Abstract
Nuclear factor-κB (NF-κB) is constitutively activated in diverse human malignancies by mechanisms that are not understood1,2. The MUC1 oncoprotein is aberrantly overexpressed by most human carcinomas and, similarly to NF-κB, blocks apoptosis and induces transformation3–6. This study demonstrates that overexpression of MUC1 in human carcinoma cells is associated with constitutive activation of NF-κB p65. We show that MUC1 interacts with the high-molecular-weight IκB kinase (IKK) complex in vivo and that the MUC1 cytoplasmic domain binds directly to IKKβ and IKKγ. Interaction of MUC1 with both IKKβ and IKKγ is necessary for IKKβ activation, resulting in phosphorylation and degradation of IκBα. Studies in non-malignant epithelial cells show that MUC1 is recruited to the TNF-R1 complex and interacts with IKKβ–IKKγ in response to TNFα stimulation. TNFα-induced recruitment of MUC1 is dependent on TRADD and TRAF2, but not the death-domain kinase RIP1. In addition, MUC1-mediated activation of IKKβ is dependent on TAK1 and TAB2. These findings indicate that MUC1 is important for physiological activation of IKKβ and that overexpression of MUC1, as found in human cancers, confers sustained induction of the IKKβ–NF-κB p65 pathway.
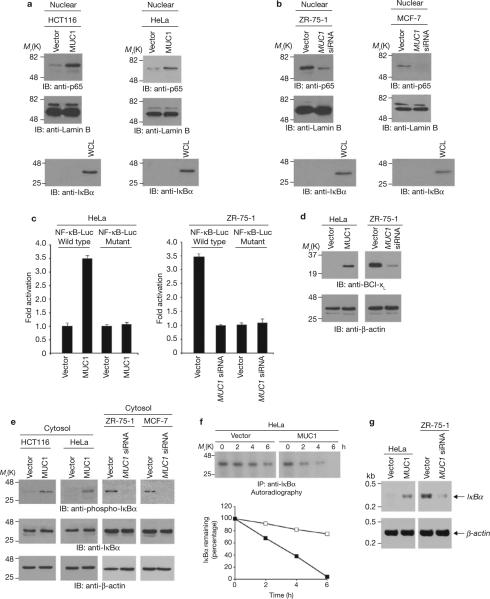
Nuclear localization of NF-κB p65 was studied in HCT116 colon cancer and HeLa cervical cancer cells that stably express either an empty vector or MUC1 (ref. 4, also see Supplementary Information, Fig. S1a). Levels of nuclear NF-κB p65 were lower in vector cells than in cells expressing MUC1 (Fig. 1a). Human ZR-75-1 and MCF-7 breast cancer cells that express endogenous MUC1 were stably transfected to express either an empty vector or a MUC1 siRNA4 (Supplementary Information, Fig. S1a). Silencing of MUC1 in ZR-75-1 (ref. 4) and MCF-7 cells7 decreased nuclear NF-κB p65 (Fig. 1b). MUC1 expression was also associated with a decrease in cytosolic NF-κB p65 levels in HeLa and ZR-75-1 cells (Supplementary Information, Fig. S1b). To determine whether MUC1 is associated with activation of the NF-κB p65 transcription function, HeLa and ZR-75-1 cells were transfected with a construct containing a NF-κB-binding site upstream of the luciferase reporter (pNF-κB-Luc). MUC1 expression was associated with activation of pNF-κB-Luc (Fig. 1c). In contrast, MUC1 had no effect on activation of a pNF-κB-Luc construct that was mutated at the NF-κB p65 binding site (Fig. 1c). In addition, expression of BclxL, a gene activated by NF-κB, was higher in cells expressing MUC1 (Fig. 1d). To determine whether MUC1 affects IκBα phosphorylation (as phosphorylated IκBα is targeted for ubiquitination and proteosomal degradation) cytosolic lysates were immunoblotted with an anti-phospho-IκBα antibody. Indeed, phospho-IκBα levels were significantly higher in cells expressing MUC1 (Fig. 1e). Assessment of IκBα stability indicated that MUC1 expression increases degradation of IκBα (Fig. 1f). The half-lives of IκBα in the absence and presence of MUC1, were 6.7 ± 0.5 h and 3.8 ± 0.3 h (mean ± s.d., n = 3), respectively. Similar results were obtained in ZR-75-1 cells (data not shown), indicating that MUC1-induced increases in phosphorylation of IκBα are associated with increases in IκBα degradation. Targeting of NF-κB p65 to the nucleus activates IκBα gene transcription in an inducible, autoregulatory pathway that replenishes IκBα levels1,2. Consistent with this autoregulatory loop, RT-PCR analysis demonstrated that MUC1-induced increases in nuclear NF-κB p65 are associated with upregulation of IκBα mRNA levels (Fig. 1g). These findings indicate that MUC1 contributes to IκBα degradation, resulting in activation of NF-κB p65.
Figure 1.
MUC1 targets NF-κB p65 to the nucleus by inducing phosphorylation and degradation of IκBα. (a) and (b) Nuclear lysates from the indicated cells were subjected to immunoblotting with anti-p65, anti-lamin B and anti-IκBα antibodies. Whole cell lysate (WCL) prepared from HCT116-vector cells was used as a control for anti-IκBα reactivity. Immunoblot analysis of the nuclear lysates with antibodies against nuclear lamin B and cytosolic IκBα confirmed equal loading of the lanes and lack of cytoplasmic contamination. (c) The indicated cells were transfected with a pNF-κB-Luc reporter plasmid or a mutant at the NF-κB binding site and, as a control, the SV40-Renilla-Luc plasmid. Luciferase activity was measured at 48 h after transfection. The results are expressed as the fold activation (mean ± s.d., of three separate experiments) compared with that obtained in HeLa-vector (left) or ZR-75-1-MUC1 siRNA (right) cells (each assigned a value of 1). (d) Whole cell lysates from the indicated cells were immunoblotted with anti-Bcl-xL and anti-β-actin antibodies. (e) Cytosolic lysates from the indicated cells were immunoblotted with anti-phospho-IκBα, anti-IκBα and anti-β-actin antibodies. (f) HeLa-vector and HeLa-MUC1 cells were pulsed with 35S-methionine and chased for the indicated times. Anti-IκBα immunoprecipitates from equal amounts of lysate were subjected to SDS–PAGE and autoradiography (upper panels). Intensity of the IκBα signals was determined by scanning densitometry and is expressed as the percentage IκBα remaining compared with that obtained at 0 h (lower panels). Similar results were obtained in two separate experiments. (g) IκBα and β-actin mRNA levels were determined for the indicated cells by quantitative RT-PCR. Full scans of the gels in a, b, e and f are shown in Supplementary Fig. S6-1.
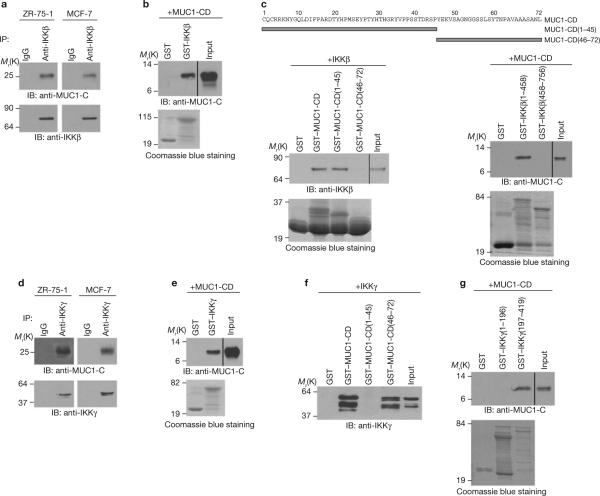
The presence of IKKβ in a complex with IKKγ is necessary and sufficient for phosphorylation of IκBα in the classical NF-κB pathway. IKKγ binds directly to IKKβ and is required for IKKβ activation. Analysis of anti-IKKβ immunoprecipitates from ZR-75-1 and MCF-7 cells showed that MUC1 carboxy-terminal subunit (MUC1-C) associates with IKKβ (Fig. 2a). In vitro studies with purified GST–IKKβ and the MUC1 cytoplasmic domain (MUC1-CD) demonstrated that these proteins interact directly with each other (Fig. 2b). This interaction was confirmed in experiments with purified GST–MUC1-CD and IKKβ (Fig. 2c, lower left). Studies with MUC1-CD amino acid fragments 1–45 and 46–72 demonstrated that MUC1-CD(1–45) confers binding to IKKβ (Fig. 2c, left). Studies with IKKβ(1–458) and IKKβ(458–756) further demonstrated that MUC1-CD binds directly to the IKKβ-amino-terminal region (Fig. 2c, lower right). The IKKβ-carboxy-terminal region associates with the N-terminal region of IKKγ. Consistent with the formation of IKKβ–IKKγ complexes and binding of MUC1 to IKKβ, we found that MUC1-C co-precipitates with IKKγ (Fig. 2d). In vitro studies with GST–IKKγ demonstrated that MUC1-CD binds to purified IKKγ (Fig. 2e). In contrast to the interaction with IKKβ, MUC1-CD(46–72) but not MUC1-CD(1–45) binds to IKKγ (Fig. 2f) and at the C-terminal region of IKKγ (Fig. 2g). To further assess binding of MUC1-C to IKKβ and IKKγ in vivo, MUC1-C was immunodepleted from HeLa-MUC1 cell lysates by precipitation with increasing amounts of an anti-MUC1-C antibody (Supplementary Information, Fig. S1c, left). As a control, the lysates were incubated with a non-immune IgG (Supplementary Information, Fig. S1c, right). Immunoblot analysis of the lysates demonstrated that depletion of MUC1-C is associated with decreases in IKKβ and IKKγ (Supplementary Fig. S1c). These findings indicate that MUC1 binds directly to IKKβ and IKKγ, and potentially to both proteins in IKKβ–IKKγ complexes (Supplementary Information, Fig. S1d).
Figure 2.
MUC1-CD binds directly to IKKβ and IKKγ. (a) Lysates from the indicated cells were subjected to immunoprecipitation with a control IgG or anti-IKKβ antibody. The precipitates were immunoblotted with the indicated antibodies. (b) GST and GST–IKKβ bound to glutathione–agarose beads were incubated with purified MUC1-CD. The precipitates and input were immunoblotted with an anti-MUC1-C antibody. Input of the GST and GST–IKKβ proteins was assessed by Coomassie blue staining. (c) Amino-acid sequence of MUC1-CD (upper panel). GST and the indicated GST–MUC1-CD fusion proteins bound to glutathione beads were incubated with purified IKKβ. The precipitates and input were immunoblotted with an anti-IKKβ antibody (lower left). GST and the indicated GST–IKKβ fusion proteins bound to glutathione beads were incubated with MUC1-CD. The precipitates and input were immunoblotted with an anti-MUC1-C antibody (lower right). Input of GST and GST– fusion proteins was assessed by Coomassie blue staining. (d) Lysates from the indicated cells were subjected to immunoprecipitation with a control IgG or an anti-IKKγ antibody. The precipitates were immunoblotted with the indicated antibodies. (e) GST and GST–IKKγ bound to glutathione beads were incubated with purified MUC1-CD. The precipitates and input were immunoblotted with an anti-MUC1-C antibody. (f) GST and the indicated GST–MUC1-CD fusion proteins bound to glutathione beads were incubated with purified IKKγ. The precipitates and input were immunoblotted with an anti-IKKγ antibody. Input of the GST and GST– fusion proteins is shown in Fig. 2c, left. (g) GST and the indicated GST–IKKγ fusion proteins bound to glutathione beads were incubated with MUC1-CD. The precipitates and input were immunoblotted with an anti-MUC1-C antibody. Full scans of the gels in a, b, c, e and f and g are shown in Supplementary Fig. S6-2.
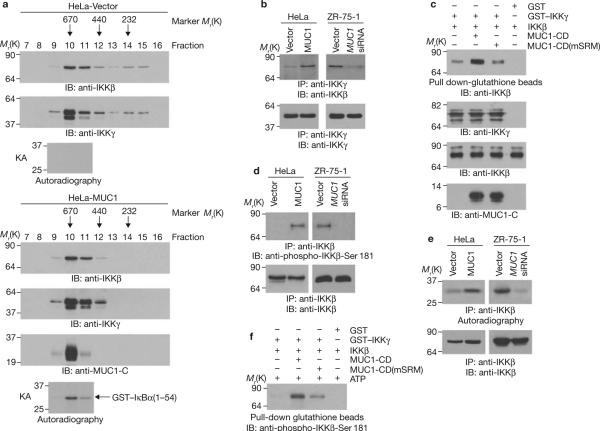
To determine whether MUC1-C associates with the IKK complex, lysates from HeLa-vector and HeLa-MUC1 cells were subjected to gel filtration chromatography followed by immunoblotting of the fractions with anti-IKKβ and anti-IKKγ antibodies. Consistent with previous findings8, analysis of HeLa-vector cells showed that IKKβ and IKKγ are detectable in a prominent pool with a relative molecular mass (Mr) of 700,000 (fractions 10–12) and also in a pool of ~300 K (fractions 14 and 15; Fig. 3a). Analysis of HeLa-MUC1 cells showed that IKKβ and IKKγ are detectable in fractions 10–12 but not in fractions 14 and 15 (Fig. 3a). MUC1-C was detected mainly in fraction 10, consistent with binding to the large ~700-K complex (Fig. 3a). The ~700-K IKKβ–IKKγ complexes isolated from HeLa-MUC1, but not HeLa-vector cells, exhibited constitutive activation (Fig. 3a). The HeLa-MUC1 lysates were also immuno-precipitated with an anti-MUC1-C antibody and the precipitates were released by adding MUC1-C peptide before gel filtration chromatography. Immunoblot analysis of the fractions confirmed that MUC1-C associates with the large IKKβ–IKKγ complexes (Supplementary Information, Fig. S2a). MUC1 expression was associated with increased binding of IKKβ to IKKγ (Fig. 3b). Moreover, incubation of purified IKKβ with IKKγ in vitro demonstrated that MUC1-CD increases the interaction between IKKβ and IKKγ (Fig. 3c). MUC1-CD(1–45) binds to IKKβ and MUC1-CD(46–72), which contains a serine-rich SAGNGGSSLS motif (SRM; amino acids 50–59), binds to IKKγ (Fig. 2). Mutation of the SRM to AAGNGGAAAA (mSRM) had no effect on the interaction between MUC1-CD and IKKβ (Supplementary Information, Fig. S2b), but attenuated binding to IKKγ (Supplementary Information, Fig. S2c). MUC1-CD(mSRM) was substantially less effective than MUC1-CD in inducing the association of IKKβ and IKKγ (Fig. 3c), indicating that this response is dependent on binding of both IKKs to MUC1-CD. Phosphorylation of IKKβ on Ser 181 in the activation loop, perhaps by a trans-autophosphorylation mechanism, is required for induction of IKKβ activity. Immunoblot analysis with an anti-phospho-IKKβ-Ser 181 antibody showed that IKKβ is phosphorylated on Ser 181 by a MUC1-dependent mechanism (Fig. 3d). Analysis of anti-IKKβ precipitates for phosphorylation of IκBα further demonstrated that MUC1 stimulates the IKKβ kinase function (Fig. 3e). Incubation of IKKβ and IKKγ with ATP in vitro resulted in phosphorylation of IKKβ on Ser 181 (Fig. 3f). The extent of IKKβ phosphorylation was increased significantly by adding MUC1-CD to the reaction (Fig. 3f). However, this effect of MUC1-CD was attenuated by mutation of the SRM (Fig. 3f).
Figure 3.
MUC1 activates the IKKβ–IKKγ complex. (a) HeLa-vector and HeLa-MUC1 cell lysates were separated on a Sephacryl S-200 HR column. The indicated fractions were analysed by immunoblotting with the indicated antibodies and for phosphorylation of GST–IκBα in kinase assays (KAs). (b) Anti-IKKγ immunoprecipitates from the indicated cells were immunoblotted with anti-IKKβ and anti-IKKγ antibodies. (c) GST or GST–IKKγ bound to glutathione beads was incubated with IKKβ in the absence and presence of MUC1-CD or MUC1-CD(mSRM). The precipitates were immunoblotted with an anti-IKKβ antibody (upper panel). Input of the proteins was assessed by immunoblotting with the indicated antibodies (lower 3 panels). (d) Anti-IKKβ precipitates from the indicated cells were immunoblotted with anti-phospho- IKKβ-Ser 181 and anti-IKKβ antibodies. (e) Anti-IKKβ precipitates from the indicated cells were incubated with GST-IκBα(1–54) and γ-32P-ATP. The reaction products were analysed by SDS–PAGE and autoradiography (upper panels). The precipitates were also immunoblotted with an anti-IKKβ antibody (lower panels). (f) GST or GST–IKKγ bound to glutathione beads was incubated with IKKβ in the absence and presence of MUC1-CD or MUC1- CD(mSRM). The precipitated complexes were suspended in kinase buffer containing ATP and incubated for 30 min at 30 °C. The reaction products were immunoblotted with an anti-phospho-IKKβ-Ser 181 antibody. Input of the GST–IKKγ, IKKβ and MUC1-CD proteins is shown in Fig. 3c. Full scans of the gels in a, b, c and f are shown in Supplementary Fig. S6-3.
To determine whether the association between MUC1-CD and the IKKβ–IKKγ complex is sufficient to activate the NF-κB p65 pathway, ZR-75-1-MUC1 siRNA cells were transfected to express Flag-tagged MUC1-CD or MUC1-CD (mSRM) (Supplementary Information, Fig. S3a). When compared with ZR-75-1-MUC1 siRNA cells transfected with the empty vector, expression of MUC1-CD was associated with an increase in the formation of IKKβ–IKKγ complexes (Supplementary Information, Fig. S3b), increased phosphorylation of IKKβ on Ser 181 and of IκBα (Supplementary Information, Figs. S3c and S3d) and increased targeting of NF-κB p65 to the nucleus (Supplementary Fig. S3e). Expression of MUC1-CD (mSRM) was less effective than MUC1-CD in inducing the formation of IKKβ–IKKγ complexes (Supplementary Information, Fig. S3b). MUC1-CD-induced phosphorylation of IKKβ on Ser 181, phosphorylation of IκBα and targeting of NF-κB p65 to the nucleus were each attenuated by mutation of the SRM (Supplementary Information, Figs. S3c-e). These findings indicate that MUC1-CD is sufficient to activate the classical IKKβ–NF-κB p65 pathway.
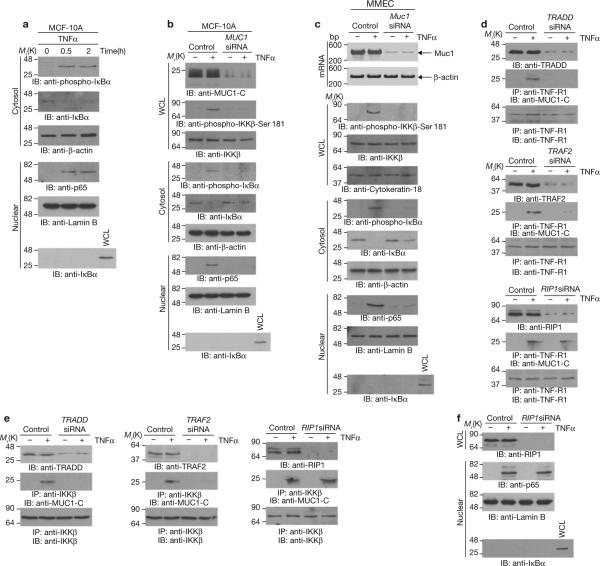
Binding of MUC1-C to the IKKβ–IKKγ complex may represent a physiological response in non-transformed cells that is constitutively activated by the overexpression of MUC1 in carcinoma cells. To address this hypothesis, studies were performed on the non-transformed mammary epithelial cell line, MCF-10A9,10, in which MUC1 is expressed at levels lower than those found in MCF-7 and ZR-75-1 cells (Supplementary Information, Fig. S4a) and where binding of MUC1-C to IKKβ or IKKγ is minimal (Supplementary Information, Fig. S4b). IKKβ and IKKγ are responsible for signalling to NF-κB in the response to tumour necrosis factor alpha (TNFα) and other pro-inflammatory cytokines. Significantly, stimulation of the MCF-10A cells with TNFα was associated with increased binding of MUC1-C to IKKβ and IKKγ (Supplementary Information, Fig. S4b). In contrast, TNFα had no apparent effect on binding of MUC1-C to IKKβ or IKKγ in MCF-7 cells (data not shown). Treatment of MCF-10A cells with TNFα was also associated with phosphorylation and degradation of IκBα (Fig. 4a), and targeting of NF-κB p65 to the nucleus (Fig. 4a). Transient silencing of MUC1 attenuated phosphorylation of IKKβ on Ser 181, phosphorylation of IκBα and nuclear targeting of NF-κB p65 in the response to TNFα stimulation (Fig. 4b). Previous work has demonstrated that the effects of TNFα on mammary epithelial cells are of physiological importance for mammary gland morphogenesis during puberty and pregnancy11–15. To determine whether these effects of TNFα are regulated by MUC1, primary mouse mammary epithelial cells (MMECs) were transfected with control or mouse-specific Muc1 siRNAs (Fig. 4c). Silencing of Muc1 in MMECs blocked TNFα-induced phosphorylation of IKKβ and IκBα (Fig. 4c). Muc1 was also necessary for TNFα-induced targeting of NF-κB p65 to the nucleus (Fig. 4c).
Figure 4.
MUC1-C contributes to NF-κB activation in the response of MCF-10A cells to TNFα. (a) MCF-10A cells were left untreated or stimulated with 20 ng ml-1 TNFα for the indicated times. Cytosolic and nuclear fractions were immunoblotted with the indicated antibodies. (b) MCF-10A cells were transfected with control siRNA or MUC1 siRNA pools for 72 h. Whole cell lysates, cytosolic fractions and nuclear fractions were immunoblotted with the indicated antibodies. (c) Primary mouse mammary epithelial cells were transfected with control or Muc1 siRNA pools for 72 h. Total RNA was isolated for determination of Muc1 and β-actin mRNA levels by RT-PCR (upper panel). Whole cell lysates and cytoplasmic and nuclear fractions were immunoblotted with the indicated antibodies. (d, e) MCF-10A cells were transfected with control siRNA, TRADD siRNA, TRAF2 siRNA or RIP1 siRNA pools for 72 h. The transfected cells were left untreated or stimulated with TNFα for 30 min. Lysates were directly immunoblotted with the indicated antibodies. Lysates were also precipitated with an anti-TNF-R1 antibody (d) or an anti-IKKβ antibody (e) and the precipitates were immunoblotted with the indicated antibodies. (f) MCF-10A cells were transfected with control siRNA or RIP1 siRNA pools for 72 h and then stimulated with TNFα. Whole-cell- or nuclear-lysates were immunoblotted with the indicated antibodies. Full scans of the gels in a, b, d and f are shown in Supplementary Fig. S6-4.
Binding of TNFα to TNF-R1 is associated with the recruitment of TNF-R1-associated death domain protein (TRADD) and TNF receptor-associated factor 2 (TRAF2) to the receptor complex2. In turn, TRAF2 recruits IKKs to the complex. Stimulation of MCF-10A cells with TNFα induced the association of MUC1-C with TNF-R1, TRADD and TRAF2 (Supplementary Information, Fig. S4c), indicating that MUC1-C is recruited to the TNF-R1 complex. Silencing of TRADD or TRAF2 demonstrated that both of these proteins are required for TNFα-induced recruitment of MUC1-C (Fig. 4d). The death-domain kinase receptor-interacting protein 1 (RIP1) is also recruited to the TNF-R1 complex, where it functions as a scaffold for IKK activation2. Notably, silencing of RIP1 had little, if any, effect on the recruitment of MUC1-C (Fig. 4d). Silencing of TRADD or TRAF2, but not RIP1, also blocked the TNFα-induced association of MUC1-C with IKKβ (Fig. 4e). In TNFα-treated fibroblasts, HeLa cells and Jurkat cells16–20, which are null for MUC1 expression6,21,22, RIP1 mediates IKK activation by binding directly to IKKγ and is essential for induction of NF-κB signalling. In contrast, we found that silencing of RIP1 in MCF-10A cells had little effect on TNFα-induced activation of NF-κB p65 (Fig. 4f). These results indicate that in MCF-10A cells, TNFα recruits MUC1-C to the TNF-R1 complex by a TRADD- and TRAF2-dependent mechanism and that MUC1-C functions independently of RIP1 in the activation of IKKβ.
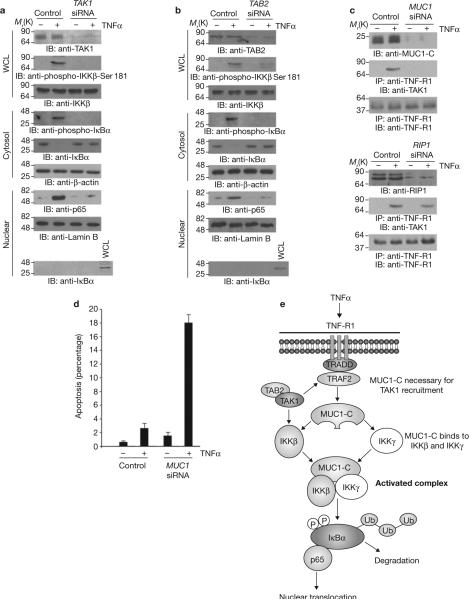
To determine whether MUC1-dependent phosphorylation of IKKβ on Ser 181 is mediated by transforming growth factor-β-activated kinase 1 (TAK1), which phosphorylates IKKβ on Ser 181 and activates the NF-κB pathway in response to TNFα stimulation23,24, TAK1 was silenced in MCF-10A cells (Fig. 5a). These results demonstrate that TNFα stimulation is associated with phosphorylation of IKKβ on Ser 181 by a TAK1-mediated mechanism (Fig. 5a). TNFα-induced phosphorylation of IκBα and targeting of NF-κB p65 to the nucleus were also dependent on TAK1 (Fig. 5a). The TAK1-binding proteins TAB1, TAB2 and TAB3 function as adaptor molecules in linking TRAFs to TAK1 activation23,25,26. Silencing of TAB1 in MCF-10A cells had no effect on TNFα-induced phosphor-ylation of IKKβ on Ser 181, phosphorylation and degradation of IκBα, or targeting of NF-κB p65 to the nucleus (Supplementary Information, Fig. S5). However, silencing of TAB2 (Fig. 5b), but not its homologue TAB3 (ref. 26 and data not shown), was associated with attenuation of TNFα-induced phosphorylation of IKKβ on Ser 181 and activation of the NF-κB p65 pathway. In this regard, TAB2 but not TAB1, is critical for TNFα signalling23,27,28. Together with our other findings, these results support a model in which MUC1-C is necessary for TNFα-induced recruitment of TAK1 to the TNF-R1 complex. Consistent with such a model, silencing of MUC1 blocked recruitment of TAK1 to TNF-R1 in TNFα-stimulated MCF-10A cells (Fig. 5c, upper). Notably, studies in Jurkat cells have shown that RIP1 is necessary for recruitment of TAK1 to the TNF-R1 complex29. Our studies showed that silencing of RIP1 in MCF-10A cells had little, if any effect, on recruitment of TAK1 to TNF-R1 (Fig. 5c, lower). This indicates that MUC1, but not RIP1, is essential for TNFα-induced TAK1 phosphorylation of IKKβ. Silencing of MUC1 in MCF-10A cells was also associated with TNFα-induced apoptosis (Fig. 5d), a response blocked by activation of the NF-κB pathway30. These findings indicate that MUC1 is involved in activation of TAK1–IKKβ–NF-κB p65 signalling by TNFα and thereby attenuation of the apoptotic response.
Figure 5.
MUC1 is necessary for TNFα-induced recruitment of TAK1 to the TNFR1 complex. (a–c) MCF-10A cells were transfected with control siRNA or TAK1 (a), TAB2 (b), MUC1 (c, upper) or RIP1 (c, lower) siRNA pools for 72 h and then stimulated with TNFα. Whole cell lysates, cytosolic fractions and nuclear fractions were immunoblotted with the indicated antibodies (a,b). Whole cell lysates were precipitated with anti-TNF-R1. The precipitates and lysates not subjected to immunoprecipitation were blotted with the indicated antibodies (c). (d) MCF-10A cells were transfected with control siRNA or MUC1 siRNA pools for 72 h. The transfected cells were left untreated or treated with TNFα for 24 h and then monitored for DNA content. The results are expressed as percentage apoptotic cells (mean ± s.d., n = 3) with sub-G1 DNA. (e) Proposed model for the effects of MUC1 on activation of the IKKβ–IKKγ complex and the NF-κB p65 pathway. Full scans of the gels in a and b are shown in Supplementary Fig. S6-5
The present studies support a model in which MUC1-C binds directly to IKKβ and IKKγ to activate the IKK complex (Fig. 5e). As shown for heat shock proteins, which associate with multiple targets31, the evidence to date indicates that MUC1-C maintains effectors of growth and survival pathways (β-catenin, p53) and nuclear hormone receptors (ERα) in stabilized and active configurations4–7. We found that expression of MUC1 in carcinoma cells constitutively induces multiple processes, including the formation of IKKβ–IKKγ complexes, phosphorylation of IKKβ on Ser 181, IKKβ-mediated phosphorylation of IκBα and targeting of NF-κB p65 to the nucleus (Fig. 5e). A variety of human tumours aberrantly overexpress MUC1 and exhibit constitutive activation of the NF-κB pathway21,32–34. The present findings indicate that MUC1 may contribute, at least in part, to activation of NF-κB in these tumours to promote cell survival. Studies performed in non-malignant MCF-10A mammary epithelial cells demonstrate that MUC1-C associates with the TNF-R1 complex in the response to TNFα stimulation and functions in the recruitment of TAK1, TAK1-mediated phosphorylation of IKKβ, assembly of the IKKβ–IKKγ complex and trans-autophosphorylation of IKKβ. Our results also indicate that TAB2 is necessary for MUC1-C function, whereas RIP1 is not required for TNFα-induced recruitment of MUC1-C to the TNF-R1 complex, binding of MUC1-C to IKKβ or activation of NF-κB. Moreover, MUC1, and not RIP1, was shown to be necessary for recruitment of TAK1 to the TNF-R1 complex. These findings indicate that MUC1-C functions independently of RIP1 in TNFα-induced IKKβ activation. The findings further indicate that there may be more than one IKK complex, depending on cell type, and that the binding partner (for example RIP1 or MUC1-C) may vary, depending on availability and affinity for the IKKs. In summary, our findings indicate that overexpression of MUC1 as found in human tumours is important for sustained IKKβ–NF-κB signalling and that MUC1-mediated activation of this pathway may be exploited by malignant cells for survival under adverse conditions.
METHODS
Cell culture
Human HCT116 colon carcinoma cells, HeLa cervical carcinoma cells and MCF-7 breast-cancer cells were grown in Dulbecco's modified Eagle's medium with 10% heat-inactivated fetal bovine serum, 100 U ml−1 penicillin, 100 μg ml−1 streptomycin and 2 mM L-glutamine. Human ZR-75-1 breast-cancer cells were grown in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum, antibiotics and L-glutamine (Mediatech, Herndon, VA). Transfection and selection of stable clones has been described for the HCT116 (ref. 4), HeLa6, MCF-7 (ref. 7) and ZR-75-1(ref. 4) cells. Human MCF-10A breast epithelial cells were grown in mammary epithelial cell growth medium (MEGM; Lonza, Walkersville, MD). Transfection of the MCF-10A cells with siRNA pools (Dharmacon, Lafayette, CO, see Supplementary Information for primer sequences) was performed in the presence of Lipofectamine 2000 (Invitrogen, Carlsbad, CA).
Subcellular fractionation
Nuclear and cytosolic fractions were prepared as previously described3–6.
Immunoprecipitation and immunoblotting
Lysates from sub-confluent cells were prepared as previously described4. Soluble proteins were incubated with anti-IKKβ (Cell Signaling Technology, Danvers, MA), anti-IKKγ, anti-TNF-R1, anti-TRADD or anti-TRAF2 (Santa Cruz Biotechnology, Santa Cruz, CA) antibodies for 2 h at 4 °C. Immune complexes and cell lysates were subjected to immunoblotting with anti-MUC1-C (Ab5; Lab Vision, Fremont, CA), anti-β-actin (Sigma, St Louis, MO), anti-NF-κB p65 (Santa Cruz Biotechnology), anti-lamin B (Calbiochem, San Diego, CA), anti-Bcl-xL (Santa Cruz Biotechnology), anti-phospho-IκBα (Cell Signaling Technology), anti-IκBα (Santa Cruz Biotechnology), anti-IKKβ (Cell Signaling Technology), anti-IKKγ (Santa Cruz Biotechnology), anti-phospho-IKKβ (Cell Signaling Technology), anti-RIP1 (Santa Cruz Biotechnology), anti-TAK1 (Cell Signaling Technology), anti-TAB1, anti-TAB2, anti-TAB3 (Santa Cruz Biotechnology) and anti-cytokeratin-18 (Abcam, Cambridge, MA) antibodies. The immune complexes were detected with horse-radish peroxidase-conjugated secondary antibodies (GE Healthcare Biosciences, Piscataway, NJ) and enhanced chemiluminescence (ECL; GE Healthcare). For immunodepletion studies, cell lysates were incubated with increasing amounts of an anti-MUC1-C antibody or a control IgG for 2 h at 4 °C. MUC1-C complexes were precipitated with protein G beads. The immune complexes and the immunodepleted supernatant were subjected to immunoblotting.
Luciferase assays
Cells were transfected with wild-type or mutant pNF-κB-Luc and SV-40-Renilla-Luc (Promega, Madison, WI) in the presence of Lipofectamine. After 48 h, cells were lysed in passive lysis buffer. Lysates were analysed for firefly and Renilla luciferase activities using the dual luciferase assay kit (Promega).
Pulse-chase analysis
Cells were cultured in methionine-free medium containing 35S-labelled methionine (200 μCi ml−1; Perkin-Elmer Life Sciences, Waltham, MA) for 1 h, washed and then chased in the presence of complete medium. Anti-IκBα precipitates were subjected to SDS–PAGE and autoradiography. Intensity of the signals was determined by densitometric scanning.
RT-PCR
Total cellular RNA was extracted with the High Pure RNA Isolation kit (Roche, Indianapolis, IN). IκBα-specific (5’-AGTCCTGCACCACCCCGCACC-3’ and 3’-TCATAACGTCAGACGCTGGCCTC-5’), mouse Muc1-specific (5’-CCACCTCACACACGGAGCGC-3 and 3’-GTCATCAGGTGTCACCGTGG-5), human β-actin and mouse β-actin (5’-CTGTCGAGTCGCGTCCACCC-3’ and 3’-TGGTGTCCGTAACACTACCT-5’) primers were used for reverse transcription and amplification (SuperScript One-Step RT-PCR with Platinum Taq; Invitrogen). Amplified fragments were analysed by electrophoresis in 2% agarose gels.
In vitro binding assays
Purified GST–MUC1-CD was cleaved with thrombin to remove the GST moiety. GST, GST–IKKβ, GST–IKKβ(1–458), GST–IKKβ(458– 756), GST–IKKγ, GST–IKKγ(1–196) or GST–IKKγ(197–419) was then incubated with the MUC1-CD for 1 h at 25 °C. In other experiments, GST, GST–MUC1-CD, GST–MUC1-CD(1–45), GST–MUC1-CD(46–72) or GST–MUC1-CD(mSRM) were incubated with purified IKKβ or IKKγ. Adsorbates to glutathione-conjugated beads were analysed by immunoblotting.
Protein gel filtration chromatography
HeLa-vector and HeLa-MUC1 cells were lysed in 50 mM Tris-HCl, at pH 7.5, 150 mM NaCl, 1 mM NaVO3, 1 mM phenylmethylsulphonyl fluoride (PMSF), 1 mM dithiothreitol (DTT), 10 mM NaF, 10 μg μl−1 aprotinin and 10 μg μl−1 leupeptin for 15 min at 4 °C. The lysates were sedimented at 14,000 g for 15 min to remove the insoluble fraction. Soluble protein (500 mg) was injected into a Sephacryl S-200 HR column and separated by fast protein liquid chromatography (FPLC) using the lysis buffer. Thirty fractions of 4 ml each were collected and 40 μl aliquots were subjected to immunoblot analysis.
In vitro kinase assays
IKK complexes were immunoprecipitated with anti-IKKβ antibody. The precipitates were incubated in 50 mM HEPES (pH 7.4), 10 mM MgCl2, 10 mM MnCl2, 2 mM DTT, 0.1 mM NaF, 10 μM ATP, 0.4 μCi μl−1 32PATP (Perkin-Elmer Life Sciences) and 0.1 μg μl−1 purified GST–IκBα(1–54) for 30 min at 30 °C. The reaction products were analysed by SDS–PAGE and auto-radiography.
Isolation of mouse mammary epithelial cells (MMECs)
The fourth and fifth mammary glands were resected from 8-week-old virgin C57BL/6 female mice, minced and digested in 0.2% collagenase I, 0.2% trypsin and 5% fetal bovine serum in mammary epithelial basal medium (Lonza) for 2 h at 37 °C as described35. The cells were pelleted at 469 g, washed in mammary epithelial basal medium (Lonza) for 10 cycles and then seeded in MEGM (Lonza).
Transfection of MMECs
Cells were transfected with control and mouse-specific Muc1 siRNAs (Dharmacon, see Supplementary Information for primer sequences) in the presence of PrimeFect siRNA transfection reagent (Lonza) for 72 h.
Apoptosis assays
Cells were fixed in 70% ethanol and incubated in PBS containing 50 μg ml−1 RNase and 2.5 μg ml−1 propidium iodide as previously described4. DNA content was analysed by flow cytometry. The percentage of cells with sub-G1 DNA was determined by the MODFIT LT Program (Verity Software, Topsham, ME).
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by Grant CA97098, CA42802 and CA100707 awarded by the National Cancer Institute (Bethesda, MD). The authors thank Michael Karin (University of California San Diego, CA) for the GST–IKKβ plasmid, Richard Gaynor (Lilly Research Laboratories, Indianapolis, IN) for the GST–IKKγ plasmid, and Al Baldwin (University of North Carolina, Chapel Hill, NC) for the wild-type and mutant pNF-κB-Luc reporters. Kamal Chauhan is acknowledged for technical support.
Footnotes
Supplementary Information is available on the Nature Cell Biology website.
References
- 1.Yamamoto Y, Gaynor R. IκB kinases: key regulators of the NF-κB pathway. Trends Biochem. Sci. 2003;29:72–79. doi: 10.1016/j.tibs.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Hayden MS, Ghosh S. Signaling to NF-κB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Liu D, Chen D, Kharbanda S, Kufe D. Human DF3/MUC1 carcinoma-associated protein functions as an oncogene. Oncogene. 2003;22:6107–6110. doi: 10.1038/sj.onc.1206732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ren J, et al. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anti-cancer agents. Cancer Cell. 2004;5:163–175. doi: 10.1016/s1535-6108(04)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei X, Xu H, Kufe D. Human MUC1 oncoprotein regulates p53-responsive gene transcription in the genotoxic stress response. Cancer Cell. 2005;7:167–178. doi: 10.1016/j.ccr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Huang L, et al. MUC1 oncoprotein blocks GSK3β-mediated phosphorylation and degradation of β-catenin. Cancer Res. 2005;65:10413–10422. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 7.Wei X, Xu H, Kufe D. MUC1 oncoprotein stabilizes and activates estrogen receptor alpha. Mol. Cell. 2006;21:295–305. doi: 10.1016/j.molcel.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 8.Mercurio F, et al. IκB kinase (IKK)-associated protein 1, a common component of the heterogeneous IKK complex. Mol. Cell Biol. 1999;19:1526–1538. doi: 10.1128/mcb.19.2.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soule HD, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- 10.Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nature Cell Biol. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varela LM, Ip MM. Tumor necrosis factor-α: a multifunctional regulator of mammary gland development. Endocrinology. 1996;137:4915–4924. doi: 10.1210/endo.137.11.8895364. [DOI] [PubMed] [Google Scholar]
- 12.Varela LM, Darcy KM, Ip MM. The epidermal growth factor receptor is not required for tumor necrosis factor-α action in normal mammary epithelial cells. Endocrinology. 1997;138:3891–3900. doi: 10.1210/endo.138.9.5389. [DOI] [PubMed] [Google Scholar]
- 13.Lee PP, Hwang JJ, Murphy G, Ip MM. Functional significance of MMP-9 in tumor necrosis factor-induced proliferation and branching morphogenesis of mammary epithelial cells. Endocrinology. 2000;141:3764–3773. doi: 10.1210/endo.141.10.7697. [DOI] [PubMed] [Google Scholar]
- 14.Brantley DM, et al. Dynamic expression and activity of NF-κB during post-natal mammary gland morphogenesis. Mech. Dev. 2000;97:149–155. doi: 10.1016/s0925-4773(00)00405-6. [DOI] [PubMed] [Google Scholar]
- 15.Shea-Eaton WK, Lee PP, Ip MM. Regulation of milk protein gene expression in normal mammary epithelial cells by tumor necrosis factor. Endocrinology. 2001;142:2558–2568. doi: 10.1210/endo.142.6.8199. [DOI] [PubMed] [Google Scholar]
- 16.Ting AT, Pimentel-Muinos FX, Seed B. RIP mediates tumor necrosis factor receptor 1 activation of NF-κB but not Fas/APO-1-initiated apoptosis. EMBO J. 1996;15:6189–6196. [PMC free article] [PubMed] [Google Scholar]
- 17.Kelliher MA, et al. The death domain kinase RIP mediates the TNF-induced NF-κB signal. Immunity. 1998;8:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- 18.Devin A, et al. The alpha and beta subunits of IκB kinase (IKK) mediate TRAF2-dependent IKK recruitment to tumor necrosis factor (TNF) receptor 1 in response to TNF. Mol. Cell Biol. 2001;21:3986–3994. doi: 10.1128/MCB.21.12.3986-3994.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu H, Shu H-B, Pan M-G, Goeddel DV. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 20.Delhase M, Hayakawa M, Chen Y, Karin M. Positive and negative regulation of IκB kinase activity through IKKβ subunit phosphorylation. Science. 1999;284:309–313. doi: 10.1126/science.284.5412.309. [DOI] [PubMed] [Google Scholar]
- 21.Kufe D, et al. Differential reactivity of a novel monoclonal antibody (DF3) with human malignant versus benign breast tumors. Hybridoma. 1984;3:223–232. doi: 10.1089/hyb.1984.3.223. [DOI] [PubMed] [Google Scholar]
- 22.Li Q, Ren J, Kufe D. Interaction of human MUC1 and β-catenin is regulated by Lck and ZAP-70 in activated Jurkat T cells. Biochem. Biophys. Res. Commun. 2004;315:471–476. doi: 10.1016/j.bbrc.2004.01.075. [DOI] [PubMed] [Google Scholar]
- 23.Wang C, et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 24.Takaesu G, et al. TAK1 is critical for IκB kinase-mediated activation of the NF-κB pathway. J. Mol. Biol. 2003;326:105–115. doi: 10.1016/s0022-2836(02)01404-3. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Q, Lee FS. Mitogen-activated protein kinase/ERK kinase kinases-2 and 3 activate nuclear factor-kB through IκB kinase-α and IκB kinase-β. J.Biol.Chem. 1999;274:8355–8358. doi: 10.1074/jbc.274.13.8355. [DOI] [PubMed] [Google Scholar]
- 26.Ishitani T, et al. Role of the TAB2-related protein TAB3 in IL-1 and TNF signaling. EMBO J. 2003;22:6277–6288. doi: 10.1093/emboj/cdg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanayama A, et al. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol. Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 28.Komatsu Y, et al. Targeted disruption of the Tab1 gene causes embryonic lethality and defects in cardiovascular and lung morphogenesis. Mech. Dev. 2002;119:239–249. doi: 10.1016/s0925-4773(02)00391-x. [DOI] [PubMed] [Google Scholar]
- 29.Blonska M, et al. TAK1 is recruited to the tumor necrosis factor-alpha (TNF-α) receptor 1 complex in a receptor-interacting protein (RIP)-dependent manner and cooperates with MEKK3 leading to NF-κB activation. J. Biol. Chem. 2005;280:43056–43063. doi: 10.1074/jbc.M507807200. [DOI] [PubMed] [Google Scholar]
- 30.Karin M. Nuclear factor-κB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 31.Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem. Sci. 2006;31:164–172. doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 32.Dyomin VG, et al. MUC1 is activated in a B-cell lymphoma by the t(1;14)(q21;q32) translocation and is rearranged and amplified in B-cell lymphoma subsets. Blood. 2000;95:2666–2671. [PubMed] [Google Scholar]
- 33.Baldwin AS. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-κB. J. Clin. Invest. 2001;107:241–246. doi: 10.1172/JCI11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakshatri H, Bhat-Nakshatri P, Martin DA, Goulet RJ, Jr., Sledge GW., Jr Constitutive activation of NF-κB during progression of breast cancer to hormone-independent growth. Mol. Cell Biol. 1997;17:3629–3639. doi: 10.1128/mcb.17.7.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sotgia F, et al. Caveolin-1 deficiency (−/−) conveys premalignant alterations in mammary epithelia, with abnormal lumen formation, growth factor independence, and cell invasiveness. Am. J. Pathol. 2006;168:292–309. doi: 10.2353/ajpath.2006.050429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.