Abstract
MicroRNA plays an important role in multiple processes of cancer development. Aberrant expression of miR-183 has been frequently reported in a variety of cancer types; however, the roles and mechanisms of miR-183 in gastric cancer are largely unknown. Here, we report that miR-183 is significantly up-regulated in human gastric tumor tissues compared to the adjacent normal tissues. Up-regulation of miR-183 is associated with advanced clinical stage, positive lymph node, deep stromal invasion, and distant metastasis in gastric cancer patients. We further demonstrated that miR-183 promotes gastric cancer cell growth in vitro by inhibition of apoptosis. Moreover, overexpression of miR-183 enhances gastric cancer cell migration and invasion. Mechanistically, we demonstrated that overexpression of miR-183 decreased, and inhibition of miR-183 increased the expression of PDCD4, a tumor suppressor, at both mRNA and protein levels. Taken together, our results suggest that miR-183 may modulate progression and metastatic potential of gastric cancer through inhibition of PDCD4 expression. miR-183 could serve as a potential biomarker for gastric cancer progression and a novel therapeutic target for gastric cancer treatment.
Keywords: Gastric cancer, miR-183, PDCD4, metastasis
Introduction
Gastric cancer is one of the most common malignant tumors of the digestive system [1]. Due to its genomic instability and the resultant altered gene expression, gastric cancers are of aggressive growth and metastasis ability, and thus remains the leading cause of cancer death all over the world. Although the cytogenetic and molecular aspects of gastric cancer have been investigated intensively, the molecular mechanisms controlling carcinogenesis and progression of gastric cancer remain to be elucidated.
Recently, the classical categories of oncogenes and tumor suppressor genes have been expanded to include a new family of RNAs known as microRNAs (miRNAs), which are small 21-25 nucleotides of RNA that participate in the regulation of cell differentiation, cell cycle progression, apoptosis, and tumorigenesis [2-7]. The study of microRNA has been extended to many types of cancers. miRNAs target protein-coding mRNAs at the post-transcriptional level by direct cleavage of the mRNA or by inhibition of protein synthesis [8,9]. Accumulating evidence shows that miR-183 is one of the oncogenic RNAs, as a member of an evolutionarily conserved miRNA cluster (miR-183, miR-96, and miR-182). miR-183 is located in human chromosome 7 and has been implicated in key cellular functions such as apoptosis, proliferation, and tumorigenesis [10-12]. Likewise, miR-183 expression was revealed to be deregulated in several types of human malignant solid tumors, including breast cancer, osteosarcoma, hepatocellular carcinoma, and colorectal cancer [13-16]. However, the association between the miR-183 expression, clinicopathological characteristics and the function and molecular mechanism of miR-183 in gastric cancer has not yet been reported.
miRNAs exert their biological effects by targeting specific mRNAs. Programmed cell death4 (PDCD4) is a novel tumor suppressor gene whose protein product plays a role in the suppression of tumorigenesis, tumor progression, and invasion [17-19]. Lost expression of PDCD4 mRNA and protein has been identified in a number of different human cancers including lung, breast, colon, prostate, brain, and liver cancer [20-22]. Reduced expression of miR-183 is associated with higher grades of pathology and worse prognosis [23]. In gastric cancer, PDCD4 plays a significant tumor suppressor role. It is suggested that PDCD4 expression in gastric cancer can be employed to indicate a favorable prognosis for the disease outcome [24]. Previous studies show that PDCD4 is one of the target genes of miR-183 and miR-183 can inhibit apoptosis in human hepatocellular carcinoma cells by repressing the PDCD4 expression [25]. To our knowledge, there are no reports on a possible correlation between PDCD4 and miR-183 in gastric cancer.
In the present study, we found that miR-183 was significantly increased in gastric cancer tissues and high expression of miR-183 correlated with advanced clinical stage (TNM stage), deep stromal invasion and positive lymph node and distant metastasis. Ectopic expression of miR-183 increased cell proliferation, migration, invasion, and inhibited apoptosis of gastric cells in vitro. We also found that PDCD4 was a target gene of miR-183 in gastric cancer and miR-183 is capable of repressing the expression of PDCD4. These results suggest that miR-183 might play an important role in the development of gastric cancer by modulating cell proliferation and apoptosis by targeting PDCD4 expression.
Material and methods
Patient and tissue samples
Eighty fresh frozen surgical specimens of gastric cancer were collected at Ruijin Hospital, Luwan Branch, Shanghai Jiaotong University, School of Medicine, in China from 2011 to 2013. Non-tumor gastric tissues were randomly selected from 20 of these patients and used as controls (5 cm away from the tumor border). All samples were derived from patients who did not receive adjuvant treatment including radiotherapy or chemotherapy prior to surgery in order to eliminate potential treatment-induced changes to gene expression profiles. Immediately following the surgical resection, tissues were frozen in liquid nitrogen and kept at -80°C until RNA extraction. Sections from each specimen were independently examined by two pathologists, and histological typing was performed using Lauren’s classification. TNM classification of malignant tumors was assigned in accordance to The International Union Against Cancer. This study was approved by The Research Ethics Committee of Ruijin Hospital, Luwan Branch, Shanghai Jiaotong University School of medicine, China. Also, written informed consent was obtained from all patients. All specimens were handled and made anonymous in accordance with ethical and legal standards.
Cell lines and culture condition
The human gastric cancer cell line, SGC-7901, was obtained from the KeyGEN BioTECH (Nanjing, China) and cultured in Dulbecco’s Modified Eagle’s Medium (Sigma-Aldrich, St. Louis, MO, USA) containing 10% fetal bovine serum, at 37°C in a humidified 5% CO2 incubator.
miRNA mimic and inhibitor transfection
Gastric cancer cells were transfected with miR-183 mimic, miR-183 inhibitor (Applied Biosystems), or scrambled sequence (GenePharma) by using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. A final concentration of 50 nM of RNA mimics or 150 nM of inhibitor and their respective negative controls were used for each transfection. At 48 hours after transfection, cells were harvested for Western blotting or qRT-PCR analyses.
RNA extraction and qRT-PCR analyses
Total RNA was extracted from tissues or cultured cells with TRIzol reagent (Invitrogen). For Quantitative Real-time PCR (qRT-PCR), RNA was reverse transcribed to cDNA from 1 mg of total RNA by using a Reverse Transcription Kit (Takara). qRT-PCR analyses were conducted with Power SYBR Green (Takara). All protocols were carried out according to the manufacturer’s instructions. Results were normalized to the expression of U6 or glyceraldehyde-3-phos-phatedehydrogenase (GAPDH). The primer sequence: PDCD4-F 5’-AAA GGG AAG GTT GCT GGA TA-3’, PDCD4-R 5’-TCC ACC TCC TCC ACA TCA TA-3’, GAPDH-F 5’-CAT CTT CTT TTG CGT CGC CA-3’, GAPDH-R 5’-TTA AAA GCA GCC CTG GTG ACC-3’, hsa-miR-183-5p-RT primer: 5’-GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACA GTG AA-3’, has-U6 snRNA-RT primer: 5’-GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACA AAA TA-3’, Common reverse primer: 5’-GTG CAG GGT CCG AGG T-3’.
Cell proliferation assays (MTT proliferation assay)
The capacity for cellular proliferation was measured with a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. At 24 h after RNA transfection, cells (approximately 5 × 104 cells/well) were seeded into 96-well culture plates for 24, 48, and 72 h. The cells were then incubated with 20 μL of MTT (5 mg/ml) for 4h at 37°C and150 μL of DMSO was added to solubilize the crystals for 20 min at room temperature. The optical density was determined at a wavelength of 490 nm. Tests were performed in triplicate. The data was presented as the inhibition rate. Inhibition rate (%) = (Negative control group-Experimental group)/Negative control group ×100%.
Apoptosis analysis
The cells were seeded into 6-well plates and then transfected with miR-183 mimic or inhibitor and their negative controls. After 48 h, the cells were harvested, and the ratio of apoptosis was determined using the Annexin V Apoptosis Detection Kit according to the manufacturer’s instructions. Apoptosis cells were examined and quantified by flow cytometry (FCM). Tests were performed in triplicate.
Scratch-wound assay
Cells were transfected and cultured to confluence or near (> 90%) confluence in 6-well dishes. A sterile 200 μl pipette tip was used to scratch a straight wound through the cells. Subsequently the medium was removed and replaced with fresh medium. Images were taken at 0 and 24 h after scratching, respectively. Two parallel lines were drawn at the border of the wound, and the distance between the lines were measured. Tests were performed in triplicate.
Transwell assay
Transiently transfected cells were trypsinized and suspended without serum-free RPMI-1640 culture medium. The suspended cells were seeded in the upper chamber of the Transwell® insert and RPMI-1640 medium containing 20% FBS (600 μl) was added to the lower chamber of the Transwell insert. Following culturing at 37°C in a humidified 5% CO2 incubator for 24 h, the inserts were washed with PBS. A cotton swab was used to remove adherent cells on the inner side of the upper chamber membrane, then the chamber membranes were fixed in paraformaldehyde (4%) for 10 min. Coomassie Blue (600 μl) was added into each well and incubated at room temperature for 15 min. The inserts were washed again and the upper chamber was left to dry naturally. Visual fields (n = 15) of each insert were randomly counted under an upright light microscope (BX51, Olympus) and the average value was calculated. Tests were performed in triplicate.
Western blotting
Total protein was extracted from tumor tissue samples and corresponding normal tissues or GC cell lines. The concentration of proteins was measured by BCA protein assay kit (Pierce, IL, USA). Proteins were separated by 10% SDS-PAGE gel and transferred to polyvinylidene fluoride (PVDF) membrane (Millipore, MA, USA). The membrane was first incubated with specific primary antibodies, then with secondary antibodies labeled with HRP and detected by ECL. The signal intensity was determined by Image J software.
Statistical analysis
The relationships between the miR-183 expression level and clinicopathological parameters were analyzed using the Pearson χ2 test. For comparisons between two different groups, statistical significance was determined using the Student’s t-test. All statistical analyses were performed using the SPSS 13.0 software package. A two-tailed value of p < 0.05 was considered statistically significant.
Results
Expression levels of miR-183 associated with advanced gastric cancer
We first measured the expression levels of miR-183 in a total of 100 gastric tissue samples including 20 non-tumorous gastric mucosa samples and 80 primary gastric tumor tissue samples by qRT-PCR. We found that the expression levels of miR-183 are up-regulated in tumor tissues than in the normal gastric tissues (Figure 1A). The mean (± SE) expression level of miR-183 in the tumor tissues (2.12 ± 0.24) was significantly higher than that in the non-tumor tissues (1.04 ± 0.15) (p < 0.01). Furthermore, there is a significant increase of miR-183 expression levels in metastatic tumors (lymph node metastasis and liver metastasis) than in localized gastric tumors (Figure 1A).
Figure 1.
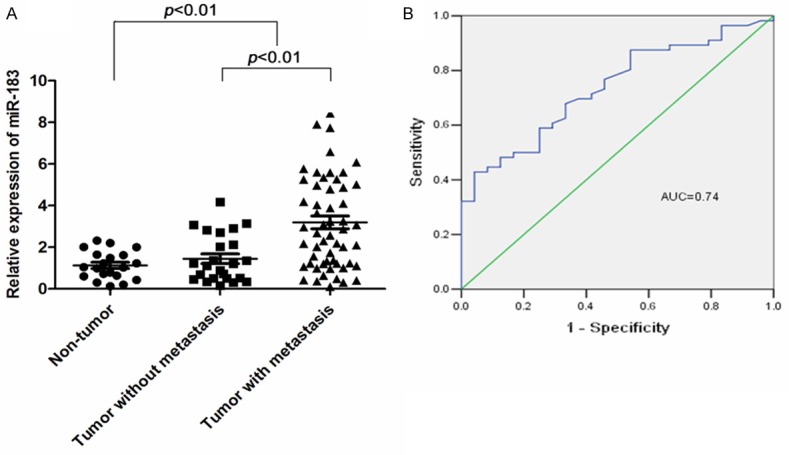
miR-183 is up-regulated in gastric cancer and as a novel biomarker for metastasis. A. Comparison of levels of miR-183 in samples from gastric cancer patients without metastasis (n = 24), gastric cancer with metastasis (lymph node and distant metastasis) (n = 56) and normal controls (n = 20). Data are expressed as the mean ± standard error (SE). Expression of miR-183 in tumor tissues was significantly higher than that in normal tissues (p < 0.001), especially higher in metastatic tissues (p < 0.001). The miR-183 expression levels were tested by qRT-PCR and normalized to U6. Statistical analysis was performed using the paired t-test. B. To test the ability of miR-183 in gastric cancer as a biomarker for metastasis, ROC curves were established. We observed clear separations between the patients with and without lymph node and distant metastasis, with an AUC of 0.74, 95% CI, 0.63-0.85.
To evaluate the correlation between miR-183 expression levels and clinicpathological characteristics, 80 patients with gastric cancer were divided into two groups (high or low) based on the expression levels of miR-183. The cut off levels for miR-183 were determined as mean levels (2.67) of relative quantity (2-ΔΔCt) according to a previous publication [26]. As shown in Table 1, high level expression of miR-183 was significantly correlated with advanced clinical stage (TNM stage) positive lymph node, distant metastases, and depth of cancer cell invasion (p < 0.05).
Table 1.
Correlation between the clinicopathologic characteristics and expression of miR-183 in gastric cancer
| Variable | miR-183 | |||
|---|---|---|---|---|
|
|
||||
| All case (n = 80) | Low expression (n = 45) | High expression (n = 35) | p value* | |
| Age (years) | ||||
| ≤ 60 | 37 | 23 | 14 | 0.323 |
| > 60 | 43 | 22 | 21 | |
| Gender | ||||
| Male | 45 | 25 | 20 | 0.887 |
| Female | 35 | 20 | 15 | |
| Tumor size (cm) | ||||
| < 5 | 46 | 29 | 17 | 0.154 |
| ≥ 5 | 34 | 16 | 28 | |
| Histological type | ||||
| Well-differentiated | 28 | 19 | 9 | 0.125 |
| Poorly-differentiated | 52 | 26 | 26 | |
| Depth of invasion | ||||
| T1, T2 | 21 | 16 | 5 | 0.032* |
| T3, T4 | 59 | 29 | 30 | |
| Lymph node metastasis | ||||
| Absent | 24 | 18 | 6 | 0.027* |
| Present | 56 | 27 | 29 | |
| Distant metastasis | ||||
| Absent | 73 | 44 | 29 | 0.039* |
| Present | 7 | 1 | 6 | |
| TNM stage | ||||
| I, II | 36 | 27 | 9 | 0.002** |
| III, IV | 44 | 18 | 26 | |
p < 0.05;
p < 0.01.
To evaluate miR-183 expression in gastric cancer as a new biomarker for metastasis, ROC curves were established. We observed clear separations between the metastatic tumors and localized gastric tumors with an AUC of 0.74 (Figure 1B, 95% CI, 0.63-0.85).
miR-183 represses the expression of PDCD4
To determine the functions of miR-183 in gastric cancer, we searched for putative target genes of miR-183 by using the Target Scan (http://www.targetscan.org/ and http://www.microrna.org/) bioinformatics tools. A putative miR-183 binding site at 3’-UTR of the PDCD4 gene was identified, which is conserved across various species. The seed for miR-183 to PDCD4 3’ UTR is shown in Figure 2A. Given the potential role of miR-183 being oncogenic in gastric cancer, we selected the programmed cell death 4 (PDCD4), a tumor suppressor, as the putative miR-183 effector in gastric cancer for further validation.
Figure 2.
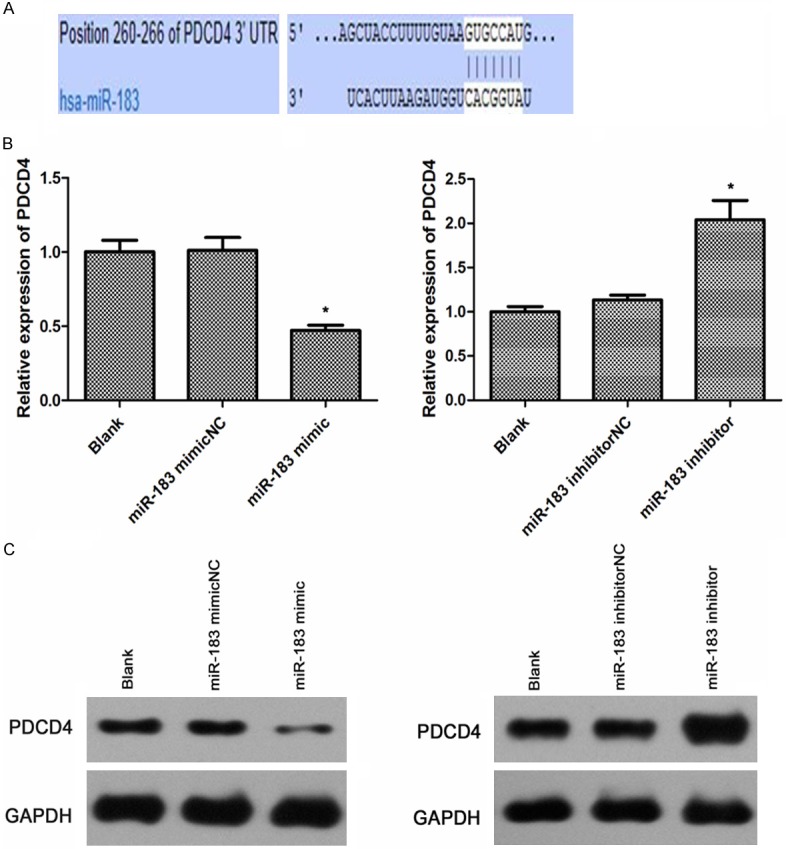
miR-183 inhibits the mRNA and protein expression of PDCD4. A. PDCD4 was predicted to be a potential target of miR-183. B. The mRNA levels of PDCD4 in the SGC-7901 cells of miR-183 mimic group and miR-183 inhibitor group. C. The protein levels of PDCD4 in the SGC-7901 cells of miR-183 mimic group and miR-183 inhibitor group. GAPDH was used as an internal loading control. Statistical analysis was performed using the student’s t-test. *p < 0.05.
The relationship between miR-183 level and PDCD4 expression was analyzed in SGC-7901 cells. qRT-PCR was performed to detect the mRNA levels of PDCD4 and showed that PDCD4 expression was down-regulated in SGC-7901 cells transfected with miR-183-mimics when compared with negative control and blank, whereas the expression levels of miR-183-inhabitor was up-regulated (Figure 2B).
The level of PDCD4 protein was also detected by western blotting. As shown in Figure 2C, there was a significant inverse correlation between miR-183 and PDCD4 protein level in the miR-183-mimic group versus miR-183-mimicNC group (p < 0.05), while there was no obvious difference between blank group and miR-183NC group. In contrast, the miR-183-inhibitor played the opposite effect of miR-183-mimic with an increasing level of PDCD4 protein expression versus either miR-183-mimic or miR-183-inhibitorNC group (p < 0.05). All of these results indicate that miR-183 post-transcriptionally inhibits PDCD4 expression and miR-183 is negatively correlated with mRNA and protein level of PDCD4 in gastric cancer cells.
miR-183 promoted cellular proliferation of gastric cancer
Given the up-regulation of miR-183 in gastric cancer, we speculate that miR-183 may influence gastric cancer cell proliferation, MTT assay was performed in SGC-7901 cells. The result demonstrated that SGC-7901 cells transfected with miR-183-mimic exhibited a significant increase of cellular viability when compared to cells treated by miR-183-mimicNC (p < 0.05) (Figure 3A). Furthermore, there was no significant difference between blank group and miR-183-mimicNC group. On the contrary, miR-183-inhabitor decreased cellular proliferation in SGC-7901 cells compared with miR-183-inhabitorNC group (p < 0.05) (Figure 3B), suggesting that miR-183 promotes gastric cell proliferation.
Figure 3.
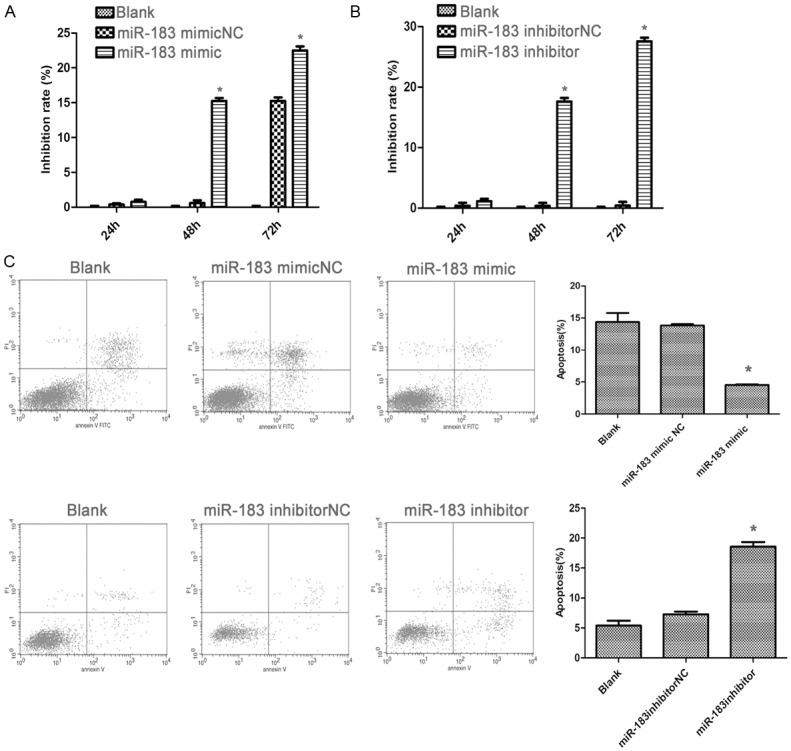
miR-183 promoted gastric cancer cell lines proliferation. A and B. MTT assay showed growth promotion in SGC-7901 cells transfected with miR-183 mimic, inhibitor, or negative controls at different time points. C. Cell apoptosis was analyzed by FCM in SGC-7901 cells 48 h after transfection of miR-183 mimic, inhibitor, or negative controls. *p < 0.05.
miR-183 inhibited the apoptosis of gastric cancer
To confirm that the over-expression of miR-183 was associated with apoptosis, we examined the apoptosis of the cells by flow cytometry 48 h after transfection. The results obviously demonstrated that there was a significant decrease of apoptosis in gastric cancer cells transfected with miR-183-mimic compared with the cells transfected with miR-183-mimicNC (p < 0.05). In contrast, the gastric cells transected with miR-183-inhabitor showed an increase of apoptosis compared with miR-183-inhabitorNC and miR-183-mimic groups (p < 0.05) (Figure 3C), suggesting that miR-183 plays a pro-apoptotic role in gastric cells.
miR-183 increased gastric cancer cell migration and invasion
Since the expression level of miR-183 correlates with advanced gastric cancer, we further assessed the effects of miR-183 on cell migration and invasion, a key determinant of malignant progression and metastasis. The scratch-wound assay was performed to measure and compare the scratch breadth at 24 hours following transfection. As shown in Figure 4A, miR-183-mimic group scratch wound-healing motility was faster compared to the control group; miR-183-inhibitor led to significantly decreased migration of SGC-7901 cells. Furthermore, the Transwell assay was used to detect the cell invasion ability. As shown in Figure 4B, the numbers of cells that had traversed the membrane were counted following transfection of miR-183 for 48 hours. When evaluated against the control group, the miR-183-mimic group significantly increased the invasion ability of GC cells. By contrast, the miR-183-inhibitor group significantly reduced the invasion ability (p < 0.05).
Figure 4.
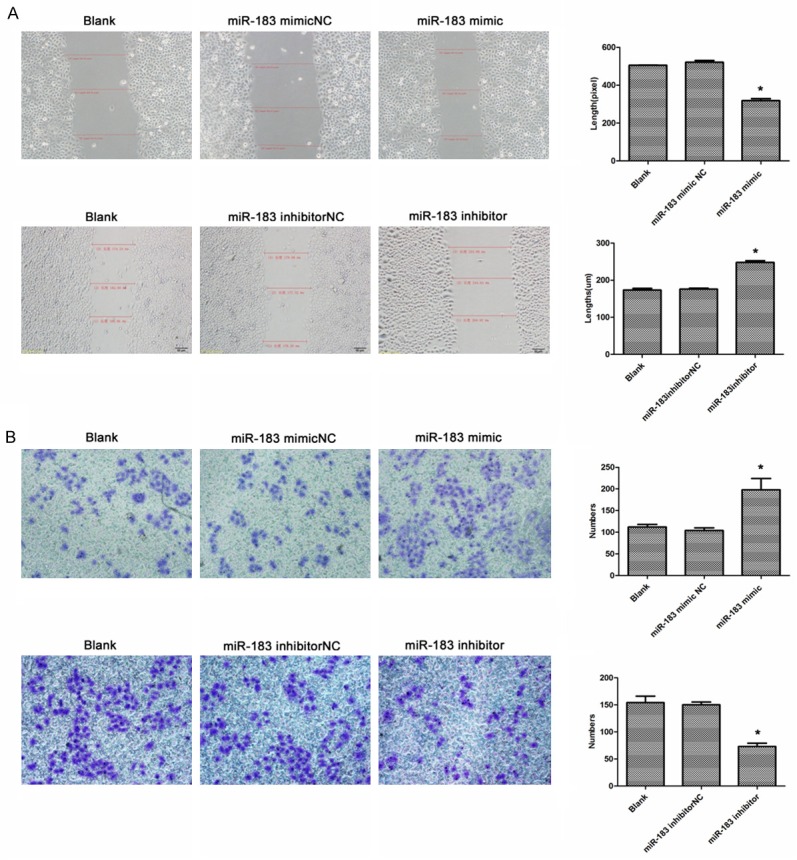
miR-183 promoted the migration and invasion of gastric cancer. A. Analysis the effect of miR-183 on the migration of SGC-7901 cells after transfection with mimic, inhibitor, or negative controls by scratch-wound assay. B. Analysis the effect of miR-183 on the invasion of SGC-7901 cells after transfection with mimic, inhibitor, or negative controls by transwell assay. *p < 0.05.
These results propose a functional role for miR-183 in mediating cell migration and invasion in gastric cancer and suggest a mechanism by which up-regulation of miR-183 potentially contributes to tumor metastasis in gastric cancer.
Discussion
Overexpression of miR-183 has been reported in a variety of human cancer types including bladder, breast, liver, and colorectal cancers [27-30]. It confers invasive potential to liver, lung, and prostate cancer cells [25,31,32]; however, the role of miR-183 in gastric cancer is yet to be elucidated. In the present study, we observed significant up-regulation of miR-183 in gastric tumors. The expression level of miR-183 increased about one-fold in metastatic gastric tumors compared to localized gastric tumors (Figure 1). Detailed analysis of the expression levels of miR-183 and the clinicopathological characteristics of the gastric tumors showed statistical significant correlation between high level expression of miR-183 and metastatic phenotypes of gastric cancers. Our data provides evidence that high level expression of miR-183 is clinically relevant and suggest that miR-183 may be a potent biomarker for advanced gastric cancer.
In this study, we further demonstrated the biological significance of miR-183 expression in gastric cancer in vitro. We showed that overexpression of miR-183 in human gastric cell line SGC-790 promotes cell proliferation and inhibits apoptosis whereas inhibition of miR-183 causes reverse effects. Since increased cell migration and invasion are required for tumor metastasis, we examined the effect of miR-183 on gastric cancer cell migration and invasion. We also demonstrated that miR-183 enhances migration and invasion activities of gastric cancer cells, which was verified by the evidence that miR-183 accelerates wound healing of cultured cells and promotes invasion of cells to pass through the simulated extra cellular matrix. Contrarily, inhibition of miR-183 reduced cancer cell migration and invasion. These data suggest that miR-183 has an oncogenic and prometastatic function in gastric cancer cells through inhibition of apoptosis and promotion of cancer proliferation.
To date, several miR-183 direct targets have been reported including Ezrin in lung cancer, FOXO1 in endometrial cancer, and EGR1in synovial sarcoma [31,33,34]. The direct target of miR-183 in gastric cancer is unknown. We identified programmed cell death 4 (PDCD4) as a candidate direct target gene of miR-183 in gastric cancer using the bioinformatic algorithms. PDCD4 is a tumor suppressor gene to suppress transformation [35,36], tumorigenesis and progression [17], invasion and metalloproteinase activation [19], and to induce apoptosis [37,38]. It was also reported that PDCD4 mRNA/protein expression is lower in primary gastric tumor tissues compared to corresponding normal tissues and in cell culture models [39]. Given that miR-183 binding to the 3’ UTR of PDCD4 and over expression of miR-183 down-regulates PDCD4 at both the mRNA and protein level, our data reported here support the notion that down-regulation of PDCD4 observed in gastric cancer is very likely due to the up-regulation of miR-183. miR-183 may attenuate PDCD4 in gastric cancer for progression and metastasis.
In summary, we demonstrated for the first time that miR-183 promotes gastric cancer cell proliferation and invasion through repression of PDCD4, a tumor suppressor critical for inhibition of cancer cell proliferation and invasion. Given that miR-183 is significantly up-regulated in gastric cancer and associates with metastatic phenotype of gastric cancer, miR-183 may be a potent biomarker for advanced gastric cancer and a potential therapeutic target for treatment of gastric cancer in future.
Acknowledgements
This research was made possible with financial support from Shanghai Municipal Health Bureau (to Mei-Jie Hu) (No. 2011308). We appreciate the valuable comments and help from Dr. Wanguo Liu of Louisiana State University Health Sciences Center New Orleans.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Medina PP, Slack FJ. microRNA and cancer: an overview. Cell Cycle. 2008;7:2485–2492. doi: 10.4161/cc.7.16.6453. [DOI] [PubMed] [Google Scholar]
- 3.Paranjape T, Slack FJ, Weidhaas JB. MicroRNAs: tools for cancer diagnostics. Gut. 2009;58:1546–1554. doi: 10.1136/gut.2009.179531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 5.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 6.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 7.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang H, Bian Y, Tu C, Wang Z, Yu Z, Liu Q, Xu G, Wu M, Li G. The miR-183/96/182 cluster regulates oxidative apoptosis and sensitizes cells to chemotherapy in gliomas. Curr Cancer Drug Targets. 2013;13:221–231. doi: 10.2174/1568009611313020010. [DOI] [PubMed] [Google Scholar]
- 11.Weeraratne SD, Amani V, Teider N, Pierre-Francois J, Winter D, Kye MJ, Sengupta S, Archer T, Remke M, Bai AH, Warren P, Pfister SM, Steen JA, Pomeroy SL, Cho YJ. Pleiotropic effects of miR-183 ~96~182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol. 2012;123:539–552. doi: 10.1007/s00401-012-0969-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ueno K, Hirata H, Shahryari V, Deng G, Tanaka Y, Tabatabai ZL, Hinoda Y, Dahiya R. microRNA-183 is an oncogene targeting Dkk-3 and SMAD4 in prostate cancer. Br J Cancer. 2013;108:1659–1667. doi: 10.1038/bjc.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, Zucman-Rossi J. MicroRNA profiling in hepatocellular tumorsis associated with clinical features and oncogene/tumorsuppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 14.Lowery AJ, Miller N, Dwyer RM, Kerin MJ. Dysregulated miR-183 inhibits migration in breast cancer cells. BMC Cancer. 2010;10:502. doi: 10.1186/1471-2407-10-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao H, Guo M, Zhao G, Ma Q, Ma B, Qiu X, Fan Q. miR-183 inhibits the metastasis of osteosarcoma via downregulation of the expression of Ezrin in F5M2 cells. Int J Mol Med. 2012;30:1013–1020. doi: 10.3892/ijmm.2012.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, Uetake H, Sugihara K, Mori M. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol. 2009;34:1069–1075. doi: 10.3892/ijo_00000233. [DOI] [PubMed] [Google Scholar]
- 17.Jansen AP, Camalier CE, Colburn NH. Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorgenesis. Cancer Res. 2005;65:6034–6041. doi: 10.1158/0008-5472.CAN-04-2119. [DOI] [PubMed] [Google Scholar]
- 18.Hilliard A, Hilliard B, Zheng SJ, Sun H, Miwa T, Song W, Göke R, Chen YH. Tanslational regulation of autoimmune inflammation and lymphomagenesis by programmed cell death 4. J Immunol. 2006;177:8095–8102. doi: 10.4049/jimmunol.177.11.8095. [DOI] [PubMed] [Google Scholar]
- 19.Yang HS, Matthews CP, Clair T, Wang Q, Baker AR, Li CC, Tan TH, Colburn NH. Tumorrigenesis suppressor Pdcd4 down regulates mitogen-avtivated protein kinase kinase kinase kinase 1expression to suppress colon carcinoma cell invasion. Mol Cell Biol. 2006;26:1297–1306. doi: 10.1128/MCB.26.4.1297-1306.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Göke R, Barth P, Schmidt A, Samans B, Lankat-Buttgereit B. Programmed cell death 4 supresses CDK1/cdc2via induction of p21(Waf1/Cip1) Am J Physiol Cell Physiol. 2004;287:C1541–C1546. doi: 10.1152/ajpcell.00025.2004. [DOI] [PubMed] [Google Scholar]
- 21.Gao F, Zhang P, Zhou C, Li J, Wang Q, Zhu F, Ma C, Sun W, Zhang L. Frequent loss of Pdcd4 expression in human glioma: Possible role in the tumorigenesis of glioma. Oncol Rep. 2007;17:123–128. [PubMed] [Google Scholar]
- 22.Zhang H, Ozaki I, Mizuta T, Hamajima H, Yasutake T, Eguchi Y, Ideguchi H, Yamamoto K, Matsuhashi S. Involvement of programmed cell death 4 in tansforming growth factor-betal-induced apoptosis in human hepatocellular carcinoma. Oncogene. 2006;25:6101–6112. doi: 10.1038/sj.onc.1209634. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Knösel T, Kristiansen G, Pietas A, Garber ME, Matsuhashi S, Ozaki I, Petersen I. Loss of PDCD4 exprossion in human lung cancer correlates with tumor progression and prognosis. J Pathol. 2003;200:640–646. doi: 10.1002/path.1378. [DOI] [PubMed] [Google Scholar]
- 24.Guo PT, Yang D, Sun Z, Xu HM. PDCD4 functions as a suppressor for pT2a and pT2b stage gastric cancer. Oncol Rep. 2013;29:1007–1012. doi: 10.3892/or.2013.2232. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Fu H, Xu C, Tie Y, Xing R, Zhu J, Qin Y, Sun Z, Zheng X. miR-183 inhibits TGF-beta1-induced apoptosis by down regulation of PDCD4 expression in human hepatocellular carcinoma cells. BMC Cancer. 2010;10:354. doi: 10.1186/1471-2407-10-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology. 2010;79:313–320. doi: 10.1159/000323283. [DOI] [PubMed] [Google Scholar]
- 27.Han Y, Chen J, Zhao X, Liang C, Wang Y, Sun L, Jiang Z, Zhang Z, Yang R, Chen J, Li Z, Tang A, Li X, Ye J, Guan Z, Gui Y, Cai Z. MicroRNA expression signatures of bladder cancer revealed by deep sequencing. PLoS One. 2011;6:e18286. doi: 10.1371/journal.pone.0018286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehmann U, Streichert T, Otto B, Albat C, Hasemeier B, Christgen H, Schipper E, Hille U, Kreipe HH, Länger F. Identification of differentially expressed microRNAs in human male breast cancer. BMC Cancer. 2010;10:109. doi: 10.1186/1471-2407-10-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang Z, Gao Y, Shi W, Zhai D, Li S, Jing L, Guo H, Liu T, Wang Y, Du Z. Expression and significance of microRNA-183 in hepatocellular carcinoma. ScientificWorldJournal. 2013;2013:381874. doi: 10.1155/2013/381874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou T, Zhang GJ, Zhou H, Xiao HX, Li Y. Overexpression of microRNA-183 in human colorectal cancer and its clinical significance. Eur J Gastroenterol Hepatol. 2014;26:229–233. doi: 10.1097/MEG.0000000000000002. [DOI] [PubMed] [Google Scholar]
- 31.Wang G, Mao W, Zheng S. MicroRNA-183 regulates Ezrin expression in lung cancer cells. FEBS Lett. 2008;582:3663–3668. doi: 10.1016/j.febslet.2008.09.051. [DOI] [PubMed] [Google Scholar]
- 32.Mihelich BL, Khramtsova EA, Arva N, Vaishnav A, Johnson DN, Giangreco AA, Martens-Uzunova E, Bagasra O, Kajdacsy-Balla A, Nonn L. miR-183-96-182 cluster is overexpressed in prostate tissue and regulates zinc homeostasis in prostate cells. J Biol Chem. 2011;286:44503–44511. doi: 10.1074/jbc.M111.262915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myatt SS, Wang J, Monteiro LJ, Christian M, Ho KK, Fusi L, Dina RE, Brosens JJ, Ghaem-Maghami S, Lam EW. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res. 2010;70:367–377. doi: 10.1158/0008-5472.CAN-09-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 2010;70:9570–9580. doi: 10.1158/0008-5472.CAN-10-2074. [DOI] [PubMed] [Google Scholar]
- 35.Cmarik JL, Min H, Hegamyer G, Zhan S, Kulesz-Martin M, Yoshinaga H, Matsuhashi S, Colburn NH. Differentially expressed protein PDCD4 inhibits tumor promoter-induced neoplastic transformation. Proc Nat Acad Sci U S A. 1999;96:14037–14042. doi: 10.1073/pnas.96.24.14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang HS, Jansen AP, Nair R, Shibahara K, Verma AK, Cmarik JL, Colburn NH. A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB or ODC transactivation. Oncogene. 2001;20:669–676. doi: 10.1038/sj.onc.1204137. [DOI] [PubMed] [Google Scholar]
- 37.Shibahara K, Asano M, Ishida Y, Aoki T, Koike T, Honjo T. Isolation of a novel mouse gene MA-3 that is induced upon programmed cell death. Gene. 1995;166:297–301. doi: 10.1016/0378-1119(95)00607-9. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Z, DuBois RN. Detection of differentially expressed genes in human colon carcinoma cells treated with a selective COX-2 inhibitor. Oncogene. 2001;20:4450–4456. doi: 10.1038/sj.onc.1204588. [DOI] [PubMed] [Google Scholar]
- 39.Motoyama K, Inoue H, Mimori K, Tanaka F, Kojima K, Uetake H, Sugihara K, Mori M. Clinicopathlogical and prognostic significance of PDCD4 and microRNA-21 in human gastric cancer. Int J Oncol. 2010;36:1089–1095. doi: 10.3892/ijo_00000590. [DOI] [PubMed] [Google Scholar]


