–521. doi: 10.1086/659002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC, Rep M. Pieterse CMJ. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- van Noordwijk AJ. de Jong G. Acquisition and allocation of resources: their influence in variation in life history tactics. Am. Nat. 1986;128:137–142. [Google Scholar]
- Wolinska J. King KC. Environment can alter selection in host-parasite interactions. TRENDS Parasitol. 2009;25:236–244. doi: 10.1016/j.pt.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Yarwood CE. History and taxonomy of powdery mildews. In: Spencer MD, editor. The powdery mildews. London: Academic Press; 1978. pp. 1–32. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Occurrence of Podosphaera plantaginis in the Åland metapopulation in 2010.
Table S1. Coordinates of the pathogen populations sampled for the inoculation study, and of the allopatric host populations.
Table S2. Random estimates of the GLMMs used to analyze life-history correlations in sympatry and allopatry.