Significance
Long-range organ-to-organ communications are important for the coordination of development and environmental adaptation in multicellular organisms, particularly plants that continuously produce postembryonic lateral organs in various environmental conditions. The substance of homeostatic regulation of organ development via long-distance signals has not yet been identified, however. Legumes use an autoregulatory negative-feedback system involving root–shoot communication to maintain optimal numbers of nodules by systemically suppressing nodulation. We show that a transcription factor, NODULE INCEPTION (NIN), an essential inducer for nodule primordium formation, directly activates genes encoding small peptides that act as root-derived long-distance mobile signals, leading to repression of endogenous NIN though the root–shoot communication and resulting in systemic suppression of nodulation. We demonstrate that an autoregulatory negative-feedback loop homeostatically regulates nodule production via this long-range signaling.
Abstract
Autoregulatory negative-feedback loops play important roles in fine-balancing tissue and organ development. Such loops are composed of short-range intercellular signaling pathways via cell–cell communications. On the other hand, leguminous plants use a long-distance negative-feedback system involving root–shoot communication to control the number of root nodules, root lateral organs that harbor symbiotic nitrogen-fixing bacteria known as rhizobia. This feedback system, known as autoregulation of nodulation (AON), consists of two long-distance mobile signals: root-derived and shoot-derived signals. Two Lotus japonicus CLAVATA3/ENDOSPERM SURROUNDING REGION (CLE)-related small peptides, CLE ROOT SIGNAL1 (CLE-RS1) and CLE-RS2, function as root-derived signals and are perceived by a shoot-acting AON factor, the HYPERNODULATION ABERRANT ROOT FORMATION1 (HAR1) receptor protein, an ortholog of Arabidopsis CLAVATA1, which is responsible for shoot apical meristem homeostasis. This peptide–receptor interaction is necessary for systemic suppression of nodulation. How the onset of nodulation activates AON and how optimal nodule numbers are maintained remain unknown, however. Here we show that an RWP-RK–containing transcription factor, NODULE INCEPTION (NIN), which induces nodule-like structures without rhizobial infection when expressed ectopically, directly targets CLE-RS1 and CLE-RS2. Roots constitutively expressing NIN systemically repress activation of endogenous NIN expression in untransformed roots of the same plant in a HAR1-dependent manner, leading to systemic suppression of nodulation and down-regulation of CLE expression. Our findings provide, to our knowledge, the first molecular evidence of a long-distance autoregulatory negative-feedback loop that homeostatically regulates nodule organ formation.
Long-distance organ-to-organ communications are generally critical for coordinating development and environmental adaptation in multicellular organisms, particularly plants that continuously produce postembryonic organs in various environmental conditions (1, 2). Autoregulatory negative-feedback loops play important roles in fine-balancing tissue and organ development. Such feedback loops include short-range intercellular signaling via cell–cell communication. In Arabidopsis, shoot apical meristem (SAM) regulation involves a well-characterized short-range negative-feedback loop that maintains the homeostasis of plant organ development (3–6). The molecular substance of long-distance negative-feedback loops that homeostatically regulate organ production and development remains largely unknown, however.
Leguminous plants use long-distance autoregulatory negative-feedback systems involving root–shoot communications to control the number of root nodules, symbiotic root lateral organs formed as consequence of successful interaction with nitrogen-fixing bacteria, collectively known as rhizobia. This nodule symbiosis is beneficial to host plants, because rhizobia accommodated in nodules convert atmospheric nitrogen to ammonium, a usable nitrogen source for host plants; however, excessive nodulation interferes with plant growth, likely because of the high energy cost of nitrogen fixation. Thus, legumes have developed negative-feedback pathways to maintain total nodule numbers and mass in a single plant. Autoregulation of nodulation (AON) is a major long-distance negative-feedback pathway, consisting of root-derived and shoot-derived long-distance mobile signals, which restricts the production of root nodules (7). The root-derived signal generated during early nodulation processes is translocated to the shoot, where it activates shoot-acting AON factors to produce the shoot-derived inhibitor, which is transported down to the root and inhibits nodulation. Thus, AON activated by rhizobial infection systemically prevents nodule formation stimulated by subsequent infection.
Lotus japonicus CLAVATA3/ENDOSPERM SURROUNDING REGION (CLE) peptides, CLE ROOT SIGNAL1 (CLE-RS1) and CLE-RS2, act as root-derived mobile signals and are perceived by a shoot-acting AON factor, the HYPERNODULATION ABERRANT ROOT FORMATION1 (HAR1) receptor protein kinase, an ortholog of Arabidopsis CLAVATA1 (CLV1), which is involved in the short-range feedback loops regulating SAM homeostasis and restricts meristem sizes through interaction with CLV3 peptide (6, 8–10). The interaction of root-derived CLE peptides with the shoot-acting receptor is required for systemic suppression of nodulation.
AON starts with the rhizobia-induced up-regulation of CLE genes encoding the root-derived mobile signals (11). Although the transcriptional regulation of CLE genes is important for the elicitation and attenuation of AON to maintain optimal nodule numbers, how CLE-RS genes are activated, and the molecular mechanism through which optimal nodule numbers are maintained, have not yet been elucidated. Caetano-Anollés and Gresshoff (12) have shown that subepidermal cell division at early stages of nodule organogenesis is required for the elicitation of AON through approach-graft experiments between WT soybean and symbiotic mutants exhibiting different phenotypes at early stages of nodulation. Expression of Medicago truncatula CLE genes (MtCLE12 and MtCLE13) and of soybean RIC1 and RIC2, which systemically suppress nodule primordium formation similar to CLE-RS1 and CLE-RS2, is associated with nodule primordia (11, 13, 14). These results imply that expression of CLE genes is up-regulated by a transcription factor that is necessary for nodule primordium formation.
NODULE INCEPTION (NIN) encodes an RWP-RK–containing transcription factor essential for cortical cell division, an initial step in nodule organogenesis (15, 16). Ectopic expression of NIN induces nodule primordium-like structures in the absence of rhizobia (17). We postulated that CLE-RS1 and CLE–RS2 expression is associated with NIN activity in L. japonicus, and investigated the activity of NIN in the systemic suppression of nodulation in response to rhizobial infection through activation of AON.
Results
Requirement of NIN for CLE-RS1 and CLE-RS2 Expression.
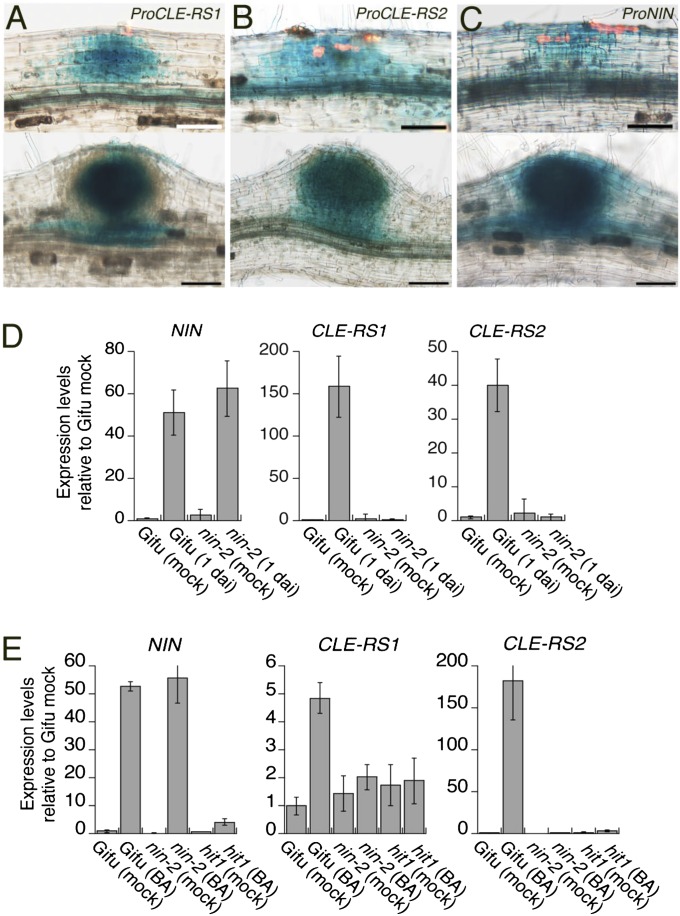
We investigated the spatial expression patterns of CLE-RS1, CLE-RS2, and NIN in L. japonicus using GUS reporter constructs (17). A 5.5-kb fragment from the putative translation initiation codon of either CLE-RS1 or CLE-RS2 was inserted upstream of the GUS gene. These reporters were introduced into L. japonicus roots using the Agrobacterium rhizogenes-mediated root transformation method. Transformed roots were inoculated with DsRed-labeled Mesorhizobium loti, a bacterial symbiont of L. japonicus, to visualize infection events. GUS expression from the promoters of CLE-RS1 and CLE-RS2 was detected in nodule primordia generated beneath root hairs infected by M. loti, as well as in developing nodules (Fig. 1 A and B). This expression coincided with that controlled by the NIN promoter during nodule primordium formation (Fig. 1C). Thus, the transcriptional activities of CLE-RS1, CLE-RS2, and NIN are coregulated and associated with the development of nodule primordia.
Fig. 1.
Expression analysis of CLE-RS1, CLE-RS2, and NIN. (A–C) GUS expression controlled by the CLE-RS1 (A), CLE-RS2 (B), and NIN (C) promoters in nodule primordia (Upper) and developing nodules (Lower). DsRed fluorescence in the upper panels represents infection foci. (Scale bars: 50 µm in Upper; 100 µm in Lower). (D and E) RT-PCR analyses of gene expression in roots. (D) Gifu B-129 (WT) and nin-2 were inoculated with (1 dai) or without (mock) M. loti. (E) Gifu B-129, nin-2, and hit1 were incubated in liquid media supplemented with (BA) or without (mock) 50 nM benzyladenin for 20 h. Expression levels were normalized to polyubiquitin expression. Data are mean ± SD of three biological repeats.
We examined the possible requirement of NIN for the activation of CLE-RS1 and CLE-RS2 in response to rhizobial infection and exogenous cytokinin, which triggers the formation of nodule-like structures in the absence of rhizobia (18). Cytokinin signaling is involved in nodule organogenesis after rhizobial infection (19–21). Expression of CLE-RS genes as well as NIN was induced in WT roots within 1 d after inoculation (dai) with M. loti (Fig. 1D). At this point, the symbiotic epidermal response can be recognized as a deformation of root hairs, with cortical cell division not yet occurring. The response of the CLE genes to M. loti inoculation occurred before cortical cell division, similar to that of NIN. Cytokinin also induced expression of the CLE genes and of NIN within 20 h (Fig. 1E). Although the nin-2 mutation did not affect expression of the NIN carrying the mutation after inoculation and cytokinin treatment, expression of both CLE genes was not up-regulated in the mutants. The induction of CLE genes and NIN in response to cytokinin was diminished in loss-of-function mutants (hit1) of the LHK1 cytokinin receptor. The findings indicate that CLE-RS1 and CLE-RS2 expression is mediated by cytokinin signaling, and that NIN expression is a prerequisite for infection and cytokinin-induced CLE expression, consistent with previous observations on the expression of MtCLE12 and MtCLE13 (22).
CLE-RS1 and CLR-RS2 Are Direct Targets of NIN.
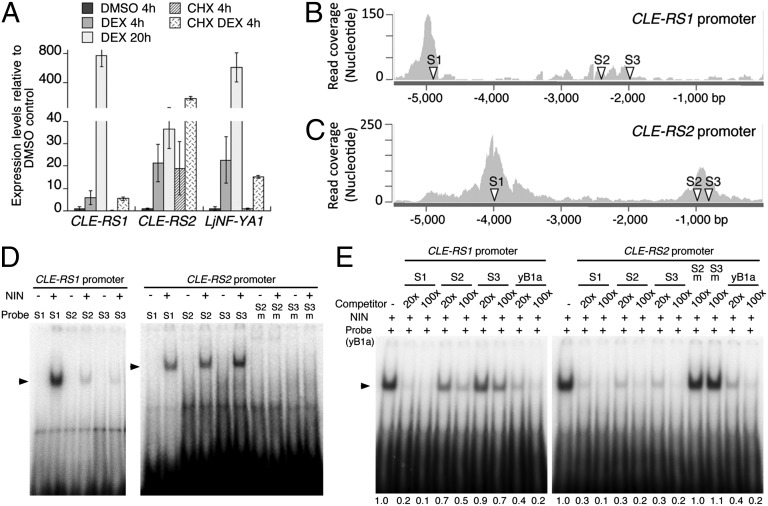
We next examined whether NIN actively induces the expression of CLE-RS1 and CLE-RS2. NIN fused to the glucocorticoid receptor (NIN-GR) was overexpressed under the CaMV35S promoter in L. japonicus roots (17). Expression of CLE-RS1 and CLE-RS2 was induced within 4 h of treatment with dexamethasone (DEX) (Fig. 2A), followed by steady increases in transcript levels over the next 16 h, indicating that NIN expression is sufficient to activate the CLE genes. Cycloheximide (CHX) together with DEX did not repress the DEX-inducible expression, demonstrating that de novo protein synthesis is not required for activation of the CLE genes. The expression pattern was similar to that of LjNF-YA1, a direct target of NIN involved in the regulation of cortical cell division (17). These results suggest that NIN might directly target CLE-RS1 and CLE-RS2. Supporting this notion, co-overexpression of LjNF-YA1 and LjNF-YB1, which stimulates cell division downstream of NIN (17), did not up-regulate expression of the CLE genes (Fig. S1).
Fig. 2.
NIN directly targets CLE-RS1 and CLE-RS2. (A) RT-PCR analysis of gene expression in Pro35S-NIN-GR roots. Roots were preincubated in liquid medium supplemented with CHX or DMSO for 30 min before DEX addition, and then incubated further for the indicated durations. Expression levels were normalized to polyubiquitin expression. Data are mean ± SD of three biological repeats. (B and C) Read coverage of the CLE-RS1 (B) and CLE-RS2 (C) promoters obtained by ChIP-seq analysis. Arrowheads (S1–S3) indicate positions of possible NBSs. (D) NIN binding with oligo-DNAs containing possible NBSs. 32P-labeled probes were incubated with NIN(520-878)-myc (+) or in vitro translation products without template (−). The CLE-RS2 promoter S2m and S3m contain mutations in S2 and S3, respectively (Fig. S4A). (E) Competition analyses. NIN(520-878)-myc and a labeled probe, NBS-yB1a (yB1a), were incubated with 20-fold and 100-fold amounts of competitors. Relative intensities of shifted bands are shown at the bottom.
We performed ChIP-seq analysis to identify NIN-binding regions in L. japonicus genomic nucleotide sequences. NIN tagged with myc epitopes was expressed under the control of an L. japonicus polyubiquitin promoter (ProLjUb-NIN-myc) in roots (17). As a control, roots were transformed with an empty vector. DNA fragments were coimmunoprecipitated with polyclonal anti-myc antibodies and sequenced with Illumina Hi-seq1000. Sequence reads were enriched around NIN-binding nucleotide sequences (NBSs) that were identified in a previous study (17), indicating that our ChIP-seq analysis successfully detected NIN-binding regions in the genomic sequences (Fig. S2A and Dataset S1). Read coverage in the promoter regions of CLE-RS1 and CLE-RS2 showed that two separate regions were enriched in the immunoprecipitate (−5,193 to −4,631 bp and −2,368 to −2,187 bp from the putative initiation codon of CLE-RS1, and −4,358 to −3,545 bp and −1,052 to −709 bp from that of CLE-RS2) (Fig. 2 B and C). ChIP-PCR analysis confirmed the enrichment in these regions (Fig. S2 B and C). These results indicate that NIN binds to the CLE-RS1 and CLE-RS2 promoters in vivo.
The binding of NIN to the promoter regions was further confirmed by electrophoresis mobility shift assay (EMSA). We synthesized eight DNA fragments covering the promoter regions identified by ChIP (Fig. S3A). Specific bands were detected when CLE-RS1 probes (2, 4, 7) and CLE-RS2 probes (2, 5, 6) were incubated with the carboxyl-terminal half of the NIN recombinant protein, NIN(520-878)-myc (17), which contains the RWP-RK domain responsible for DNA binding (Fig. S3B). Supershift analyses and competition assays confirmed that NIN bound specifically with these promoter regions (Fig. S3 C and D).
We also searched nucleotide sequences similar to that of NBS-yB1a in the promoter fragments bound by NIN. NBS-yB1a is an NBS identified in the LjNF-YB1 promoter (17). We found possible NBSs in each DNA fragment (Fig. 2 B and C and Fig. S4 A and B). NIN bound with oligo-DNA probes containing these possible NBSs (Fig. 2D). Competition analyses showed that double-stranded oligo-DNAs containing the possible NBSs reduced the intensities of shifted bands caused by binding of the probe NBS-yB1a with the NIN protein (Fig. 2E). Mutations in possible NBSs, CLE-RS2 S2 and S3, diminished the binding affinity with the NIN protein (Fig. 2 D and E and Fig. S4A). CLE-RS1 S1 and CLE-RS2 S1, S2, and S3 competed out the shifted band comparably to or more efficiently than NBS-yB1a.
To examine whether NBSs identified in the CLE-RS1 and CLE-RS2 promoters are involved in NIN-mediated transcriptional activation, we inserted promoter fragments containing NBSs upstream of the CaMV35S minimal promoter, followed by the GUS reporter (Fig. S4C). These reporter constructs were introduced into Nicotiana benthamiana leaves by Agrobacterium infiltration, and GUS activities were quantified. These promoter fragments activated GUS expression when coexpressed with NIN, whereas GUS activity was significantly reduced by NBS mutations in each promoter fragments (Fig. S4D). Moreover, these reporters were expressed in nodule primordia depending on the NBSs in each promoter fragment (Fig. S4E). These results indicate that the NBSs are required for activation of the reporter. Taken together, our data demonstrate that NIN directly targets CLE-RS1 and CLE-RS2 promoters to activate gene expression.
NIN Ectopic Expression Systemically Suppresses Nodulation Through AON.
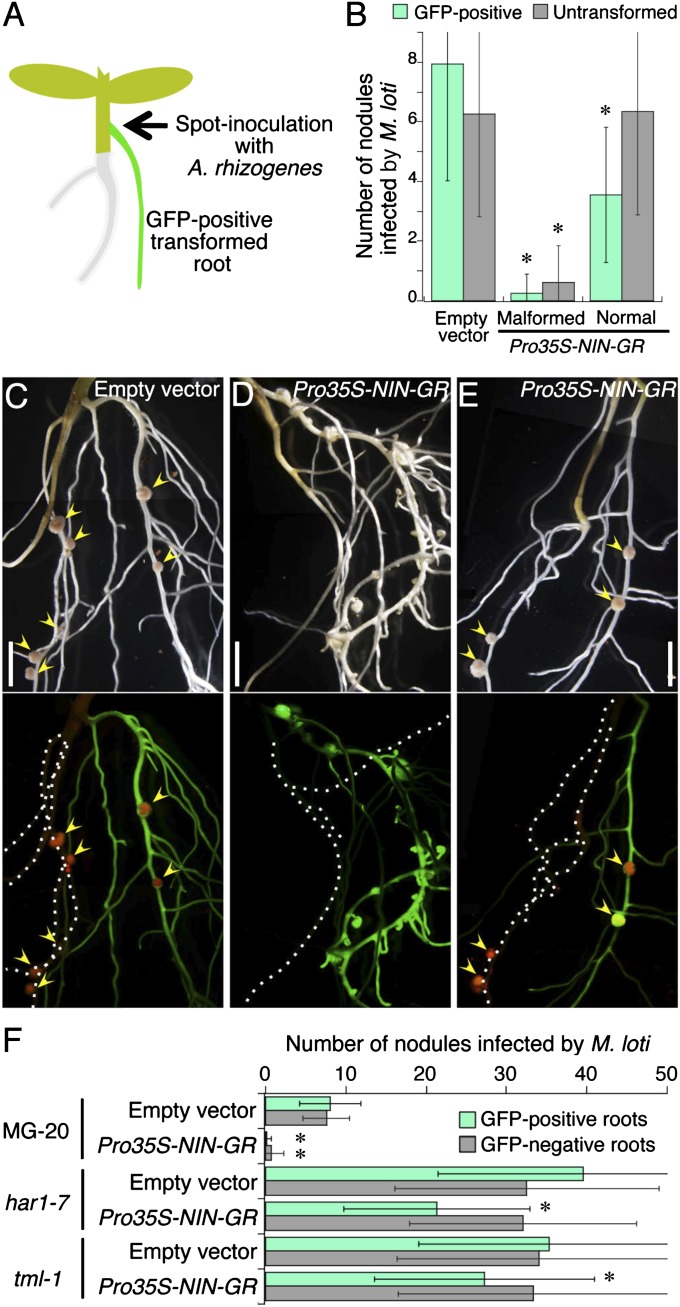
To test a hypothesis that NIN ectopic expression systemically suppresses nodulation by promoting the production of CLE-RS1 and CLE-RS2 peptides, we spot-inoculated hypocotyls of intact seedlings with A. rhizogenes strains containing a binary vector carrying Pro35S-NIN-GR or an empty control vector (Fig. 3A). We used the CaMV35S promoter to constitutively overexpress NIN-GR in transformed roots. The posttranslational induction system was used to avoid inhibiting the growth of roots that were strongly affected by NIN overexpression before inoculation with rhizobia (see below). Transformed roots were identified by GFP fluorescence, which served as a transformation marker. Plants that generated hairy roots with GFP fluorescence were treated with DEX for 5 d to activate the NIN-mediated pathway, and then inoculated with DsRed-labeled M. loti in soil supplemented with DEX. Nodules that formed on untransformed and GFP-positive transformed roots were counted separately to assess the systemic effect of NIN ectopic expression at 14 dai.
Fig. 3.
Systemic inhibition of nodulation by ectopic expression of NIN. (A) Illustration of the Agrobacterium spot-inoculation method. (B) Number of nodules formed on untransformed or GFP-positive roots. Roots were transformed with Pro35S-NIN-GR or the empty vector by the spot-inoculation method. Data are mean ± SD. More than 50 plants were analyzed in each individual experiment. Asterisks indicate significant differences from controls according to the Student t test (P < 0.01). (C–E) Nodulation on untransformed roots and GFP-positive roots generated by spot-inoculation with A. rhizogenes carrying either the empty vector (C) or the binary vector with Pro35S-NIN-GR (D and E). A Pro35S-NIN-GR root in (D) displays the malformed structure. (Upper) Composite bright-field images. (Lower) Corresponding fluorescent images with GFP as a transformation marker for roots and DsRed expressed in M. loti. Arrowheads indicate infected nodules. Broken lines in the lower panels represent GFP-negative roots. (Scale bars: 5 mm.) (F) Number of nodules on GFP-negative and -positive roots that were transformed with the empty vector or Pro35S-NIN-GR. MG-20, har1-7, and tml-1 were inoculated with M. loti after 5 d of DEX treatment. Nodule numbers were counted at 14 dai. Data are mean ± SD. More than 60 plants were analyzed in each experiment. Asterisks indicate significant differences from empty vector controls according to the Student t test (P < 0.01).
Roots that were transformed with Pro35S-NIN-GR exhibited two morphological phenotypes: malformed structures (Fig. 3D and Fig. S5B) and an apparently normal architecture (Fig. 3E and Fig. S5C). These malformed roots were found regardless of M. loti inoculation and had enlarged root tips owing to extra cell divisions (17). Plants that generated only Pro35S-NIN-GR roots with normal architecture produced nodules on untransformed roots as well as on GFP-positive transformed roots, as was also seen in empty vector control plants (Fig. 3 B, C, and E). In contrast, nodulation in plants with malformed Pro35S-NIN-GR roots was significantly suppressed in both untransformed and GFP-positive roots (Fig. 3 B and D). RT-PCR analysis showed that CLE-RS1 and CLE-RS2 were more strongly expressed in malformed roots compared with normal roots in the absence of rhizobia (Fig. S5A). Our results indicate that NIN ectopic expression systemically inhibits nodulation, an effect correlated with CLE expression levels.
HAR1 and TOO MUCH LOVE (TML) are required for AON to inhibit nodulation. TML encodes an F-box protein that acts downstream of HAR1 in the root (23). CLE-RS1 and CLE-RS2 suppress nodulation in an HAR1- and TML-dependent fashion (11, 23); har1 and tml mutants produce excessive numbers of nodules because AON is not operational. To investigate whether the systemic effects of NIN-GR are related to activation of AON, we introduced Pro35S-NIN-GR into roots of har1-7 and tml-1 mutants, as well as WT plants. Although Agrobacterium spot-inoculation is a reliable method for analyzing systemic effects, it has difficulty maintaining untransformed roots during the transformation procedure; thus, thereafter, roots were transformed by the standard A. rhizogenes-mediated root transformation method (Fig. S5D). Plants were inoculated with A. rhizogenes after roots were cut off from hypocotyls. Several GFP-positive and GFP-negative roots were generated from different positions of the same hypocotyl. Transformed roots were distinguishable by the fluorescence of the GFP marker. Most GFP-negative roots were untransformed.
The mean numbers of nodules formed on GFP-positive Pro35S-NIN-GR roots and on GFP-negative roots were significantly reduced compared with control plants whose roots were transformed with the empty vector (Fig. 3F and Fig. S5E), confirming the systemic inhibitory effect. This effect was absent in the har1-7 and tml-1 mutant backgrounds (Fig. 3F and Fig. S5 F and G). These findings indicate that NIN-GR activates AON to systemically inhibit nodulation. The number of nodules in GFP-positive Pro35S-NIN-GR roots tended to be reduced in these mutants, likely related to local inhibitory effects of NIN expression on rhizobial infection (24). Alternatively, morphological alteration of roots caused by NIN overexpression might affect infection frequency. The former possibility seems more likely, given that this tendency was observed in Pro35S-NIN-GR roots with normal architecture as well (Fig. 3B), likely because of constitutive expression of the exogenous NIN at levels that did not affect the root structure.
A Negative-Feedback Loop Attenuates NIN Activity and Suppresses Nodulation.
Based on the foregoing results supporting NIN’s systemic function in controlling nodulation, we focused our subsequent analyses on NIN–CLE interactions in this long-distance signaling pathway, specifically analyzing roots influenced by the systemic effects of NIN constitutive expression. We investigated the expression of CLE-RS genes in these roots. Because NIN systemically suppresses nodulation, we assumed that the systemic effect represses CLE-RS1 and CLE-RS2, which are expressed in nodule primordia. To examine the effects of NIN on CLE-RS2 expression, we cointroduced Pro35S-NIN-GR with ProCLE-RS2-GUS into roots. Shoots of 3-d-old seedlings were inoculated with an A. rhizogenes strain carrying the ProCLE-RS2-GUS binary vector and a strain carrying either Pro35S-NIN-GR or its empty vector. The former vector carries Pro35S-GFP as a transformation marker, and the latter two vectors carry Pro35S-DsRed. We selected plants exhibiting GFP and DsRed fluorescence in different roots or the same root (Fig. S6A) and inoculated them with M. loti after 5 d of treatment with DEX. GUS expression was examined in DsRed-negative roots at 4 dai.
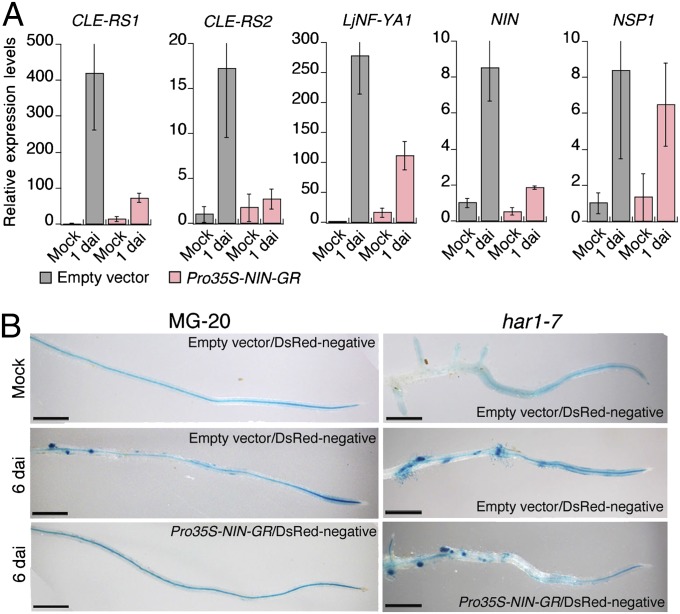
In control plants (whose roots were transformed with ProCLE-RS2-GUS and the empty vector), GUS expression with spot-staining patterns was detected on DsRed-negative roots (Fig. S6B). GUS staining in the stele was occasionally observed regardless of the presence or absence of rhizobia. Divided cortical cells were found in 55% of the GUS spots in the DsRed-negative control roots (Fig. S6 C–F). There were fivefold fewer GUS spots in DsRed-negative roots of plants with malformed Pro35S-NIN-GR roots compared with DsRed-negative control roots (Fig. S6E). The proportion of GUS spots accompanied by cortical cell divisions was twofold lower than that in the control roots (Fig. S6F). These findings suggest that NIN ectopic expression systemically represses CLE-RS2 and inhibits cortical cell division.
Given that the locations of the GUS staining spots indicate where the endogenous NIN gene is activated, the repression of CLE-RS2 in DsRed-negative roots of plants with malformed Pro35S-NIN-GR roots appears to be related to attenuation of NIN activity. RT-PCR analysis showed that whereas inoculation with M. loti activated NIN and its transcriptional targets CLE-RS1, CLE-RS2, and LjNF-YA1 in DsRed-negative control roots within 1 d, there was no significant induction of these genes in DsRed-negative roots of plants with malformed Pro35S-NIN-GR roots (Fig. 4A). In contrast, expression of NSP1, NSP2, CCaMK, and CYCLOPS, which are involved in NIN activation in symbiotic responses (25, 26), was not significantly altered by the systemic effect of NIN expression (Fig. 4A and Fig. S6G). Expression patterns of these genes resembled those seen in roots overexpressing CLE-RS1 (Fig. S6H).
Fig. 4.
NIN ectopic expression down-regulates NIN activity. (A) RT-PCR analysis of gene expression in DsRed-negative control roots and DsRed-negative roots of plants that also generated DsRed-positive 35S-NIN-GR malformed roots. Plants were inoculated with (1 dai) or without (mock) M. loti after 5 d of DEX treatment. Expression levels were normalized to polyubiquitin expression. Data are mean ± SD from three biological repeats. (B) GUS expression controlled by the NIN promoter in roots. Roots were cotransformed with ProNIN-GUS and either the empty vector or Pro35S-NIN-GR. The latter two constructs carried Pro35S-DsRed as a transformation marker. Plants were inoculated with (6 dai) or without (mock) M. loti after 5 d of DEX treatment. GUS expression in DsRed-negative roots was analyzed at 6 dai. (Scale bars: 1 mm.)
We further characterized the down-regulation of endogenous NIN expression by the systemic effects of NIN overexpression by studying ProNIN-GUS expression. Roots were transformed with ProNIN-GUS and Pro35S-NIN-GR by the same method shown in Fig. S6A. The GUS expression typical of infected roots was observed in DsRed-negative control roots that were cogenerated with hairy roots transformed with the empty vector (Fig. 4B) (15, 27). This expression was repressed in DsRed-negative roots of plants with malformed Pro35S-NIN-GR roots (Fig. 4B). The systemic and negative effects on GUS expression were suppressed in har1-7 mutants (Fig. 4B). These findings indicate that AON negatively regulates NIN expression at the transcription level. This negative feedback results in the repression of NIN target genes and prevention of cortical cell division in response to rhizobial infection.
Finally, we investigated the effects of NIN ectopic expression on cortical cell division in har1 and tml mutants. Pro35S-NIN-GR was introduced into roots of har1-7 and tml-4 mutants, and sites in which cortical cell division occurred were counted at 3 wk after DEX treatment. In the WT background, NIN ectopic expression induced cortical cell division in 25–28% of the transformed roots (Fig. S7A), with 2.2 ± 1.0 (mean ± SD) distinct locations showing cortical cell division in these roots (Fig. S7B). Ectopic cortical cell division was occasionally found in har1-7 and tml-4 roots that has been transformed with the empty vector, in accordance with previous observations of enhanced mitotic activity (28). The numbers of roots and sites on roots at which cortical cell division occurred approximately doubled in har1-7 and tml-4 roots transformed with Pro35S-NIN-GR (Fig. S7 A and B). In addition, the size of the regions showing cortical cell division was significantly increased in the mutants (Fig. S7C). These results indicate that the har1-7 and tml-4 mutations enhance cortical cell division rates induced by NIN expression. This enhancement suggests that AON negatively influences NIN-mediated cortical cell division.
Discussion
In this study, we have shown how early events in nodule organogenesis activate AON at the molecular level and have demonstrated that an autoregulatory negative-feedback loop homeostatically regulates nodule development via long-distance signaling. NIN is the transcription factor that targets CLE genes whose expression is regulated by exogenous stimuli and influences organ development. NIN activates AON by directly regulating the expression of CLE-RS1 and CLE-RS2. AON elicited by CLE-RS1 and CLE-RS2 expression down-regulates NIN activity, leading to the suppression of nodulation and repression of the CLE genes, thereby allowing recovery from the AON-mediated suppression of nodulation. Thus, NIN plays a central role in the integration of the nodulation signaling pathway and AON.
This shoot-mediated signaling is a reasonable strategy for the systemic regulation of nodulation, because roots that were adventitiously generated from the hypocotyl or stem are not directly connected to other roots. It takes a few days for shoot-derived AON signals to return to the root (29). This interval enables AON to suppress nodulation stimulated by subsequent infection through repression of this gene essential to nodule organogenesis. In addition, NIN expression in the cortex also locally inhibits rhizobial infection, whereas this gene is essential for infection thread development and for cortical cell division to generate nodule primordia (24). This report and our data show that NIN has multiple functions in nodulation. Recruitment of NIN in the nodule symbiosis is crucial for proper regulation of nodule formation.
Enhancement of the NIN-mediated cortical cell division by har1 and tml mutations implies that AON may posttranslationally influence NIN activity in addition to its transcription-regulating function, given that NIN is expressed under the control of the constitutively active CaMV35S promoter. Alternatively, AON may regulate factors other than NIN involved in cortical cell division, factors that influence endogenous NIN expression. TML F-box protein acts in roots downstream of shoot-derived mobile inhibitors that are produced in a HAR1-dependent manner and inhibit nodulation. Identification of factors targeted by TML is important for understanding how NIN expression is regulated by AON. Legumes have coopted receptor proteins responsible for the SAM homeostatic regulation as shoot-acting AON factors (8, 9, 30, 31). Cytokinin signaling is commonly implicated in nodule organogenesis as well as in the SAM homeostatic regulation and protoxylem vessel formation regulated by CLE10 (32-34). NIN expression is not up-regulated in the hit1 cytokinin-receptor mutant. Thus, factors regulated by cytokinin signaling may be targeted by CLE peptide-mediated AON.
NIN is evolved from a member of NIN-like proteins (NLPs), which regulate the expression of primary nitrate-responsive genes (35, 36), suggesting integration of a part of nitrate-responsive pathway in the nodule symbiosis. Indeed, CLE-RS2 and GmNIC1 are activated by nitrate, which also has an inhibitory effect on nodulation (11, 37, 38). Intriguingly, nitrate systemically regulates root architecture in Arabidopsis via root–shoot communication (39, 40). There may be an evolutionary link between pathways for nitrate responses and AON.
Materials and Methods
Plant Materials and Bacterial Strains.
We used two L. japonicus accessions, Gifu B-129 and MG-20, as the WT, along with five symbiotic mutants, nin-2, hit1, har1-7, tml-1, and tml-4 (15, 19, 23, 41). Unless stated otherwise in a figure legend, MG-20 served as the WT. Root transformation with A. rhizogenes and inoculation with M. loti MAFF303099 and a line constitutively expressing DsRed were performed as described previously (11, 17). Transformation of tobacco leaf cells was performed as described previously (17).
Plasmids.
Detailed information is provided in SI Materials and Methods.
Expression Analysis.
Total RNA was isolated from roots for RT-PCR analysis. First-strand cDNAs were synthesized using the QuantiTect Reverse-Transcription kit (Qiagen). RT-PCR was performed in a LightCycler with LightCycler FastStart DNA Master SYBR Green I reaction mix (Roche Applied Science). Expression levels were normalized using polyubiquitin transcripts. Primers used for RT-PCR were synthesized as described previously (11, 17). Histochemical GUS staining was performed as described previously (17). A detailed description of the transient expression assay in N. benthamiana is provided in SI Materials and Methods.
ChIP Analysis.
ChIP assays were performed as described previously (17). Primers used for ChIP-PCR are listed in Table S1. Detailed information on ChIP-seq analysis is provided in SI Materials and Methods.
EMSA.
EMSA was performed as described previously (17). Primers used for probe synthesis are listed in Table S1. The in vitro translation product containing NIN(520-878)-myc was incubated with probes for 30 min at 27 °C. For supershift analysis, the reaction mixture was incubated for another 1 h after the addition of polyclonal anti-myc antibodies (Santa Cruz Biotechnology).
Supplementary Material
Acknowledgments
We thank the National Institute for Basic Biology’s Functional Genomics Facility for technical support. This research was supported by the Funding Program for Next-Generation World-Leading Researchers from the Japan Society for the Promotion of Science and by the National Institute of Agrobiological Sciences Strategic Research Fund (to M.H.), and by Grants-in-Aid for Scientific Research 22128006 and 25291066 from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412716111/-/DCSupplemental.
References
- 1.Leyser O. Auxin, self-organisation, and the colonial nature of plants. Curr Biol. 2011;21(9):R331–R337. doi: 10.1016/j.cub.2011.02.031. [DOI] [PubMed] [Google Scholar]
- 2.Puig J, Pauluzzi G, Guiderdoni E, Gantet P. Regulation of shoot and root development through mutual signaling. Mol Plant. 2012;5(5):974–983. doi: 10.1093/mp/sss047. [DOI] [PubMed] [Google Scholar]
- 3.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289(5479):617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- 4.Schoof H, et al. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100(6):635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- 5.Yadav RK, et al. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011;25(19):2025–2030. doi: 10.1101/gad.17258511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol. 2009;5(8):578–580. doi: 10.1038/nchembio.182. [DOI] [PubMed] [Google Scholar]
- 7.Kosslak RM, Bohlool BB. Suppression of nodule development of one side of a split-root system of soybeans caused by prior inoculation of the other side. Plant Physiol. 1984;75(1):125–130. doi: 10.1104/pp.75.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krusell L, et al. Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature. 2002;420(6914):422–426. doi: 10.1038/nature01207. [DOI] [PubMed] [Google Scholar]
- 9.Nishimura R, et al. HAR1 mediates systemic regulation of symbiotic organ development. Nature. 2002;420(6914):426–429. doi: 10.1038/nature01231. [DOI] [PubMed] [Google Scholar]
- 10.Okamoto S, Shinohara H, Mori T, Matsubayashi Y, Kawaguchi M. Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat Commun. 2013;4:2191. doi: 10.1038/ncomms3191. [DOI] [PubMed] [Google Scholar]
- 11.Okamoto S, et al. Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol. 2009;50(1):67–77. doi: 10.1093/pcp/pcn194. [DOI] [PubMed] [Google Scholar]
- 12.Caetano-Anollés G, Gresshoff PM. Early induction of feedback regulatory responses governing nodulation in soybean. Plant Sci. 1990;71:69–81. [Google Scholar]
- 13.Mortier V, et al. CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 2010;153(1):222–237. doi: 10.1104/pp.110.153718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim CW, Lee YW, Hwang CH. Soybean nodule-enhanced CLE peptides in roots act as signals in GmNARK-mediated nodulation suppression. Plant Cell Physiol. 2011;52(9):1613–1627. doi: 10.1093/pcp/pcr091. [DOI] [PubMed] [Google Scholar]
- 15.Schauser L, Roussis A, Stiller J, Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402(6758):191–195. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- 16.Marsh JF, et al. Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 2007;144(1):324–335. doi: 10.1104/pp.106.093021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soyano T, Kouchi H, Hirota A, Hayashi M. Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 2013;9(3):e1003352. doi: 10.1371/journal.pgen.1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heckmann AB, et al. Cytokinin induction of root nodule primordia in Lotus japonicus is regulated by a mechanism operating in the root cortex. Mol Plant Microbe Interact. 2011;24(11):1385–1395. doi: 10.1094/MPMI-05-11-0142. [DOI] [PubMed] [Google Scholar]
- 19.Murray JD, et al. A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science. 2007;315(5808):101–104. doi: 10.1126/science.1132514. [DOI] [PubMed] [Google Scholar]
- 20.Tirichine L, et al. A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science. 2007;315(5808):104–107. doi: 10.1126/science.1132397. [DOI] [PubMed] [Google Scholar]
- 21.Plet J, et al. MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J. 2011;65(4):622–633. doi: 10.1111/j.1365-313X.2010.04447.x. [DOI] [PubMed] [Google Scholar]
- 22.Mortier V, De Wever E, Vuylsteke M, Holsters M, Goormachtig S. Nodule numbers are governed by interaction between CLE peptides and cytokinin signaling. Plant J. 2012;70(3):367–376. doi: 10.1111/j.1365-313X.2011.04881.x. [DOI] [PubMed] [Google Scholar]
- 23.Takahara M, et al. Too much love, a novel Kelch repeat-containing F-box protein, functions in the long-distance regulation of the legume-Rhizobium symbiosis. Plant Cell Physiol. 2013;54(4):433–447. doi: 10.1093/pcp/pct022. [DOI] [PubMed] [Google Scholar]
- 24.Yoro E, et al. A positive regulator of nodule organogenesis, NODULE INCEPTION, acts as a negative regulator of rhizobial infection in Lotus japonicus. Plant Physiol. 2014;165(2):747–758. doi: 10.1104/pp.113.233379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirsch S, et al. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell. 2009;21(2):545–557. doi: 10.1105/tpc.108.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh S, Katzer K, Lambert J, Cerri M, Parniske M. CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe. 2014;15(2):139–152. doi: 10.1016/j.chom.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Kosuta S, et al. Lotus japonicus symRK-14 uncouples the cortical and epidermal symbiotic program. Plant J. 2011;67(5):929–940. doi: 10.1111/j.1365-313X.2011.04645.x. [DOI] [PubMed] [Google Scholar]
- 28.Wopereis J, et al. Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. Plant J. 2000;23(1):97–114. doi: 10.1046/j.1365-313x.2000.00799.x. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki A, et al. Split-root study of autoregulation of nodulation in the model legume Lotus japonicus. J Plant Res. 2008;121(2):245–249. doi: 10.1007/s10265-007-0145-5. [DOI] [PubMed] [Google Scholar]
- 30.Miyazawa H, et al. The receptor-like kinase KLAVIER mediates systemic regulation of nodulation and non-symbiotic shoot development in Lotus japonicus. Development. 2010;137(24):4317–4325. doi: 10.1242/dev.058891. [DOI] [PubMed] [Google Scholar]
- 31.Krusell L, et al. The Clavata2 genes of pea and Lotus japonicus affect autoregulation of nodulation. Plant J. 2011;65(6):861–871. doi: 10.1111/j.1365-313X.2010.04474.x. [DOI] [PubMed] [Google Scholar]
- 32.Leibfried A, et al. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature. 2005;438(7071):1172–1175. doi: 10.1038/nature04270. [DOI] [PubMed] [Google Scholar]
- 33.Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci USA. 2009;106(38):16529–16534. doi: 10.1073/pnas.0908122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kondo Y, Hirakawa Y, Kieber JJ, Fukuda H. CLE peptides can negatively regulate protoxylem vessel formation via cytokinin signaling. Plant Cell Physiol. 2011;52(1):37–48. doi: 10.1093/pcp/pcq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konishi M, Yanagisawa S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat Commun. 2013;4:1617. doi: 10.1038/ncomms2621. [DOI] [PubMed] [Google Scholar]
- 36.Marchive C, et al. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat Commun. 2013;4:1713. doi: 10.1038/ncomms2650. [DOI] [PubMed] [Google Scholar]
- 37.Reid DE, Ferguson BJ, Gresshoff PM. Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Mol Plant Microbe Interact. 2011;24(5):606–618. doi: 10.1094/MPMI-09-10-0207. [DOI] [PubMed] [Google Scholar]
- 38.Carroll BJ, McNeil DL, Gresshoff PM. Isolation and properties of soybean [Glycine max (L.) Merr.] mutants that nodulate in the presence of high nitrate concentrations. Proc Natl Acad Sci USA. 1985;82(12):4162–4166. doi: 10.1073/pnas.82.12.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, Jennings A, Barlow PW, Forde BG. Dual pathways for regulation of root branching by nitrate. Proc Natl Acad Sci USA. 1999;96(11):6529–6534. doi: 10.1073/pnas.96.11.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruffel S, et al. Nitrogen economics of root foraging: Transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc Natl Acad Sci USA. 2011;108(45):18524–18529. doi: 10.1073/pnas.1108684108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Magori S, et al. Too much love, a root regulator associated with the long-distance control of nodulation in Lotus japonicus. Mol Plant Microbe Interact. 2009;22(3):259–268. doi: 10.1094/MPMI-22-3-0259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.