Abstract
The molecular events that control cell fate determination in cardiac and smooth muscle lineages remain elusive. Myocardin is an important transcription co-factor that regulates cell proliferation, differentiation and development of the cardiovascular system. Here, we describe the construction and analysis of a dual Cre and Enhanced Green Fluorescent Protein (EGFP) knock-in mouse line in the Myocardin locus (MyocdKI). We report that the MyocdKI allele expresses the Cre enzyme and the EGFP in a manner that recapitulates endogenous Myocardin expression patterns. We show that Myocardin expression marks the earliest cardiac and smooth muscle lineages. Furthermore, this genetic model allows for the identification of a cardiac cell population which maintains both Myocardin and Isl-1 expression, in E7.75 - E8.0 embryos, highlighting the contributions and merge of the first and second heart fields during cardiogenesis. Therefore, the MyocdKI allele is a unique tool for studying cardiovascular development and lineage-specific gene manipulation.
Keywords: Myocardin, Cre, EGFP, heart, smooth muscle
The heart is the first functional organ of vertebrate animals, which evolves from a peristaltic linear tube to a complex four chamber pump (Moorman et al., 2003; Virágh and Challice, 1973). As the heart develops the concomitant vasculature tree forms, grows and branches out from large vessels to tiny capillaries directing the blood flow and carrying nutrients and oxygen within (Baldwin, 1996). Vasculature inner pressure is generated by the heartbeat, and its magnitude is greater the closer a vessel is to the heart. Thus, to protect vessels against possible pressure damage, a smooth muscle sheath confers elasticity to aid in vessel stretching and strength to avoid vessel collapse (Owens et al., 2004). Smooth muscle is not only found sheathing vessels, but also around or within organs which expand as a result of positive pressure, such as the trachea, lungs, bladder, lymphatic vessels, uterus, etc (Owens et al., 2004). Interestingly, early cardiac tube gene expression resembles that of smooth muscle, expressing SM22α, and SM α-actin genes along with cardiac-specific genes (Li et al., 1996; Owens et al., 2004). However, the molecular mechanisms that control cardiac and smooth muscle gene expression and cardiac and smooth muscle lineage specification are not fully understood.
Myocardin was identified through an in silico screening of a cardiac specific Expressed Sequenced Tagged (EST)-library (Wang et al., 2001) and was characterized as a Serum Response Factor (SRF) transcription co-factor. Myocardin physically interacts with SRF and stabilizes a multimeric complex in which SRF ultimately binds to at least two SRF response elements located within the promoter/enhancers of many cardiac and smooth muscle genes. Thus, Myocardin expression and activity is necessary to induce or to enhance the expression of cardiac and smooth muscle genes such as Atrial Natriuretic Factor (ANF), Transgelin (Tlgn, SM22α), smoothelins, etc. (Wang et al., 2001; Wang et al., 2004; Wang et al., 2003). Myocardin mRNA expression was revealed to be restricted to cardiac progenitor tissue at early stages of development, while also being expressed in the developing and adult smooth muscle tissues present in different organs. Myocardin mRNA expression pattern during embryogenesis suggested its important role during cardiac and smooth muscle development (Wang et al., 2001; Wang et al., 2004; Wang et al., 2003). Indeed, Myocardin homozygous mutation results in embryonic lethality at around embryonic day (E)9.5 displaying cardiac malformations and lack of smooth muscle formation (Li et al., 2003). On the other hand, conditional cardiac ablation of Myocardin at late stages of development results in a post-natal cardiac physiology imbalance, an increase of fibrotic tissue and an increase of cell death leading to cardiac enlargement (Huang et al., 2009). Conversely, Myocardin over-expression in primary human mesenchymal stem cells or human vascular smooth muscle cells results in a reduction of cell proliferation and a forced expression of cardiac and smooth muscle molecular markers (Chen et al., 2011; Tang et al., 2008; van Tuyn et al., 2005; Wang et al., 2003; Wystub et al., 2013). Further, Myocardin over-expression induces cardiac hypertrophy in neonatal rat cardiomyocytes as well as in transgenic mice (Wystub et al., 2013; Xing et al., 2006).
As noted above, the mRNA expression of Myocardin was revealed to be restricted to cardiac forming tissues from E7.5. Later on, it is also expressed in the smooth muscle cells from E9.5 at the time of vasculature formation during mouse embryogenesis (Wang et al., 2001). Myocardin expression is regulated by several transcription factors and signaling regulators in a complex genetic network. More specifically the homeodomain protein Nkx2-5 (Ueyama et al., 2003), the MADS box transcription factor Mef2c, the transcription enhancer factor Tead1, the forkhead box binding protein Foxo4, the nuclear factor of activated T cells Nfatc3, as well as Myocardin itself, have been shown to interact with specific cis-acting elements, present within a 10Kbp promoter/enhancer DNA fragment upstream of the Myocardin transcriptional start site, to regulate the transcription of the Myocardin gene (Creemers et al., 2006; Liu et al., 2014; Wang et al., 2010). Additionally, the transcription factor Smad3, a transforming growth factor TGF-β signal transducer, has been shown to repress Myocardin expression in the early stages of smooth muscle differentiation by sequestering the phosphoinositol-3 (PI3) kinase-activated Nkx2-5 (Xie et al., 2011). Together, these studies suggest that the combination of different factors at a given time results in cardiac and/or smooth muscle specific Myocardin expression (Creemers et al., 2006).
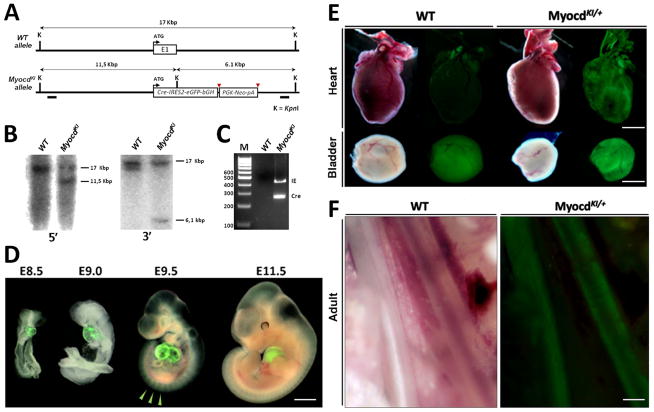
In this regard, we decided to knock-in the Cre enzyme and the Enhanced Green Fluorescent Protein (EGFP) reporters into the Myocardin locus in order to facilitate the investigation of Myocardin expression and function. In this model the first exon of the Myocardin coding sequence was replaced with a Cre-IRES2-EGFP-bGHpA cassette followed by an Frt-flanked Neomycin resistance gene (Fig. 1A). The final construct, named Myocd-KI was linearized, purified and electroporated into 129SJ1;C57BL/6 hybrid mouse embryonic (ES) stem cells. Targeted ES cells were screened by PCR and Southern blot (Fig. 1B–C). Seven clones were identified to have the correct homologous recombination and one was used for blastocyst injection. Six male chimera mice were obtained, all lead to germline transmission and viable heterozygous mutant offspring.
Figure 1. Generation of MyocardinKI allele.
A) Diagram depicting the genomic structure of Myocardin wild type (WT) and mutant (MyocdKI) alleles. Myocardin mutant allele contains a Cre-IRES2-EGFP-bGHpA cassette followed by an Frt-flanked positive selection Neomycin cassette which replaces the first Exon (E1) of the gene reading frame. B) Southern blots with 5′ and 3′ probes (black boxes in genomic locus) showing the specificity of the genomic recombination. C) PCR genotyping indicates the presence of the Cre-IRES2-EGFP-bGHpA cassette only in DNA obtained from mutant samples. D) Cell fate determination by observation of EGFP expression from early stages of embryonic development. EGFP is detected in developing hearts. EGFP is also observed in somites (arrowheads) (Scale bar, 1mm). E) EGFP expression tissues such as the heart and the bladder. Note the EGFP signal in the bladder of WT mice is an auto-fluorescence signal due to urine content (Scale bar; hearts, 2mm; bladders, 1mm). F) EGFP expression in postnatal day 2 (P2) tissues in tissues containing smooth muscle such as the Esophagus (E) and Dorsal Aorta (DAo) (Scale bar, 100μm).
Analysis of EGFP expression under UV light at different stages of mouse embryonic development shows a recapitulation of Myocardin expression pattern. EGFP expression is observed from early stages of development and is restricted to the cardiac crescent at E7.5, and to cardiac tissue throughout embryogenesis in MyocdKI/+ mice (Fig. 1D, Supplementary Fig. 1). Also, a faint EGFP expression is observed in the forming somites from E8.0 until E11.5; however, this faint expression is no longer visible in embryos beyond E11.5, indicating a transient expression of Myocardin in this region (Fig. 1D). Nevertheless, upon dissection, EGFP expression in adult tissues was readily observed in the heart as well as in tissues which contain smooth muscle cells such as the bladder, intestines, veins, arteries and other large blood vessels (Fig. 1E, F). Homozygocity of the MyocdKI allele results in embryonic lethality showing cardiac edema, a sign of cardiac dysfunction, and overall developmental delay (Fig. 2A). This phenotype is consistent with what has been reported from an independent Myocardin mutant mouse line (Li et al., 2003). Interestingly, cardiac EGFP expression is maintained in MyocdKI/KI embryos indicating that the Myocardin promoter is still active even during cardiac abnormalities at this developmental stage. Further, RNA was isolated from E9.5 embryonic hearts and used in qPCR assays; results show that Myocardin expression is abolished in MyocdKI/KI embryos when compared to wild type controls (Fig. 2B). Taken together, these results confirm that our knock-in strategy results in a true Myocardin null mutation.
Figure 2. MyocdKI allele is a true Myocd null allele.
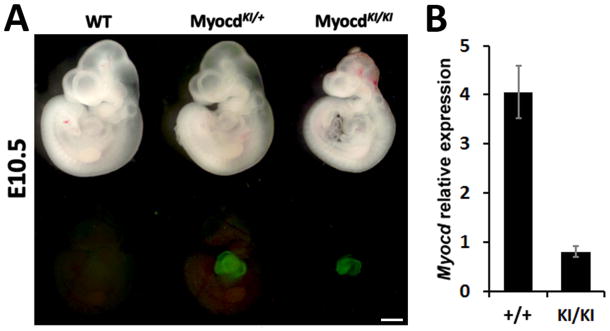
A) EGFP expression in cardiac tissue of E10.5 Myocd wild type (WT), heterozygote (MyocdKI/+) and null mutant (MyocdKI/KI) embryos. The MyocdKI/KI embryo shows cardiac defects and embryonic development delay (Scale bar, 1mm). B) Myocardin relative RNA expression levels by qPCR in Myocd wild type (WT) and null mutant (MyocdKI/KI) hearts.
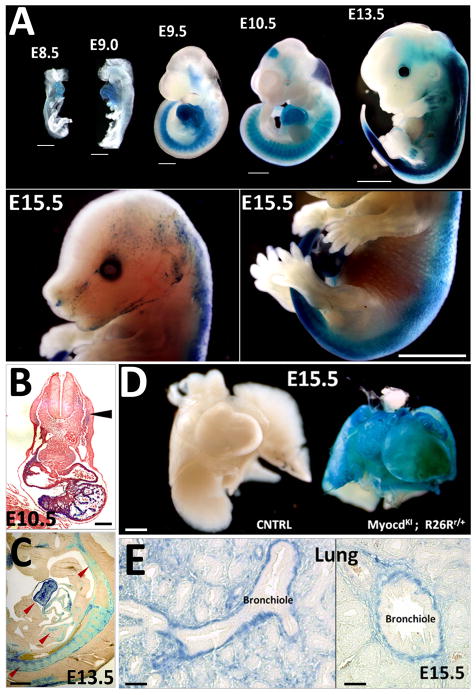
In order to confirm the efficacy of the Cre enzyme present in our genetic model, lineage tracing analysis was performed utilizing the R26R LacZ conditional reporter mouse line. Indeed, LacZ expression recapitulates Myocardin expression in double heterozygous embryos (MyocdKI/+; R26Rr/+) (Fig. 3A–E). In particular, LacZ expression is observed in the developing heart from early stages of development, similar to that of EGFP expression (Fig. 3A). Stronger LacZ staining was detected in the ventricles of the developing heart when compared to that of the atria (Fig. 3B, C). Additionally, LacZ positive signal was observed in the somites and midbrain of E9.5 and E10.5 embryos, indicating the transient expression of Myocardin in these tissues (Fig. 3A). At E13.5, LacZ expression is observed to extend to the limbs and at E15.5 LacZ staining appears to be restricted to the dorsal side of embryo (Fig. 3A). However, closer inspection of the internal organs shows LacZ expression in the smooth muscle layers of other organs such as the lung and intestines, in addition to the heart (Figs. 3C–E, Supplementary Fig. 2); and LacZ staining is clearly detected in the smooth muscle layer of bronchioles in the developing lung (Fig. 3E).
Figure 3. Myocardin expression pattern in MyocdKI/+; R26Rr/+ reporter mice.
A) X-gal staining at different stages of developing mouse embryos. LacZ expression is observed in the developing heart, somites and mid-brain. Note that LacZ expression expands to proximal limb internal structures (from E13.5) (Scale bars, 250μm). B) Transversal section of an E10.5 embryo showing cardiac specific expression of LacZ, somite expression is low compared to the heart (arrowhead) (Scale bar, 500μm). C) X-gal staining on a sagital section of an E13.5 embryo reveals LacZ expression in the heart, somite derivatives and forming vasculature (arrowhead) (Scale bar, 500μm). D) LacZ staining in an E15.5 MyocdKI/+;R26Rr/+ embryo heart and lung. A wild type littermate embryo was used as a control (Scale bar, 500μm). E) Cross sections of a MyocdKI/+;R26Rr/+ E15.5 embryo lung after X-gal staining showing LacZ expression in the smooth muscle layer surrounding the forming bronchioles (Scale bars, 125μm).
In order to confirm the above observations, we utilized another independent conditional reporter mouse line, the Tomato/EGFP (mTmG) in which expression of EGFP (mG) is achieved conditionally by a Cre-induced excision of an upstream loxP-flanked actively expressed red fluorescent tdTomato gene (mT). Indeed, the defined expression domains of Myocardin, such as cardiac, somite and midbrain were further confirmed in double heterozygote MyocdKI/+;mTmGm/+ mice (Fig. 4A). Additionally, EGFP expression was also observed in skeletal muscle, foot pad, tongue, bladder, lung and other smooth muscle containing tissues (Fig. 4B, C; Supplementary Figure 3). As a result, lineage tracing analyses reveal a larger and broader Myocardin expression domain and pattern when compared to earlier in situ hybridization reports (Wang et al., 2001). Together, our results are in accord with other reports obtained with a similarly engineered Cre knock-in mouse line (Long et al., 2007).
Figure 4. EGFP expression pattern in a MyocdKI/+;mTmGm/+ double heterozygote E10.5 embryo.
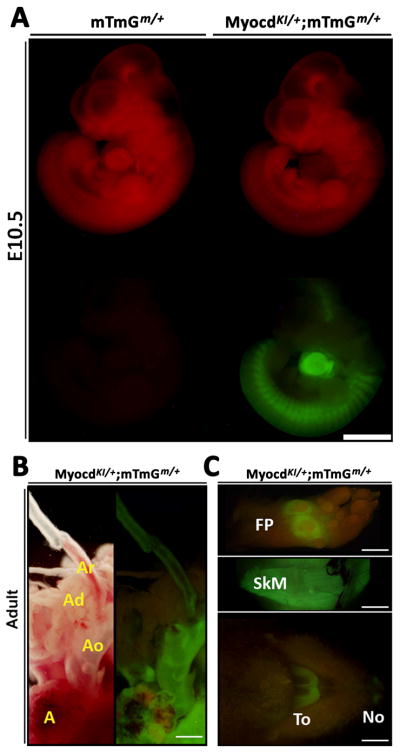
A) EGFP expression in the heart, somites and mid-brain of a MyocdKI/+;mTmGm/+ embryo. Notice that red fluorescence is diminished in regions in which EGFP is expressed. A mTmGm/+ single heterozygote embryo was included as a control (Scale bar, 1mm). B) EGFP expression in a 2 month old adult MyocdKI/+;mTmGm/+ mouse cardiovascular tissues such as the Atrium (A), Aorta (Ao), and in the left common carotid aortic branch (Ar), while EGFP is not observed in Adipose tissue (Ad) of (Scale bar, 2mm). C) EGFP expression in the foot pad (FP), skeletal muscle (SkM), tongue (To) and nostrils (No) of a 2 month old adult MyocdKI/+;mTmGm/+ mouse (Scale bar, 4mm).
The formation of the embryonic heart is a very dynamic process with the involvement of at least two cardiac progenitor cell populations termed first (primary) and second heart fields (Vincent et al., 2010). These cardiac fields, which display distinct molecular signatures, give rise to different cardiac regions of the mature heart (Cai et al., 2003; Lyons et al., 1995; Meilhac et al., 2004; Tanaka et al., 1999). Indeed, the first heart field is molecularly primed by the expression of the homeodomain transcription factor Nkx2-5 that regulates a downstream cascade of molecular signals to warrant the formation of the left ventricle and parts of both atria (Lyons et al., 1995; Tanaka et al., 1999). The second heart field was characterized by the specific and restricted expression of the Lim/homeodomain transcription factor Isl-1 which warrants the formation of the right ventricle, outflow tract and portions of both atria and inflow tract (Cai et al., 2003). Both Nkx2-5 and Isl-1 genes are expressed at the earliest stages of cardiac development; however, the expression of Isl-1 becomes restricted to the dorsal side and to the cranial and caudal poles of the forming tubular heart (Cai et al., 2003; Zhuang et al., 2013). Even though Nkx2-5 is one of the earliest markers of cardiac lineage, its expression domain is not restricted to the primary heart field as its expression domain expands to almost the entire myocardium at later developmental stages (Espinoza-Lewis et al., 2011; Kasahara et al., 1998).
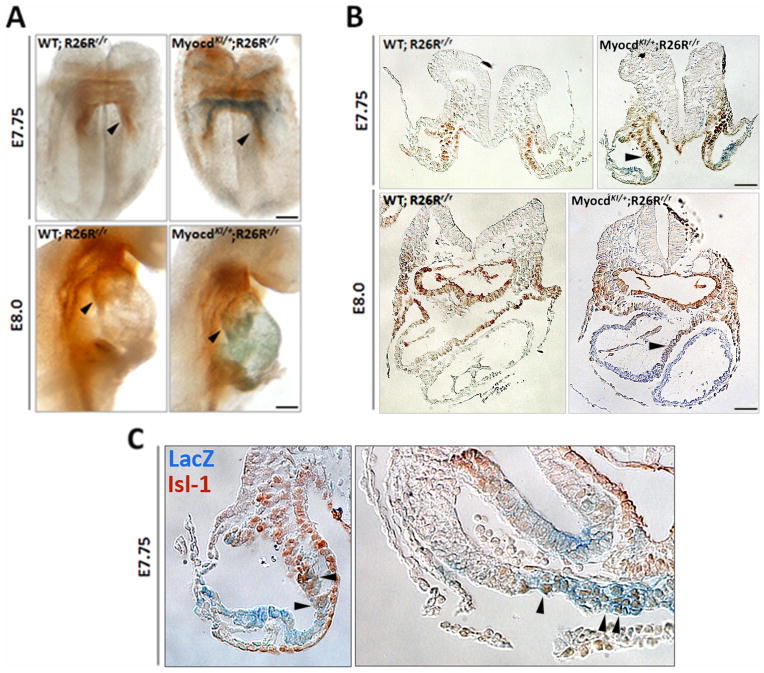
It has proven challenging to find a cardiac molecular marker that is unique to the first heart field. It was only until very recently that the Hyperpolarization-activated cation-gated channel 4 (Hcn4), a specific sinoatrial node and pacemaker molecular marker (Garcia-Frigola et al., 2003; Stieber et al., 2003), was described as a unique first heart field molecular marker (Liang et al., 2013; Später et al., 2013). However, the scarcity of first heart field molecular markers continues. Thus, we asked if Myocardin could also be a first heart field marker. We obtained MyocdKI/+; R26Rr/+ embryos from E7.5 up to E8.0 and subjected them to X-gal staining, which recapitulates the expression of Myocardin, followed by Isl-1 immunostaining. Whole mount staining reveals that LacZ (Myocardin) expression and Isl-1 protein expression domains are mostly mutually exclusive (Fig. 5A). Indeed, transversal sections in these embryos show that Myocardin and Isl-1 expression domains are complementary; indicating that Myocardin is not expressed in the second heart field. However, there is significant expression overlapping at the edges of both genes expression domains (E8.0) (Fig. 5A–C, arrowheads). Regions of overlapping are located in both the primordial outflow and inflow tract. These results indicate that there is a cell population that still maintains Isl-1 protein expression, and indicate that the switch from cardiac progenitor cell to a differentiated cardiomyocyte is a progressive event and not immediate. Prior reports addressing this issue utilize Hcn4, Isl-1, and MLC2a as markers for cardiac progenitors of the first heart field, second heart field and differentiated cardiomyocytes, respectively. Fluorescent in situ hybridizations show that the expression patterns of Hcn4 and Isl-1 do not overlap with that of MLC2a and are clearly discernible indicating a clear cellular phenotype difference (Cai et al., 2003; Liang et al., 2013; Später et al., 2013). Interestingly, our studies suggest that myocardin expression marks first heart field during early embryogenesis. Additionally, Myocardin expression highlights a cell population which also transiently expresses Isl-1, indicating a transitional phenotype between progenitor and differentiated cell and the merge of both first and second heart fields.
Figure 5. Myocardin is expressed in the first heart field.
A) Whole mount X-gal staining and Isl-1 immunostaining at early developmental staged embryos (E7.75 and E8.0) of MyocdKI/+;R26Rr/+ mice. At E7.75 LacZ expression appears ventral to that of Isl-1 staining (arrowheads) in the primordial forming heart. At E8.0 LacZ expression is present in the lineal heart tube while Isl-1 expression remains dorsal to that of LacZ and at the poles of the forming tubular heart (arrowheads). WT;R26Rr/+ were included as controls. B) Transversal sections of E7.75 and E8.0 embryos stained for LacZ followed by Isl-1 immunostaining showing the different expression domains. Arrowheads point to a cell population which seems to express both markers, LacZ and Isl-1. C) Higher magnification pictures of the transversal sections of the E7.75 MyocdKI/+;R26Rr/r embryo depicted in A) and B). Images correspond to the horns and to the medial arc of the forming cardiac tube respectively. Arrowheads point to cells and regions where double staining is observed (Scale bars, 100 μm.).
Taken together, here we describe the generation and characterization of a Myocardin-Cre-EGFP knock-in mouse model as a highly useful biological and genetic tool. This mouse model recapitulates Myocardin expression and is a true null allele; additionally, the inclusion of both the Cre as well as the EGFP genes provides a dual tool for further cell fate and lineage tracing analysis and conditional inactivation of target genes. Gene inactivation and cellular analyses are also possible even at the earliest stages of cardiac development with high accuracy and could be performed in parallel and at the same time due to the duality of the model. Finally, the presence of the EGFP gene in this model makes it useful for possible further screening experimentation to elucidate the complex regulation of cardiac and smooth muscle cell fate specification and differentiation.
Materials and Methods
Generation of the MyocdKI/+ knock-in mouse line and other mouse strains
A BAC (RP24213H11, BAC library RP24, C57BL/6 Male, Children’s Hospital Oakland Research Institute Oakland, CA) containing the Myocardin locus was purchased from used as a template to amplify by PCR a long 6.5Kbp upstream fragment and a short 4.7Kbp downstream fragment, relative to the Myocardin gene first exon. The fragments were inserted into the TOPO-pCRII vector containing a Cre-IRES2-EGFP-bGHpA cassette followed by an Frt-flanked Neomycin resistant gene and a Thymidine Kinase (TK) negative selection gene. The Cre-IRES2-EGFP-bGHpA cassette contains the DNA sequences for the Cre enzyme, the Internal Ribosome Entry, the Enhanced Fluorescence Protein and the bovine Growth Hormone polyadenylation signal (a specialized termination sequence present in several cloning vectors including the pcDNA3.1 family of vectors). The Final Myocd-KI construct was linearized with NdeI restriction endonuclease, purified by Phenol:Chloroform:IAA (25:24:1) and electroporated into 129SJ1;C57BL/6 hybrid mouse stem cells. Stem cells were cultured under G418 conditions. Surviving cell clones were further screened by PCR and southern blot. Seven clones were identified as having the correct genomic homologous recombination and one clone was selected for blastocyst injection. Six male and one female chimera were obtained. All males were fertile and produced MyocdKI/+ offspring, all displaying expected myocardin expression pattern. One mouse line was kept for further analysis and experimentation. This mouse line is available upon request.
The ROSA26R conditional LacZ (B6.129S4-Gt(ROSA)26Sortm1Sor/J) (hitherto termed R26R) and the ROSA26mT/mG (Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J) (hitherto termed mTmG) reporter mouse lines were obtained from The Jackson Laboratory.
X-gal staining
Tissues and mouse embryos at different developmental stages were dissected in PBS, fixed in 4% Paraformaldehyde (PFA) in PBS at various times at room temperature or at 4°C overnight. They were rinsed in X-gal washing buffer (PBS, 2mM magnesium chloride, 0.02% NP-40, 0.01% deoxycholate) 3 times, 15 min each at room temperature or on ice. Specimens were incubated in X-gal staining solution (5–16mM ferricyanide, 5–16mM ferrocyanide, 2mM MgCl2, 1mg/ml X-gal in PBS) for various times at room temperature or at 4°C overnight.
Immunostaining
Early stage embryos were dissected and rinsed in PBS at room temperature, fixed in 4% PFA for 2h at room temperature or at 4°C overnight. Embryos were rinsed in PBS 3 times, 15 min each in PBS, dehydrated in a series of methanol, post-fixed in a Methanol:DMSO (4:1) solution at 4°C overnight, bleached with a Methanol:DMSO:H2O2 (4:1:1) solution for 5h at room temperature, rinsed in methanol and rehydrated to PBS. Embryos were incubated with primary antibody (mouse anti Isl-1, DHSB 39.4D5c, 1:500) and secondary antibody (HRP-conjugated goat anti-mouse IgG2b, 1:200) in PBSMT (PBS, 2% non-fat dried milk, 0.5% triton X-100) for 48h and 24h respectively at 4°C. Rinsing with PBTx (PBS pH 7.2, 2% BSA, 0.5% Triton X-100) was performed 3 times, 1h each at room temperature prior to incubation in DBA (100ng/ml in PBS pH7.2) for 30min at room temperature followed by color reaction solution (DBA, H2O2 1:150). Color reaction was observed under microscope and stopped by the addition of 3 volumes of distilled water. Embryos were finally rinsed in distilled water, fixed in 4%PFA in PBS, and treated for paraffin embedding.
X-gal staining followed by immunostaining
Early stage embryos were dissected in PBS at room temperature, fixed in 4% PFA 15 min on ice, rinsed in PBS containing 2mM MgCl2 3 times, 10 min each on ice. Embryos were then incubated in X-gal staining solution (PBS, 2mM MgCl2, 5mM ferricyaninde, 5mM ferrocyaninde, 1mg/ml X-gal) 1h at 37°C. When X-gal staining is deemed complete embryos were washed in PBS 3 times, 10min each at room temperature and further fixed in 4% PFA for 2h at room temperature or at 4°C overnight. Immunostaining was performed as described above.
Southern Blot
For Southern blot 20μg of genomic DNA (gDNA) were digested with the respective restriction endonuclease and run in a 0.8% agarose gel overnight. The gel was depurinated (0.25N HCl, 15min at room temperature), denatured (0.5N NaOH, 15min at room temperature), neutralized (0.65M Tris base, 1M NaCl) and rinsed in distilled water. Transfer of gDNA to a nitrocellulose membrane was performed by capillary action in a 10XSSC bath overnight at room temperature. Transferred gDNA was cross linked under UV light in a cross linker oven. The membrane was prehybridized (20XSSPE, 10% PEG-8000, 7%SDS) at 65°C for 1h. Hybridization was carried with a radioactive probe added to fresh warmed prehybridization solution and incubated overnight at 65°C. Washes were performed at 65°C and film exposure was carried at −80°C for a week.
Radioactive probe labeling
Probes were amplified from genomic DNA by PCR, labeled following manufacturer’s instructions (AMERSHAM Rediprime labeling kit II), and purified following manufacturer’s instructions (Illustra NICK columns, Sephadex G-50).
Supplementary Material
Supplementary Figure 1. MyocdKI EGFP expression at early embryonic stages. EGFP expression is observed at early stages of cardiac formation and restricted to the cardiac crescent at E7.5 and early forming tubular heart at E8.0. (Scale bar, 100μm).
Supplementary Figure 2. MyocdKI LacZ expression at late embryonic stages. A) X-gal staining on E15.5 embryo sectioned through the midline. B) Transversal section showing LacZ expression in the Neointima layer (arrowhead) of the carotid artery (CA) of a Postnatal (P2) pup. (Scale bar, 1mm) C) Transversal section of the small intestine of an E15.5 embryo. LacZ expression is observed in the forming muscularis mucosae (yellow arrowhead) and muscularis externa (black arrowhead) layers. (Scales bars, 100μm).
Supplementary Figure 3. Tissue autofluorescence. Images of several adult wild type control tissues and organs showing minimal autofluorescence when exposed to UV light.
Acknowledgments
We thank members of the Wang laboratory for advice and support. This work is supported by the March of Dimes Foundation and the NIH (HL085635, HL116919). RA Espinoza-Lewis was supported by postdoctoral fellowship from the American Heart Association.
References
- Baldwin S. Early embryonic vascular development. Cardiovascular Research. 1996;31:E34–E45. [PubMed] [Google Scholar]
- Cai C-L, Liang X, Shi Y, Chu P-H, Pfaff SL, Chen J, Evans S. Isl1 Identifies a Cardiac Progenitor Population that Proliferates Prior to Differentiation and Contributes a Majority of Cells to the Heart. Developmental Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Yin H, Jiang Y, Radhakrishnan SK, Huang Z-P, Li J, Shi Z, Kilsdonk EPC, Gui Y, Wang D-Z, Zheng X-L. Induction of MicroRNA-1 by Myocardin in Smooth Muscle Cells Inhibits Cell Proliferation. Arteriosclerosis, Thrombosis, and Vascular Biology. 2011;31:368–375. doi: 10.1161/ATVBAHA.110.218149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creemers EE, Sutherland LB, McAnally J, Richardson JA, Olson EN. Myocardin is a direct transcriptional target of Mef2, Tead and Foxo proteins during cardiovascular development. Development. 2006;133:4245–4256. doi: 10.1242/dev.02610. [DOI] [PubMed] [Google Scholar]
- Espinoza-Lewis RnA, Liu H, Sun C, Chen C, Jiao K, Chen Y. Ectopic expression of Nkx2.5 suppresses the formation of the sinoatrial node in mice. Developmental Biology. 2011;356:359–369. doi: 10.1016/j.ydbio.2011.05.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Frigola C, Shi Y, Evans SM. Expression of the hyperpolarization-activated cyclic nucleotide-gated cation channel HCN4 during mouse heart development. Gene Expression Patterns. 2003;3:777–783. doi: 10.1016/s1567-133x(03)00125-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Min Lu M, Cheng L, Yuan L-J, Zhu X, Stout AL, Chen M, Li J, Parmacek MS. Myocardin is required for cardiomyocyte survival and maintenance of heart function. Proceedings of the National Academy of Sciences. 2009;106:18734–18739. doi: 10.1073/pnas.0910749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara H, Bartunkova S, Schinke M, Tanaka M, Izumo S. Cardiac and Extracardiac Expression of Csx/Nkx2.5 Homeodomain Protein. Circulation Research. 1998;82:936–946. doi: 10.1161/01.res.82.9.936. [DOI] [PubMed] [Google Scholar]
- Li L, Miano JM, Cserjesi P, Olson EN. SM22α, a Marker of Adult Smooth Muscle, Is Expressed in Multiple Myogenic Lineages During Embryogenesis. Circulation Research. 1996;78:188–195. doi: 10.1161/01.res.78.2.188. [DOI] [PubMed] [Google Scholar]
- Li S, Wang D-Z, Wang Z, Richardson JA, Olson EN. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proceedings of the National Academy of Sciences. 2003;100:9366–9370. doi: 10.1073/pnas.1233635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Wang G, Lin L, Lowe J, Zhang Q, Bu L, Chen Y-H, Chen J, Sun Y, Evans SM. HCN4 Dynamically Marks the First Heart Field and Conduction System Precursors. Circulation Research. 2013;113:399–407. doi: 10.1161/CIRCRESAHA.113.301588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wang X, Hu G, Wang Y, Zhou J. The Transcription Factor TEAD1 Represses Smooth Muscle-specific Gene Expression by Abolishing Myocardin Function. Journal of Biological Chemistry. 2014;289:3308–3316. doi: 10.1074/jbc.M113.515817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Creemers EE, Wang D-Z, Olson EN, Miano JM. Myocardin is a bifunctional switch for smooth versus skeletal muscle differentiation. Proceedings of the National Academy of Sciences. 2007;104:16570–16575. doi: 10.1073/pnas.0708253104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes & Development. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- Meilhac SnM, Esner M, Kelly RG, Nicolas J-Fo, Buckingham ME. The Clonal Origin of Myocardial Cells in Different Regions of the Embryonic Mouse Heart. Developmental Cell. 2004;6:685–698. doi: 10.1016/s1534-5807(04)00133-9. [DOI] [PubMed] [Google Scholar]
- Moorman A, Webb S, Brown NA, Lamers W, Anderson RH. DEVELOPMENT OF THE HEART: (1) FORMATION OF THE CARDIAC CHAMBERS AND ARTERIAL TRUNKS. Heart. 2003;89:806–814. doi: 10.1136/heart.89.7.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens GK, Kumar MS, Wamhoff BR. Molecular Regulation of Vascular Smooth Muscle Cell Differentiation in Development and Disease. Physiological Reviews. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- Später D, Abramczuk MK, Buac K, Zangi L, Stachel MW, Clarke J, Sahara M, Ludwig A, Chien KR. A HCN4+ cardiomyogenic progenitor derived from the first heart field and human pluripotent stem cells. Nat Cell Biol. 2013;15:1098–1106. doi: 10.1038/ncb2824. [DOI] [PubMed] [Google Scholar]
- Stieber J, Herrmann S, Feil S, Löster J, Feil R, Biel M, Hofmann F, Ludwig A. The hyperpolarization-activated channel HCN4 is required for the generation of pacemaker action potentials in the embryonic heart. Proceedings of the National Academy of Sciences. 2003;100:15235–15240. doi: 10.1073/pnas.2434235100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- Tang R-h, Zheng X-L, Callis TE, Stansfield WE, He J, Baldwin AS, Wang D-Z, Selzman CH. Myocardin inhibits cellular proliferation by inhibiting NF-κB(p65)-dependent cell cycle progression. Proceedings of the National Academy of Sciences. 2008;105:3362–3367. doi: 10.1073/pnas.0705842105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueyama T, Kasahara H, Ishiwata T, Nie Q, Izumo S. Myocardin Expression Is Regulated by Nkx2.5, and Its Function Is Required for Cardiomyogenesis. Molecular and Cellular Biology. 2003;23:9222–9232. doi: 10.1128/MCB.23.24.9222-9232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tuyn J, Knaän-Shanzer S, van de Watering MJM, de Graaf M, van der Laarse A, Schalij MJ, van der Wall EE, de Vries AAF, Atsma DE. Activation of cardiac and smooth muscle-specific genes in primary human cells after forced expression of human myocardin. Cardiovascular Research. 2005;67:245–255. doi: 10.1016/j.cardiores.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Vincent SD, Buckingham ME, Peter K. Current Topics in Developmental Biology. Academic Press; 2010. Chapter One - How to Make a Heart: The Origin and Regulation of Cardiac Progenitor Cells; pp. 1–41. [DOI] [PubMed] [Google Scholar]
- Virágh S, Challice CE. Origin and differentiation of cardiac muscle cells in the mouse. Journal of Ultrastructure Research. 1973;42:1–24. doi: 10.1016/s0022-5320(73)80002-4. [DOI] [PubMed] [Google Scholar]
- Wang D-Z, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of Cardiac Gene Expression by Myocardin, a Transcriptional Cofactor for Serum Response Factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- Wang K, Long B, Zhou J, Li P-F. miR-9 and NFATc3 Regulate Myocardin in Cardiac Hypertrophy. Journal of Biological Chemistry. 2010;285:11903–11912. doi: 10.1074/jbc.M109.098004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wang D-Z, Hockemeyer D, McAnally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang D-Z, Pipes GCT, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proceedings of the National Academy of Sciences. 2003;100:7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wystub K, Besser J, Bachmann A, Boettger T, Braun T. miR-1/133a Clusters Cooperatively Specify the Cardiomyogenic Lineage by Adjustment of Myocardin Levels during Embryonic Heart Development. PLoS Genet. 2013;9:e1003793. doi: 10.1371/journal.pgen.1003793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W-B, Li Z, Miano JM, Long X, Chen S-Y. Smad3-mediated Myocardin Silencing: A Novel Mechanism governing the initiation of smooth muscle differentiation. Journal of Biological Chemistry. 2011;286:15050–15057. doi: 10.1074/jbc.M110.202747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing W, Zhang T-C, Cao D, Wang Z, Antos CL, Li S, Wang Y, Olson EN, Wang D-Z. Myocardin Induces Cardiomyocyte Hypertrophy. Circulation Research. 2006;98:1089–1097. doi: 10.1161/01.RES.0000218781.23144.3e. [DOI] [PubMed] [Google Scholar]
- Zhuang S, Zhang Q, Zhuang T, Evans SM, Liang X, Sun Y. Expression of Isl1 during mouse development. Gene Expression Patterns. 2013;13:407–412. doi: 10.1016/j.gep.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. MyocdKI EGFP expression at early embryonic stages. EGFP expression is observed at early stages of cardiac formation and restricted to the cardiac crescent at E7.5 and early forming tubular heart at E8.0. (Scale bar, 100μm).
Supplementary Figure 2. MyocdKI LacZ expression at late embryonic stages. A) X-gal staining on E15.5 embryo sectioned through the midline. B) Transversal section showing LacZ expression in the Neointima layer (arrowhead) of the carotid artery (CA) of a Postnatal (P2) pup. (Scale bar, 1mm) C) Transversal section of the small intestine of an E15.5 embryo. LacZ expression is observed in the forming muscularis mucosae (yellow arrowhead) and muscularis externa (black arrowhead) layers. (Scales bars, 100μm).
Supplementary Figure 3. Tissue autofluorescence. Images of several adult wild type control tissues and organs showing minimal autofluorescence when exposed to UV light.