Abstract
Mo-CBP3 is a chitin-binding protein purified from Moringa oleifera Lam. seeds that displays inhibitory activity against phytopathogenic fungi. This study investigated the structural properties and the antifungal mode of action of this protein. To this end, circular dichroism spectroscopy, antifungal assays, measurements of the production of reactive oxygen species and microscopic analyses were utilized. Mo-CBP3 is composed of 30.3% α-helices, 16.3% β-sheets, 22.3% turns and 30.4% unordered forms. The Mo-CBP3 structure is highly stable and retains its antifungal activity regardless of temperature and pH. Fusarium solani was used as a model organism for studying the mechanisms by which this protein acts as an antifungal agent. Mo-CBP3 significantly inhibited spore germination and mycelial growth at 0.05 mg.mL−1. Mo-CBP3 has both fungistatic and fungicidal effects, depending on the concentration used. Binding of Mo-CBP3 to the fungal cell surface is achieved, at least in part, via electrostatic interactions, as salt was able to reduce its inhibitory effect. Mo-CBP3 induced the production of ROS and caused disorganization of both the cytoplasm and the plasma membrane in F. solani cells. Based on its high stability and specific toxicity, with broad-spectrum efficacy against important phytopathogenic fungi at low inhibitory concentrations but not to human cells, Mo-CBP3 has great potential for the development of new antifungal drugs or transgenic crops with enhanced resistance to phytopathogens.
Introduction
Plants use several strategies to overcome fungal attacks, including the production of antimicrobial peptides and proteins [1], [2]. Much effort has been dedicated to researching these bioactive constituents, particularly because the chemically-synthesized antifungal compounds used to prevent and contain these pathogens comprise a potential environmental threat [3], [4], [5]. In general, these defense-related proteins interfere with the fungal life cycle by either impairing growth or killing the pathogen [6], [7]. The antifungal properties of these proteins may be exploited for use in the development of transgenic crops that have enhanced resistance to phytopathogenic fungi [8].
Chitin-binding proteins (CBPs) represent a group of proteins also found in plants that often have a basic pI, a molecular mass ranging from 3.1 kDa up to 20 kDa, and high resistance to both extreme pH changes and proteolysis. Some CBPs have the ability to inhibit fungal growth [9], as they bind to and disrupt the proper function of chitin, a key component of the fungal cell wall [10]. It has been suggested that the binding of these proteins to chitin in filamentous fungi leads to the disruption of both cell wall biogenesis and cell polarity [11], [12].
Recently, our research group isolated a chitin-binding protein named Mo-CBP3 from Moringa oleifera Lam. seeds [13]. This protein is a basic glycoprotein (18 kDa by SDS-PAGE) and does not display haemagglutination, chitinase or β-1,3-glucanase activity. Mo-CBP3 presented potent antifungal activity against the phytopathogenic fungi Fusarium solani, F. oxysporum, Colletotrichum musae and C. gloeosporioides at a low concentration (0.05 mg.mL−1). The phytopathogenic effect of Mo-CBP3 against fungi was observed even when the protein was heated at 100°C for 1 h or pre-treated with 150 mM N-acetyl-D-glucosamine.
As Mo-CBP3 has a low molecular mass and is a protein with potent antifungal activity at low concentrations, it is a very promising bioactive candidate that may be explored to determine whether it can confer resistance against phytopathogenic fungi to economically and nutritionally important crops. To further test Mo-CBP3, it is essential to obtain additional knowledge about its structure and mode of action. Here, we report new structural features of Mo-CBP3 that reveal a correlation between its structural stability and its antifungal activity. In addition, to know about the mechanisms by which this protein acts as an antifungal agent, its ability to induce the endogenous production of reactive oxygen species and to trigger morphologic and ultrastructural alterations were analyzed using F. solani as a model. F. solani is an easy-to-handle and fast-developing species, making it ideal for in vitro assays, and it holds relevance as a phytopathogenic fungus that attacks economically important crop plants. Furthermore, to have a preliminary clue whether Mo-CBP3, as a chemical agent against fungi, displays cytotoxicity, the level of lysis of the human red blood cells was examined.
Materials and Methods
Biological materials and reagents
M. oleifera seeds were obtained from trees at the Campus do Pici of the Federal University of Ceará (UFC), Fortaleza, Brazil. A voucher specimen (No. EAC34591) was deposited in the Prisco Bezerra Herbarium, UFC. The filamentous fungus F. solani (URM 3708) was provided by the Departamento de Micologia of the Universidade Rural de Pernambuco, Recife, Brazil. All chemicals used were of analytical grade.
Mo-CBP3 preparation
A highly purified Mo-CBP3 preparation devoid of contaminating proteins was obtained according to Gifoni et al. [13]. Mature seeds were ground in a coffee grinder, and the resulting flour was treated with n-hexane. Defatted flour was extracted with 50 mM Tris-HCl, pH 8.0, containing 150 mM NaCl (1∶10 w/v), for 4 h at 4°C under constant stirring and was then filtered through cheesecloth. After centrifugation at 15,000 g at 4°C for 30 min, the supernatant was exhaustively dialyzed against Milli-Q grade water and centrifuged again under the same conditions. (NH4)2SO4 was added to the soluble material, denoted as the albumins, to yield 90% saturation. This protein fraction (F0–90%) was then dissolved in and dialyzed against the extracting buffer and applied to a chitin column that had been equilibrated with the same buffer. After elution with the starting buffer of the unbound proteins from the column, the chitin-bound proteins, named PNAG and PAC, were eluted with 100 mM N-acetyl-D-glucosamine (NAG) that was prepared in the extracting buffer and with 50 mM acetic acid (pH 5.0), respectively. The PNAG sample was dialyzed against 100 mM acetic acid and distilled water, lyophilized and applied to a cation-exchange matrix (Resource S) that had been previously equilibrated with 50 mM sodium acetate buffer, pH 5.2. Four major adsorbed protein peaks (Mo-CBP2, Mo-CBP3, Mo-CBP4, and Mo-CBP5) were recovered after being selectively desorbed by stepwise elution with 400, 500, 600, and 700 mM NaCl, respectively, included in the equilibrium buffer. As Mo-CBP3 was purified to homogeneity, had high yield and presented the highest activity against the phytopathogenic fungi Fusarium solani, Fusarium oxysporum, Colletotrichum musae and Colletotrichum gloesporioides, as previously reported by our research group [13], it was used for further analyses. The purity of Mo-CBP3 was checked by denaturing gel electrophoresis [14]. The identity of Mo-CBP3 was confirmed by N-terminal amino acid sequence analysis by Edman degradation (Shimadzu PPSQ-10A automated protein sequencer).
Protein concentration
The protein concentration was determined according to Bradford [15], using BSA as a standard. Absorbance at 280 nm was also used to monitor the protein elution profiles during chromatography.
Far-UV circular dichroism (CD) spectroscopy
CD spectra measurements were made on a JASCO J-715 spectropolarimeter (Jasco Instruments, Tokyo, Japan) in an N2 atmosphere at 25°C. Mo-CBP3 (40 µg) was dissolved in 20 mM phosphate-buffered saline (PBS) at pH 7.0 and transferred to a rectangular quartz cuvette with a 0.1 cm path length. Eight scans were performed with a scan rate of 20 nm.min−1 and a 4 s response time. CD spectra were measured from 190 to 250 nm. The contributions of the secondary structural elements of Mo-CBP3 were determined by CD spectra deconvolution analyses using the basis reference protein set SMP56 of the CDPro software [16] and applying three methods, CONTIN/LL, SELCON 3 and CDSSTR. CD spectroscopy was also used to assess Mo-CBP3 thermal stability. To do this, Mo-CBP3 (40 µg in PBS) was heated gradually in 10°C increments from 26 to 90°C in a TC-100 circulating water bath (Jasco). The samples were maintained at each temperature for 10 min, and spectra were recorded from 190 to 250 nm. To evaluate structural stability as a function of pH, Mo-CBP3 (40 µg) was incubated for 240 min in 20 mM sodium acetate/phosphate/borate buffer at different pH values (2.0, 4.0, 6.0, 8.0, 10.0, and 12.0) before recording the CD spectrum.
Effect of pH and temperature on the inhibition of the conidial germination of F. solani by Mo-CBP3
The filamentous fungus F. solani was grown in Petri dishes containing potato dextrose agar (PDA) medium for 12 days at room temperature (22°C). Fresh conidia suspensions were prepared by rinsing the surface of the 12-day-old sporulated cultures with sterile distilled water and the aid of a triangular Drigalski rod. Spore suspensions were filtered through cheesecloth in a laminar flux chamber under sterile conditions, and conidia were quantified using a Neubauer chamber under an optical microscope (Olympus System Microscope BX 60). Antifungal assays were conducted as described by Ji and Kuc [17]. For analyzing the changes in conidial germination as a function of pH, Mo-CBP3 samples, at antifungal concentration (0.1 mg.mL−1), were dissolved in 20 mM sodium acetate/phosphate/borate buffer at different pH values (2.0, 4.0, 6.0, 8.0, 10.0 and 12.0) and incubated with 10 µL of the conidia suspension (2×105.mL−1) in reticulated plates. For the non-inhibitory controls, conidia were incubated in each buffer in the absence of Mo-CBP3. The plates were placed in a plastic box maintained near 100% relative humidity at 22°C in the dark for 24 h. After this time, 50 conidia were randomly selected from each treatment and evaluated for germination under an optical microscope. A conidium that had emitted a hyphae at least twice the length of the ungerminated conidium was considered to have successfully germinated. In parallel, to assess whether the ability of Mo-CBP3 to inhibit spore germination was affected by heat treatment, Mo-CBP3 was heated in a water bath at 100°C for 60 min, cooled on ice for 10 min, and assayed as described above. Each experiment was performed in triplicate, and images were taken with a digital camera (Sony, MCV-CD350 model, 14.2 megapixels).
Effect of Mo-CBP3 on the mycelial growth and conidial viability of F. solani
A quantitative assay for fungal growth inhibition was performed following the protocol developed by Broekaert et al. [18]. A conidia suspension (2×105 cells.mL−1) was incubated in 96-well flat microplates with 100 µL of potato dextrose broth in the absence of Mo-CBP3 and allowed to germinate for up to 12 h in the dark at 37°C. Next, 100 µL of different concentrations of Mo-CBP3 (0.05, 0.1, 0.5 and 1 mg.mL−1) were added. Cell growth was also determined without the addition of Mo-CBP3. Fungal growth was monitored by turbidimetry at 630 nm from 0 to 49 h using an automated microplate reader (Model Elx800, Bio-Tek Instruments). The absorbance values taken immediately after Mo-CBP3 addition were recorded and established as zero and were discounted from every readings taken onwards. To evaluate the conidial viability of F. solani after treatment with different concentrations of Mo-CBP3, 150 µL aliquots were taken from the wells, transferred to Eppendorf tubes and centrifuged at 3,000 g for 1 min at 25°C and the supernatant discarded. The remaining conidia were washed with sterile distilled water to remove Mo-CBP3, reculturing in Petri dishes containing PDA medium and kept in an incubator at 27°C. Images of the mycelium growth were taken after 5 days. All experiments were carried out in triplicate.
Evaluation of the electrostatic interaction of Mo-CBP3 with the conidial membrane
To evaluate the presence of electrostatic interactions between Mo-CBP3 and the conidial plasma membrane, Mo-CBP3 (0.1 mg.mL−1) was first dissolved in solutions with different NaCl concentrations (25, 75 and 150 mM) and antifungal assays were performed following the methodology described in Section 2.5. In the negative, non-inhibitory controls, conidia were incubated only in 25, 75 and 150 mM NaCl, all in the absence of Mo-CBP3. As a positive inhibitory control, Mo-CBP3 was used at antifungal concentration (0.1 mg.mL−1). All experiments were carried out in triplicate.
Evaluation of reactive oxygen species (ROS) production by F. solani conidia after Mo-CBP3 treatment
To evaluate the ability of Mo-CBP3 to induce the endogenous production of ROS in F. solani conidia, the in situ assay described by Thordal-Christensen et al. [19], with some modifications [20], was conducted, using 3,3′-diaminobenzidine (DAB). F. solani conidia (2×106 cells.mL−1) were incubated with Mo-CBP3 (0.1 mg.mL−1) prepared in H2O, with only H2O or bovine serum albumin (BSA, 0.1 mg.mL−1 in H2O) used as controls, all in the presence of DAB (0.5 mg.mL−1 in H2O). After 1 h incubation, aliquots of conidial suspensions were placed on glass slides and examined under a light microscope (Olympus System Microscope BX 60).
Scanning electron microscopy (SEM)
To analyze the F. solani conidial morphology after treatment with Mo-CBP3, the fungal cells (2×106 conidia.mL−1) were incubated in either the absence or presence of Mo-CBP3 (0.05 mg.mL−1 in H2O). After 48 h incubation, the cells were harvested and fixed for 30 min at 25°C with 2.5% (v/v) fresh glutaraldehyde and 4% (v/v) paraformaldehyde prepared in 50 mM cacodylate buffer, pH 7.2. Subsequently, the materials were rinsed three times with the above buffer, post-fixed for 30 min at 25°C with 1% (m/v) osmium tetroxide (OsO4) solution diluted in the same buffer and rinsed with distilled water. After that, conidia were dehydrated in a graded acetone series (30, 50, 70, 90, and 100%; v/v), critical-point dried in CO2, coated with 20 nm gold and observed in a Zeiss 962 scanning electron microscope.
Transmission electron microscopy (TEM)
Structural changes of F. solani conidium induced by Mo-CBP3 were assessed by TEM. Conidia were grown for 48 h in water in either the presence (0.05 mg.mL−1) or absence of Mo-CBP3 and processed as for SEM analysis. After post-fixation in 1% (m/v) OsO4 and dehydration in a graded acetone series, the specimens were embedded in Epon resin (Polybeded 812). Ultrathin sections (0.1 µm) were fixed onto copper grids, stained with uranyl acetate (10 min) and lead with citrate (5 min). Visualization of cells was performed in a transmission electron microscope (Zeiss TEM 900) operating at 80 kV.
Haemolytic assay
This was carried out using human red blood cells (hRBCs) collected from healthy donors in heparinized tubes [21]. hRBCs were separated from plasma by centrifugation (3,000 g, 10 min, 25°C) and washed three times with 100 mM sodium phosphate buffer, pH 7.4, containing 150 mM NaCl (PBS). A 1% (v/v) suspension was prepared and incubated for 1 h at 37°C with serial dilutions of Mo-CBP3 (from 280 to 0.137 µM) in PBS. After the incubation period, suspensions were centrifuged at 3,000 g for 10 min at 25°C, aliquots of the supernatants were transferred to Eppendorf tubes and the absorbances taken at 405 nm (spectrophotometer Novaspec II, Pharmacia) to monitor the release of haemoglobin. Triton X-100 and PBS were used as positive (100% haemolysis) and negative controls, respectively. The haemolysis percentage was calculated using the following equation: Haemolysis (%) = [Aprotein–APBS]/[ATriton–APBS], where A means absorbance at 405 nm.
Results and Discussion
Mo-CBP3 preparation
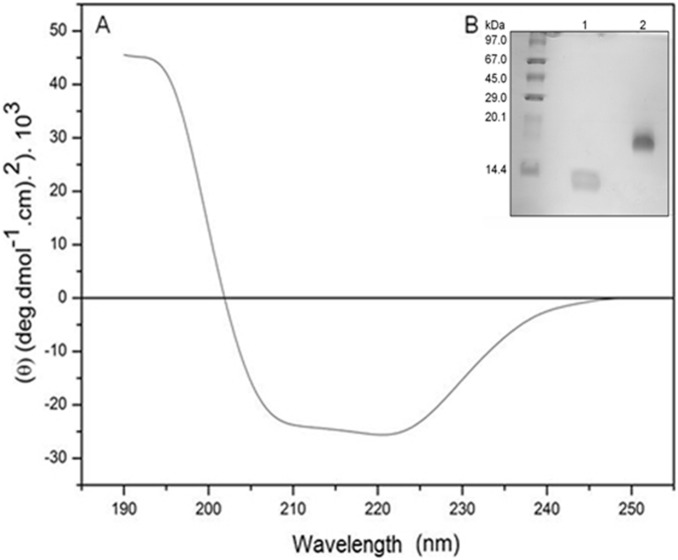
The Mo-CBP3 preparations used in the present study were confirmed to be homogeneous and free of contaminants, as shown in a representative Figure 1. Mo-CBP3 is an 18 kDa protein that inhibits the conidia germination of F. solani, F. oxysporum, C. musae and C. gloeosporioides, confirming the findings of Gifoni et al. [13]. The purity of Mo-CBP3 was further proved by N-terminal sequencing analysis.
Figure 1. Structural properties of Mo-CBP3.
(A) Circular dichroism spectra (Far-UV) of Mo-CBP3 (2.22 mM) in 20 mM sodium phosphate buffer, pH 7.0, using a rectangular quartz cuvette with a 0.1 cm path length. (B) Denaturing polyacrilamide gel electrophoresis (SDS-PAGE - 15% acrylamide gel) of Mo-CBP3. Molecular mass standards are shown (in kDa) on the left; Lanes 1 and 2, Mo-CBP3 (20 µg) in reducing (4 kDa and 8 kDa subunits) and non-reducing conditions (18 kDa), respectively.
Structure-antifungal activity relationships
The far-UV CD spectra of native Mo-CBP3 showed minima at approximately 208 and 222 nm (Figure 1). The deconvolution of the CD spectra performed using the CDPro package [16] revealed the following content of the secondary structure fraction: 30.3% α-helices, 16.3% β-sheets, 22.3% turns and 30.4% unordered forms. Therefore, Mo-CBP3 can be classified as an alpha-beta protein [22].
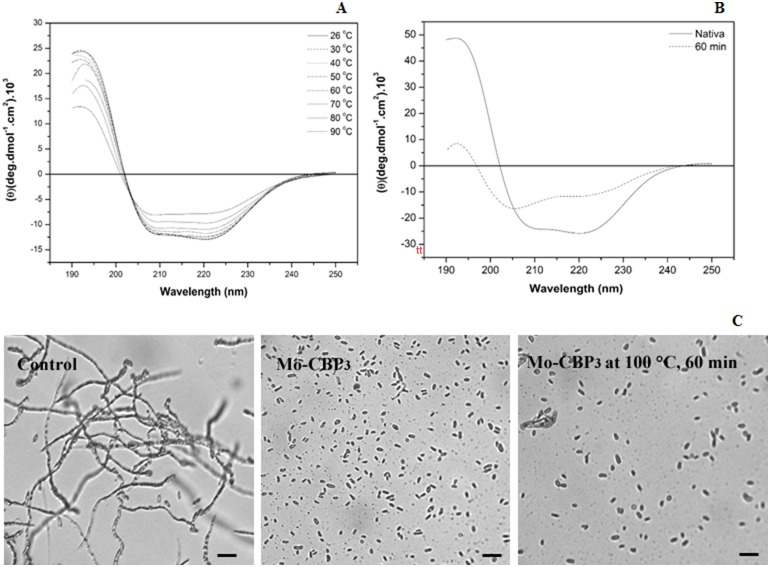
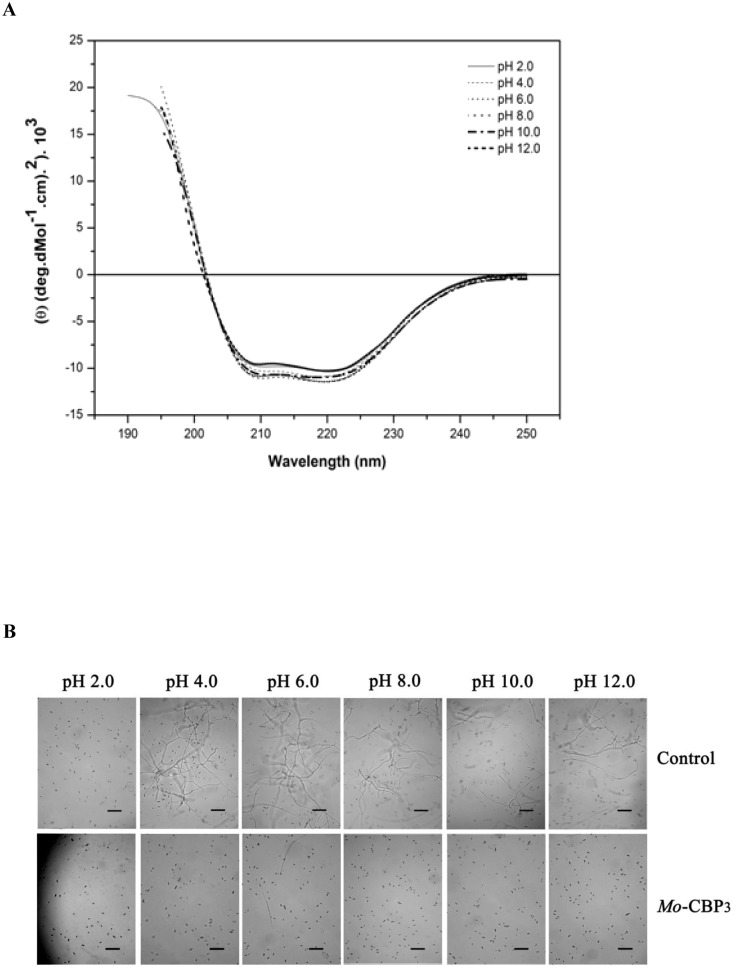
To further characterize Mo-CBP3, the effects of temperature and pH on its secondary structure and antifungal activity were evaluated. No significant changes in the CD spectra of Mo-CBP3 were observed after heat treatment at 90°C for 10 min (Figure 2A). After heat treatment at 100°C for 60 min, the CD spectra of Mo-CBP3 demonstrated only a discrete alteration (Figure 2B). Similarly, Mo-CBP3 was still able to inhibit the spore germination of F. solani after heating at 100°C for 60 min. Additionally, the CD spectral shape did not change from pH 2.0 to pH 12.0 (Figure 3A), suggesting that the protein structure is maintained and that even the pH extremes were insufficient to alter the net charge of Mo-CBP3 in a way to cause electrostatic repulsion, with later rupture of the hydrogen bonds. To correlate the structure of Mo-CBP3 to its antifungal activity, Mo-CBP3 was dissolved in 20 mM sodium acetate-borate-phosphate buffer at different pH values, and the antifungal activity on F. solani spore germination was tested. The inhibitory activity of Mo-CBP3 was similar at all pH ranges tested (4.0, 6.0, 8.0, 10 and 12.0) (Figure 3B). However, it was not possible to evaluate the behavior of the protein at pH 2.0, as spore germination did not occur even in the control, most likely because this pH is very acidic and has an adverse effect on the development of F. solani. These data demonstrate that Mo-CBP3 exhibits high resistance to both temperature and pH changes, thus retaining its antifungal activity. The elevated structural and functional stability of Mo-CBP3 can be attributed to the presence of cysteine residues in its structure. Mo-CBP3 is a chitin-binding protein, and many proteins for which the amino acid sequences are known that possess this property share a common structural domain composed of 43 amino acids, with many cysteine and glycine residues in conserved positions [23]. In fact, of the 22 identified residues from the N-terminus of Mo-CBP3, 27.3% were cysteines [13]. The presence of such residues can lead to formation of disulphide bridges, making these proteins more resistant to denaturation [9], [24]. In fact, the reduction of these disulphide bridges abolished the antifungal activity of Mo-CBP3. Several disulphide bonds are present in osmotins and thaumatin-like proteins, and it is thought that they contribute to the high structural stability of these proteins [25].
Figure 2. Effect of temperature on the conformation and antifungal activity of Mo-CBP3.
(A) Far-UV CD spectra of Mo-CBP3 (2.22 mM) at various temperatures. (B) CD spectra of Mo-CBP3 (2.22 mM) after heating at 100°C for 60 min. (C) Light micrographs of F. solani spores in either the culture medium (control) or incubated with Mo-CBP3 (0.1 mg.mL−1) and either unheated or previously heated at 100°C for 60 min in a water bath. Trials were conducted for 24 h at 22°C in the dark. Bars: 2.5 µm.
Figure 3. Effect of pH on the conformation and antifungal activity of Mo-CBP3.
(A) Far-UV CD spectra of Mo-CBP3 (2.22 mM) at various pH values. (B) Light micrographs of F. solani spores either in 20 mM sodium acetate-borate-phosphate buffer at different pH values (control) or incubated with Mo-CBP3 (0.1 mg.mL−1) prepared in these buffers. Trials were conducted for 24 h at 22°C in the dark. Bars: 2.5 µm.
Effect of Mo-CBP3 on the mycelial growth and conidial viability of F. solani
In addition to inhibiting spore germination, the ability of Mo-CBP3 to affect fungal growth was analyzed. Mo-CBP3 was inhibitory to the mycelial mass development of F. solani in comparison to the control incubated in the absence of Mo-CBP3 (Figure 4A). Mo-CBP3 displayed a significant inhibitory effect on fungus growth at a concentration of only 0.05 mg.mL−1, with close to 62% inhibition within 49 h. At higher concentrations (0.5 and 1.0 mg.mL−1), the antifungal effect was observed at earlier stages. For example, at 1.0 mg.mL−1, Mo-CBP3 inhibited 94% of the growth of F. solani 26 h post-incubation. In reality, Mo-CBP3 behaves both as fungistatic and fungicidal protein, depending on its concentration and the stage of fungus development. Pre-incubation of F. solani spores with Mo-CBP3 at concentrations up to 0.5 mg.mL−1 for 49 h followed by removal of the protein restored the mycelial growth capacity of the fungus, indicating the fungistatic effect of Mo-CBP3 at low concentrations. In contrast, prior incubation of F. solani spores with 1.0 mg.mL−1 Mo-CBP3 followed by removal of the protein abolished (100% inhibition) the fungus viability, as mycelial growth was inhibited (Figure 4B), which characterizes Mo-CBP3 fungicidal action. It is well known that several chitin-binding proteins have antifungal activity [26]. A chitin-binding lectin from Setcreasea purpurea (SPL) causes inhibition of Rhizoctonia solani, Penicillium italicum, Sclerotinia sclerotiorum, and Helminthosporium maydis at 1.51 mg.mL−1 [27]. However, it is remarkable that the inhibitory effects of Mo-CBP3 on F. solani growth were more pronounced in this study than those observed by Gifoni et al. [13]. The differences observed are presumably due to the different protocols used. For example, the growth inhibition assays in the present study were performed in liquid medium, differing from the previous study, which was made on solid medium (Petri dish). In addition, it is worth noting that the cultivation medium has a great influence on the antifungal activity of Mo-CBP3, as F. solani growth inhibition could be detected only when yeast extract was not present in the medium composition. Based on these data, it is plausible to speculate that in the presence of yeast extract, which contains cell wall fragments and negatively charged mannan [28], the interaction of Mo-CBP3 with the filamentous fungus would be compromised, affecting its antifungal activity.
Figure 4. Effect of Mo-CBP3 on the mycelial growth and conidial viability of F. solani.
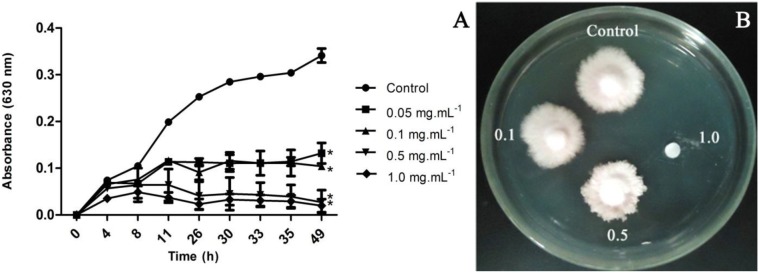
(A) Mycelial growth of F. solani in the presence of Mo-CBP3. A conidium suspension (2×105 cells.mL−1) was incubated in the absence of Mo-CBP3 and allowed to germinate for up to 12 h in the dark at 37°C. Next, 100 µL of different concentrations of Mo-CBP3 were added. The fungal culture in the absence of Mo-CBP3 was used as control. Each point is the mean of three estimates. The values are means (± SD) of triplicates. Asterisks indicate significant differences (P<0.05) compared to control group (Tukey’s Test). (B) Effects of Mo-CBP3 (0.1, 0.5 and 1.0 mg.mL−1) on the conidium viability of F. solani after inhibition growth assay.
Mode of action of Mo-CBP3 upon fungal cell
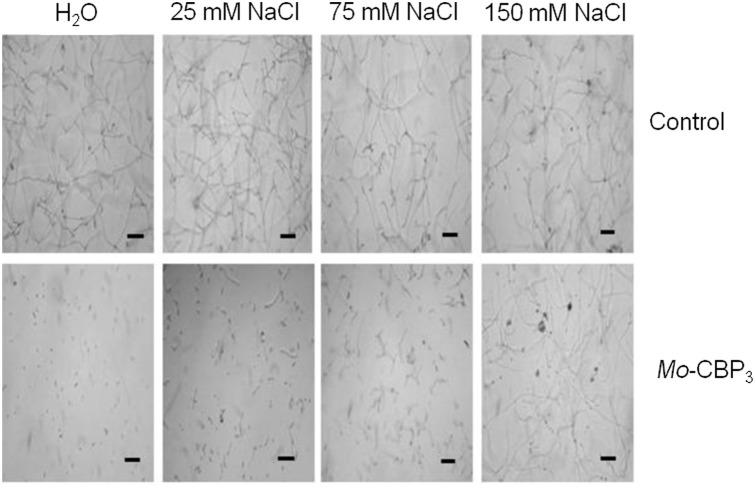
Antimicrobial molecules possess several features to fulfill their role in plant defense mechanisms. For rapid killing, antimicrobial molecules often act at the cell surface rather than the cell interior [29]. Therefore, it was hypothesized that besides the binding of Mo-CBP3 to the fungus chitin, since it is a chitin-binding protein as established by affinity chromatography [13], Mo-CBP3 could also bind to F. solani cell membrane components, at least in part, via electrostatic interactions. This prediction is supported by the observation that NaCl at 25, 75 and 150 mM reduced the inhibitory effect of Mo-CBP3 (0.1 mg.mL−1) on F. solani spore germination in comparison to the controls in the absence of Mo-CBP3 and presence of the same above concentrations of NaCl (Figure 5). Sensitivity to ionic strength with loss of antimicrobial activity has been described for other basic antimicrobial proteins and peptides that are thought to act via electrostatic interactions with negatively charged membrane components, as well as for those that bind to specific receptors [30], [31]. In the F. solani membrane, such electrostatic interactions of Mo-CBP3, which is a cationic protein, most likely occur with the negatively charged phospholipid phosphatidylinositol [32].
Figure 5. Effect of NaCl on the antifungal activity of Mo-CBP3.
Light micrographs of F. solani spores in either H2O or different NaCl concentrations (control), with or without incubation with Mo-CBP3 (0.1 mg.mL−1) prepared in these solutions. Trials were conducted for 24 h at 22°C in the dark. Bars: 2.5 µm.
After this initial binding to components of the fungal membrane, secondary effects that are induced internally in the cell were investigated. Figure 6C shows that Mo-CBP3 (0.1 mg.mL−1) induced ROS production as reveled by the presence of a reddish-brown pellet inside the F. solani spores, in contrast to the negative controls, H2O and BSA (0.1 mg.mL−1) (Figures 6A and 6B, respectively). The ROS induction capacity of various antifungal peptides and proteins has been previously reported. Similar to Mo-CBP3, the defensin from Phaseolus vulgaris (PvD1) causes ROS induction in F. solani cells at 0.1 mg.mL−1 [33]. Another example is the Raphanus sativus antifungal peptide 2 (Rs-AFP2), which is able to stimulate ROS production in Candida albicans in a dose-dependent manner, but is unable to do so in an Rs-AFP2-resistant Δgcs C. albicans mutant that lacks the Rs-AFP2-binding site in its membranes [34]. This finding suggests that upstream binding of the macromolecule is needed for ROS production. An increase in the generation of ROS that exceeds the cellular neutralization capacity of the fungus promotes oxidative stress and may cause the hyperoxidation of proteins, lipids and nucleic acids and consequently cell death [35].
Figure 6. Induction of reactive oxygen species (ROS) in F. solani spores.
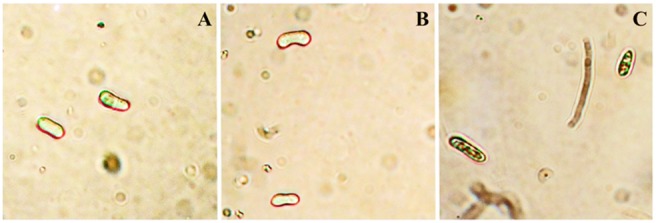
Cells were treated with 3,3′-diaminobenzidine (DAB) for ROS detection. Cells were previously incubated with (A) H2O, (B) BSA (0.1 mg.mL−1) or (C) Mo-CBP3 (0.1 mg.mL−1). Uptake of DAB is confirmed by the dark staining (reddish-brown) reaction in conidia, as indicated by arrows. Bars: 2.5 µm (A–C).
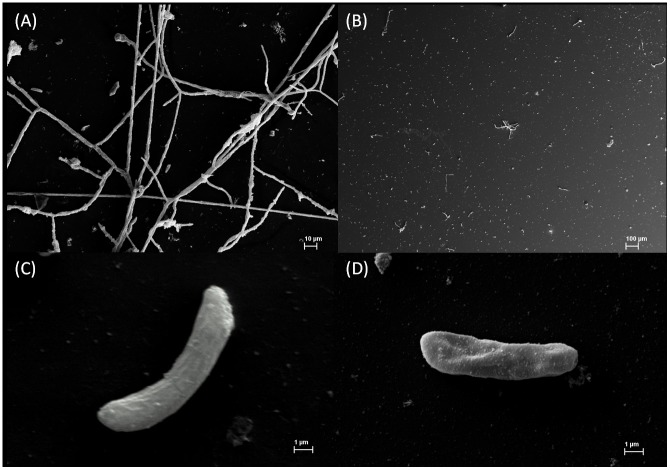
SEM was employed to allow visualization of any morphological changes promoted by Mo-CBP3 on F. solani cells. Photomicrographs of F. solani conidia were taken 48 h after growth in the presence or absence of Mo-CBP3 (0.05 mg.mL−1). Normal hyphal growth was observed in the control cells (Figure 7A), but not in in the cells treated with Mo-CBP3 (Figure 7B). Closer examination of F. solani cells treated with Mo-CBP3 revealed loss of asymmetry, deformations and wrinkles in comparison to control cells, as represented in Figures 7D and 7C, respectively. Similar alterations were detected in S. cerevisiae cells after incubation with a 2S albumin-homologous protein (0.1 mg.mL−1) from passion fruit seeds [36].
Figure 7. Scanning electron microscopy of F. solani cells.
The cells were cultured either in the absence (A, C) and presence (B, D) of Mo-CBP3 (0.05 mg.mL−1). In (A) the fungus cell has typical growth and developed hyphae in contrast with (B) which shows ungerminated spores and spores that emitted the germination peg, but not developed further. The zooming image of a Mo-CBP3 treated spore (D) shows typical alterations in the cell surface morphology in contrast with control spore (C).
Ultrastructural analysis of F. solani cells also revealed alterations in the presence of Mo-CBP3 (0.05 mg.mL−1). It was observed condensation of the cytosol content, vacuolation and shrinkage of the cell wall (Figures 8B and C) when compared with control cells (Figure 8A). Vacuoles serve as compartments either for storage of resources or for detoxification purposes [37]. Thus, possibly the increased vacuole formation in the fungus cell might be related to a defense response of F. solani to the toxic effects of Mo-CBP3. In addition to these above changes observed, notable accumulation of electrodense granular material was observed in the cytosol of the cells incubated with Mo-CBP3 (Figure 8C). It is plausible to speculate that the electrodense granular material observed might result from the electrostatic interactions of the cationic Mo-CBP3 with negatively charged primary or secondary metabolites present into the fungus cell, based on the coagulant (flocculent) properties of this protein as previously reported [13]. These alterations in F. solani morphology as visualized by TEM are typically found in cells that have undergone apoptosis [38]. This is in agreement with the results shown above, as ROS are classical apoptotic markers [39]. Thus, these data together suggest that the antifungal properties of Mo-CBP3 are triggered by alterations in the cell surface. Brul et al. [40] found that the filamentous fungi Penicillium roqueforti, Trichoderma harzianum, Paecilomyces variotii, Aspergillus niger, and A. nidulans allow molecules up to 150 kDa to cross the cell wall. Thus, it cannot be ruled out that Mo-CBP3 may eventually pass through the cell wall barrier, interact with the cell membrane receptors and induce secondary effects internally in F. solani to promote cell death.
Figure 8. Transmission electron microscopy of F. solani cells.
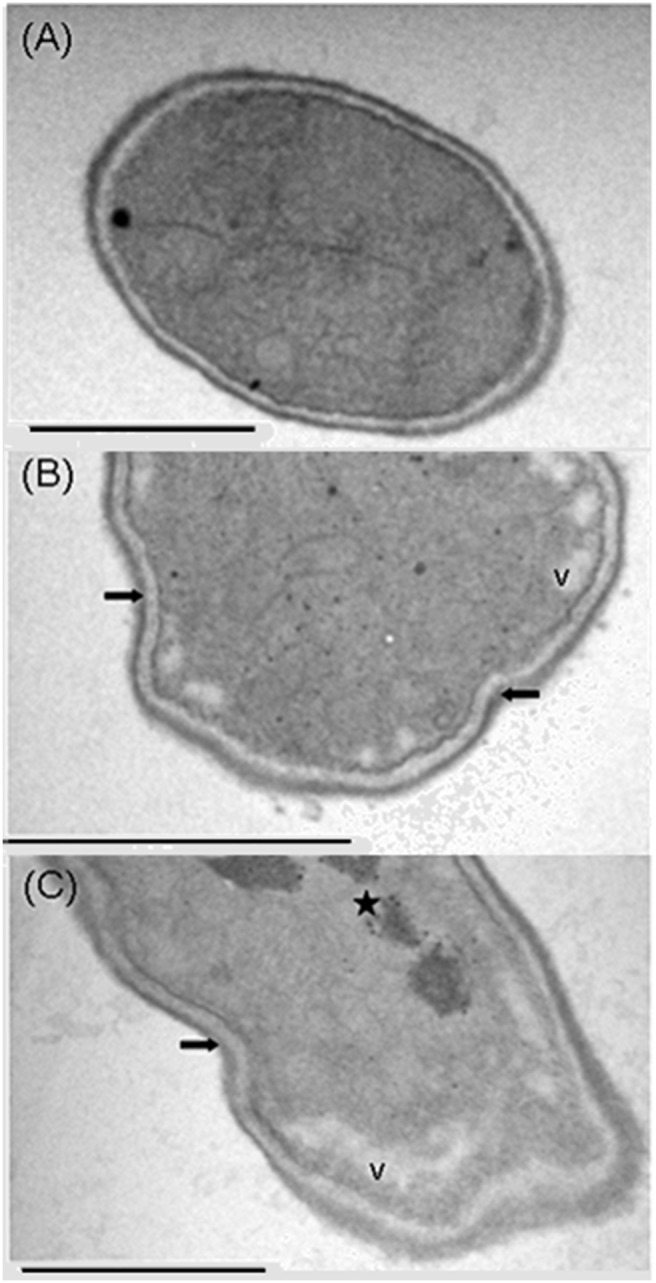
The cells were cultured either in the absence (A) and presence (B and C) of Mo-CBP3 (0.05 mg.mL−1). Star indicates condensation of the cytosolic content. Vacuole condensation (V) is also shown. Arrows indicate shrinkage of the cell wall. Bars: 0.5 µm (A–C).
Evaluation of cytotoxicity effects of Mo-CBP3
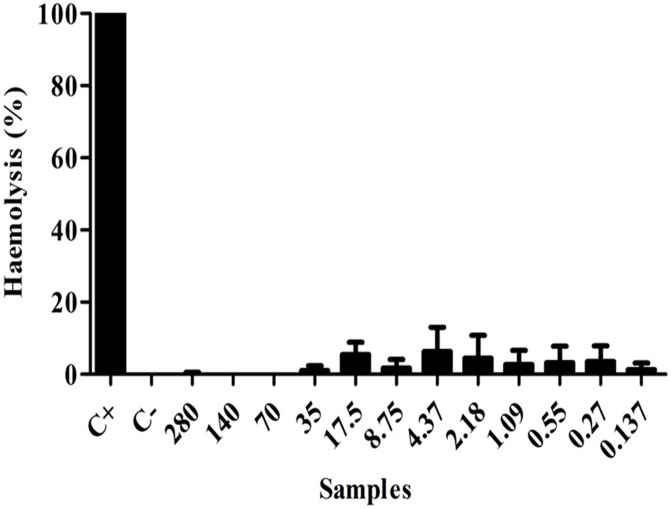
Many antimicrobial proteins also exhibit toxic potential on eukaryotic cells. In this context, the mechanical stability of the membrane of red blood cells is a good indicator to evaluate in vitro the effects of various compounds when screening for cytotoxicity [41]. These cells may undergo a loss of membrane integrity and die rapidly as a result of cell lysis. Thus, to evaluate whether Mo-CBP3 causes cytotoxicity, haemolytic assay was utilized by measuring the release of haemoglobin at different Mo-CBP3 concentrations. Mo-CBP3 was compared with the detergent Triton X-100, whose relative haemoglobin release was set at 100%. For all concentrations tested (from 0.137 a 280 µM) Mo-CBP3 did not show haemolytic activity (Figure 9), suggesting that the antifungal action of this protein occurs via a selective interaction with the fungal membrane. This result shows that although Mo-CBP3 displayed remarkable antifungal activity against phytopathogenic fungi, it shows no haemolytic activity. Similar results were found for an antifungal peptide (AFP-J) purified from potato tubers (Solanum tuberosum cv. L Jopung) [42].
Figure 9. Evaluation of the cytotoxic effect of Mo-CBP3.
In vitro haemolytic activity of Mo-CBP3 on human erythrocytes using concentrations ranging from 280 to 0.137 µM. Positive control (C+): 1% Triton X-100. Negative control (C–): 100 mM sodium phosphate buffer, pH 7.4, 150 mM NaCl.
In conclusion, this study reinforces previous data on the antifungal properties of Mo-CBP3 and reports new information about its structural features and mode of action. The CD spectral data from different temperature and pH conditions indicate that the high structural stability of Mo-CBP3 results in the effectiveness of its antifungal activity through interactions with the cell membrane, which causes prominent morphological changes followed by the induction of oxidative stress, eventually leading to cell death. Considering its elevated stability and specific toxicity, with broad-spectrum efficacy against important phytopathogenic fungi at low inhibitory concentrations but not to human cells, Mo-CBP3 has great potential for the development of new antifungal drugs or transgenic crops with enhanced resistance to phytopathogens.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
This work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq (www.cnpq.br), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - CAPES (www.capes.gov.br), and Fundação Cearense de Apoio ao Desenvolvimento Científico e Tecnológico - FUNCAP (www.funcap.ce.gov.br). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Duan XH, Jiang R, Wen YJ, Bin JH (2013) Some 2S albumin from peanut seeds exhibits inhibitory activity against Aspergillus flavus . Plant Physiol Biochem 66: 84–90. [DOI] [PubMed] [Google Scholar]
- 2. Zottich U, Da Cunha M, Carvalho AO, Dias GB, Silva NC, et al. (2011) Purification, biochemical characterization and antifungal activity of a new lipid transfer protein (LTP) from Coffea canephora seeds with alfa-amylase inhibitor properties. Biochim Biophys Acta 1810: 375–383. [DOI] [PubMed] [Google Scholar]
- 3. Selitrennikoff CP (2001) Antifungal proteins. Appl Environm Microbiol 67: 2883–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Borges AF, Ferreira RB, Monteiro S (2013) Transcriptomic changes following the compatible interaction Vitis vinifera-Erysiphe necator. Paving the way towards an enantioselective role in plant defence modulation. Plant Physiol Biochem 68: 71–80. [DOI] [PubMed] [Google Scholar]
- 5. Wong JH, Ip DCW, Ng TB, Chan YS, Fang F, et al. (2012) A defensin-like peptide from Phaseolus vulgaris cv. ‘King Pole Bean’. Food Chem 135: 408–414. [DOI] [PubMed] [Google Scholar]
- 6. Morais JKS, Gomes VM, Oliveira JTA, Santos IS, Da Cunha M, et al. (2010) Soybean toxin (SBTX), a protein from soybeans that inhibits the life cycle of plant and human pathogenic fungi. J Agric Food Chem 58: 10356–10363. [DOI] [PubMed] [Google Scholar]
- 7. Choi H, Cho J, Jin Q, Woo ER, Lee DG (2012) Antifungal property of dihydrodehydrodiconiferyl alcohol 9′-O-β-d-glucoside and its pore-forming action in plasma membrane of Candida albicans . Biochim Biophys Acta 1818: 1648–1655. [DOI] [PubMed] [Google Scholar]
- 8. Deo Prasad B, Jha S, Chattoo BB (2008) Transgenic indica rice expressing Mirabilis jalapa antimicrobial protein (Mj-AMP2) shows enhanced resistance to the rice blast fungus Magnaporthe oryzae . Plant Sci 175: 364–371. [Google Scholar]
- 9. Trindade MB, Lopes JLS, Soares-Costa A, Monteiro-Moreira AC, Moreira RA, et al. (2006) Structural characterization of novel chitin-binding lectins from the genus Artocarpus and their antifungal activity. Biochim Biophys Acta 1764: 146–152. [DOI] [PubMed] [Google Scholar]
- 10. Bindschedler LV, Whitelegge JP, Millar DJ, Bowell GP (2006) A two component chitin-binding protein from French bean – Association of a proline-rich protein with a cysteine-rich polypeptide. FEBS Lett 580: 1541–1546. [DOI] [PubMed] [Google Scholar]
- 11. Bormann C, Baier D, Hörr I, Raps C, Berger J, et al. (1999) Characterization of a novel, antifungal, chitin-binding protein from Streptomyces tendae Tü901 that interferes with growth polarity. J Bacteriol 181: 7421–7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yao Q, Wu CF, Luo P, Xiang XC, Liu JJ, et al. (2010) A new chitin-binding lectin from rhizome of Setcreasea purpurea with antifungal, antiviral and apoptosis-inducing activities. Proc Biochem 45: 1477–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gifoni JM, Oliveira JTA, Oliveira HD, Batista AB, Pereira ML, et al. (2012) A novel chitin-binding protein from Moringa oleifera seed with potential for plant disease control. Biopolymers 98: 406–415. [DOI] [PubMed] [Google Scholar]
- 14. Laemmli UK (1970) Cleavage of structural proteins during the assembly of bacteriophage T4. Nature 277: 685–689. [DOI] [PubMed] [Google Scholar]
- 15. Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- 16. Sreerama N, Woody RW (2000) Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON and CDSSTR methods with an expanded reference set. Anal Biochem 287: 252–260. [DOI] [PubMed] [Google Scholar]
- 17. Ji C, Kuc J (1996) Antifungal activity of cucumber β-1,3-glucanase and chitinase. J Physiol Mol Plant Pathol 49: 257–265. [Google Scholar]
- 18. Broekaert WF, Terras FRG, Cammue BPA, Vanderleyden J (1990) An automated quantitative assay for fungal growth inhibition. FEMS Microbiol Lett 69: 55–59. [Google Scholar]
- 19. Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB (1997) Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11: 1187–1194. [Google Scholar]
- 20. Mendieta JR, Pagano MR, Muñoz FF, Daleo GR, Guevara MG (2006) Antimicrobial activity of potato aspartic proteases (StAPs) involves membrane permeabilization. Microbiology 152: 2039–2047. [DOI] [PubMed] [Google Scholar]
- 21. Bignami GS (1993) A rapid and sensitive hemolysis neutralization assay for palytoxin. Toxicon 31: 817–820. [DOI] [PubMed] [Google Scholar]
- 22. Ranjbar B, Gill P (2009) Circular dichroism techniques: biomolecular and nanostructural analyses. Chem Biol Drug Des 74: 101–120. [DOI] [PubMed] [Google Scholar]
- 23. Beintema JJ (1994) Structural features of plant chitinases and chitin-binding proteins. FEBS Lett 350: 159–163. [DOI] [PubMed] [Google Scholar]
- 24. Asensio JL, Cañada FJ, Siebert HC, Laynez J, Poveda A, et al. (2000) Structural basis for chitin recognition by defense proteins: GlcNAc residues are bound in a multivalent fashion by extended binding sites in hevein domains. Chem Biol 7: 529–543. [DOI] [PubMed] [Google Scholar]
- 25. Freitas CDT, Lopes JLS, Beltramini LM, Oliveira RSB, Oliveira JTA, et al. (2011) Osmotin from Calotropis procera latex: New insights into structure and antifungal properties. Biochim Biophys Acta 1808: 2501–2507. [DOI] [PubMed] [Google Scholar]
- 26. Broekaert WF, Marien W, Terras FRG, De Bolle MFC, Proost P, et al. (1992) Antimicrobial peptides from Amaranthus caudatus seeds with sequence homology to the cysteine/glycine-rich domain of chitin-binding proteins. Biochemistry 31: 4308–4314. [DOI] [PubMed] [Google Scholar]
- 27. Yao Q, Wu C, Luo P, Xiang X, Liu J, et al. (2010) A new chitin-binding lectin from rhizome of Setcreasea purpurea with antifungal, antiviral and apoptosis-inducing activities. Proc Biochem 45: 1477–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lindquist W (1953) On the mechanism of yeast flocculation. J Inst Brew 59: 59–61. [Google Scholar]
- 29. Jenssen H, Hamill P, Hancock RE (2006) Peptide antimicrobial agents. Clin Microbiol Rev 19: 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Theis T, Wedde M, Meyer V, Stahl U (2003) The antifungal protein from Aspergillus giganteus causes membrane permeabilization. Antimicrob Agents Chemoter 47: 588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carvalho AO, Gomes VM (2011) Plant defensins and defensin-like peptides – biological activities and biotechnological applications. Curr Pharm Des 17: 4270–4293. [DOI] [PubMed] [Google Scholar]
- 32. Koka LT, Norris DM (1972) Comparative phospholipid compositions of adult female Xyleborus ferrugineus and its mutualistic fungal ectosymbionts. Comp Biochem Physiol B 42: 245–254. [Google Scholar]
- 33. Mello EO, Ribeiro SFF, Carvalho AO, Santos IS, Da Cunha M, et al. (2011) Antifungal activity of PvD1 defensin involves plasma membrane permeabilization, inhibition of medium acidification, and induction of ROS in fungi cells. Curr Microbiol 62: 1209–1217. [DOI] [PubMed] [Google Scholar]
- 34. Aerts AM, François IEJA, Meert EMK, Li QT, Cammue BPA, et al. (2007) The antifungal activity of Rs-AFP2, a plant defensin from Raphanus sativus, involves the induction of reactive oxygen species in Candida albicans . J Mol Microbiol Biotechnol 13: 243–247. [DOI] [PubMed] [Google Scholar]
- 35. Gessler NN, Aver’yanov AA, Belozerskaya TA (2007) Reactive oxygen species in regulation of fungal development. Biochemistry 72: 1091–1109. [DOI] [PubMed] [Google Scholar]
- 36. Agizzio AP, Da Cunha M, Carvalho AO, Oliveira MA, Ribeiro SFF, et al. (2006) The antifungal properties of a 2S albumin-homologous protein from passion fruit seeds involve plasma membrane permeabilization and ultrastructural alterations in yeast cells. Plant Sci 171: 515–522. [DOI] [PubMed] [Google Scholar]
- 37. Richards A, Veses V, Gow NAR (2010) Vacuole dynamics in fungi. Fungal Biol Rev 24: 93–105. [Google Scholar]
- 38. Narasimham ML, Damsz B, Coca MA, Ibeas JI, Yun DJ, et al. (2001) A plant defense response effector induces microbial apoptosis. Mol Cell 8: 921–930. [DOI] [PubMed] [Google Scholar]
- 39. Qi G, Zhu F, Du P, Yang X, Qiu D, et al. (2010) Lipopeptide induces apoptosis in fungal cells by a mitochondria-dependent pathway. Peptides 31: 1978–1986. [DOI] [PubMed] [Google Scholar]
- 40. Brul S, Nussbaum J, Dielbandhoesing SK (1997) Fluorescent probes for wall porosity and membrane integrity in filamentous fungi. J Microbiol Methods 28: 169–178. [Google Scholar]
- 41. Riaz M, Rasool N, Bukhari IH, Shahid M, Zubair M, et al. (2012) In vitro antimicrobial, antioxidant, cytotoxicity and GC-MS analysis of Mazus goodenifolius . Molecules 17: 14275–14287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee J-K, Gopal R, Seo CH, Cheong H, Park Y (2012) Isolation and purification of a novel deca-antifungal peptide from potato (Solanum tuberosum L cv. Jopung) against Candida albicans . Int J Mol Sci 13: 4021–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.