Significance
Duplication of genomes following hybridization (allopolyploidy) is common among flowering plants, particularly in the grasses that cover vast areas of the world and provide food and fuel. Here, we find that genome duplication has occurred at a remarkable rate, accounting for at least a third of all speciation events in a group of about 1,200 species. Much of this genome duplication occurred during the expansion of the C4 grasslands in the Late Miocene. We find no evidence that allopolyploidy leads directly to a change in the net rate of diversification or correlates with the origin of novel morphological characters. However, as a mode of speciation, the frequency of allopolyploidization is surprisingly high.
Abstract
The role of polyploidy, particularly allopolyploidy, in plant diversification is a subject of debate. Whole-genome duplications precede the origins of many major clades (e.g., angiosperms, Brassicaceae, Poaceae), suggesting that polyploidy drives diversification. However, theoretical arguments and empirical studies suggest that polyploid lineages may actually have lower speciation rates and higher extinction rates than diploid lineages. We focus here on the grass tribe Andropogoneae, an economically and ecologically important group of C4 species with a high frequency of polyploids. A phylogeny was constructed for ca. 10% of the species of the clade, based on sequences of four concatenated low-copy nuclear loci. Genetic allopolyploidy was documented using the characteristic pattern of double-labeled gene trees. At least 32% of the species sampled are the result of genetic allopolyploidy and result from 28 distinct tetraploidy events plus an additional six hexaploidy events. This number is a minimum, and the actual frequency could be considerably higher. The parental genomes of most Andropogoneae polyploids diverged in the Late Miocene coincident with the expansion of the major C4 grasslands that dominate the earth today. The well-documented whole-genome duplication in Zea mays ssp. mays occurred after the divergence of Zea and Sorghum. We find no evidence that polyploidization is followed by an increase in net diversification rate; nonetheless, allopolyploidy itself is a major mode of speciation.
Polyploidy (whole-genome duplication) is often linked with the acquisition of new traits and subsequent species diversification, particularly in plants (1, 2). Ancient polyploidy correlates with major land-plant radiations (3) and the origins of orders, large families, and major clades (4–7) although, in many cases, sharp changes in diversification rates are delayed for millions of years after the polyploidization event (1). This phylogenetic pattern has led to the hypothesis that polyploidy causes or promotes diversification. Good mechanistic reasons support such a hypothesis. Studies of naturally occurring and synthetic polyploids find changes in gene expression, gene loss, release of transposons, and changes in morphology and physiology immediately after polyploidy (4, 8–12). This pattern is particularly true for allopolyploids, which originate from a cross between genetically distinct parents often representing different species; in such cases, the biological changes seen after polyploidization may not reflect the effects of genome doubling per se, but rather the effect of hybridization between distantly related progenitors (4).
Despite the appeal of the hypothesis that polyploidy causes diversification, there is evidence to the contrary. As noted by Stebbins (13), “polyploidy has been important in the diversification of species and genera within families, but not in the origin of the families and orders themselves,” implying that polyploidy is only a minor force in diversification (see also ref. 14). Surveys of angiosperms and ferns have found no evidence for increased speciation after polyploidization (15, 16), supporting Stebbins's hypothesis. However, because of the large scale of the analyses, these studies necessarily had to rely on inference of polyploidization events from chromosome numbers. Although unavoidable, inferences from chromosome numbers require assumptions about which numbers represent polyploids and when the polyploids arose.
Phylogenetic trees of nuclear genes can reliably identify allopolyploidization events (17–21) because they produce characteristic double-labeled tree topologies in which the polyploid species appears twice (Fig. S1). In such trees, allopolyploids are readily identified and can be discovered even when no chromosome counts are available (e.g., ref. 22). This tree-based method will recover genetic allopolyploids; following the terminology of Doyle and Egan (23), these polyploids may be descendants of two species (taxonomic allopolyploids) or one (taxonomic autopolyploids). The gene-tree approach thus provides unambiguous evidence of allopolyploidization and provides an estimate of relative timing that does not require calibration of a tree. The approach does require identification of all or most paralogues, which can be accomplished by investigating genes in categories that tend to be retained after whole-genome duplication, such as transcription factors.
The grass family is a powerful system in which to study allopolyploidization. About 80% of its species are polyploid (24), and Hunziker and Stebbins (25) have noted that the grasses are the “only large family in which high frequency of polyploids prevails throughout the family.” The polyploids that Stebbins refers to and that we discuss in this paper all formed after the well-documented whole-genome duplication at the origin of the family (26, 27); the grass WGD (rho) occurred about 80–90 million years ago (Mya) (28, 29).
Here, we focus on the tribe Andropogoneae (subfamily Panicoideae), a group of about 1,200 species in 90 genera. The tribe is morphologically diverse and contains some of our most economically important crop plants [maize (Zea mays), sorghum, and sugarcane (Saccharum officinarum)]. In addition, members of the tribe are ecologically dominant species of temperate and tropical grasslands (e.g., Andropogon gerardii, Schizachyrium scoparium, and Sorghastrum nutans of the North American tall grass prairie; Themeda triandra of East African and Australian grasslands), some of the most troublesome agricultural weeds (Imperata cylindrica, Sorghum halepense, Ischaemum spp.), many important forage crops (Dichanthium spp., Bothriochloa spp., Andropogon spp.), and some burgeoning biofuels (Miscanthus and Saccharum). In short, the Andropogoneae feed, and increasingly fuel, the planet.
Of the species listed in the preceding paragraph, all but sorghum are allopolyploid. We therefore sought to test whether each allopolyploidy event leads only to one or a few species (the Stebbins hypothesis), in which case, the allopolyploids should appear in the phylogeny as the result of numerous independent polyploidy events. Alternatively, allopolyploidy could correlate with diversification of a major clade or even the entire tribe (the diversification hypothesis), in which case most allopolyploids would be the product of a single polyploidy event followed by radiation at the tetraploid or hexaploid level; we would see only a handful of polyploidization events deep in the tree (Fig. S1, second row).
Previous attempts to resolve relationships within Andropogoneae, using both chloroplast and nuclear markers, have produced phylogenies that are poorly supported, with intergeneric relationships unresolved (30–36). The pattern apparently indicates a rapid radiation. One reason for the difficulty of resolving the phylogeny may be a failure to account for polyploidy. Mixing of orthologues and paralogues in gene trees, combined with extensive reticulation in the history of the genera, could easily destroy any phylogenetic signal. The apparent rapid radiation of the tribe might also reflect diversification following a single polyploidy event.
In this paper, we used a phylogenetic approach, supported by data on chromosome number and genome size. We found a remarkable number of allopolyploidization events, none of which correlate with shifts in diversification rate or with the origin of unique morphological characters. Repeated formation of polyploids correlates with expansion of the C4 grasslands in the Late Miocene.
Results
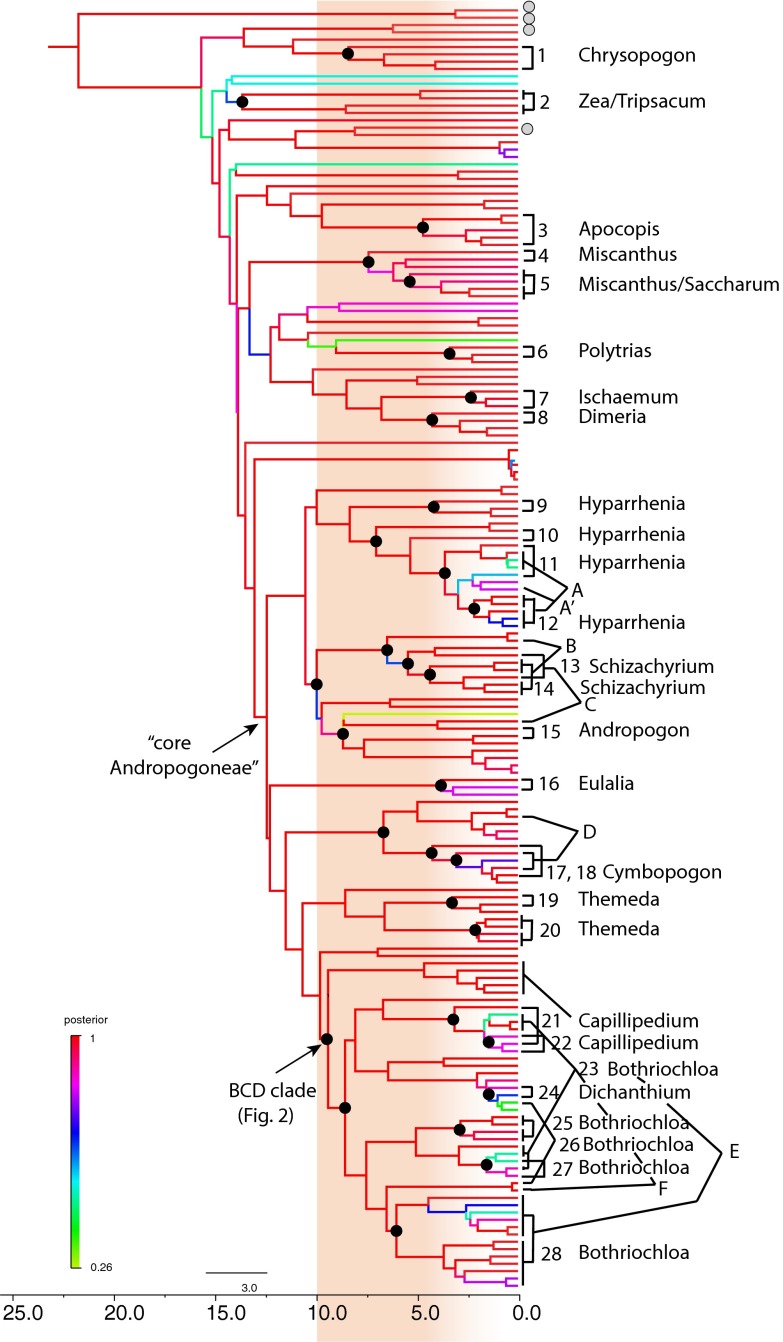
Allopolyploidy occurs repeatedly throughout the Andropogoneae, accounting for at least a third of the speciation events. Based on our phylogenetic criteria, 28 groups (numbered in Fig. 1 and Figs. S2 and S3) show the phylogenetic signature of allotetraploidization, with an additional six hexaploid events (letters in Fig. 2). Together, these events have given rise to 32 of the 100 sampled species, with a few species forming repeatedly, from more than one independent event.
Fig. 1.
Chronogram of the phylogeny of Andropogoneae produced in BEAST (57). Branch colors indicate Bayesian posterior probability with red highest, green lowest. Numbers, allotetraploidization events; letters, allohexaploidization events; black dots, common ancestors of the parental genomes of the polyploids; gray dots, accessions with large, likely polyploid, genome sizes but without evidence of genetic allopolyploidy; shading, approximate time of the Miocene grassland expansion. The order in which the hexaploid genomes came together is unknown; for clarity, they are drawn as though a tetraploid formed from the most closely related parents, and the hexaploidy event added the more distant one, but this is merely a graphical convention.
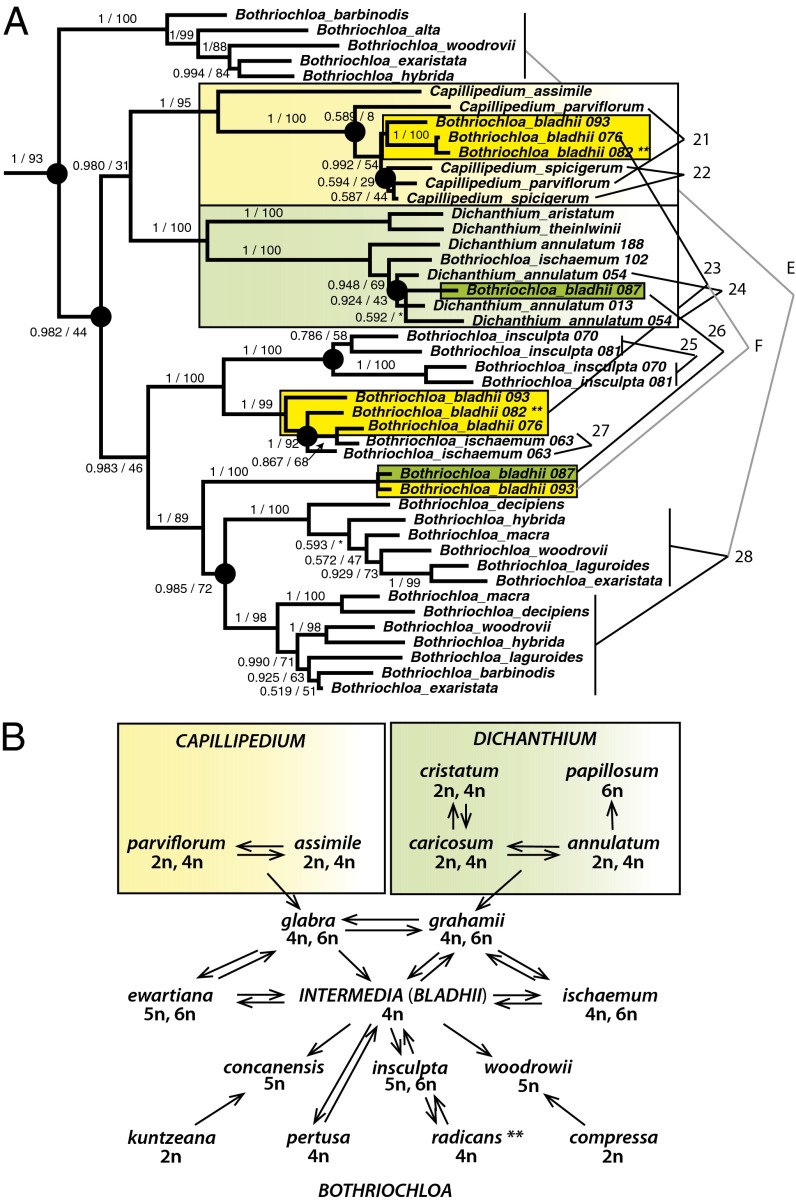
Fig. 2.
(A) Phylogeny of the Bothriochloa-Capillipedium-Dichanthium clade, showing the complex history of B. bladhii, which combines genomes of all three genera in plants that are morphologically placed in Bothriochloa. Numbered polyploidization events correspond to those in Fig. 1 and Figs. S2 and S3. For species with more than one accession, accession numbers follow the species name. Black lines connect genomes of tetraploids, and gray lines connect hexaploids. (B) Summary of data from extensive crossing experiments and chromosome pairing studies [redrawn from de Wet and Harlan (42)]. The genomic history inferred from cytogenetics is congruent with that in our phylogenetic study. Green shading, Dichanthium genomes; yellow shading, Capillipedium genomes; dark green and dark yellow shading, Bothriochloa accessions bearing Dichanthium and Capillipedium genomes, respectively.
Most allopolyploidization events have occurred within genera or species. For example, T. triandra is the product of an event (event 20) separate from the tetraploidy that gave rise to T. villosa and T. arundinacea (event 19) (Fig. 1 and Fig. S3). Hyparrhenia hirta, Hyparrhenia rufa, and Hyparrhenia diplandra appear to be the products of four separate events (events 9–12), with additional rounds of polyploidy leading to hexaploids in H. hirta (events A and A′) (Fig. 1 and Fig. S3).
We looked for shifts in net diversification rate (speciation minus extinction) using Bayesian Analysis of Macroevolutionary Mixtures (BAMM) (37). A model with no shift of evolutionary rates had a slightly higher posterior probability than one with one shift (odds ratio 1.0395); in the model with one shift, the position of the shift cannot be determined with certainty (Fig. S4). However, there is no evidence of an increase in diversification in polyploid clades.
To test net diversification rates in diploids versus polyploids directly, we used a nonparametric comparison between the numbers of species in diploid clades and those in their sister polyploid clades and found no significant difference (P = 0.637). We then used the Binary State Speciation and Extinction (BiSSE) model (38) and found that net diversification is significantly higher in diploids, assuming a sampling fraction of 8% (likelihood ratio test, P < 0.02) (Fig. S5); the pattern is similar assuming 50% sampling, but the difference is not significant. Although estimated speciation rates are higher in polyploids, estimated extinction rates are also higher (K-S test, P < 2.2e−16), leading to lower net diversification.
Only in 2 of the 32 allopolyploid lineages can we be certain that polyploidy is followed by diversification to produce more than one or two species. Event 2 represents a hybridization and polyploidy event that led to Z. mays plus Tripsacum dactyloides: i.e., the allopolyploidy apparently preceded the origin of the genera, which together include 17 species. Tetraploidy 28 and hexaploidy E precede the diversification of a clade of Bothriochloa species. Because this clade includes all of the South American species that we sampled (in addition to a few species from the Old World) and because all South American species are thought to be hexaploid or higher, we suspect that all of the ca. 13 species will fall into this clade. Thus, we identify one event leading to 17 species and the other to at least 13 species; both radiations occur at approximately the average rate of speciation for the entire clade.
In addition to the events identified by gene-tree patterns, we found several species with large genome sizes (>1,800 Mbp) but no double-labeling in the gene trees (gray dots; Fig. 1 and Fig. S2); assuming an approximate diploid genome size for the tribe of about 900 Mbp, any plant with a number larger than 1,800 is likely to be polyploid. These plants may be genetic autopolyploids (sensu ref. 23), allopolyploids for which we have not sequenced deeply enough to detect paralogues, or simply diploids in which transposons have amplified to produce large genomes. If these four species are indeed polyploids, then the total for the tribe is a minimum of 38%.
The most complex history of hybridization is in the clade including Bothriochloa, Capillipedium, and Dichanthium (the BCD clade) (Fig. 2A). Tetraploid and hexaploid plants that are morphologically classified as Bothriochloa bladhii have some sequences derived from Bothriochloa-like clades and some derived from Capillipedium (tetraploidy events 24 and hexaploidy F). Other plants that are morphologically B. bladhii have sequences derived from Dichanthium-like relatives (e.g., tetraploidy 27). Thus, the B. bladhii morphology is retained even in crosses involving distinct ancestral species.
The Zea-Tripsacum clade is the only unambiguous example of polyploidization preceding diversification of genera. In contrast, we find five instances in which polyploidization has occurred after divergence of morphologically defined genera. Event 3 involves Apocopis intermedius, in which one parent is classified as Apocopis and the other as Germainia. Event 5 precedes the divergence of Miscanthus eckloni and Saccharum narenga but postdates the divergence of the genus Miscanthus. Events 23 and 26 in the BCD clade involve all three genera. Two sequences of Schizachyrium hirtiflorum appear to be from disparate ancestors within Schizachyrium (tetraploidy event 13) whereas the third is derived from species of Andropogon (hexaploidy C).
The precise number of diploids in our sample is unclear; although we are confident that 38% of the species are the product of allopolyploidization, it is unlikely that all—or even most—of the remaining 62% are diploid. We infer diploidy if our accession has a genome size less than ca. 1 Gbp, published counts for the species are all diploid, and we find only a single gene copy for each locus; all three criteria apply, for example, to Sorghum bicolor, Diheteropogon amplectens, Pseudosorghum fasciculare, and Andropogon virginicus. The latter species is a member of a clade of four species that is sister to one of the big bluestem (A. gerardii) genomes (Fig. S3); A. virginicus is, like A. gerardii, a native of the North American prairie and thus is a plausible model for one genome of big bluestem. For most other species, however, chromosome counts are unavailable or published counts include several ploidy levels and are often poorly documented. Genome-size estimates may be lacking for field-collected specimens and are in any case not definitive. Discovering only one gene copy is weak evidence because our PCR-based approach may not have uncovered all paralogues. We do not think gene loss is a major concern because we have always been able to locate paralogues in known polyploids given sufficient depth of sequencing and because we deliberately chose gene classes that are generally retained in duplicate after polyploidization events.
Most allopolyploids formed in the Late Miocene or more recently. Of the documented allopolyploidization events, only three could have occurred more than 9 Mya (Table 1). Gene-tree data provide an estimate of the divergence time of the parental diploid genomes but cannot determine exactly when the hybridization and genome-doubling events occurred (23); dates in Table 1 are thus the oldest possible dates for polyploidization. However, if speciation has occurred after allopolyploidization, as in Themeda villosa-arundinacea, or Zea-Tripsacum, the date of the earliest tetraploid speciation event provides the most recent possible date. Thus, the Zea-Tripsacum event occurred after 13 Mya (the divergence of the parental genomes) but before 8.5 Mya (the earliest estimate for divergence of the two genera).
Table 1.
Divergence dates of parental genomes of allopolyploids, and 95% highest posterior density (HPD) values
| Labeled event | Taxon | Divergence of parental genomes, Mya | 95% HPD | |
| 1 | Chrysopogon gryllus | 8.4 | 10.6 | 4.1 |
| 2 | Zea-Tripsacum | 13.6 | 18.2 | 9.1 |
| 3 | Apocopis intermedius | 4.7 | 6.3 | 2.4 |
| 4 | Miscanthus sinensis | 7.4 | 9.4 | 3.7 |
| 5 | Miscanthus-Saccharum | 3.8 | 6.2 | 2.0 |
| 6 | Polytrias indica | 3.4 | 5.8 | 1.9 |
| 7 | Ischaemum santapaui | 2.3 | 3.4 | 0.9 |
| 8 | Dimeria fuscescens | 4.2 | 7.9 | 3.4 |
| 9 | Hyparrhenia diplandra | 4.2 | 6.1 | 1.9 |
| 10 | Hyparrhenia rufa (153) | 7.0 | 8.3 | 3.8 |
| 11 | Hyparrhenia rufa (108) | 7.0 | 8.3 | 3.8 |
| 12 | Hyparrhenia hirta (018, 185) | 2.2 | 3.0 | 1.1 |
| 13 | Schizachyrium sanguineum var. hirtiflorum | 5.5 | 7.2 | 3.0 |
| 14 | Schizachyrium sanguineum | 4.3 | 6.3 | 2.4 |
| 15 | Andropogon gerardii | 8.6 | 10.2 | 4.7 |
| 16 | Eulalia quadrinevis | 3.8 | 8.6 | 2.9 |
| 17 | Cymbopogon citratus | 3.0 | 3.6 | 1.5 |
| 18 | Cymbopogon parkeri | 4.2 | 4.7 | 1.9 |
| 19 | Themeda villosa | 3.3 | 3.8 | 0.8 |
| 20 | Themeda triandra | 2.0 | 3.3 | 1.0 |
| 21 | Capillipedium parviflorum | 3.1 | 3.6 | 1.2 |
| 22 | Capillipedium spicigerum | 1.4 | 1.6 | 0.4 |
| 23 | Bothriochloa bladhii (093, 082, 076) | 7.5 | 9.8 | 5.0 |
| 24 | Dichanthium annulatum | 1.4 | 2.2 | 0.8 |
| 25 | Bothriochloa insculpta | 2.8 | 4.6 | 1.8 |
| 26 | Bothriochloa bladhii (087) | 8.5 | 11.1 | 5.7 |
| 27 | Bothriochloa ischemum (063) | 1.5 | 2.7 | 0.7 |
| 28 | Bothriochloa spp. | 6.0 | 7.3 | 3.4 |
| A | Hyparrhenia hirta (185, 189, 018) | 3.6 | 5.4 | 2.3 |
| A′ | Hyparrhenia hirta (018) | 3.0 | 4.1 | 1.7 |
| B | Schizachyrium sangineum | 6.5 | 8.3 | 3.5 |
| C | S. sanguineum var. hirtiflorum | 9.9 | 11.8 | 5.8 |
| D | Cymbopogon citratus | 6.6 | 7.9 | 3.3 |
| E | Bothriochloa spp. | 9.4 | 11.8 | 6.1 |
| F | Bothriochloa bladhii (093) | 8.5 | 11.1 | 5.7 |
Numbered events indicate allotetraploids; letters indicate allohexaploids. Accession numbers in parentheses are for species represented by more than one accession.
Discussion
The number of independent allopolyploidy events in Andropogoneae is remarkably and unexpectedly high. Thirty-four unambiguous allopolyploid events can be identified easily from the tree, and this is certainly an underestimate of the true number. Although we do not know all mechanisms of speciation in the tribe, polyploidy may be as important as any other single mechanism. Major clades with distinctive morphology were established before the allopolyploidy events, which also do not correlate with obvious habitat shifts, or subsequent bursts of speciation. No polyploidization event encompasses the base of the tree; we thus reject the most extreme diversification hypothesis—that allopolyploidy precedes the radiation of the tribe. Instead, allopolyploidy events have occurred repeatedly postdiversification.
Simple inspection of the tree, plus results of the BAMM and BiSEE analyses, is consistent with the Stebbins hypothesis—that polyploidization is common but not accompanied by an increase in net speciation rate (13, 15, 16, 39). In fact, we found a modest net decrease in diversification, similar to that modeled by Arrigo and Barker (39). The polyploidization events we identify are old enough that there has been enough time for subsequent speciation (Table 1). The tree includes speciation events as young as 0.5 My. Generation time in these plants is 1–2 y, so 0.5 My represents at least 250,000 generations, presumably long enough for detectable evolutionary change.
Our data also fit with the Radiation Time-lag Hypothesis—that polyploidy has no immediate effect on diversification at all but rather that the effects are delayed (1). For example, nearly 20 My elapsed between the genome duplication in the common ancestor of the grasses and the major radiation of species (28), and a similar time lag occurred after the duplication at the origin of Brassicaceae (40). The time-lag hypothesis provides one possible way to reconcile the diversification hypothesis and the Stebbins hypothesis—polyploidy may have no effect that can be detected over the first 10–20 million years, and any burst of speciation occurs later. However, the time-lag hypothesis also posits that genome duplication correlates with the origin of a novel and possibly adaptive trait. If that has occurred in any of our taxa, we have not been able to identify it.
Unlike most previous studies, our estimate of polyploidization is drawn solely from gene-tree structure although we bolster the conclusion with available data on genome size and in a few cases chromosome number. Although we could compare the speciation rate in a clade of polyploids sister to a clade of diploids, the polyploids may have formed (as in Hyparrhenia) by repeated independent polyploidization events; thus, the comparison does not directly assess diversification after polyploidization.
Although many lineages have failed to diversify after allopolyploidy, there are exceptions. The 17 species in Zea-Tripsacum (event 2) and 13 species of New World Bothriochloa (events 28 and E) clearly diversified after polyploidization. Without exhaustively sampling all 1,200 species in the tribe, we cannot be certain exactly how many other such groups exist, but it is possible to estimate an upper bound on the number. For example, the three sampled species of Hyparrhenia result from at least four separate tetraploidization events (events 9–12). About 75% of the 54 species in the genus are polyploid, or about 40; if they all descended from one of the identified allopolyploid events, then the largest possible radiation would be about 37 species. However, it is unlikely that we accidentally found the only polyploidization events in the genus and that all remaining polyploids are descended from one of them, so the total number following a single allopolyploidization event is likely to be much lower. Similar arguments can be made regarding other genera.
Most of the allopolyploidization events identified here occurred in the Late Miocene, simultaneous with or following the well-documented expansion of the C4 grasslands (41). The dominant species of modern C4 grasslands are members of Andropogoneae, and most are allopolyploid. Unexpectedly, we found that many of these ecological dominants in the genera Andropogon, Schizachyrium, and Hyparrhenia form a single, strongly supported clade [Bayesian posterior probability (pp) 1.0, Maximum Likelihood bootstrap (ML) 100] whose origin is dated to about 10.5 [13.5–7.5, 95% highest posterior density (HPD)] Mya. This date correlates closely with the date when C4 species came to dominate grasslands in Africa and Southern Asia (Pakistan), also estimated about 10–11 Mya; the expansion in North America is dated about 7 Mya (41). Allopolyploidy is thus correlated with ecological success, whether or not it leads to diversification.
Polyploid formation and grassland expansion are correlated in time but may or may not be causally connected. Allopolyploids might tolerate more diverse environments than their diploid ancestors, such that repeated formation of polyploids could have helped drive the formation of the grasslands. Alternatively, expansion of the grasslands might have led to expansion of the ranges of progenitor diploids; if these diploids then came into contact with diverged species, hybridized and formed allopolyploids, then the causation is the opposite—expansion of the grasslands drove polyploidization. The third possibility, of course, is that the correlation of polyploidy and grassland expansion is simply fortuitous and neither caused the other.
The complex phylogenetic structure of the BCD clade confirms and extends data collected over 40 y ago by de Wet and Harlan (42) (Fig. 2B). In a substantial set of crossing and cytogenetic studies, they demonstrated that the three genera, which are easily distinguished morphologically, are intersterile at the diploid level. However, at the tetraploid level, fertility barriers are weak, and intergeneric crosses and allopolyploidy are rampant. Many of the species are also apomictic, which is known to correlate strongly with polyploidy (43). At the center of the complex is a species that was then called Bothriochloa intermedia, now B. bladhii. This species can be clearly identified as a Bothriochloa, despite the acquisition of genomes from Capillipedium and Dichanthium. Harlan and de Wet (44) coined the term “compilospecies” for B. bladhii to indicate its ability to “absorb” genomes from disparate sources.
Some accessions of B. bladhii indeed have a Bothriochloa genome plus another that is part of a clade with Capillipedium (event 23); other Bothriochloa accessions have Bothriochloa plus Dichanthium genomes (event 26). These accessions were all grown to flowering for this study and are unmistakably morphologically Bothriochloa despite their parentage. Thus, the B. bladhii morphology appears to be dominant in some way, despite the disparate genomes from the other two species.
Our data do not identify autopolyploids, which are defined taxonomically as polyploids in which the parents are the same species, or genetically as plants exhibiting polysomic inheritance (23, 45, 46). In most cases, the lack of diploids makes it impossible to assess whether the parents are conspecific with the polyploids or not. For example, Bothriochloa insculpta (event 25) and T. triandra (event 20) are tetraploids with two distinct genomes (genetic allopolyploids), but we do not know whether the genome donors would both be classified in the same species. Our data also suggest a need for caution in inference of taxonomic autopolyploidy. Each accession of B. bladhii includes genomes from Bothriochloa and from either Capillipedium and Dichanthium, but the latter genomes are morphologically “invisible.” Likewise, A. intermedius has one parent classified as Apocopis and one as Germainia. Going by morphology alone, plants of B. bladhii or A. intermedius with different chromosome numbers would be interpreted as taxonomically autopolyploid whereas, in fact, they are both taxonomically and genetically allopolyploid. Morphology gives little or no hint of the allopolyploid origin of the species.
The phylogeny presented here (Fig. 1 and Figs. S2 and S3) is consistent with previous studies of Andropogoneae but considerably better resolved. Like previous workers, we found that the sister genus to the rest of the tribe is Arundinella (formerly in Arundinelleae), that Andropogoneae is monophyletic, and that the early diversification appears to have been rapid. Novel results are too numerous to list here but include a moderately well supported clade (1.0 pp, 71% ML) that we call “core Andropogoneae,” which includes over half the species in the tribe and within which relationships are mostly strongly supported. This clade is considered to be subtribe Andropogoninae by Kellogg (47), an interpretation that is broader than that of Clayton and Renvoize (48). The ecologically dominant genera Andropogon, Schizachyrium, and Hyparrhenia together form a strongly supported clade. Saccharum and Miscanthus are closely related, consistent with previous work (49).
Zea is a well-documented allotetraploid, and the relationships of its genomes to sorghum have long been of interest (50, 51). A previous study (52) concluded that the divergence of sorghum and the maize genomes was nearly simultaneous and occurred about 11.9 Mya. We provide a slightly earlier date, along with strong evidence that maize allopolyploidy occurred after the divergence from sorghum. In addition, we confirm that polyploidy of Miscanthus sinensis also occurred after the Miscanthus-Sorghum divergence, consistent with recent mapping studies (53–55).
The methods used here provide a robust minimum estimate of the number of allopolyploidization events in the clade and do not require the same assumptions as other approaches. Specifically, assessment of polyploidy from chromosome number or genome size requires an assumption of what the diploid value is likely to be; because both chromosome number and genome size can increase by mechanisms unconnected to polyploidy, they are helpful but not definitive. Likewise, the frequency distribution of synonymous sites (Ks plot) is an effective method for inferring polyploidy but does not have the power to determine whether near-simultaneous events are independent or not, or to distinguish allopolyploidy from autopolyploidy. As increasing numbers of nuclear genomes become available, we suggest that the gene-tree signature of allopolyploidy may provide valuable estimates of the precise timing and independence of allopolyploidization events.
Finally, we note that our formal tests for diversification, like those in the literature, assume an underlying bifurcating model of evolution whereas the question addressed here (and elsewhere) concerns the result of reticulating evolution, for which we have no appropriate null model (56). For the analyses in BAMM and BiSSE, we included only one genome of the polyploids, thereby forcing the tree to be bifurcating. In addition, BiSSE simply tests whether there is an elevated rate of net diversification in clades that contain polyploids, confounding the numbers of species that are formed by the polyploidization event itself and species formed by divergent evolution after the reticulation (Fig. S1). Rigorous determination of the role of polyploidy in subsequent diversification awaits development of new methods.
In summary, allopolyploidization is not clearly linked to increases in net diversification rate over a time scale of ca. 20 million y (the depth of the current phylogeny) although, in a few lineages, there has been some speciation following the polyploidization event. Nonetheless, allopolyploidization itself is surprisingly common and clearly produces new combinations of genomes repeatedly and frequently. Whatever their long-term fate, the resulting allopolyploids have come to dominate the major grassland ecosystems of the planet.
Materials and Methods
Taxon sampling included 114 accessions; 2 outgroup species, 2 species of Arundinella, and 100 species of Andropogoneae in 40 genera. Vouchers and sources of material are listed in Table S1. Trees were constructed from five regions of four nuclear loci, following Estep et al. (57) (SI Materials and Methods). We found no evidence that these loci are lost after polyploidization; in known polyploids, paralogues are found consistently as long as sequence depth is sufficient. Sequences were concatenated to produce a single data matrix with a minimum of three out of five loci for each taxon. Concatenated trees were reconstructed using both maximum likelihood (ML) and Bayesian approaches. Divergence times were estimated using BEAST 1.7.5 (58). Tests for shifts in the underlying model of diversification were conducted using BAMM (37), on a tree constructed by pruning the BEAST tree to leave only a single paralogue (genome) per species. Differences in speciation rate for polyploids versus diploids were estimated using the Binary State Speciation and Extinction (BiSSE) model (38) as implemented in Mesquite (59) and diversitree (60). See SI Materials and Methods for more details. Genome size was measured by flow cytometry a minimum of four times using standard methods, and the values were averaged (61). To obtain a minimum estimate of the number of allopolyploidy events, we looked for a clear phylogenetic signal of allopolyploidy, in the form of a multiple-labeled gene tree (Fig. S1).
Supplementary Material
Acknowledgments
This study was supported National Science Foundation Grants DBI-0604923 and DEB-11456884 (to E.A.K.) and by the E. Desmond Lee and Family Endowment at the University of Missouri–St. Louis.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database. For a list of accession numbers, see Table S1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404177111/-/DCSupplemental.
References
- 1.Schranz ME, Mohammadin S, Edger PP. Ancient whole genome duplications, novelty and diversification: The WGD Radiation Lag-Time Model. Curr Opin Plant Biol. 2012;15(2):147–153. doi: 10.1016/j.pbi.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Edger PP, Pires JC. Gene and genome duplications: The impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 2009;17(5):699–717. doi: 10.1007/s10577-009-9055-9. [DOI] [PubMed] [Google Scholar]
- 3.Jiao Y, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473(7345):97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 4.Doyle JJ, et al. Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet. 2008;42:443–461. doi: 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- 5.Soltis DE, et al. Polyploidy and angiosperm diversification. Am J Bot. 2009;96(1):336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- 6.Barker MS, Vogel H, Schranz ME. Paleopolyploidy in the Brassicales: Analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biol Evol. 2009;1:391–399. doi: 10.1093/gbe/evp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barker MS, et al. Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years. Mol Biol Evol. 2008;25(11):2445–2455. doi: 10.1093/molbev/msn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flagel LE, Wendel JF. Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol. 2010;186(1):184–193. doi: 10.1111/j.1469-8137.2009.03107.x. [DOI] [PubMed] [Google Scholar]
- 9.Rapp RA, Udall JA, Wendel JF. Genomic expression dominance in allopolyploids. BMC Biol. 2009;7:18. doi: 10.1186/1741-7007-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bekaert M, Edger PP, Pires JC, Conant GC. Two-phase resolution of polyploidy in the Arabidopsis metabolic network gives rise to relative and absolute dosage constraints. Plant Cell. 2011;23(5):1719–1728. doi: 10.1105/tpc.110.081281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pires JC, Gaeta RT. 2011. Structural and functional evolution of resynthesized polyploids. Genetics and Genomics of the Brassicaceae, Plant Genetics and Genomics: Crops and Models, eds Schmidt R, Bancroft I (Springer, New York), Vol 9, pp 195–214.
- 12.Pires JC, et al. Molecular cytogenetic analysis of recently evolved Tragopogon (Asteraceae) allopolyploids reveal a karyotype that is additive of the diploid progenitors. Am J Bot. 2004;91(7):1022–1035. doi: 10.3732/ajb.91.7.1022. [DOI] [PubMed] [Google Scholar]
- 13.Stebbins GL. Chromosomal Evolution in Higher Plants. Edward Arnold; London: 1971. [Google Scholar]
- 14.Stebbins GL. Variation and Evolution in Plants. Columbia Univ Press; New York: 1950. [Google Scholar]
- 15.Mayrose I, et al. Recently formed polyploid plants diversify at lower rates. Science. 2011;333(6047):1257. doi: 10.1126/science.1207205. [DOI] [PubMed] [Google Scholar]
- 16.Wood TE, et al. The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci USA. 2009;106(33):13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mason-Gamer RJ, Burns MM, Naum M. Phylogenetic relationships and reticulation among Asian Elymus (Poaceae) allotetraploids: Analyses of three nuclear gene trees. Mol Phylogenet Evol. 2010;54(1):10–22. doi: 10.1016/j.ympev.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Mason-Gamer RJ, Burns MM, Naum M. Reticulate evolutionary history of a complex group of grasses: Phylogeny of Elymus StStHH allotetraploids based on three nuclear genes. PLoS ONE. 2010;5(6):e10989. doi: 10.1371/journal.pone.0010989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Q, Triplett JK, Wen J, Peterson PM. Allotetraploid origin and divergence in Eleusine (Chloridoideae, Poaceae): Evidence from low-copy nuclear gene phylogenies and a plastid gene chronogram. Ann Bot (Lond) 2011;108(7):1287–1298. doi: 10.1093/aob/mcr231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Triplett JK, Wang Y, Zhong J, Kellogg EA. Five nuclear loci resolve the polyploid history of switchgrass (Panicum virgatum L.) and relatives. PLoS ONE. 2012;7(6):e38702. doi: 10.1371/journal.pone.0038702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marcussen T, et al. Inferring species networks from gene trees in high-polyploid North American and Hawaiian violets (Viola, Violaceae) Syst Biol. 2012;61(1):107–126. doi: 10.1093/sysbio/syr096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doust AN, Penly AM, Jacobs SWL, Kellogg EA. Congruence, conflict and polyploidization shown by nuclear and chloroplast markers in the monophyletic “bristle clade” (Paniceae, Panicoideae, Poaceae) Syst Bot. 2007;32:531–544. [Google Scholar]
- 23.Doyle JJ, Egan AN. Dating the origins of polyploidy events. New Phytol. 2010;186(1):73–85. doi: 10.1111/j.1469-8137.2009.03118.x. [DOI] [PubMed] [Google Scholar]
- 24.Stebbins GL. Polyploidy, hybridization and the invasion of new habitats. Ann Mo Bot Gard. 1985;72:824–832. [Google Scholar]
- 25.Hunziker JH, Stebbins GL. Chromosomal evolution in the Gramineae. In: Soderstrom TR, Hilu KW, Campbell CS, Barkworth ME, editors. Grass Systematics and Evolution. Smithsonian Institution; Washington, DC: 1987. pp. 179–187. [Google Scholar]
- 26.Paterson AH, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457(7229):551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 27.Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA. 2004;101(26):9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vicentini A, Barber JC, Giussani LM, Aliscioni SS, Kellogg EA. Multiple coincident origins of C4 photosynthesis in the Mid- to Late Miocene. Glob Change Biol. 2008;14:2963–2977. [Google Scholar]
- 29.Tang H, Bowers JE, Wang X, Paterson AH. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc Natl Acad Sci USA. 2010;107(1):472–477. doi: 10.1073/pnas.0908007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathews S, Spangler RE, Mason-Gamer RJ, Kellogg EA. Phylogeny of Andropogoneae inferred from phytochrome B, GBSSI, and ndhF. Int J Plant Sci. 2002;163:441–450. [Google Scholar]
- 31.Spangler R, Zaitchik B, Russo E, Kellogg E. Andropogoneae evolution and generic limits in Sorghum (Poaceae) using ndhF sequences. Syst Bot. 1999;24:267–281. [Google Scholar]
- 32.Bomblies K, Doebley JF. Molecular evolution of FLORICAULA/LEAFY orthologs in the Andropogoneae (Poaceae) Mol Biol Evol. 2005;22(4):1082–1094. doi: 10.1093/molbev/msi095. [DOI] [PubMed] [Google Scholar]
- 33.Lukens L, Doebley J. Molecular evolution of the teosinte branched gene among maize and related grasses. Mol Biol Evol. 2001;18(4):627–638. doi: 10.1093/oxfordjournals.molbev.a003843. [DOI] [PubMed] [Google Scholar]
- 34.Hodkinson TR, Chase MW, Lledó MD, Salamin N, Renvoize SA. Phylogenetics of Miscanthus, Saccharum and related genera (Saccharinae, Andropogoneae, Poaceae) based on DNA sequences from ITS nuclear ribosomal DNA and plastid trnLintron and trnL-F intergenic spacers. J Plant Res. 2002;115(5):381–392. doi: 10.1007/s10265-002-0049-3. [DOI] [PubMed] [Google Scholar]
- 35.Teerawatananon A, Jacobs SWL, Hodkinson TR. Phylogenetics of Panicoideae (Poaceae) based on chloroplast and nuclear DNA sequences. Telopea (Syd) 2011;13:115–142. [Google Scholar]
- 36.Skendzic EM, Columbus JT, Cerros-Tlatilpa R. Phylogenetics of Andropogoneae (Poaceae: Panicoideae) based on nuclear ribosomal internal transcribed spacer and chloroplast trnL-F sequences. Aliso. 2007;23:530–544. [Google Scholar]
- 37.Rabosky DL. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS ONE. 2014;9(2):e89543. doi: 10.1371/journal.pone.0089543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maddison WP, Midford PE, Otto SP. Estimating a binary character’s effect on speciation and extinction. Syst Biol. 2007;56(5):701–710. doi: 10.1080/10635150701607033. [DOI] [PubMed] [Google Scholar]
- 39.Arrigo N, Barker MS. Rarely successful polyploids and their legacy in plant genomes. Curr Opin Plant Biol. 2012;15(2):140–146. doi: 10.1016/j.pbi.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 40.Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107(43):18724–18728. doi: 10.1073/pnas.0909766107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edwards EJ, et al. C4 Grasses Consortium The origins of C4 grasslands: Integrating evolutionary and ecosystem science. Science. 2010;328(5978):587–591. doi: 10.1126/science.1177216. [DOI] [PubMed] [Google Scholar]
- 42.de Wet JMJ, Harlan JR. Apomixis, polyploidy, and speciation in Dichanthium. Evolution. 1970;24:270–277. doi: 10.1111/j.1558-5646.1970.tb01760.x. [DOI] [PubMed] [Google Scholar]
- 43.Ozias-Akins P. Apomixis: Developmental characteristics and genetics. Crit Rev Plant Sci. 2006;25:199–214. [Google Scholar]
- 44.Harlan JR, de Wet JMJ. The compilospecies concept. Evolution. 1963;17:497–501. [Google Scholar]
- 45.Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6(11):836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- 46.Parisod C, Holderegger R, Brochmann C. Evolutionary consequences of autopolyploidy. New Phytol. 2010;186(1):5–17. doi: 10.1111/j.1469-8137.2009.03142.x. [DOI] [PubMed] [Google Scholar]
- 47.Kellogg EA. Poaceae. In: Kubtizki K, editor. The Families and Genera of Vascular Plants. Springer; New York: 2014. in press. [Google Scholar]
- 48.Clayton WD, Renvoize SA. Genera Graminum: Grasses of the World. Her Majesty's Stationery Office; London: 1986. [Google Scholar]
- 49.Hodkinson TR, et al. The use of DNA sequencing (ITS and trnL-F), AFLP, and fluorescent in situ hybridization to study allopolyploid Miscanthus (Poaceae) Am J Bot. 2002;89(2):279–286. doi: 10.3732/ajb.89.2.279. [DOI] [PubMed] [Google Scholar]
- 50.Schnable PS, et al. The B73 maize genome: Complexity, diversity, and dynamics. Science. 2009;326(5956):1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 51.Schnable JC, Springer NM, Freeling M. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci USA. 2011;108(10):4069–4074. doi: 10.1073/pnas.1101368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swigonová Z, et al. Close split of sorghum and maize genome progenitors. Genome Res. 2004;14(10A):1916–1923. doi: 10.1101/gr.2332504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swaminathan K, et al. A framework genetic map for Miscanthus sinensis from RNAseq-based markers shows recent tetraploidy. BMC Genomics. 2012;13:142. doi: 10.1186/1471-2164-13-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim C, et al. SSR-based genetic maps of Miscanthus sinensis and M. sacchariflorus, and their comparison to sorghum. Theor Appl Genet. 2012;124(7):1325–1338. doi: 10.1007/s00122-012-1790-1. [DOI] [PubMed] [Google Scholar]
- 55.Ma XF, et al. High resolution genetic mapping by genome sequencing reveals genome duplication and tetraploid genetic structure of the diploid Miscanthus sinensis. PLoS ONE. 2012;7(3):e33821. doi: 10.1371/journal.pone.0033821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soltis DE, et al. Are polyploids really evolutionary dead-ends (again)? A critical reappraisal of Mayrose et al. (2011) New Phytol. 2014;202(4):1105–1117. doi: 10.1111/nph.12756. [DOI] [PubMed] [Google Scholar]
- 57.Estep MC, Vela Diaz DM, Zhong J, Kellogg EA. Eleven diverse nuclear-encoded phylogenetic markers for the subfamily Panicoideae (Poaceae) Am J Bot. 2012;99(11):e443–e446. doi: 10.3732/ajb.1200186. [DOI] [PubMed] [Google Scholar]
- 58.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maddison WP, Maddison DR. 2011 Mesquite: A Modular System for Evolutionary Analysis, Version 2.75. Available at mesquiteproject.org.
- 60.FitzJohn RG. Diversitree: Comparative phylogenetic analysis of diversification in R. Methods Ecol. Evol. 2012;3:1084–1092. [Google Scholar]
- 61.Arumuganathan K, Earle ED. Estimation of nuclear DNA amounts of plants by flow cytometry. Plant Mol Biol Rep. 1991;9:229–241. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.