Significance
Nitric oxide (NO) influences diverse biological processes, ranging from vasodilation in mammals to communal behavior in bacteria. Heme-nitric oxide/oxygen (H-NOX) binding domains, a recently discovered family of heme-based gas sensor proteins, have been implicated as regulators of these processes. Crucial to NO-dependent activation of H-NOX proteins is rupture of the heme–histidine bond and formation of a five-coordinate NO complex. To delineate the molecular details of NO binding, high-resolution crystal structures of a bacterial H-NOX protein in the unligated and intermediate six- and five-coordinate NO-bound states are reported. From these structures, it is evident that NO-induced scission of the heme–histidine bond elicits a pronounced conformational change in the protein as a result of structural rearrangements in the heme pocket.
Keywords: gas sensing, hemoprotein
Abstract
Heme-nitric oxide/oxygen (H-NOX) binding domains are a recently discovered family of heme-based gas sensor proteins that are conserved across eukaryotes and bacteria. Nitric oxide (NO) binding to the heme cofactor of H-NOX proteins has been implicated as a regulatory mechanism for processes ranging from vasodilation in mammals to communal behavior in bacteria. A key molecular event during NO-dependent activation of H-NOX proteins is rupture of the heme–histidine bond and formation of a five-coordinate nitrosyl complex. Although extensive biochemical studies have provided insight into the NO activation mechanism, precise molecular-level details have remained elusive. In the present study, high-resolution crystal structures of the H-NOX protein from Shewanella oneidensis in the unligated, intermediate six-coordinate and activated five-coordinate, NO-bound states are reported. From these structures, it is evident that several structural features in the heme pocket of the unligated protein function to maintain the heme distorted from planarity. NO-induced scission of the iron–histidine bond triggers structural rearrangements in the heme pocket that permit the heme to relax toward planarity, yielding the signaling-competent NO-bound conformation. Here, we also provide characterization of a nonheme metal coordination site occupied by zinc in an H-NOX protein.
Nitric oxide (NO) is a freely diffusible radical gas that is critical to numerous eukaryotic processes, such as blood vessel homeostasis and neurotransmission (1, 2). An integral component of mammalian NO signaling is soluble guanylate cyclase (sGC), the primary receptor for NO (3–6). The binding of NO to the heme cofactor of sGC increases guanylate cyclase activity, resulting in activation of cGMP-dependent signaling pathways. The interaction of NO with the histidyl-ligated heme cofactor of sGC has been extensively studied through biochemical and spectroscopic means, providing an overarching model of NO-induced activation. NO binds to the ferrous (FeII) heme of sGC at the open coordination site, axial to the heme-ligating histidine, to form a transient six-coordinate nitrosyl complex. NO association severely weakens the iron–histidine bond, resulting in histidine dissociation and the formation of an activated five-coordinate, NO-bound heme (7–10) (Fig. 1A). Rupture of the iron–histidine bond is the key molecular event that stimulates sGC activity (reviewed in ref. 6). However, due to limited structural data, a detailed molecular model regarding the binding of NO to the heme and propagation of this signal has remained elusive.
Fig. 1.
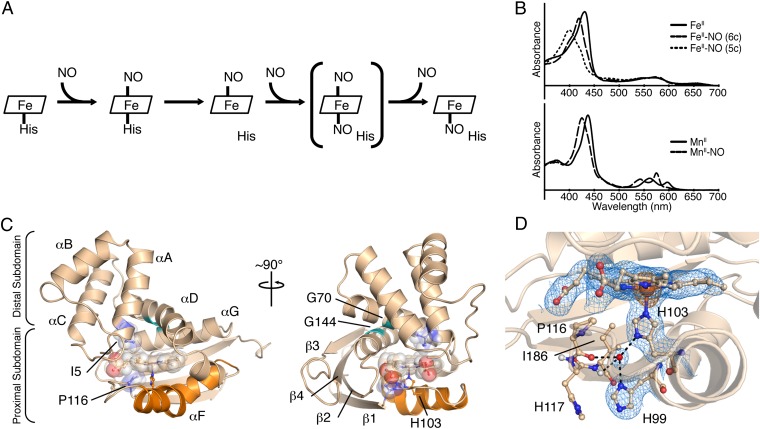
NO binding in H-NOX proteins and structural features of FeII-unligated So H-NOX. (A) Reaction scheme of NO binding to H-NOX proteins. The putative dinitrosyl intermediate is shown in brackets. (B) UV-visible absorption spectra for FeII (Upper) and MnII (Lower) nitrosyl complexes. The six-coordinate (6c) FeII nitrosyl intermediate spectrum (Upper, thick dashed line) was obtained via stopped-flow UV-visible spectroscopy (5c, five coordinate). (C) Cartoon representation of the FeII-unligated So H-NOX structure [molecule A of the asymmetric unit (ASU)]. Secondary structure elements and important structural features are highlighted. The signaling helix, αF, is displayed in orange; residues I5 and P116 and the heme are shown as transparent spheres; and conserved glycines G70 and G144 are shown in green. (D) Detailed view of the FeII-unligated H-NOX heme pocket showing a strongly hydrogen-bonded water molecule (shown as a red sphere) with residues in the proximal heme pocket. The 2mFo-DFc electron density (blue mesh, 1σ) and the anomalous difference density (orange mesh, 5σ) are displayed.
Significant advances in understanding the mechanism of sGC activation have resulted from studies involving homologous prokaryotic proteins from the heme-nitric oxide/oxygen (H-NOX) binding family of gas receptors (11–14). These bacterial homologs share similar spectroscopic properties, high sequence homology (18–40%), and key conserved residues with the sGC heme pocket, and therefore have served as important models of the sGC heme domain (15, 16).
Previous structural studies to probe the NO activation mechanism in H-NOX proteins have relied on structural mimics, wherein the heme-ligating histidine residue had been substituted with a glycine and heme binding had been rescued using supplemented imidazole as a surrogate for the histidine side chain (13, 14). Analyses of these structural mimics demonstrated that artificially “cleaving” the iron–histidine bond elicits a conformational change involving a rotation of the distal subdomain relative to the proximal subdomain, centered about the heme cofactor. Further analysis showed that alongside this conformational change, severing the iron–histidine bond resulted in relaxation of the heme toward planarity. The relationship between heme and protein conformation was first noted in the H-NOX crystal structure from Thermoanaerobacter tengcongensis (Tt H-NOX), wherein the two distinct protein conformations observed crystallographically correlate well with the overall degree of heme distortion (11). The pronounced distortion of the heme from planarity is the result of van der Waals interactions with residues I5 and P115 that flank the distal and proximal faces of the heme, respectively. Removal of the steric bulk imparted by P115, a residue that is strictly conserved in H-NOX proteins, resulted in a significantly flattened heme (17) and the same conformational change that was observed upon mimicking heme–histidine cleavage (13, 14).
From these studies, a heme-strain model of NO activation was proposed. In this model, in the basal, FeII-unligated state, the heme is distorted from planarity due to steric interactions with key conserved residues in the heme pocket. NO binding and subsequent scission of the iron–histidine bond release the steric strain imparted on the heme, allowing it to relax toward planarity and, via intimate interactions of the protein and heme, to elicit a rotation of the distal subdomain with respect to the proximal subdomain. This conformational change thus is hypothesized to communicate NO binding to downstream signaling partners (13, 14, 17).
Although these prior studies provided a structural description of conformational changes associated with rupture of the iron–histidine bond, structure determination used heme ligands and mutations within the αF, or signaling helix, that do not accurately represent the native H-NOX signaling species. Importantly, crucial structural changes involving the heme-ligating histidine were absent. The present study provides molecular-level insight into NO-induced activation of H-NOX proteins through high-resolution crystal structures of distinct states along the NO activation pathway. Reported structures include the inactive FeII-unligated state, as well as both the intermediate six-coordinate and activated five-coordinate NO-bound states, of the H-NOX protein from Shewanella oneidensis (MR-1), which has high homology and similar ligand-binding properties to sGC. Structures presented here provide high-resolution snapshots of key steps in NO activation and expand previous work on structural changes that propagate the NO-binding event. Together, these data show that several structural features in the heme pocket of the FeII-unligated H-NOX maintain a distorted heme that, following NO-induced scission of the iron–histidine bond and subsequent structural rearrangements in the proximal heme pocket, relaxes toward planarity to yield the active H-NOX protein conformation. In addition, the characterization of a nonheme metal coordination site occupied by zinc in an H-NOX protein is presented.
Results and Discussion
Structural Features of the FeII-Unligated Structure.
The H-NOX protein from the facultative anaerobe Shewanella oneidensis MR-1 (SO2144; So H-NOX) was chosen for characterization of the structural changes associated with NO activation in H-NOX proteins because of its high (among bacterial H-NOX proteins) sequence identity (30%) and similar ligand-binding properties to sGC, namely, no measurable affinity for O2 and formation of a stable, five-coordinate complex with NO (λmax = 399 nm) that occurs through a transient, six-coordinate intermediate (λmax = 420 nm) (7, 8) (Fig. 1B, Upper). Importantly, this latter feature differs from the previously reported crystal structure of the non–O2-binding H-NOX domain from the facultative anaerobe Nostoc sp. PCC 7120 (Ns H-NOX), which was crystallized as a six-coordinate complex with NO (12).
The FeII-unligated structure of So H-NOX was determined to a resolution of 2.00 Å (Table 1) through the employment of surface entropy reduction (18) (Materials and Methods). As with other previously crystallized H-NOX proteins, So H-NOX contains a heme cofactor that is highly distorted from planarity (rmsd from planarity = 0.312 Å) (19). Although the degree and modes of heme distortion (saddling, ruffling, and doming) vary among the reported H-NOX structures, the heme pocket residues that impart distortion, a branched hydrophobic residue in the distal pocket (position 5 in helix αA) and a proline in the proximal pocket, are conserved (11, 12, 15, 17, 20). Consistent with these observations, the FeII-unligated structure of So H-NOX displays a highly distorted heme due to a van der Waals interaction between the conserved proline 116 and pyrrole A (Fig. 1 C and D). This interaction results in a hinge-like displacement of pyrroles A and D from planarity, with the bending axis centered about the heme β-δ methine positions (Fig. 1D). Isoleucine 5, located in helix αA within the distal pocket, abuts the heme opposite to proline 116, and thereby serves to communicate the upward displacement of pyrrole D from the heme plane to the distal subdomain (residues 1–83) (Fig. 1C and Fig. S1). In agreement with studies on other H-NOX family members, the orientation of the distal subdomain with respect to the proximal subdomain appears to be linked to the conformation of the heme (13, 14, 17).
Table 1.
X-ray data collection and refinement statistics for So H-NOX
| FeII | MnII | ||||
| Unligated* | NO | CO* | Unligated* | NO* | |
| Data collection | |||||
| Wavelength, Å | 1.00 | 1.11 | 1.00 | 1.00 | 1.00 |
| Space group | P6322 | P21212 | P6322 | P6322 | P6322 |
| Cell dimensions | |||||
| a, b, c; Å | 164.0, 164.0, 101.7 | 67.6, 86.7, 33.8 | 164.2, 164.2, 102.4 | 163.8, 163.8, 102.0 | 164.3, 164.3, 101.7 |
| α, β, γ; ° | 90, 90, 120 | 90, 90, 90 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Resolution, Å | 47.88–2.00 | 33.82–1.65 | 43.01–2.25 | 48.02–2.10 | 47.88–2.45 |
| (2.05–2.00) | (1.74–1.65) | (2.32–2.25) | (2.16–2.10) | (2.55–2.45) | |
| Redundancy | 7.6 (6.0) | 3.9 (3.3) | 11.1 (11.1) | 4.8 (2.4) | 10.7 (11.0) |
| Completeness, % | 99.6 (97.9) | 99.7 (99.5) | 97.4 (98.1) | 98.5 (98.6) | 100.0 (100.0) |
| Rp.i.m.,† % | 2.1 (61.8) | 2.8 (41.8) | 2.8 (44.6) | 3.2 (65.5) | 3.2 (66.3) |
| I/σ | 20.8 (1.4) | 14.4 (1.7) | 20.2 (1.8) | 12.8 (1.1) | 19.3 (1.6) |
| CC1/2 | 99.9 (55.5) | 99.9 (68.7) | 99.9 (64.4) | 99.9 (74.2) | 99.9 (52.7) |
| Refinement | |||||
| Molecules in ASU | 2 | 1 | 2 | 2 | 2 |
| Rwork/Rfree, % | 17.2/19.1 | 16.8/20.3 | 18.2/19.3 | 17.1/19.7 | 18.2/20.7 |
| Ramachandran | |||||
| Favored, % | 96.8 | 97.2 | 96.2 | 96.8 | 96.0 |
| Outliers, % | 0 | 0 | 0 | 0 | 0 |
| B factors, Å2 | |||||
| Overall | 61.65 | 34.52 | 66.21 | 63.34 | 73.76 |
| Protein | 56.60 | 31.14 | 60.81 | 58.14 | 67.54 |
| Heme/MnPPIX‡ | 37.06 (A) | 20.69 | 41.86 (A) | 41.41 (A) | 48.06 (A) |
| 50.05 (B) | 52.26 (B) | 54.43 (B) | 63.32 (B) | ||
| Gas ligands§ | — | 26.58 (N) | 58.71 (C) | — | 47.78 (N) |
| 26.04 (O) | 67.34 (O) | 70.23 (O) | |||
| Zinc‡ | 35.58 (A) | 20.46 | 39.37 (A) | 41.91 (A) | 47.63 (A) |
| 44.79 (B) | 48.08 (B) | 51.83 (B) | 55.99 (B) | ||
| Gas ligand RSR¶ | — | 0.12 (0.11) | 0.25 (0.16) | — | 0.25 (0.17) |
| rmsd | |||||
| Bond lengths, Å | 0.005 | 0.019 | 0.009 | 0.021 | 0.022 |
| Bond angles, ° | 0.865 | 0.864 | 0.887 | 1.650 | 1.643 |
| MolProbity score | 1.03 | 1.20 | 1.41 | 1.34 | 1.31 |
Unless otherwise noted, values in parentheses correspond to the highest resolution shell. CC1/2, Pearson's correlation coefficient between average intensities of random half datasets for each unique reflection (51).
Denotes the 154AAA mutant of So H-NOX.
Calculated using .
Corresponding molecule in asymmetric unit (ASU) is listed in parentheses.
Corresponding gas atom is listed in parentheses.
Real-space R-value (RSR) for the gas ligand and porphyrin (listed in parentheses).
In addition, the FeII-unligated structure of So H-NOX contains a hydrogen-bonding network that intimately links crucial secondary structure elements in the proximal heme pocket. Specifically, the proximal pocket contains an ordered water molecule that hydrogen-bonds with the Nδ1 nitrogen of the heme-ligating histidine residue (H103), the Nδ1 nitrogen of H99, the carbonyl oxygen (C=O) of P116, and the amide hydrogen (H-N) of I118 (Fig. 1D). As a result, this hydrogen-bonding network appears to link the conformation of the P116-containing αF-β1 loop to the conformation of the αF helix. Based on this observation, it is hypothesized that heme-histidine ligation serves to maintain P116 in van der Waals contact with the heme through hydrogen-bonding interactions within the proximal pocket. Notably, due to the coupling of the heme and protein conformations, it is then postulated that this proximal pocket hydrogen-bonding network aids in maintaining the protein in the basal state conformation. Furthermore, a similar mode of water coordination in the proximal pocket was also observed in Ns H-NOX (12, 21) and has been hypothesized to be an important feature in H-NOX activation (22).
Analysis of a Structural Zinc Site in So H-NOX.
Analysis of the electron density in the So H-NOX structure following molecular replacement showed a very strong peak in the anomalous difference map centered between residues C139, H161, C164, and C172 (Fig. S2A). Because histidine and cysteine residues are commonly observed in zinc or copper coordination sites, it was reasoned that So H-NOX possesses a previously unidentified nonheme metal-binding site. To unambiguously identify the bound metal, purified So H-NOX used for crystallization was subjected to inductively coupled plasma absorption emission spectroscopy (ICP-AES) (SI Materials and Methods). ICP-AES analysis revealed near-stoichiometric amounts of both iron (originating from the bound heme) and zinc (Fig. S2B). Negligible amounts of copper, manganese, magnesium, nickel, or cobalt were detected. Following this analysis, zinc was modeled into the FeII-unligated crystal structure and, after refinement with loose distance restraints, was found to adopt geometric values that correlate well with published statistics for tetrahedral zinc coordination sites (23).
The unprecedented observation of a nonheme metal coordination site in an H-NOX protein garnered curiosity about its prevalence within the H-NOX family. Analysis of a sequence alignment containing all available bacterial H-NOX sequences indicates that the residues involved in zinc coordination are almost exclusively found in gammaproteobacteria. Only a few examples were observed in other classes of bacteria. Furthermore, 78 of 158 known gammaproteobacteria sequences possess putative zinc-binding residues. Of those gammaproteobacteria sequences, 34% contain histidine as the fourth ligand, whereas 66% contain glutamine, both of which are capable of binding zinc (23). Also included in the sequence alignment analysis are sequences derived from rat and human sGCs, neither of which possesses the determined zinc-binding residues. As such, the observation of a zinc coordination site is limited predominantly to a select group of H-NOX proteins from gammaproteobacteria (Fig. S2C).
To confirm that those H-NOX proteins containing glutamine as the fourth ligand also coordinate zinc, the H-NOX from Vibrio cholerae (Vc H-NOX), which possesses a glutamine in place of H161 (So H-NOX numbering), was purified and subjected to ICP-AES analysis. Tt H-NOX, not predicted to bind zinc, was also analyzed by ICP-AES. These analyses indicate that So and Vc H-NOX proteins possess stoichiometric iron and zinc, whereas Tt H-NOX, as expected, does not harbor any other metals besides the iron derived from the heme cofactor (Fig. S2B).
Mutational analysis of the zinc site in So H-NOX identifies this feature as a crucial component in maintaining the structural integrity of the protein. Specifically, mutation of any one of the zinc-coordinating cysteine residues to serine or H161 to an alanine completely abolished soluble protein expression, presumably as a result of the inability of the protein to incorporate zinc and fold properly (Fig. S3C). However, following the observation that Vc H-NOX is capable of binding zinc, H161 in So H-NOX was replaced with a glutamine and purified. ICP-AES analysis determined that H161Q So H-NOX bound a stoichiometric amount of zinc and displayed similar stability and ligand-binding properties as the WT protein (Fig. S3). It is postulated then that zinc coordination in H-NOX proteins serves a structural role, a feature common to sites of tetrahedral zinc coordination (24), and links the β3-β4 loop to helix αG within the proximal subdomain of zinc-binding H-NOX proteins.
Mimicking the Transient Six-Coordinate FeII-NO Intermediate.
H-NOX proteins from facultative anaerobes and mammalian sGC initially bind NO as a six-coordinate complex (Fig. 1A). NO binding severely weakens the iron–histidine bond due to the strong trans-effect of NO, resulting in the rapid formation of the five-coordinate NO-bound species following histidine dissociation (7–10). Although the six-coordinate NO complex is a transient intermediate, structural changes within the heme pocket concomitant with NO binding may aid in histidine dissociation. To elucidate structural changes associated with the formation of a six-coordinate NO complex, a structural analog was sought due to the short lifetime of this species.
Previous work has shown that sGC reconstituted with manganese (II) (MnII) protoporphyrin IX (MnPPIX) remains in a stable, six-coordinate NO-bound state due to lack of scission of the proximal histidine bond, and thus does not show NO-dependent activation (25). Similar to sGC, binding of NO to MnII-unligated So H-NOX (λmax = 437 nm) was found to yield a stable, six-coordinate NO complex (λmax = 425 nm) (Fig. 1B, Lower). Therefore, it was hypothesized that the structure of MnII-NO So H-NOX could accurately mimic the transient six-coordinate intermediate in the native protein.
Because the crystal structure of MnII-NO So H-NOX would be determined through crystal derivatization of the parent MnII-substituted So H-NOX, the structure of MnII-unligated So H-NOX was first pursued to establish feasibility. Crystals of MnII-unligated So H-NOX were obtained under similar conditions as FeII So H-NOX (Materials and Methods). The crystals were found to diffract to a comparable resolution (2.10 Å vs. 2.00 Å) in the same space group, with similar unit cell parameters (Table 1). The FeII-unligated and MnII-unligated structures were found to superimpose to a high degree overall with an rmsd of 0.119 Å (Cα-Cα alignment) (Fig. 2A), indicating that replacement of the heme Fe with Mn does not lead to any significant alterations in the protein conformation.
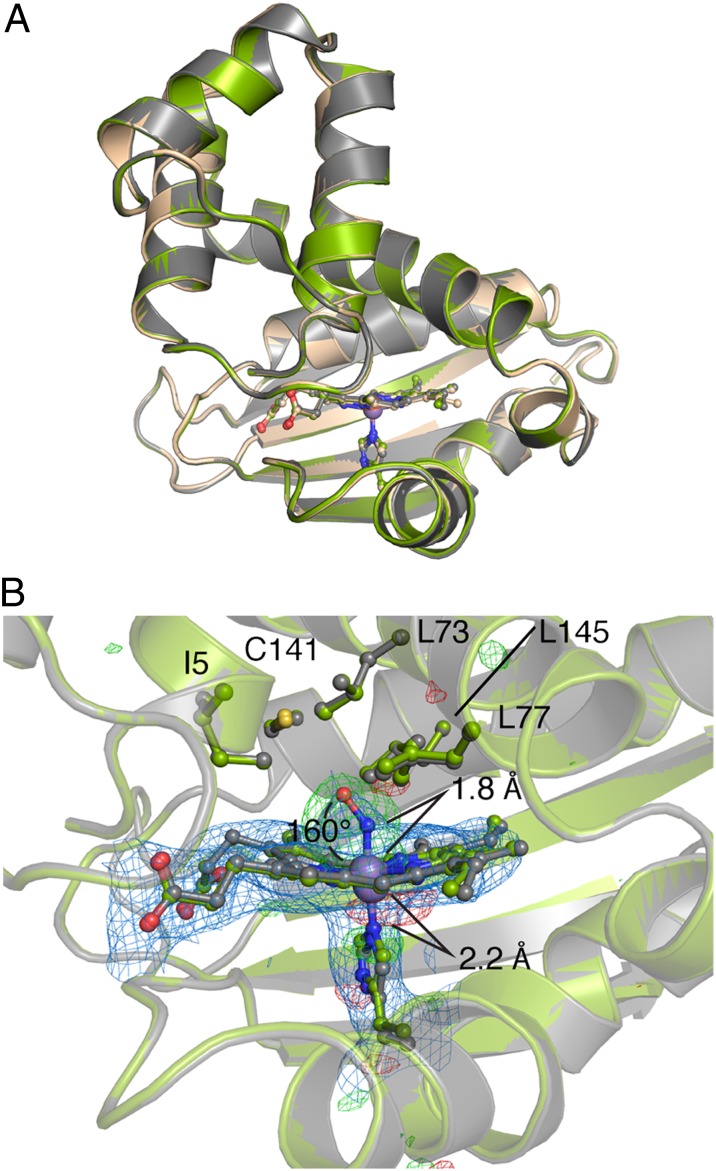
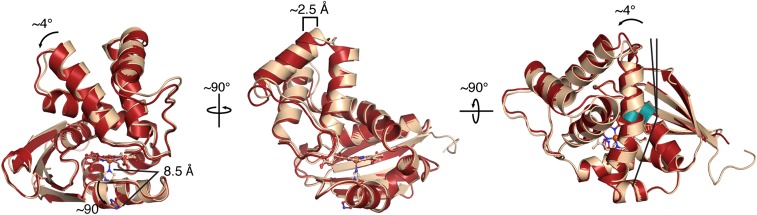
Fig. 2.
Structural comparison of FeII, MnII, and MnII-NO So H-NOX. (A) Overall structural alignment of FeII (wheat), MnII (gray), and MnII-NO (green) So H-NOX structures (molecules A of the ASU). (B) Detailed comparison of MnII and MnII-NO heme pockets showing that the six-coordinate NO complex minimally perturbs residues in the distal pocket. The 2mFo-DFc electron density (blue mesh, 1σ) and the isomorphous Fo-Fo (NO − native) difference density (positive, green mesh; negative, red mesh; 5σ) (within a 10-Å radius of the MnII metal) are displayed.
To determine the structure of the six-coordinate MnII–NO complex, crystals of the MnII-unligated protein were soaked with NO (Materials and Methods). Analysis of the MnII-NO–MnII-unligated Fo-Fo isomorphous difference map demonstrated strong positive density distal to H103 in molecule A of the asymmetric unit, indicative of NO binding (Fig. 2B). NO was not present in molecule B (Fig. S4). NO was observed to bind in a bent geometry (Mn-N-O angle of ∼160°), staggered between pyrroles A and D, and was refined to full occupancy. However, weak electron density for the oxygen atom of the NO ligand indicates that there might be a degree of disorder of the Mn-N-O angle. The orientation of the NO ligand toward the propionate groups appears to be due to steric restraints imposed by distal pocket residues L73, L77, and L145 (Fig. 2B and Fig. S5). Based on comparison with the MnII structure, NO binding is associated with a slight movement of the MnII atom within the porphyrin pyrrole nitrogen plane toward the nitrogen atom of the NO ligand (∼0.5 Å), along with a slight movement of H103 toward the porphyrin (∼0.4 Å) (Fig. 2B). Movement of H103 maintains a constant Mn–histidine bond length of ∼2.2 Å. However, in the six-coordinate FeII–nitrosyl complex, movement of H103 is not expected to occur with movement of the metal within the plane of the porphyrin, thereby leading to an elongated, and thus weaker, iron–histidine bond (26).
Superimposition of the MnII-unligated and MnII-NO structures yields an rmsd of 0.099 Å (Cα-Cα alignment), indicating that formation of the six-coordinate NO complex does not induce an appreciable conformational change (Fig. 2A). Furthermore, formation of the six-coordinate NO complex occurs with minimal displacement of distal heme pocket residues (Fig. 2B) as a result of local interactions with surrounding residues. These observations indicate that the primary determinant of iron–histidine bond cleavage appears to be the trans-effect exhibited by NO. Additionally, these findings are in agreement with the body of biochemical evidence suggesting that scission of the iron–histidine bond is a primary contributor to conformational activation of H-NOX proteins, and not simply NO bound to the heme (6, 25).
NO in Excess Binds to the Proximal Face of the Heme.
Initial attempts to obtain a five-coordinate NO structure were performed by soaking FeII-unligated crystals of So H-NOX with various concentrations of NO following the same protocol used to obtain the MnII-NO structure. However, pronounced crystal cracking, splitting of diffraction spots, and/or significant loss of diffraction was observed even during brief exposure to low concentrations of NO. Thus, it was reasoned that the conformational change associated with formation of the five-coordinate NO complex was not suitable with the lattice constraints induced by crystallographic contacts and efforts should be focused on crystallization of a preformed FeII–NO complex. Briefly, immediately before crystallization, the FeII-unligated protein was mixed with an excess of quick-releasing, PROLI-NONOate [t1/2 of ∼5 s at room temperature (RT)] and slow-releasing DETA-NONOate (t1/2 of ∼56 h at RT) NO donors (equal mixture) to ensure immediate formation of the NO complex and to maintain adequate NO concentration following drop equilibration, respectively. Because NO is labile in the presence of oxygen, all protein manipulation and crystallization procedures were performed under an argon atmosphere (Materials and Methods).
Crystals of preformed FeII-NO So H-NOX diffracted to 1.65 Å and were assigned to a different space group than the FeII-unligated crystals (P21212 vs. P6322), consistent with the hypothesis that the FeII-unligated crystal lattice constraints were not conducive to the FeII-NO structure. In agreement with prior spectroscopic studies, the NO ligand in the FeII-NO structure was found to bind as a five-coordinate adduct (7–9, 27), wherein the heme-ligating histidine was displaced ∼8.5 Å from the heme iron following rupture of the iron–histidine bond. Furthermore, NO was found to adopt a bent geometry (Fe-N-O angle of ∼124°) with the NO oxygen oriented between pyrroles A and B (Fig. 3A and Fig. S5).
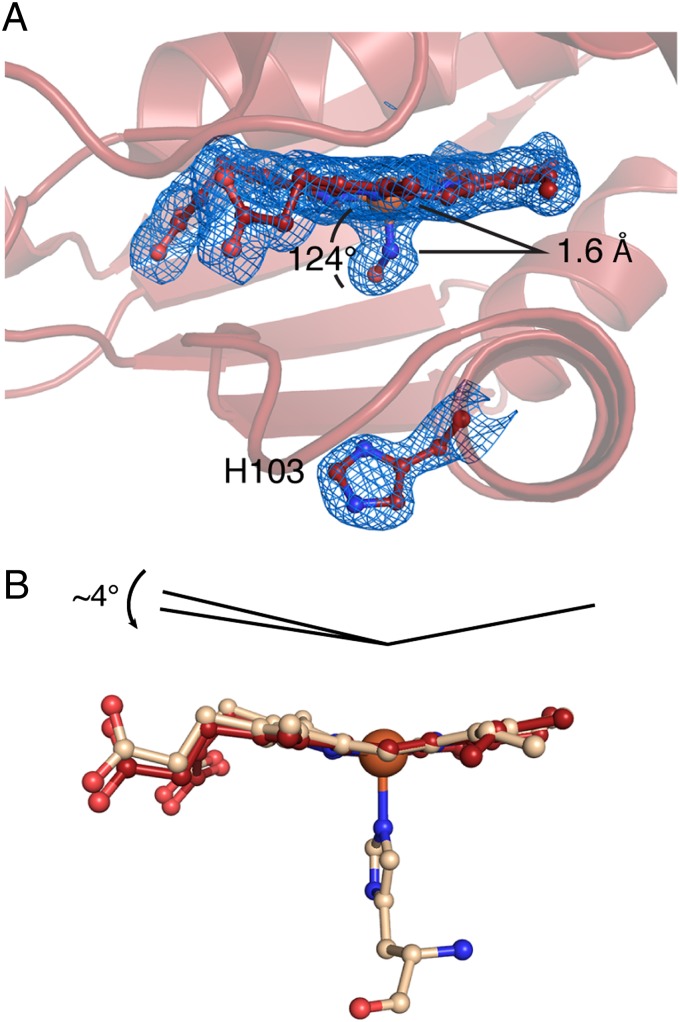
Fig. 3.
Proximal-bound, five-coordinate NO complex and NO-induced heme flattening. (A) Zoomed-in view of the FeII-NO So H-NOX heme pocket shows NO bound in a bent geometry to the proximal heme face, with the heme-ligating histidine (residue 103) considerably displaced from the heme iron. The 2mFo-DFc electron density (1σ) is displayed in blue mesh. (B) Alignment of the FeII and FeII-NO hemes (aligned using pyrroles B and C; molecules A of the ASU) shows an increase in heme planarity upon histidine dissociation.
Notably, NO was observed to bind to the proximal face of the heme (Fig. 3A and Fig. S5A). This observation was previously reported for cytochrome c′ from Alcaligenes xylosoxidans (Axcyt c′) (28), which, similar to So H-NOX and sGC, also reacts with NO to form a five-coordinate complex via a six-coordinate intermediate (29). Previous studies on NO binding in H-NOX proteins have revealed that both the rates of initial NO binding and subsequent dissociation of the histidine to form the five-coordinate NO complex exhibit NO concentration dependence. Therefore, it was suggested that under conditions where there are at least 2 eq of NO per H-NOX protein, a second NO could bind to the proximal face of the heme in place of the dissociated histidine to form a putative dinitrosyl intermediate, from which the original distal NO then dissociates to leave a five-coordinate NO complex with the NO residing in the proximal pocket (7–10, 30) (Fig. 1A). Additional support for this hypothesis was obtained from studies using double-isotope sequential stopped-flow and freeze-quench EPR experiments, where a second NO-binding event to the proximal heme face could be discerned (10). Further detailed in vitro investigations into the involvement of additional equivalents of NO in the activation of sGC provided evidence that two distinct, spectrally identical, NO-bound states of the enzyme exist. Importantly, these states were hypothesized to be a result of either (i) two five-coordinate NO-bound species, with NO located in either the distal or proximal pocket, or (ii) a five-coordinate NO-bound heme with the additional NO equivalent modifying a cysteine residue to generate an NO-addition complex (4, 31–34). The FeII-NO structure presented here provides crystallographic evidence for the formation of proximal NO in H-NOX proteins. It must be noted, however, that the conditions used to obtain this structure utilized a large excess of NO to maintain a sufficient concentration of NO during crystallization. Although it is doubtful that an H-NOX signaling system would encounter these concentrations of NO in vivo, it would seem that an excess of NO favors binding to the proximal heme face. Furthermore, it must be noted that the observation of a proximal NO species in an H-NOX protein does not eliminate the possibility of the involvement of a cysteine residue in the full activation of sGC, because these species are not mutually exclusive.
It was proposed for Axcyt c′ that steric repulsion imposed by a leucine (L16), located in the distal pocket and directly above the heme iron, decreased the stability of ligands bound in the distal pocket to favor NO binding in the proximal pocket. Support for this hypothesis was grounded in the observation that carbon monoxide (CO) in the CO-bound Axcyt c′ structure was located in the same position previously occupied by L16 and possessed higher thermal parameters than the heme, indicative of partially occupied or weakly bound CO. It was reasoned that this steric hindrance destabilized distal-bound ligands, contributing to the transient nature of the six-coordinate nitrosyl complex and favoring binding of NO to the proximal heme face (28, 35).
To obtain further information into whether steric crowding in the distal pocket potentially favors proximal binding of NO in So H-NOX, a crystal structure of the FeII–CO complex was determined to 2.25 Å (Table 1). Briefly, crystals of FeII were subjected to the same derivatization protocol used for MnII-NO, except with replacement of the NO solution with CO-saturated cryoprotectant (Materials and Methods). Analysis of the FeII-CO structure reveals that CO binds in a linear geometry (Fe-C-O angle of ∼176°) at full occupancy to molecule A of the asymmetric unit. No ligand was observed for molecule B. Superimposition of the FeII and FeII-CO structures resulted in an rmsd of 0.075 Å (Cα-Cα alignment) (Fig. S6A), indicating very modest structural differences. Analysis of the distal pocket reveals that the side chain of L77, located above pyrrole C, moves ∼0.6 Å upon CO binding as a result of a van der Waals interaction with CO. This interaction results in minor displacement of the CO ligand from linearity, with a slight movement of the CO toward pyrrole D and away from L77 (Fig. S6B). However, the structural changes are modest and do not result in significant elevation of the B-factors for the CO ligand above adjacent heme atoms (pyrrole nitrogens and iron) or an unusual binding geometry. Similarly, the MnII-NO structure possessed a distal-bound NO without noticeable alterations in the heme pocket. As such, it is reasoned that because So H-NOX is able to accommodate ligands in the distal pocket with little structural change to distal pocket residues or unusual ligand geometries, other factors may contribute to preferential binding of NO to the proximal pocket in H-NOX proteins.
Histidine Dissociation and Structural Changes in the Signaling Helix.
Comparison of the FeII-unligated and five-coordinate FeII-nitrosyl structures revealed that several interconnected structural changes occur within the proximal subdomain following scission of the iron–histidine bond. Most pronounced are structural changes in the signaling helix αF. These changes, predominantly localized to residues 99–106 and centered about heme-ligating histidine H103, involve a rigid rotation of ∼45° of these residues about an axis centered along the length of the helix. In the FeII-unligated structure, the backbone carbonyl oxygen atoms of residues H99 and D100 lack hydrogen-bonding partners, resulting in a pronounced bulge within helix αF. Upon iron–histidine bond cleavage and rotation of the αF helix, the carbonyl oxygen of D100 rotates into hydrogen-bonding distance of the amide hydrogen of L104 (C=O – H-N distance of 4.6 Å vs. 2.4 Å) to contribute a new backbone hydrogen bond within helix αF that serves to stabilize the new conformation. Concomitant with rotation of this helix is the displacement of the side chain of H103 by ∼8.5 Å (H103 Nε2-Nε2 distance between structures). This displacement is a result of movement of H103 backbone atoms ∼1 Å away from the heme, due to rotation of the αF helix, alongside a change in the rotamer of H103 (Δχ1 of ∼90°) (Figs. 3A and 4). Therefore, it appears that rotation of the αF helix and formation of a new backbone hydrogen bond between residues D100 (C=O) and L104 (H-N) aid in stabilization of the αF helix with H103 in the out conformation.
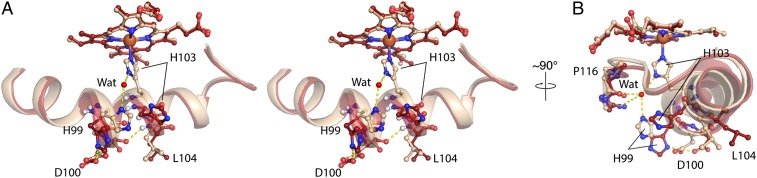
Fig. 4.
Structural changes within the signaling helix upon NO binding. (A) Stereoview of the proximal pocket of FeII (wheat) and FeII-NO (red) (molecules A of the ASU) focusing on important structural changes of the signaling helix (αF) and the αF-β1 loop (residues 91–118). NO has been omitted for clarity. (B) Rotation of 90° of A. The ordered water molecule in the proximal pocket is shown as a red sphere. Hydrogen bonds are shown as yellow dashes.
Histidine dissociation and outward displacement result in loss of the ordered water that was observed between residues H103, H99, P116, and L118 in the FeII-unligated structure. Specifically, the side chain of H99 rotates ∼180° about χ1 to reorient Nδ1 from a location capable of interacting with the proximal pocket water to a conformation that now hydrogen-bonds with one of the side chain oxygen atoms of D100 (Fig. 4). This new bonding interaction, alongside the additional backbone hydrogen bond, places D100 within a contact network involving a backbone hydrogen bond with L104 and a side chain hydrogen bond with H99. It is hypothesized that this D100-centric contact network contributes significantly to stabilization of the H103 out conformation. It must also be noted that movements of histidines 99 and 103 create a large, solvent-accessible void that exposes the proximal face of the heme, which may potentially serve a role in the formation of proximal NO.
Additionally, the loss of the proximal water molecule eliminates the interactions between helix αF and the αF-β1 loop to favor interactions solely within the αF helix. As such, contacts previously anchoring the αF-β1 loop are released, thereby allowing P116 to translate away from the heme and relieves the heme strain imparted by P116. These movements allow for subsequent relaxation of the heme toward planarity [0.312 (FeII) vs. 0.269 (FeII-NO) Å rmsd from planarity] that is a result of a piano hinge-type bending centered about the heme β-δ methine positions (aligned using pyrroles B and C) (Fig. 3B). This flattening involves movement of pyrroles A and D by ∼4° toward the plane of pyrroles B and C. This observation is consistent with the hypothesis that NO-induced scission of the iron–histidine bond results in activation of H-NOX proteins through relaxation of the heme toward planarity.
An important feature in the basal-to-active conformational switch in So H-NOX is loss of the proximal pocket water molecule. This water molecule has also previously been observed in Ns H-NOX and is postulated to be a general feature of H-NOX proteins from facultative anaerobes and sGC (12, 21). Evidence has been provided for this water molecule serving a similar role in sGC, wherein mutation of the proposed water-coordinating residue in sGC, D102, to an alanine drastically affected heme binding and activation of the enzyme. As such, it was reasoned that loss of one of the amino acids that stabilize the proximal pocket water molecule destabilized interactions in the heme region to such an extent that the integrity of the enzyme was severely compromised. In sharp contrast to the D102A mutation, replacement of D102 with asparagine resulted in heme binding and formation of a stable, five-coordinate NO complex. However, activation by NO was drastically reduced, indicating that breakage of the iron–histidine bond and NO activation had been uncoupled (22).
Conformational Changes Associated with NO Binding.
Superimposition of molecules A from the FeII-unligated and FeII-NO structures via their proximal subdomains (residues 95–180) demonstrates that the distal subdomain (residues 1–83) of the FeII-NO structure undergoes a rigid body displacement of ∼2.5 Å that is the result of a ∼4° rotation about the heme with a pivot point located within helix αD (Fig. 5). The pronounced kink within helix αD, located at G70, appears to accommodate this rotation by displacing the N terminus of helix αD by ∼4°. This pivot point is of particular interest because the strictly conserved G70 residue makes van der Waals contact with another conserved glycine, G144, located in helix αG (Figs. 1C and 5). Glycine residues at the interfaces of crossing helices have been observed within polytopic membrane proteins as a means to allow for close helix proximity as well as to provide crossing helices with the ability to sample a wider range of crossing angles without disrupting their interface, neither of which would be possible with larger amino acid side chains at helical interfaces (24, 36). Taken together, it is postulated that H-NOX proteins possess glycine residues at these positions, termed the “glycine hinge,” to maintain the αD-αG helical interface, while allowing for displacement of the distal subdomain about helix αD during NO-induced activation.
Fig. 5.
Structural comparison of FeII and FeII-NO structures. Overlay of FeII (wheat) and FeII-NO (red) (molecules A of the ASU) crystal structures following alignment of the proximal subdomains (residues 95–180) showing displacement of the distal subdomain upon NO binding and breakage of the iron–histidine bond. Conserved glycines (G70 and G144, highlighted in green) in helices αD and αG serve as a hinge point for subdomain rotation within So H-NOX.
Importantly, the degree by which the distal subdomain rotates about the proximal subdomain (∼4°) in formation of the five-coordinate NO structure can be correlated with the degree of pyrrole A and D rotation toward planarity (∼4°). Therefore, upon iron–histidine bond cleavage and release of the proximal pocket restraints distorting the heme, residues in the distal pocket that make van der Waals contact with the distal face of the heme, primarily I5, move with the trajectory of the heme toward planarity. This movement results in displacement of the distal subdomain with respect to the proximal subdomain to yield the active H-NOX conformation. These observations are in agreement with previous structural studies on H-NOX proteins that related the degree of heme distortion to distal subdomain displacement (13, 14, 17).
Rotational displacement of the distal subdomain serves as a probable mechanism for communicating NO binding to the H-NOX protein to associated signaling effector proteins. Conceivably, the effector protein shares an interaction surface with both the distal and proximal H-NOX subdomains in such a manner that following NO binding and distal subdomain rotation, the change in relative subdomain orientation imparts a structural change in the effector protein. In support of this hypothesis, interface mapping in sGC indicates that this interface potentially lies across the distal and proximal subdomains involving the αA and αF helices, as well as the αB-αC and αF-β1 loops (37).
Conclusion
To provide molecular insight into the mechanism of NO-dependent activation of H-NOX proteins, crystal structures of the H-NOX protein from So in the FeII-unligated and both the intermediate six-coordinate and active five-coordinate NO-bound states were determined. Analysis of these structures reveal that NO binding and subsequent cleavage of the iron–histidine bond yield the active H-NOX conformation via several, interconnected structural changes. An integrated heme-helix strain model for NO activation has emerged that couples conformational changes within the signaling αF helix to flattening of the heme and rotational displacement of the distal subdomain.
An integral feature of the heme-helix strain model proposed here (Fig. 6) is the stored potential energy within the porphyrin macrocycle, analogous to a loaded spring, as a result of its deformation from planarity in the unligated structure. In the basal state (shown in gold in Fig. 6), a proximal pocket hydrogen-bonding network linking the signaling helix to a conserved proline in the αF-β1 loop, places this proline in van der Waals contact with the heme, thereby distorting it from planarity. Following NO binding and scission of the iron–histidine bond, several rearrangements in the signaling helix act to stabilize the heme-ligating histidine in an outward conformation. As such, these rearrangements eliminate the proximal pocket hydrogen-bonding network and release the proline from its restricted position, allowing it to translate away from the heme, and subsequently allowing the porphyrin to relax back toward planarity. Residues within the distal pocket contacting the heme opposite to the proline translate heme relaxation to the distal subdomain to drive rotational displacement of the distal subdomain about the proximal subdomain, yielding the active state (shown in red in Fig. 6). This conformational change communicates NO binding to the effector protein via a binding interface that presumably spans the distal and proximal subdomains. Following NO dissociation back to the unligated state, the system is reset, effectively “reloading” the spring and returning the signaling pathway back to the basal state.
Fig. 6.
Heme-helix strain model for NO-induced activation of H-NOX proteins. In the basal, FeII-unligated (gold) state, the heme (orange square) is distorted from planarity from a coordination network in the proximal involving proline 116 (green pentagon) and the heme-ligating histidine 103 (blue pentagon). Upon binding of NO (blue and red circles) the iron–histidine bond is severed, allowing for histidine 103 dissociation and movement of proline 116 away from the heme. These movements result in a more planar heme, structural changes in the signaling helix (αF; pink cylinder) to stabilize H103 in the outward conformation, and rotation of the distal subdomain with respect to the proximal subdomain. The resulting active conformation communicates NO binding to the signaling effector domain (shown as a gray rectangle) through contacts bridging the distal and proximal subdomains.
Materials and Methods
Protein Expression and Purification.
All H-NOX proteins were expressed in the RP523(DE3) Escherichia coli strain (38). Vc H-NOX and Tt H-NOX were purified as described previously (16). All So H-NOX variants were purified using nickel affinity chromatography followed by size-exclusion chromatography (SI Materials and Methods).
Protein Crystallization.
Following gel filtration, all protein manipulation and crystallization procedures were performed in an anoxic glove box (Plas Labs) under an argon/hydrogen (95:5) environment at ∼25 °C with 40–60% humidity. In addition, all samples containing MnPPIX were handled with minimized light exposure or under illumination of a red lamp.
Before crystallization, heme-bound So H-NOX was exchanged into buffer C lacking DTT, fully oxidized (λmax = 403 nm) using 1 to 2 eq of potassium ferricyanide, exchanged into fresh buffer C, and reduced (λmax = 430 nm; FeII) using 1 mM sodium dithionite (final). Residual dithionite was removed by exchanging the protein into fresh buffer C. Crystallization of FeII-unligated H-NOX was performed immediately.
Exhaustive crystallization attempts of WT So H-NOX in the FeII oxidation state were fruitless. WT So H-NOX was submitted to the Surface Entropy Reduction prediction (SERp) server (18), which identified residues 154–156 (QQK), located in the solvent-exposed αG-β1 loop, as candidates for mutation. Substitutions with alanine at all three positions (154AAA) yielded multiple crystal hits following sparse-matrix screening. It should be noted that crystallization attempts of 155AA and 154ASA mutants were unsuccessful. Crystals of FeII-unligated and MnII-unligated 154AAA So H-NOX for diffraction studies were obtained using sitting drop vapor diffusion by equilibrating a 2-μL drop of 1:1 protein/reservoir against a 700-μL reservoir containing 1.6–1.9 M dl-malic acid (pH 7.3). For cryoprotection, 2 μL of mother liquor containing 10% (vol/vol) glycerol was added directly to the drop, and crystals were serial-transferred into mother liquor solution containing 5%, 7.5%, and 10% (vol/vol) glycerol before flash-freezing in liquid nitrogen.
MnIIPPIX-substituted 154AAA So H-NOX was purified in the MnII-unligated state (λmax = 437 nm) and needed no further modification before crystallization. Crystallization was performed as described for 154AAA FeII-unligated So H-NOX.
WT FeII-NO So H-NOX was reduced using the same protocol as for 154AAA FeII-unligated So H-NOX. Immediately before crystallization, reduced protein (25–30 mg/mL) was mixed with 500 μM (final) quick-releasing PROLI-NONOate (t1/2 of ∼5 s at RT), and slow-releasing DETA-NONOate (t1/2 of ∼56 h at RT) NO donors (equal mixture) to ensure immediate formation of the NO complex and to maintain adequate NO levels following drop equilibration, respectively. UV-visible spectra confirmed formation of a five-coordinate nitrosyl complex (λmax = 399 nm). Crystals of WT FeII-NO H-NOX for diffraction studies were crystallized using sitting drop vapor diffusion by equilibrating a 2- or 4-μL drop of 1:1 protein/reservoir against a 700-μL reservoir containing 0.7 M NaH2PO4 and 0.9 M K2HPO4. Cryoprotection was achieved by carefully adding an equal drop volume of mother liquor containing 50% (vol/vol) glycerol. Crystals were then transferred into mother liquor containing 30% (vol/vol) glycerol and flash-frozen in liquid nitrogen.
NO and CO Derivatization.
Crystals of MnII-NO were obtained by transferring cryoprotected MnII-unligated crystals into 10 μL of cryoprotectant in a depression well. Crystals were then covered in 700 μL of silicon oil. Using a gas-tight syringe (Hamilton), 200 μL of anaerobic cryoprotectant containing 2 mM PROLI-NONOate (Cayman Chemical) was added under the layer of oil. Crystals were allowed to incubate for 15–45 min before freezing in liquid nitrogen.
FeII-CO crystals were generated using the same protocol for NO derivatization, with the exception that CO-saturated cryoprotectant was used in place of the NO solution. CO-saturated cryoprotectant was generated by sparging the headspace of a septum-sealed Reacti-Vial (Pierce) containing anaerobic cryoprotectant with CO (99.99%; Praxair, Inc.) for 5–15 min.
X-Ray Data Collection and Structure Refinement.
X-ray data were collected using synchrotron radiation at beamline 8.2.1, 8.2.2, or 8.3.1 at the Advanced Light Source, Lawrence Berkeley National Laboratory. Diffraction images were collected at 100 K at a wavelength of λ = 1.00 or 1.11 Å using 0.5° to 1.0° oscillations per frame. Data were indexed and integrated using XDS (39) and scaled using Aimless (40) from the CCP4 program suite (41, 42). Phasing of the FeII data was carried out by molecular replacement with the program Phaser (43) using a CHAINSAW (44)-prepared model of Ns H-NOX [Protein Data Bank (PDB) ID code 3TF8] (12) that was further split into two subdomains (residues 1–83 and 84–180). Note that the NMR structure of So H-NOX (PDB ID code 2KII) (13) did not yield a molecular replacement solution; thus, Ns H-NOX was used. The placed model was then subjected to density modification using Parrot (45), followed by extensive manual model building using the program Coot (26). The entire model was rebuilt using a combination of outputs from Buccaneer (46) and ARP/wARP (47). Phasing of the MnII-unligated structure was performed using molecular replacement with the FeII-unligated structure as the search model. The FeII-CO and MnII-NO structures were solved by rigid body refinement of the parent structures, FeII-unligated and MnII-unligated, respectively, against the CO- and NO-derivatized X-ray datasets. Phasing of the FeII-NO structure was achieved using molecular replacement with two subdomains (residues 1–83 and 84–180) from the FeII structure as the search models. Gas ligands were initially placed into overlapping Fo-Fc, 2mFo-DFc, and Fo-Fo densities with an occupancy of 0.5 and temperature factors approximately equal to the average of the four pyrrole nitrogen atoms. Ligand occupancy, position, and B factors were refined simultaneously using only loose distance restraints between the porphyrin metal and bonded gas atom. All structures were refined using PHENIX (48) with TLS refinement parameters incorporated. MnPPIX (MNH) restraints were generated using eLBOW within the PHENIX suite (49). Stereochemical properties were assessed by MOLPROBITY (50). Data collection and refinement statistics are listed in Table 1. Coordinates are deposited in the Research Collaboratory for Structural Bioinformatics (RCSB) PDB as entries 4U99 (FeII-unligated 154AAA So H-NOX), 4U9B (WT FeII-NO So H-NOX), 4U9G (FeII-CO 154AAA So H-NOX), 4U9J (MnII-unligated 154AAA So H-NOX), and 4U9K (MnII-NO 154AAA So H-NOX).
Supplementary Material
Acknowledgments
We thank the 8.2.1, 8.2.2, and 8.3.1 beamline scientists at Lawrence Berkeley National Laboratory for use of equipment and technical assistance. We are grateful to Dr. Peter Goldman and Prof. Catherine M. Drennan at the Massachusetts Institute of Technology for their assistance and use of anaerobic, high-throughput crystallization screening equipment. Finally, we thank current and former members of the M.A.M. and J.K. laboratories (particularly Dr. Michael Winter, Dr. Tiago Barros, Dr. Jonathan Winger, Dr. Christine M. Phillips-Piro, and Dr. Jeff Iwig) for frequent helpful discussions and their invaluable insight. This study was supported in part by the American Heart Association (Western States Affiliate) Predoctoral Fellowship Award 11PRE7370086 (to M.A.H.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Coordinates are deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank, www.rcsb.org/pdb/home/home.do, as entries 4U99 (FeII-unligated So H-NOX), 4U9B (WT FeII-NO So H-NOX), 4U9G (FeII-CO 154AAA So H-NOX), 4U9J (MnII-unligated 154AAA So H-NOX), and 4U9K (MnII-NO 154AAA So H-NOX).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416936111/-/DCSupplemental.
References
- 1.Ignarro LJ, Cirino G, Casini A, Napoli C. Nitric oxide as a signaling molecule in the vascular system: An overview. J Cardiovasc Pharmacol. 1999;34(6):879–886. doi: 10.1097/00005344-199912000-00016. [DOI] [PubMed] [Google Scholar]
- 2.Bredt DS, Snyder SH. Nitric oxide, a novel neuronal messenger. Neuron. 1992;8(1):3–11. doi: 10.1016/0896-6273(92)90104-l. [DOI] [PubMed] [Google Scholar]
- 3.Denninger JW, Marletta MA. Guanylate cyclase and the .NO/cGMP signaling pathway. Biochim Biophys Acta. 1999;1411(2-3):334–350. doi: 10.1016/s0005-2728(99)00024-9. [DOI] [PubMed] [Google Scholar]
- 4.Russwurm M, Koesling D. NO activation of guanylyl cyclase. EMBO J. 2004;23(22):4443–4450. doi: 10.1038/sj.emboj.7600422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poulos TL. Soluble guanylate cyclase. Curr Opin Struct Biol. 2006;16(6):736–743. doi: 10.1016/j.sbi.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Derbyshire ER, Marletta MA. Structure and regulation of soluble guanylate cyclase. Annu Rev Biochem. 2012;81:533–559. doi: 10.1146/annurev-biochem-050410-100030. [DOI] [PubMed] [Google Scholar]
- 7.Stone JR, Marletta MA. Spectral and kinetic studies on the activation of soluble guanylate cyclase by nitric oxide. Biochemistry. 1996;35(4):1093–1099. doi: 10.1021/bi9519718. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Y, Brandish PE, Ballou DP, Marletta MA. A molecular basis for nitric oxide sensing by soluble guanylate cyclase. Proc Natl Acad Sci USA. 1999;96(26):14753–14758. doi: 10.1073/pnas.96.26.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballou DP, Zhao Y, Brandish PE, Marletta MA. Revisiting the kinetics of nitric oxide (NO) binding to soluble guanylate cyclase: The simple NO-binding model is incorrect. Proc Natl Acad Sci USA. 2002;99(19):12097–12101. doi: 10.1073/pnas.192209799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin E, Berka V, Sharina I, Tsai A-L. Mechanism of binding of NO to soluble guanylyl cyclase: Implication for the second NO binding to the heme proximal site. Biochemistry. 2012;51(13):2737–2746. doi: 10.1021/bi300105s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pellicena P, Karow DS, Boon EM, Marletta MA, Kuriyan J. Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases. Proc Natl Acad Sci USA. 2004;101(35):12854–12859. doi: 10.1073/pnas.0405188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma X, Sayed N, Beuve A, van den Akker F. NO and CO differentially activate soluble guanylyl cyclase via a heme pivot-bend mechanism. EMBO J. 2007;26(2):578–588. doi: 10.1038/sj.emboj.7601521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erbil WK, Price MS, Wemmer DE, Marletta MA. A structural basis for H-NOX signaling in Shewanella oneidensis by trapping a histidine kinase inhibitory conformation. Proc Natl Acad Sci USA. 2009;106(47):19753–19760. doi: 10.1073/pnas.0911645106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olea C, Jr, Herzik MA, Jr, Kuriyan J, Marletta MA. Structural insights into the molecular mechanism of H-NOX activation. Protein Sci. 2010;19(4):881–887. doi: 10.1002/pro.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iyer LM, Anantharaman V, Aravind L. Ancient conserved domains shared by animal soluble guanylyl cyclases and bacterial signaling proteins. BMC Genomics. 2003;4(1):5. doi: 10.1186/1471-2164-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karow DS, et al. Spectroscopic characterization of the soluble guanylate cyclase-like heme domains from Vibrio cholerae and Thermoanaerobacter tengcongensis. Biochemistry. 2004;43(31):10203–10211. doi: 10.1021/bi049374l. [DOI] [PubMed] [Google Scholar]
- 17.Olea C, Boon EM, Pellicena P, Kuriyan J, Marletta MA. Probing the function of heme distortion in the H-NOX family. ACS Chem Biol. 2008;3(11):703–710. doi: 10.1021/cb800185h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldschmidt L, Cooper DR, Derewenda ZS, Eisenberg D. Toward rational protein crystallization: A Web server for the design of crystallizable protein variants. Protein Sci. 2007;16(8):1569–1576. doi: 10.1110/ps.072914007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jentzen W, Ma J-G, Shelnutt JA. Conservation of the conformation of the porphyrin macrocycle in hemoproteins. Biophys J. 1998;74(2 Pt 1):753–763. doi: 10.1016/S0006-3495(98)74000-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran R, Boon EM, Marletta MA, Mathies RA. Resonance Raman spectra of an O2-binding H-NOX domain reveal heme relaxation upon mutation. Biochemistry. 2009;48(36):8568–8577. doi: 10.1021/bi900563g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winter MB, Herzik MA, Jr, Kuriyan J, Marletta MA. Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain. Proc Natl Acad Sci USA. 2011;108(43):E881–E889. doi: 10.1073/pnas.1114038108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baskaran P, Heckler EJ, van den Akker F, Beuve A. Aspartate 102 in the heme domain of soluble guanylyl cyclase has a key role in NO activation. Biochemistry. 2011;50(20):4291–4297. doi: 10.1021/bi2004087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harding MM, Nowicki MW, Walkinshaw MD. Metals in protein structures: A review of their principal features. Crystallogr Rev. 2010;16(4):247–302. [Google Scholar]
- 24.Russ WP, Engelman DM. The GxxxG motif: A framework for transmembrane helix-helix association. J Mol Biol. 2000;296(3):911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 25.Dierks EA, et al. Demonstration of the role of scission of the proximal histidine−iron bond in the activation of soluble guanylyl cyclase through metalloporphyrin substitution studies. J Am Chem Soc. 1997;119(31):7316–7323. [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Price MS, Chao LY, Marletta MA. Shewanella oneidensis MR-1 H-NOX regulation of a histidine kinase by nitric oxide. Biochemistry. 2007;46(48):13677–13683. doi: 10.1021/bi7019035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawson DM, Stevenson CEM, Andrew CR, Eady RR. Unprecedented proximal binding of nitric oxide to heme: Implications for guanylate cyclase. EMBO J. 2000;19(21):5661–5671. doi: 10.1093/emboj/19.21.5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrew CR, Green EL, Lawson DM, Eady RR. Resonance Raman studies of cytochrome c′ support the binding of NO and CO to opposite sides of the heme: Implications for ligand discrimination in heme-based sensors. Biochemistry. 2001;40(13):4115–4122. doi: 10.1021/bi0023652. [DOI] [PubMed] [Google Scholar]
- 30.Wu G, Liu W, Berka V, Tsai A-L. The selectivity of Vibrio cholerae H-NOX for gaseous ligands follows the “sliding scale rule” hypothesis. Ligand interactions with both ferrous and ferric Vc H-NOX. Biochemistry. 2013;52(52):9432–9446. doi: 10.1021/bi401408x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cary SPL, Winger JA, Marletta MA. Tonic and acute nitric oxide signaling through soluble guanylate cyclase is mediated by nonheme nitric oxide, ATP, and GTP. Proc Natl Acad Sci USA. 2005;102(37):13064–13069. doi: 10.1073/pnas.0506289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Derbyshire ER, et al. Characterization of two different five-coordinate soluble guanylate cyclase ferrous-nitrosyl complexes. Biochemistry. 2008;47(12):3892–3899. doi: 10.1021/bi7022943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernhoff NB, Derbyshire ER, Marletta MA. A nitric oxide/cysteine interaction mediates the activation of soluble guanylate cyclase. Proc Natl Acad Sci USA. 2009;106(51):21602–21607. doi: 10.1073/pnas.0911083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith BC, Marletta MA. Mechanisms of S-nitrosothiol formation and selectivity in nitric oxide signaling. Curr Opin Chem Biol. 2012;16(5-6):498–506. doi: 10.1016/j.cbpa.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawson DM, Stevenson CEM, Andrew CR, George SJ, Eady RR. A two-faced molecule offers NO explanation: The proximal binding of nitric oxide to haem. Biochem Soc Trans. 2003;31(Pt 3):553–557. doi: 10.1042/bst0310553. [DOI] [PubMed] [Google Scholar]
- 36.Eilers M, Shekar SC, Shieh T, Smith SO, Fleming PJ. Internal packing of helical membrane proteins. Proc Natl Acad Sci USA. 2000;97(11):5796–5801. doi: 10.1073/pnas.97.11.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Underbakke ES, Iavarone AT, Marletta MA. Higher-order interactions bridge the nitric oxide receptor and catalytic domains of soluble guanylate cyclase. Proc Natl Acad Sci USA. 2013;110(17):6777–6782. doi: 10.1073/pnas.1301934110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winter MB, Woodward JJ, Marletta MA. Cytochrome P450 Protocols, Methods in Molecular Biology. Humana Press; Totowa, NJ: 2013. pp. 95–106. [Google Scholar]
- 39.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 7):1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morin A, et al. Collaboration gets the most out of software. eLife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 1):32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stein N. CHAINSAW: A program for mutating pdb files used as templates in molecular replacement. J Appl Crystallogr. 2008;41:641–643. [Google Scholar]
- 45.Cowtan K. Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cowtan K. Completion of autobuilt protein models using a database of protein fragments. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):328–335. doi: 10.1107/S0907444911039655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3(7):1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moriarty NW, Grosse-Kunstleve RW, Adams PD. electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 10):1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.