Significance
During rapid adaptation, populations start in hostile conditions and must evolve new traits to survive. Development of cancer from a population of precancerous cells within a body is an example of such rapid adaptation. New traits required for cancer progression are acquired by driver mutations in a few key genes. Most mutations, however, are unimportant for progression and can be damaging to cancer cells, termed “passengers.” The role these damaging passengers play in cancer and other adaptive processes is unknown. Here we show that driver mutations engage in a tug-of-war with damaging passengers. This tug-of-war explains many phenomena in oncology, suggesting how to develop new therapies and target existing therapies to exploit damaging passengers.
Keywords: evolution, mathematical modeling, simulations, cancer genomics, chemotherapy
Abstract
Cancer progression is an example of a rapid adaptive process where evolving new traits is essential for survival and requires a high mutation rate. Precancerous cells acquire a few key mutations that drive rapid population growth and carcinogenesis. Cancer genomics demonstrates that these few driver mutations occur alongside thousands of random passenger mutations—a natural consequence of cancer’s elevated mutation rate. Some passengers are deleterious to cancer cells, yet have been largely ignored in cancer research. In population genetics, however, the accumulation of mildly deleterious mutations has been shown to cause population meltdown. Here we develop a stochastic population model where beneficial drivers engage in a tug-of-war with frequent mildly deleterious passengers. These passengers present a barrier to cancer progression describable by a critical population size, below which most lesions fail to progress, and a critical mutation rate, above which cancers melt down. We find support for this model in cancer age–incidence and cancer genomics data that also allow us to estimate the fitness advantage of drivers and fitness costs of passengers. We identify two regimes of adaptive evolutionary dynamics and use these regimes to understand successes and failures of different treatment strategies. A tumor’s load of deleterious passengers can explain previously paradoxical treatment outcomes and suggest that it could potentially serve as a biomarker of response to mutagenic therapies. The collective deleterious effect of passengers is currently an unexploited therapeutic target. We discuss how their effects might be exacerbated by current and future therapies.
Although many populations evolve new traits via a gradual accumulation of changes, some adapt very rapidly. Examples include viral adaptation during infection (1), the emergence of antibiotic resistance (2), artificial selection in biotechnology (3), and cancer (4). Rapid adaptation is characterized by three key features: (i) the availability of strongly advantageous traits accessible by rare mutations, (ii) an elevated mutation rate (1), and (iii) a dynamic population size (5). Because traditional theories of gradual adaptation are not applicable under these conditions, new approaches are needed.
Cancer progression is an example of a rapidly adapting population: cancers develop as many as 10 new traits (6), exhibit a high mutation rate (6–8), and rapidly change in population size (9). Progression is driven by a handful of mutations (10) and chromosomal abnormalities (11) in cancer-related genes (oncogenes and tumor suppressors), collectively called “drivers.” Drivers are beneficial to cancer cells as they facilitate uncontrolled proliferation and other hallmarks of cancer (6). Drivers, however, arise alongside thousands of other mutations/alterations, called “passengers,” that are randomly dispersed throughout the genome, are nonrecurrent in patients, and have no immediate beneficial effect (10).
Passengers have previously been assumed to be neutral and largely ignored in cancer research, yet growing evidence suggests that they can be deleterious to cancer cells and play an important role in both cancer progression and clinical outcomes. Previously, we showed that deleterious passengers readily accumulate during tumor progression and exhibit signatures of damaging mutations (12). Passenger mutations and chromosomal abnormalities, including aneuploidy, can be deleterious via a variety of mechanisms such as direct loss-of-function (13), cytotoxicity from protein disbalance and aggregation (14), or by eliciting an immune response (15).
Although the role of deleterious mutations in cancer is largely unknown, their effects on natural populations have been extensively studied in genetics (16–18). The accumulation of deleterious mutations can cause population extinction by Muller’s ratchet and mutational meltdown (16, 19). How this applies to rapidly adapting populations with a varying size and advantageous mutations, and specifically to cancer, remains unknown.
A rapidly adapting population faces a double bind: it must quickly acquire often exceedingly rare, adaptive mutations, yet also avoid mutational meltdown. As a result, adaptive processes frequently fail. Indeed, less than 0.1% of species on Earth have adapted fast enough to avoid extinction (20), and similarly, only ∼0.1% of precancerous lesions ever advance to cancer (21). Evolutionary properties of extinction may be exploitable in evolving tumors (22).
Here we investigate how asexual populations such as cancer rapidly evolve new traits while avoiding mutational meltdown. We observed a tug-of-war between beneficial drivers and deleterious passengers that creates two major regimes of population dynamics: an adaptive regime, where the probability of adaptation (cancer) is high, and a nonadaptive regime, where adaptation (cancer) is exceedingly rare. These regimes are separated by an effective barrier, which makes cancer progression an unlikely event. Our model is consistent with cancer genomic and age–incidence data, offers a new interpretation of cancer treatment strategies, and explains a previously paradoxical relationship between cancer mutation rates and clinical outcomes. Most importantly, it suggests that deleterious passengers offer a new, unexploited avenue of cancer therapy.
Results
Model.
In simulations, each cell can divide, mutate (in a general sense, i.e., including copy number alterations, chromosomal gains and losses, epigenetic changes, etc.), and die stochastically. Mutations occur during cell divisions with a per-locus rate μ. The frequency of driver mutations per cell division is the overall mutation rate times the number of driver loci in the genome (i.e., target size) Td; hence, μd = μTd. The frequency of passenger mutations is μp = μTp. Because there are many more passenger loci than driver loci, Tp ≫ Td (SI Appendix).
A single cell’s fitness w is determined by its number of accumulated drivers nd that each increase its fitness by sd, and its number of accumulated passengers np that each decrease fitness by sp: . This form assumes that mutations act independently, i.e., no epistasis. In SI Appendix and Discussion, we consider a simple two-hit form of epistasis. Constant values of sd and sp are used here because previous work showed that sampling s from various distributions exhibits qualitatively similar dynamics (12). We assume here and estimate from data below that sd > sp.
Cell fitness w and the population size N determine the birth and the death rate of that cell. The death rate increases with N according to a Gompertzian function often used to describe cancer dynamics (9). Stochastic cell division (with mutations) and death were modeled using a Gillespie algorithm. To develop the most tractable and foundational model, we assume that all parameters stay constant in time.
Adaptive processes occur within a broad range of evolutionary parameters. For example, μ varies dramatically across cancers (8), whereas estimates of sd range from 0.0001 (4) to 0.58 (23). Hence, we varied each parameter by 1,000-fold (SI Appendix, Table S1) and found that dynamics varied considerably over this range but fell into two broad categories: adaptation (cancer) and extinction (no progression). Previous studies have concluded that passengers minimally affect progression when lethal (7) or if only deleterious in a few housekeeping genes (24); however, we previously presented genomic evidence that cancers accumulate myriad mildly deleterious passengers (12).
A Critical Population Size.
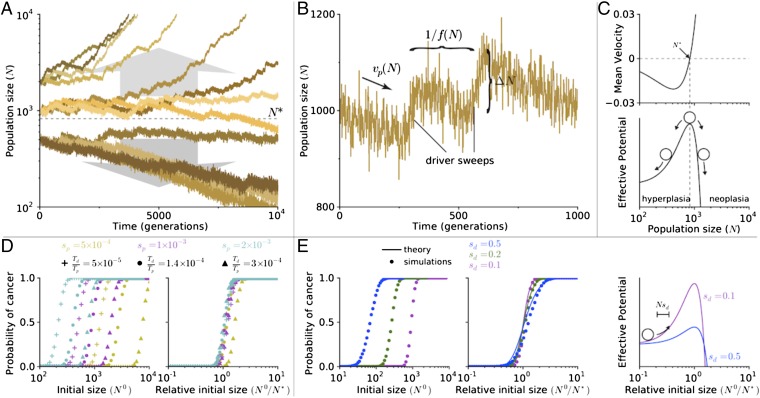
Fig. 1A shows the dynamics N(t) of individual populations starting at different initial sizes N0, which correspond to different potential hyperplasia sizes (trajectories begin immediately after a stem cell acquires its first driver; see SI Appendix for a discussion of dynamics before this point). Populations exhibit two ultimate outcomes, growth to a macroscopic size (i.e., cancer progression) or extinction, which depend on a critical population size N*. Larger populations (N > N*) generally commit to rapid growth, whereas smaller populations (N < N*) generally commit to extinction.
Fig. 1.
Tug-of-war between drivers and passengers leads to a critical population size. (A) Population size verses time of simulations initiated at various sizes (N0 = 500, 1,000, or 2,000). For all simulations presented in this paper, μ = 10−8, Td = 1,400, Tp = 107, sd = 0.1, and sp = 0.001 (SI Appendix, Table S1), unless specified otherwise. (B) A segment of a trajectory shows periods of rapid growth and gradual decline. New drivers arrive with a frequency f(N), abruptly increasing the population size by an amount ΔN. Passenger accumulation causes a gradual decline with rate vp. (C) Analytically computed mean velocity of population growth (Upper) and an effective barrier (Lower) as a function of population size N. Velocity is negative below N* and positive above it. (D) The probability of adaptation (cancer) as a function of initial population size N (Left) and a relative initial population size (N/N*; Right) for nine sets of evolutionary parameters. Curves collapse and behave similarly when plotted relative to N*. (E) Same as in D for simulations and theory but for different values of sd. Higher values of sd leads to a more gradual transition from nonadaptive to adaptive regime. In our formalism, an increase in sd results in a larger jump size ΔN and lower potential barrier, allowing more populations to overcome the barrier (Right).
To understand the origin of this critical population size N*, we examined the short-term dynamics of populations. All trajectories follow a reversed sawtoothed pattern (Fig. 1B), resulting from a tug-of-war between drivers and passengers (12). When a new driver arises and fixates in the population, the population size increases to a new stationary value. In between these rare driver events, the population size gradually declines due to passenger accumulation. The relative rates of these competing processes determine whether a population grows rapidly or goes extinct.
The value of N* can be identified by considering the average change in population size over time (〈dN/dt〉), which is the average population growth due to accumulating drivers (vd) minus the population decline due to accumulating passengers (vp). When a new driver fixates, the population size immediately increases by ΔN = Nsd. These jumps occur randomly at a nearly constant rate f = μdNsd [a product of the driver occurrence rate μdN and its probability of fixation sd/(1 + sd) ≈ sd]. Thus, the velocity due to drivers is . Similarly, passengers’ velocity vp = μpNsp is a product of their rate of occurrence μpN, effect on population size Nsp, and fixation probability (near-neutral ∼1/N; see SI Appendix for a more precise estimate). Thus, we obtain
| [1] |
where is the critical population size. In this equation, a population’s mean velocity is negative below N* and positive above N*, explaining why populations above N* grow rapidly and populations below N* continually decline.
These dynamics suggests that N* constitutes an effective barrier to cancer, which can prevent most cancers from progressing (Fig. 1C). Simulations support this conclusion because they exhibit a sharp transition in the probability of progression at N* (Fig. 1D). Indeed, drastically different probability curves collapse onto a single curve when N0 is rescaled by N*.
By considering only the average dynamics, we miss the variability of outcomes in adapting populations. In SI Appendix, we formulate and solve a stochastic generalization of Eq. 1 that explains the variability of outcomes well (Fig. 1E). Outcome variability depends only upon N* and the strength of drivers sd. Higher values of sd lead to larger stochastic jumps ΔN, which increases variability and leads to more gradual changes in the probability of cancer across N0.
The effects of N* and sd on cancer progression can be intuitively understood using a simple random-walk analogy. The population’s reverse sawtoothed path abides by a one-dimensional random walk in an effective potential (Fig. 1C). Akin to chemical reactions activated by thermal energy, cancer progression is a rare event that transitions through an unstable state, requiring a quick succession of driver fixations.
Our analytical formalism indicates that population dynamics depend entirely on two dimensionless parameters: a deterministic mean velocity dependent only upon N/N* and a stochastic step size of ∼sd. By reducing the complexity of our evolutionary system to two parameters, we were next able to infer their values from real cancers without overfitting.
Model Validation Using Cancer Incidence and Genomics Data.
Our model of cancer progression predicts several properties of cancer: the presence of an effective barrier that small lesions seldom overcome, a broad distribution in the number of accumulated drivers and passengers in tumors, and a tug-of-war between drivers and passengers in individual cancers. We looked for evidence of these phenomena in age–incidence data (25) and recent cancer genomics data, which also allowed us to estimate sd, sp, and the probability of cancer progression.
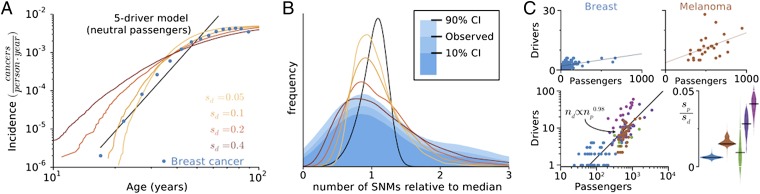
Fig. 2A presents the incidence rate of breast cancer versus age (25) alongside the predictions from our model and a classic driver-only model (SI Appendix). The incidence rate was calculated by assuming that precancerous lesions arise with a constant rate r beginning at birth. Lesions then progress to cancer in time τ with probability P(τ), determined from simulations (SI Appendix, Fig. S1). The age–incidence rate I(t) is then the convolution of P(τ) with the lesion initiation rate r. Because many lesions never progress and go extinct in our model, the incidence rate saturates at old age: , where P∞ is the probability that a lesion ever progresses to cancer.
Fig. 2.
Signatures of balance between drivers and passengers in incidence and genomics data. (A) Predicted and observed breast cancer incidence rates verses age. Incidence rates in our model and the data both plateau at old age, but a traditional driver-only model (I ∝ tk) does not. (B) Histogram (blue) of the collective number of protein-coding mutations (SNMs) in breast cancer, alongside predicted distributions (lines colors as in A). Our model captures the width and asymmetry of the distribution when sd = 0.4, whereas a driver-only model predicts a narrower, less-skewed distribution. (C) (Upper) The total number of aggregate SCNA and SNM drivers verses the total number of passengers in sequenced tumors (points) fitted by linear regression. (Lower Left) Aggregated cancer genomics data plotted on log axes, with the y intercept from each subtype’s linear fit subtracted [lung cancer (green), colorectal cancer (MIN−, dark purple; MIN+, light purple)]. (Lower Right) The distribution of slope values obtained by bootstrapping 10,000 samples of each tumor type. All cancers exhibit positive slopes (P < 0.08 − 10−5), suggesting estimates of sp/sd of breast, 0.0060 ± 0.0010; melanoma, 0.016 ± 0.003; lung, 0.0094 ± 0.0093; colorectal, MIN− 0.028 ± 0.007 and MIN+ 0.041 ± 0.006.
Observed age–incidence rates saturate with age, allowing us to estimate the efficiency of cancer progression. We estimate the rate of lesion formation in breast cancer r ≥ 10 per year, deducible in two ways: by multiplying the number of human breast epithelial stem cells by their rate of mutation into a lesion (SI Appendix) or by considering the number of lesions observed in normal breasts (21). By comparing this limit (r ∼ 10 y−1) to the maximum observed breast cancer incidence rate Imax ∼ 10−2cancers⋅year−1, we find that P∞ ∼ 10−3, or only ∼0.1% of lesions ever progress. Likewise, most other cancer subtypes (84%) plateau at old age, indicating that inefficient progression is common (SI Appendix, Fig. S1). These findings are consistent with medical observations that very few lesions ever progress to cancer (26, 27). Like our model, these studies also find that most lesions regress to undetectable size. Conversely, in a driver-only model (SI Appendix), every lesion eventually progresses to cancer after sufficient time (i.e., P∞ = 1), and incidence rates plateau only if the lesion formation rate is unrealistically low (0.01 per year). Good agreement between age–incidence data and our model is obtained for sd ≈ 0.1−0.2.
Recent cancer genomics data offer a new opportunity to validate our model. Specifically, we looked at somatic nonsynonymous mutations (SNMs) and somatic copy-number alterations (SCNAs) from over 700 individual cancer–normal sample pairs (SI Appendix, Table S2). Analyzing SNMs and SCNAs separately and in aggregation yielded similar results (SI Appendix, Fig. S2 and Table S3). Fig. 2B shows a wide and asymmetric distribution of the total number of SNMs in breast cancer. Our model predicts a similarly wide distribution of total SNMs due to the stochastic period that cancers linger at the critical population size N* while accumulating mutations. To compare these data with various models we normalized the total number of mutations by their median (27) because several evolutionary parameters and sequencing decisions can alter these distributions by a multiplicative factor (SI Appendix). Our model agrees with the data when the effect size of drivers is large (sd ≈ 0.4). In contrast, a traditional five-driver model (SI Appendix) which neglects deleterious passengers and, thus, a critical barrier yields a narrower, less-skewed distribution than observed.
Overall, our model agrees with the 11 most sequenced cancer subtypes best when sd ≈ 0.1−0.6 (SI Appendix, Fig. S3 and Table S4). These estimates agree with sd = 0.16−0.58 experimentally measured as growth rate changes of mouse cells upon mutations in p53, APC, or Kras (23). These experimental measurements and our estimates are also considerably larger than previous theoretical estimates of sd = 0.004 (4), where cancer progression was modeled as an exponential growth unaffected by passengers. Such a driver-only model fits SNM distributions poorly, and fits suggest that just one to two drivers are needed for cancer (SI Appendix, Table S4), which is inconsistent with known biology. Taken together, cancer genomics data and recent experiments (23) strongly support our model and refute the driver-only model.
We then compared the number of drivers and the number of passengers observed in individual cancer samples to our model. Lesions that linger around N* in our model acquire additional passengers and additional counterbalancing drivers, whereas lesions that progress quickly acquire fewer of each. This predicts a linear relationship: nd = sp/sd ⋅ np + constant (SI Appendix), where the slope provides an estimate of sp/sd. We indeed observed a positive linear relationship between nd and np in all tumors studied (Fig. 2C and SI Appendix, Table S3; P < 0.08−10−6). Linearity was confirmed by regressing the aggregated data in log–log axes (Fig. 2C), which yielded , consistent with nd ∼ np. Regressing nd on np and using 104 bootstrapped samples to estimate the confidence, we obtain an sp/sd ∼ 0.005−0.05 (Fig. 2C). Using our estimate sd ≈ 0.1−0.6, we obtain a damaging effect of a passenger mutation sp ≈ 5 ⋅ (10−4−10−2). These estimates are consistent with effects of germ-line SNMs in humans where 64% of mutations decrease fitness by 10−5−10−2 (28). In summary, this analysis shows that passengers are indeed deleterious and ∼100× weaker than drivers.
We considered and refuted several alternative explanations for the observed positive linear relationship between drivers and passengers. First, variation in the tumor stage or the rate/mechanism of mutagenesis cannot explain the observed relationship (SI Appendix, Table S3). Second, SCNAs and SNMs are uncorrelated, so the linear relationship could not result from their differing effect sizes (SI Appendix, Fig. S2 and Table S3). Because the data disagree with these alternate hypotheses, we conclude that cancer indeed proceeds as a tug-of-war between drivers and passengers.
A Critical Mutation Rate.
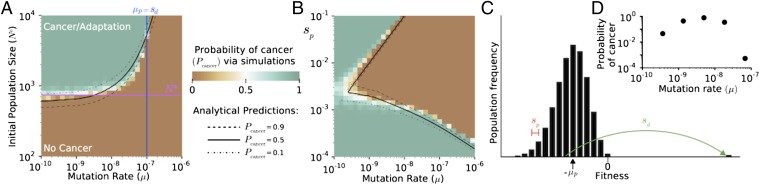
By simulating cancer progression over a broad range of evolutionary parameters (SI Appendix, Fig. S4), we observed another barrier to cancer: a critical mutation rate, above which the probability of cancer is exceedingly low (Fig. 3A). Through further analytical treatment (SI Appendix), we found that this mutational barrier is created and determined by the load of segregating (unfixed) passengers in the population. The origin of a critical mutation rate can be understood by considering the number of segregating passengers per cell, previously shown to be Poisson distributed with mean μp/sp, during mutation–selection balance (29). The average fitness reduction of a cell due to this mutational load is then μp. If this fitness reduction exceeds the benefit of a new driver (μp > sd), then drivers seldom fixate (17, 30) (Fig. 3C). Hence, cancer is extremely rare above the critical mutations rate μ* = sd/Tp.
Fig. 3.
Effect of mutation rate on cancer dynamics. (A) The probability of cancer (adaptation) computed by simulations across mutation rates and initial population sizes. Evolutionary parameters partition adaptation into a regime where cancer is almost certain (green) and a regime where it is exceedingly rare (brown), accurately predicted by theoretical estimates of N* (magenta) and μ* (blue). A theory incorporating passenger interference with driver sweeps and selection against passengers explains simulations well (black lines). (B) Cancer probability as a function of μ and sp (N0 = 103). Theory (black lines) accurately reproduces the complex transition between regimes. (C) Diagram illustrating how the load of segregating (unfixed) passengers influences relative cell fitness and the probability of a driver fixating. Hitchhiking passengers reduce a driver’s fitness benefit and probability of fixation. (D) Probability of cancer constrained to grow within a human lifespan ∼60 years, 104 generations, with N0 = 103 for various mutation rates exhibits an optimum mutation rate.
Fig. 3A shows that this simple argument accurately predicts the location of the critical mutation rate. By incorporating more details about segregating passengers and selection against them (SI Appendix), we explain observed simulation dynamics well across the entire phase space (Fig. 3 A and B and SI Appendix, Fig. S4).
This analysis offers a new mode by which mutational meltdown operates. Whereas prior models of mutational meltdown consider deleterious mutations in isolation (16), we find that mutational meltdown can occur when deleterious mutations inhibit the accumulation of advantageous mutations.
When cancer progression is constrained to develop within a human lifetime, we observe an optimum mutation rate for the probability of cancer (μopt = 10−9−10−8 nucleotide−1⋅generation−1; Fig. 3D), similar to experimentally measured rates in cancer of 10−8 (7). Above μopt, population meltdown is very common, whereas below μopt, progression is too slow.
The Adaptive Barrier and Critical Mutation Rate Explain Cancer Treatment Outcomes.
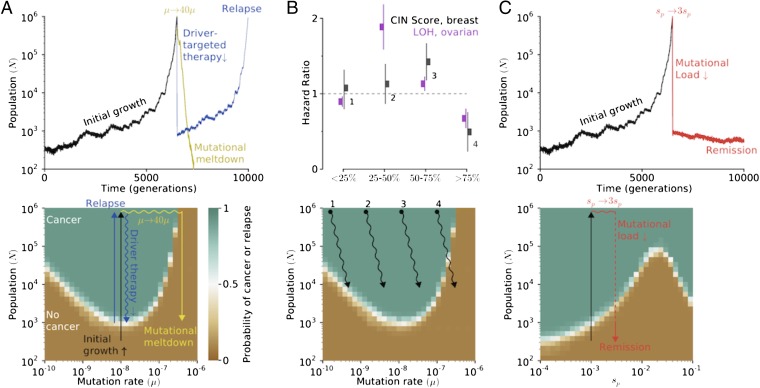
Chemotherapy and radiation are valued for their ability to kill rapidly dividing cells; however, our model shows that the elevation of mutation rates (including SCNAs and aneuploidy) by these therapies dramatically affects cancer survival. We use the phase diagrams from Fig. 3 to rationalize outcomes of these and other treatments.
In Fig. 4A, we present evolutionary paths of cancers—from hyperplasia, to cancer, to treatment, and to relapse or remission—on top of the phase diagrams described above. Treatments succeed if they push cancer into the nonadaptive regime (where the probability of growth is low) and fail if they do not. Our model suggests that chemotherapy succeeds, in part, because it moves cancers across the mutational threshold μ*. Above this threshold, drivers seldom overpower the load of segregating passengers, making readaptation difficult. Driver-targeted therapies (that eliminate an oncogene’s benefit) must bring n < N* to succeed.
Fig. 4.
Mapping and interpreting treatment outcomes. (A, Upper) An adapted population (cancer) can be reverted to extinction by increasing the mutation rate (mutagenic chemotherapy) or by decreasing the population size (e.g., surgery or cytotoxic chemotherapy). (Lower) Our phase diagrams explain therapeutic outcomes: therapies that alter evolutionary parameters enough to push it outside of the adaptive regime cause continued population collapse; those that do not readapt and relapse. (B, Upper) Cancers with intermediate mutational loads are the most aggressive (31, 33), whereas patients with very high level of chromosomal instability are most effectively treated. (Lower) This result is well explained by our phase diagrams, where cancers with high mutation rates are susceptible to mutational meltdown, yet paradoxical for all previous evolutionary models of cancer. We believe traditional therapies decrease population size and may increase the mutation rate. (C) Threefold increase in the effect of passenger mutations leads to rapid population meltdown below N*, without relapse.
Cancers with higher loads of mutations/alterations are closer to the critical mutation rate and should be most susceptible to mutagenic meltdown. Several recent studies (31–33) found that patient survival from breast (all subtypes) and ovarian cancer was greatest when tumors harbored exceptionally high levels of chromosomal alterations. These findings are paradoxical for all previous models of cancer (31), where a greater mutation rate always accelerates cancer, yet fully consistent with our model (Fig. 4B).
Treatments exploiting cancer’s load of deleterious passengers remain unexplored. Fig. 4C shows that a relatively mild threefold to fivefold increase of the deleterious effect of passengers sp causes complete remission. Increasing sp is doubly effective because it exacerbates accumulated passengers and slows down future adaptation. Below we discuss possible treatment strategies that would increase sp.
Increases in μ work synergistically with increases in sp in our model (SI Appendix, Fig. S6). Hence, combinations of mutagenic chemotherapy with treatments elevating the cost of passengers may be most effective; these therapies should further synergize with driver-targeted therapies.
Discussion
We present an evolutionary model of rapid adaptation incorporating rare, strongly advantageous driver mutations and frequent, mildly deleterious passenger mutations. In this tug-of-war process, populations either succeed and adapt or fail and go extinct. Simulations and theory identify two regimes of dynamics: one where populations almost always adapt and another where they almost always fail. The complex stochastic dynamics of this process is accurately described as simple diffusion over a potential barrier located at a critical population size separating the two regimes. This general framework for adaptive asexual populations effectively characterizes the dynamics of cancer progression and therapeutic responses.
We show that the late onset of cancer, evident in age–incidence curves, can be explained by a passenger-generated barrier to cancer and does not require more complex models of cancer progression (e.g., a specific order of mutations, or multihit model, or variable mutation rate). We also considered a commonly used two-hit model in SI Appendix, where the first driver confers no fitness benefit, whereas two drivers confer a strong cumulative effect. This two-hit model also exhibits a barrier to cancer and behaves similarly to our model (SI Appendix, Fig. S7), yet with an effectively larger N* and sd.
Our framework suggests that most normal tissues reside in a regime where cancer progression is exceedingly rare. Most lesions fail to overcome the adaptive barrier at N*. This implies that observed tumors acquired drivers faster than a mean trajectory and may be testable via phylogenetic analysis.
Clinical cancers, on the contrary, reside above the adaptive barrier in a rapidly adapting state. Successful therapies must push a cancer below this adaptive barrier N* or increase the mutation rate above the critical value μ*.
We tested our model and estimated its parameters using age–incidence curves, cancer exome sequences from ∼1,000 tumors in four cancer subtypes, and data on clinical outcomes. Age–incidence curves support our hypothesis that nearly all lesions fail to progress and allowed us to estimate the fitness benefit of a driver as sd ≈ 0.1−0.6, in good agreement with direct experimental measurements (23). Genomics data also affirmed that a damaging effect of a nonsynonymous passenger is 100 times smaller than the effect of a driver, but passengers are 100 times more numerous than drivers. Taken together, these data support the notion of a tug-of-war between rare large-effect drivers verses frequent mildly deleterious passengers.
Clinicians could exploit deleterious passengers by either increasing the mutation/alteration rate in cancers with already high mutation rates or by increasing the deleterious effect of passengers. Clinical data indeed show that cancers with a higher load of chromosomal alterations, closer to μ*, respond better to treatments (31–33). PARP inhibitors, which increase DNA damage in BRCA1/2-positive tumors, may already be curing patients by inducing mutational meltdown (34).
Passenger mutations and alterations can be deleterious by gain-of-function toxicity via proteotoxic/misfolding stress (14, 35) or by eliciting an immune response to mutated epitopes (15). Their damage to cancer cells could be magnified by (i) inhibiting unfolded protein response (UPR) pathways and proteasomes (14), (ii) hyperthermia that further destabilizes mutated proteins and clogs UPR pathways (36), or (iii) activating an immune response (37). All these strategies are in clinical trials, yet none are believed to work by exacerbating passengers’ deleterious effects. Our alternative explanation for their efficacy suggests that these therapies will be most effective in combination and in cancers with many passengers.
This study focused on the evolution of cancer, but our model should generalize to other adaptive asexual processes. Consider a small population entering a new environment. Fluctuations in its size often lead to its extinction. Occasionally, however, the population may acquire several new highly advantageous traits for this new environment, allowing it to rapidly expand its size and avert extinction. Both its evolutionary parameters (18) and behavior (38) mirror our model. Our mathematical framework further explains why these populations sometimes adapt, yet often fail.
Materials and Methods
Simulations used a first-order Gillespie algorithm (12). Drivers and passengers were classified in the studies where they were first identified. See SI Appendix for details.
Supplementary Material
Acknowledgments
We are grateful to Shamil Sunyaev, Gregory Krykov, Geoffrey Fudenberg, Maxim Imakaev, and Anton Goloborodko for many productive discussions. C.D.M. and L.A.M. were supported by National Cancer Institute Grant U54CA143874. K.S.K. was supported by the Pappalardo Fellowship in Physics at Massachusetts Institute of Technology.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404341111/-/DCSupplemental.
References
- 1.Kawashima Y, et al. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458(7238):641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Q, et al. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science. 2011;333(6050):1764–1767. doi: 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]
- 3.Wright SI, et al. The effects of artificial selection on the maize genome. Science. 2005;308(5726):1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- 4.Bozic I, et al. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci USA. 2010;107(43):18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cameron TC, O’Sullivan D, Reynolds A, Piertney SB, Benton TG. Eco-evolutionary dynamics in response to selection on life-history. Ecol Lett. 2013;16(6):754–763. doi: 10.1111/ele.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Beckman RA, Loeb LA. Negative clonal selection in tumor evolution. Genetics. 2005;171(4):2123–2131. doi: 10.1534/genetics.105.040840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domingues JS. Gompertz model: Resolution and analysis for tumors. J Math Model Appl. 2012;1(7):70–77. [Google Scholar]
- 10.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45(10):1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McFarland CD, Korolev KS, Kryukov GV, Sunyaev SR, Mirny LA. Impact of deleterious passenger mutations on cancer progression. Proc Natl Acad Sci USA. 2013;110(8):2910–2915. doi: 10.1073/pnas.1213968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacArthur DG, et al. 1000 Genomes Project Consortium A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335(6070):823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheltzer JM, Amon A. The aneuploidy paradox: Costs and benefits of an incorrect karyotype. Trends Genet. 2011;27(11):446–453. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vesely MD, Schreiber RD. Cancer immunoediting: Antigens, mechanisms, and implications to cancer immunotherapy. Ann N Y Acad Sci. 2013;1284:1–5. doi: 10.1111/nyas.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch M. The cellular, developmental and population-genetic determinants of mutation-rate evolution. Genetics. 2008;180(2):933–943. doi: 10.1534/genetics.108.090456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson T, Barton NH. The effect of deleterious alleles on adaptation in asexual populations. Genetics. 2002;162(1):395–411. doi: 10.1093/genetics/162.1.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goyal S, et al. Dynamic mutation-selection balance as an evolutionary attractor. Genetics. 2012;191(4):1309–1319. doi: 10.1534/genetics.112.141291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neher RA, Shraiman BI. Fluctuations of fitness distributions and the rate of Muller’s ratchet. Genetics. 2012;191(4):1283–1293. doi: 10.1534/genetics.112.141325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.May R, Lawton J, Stork N. Assessing Extinction Rates. Oxford Univ Press; Oxford: 1995. [Google Scholar]
- 21.Page DL, Dupont WD, Rogers LW, Rados MS. Atypical hyperplastic lesions of the female breast. A long-term follow-up study. Cancer. 1985;55(11):2698–2708. doi: 10.1002/1097-0142(19850601)55:11<2698::aid-cncr2820551127>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 22.Korolev KS, Xavier JB, Gore J. Turning ecology and evolution against cancer. Nat Rev Cancer. 2014;14(5):371–380. doi: 10.1038/nrc3712. [DOI] [PubMed] [Google Scholar]
- 23.Vermeulen L, et al. Defining stem cell dynamics in models of intestinal tumor initiation. Science. 2013;342(6161):995–998. doi: 10.1126/science.1243148. [DOI] [PubMed] [Google Scholar]
- 24.S Datta R, Gutteridge A, Swanton C, Maley CC, Graham TA. Modelling the evolution of genetic instability during tumour progression. Evol Appl. 2013;6(1):20–33. doi: 10.1111/eva.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. National Cancer Institute (2014) Surveillance, Epidemiology, and End Results (SEER) Program Research Data (1973–2011). Available at http://seer.cancer.gov/archive/csr/1975_2009_pops09/results_single/sect_01_table.10_2pgs.pdf. Accessed March 7, 2014.
- 26.Bota S, et al. Follow-up of bronchial precancerous lesions and carcinoma in situ using fluorescence endoscopy. Am J Respir Crit Care Med. 2001;164(9):1688–1693. doi: 10.1164/ajrccm.164.9.2012147. [DOI] [PubMed] [Google Scholar]
- 27.Tong WWY, et al. Progression to and spontaneous regression of high-grade anal squamous intraepithelial lesions in HIV-infected and uninfected men. AIDS. 2013;27(14):2233–2243. doi: 10.1097/QAD.0b013e3283633111. [DOI] [PubMed] [Google Scholar]
- 28.Boyko AR, et al. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4(5):e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haigh J. The accumulation of deleterious genes in a population—Muller’s Ratchet. Theor Popul Biol. 1978;14(2):251–267. doi: 10.1016/0040-5809(78)90027-8. [DOI] [PubMed] [Google Scholar]
- 30.Peck JR. A ruby in the rubbish: Beneficial mutations, deleterious mutations and the evolution of sex. Genetics. 1994;137(2):597–606. doi: 10.1093/genetics/137.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Birkbak NJ, et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011;71(10):3447–3452. doi: 10.1158/0008-5472.CAN-10-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciriello G, et al. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45(10):1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang ZC, et al. Australian Ovarian Cancer Study Group Profiles of genomic instability in high-grade serous ovarian cancer predict treatment outcome. Clin Cancer Res. 2012;18(20):5806–5815. doi: 10.1158/1078-0432.CCR-12-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee AJX, Swanton C. Tumour heterogeneity and drug resistance: Personalising cancer medicine through functional genomics. Biochem Pharmacol. 2012;83(8):1013–1020. doi: 10.1016/j.bcp.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 35.Jordan DM, Ramensky VE, Sunyaev SR. Human allelic variation: Perspective from protein function, structure, and evolution. Curr Opin Struct Biol. 2010;20(3):342–350. doi: 10.1016/j.sbi.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossi-Fanelli A, Cavaliere R, Mondov B, Moricca G. Selective Heat Sensitivity of Cancer Cells. Springer; Berlin: 1977. pp. 1–192. [Google Scholar]
- 37.Segal NH, et al. Epitope landscape in breast and colorectal cancer. Cancer Res. 2008;68(3):889–892. doi: 10.1158/0008-5472.CAN-07-3095. [DOI] [PubMed] [Google Scholar]
- 38.Bell G, Gonzalez A. Evolutionary rescue can prevent extinction following environmental change. Ecol Lett. 2009;12(9):942–948. doi: 10.1111/j.1461-0248.2009.01350.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.