Abstract
Aim
This study aimed to evaluate the relationship between a human GRIK4 gene polymorphism (rs1954787) and responsiveness to antidepressant treatment in depressed patients.
Methods
A meta-analysis was carried out on five studies. Pooled odds ratios (ORs), 95% CIs and a χ2 test measuring heterogeneity were calculated. A test of publication bias was also conducted.
Results
Alleles and genotypes from a total of 2169 depressed patients were analyzed. The results showed that the C allele appeared more frequently than the T allele in responders to treatment (OR: 1.22; 95% CI: 1.035–1.445; z = 2.36; p = 0.018). Similarly, CC homozygotes were more likely than TT homozygotes to respond to treatment (OR: 1.45; 95% CI: 1.107–1.913; z = 2.69; p = 0.007). No evidence of publication bias was detected.
Conclusion
Subjects possessing the C allele or CC genotype of the GRIK4 polymorphism rs1954787 are more likely to respond to antidepressant treatment relative to subjects harboring the T allele and TT genotype. Additional replication of this result is required before this association can be considered definitive, after which it may become possible to employ this marker in conjunction with other known predictors in order to anticipate the outcomes of treatment with antidepressant medications.
Keywords: antidepressant, glutamate, GRIK4 polymorphism, major depressive disorder, meta-analysis, predictor, treatment response
Major depressive disorder is a common, often devastating psychiatric condition that affects 10–15% of the population annually, with an estimated 14 million people afflicted at any given time [1]. Despite some significant advances in the field of psychopharmacology, antidepressant treatment is still far from ideal. For instance, in the 7-year STAR*D trial, researchers observed a remission rate of only 28–33% and a response rate of less than 50% [2]. While a prior meta-analysis revealed no effect of antidepressants in comparison to placebo [3], closer scrutiny of this study suggested a possible effect of study design, which when accounted for, yielded a stronger result [4]. Nonetheless, there is clearly a degree of uncertainty that must be addressed in order to maximize the efficacy of antidepressant treatment, and considerable room for improvement in predicting outcomes.
Research from the past decade has accumulated a large amount of evidence implicating the glutamate system as a putative mediating or moderating factor in depression. Several neuroimaging studies have observed reduced levels of glutamate or glutamate metabolites in the frontal and cingulate cortices of the brain in patients suffering from depression [5] and increased levels in the occipital and parietal cortices [6]. Furthermore, clinical studies have shown that antidepressant treatment results in a correction of glutamate imbalance, and that glutamate receptor antagonists such as ketamine have markedly stronger antidepressant effects than traditional monoaminergic antidepressants [7]. Antidepressants may interact with glutamate systems in several ways, such as inducing long-term potentiation of glutamate receptors, interfering with presynaptic glutamate release and/or reducing stress-related glutamate levels [8].
The glutamatergic system has also been implicated in pharmacogenomics studies of depression treatment. Results from the STAR*D trial, for instance, identified an association of treatment response with polymorphisms in the GRIK4, GRIN2A and GRIK1 genes, each of which encodes proteins contributing to glutamatergic signaling. The aim of these trials was to discover genetic markers that facilitate the prediction of an individual’s response to pharmacological therapies in the hope that treatment can be tailored to the individual (i.e., personalized medicine). While marker discovery is always a significant event, replication of newly discovered genetic associations in subsequent studies has been more difficult to achieve, as exemplified by the results for GRIK4. An SNP in GRIK4 was initially found to be associated with nonresponse to antidepressant therapy in humans [9]; however, subsequent studies examining GRIK4 in this context have thus far been unable to replicate this result [10–12], save for one study [13]. It is unclear whether the initial association is a case of the winner’s curse, where a true association went unreplicated due to the low power of follow-up studies, or if it was simply a false-positive result.
Pharmacogenomic data regarding GRIK4 have so far been inconsistent, and arriving at any definitive conclusions has been inhibited by small sample sizes, making this an ideal candidate for a meta-analysis. To our knowledge, no meta-analysis of GRIK4 and its association with antidepressant response has been published to date. A more recent meta-analysis of three major genome-wide association studies (the GENDEP project, the MARS project and the STAR*D trial) found no notable associations with any genes [14]. Thus, there remains considerable uncertainty regarding the putative relationships between all genes (including GRIK4) and antidepressant treatment response.
Presently, we have meta-analyzed five independent studies of the allelic association of one GRIK4 polymorphism (rs1954787) that has been previously associated with antidepressant responsiveness. By performing meta-analyses, we aimed to determine whether the divergent evidence for an association among these studies was due to their low power to detect a weak but reliable association, etiologic heterogeneity or random error in the absence of a true effect.
Methods
Literature search
In order to identify candidate studies for meta-analysis, a search was performed in the National Library of Medicine’s PubMed online catalog and Google Scholar using the terms ‘GRIK4’ and ‘antidepressant OR treatment’ in articles dating from January 1966 to March 2014. Abstracts were read in order to select appropriate articles that examined human GRIK4 gene polymorphisms in association with antidepressant response. Articles were then read in order to ensure candidacy for inclusion in the meta-analysis. Article bibliographies were also scanned in order to identify any additional studies that may have been missed during the initial search. Finally, data were also sought from genome-wide association studies where GRIK4 was likely to have been included. Data from STAR*D have been included [9]. Data from the MARS project were not publicly available; however, the major depressive patients from the project made up the sample used in the included sample from one study [12]. Data from GENDEP were not publicly available or otherwise obtainable. This process resulted in 17 articles that were potentially eligible for inclusion in the meta-analysis. Five articles were removed because they studied something other than pharmacological response in depression. Three articles were reviews and did not present original data. One article studied animal models. Three articles were reviews of the STAR*D data set and presented data that were redundant with an article that was already in use in this analysis. In total, 12 articles were removed from the search pool.
Inclusion criteria
Studies must have met all of the following criteria in order to be included in the analysis: published in a peer-reviewed journal; present original data; and publish enough data for an effect size to be calculated. If data were not present or in a usable format, requests for additional information were submitted to the original authors. Requests were made and additional data were supplied by the original authors for three of the included studies [9,10,12]. The data received were genotype and allele totals divided by responders versus non-responders. All studies genotyped patients with major depressive disorder as a primary diagnosis or those who met the criteria for a major depressive episode at the time of inclusion. One study included patients who were diagnosed with bipolar disorder in its sample; however, this subsample made up less than 10% of the sample population. The remaining 90% of the sample was diagnosed with major depressive disorder [12]. After application of inclusion criteria, five studies remained [9–13].
Coding of study characteristics
In order to delineate the potential moderating influences of various sample characteristics on the size of the effects obtained in the case–control studies under consideration, each study was coded on the following variables: the ancestry of the sample; the mean age of the control group; and a gender ratio (calculated as female cases/male cases). Responders were defined using the original authors’ criteria. Original response definitions and descriptive characteristics of the studies are presented in Table 1.
Table 1.
Descriptive characteristics of studies of the association between GRIK4 and antidepressant response in depressed patients.
| Author (year) | Cases† (n) | Ancestry | Mean age of control group | Gender ratio‡ | Response | Antidepressant treatment | Ref. |
|---|---|---|---|---|---|---|---|
| Paddock et al. (2007) | 1094 | European | 40.8 | 1.61 | 50% reduction in base QIDS score | SSRI | [9] |
| Perlis et al. (2010) | 296 | European | 44.2 | 1.72 | 50% reduction in base HAM-D score | SNRI | [10] |
| Horstmann et al. (2010) | 300 | European | 49.5 | 1.35 | 50% reduction in base HAM-D score | Naturalistic§ | [12] |
| Serretti et al. (2012) | 200 | European | 50.1 | 2.70 | HAM-D score <17 after 4 weeks of treatment | Naturalistic§ | [11] |
| Pu et al. (2013) | 279 | Asian | 38.2 | 1.42 | 50% reduction in base HAM-D score | SSRI or SNRI | [13] |
For which GRIK4 was genotyped.
Gender ratio = females cases:male cases.
Individual treatment determined by doctors. All classes of antidepressants were allowed.
HAM-D: Hamilton Rating Scale for Depression; QIDS: Quick Inventory of Depressive Symptomology; SNRI: Serotonin–norepinephrine reuptake inhibitor; SSRI: Selective serotonin reuptake inhibitor.
Statistics
Data from each study were first divided into a two-by-five table by treatment response (response vs non-response) and genotype or allele (CC, CT, TT, total C and total T). The strength of the association for each study was summarized using the odds ratio (OR) statistic, where an OR >1.0 indicated a positive association with treatment response. The prior literature had implicated the C allele as increasing the odds of anti-depressant response [9], and as such is identified as the ‘risk’ allele in this meta-analysis. Thus, in our analyses, an OR >1.0 represents an increased probability of observing the C allele or CC genotype in responders.
Studies were meta-analyzed using a random-effects model according to the methods described by DerSimonian and Laird [15], with a 95% CI calculated as described by Woolf [16]. Heterogeneity of ORs was measured using a χ2 test of goodness of fit, and the significance of pooled ORs was assessed using a z-test. The influence of individual studies on the pooled OR was determined by sequentially removing each study and recalculating the pooled OR and 95% CI.
Publication bias within the group of ORs was assessed by the method in which the standard normal deviate (SND) of the OR (z score) is regressed on the precision of the OR (POR; the inverse of the standard error of the OR) [17]. Since POR increases with sample size, the regression of z on POR should run through the origin in the absence of bias (i.e., small samples with low precision have large standard errors and small SNDs, whereas large samples with high precision have small standard errors and large SNDs). The slope of the regression line indicates the size and direction of the association and, in the presence of bias, the intercept of the regression will be significantly different from zero, as determined by the t-test. The type I error rate was set at 0.05. All statistical analyses were conducted using Stata 11.0 (Stata Corp., TX, USA).
Results
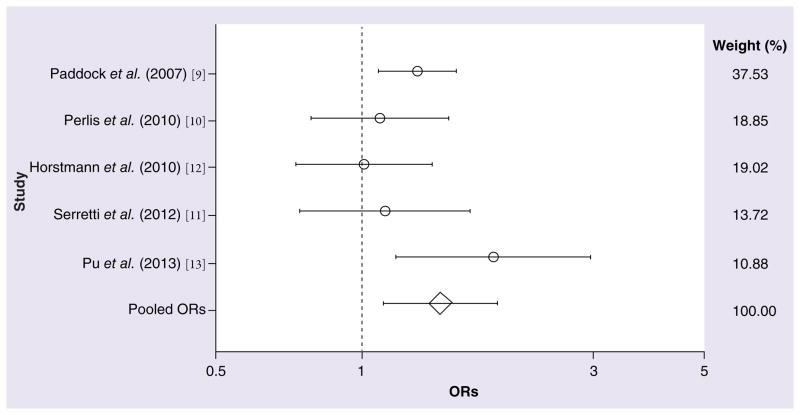
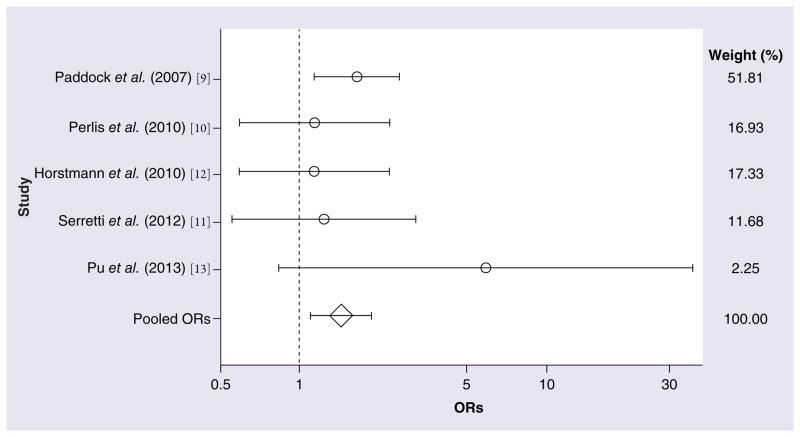
In the collective sample of 1361 responders and 808 nonresponders to antidepressant treatment (Table 1), a significant association with treatment response was observed for the C allele of rs1954787 (OR: 1.22; 95% CI: 1.035–1.445; z = 2.36; p = 0.018; Figure 1). The degree of measured heterogeneity was not significant (χ2 = 5.63; p = 0.229). The ORs and 95% CIs from each study and from the pooled analysis of all five studies are shown in Table 2. The power of each study for detecting the original effect was calculated and is also shown in Table 2 (all values <33.31%). As shown in Table 3, a significant difference in the likelihood of treatment response was also observed between CC and TT homozygotes (OR: 1.45; 95% CI: 1.107–1.913; z = 2.69; p = 0.007; Figure 2). No significant amount of heterogeneity was detected in this analysis (χ2 = 3.63; p = 0.459). For both the C allele and CC homozygosity, the direction of effect across all five studies was consistent, although the studies differed widely in their estimates of the magnitude of the effects. A model of CC homozygotes versus T carriers (TT and TC genotypes) reached marginal significance (z = 1.85; p = 0.064), but this contrast was also marked by marginally significant heterogeneity (χ2 = 8.21; p = 0.084). No significant differences in treatment response were observed between CC homozygotes and CT heterozygotes (z = 1.44; p = 0.149) or between CT heterozygotes and TT homozygotes (z = 0.82; p = 0.414).
Figure 1. Meta-analysis of five studies interrogating the C allele of the GRIK4 SNP rs1954787 in association with antidepressant responsiveness.
OR: Odds ratio.
Table 2.
Meta-analysis of case–control studies of the allelic association between the GRIK4 polymorphism rs1954787(C/T) and antidepressant response in depressed patients.
| Author (year) | Responder type | C | T | OR† | 95% CI | Power (%)‡ | Ref. |
|---|---|---|---|---|---|---|---|
| Paddock et al. (2007) | Responders Nonresponders |
874 322 |
670 322 |
1.30 | 1.09–1.57 | – | [9] |
| Perlis et al. (2010) | Responders Nonresponders |
193 132 |
153 114 |
1.09 | 0.79–1.51 | 32.53 | [10] |
| Horstmann et al. (2010) | Responders Nonresponders |
151 194 |
111 144 |
1.01 | 0.73–1.40 | 33.31 | [12] |
| Serretti et al. (2012) | Responders Nonresponders |
95 133 |
67 105 |
1.12 | 0.75–1.68 | 22.21 | [11] |
| Pu et al. (2013) | Responders Nonresponders |
349 114 |
59 36 |
1.87 | 1.18–2.98 | 25.37 | [13] |
| Pooled | Responders Nonresponders |
1662 895 |
1060 721 |
1.22 | 1.03–1.45 | – |
OR of the test of allelic effect.
For detecting the original effect observed by Paddock et al. [9].
OR: Odds ratio.
Table 3.
Meta-analysis of case–control studies of the genotypic association between the GRIK4 polymorphism rs1954787(C/T) and antidepressant response in depressed patients.
| Author (year) | Responder type | CC | TC | TT | OR† | 95% CI | Power (%)‡ | Ref. |
|---|---|---|---|---|---|---|---|---|
| Paddock et al. (2007) | Responders Nonresponders |
253 74 |
368 174 |
151 74 |
1.68 | 1.15–2.45 | – | [9] |
| Perlis et al. (2010) | Responders Nonresponders |
56 34 |
81 64 |
36 25 |
1.14 | 0.59–2.22 | 27.63 | [10] |
| Horstmann et al. (2010) | Responders Nonresponders |
42 62 |
67 70 |
22 37 |
1.14 | 0.59–2.20 | 29.49 | [12] |
| Serretti et al. (2012) | Responders Nonresponders |
29 39 |
37 55 |
15 25 |
1.24 | 0.56–2.76 | 19.41 | [11] |
| Pu et al. (2013) | Responders Nonresponders |
147 42 |
55 30 |
2 3 |
5.25 | 0.85–32.46 | 26.97 | [13] |
| Pooled | Responders Nonresponders |
527 251 |
608 393 |
226 164 |
1.46 | 1.11–1.91 | – |
OR of the test of homozygous genotypes (CC/TT).
For detecting the original effect observed by Paddock et al. [9].
OR: Odds ratio.
Figure 2. Meta-analysis of five studies interrogating the CC genotype versus the TT genotype of the GRIK4 SNP rs1954787 in association with antidepressant responsiveness.
OR: Odds ratio.
A test of influence showed that removing the original study [9] from the meta-analysis caused the pooled effect of the C allele among the remaining four studies to decrease in size (OR: 1.19; 95% CI: 0.93–1.50) and to lose significance (z = 1.40; p = 0.162). However, the magnitude of the effect decreased only minimally (original pooled OR: 1.22). Because of this, it is likely that the loss of significance could be attributed to a loss of sample size and statistical power for detecting an effect, rather than a loss of actual effect. Removing the same study also caused the pooled effect of the CC genotype against the TT genotype to decrease (OR: 1.25; 95% CI: 0.84–1.86) and to lose significance (z = 1.11; p = 0.27). No evidence of publication bias was observed in any of the meta-analyses (all p-values >0.293). Although our power to detect publication bias was small, studies aimed at replicating the original effect reported no significant result, save for one. We therefore believe that it is unlikely that publication bias played a role in publishing the included studies, given that this phenomenon would increase the rate at which studies with significant results are published.
Discussion
In this study, we aggregated and meta-analyzed the data from five studies examining the association between the GRIK4 SNP rs1954787 and antidepressant treatment response. Through meta-analysis, we are better able to detect any small but significant effects that might otherwise be masked by heterogeneity, and we are able to determine whether the general appearance of an absence of effect was due to the low power of individual studies for detecting these small effects. Results of the analysis showed that the C allele occurred more frequently among depressed patients who responded to antidepressant treatment than those who did not. Similarly, we detected a significant enrichment of CC homozygotes relative to TT homozygotes among those who responded to treatment. These results support the initial findings of STAR*D [9], which first revealed the candidacy of GRIK4 as a genetic factor in antidepressant response. In that genome-wide study, the C allele of rs1954787 was identified as the allele that was associated with improving the odds of treatment response; however, no such relationship could be observed (with statistical significance) in subsequent targeted replication efforts [10–12], save for one study [13]. The fact that we were able to ‘rediscover’ this association by pooling data suggests that the inability to consistently observe an effect may have been attributable to the relatively small sample sizes and subsequently lower power for detecting an effect in these individual efforts. This is further supported by the fact that when the largest study was removed from the analysis, no significant effects were preserved.
In addition to the allelic effect, we also observed an association of the CC genotype with treatment response, especially compared with TT homozygotes. Comparisons of each homozygote group with the heterozygote CT group were not significant, so it is premature to speculate on the genetic mechanism (additive, dominant, recessive or otherwise) by which the C allele might affect this phenotype. It is important to point out that this meta-analysis, despite being a more powerful sample than any prior individual study, could benefit from the addition of further studies and samples in order to gain power for resolving this mechanism more clearly.
Additional limitations of our analysis must also be considered. Given the nature of meta-analysis, we were unable to control for any weaknesses or design flaws that might have been embedded within any individual study; therefore, our analyses are only as strong as the data we have pooled. We were unable to detect any quality issues with any of the included studies; however, the fact remains that errors in any included study would also be represented in the meta-analysis. Furthermore, there are several variables that could not be controlled for across the studies and therefore are not consistent. For instance, treatment varied across studies by drug, dosage and trial length. The original STAR*D study treated patients exclusively with citalopram, a selective serotonin reuptake inhibitor, while one study solely utilized duloxetine, a serotonin–norepinephrine reuptake inhibitor [10]. While both types of drugs are utilized as antidepressants, they have different mechanisms of action. The remaining three studies utilized a ‘naturalistic’ approach in which the antidepressant type was determined for each individual subject. Furthermore, some studies allowed concomitant drug treatment, such as anxiolytics, mood stabilizers and (in one study) antipsychotics [12]. Finally, one study [11] defined treatment response in a different manner from the remaining four studies. Our ability to detect consistent and significant associations of treatment response with the C allele and CC homozygosity by meta-analysis is perhaps more impressive in light of these differences, which may also indicate the robustness of these effects.
While most studies included here excluded subjects with psychotic depression or any form of psychosis, one study did not [12]. This observation may be the root of the marginal heterogeneity (χ2 = 8.21; p = 0.084) observed in the analysis of CC homozygotes versus T carriers, which is supported by the fact that when the one study [12] was removed from the pool, the heterogeneity decreased considerably (χ2 = 2.70; p = 0.439). In addition, GRIK4 and other glutamatergic genes have previously been associated with the efficacy of haloperidol, an anti-psychotic [18]. Furthermore, the SNP of interest in this analysis (rs1954787) is part of a haplotype that has been associated with schizophrenia [19]. It is also worth noting, however, that due to our small sample size, our power to detect heterogeneity was low. It is therefore possible that some of our negative analyses of heterogeneity may represent a type II error.
In summary, the results of our analyses support variation in GRIK4 as a factor in determining anti-depressant response; however, more work is needed in order to clarify the genetic mechanism of action. There is an increasing amount of evidence in support of the glutamatergic system’s involvement in depression and antidepressant response, as seen most notably in the clinical use of ketamine [7]. This has focused considerable research on this system as a source of potential targets for depression treatment. Our work adds to this knowledge base, but also raises additional questions. For example, does GRIK4 variation directly and independently mediate antidepressant mechanisms or does it cooperate with variations in glutamatergic genes, such as GRIN2A, or monoamine genes, such as HTR2C? Why is GRIK4 in particular associated with antidepressant response, while other glutamatergic genes appear not to be related to the phenotype? Is the SNP implicated here functional or is it merely a marker for a nearby functional variant? Does this effect persist in ancestral groups other than those analyzed so far? Finally, will this effect continue to be supported with the addition of other sources of data? As research in this particular area continues, these are important concerns that should be addressed.
Conclusion
The results from this meta-analysis suggest that the initial findings of STAR*D [9] may not have been a type I error; rather, GRIK4 does indeed appear to be associated with antidepressant response in depressed patients. Specifically, the C allele appears more frequently than the T allele in patients who respond to antidepressant treatment, and CC homozygotes have greater odds of also responding to treatment when compared with TT homozygotes. Upon further replication, this observation may prove beneficial in the formation of future personalized antidepressant treatments.
Future perspective
Additional analyses – and cumulative meta-analyses – will allow us to build a more concrete picture of GRIK4’s role in this domain, including the genetic mechanism and potential moderators of its effect. To our knowledge, only five published studies directly examined the human GRIK4 gene in conjunction with antidepressant response, with three of which being unable to detect any significant association on their own. The results presented in this analysis suggest that GRIK4 may indeed be a genetic contributor to antidepressant response; however, additional studies need to be conducted on the topic that must include large samples of various ancestries. In the future, it is likely that pharmacogenomics will be a critical, even central, factor in the advancement of antidepressant treatment. Given the nature of the results presented here and of the recent literature surrounding the glutamatergic system in general, it would seem that GRIK4 and other glutamate-related genes may provide a target pathway for novel approaches towards the treatment of depression and other mood disorders.
Supplementary Material
Executive summary.
Aim
A GRIK4 polymorphism has been identified as a potential factor in antidepressant responsiveness. Since its discovery, several studies have sought to replicate this finding.
Results from these studies have been mixed; however, this may have been due to low statistical power or significant heterogeneity. This study aims to resolve these issues and explore whether the GRIK4 polymorphism rs1954787 is a factor in determining response to antidepressant treatment.
Methods
A literature search was conducted for all studies interrogating the GRIK4 SNP rs1954787 and its association with antidepressant responsiveness in depressed patients through PubMed and Google Scholar up until March 2014.
A meta-analysis was performed using Stata 11.0 (Stata Corp., TX, USA). Pooled odds ratios (ORs), 95% CIs and χ2 tests were calculated using a random effects model.
Results
Five studies were analyzed, totaling 2169 subjects (1361 responders and 808 nonresponders).
A significant association was observed for the C allele versus the T allele (OR: 1.22; 95% CI: 1.035–1.445; z = 2.36; p = 0.018).
A significant association was observed for the CC genotype versus the TT genotype (OR: 1.45; 95% CI: 1.107–1.913; z = 2.69; p = 0.007).
A marginal association was observed between CC genotype and T carriers (CT and TT genotypes; OR: 1.329; 95% CI: 0.983–1.797; z = 1.85; p = 0.064), but was also marked by marginal heterogeneity (χ2 = 8.21; p = 0.084).
No significant differences were found between CT genotype and TT genotype (OR: 1.108; 95% CI: 0.866–1.419; z = 0.82; p = 0.414) nor between the CC genotype and CT genotype (OR: 1.280; 95% CI: 0.915–1.791; z = 1.44; p = 0.149).
No evidence of publication bias was observed (all p-values >0.293).
Conclusion
The GRIK4 polymorphism rs1954787 may be a genetic factor in determining antidepressant responsiveness.
Individuals with the C allele and CC genotype are more likely to respond to antidepressant treatment than those with the T allele and TT genotype, respectively.
Acknowledgments
The authors would like to express their gratitude to R Perlis of the Massachusetts General Hospital, J Wood of Lilly LLC, S Paddock of Rose Li & Associates, F McMahon and A Sorant of the US NIH and E Binder of the Max-Planck Institute of Psychiatry for aiding our efforts and providing the additional data required to carry out this analysis. The authors would also like to express their gratitude to all members of the PsychGENe laboratory who contributed their time and energy to the creation of this study.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Financial & competing interests disclosure
This work was supported in part by grant R01MH085521 (SJ Glatt) from the US NIH; a Young Investigator Award, an Independent Investigator Award and the Sidney R Baer, Jr Prize for Schizophrenia Research (SJ Glatt) from NARSAD: The Brain and Behavior Research Fund; and a Research Grant from The Gerber Foundation (SJ Glatt). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest;
•• of considerable interest
- 1.Al-Harbi KS. Treatment-resistant depression: therapeutic trends, challenges, and future directions. Patient Prefer Adherence. 2012;6:369–388. doi: 10.2147/PPA.S29716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trivedi MH, Rush AJ, Wisniewski SR, et al. Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: implications for clinical practice. Am J Psychiatry. 2006;163(1):28–40. doi: 10.1176/appi.ajp.163.1.28. [DOI] [PubMed] [Google Scholar]
- 3.Kirsch I, Deacon BJ, Huedo-Medina TB, Scoboria A, Moore TJ, Johnson BT. Initial severity and antidepressant benefits: a meta-analysis of data submitted to the Food and Drug Administration. PLoS Med. 2008;5(2):e45. doi: 10.1371/journal.pmed.0050045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vohringer PA, Ghaemi SN. Solving the antidepressant efficacy question: effect sizes in major depressive disorder. Clin Ther. 2011;33(12):B49–B61. doi: 10.1016/j.clinthera.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hasler G, Van Der Veen JW, Tumonis T, Meyers N, Shen J, Drevets WC. Reduced prefrontal glutamate/glutamine and gamma-aminobutyric acid levels in major depression determined using proton magnetic resonance spectroscopy. Arch Gen Psychiatry. 2007;64(2):193–200. doi: 10.1001/archpsyc.64.2.193. [DOI] [PubMed] [Google Scholar]
- 6.Sanacora G, Gueorguieva R, Epperson CN, et al. Subtype-specific alterations of gamma-aminobutyric acid and glutamate in patients with major depression. Arch Gen Psychiatry. 2004;61(7):705–713. doi: 10.1001/archpsyc.61.7.705. [DOI] [PubMed] [Google Scholar]
- 7.Niciu MJ, Henter ID, Luckenbaugh DA, Zarate CA, Jr, Charney DS. Glutamate receptor antagonists as fast-acting therapeutic alternatives for the treatment of depression: ketamine and other compounds. Ann Rev Pharmacol Toxicol. 2014;54:119–139. doi: 10.1146/annurev-pharmtox-011613-135950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8•.Sanacora G, Treccani G, Popoli M. Towards a glutamate hypothesis of depression: an emerging frontier of neuropsychopharmacology for mood disorders. Neuropharmacology. 2012;62(1):63–77. doi: 10.1016/j.neuropharm.2011.07.036. Review summarizing the shift from the monoamine hypothesis of major depressive disorder to the glutamate neuroplasticity model. Suggests that glutamate may be the final pathway for antidepressant action. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9••.Paddock S, Laje G, Charney D, et al. Association of GRIK4 with outcome of antidepressant treatment in the STAR*D cohort. Am J Psychiatry. 2007;164(8):1181–1188. doi: 10.1176/appi.ajp.2007.06111790. Initial discovery of the GRIK4 SNP rs1954787 association with antidepressant response. Shows that the C allele may be a predictor of treatment response, which is in agreement with our results. [DOI] [PubMed] [Google Scholar]
- 10••.Perlis RH, Fijal B, Dharia S, Heinloth AN, Houston JP. Failure to replicate genetic associations with antidepressant treatment response in duloxetine-treated patients. Biol Psychiatry. 2010;67(11):1110–1113. doi: 10.1016/j.biopsych.2009.12.010. Study showing no association between the GRIK4 SNP rs1954787 and antidepressant response, which is in contrast to our results. [DOI] [PubMed] [Google Scholar]
- 11••.Serretti A, Chiesa A, Crisafulli C, et al. Failure to replicate influence of GRIK4 and GNB3 polymorphisms on treatment outcome in major depression. Neuropsychobiology. 2012;65(2):70–75. doi: 10.1159/000329553. Study finding no association between the GRIK4 SNP rs1954787 and antidepressant response, which is in contrast to our results. [DOI] [PubMed] [Google Scholar]
- 12••.Horstmann S, Lucae S, Menke A, et al. Polymorphisms in GRIK4, HTR2A, and FKBP5 show interactive effects in predicting remission to antidepressant treatment. Neuropsychopharmacology. 2010;35(3):727–740. doi: 10.1038/npp.2009.180. Study showing no association between the GRIK4 SNP rs1954787 and antidepressant response, which is in contrast to our results. This study used in-patient samples for analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Pu M, Zhang Z, Xu Z, et al. Influence of genetic polymorphisms in the glutamatergic and GABAergic systems and their interactions with environmental stressors on antidepressant response. Pharmacogenomics. 2013;14(3):277–288. doi: 10.2217/pgs.13.1. This study found that the C allele of the GRIK4 SNP rs1954787 is a predictor of good treatment response, which is in agreement with our results. [DOI] [PubMed] [Google Scholar]
- 14•.GENDEP Investigators, MARS Investigators, STAR*D Investigators. Common genetic variation and antidepressant efficacy in major depressive disorder: a meta-analysis of three genome-wide pharmacogenetic studies. Am J Psychiatry. 2013;170(2):207–217. doi: 10.1176/appi.ajp.2012.12020237. Meta-analysis of three large genome-wide association studies examining antidepressant response. No significant associations were observed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 16.Woolf B. On estimating the relation between blood group and disease. Ann Hum Genet. 1955;19:251–253. doi: 10.1111/j.1469-1809.1955.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 17.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drago A, Giegling I, Schafer M, et al. AKAP13, CACNA1, GRIK4 and GRIA1 genetic variations may be associated with haloperidol efficacy during acute treatment. Eur Neuropsychopharmacol. 2013;23(8):887–894. doi: 10.1016/j.euroneuro.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 19.Pickard BS, Malloy MP, Christoforou A, et al. Cytogenetic and genetic evidence supports a role for the kainate-type glutamate receptor gene, GRIK4, in schizophrenia and bipolar disorder. Mol Psychiatry. 2006;11(9):847–857. doi: 10.1038/sj.mp.4001867. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.